Abstract
There are two response elements for p53 in the promoter of the gene for the cyclin-dependent kinase inhibitor p21. The binding of p53 to the 5′ site was enhanced by incubation with monoclonal antibody 421, whereas the binding of p53 to the 3′ site was inhibited. Mutational analysis showed that a single-base change caused one element to behave like the other. A response element in the human cdc25C promoter is bound by p53 with properties similar to the 3′ site. These results identify two classes of p53-binding sites and suggest a mechanism for target gene selectivity by p53.
Keywords: Tumor suppressor, DNA binding, sequence specificity, p53 protein, transcriptional activation, binding sites
The tumor suppressor protein p53 has been implicated in the cellular response to DNA damage and mediates either growth arrest or apoptosis, depending on particular cellular conditions (see Gottlieb and Oren 1996; Ko and Prives 1996; Levine 1997). It has also been implicated in a spindle checkpoint (Cross et al. 1995) and in the induction of either differentiation (Shaulsky et al. 1991; Aloni-Grinstein et al. 1993; Soddu et al. 1994) or senescence (Sugrue et al. 1997). The p53 protein is a transcription factor that binds in a sequence-specific manner to particular sites in the genome and activates transcription of target genes (for review, see Gottlieb and Oren 1996; Ko and Prives 1996; Levine 1997). Utilizing immunobinding assays involving the monoclonal antibody 421, a consensus binding site for p53 has been defined and consists of four pentameric repeats of RRRCW in which R is a purine and W represents either an A or T residue (El-Deiry et al. 1992; Funk et al. 1992; Halazonetis et al. 1993). Two palindromic pentamers (half-site) are juxtaposed to a second set of two palindromic pentamers, the two half-sites being separated by no insert or insertions from 1–13 bp (El-Deiry et al. 1992). Such a consensus site is consistent with the fact that p53 exists in solution and binds to DNA as a tetramer (Friedman et al. 1993). It has been proposed that to accommodate a symmetrical tetrameric p53 on such a site, the DNA must bend (Balagurumoorthy et al. 1995; Nagaich et al. 1997a,b). Studies to date have implicated the C residue at position 4 of each pentamer as essential for the binding of p53 to DNA (Halazonetis et al. 1993; Nagaich et al. 1997b).
The structure of p53 is consistent with its role as a transcription factor with identified domains that are responsible for transcriptional activation, sequence-specific DNA binding, and oligomerization as a tetramer. Previous studies have implicated the carboxy-terminal 30 amino acids of p53 as exerting a negative regulatory effect on the DNA-binding activity of the protein. Deletion of these carboxy-terminal 30 amino acids, phosphorylation of sites within this region by casein kinase II and protein kinase C, and the binding of bacterial DnaK in this region all will activate the DNA-binding activity of p53 (Hupp et al. 1992; Takenaka et al. 1995). Consistent with this, the mAb 421, which has an epitope in this carboxy-terminal region, activates the ability of p53 to bind to DNA (Funk et al. 1992; Hupp et al. 1992; Halazonetis et al. 1993; Mundt et al. 1997). Finally, a peptide derived from the carboxyl end of p53 has also been shown to stimulate the ability of p53 to interact with DNA, although not to the same extent as the activators identified previously (Hupp et al. 1995).
DNA damage induces expression of p53 protein which, in turn, transcriptionally activates expression of particular genes, most notably those that encode the cyclin-dependent kinase inhibitor p21. Consistent with this, cells that lack p21 expression have an impaired p53-dependent response to DNA damage (Brugarolas et al. 1995; Deng et al. 1995). The human p21 promoter has been shown to contain two p53-responsive elements. Deletion analysis of reporter constructs containing the sequence of the human p21 promoter identified a distal element located 2.3–2.5 kb and a proximal element located 1.1–1.5 kb from the start site of transcription (El-Deiry et al. 1993,1995; Macleod et al. 1995).
In this report we have confirmed the existence of two p53-responsive elements in the human p21 promoter. One of these, the 3′ site, matches the consensus sequence for p53 DNA binding at 18 of 20 positions. Notably, there is a G residue in place of the C residue in the fourth position of the first pentamer (Fig. 1). In contrast to other known p53-binding sites, the binding of p53 to this 3′ site in the p21 promoter is inhibited by mAb 421. This suggests the existence of a new class of genomic sites in which the binding of p53 may be regulated differentially. Because p53 has been implicated in a variety of cellular responses, an understanding of the mechanism for selection of target genes by p53 is central to understanding its biological functions. The results presented here suggest one potential mechanism for such site selection by p53.
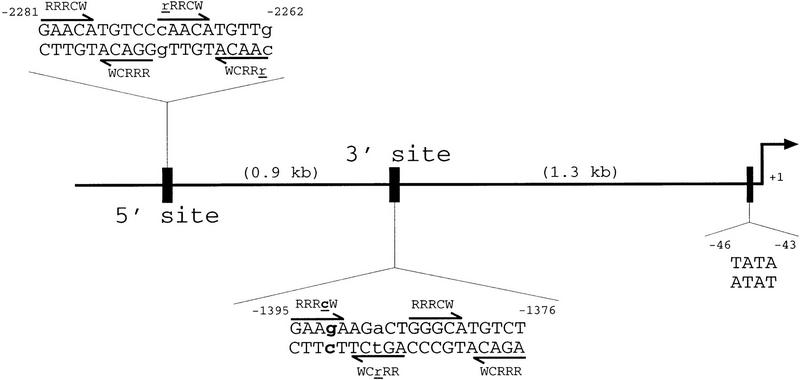
Figure 1.
Schematic of p21 promoter. Shown is 2.5 kb of the upstream sequence of the human p21 promoter; 2.2 kb from the start site of transcription is a well-documented p53-binding site at positions −2281 to −2262. It matches the published consensus sequence for a p53 DNA binding site in 18 of 20 positions; the variations from the consensus are shown by lowercase letters. A second site with similarity to the consensus sequence is 1.3 kb upstream from the start site of transcription. Contained within positions −1395 to −1376, this sequence also matches the consensus at 18 of 20 positions. Note that in this 3′ site, the fourth position of the first pentamer contains a G rather than a C residue (shown in lowercase and boldface type). The position of the TATA box of the promoter is also indicated.
Results
Two p53-responsive elements are present in the human p21 promoter
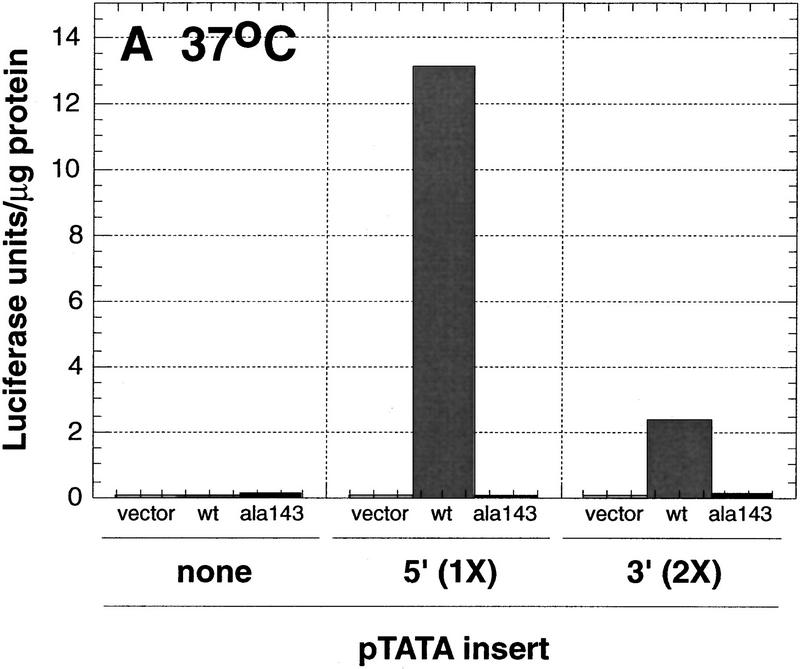
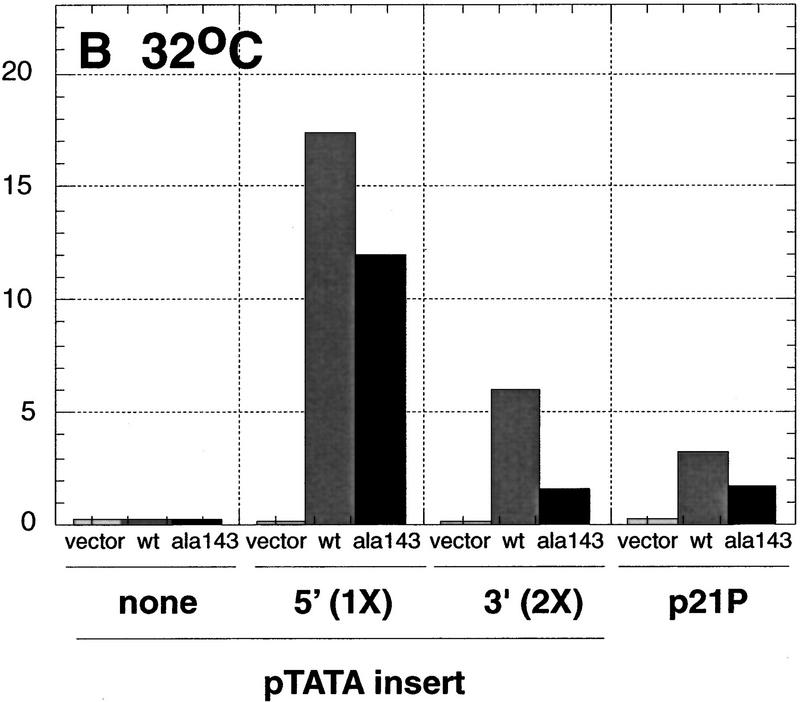
Using a series of deletion constructs, two p53-response elements had been identified previously in the human p21 promoter (Macleod et al. 1995). A well-characterized element was located 2.4 kb upstream from the start site of transcription, and a second element had been suggested to be present 1.1–1.5 kb from the start of transcription (Fig. 1). We demonstrate that both a 20-bp 5′ element, located between −2262 and −2281, and a 20-bp 3′ element, located between −1376 and −1395, are each sufficient to transactivate a reporter gene in a p53-dependent manner (Fig. 2). Double-stranded synthetic oligonucleotides containing either one copy of the 5′ site or two copies of the 3′ site were inserted into a reporter vector, pGL3–E1bTATA, containing the E1b promoter upstream of a luciferase reporter gene. Although a single copy of the 3′ site conferred p53-dependent transcriptional activation on the minimal promoter (see Fig. 5B, below), two copies of the 3′ site showed a more pronounced effect and were used in the experiments described here (Fig. 2). Each reporter construct was cotransfected into p53-negative Saos-2 cells with empty vector or a plasmid expressing either human wild-type p53 or the human temperature-sensitive mutant p53Ala143. The temperature-sensitive mutant p53Ala143 is in a mutant conformation at 37°C. At this temperature, it is unable to activate p53-response elements. However, when shifted to 32°C, this mutant can assume a wild-type conformation and has been shown to activate some p53-responsive promoters (such as p21) but not others (such as Bax) (Friedlander et al. 1996). Cells were maintained at 37°C or shifted to 32°C, 17 hr prior to lysis. At 37°C, a luciferase reporter containing a single copy of the 5′ site was activated 1730-fold by wild-type p53, but only 2-fold by p53Ala143. The reporter plasmid containing two copies of the 3′ site was activated 380-fold by wild-type p53, but again only 2-fold by p53Ala143. The reporter vector lacking either response element was minimally activated by expression of either the wild-type or mutant p53 (Fig. 2A). At 32°C, the luciferase reporter containing a single copy of the 5′ site was activated 1154-fold by wild-type p53 and 792-fold in the presence of p53Ala143. The reporter plasmid containing two copies of the 3′ site was activated 362-fold by wild-type p53 and 96-fold by the mutant p53Ala143. A luciferase reporter plasmid containing the full-length p21 promoter p21P was activated 16-fold by wild-type p53 and ninefold by the mutant p53Ala143. At 32°C, the reporter vector lacking either response element was not activated by either wild-type or mutant p53 (Fig. 2B). These data confirm that there are two p53-response elements in the human p21 promoter, each of which is sufficent to confer p53-dependent transcriptional activation on a luciferase reporter gene containing the minimal adenovirus E1b promoter. Additionally, neither the 5′ nor the 3′ site is activated by the temperature-sensitive mutant p53Ala143 at 37°C, whereas at 32°C both sites are activated, although less so than by wild-type p53.
Figure 2.
Two p53-response elements are present in the p21 promoter. Saos-2 cells were transfected as described in Materials and Methods with 2 μg of the indicated reporter constructs in the absence or presence of either 50 ng of pCMV–p53wt, an expression vector encoding human wild-type p53 under the control of the CMV promoter (light and dark bars, respectively), or 50 ng of pCMV–p53ala143, an expression vector encoding the temperature-sensitive mutant p53Ala143 (solid bars). Cells were either maintained at 37°C (A) or shifted to 32°C 17 hr prior to lysis (B) and then were assayed for luciferase activity and total protein levels as described in Materials and Methods. The indicated values are the average of three (37°C) or four (32°C) independent experiments that had been performed in duplicate.
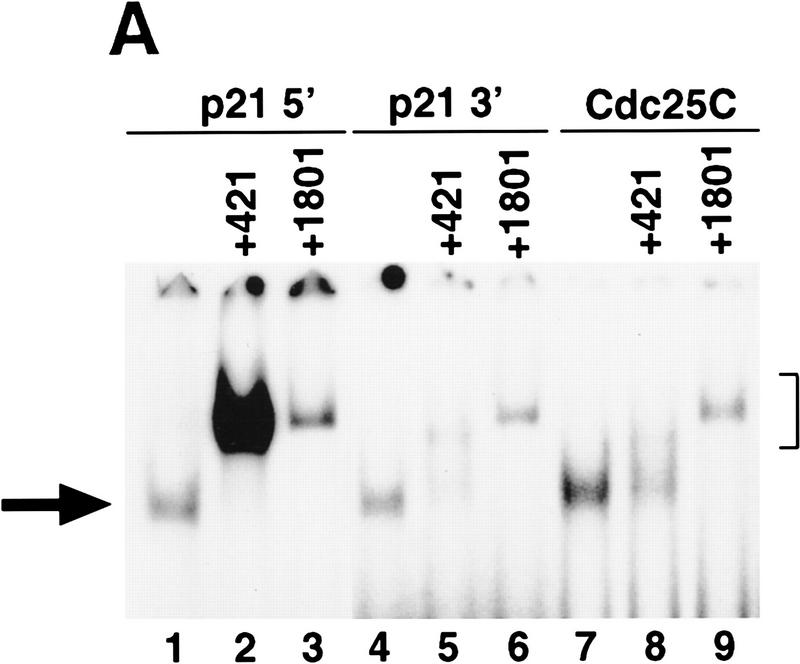
Figure 5.
The p53-response element in the promoter of the human cdc25C gene has properties similar to the 3′ site. (A) An EMSA using as radiolabeled probe the 5′ site (lanes 1–3), the 3′ site (lanes 4–6), or the site from the cdc25C promoter (lanes 7–9). The probes had an equivalent specific activity of labeling. Five nanograms of purified p53 was incubated in the absence (lanes 1,4,7) or presence of either 4 μl of mAb 421 (lanes 2,5,8) or the presence of 4 μl of mAb 1801 (lanes 3,6,9). (→) The position of the p53–DNA complex; (]) position of the supershifted p53–DNA–antibody complex. (B) Saos-2 cells were transfected as described in Materials and Methods with 2 μg of the indicated reporter constructs in the absence (open bars) or presence of increasing amounts of pCMV–p53wt (50, 100, 200, or 500 ng, shaded bars), or 500 ng of pCMV–p53ala143 (solid bars). Appropriate amounts of the vector pCMV were added to each transfection mixture to maintain a total level of plasmid DNA of 2.5 μg. Cells were maintained at 37°C and assayed for luciferase activity and total protein levels as described in Materials and Methods. The indicated values are from a representative experiment that had been performed in duplicate.
Two p53-binding sites are present in the human p21 promoter
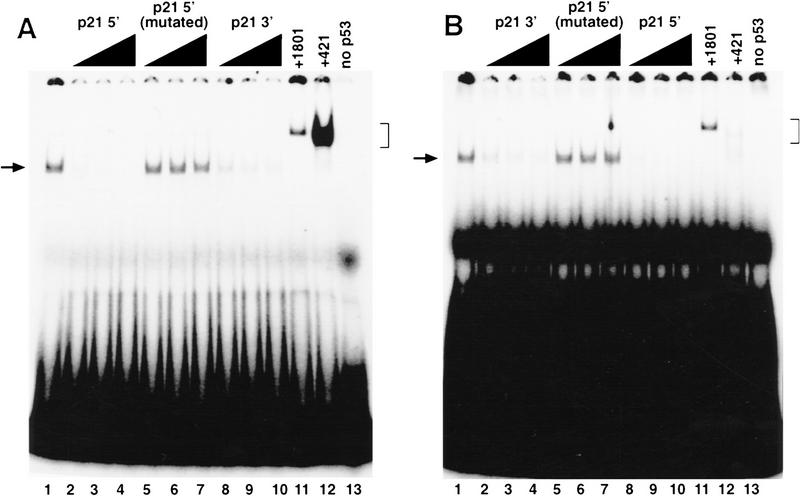
Double-stranded oligonucleotides that contain the sequence of the 5′ and 3′ sites were synthesized and used in electrophoretic mobility shift assays (EMSAs). The 5′ site was bound by purified p53, and this binding was competed by an excess of unlabeled 5′ site (Fig. 3A, lanes 2–4), as well as an excess of the 3′ site (Fig. 3A, lanes 8–10), but not by an unlabeled oligonucleotide containing a mutated sequence of the 5′ site in which the C residue in the fourth position of each pentamer has been mutated to a T residue (Fig. 3A, lanes 5–7). The 3′ site competes approximately threefold less well than the 5′ site for the binding of p53 protein to a labeled 5′ site (Fig. 3A, lanes 2–4, and 8–10). The mAb 1801 supershifts the p53–5′ site complex efficiently, demonstrating the presence of p53 in that complex (Fig. 3A, lane 11). As has been reported previously for a p53 consensus site (Funk et al. 1992), mAb 421 also efficiently supershifts the p53–5′ site complex and in so doing enhances the binding that is seen (Fig. 3A, lane 12).
Figure 3.
Monoclonal antibody enhances the binding of p53 to the 5′ site but inhibits the binding of p53 to the 3′ site. (A) An EMSA using as labeled probe the 5′ p53-binding site; (B) an EMSA using as labeled probe the 3′ p53-binding site. Five nanograms of purified p53 protein was incubated alone (lane 1), in the presence of a 17-, 33-, or 50-fold excess of each unlabeled competitor, as indicated (lanes 2–10), or in the presence of a 4 μl of mAb 1801 (lane 11) or 4 μl of mAb 421 (lane 12). p21 5′ (mutated) refers to a 5′ site in which the fourth postion of each pentamer has been mutated to a T residue. A sample that does not contain any p53 protein is shown in lane 13. (→) The position of the p53–DNA complex; (]) the position of the supershifted p53–DNA antibody complex.
The 3′ site also was bound by purified p53, and the binding of p53 to a labeled 3′ site was effectively competed by the unlabeled 3′ site (Fig. 3B, lanes 2–4), as well as the unlabeled 5′ site (Fig. 3B, lanes 8–10) but not the mutated 5′ site with all four C residues altered (Fig. 3B, lanes 5–7). Consistent with the results of the competition analysis using the labeled 5′ site, the unlabeled 5′ site competed threefold better for binding to the labeled 3′ site as the unlabeled 3′ site (Fig. 3B, lanes 2–4, and 8–10). mAb 1801 again supershifted the p53–3′ site complex (Fig. 3B, lane 11) efficiently. Surprisingly, in contrast to the result with the labeled 5′ site, mAb 421 appeared to inhibit the binding of p53 to the 3′ site and supershifted what little DNA-binding complex that was detected only poorly (Fig. 3B, lane 12). This latter result suggests the intriguing possibility that the optimal binding of p53 to each of these sites may require different conformations of the p53 tetramer.
Mutation of a C residue affects responsiveness to mAb 421
The 3′ site diverges from the published consensus for p53-binding sites in two positions. The residue in position 4 of the first pentamer is a G (instead of a C residue), and the residue in position 3 of the second pentamer is an A residue (instead of a pyrimidine). The binding of p53 to the labeled 3′ site is inhibited in the presence of mAb 421 (Fig. 4, lanes 9–12). However, when a 1-bp change, G4 to C residue, was engineered in this sequence and used as the labeled probe, the binding to p53 was enhanced in the presence of mAb 421 (Fig. 4, lanes 13–16). Thus, mutation of G4 to a C, thereby creating a site with four consensus pentamers, allowed the binding of p53 to this site to be enhanced by mAb 421.
Figure 4.
Mutational analysis demonstrates the importance of the C residue in the fourth position of a pentamer in responsiveness to mAb 421. Purified p53 (5 or 10 ng as indicated) was incubated with labeled probes containing the 5′ site (lanes 1–4), the 5′ site with the residue at position 14 mutated to a G (C14 to G) (lanes 5–8), the 3′ site (lanes 9–12), or the 3′ site with the residue at position 4 mutated to a C (G4 to C) (lanes 13–16). The labeled probes had equivalent specific activities. Reactions were performed either in the absence (lanes 1,2,5,6,9,10,13,14) or presence (lanes 3,4,7,8,11,12,15,16) of mAb 421. The arrow (→) position of the p53–DNA complex; (]) position of the supershifted p53–DNA–antibody complex.
The reverse effect could be achieved through mutagenesis of the 5′ site. This site conforms to the consensus sequence in that a C residue is present in the fourth position of each pentamer. A labeled 5′ probe bound to increasing amounts of p53 (Fig. 4, lanes 1–2) and this binding could be enhanced in the presence of mAb 421 (Fig. 4, lanes 3,4). When mutated 5′ (C14 to G) was labeled and used in this EMSA, its binding to p53 was inhibited in the presence of 421 (Fig. 4, lanes 5–8) just like the 3′ site (Fig. 4, lanes 9–12). Therefore, the primary sequence of the response element can determine whether the binding by p53 is enhanced or inhibited by antibody 421.
There is a p53-response element in the promoter of the human cdc25C gene with properties similar to the 3′ site
A search in the human genome database for other variant p53-binding sites that consist of four pentamers, only three of which contain C residues in the fourth position, was performed. A site in the promoter of the cdc25C gene, which encodes a cell cycle-regulated protein phosphatase that is necessary for progression into mitosis, was subjected to further analysis. A radiolabeled synthetic oligonucleotide containing a sequence from the human cdc25C promoter is bound by purified human p53 in an EMSA (Fig. 5A, lane 7). mAb 1801 supershifts this complex efficiently, whereas mAb 421 inhibits the binding of p53 to this site (Fig. 5A, lanes 8,9). Similar results were obtained with a radiolabeled oligonucleotide containing the sequence of the 3′ site of the p21 promoter (Fig. 5A, lanes 4–6). These results are in contrast to the ability of mAb 421 to enhance and supershift the complex of p53 with a radiolabled oligonucleotide containing the 5′ site from the p21 promoter (Fig. 5A, lanes 1–3). These results demonstrate that the site from the cdc25C promoter binds to p53 in the presence of mAb 421 with similar properties as the 3′ site from the p21 promoter.
To determine whether this p53-binding site from the cdc25C promoter can act as a p53-response element in cells, an oligonucleotide containing a single copy of the sequence of this site was inserted adjacent to the adenovirus E1b minimal promoter in a luciferase reporter plasmid. This construct was compared to constructs containing two previously characterized p53-response elements, namely one from the human bax promoter, and one of the two intronic sites found in the IGFBP3 gene, the so-called box A site (Buckbinder et al. 1995; Miyashita and Reed 1995; Friedlander et al. 1996; Ludwig et al. 1996). These reporter constructs were compared to a plasmid containing a single copy of the 3′ site from the p21 promoter and a plasmid containing a single copy of the 5′ site that contains all four C residues altered. Saos-2 cells were transfected with increasing amounts of the wild-type p53 expression vector in the presence of these various reporter constructs. Wild-type p53 activated reporters containing the Bax, IGFBP3-A, 3′ site, and Cdc25C sites, but not a reporter containing the mutated 5′ site (Fig. 5B). This demonstrates that the site from the cdc25C promoter is sufficient to confer p53-dependent transcriptional activation on a heterologous luciferase reporter construct. Thus, mAb 421 differentially affects the binding of p53 to two different genomic binding sites that can mediate p53-dependent transcriptional activation.
Discussion
The binding of the mAb 421 to p53 stimulates the ability of p53 to bind to one set of genomic sites that conform to a previously identified consensus sequence and inhibits its ability to bind to another set of genomic sites that deviate from that consensus. This ability to regulate the sequence selectivity of DNA binding by a transcription factor, even in an in vitro setting, is a novel finding. A provocative unanswered question is whether the inhibition seen in the presence of mAb 421 has a physiological counterpart in the cell such that the sequence-specific binding of p53 to elements such as the 3′ site is regulated. The relevance of the mAb 421 effect will remain an open question until cellular conditions are identified that produce selective inhibition of these variant p53-response elements. Previous studies suggest some possible mechanisms, including regulation by the coactivator p300 and phosphorylation by particular kinases. The coactivator p300 recently has been shown to stimulate the sequence-specific DNA-binding activity of p53 (Gu and Roeder 1997), and the DNA-binding activity of p53 can also be stimulated by phosphorylation by casein kinase II, protein kinase C, and cyclin-dependent kinase (Meek et al. 1990; Takenaka et al. 1995; Wang and Prives 1995). Studies to examine the role of these different kinases, as well as p300, in the regulation of the ability of p53 to interact with elements such as the 3′ site in the p21 promoter are essential to address these possibilities.
Depending on particular cellular conditions, the tumor suppressor protein p53 has been reported to induce growth arrest in both the G1 and G2 phases of the cell cycle, mediate an apoptotic response, or trigger alternatively a differentiation or a senescence pathway (see Gottlieb and Oren 1996; Ko and Prives 1996; Levine 1997; Sugrue et al. 1997). Because the DNA-binding activity of p53 appears to play a role in each of these physiological responses, the ability of p53 to select among various target genes to elicit a particular cellular response is central to the regulation of its biological function. To date, the identification of a mechanism for the regulation of target gene selectivity by p53 has been elusive. The results presented here, albeit under nonphysiological conditions, suggest one potential mechanism by which such selectivity may be achieved. It will be important to determine whether such a mechanism occurs during any of the various cellular responses to p53 and to identify the target genes that are relevant in each situation.
Materials and methods
Plasmids
The expression plasmids pCMV–p53wt and pCMV–p53Ala143, encode the indicated human p53 protein under the control of the CMV promoter. These plasmids were referred originally to as pCMV–SN3 and pCMV–CX3, respectively. The reporter plasmid, pGL3–E1bTATA, was constructed by digesting a synthetic double-stranded oligonucleotide, GCGCGGTACCCTCGAGATGCATGAATTCGCTAGCGAGCTCAGGGTATATAATGAAGCTTGGCC, with KpnI and HindIII and cloning it into the pGL3–Basic vector (Promega), which had been double-digested with KpnI and HindIII. The resulting plasmid contains a multiple cloning region with the unique restriction sites, KpnI, XhoI, NsiI, EcoRI, NheI, and SacI upstream of the minimal adenovirus E1b promoter sequence and the coding region for firefly luciferase.
The following synthetic double-stranded oligonucleotides were digested with KpnI and either NheI [5′, 3′(1×), 3′(2×), Cdc25C, and Bax] or SacI (5′ mut and IGFBP3-A) and cloned into pGL3–E1bTATA, which had been double-digested with KpnI and either NheI or SacI to produce the appropriate reporter plasmids: 5′-AATTGGTACCGAACATGTCCCAACATGTTGGCTAGCGAATT; 3′(1×)–AATTCGGTACCGAAGAAGACTGGGCATGTCTGCTAGCGAATT; 3′(2×)–AATTCGGTACCGAAGAAGACTGGGCATGTCTGAAGAAGACTGGGCATGTCTGCTAGCGAATT; 5′ mut–AATTCGGTACCGAATATATCCCAATATATTGGAGCTCGAATT; Cdc25C–AATTCGGTACCGGGCAAGTCTTACCATTTCCAGAGCAAGCACGCTAGCGAATT; Bax–AATTCGGTACCTCACAAGTTAGAGACAAGCCTGGGCGTGGGCTATATTGCTAGCGAATT; and IGFBP3-A–AATTCGGTACCAAACAAGCCACCAACATGCTTTGGAGCTCGAATT.
Transfection of reporter constructs
Saos-2 cells were transfected using the DOTAP liposomal transfection reagent (Boehringer Mannheim) according to the manufacturer’s instructions. Lysates were prepared, total protein concentration was determined, and luciferase assays were quantitated using a TD-20e Luminometer (Turner).
Purification and quantitation of human p53 protein
Sf9 cells that were infected with recombinant baculovirus were lysed in 20 mm HEPES (pH 7.4) containing 20% glycerol, 10 mm NaCl, 0.2 mm EDTA, 0.1% Triton X-100, 1 mm DTT, 1 mm PMSF, 50 μm leupeptin, and 50 μg/ml aprotinin (lysis buffer). Nuclei were pelleted by centrifugation at 2300 rpm and then resuspended in lysis buffer containing 500 mm NaCl. Extracts were diluted to 100 mm NaCl, applied to a 0.5-ml Ni-NTA-agarose column (Qiagen) that was equilibrated with 20 mm HEPES containing 100 mm NaCl, and eluted with 200 mm imidazole containing 10 mm HEPES, (pH 7.4) and 5 mm NaCl. Fractions of 0.5-ml were collected, dialyzed against 10 mm HEPES (pH 7.4), 5 mm NaCl, 0.1 mm EDTA, 20% glycerol, and 1 mm DTT, aliquoted, and stored at −70°C.
EMSAs
Complementary single-stranded oligonucleotides were annealed, and ends were filled using the Klenow fragment of DNA polymerase to produce the following double-stranded oligonucleotides: p21 5′-AATTCTCGAGGAACATGTCCCAACATGTTGCTCGAGAATT; p21 3′-AATTCTCGAGGAAGAAGACTGGGCATGTCTTCTACCTCGAGAATT; p21 5′ (mutated)–AATTCTCGAGGAATATATCTTGAATTCTTCCTCGAGAATT; p21 5′ (C14 to G)–AATTCTCGAGGAACATGTCCCAAGATGTTGCTCGAGAATT; p21 3′ (G4 to C)–AATTCTCGAGGAACAAGACTGGGCATGTCTTCTACCTCGAGAATT; and Cdc25C–AATTCTCGAGGGGCAAGTCTTACCATTTCCAGAGCAAGCACCTCGAGAATT.
Purified p53 protein, 3 ng of labeled double-stranded oligonucleotide, and hybridoma supernatant where appropriate, were incubated in a total volume of 30 μl of DNA binding buffer containing 20 mm HEPES (pH 7.5), 83 mm NaCl, 0.1 mm EDTA, 12% glycerol, 2 mm MgCl2, 2 mm spermidine, 0.7 mm DTT, 133 μg/ml BSA, and 25 μg/ml poly[d(I–C)] for 30 min at room temperature. Samples were loaded on a native 4% acrylamide gel in 0.5× TBE and electrophoresed at 4°C at 200 V for 2 hr. The gel was dried and exposed to Kodak XAR-5 film using an intensifying screen at −70°C. Bands were quantitated using the Molecular Analyst Phosphorimaging system (Bio-Rad).
Acknowledgments
We thank Bert Vogelstein for the p53 expression plasmids, and Ze’ev Ronai for the recombinant baculovirus expressing His-tagged human p53. Ron Magnusson is thanked for help with the recombinant baculovirus infections. Helpful discussions with Avrom Caplan, Robert Krauss, Jonathan Licht, Leslie Pick, Serafin Piñol-Roma, and, especially, Zhen-Qiang Pan are gratefully acknowledged. Members of the Manfredi laboratory are thanked for their help and support: Yvette Tang, Edward Thornborrow, Michelle Denberg, and Joyce Meng. These studies were supported by grants from the National Cancer Institute (CA69161) and the Breast Cancer Program of the U.S. Army Medical Research and Materiel Command (U.S. AMRMC; DAMD-17-97-1-7336 and DAMD-17-97-1-7337). S.S.C. was supported by a training grant from the Breast Cancer Program of the U.S. AMRMC (DAMD-17-94-J-4111).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL jmanfredi@smtplink.mssm.edu; FAX (212) 849-2446.
References
- Aloni-Grinstein R, Zan-Bar I, Alboum I, Goldfinger N, Rotter V. Wild type p53 functions as a control protein in the differentiation pathway of the B-cell lineage. Oncogene. 1993;8:3297–3305. [PubMed] [Google Scholar]
- Balagurumoorthy P, Sakamoto H, Lewis MS, Zambrano N, Clore GM, Gronenborn AM, Appella E, Harrington RE. Four p53 DNA-binding domain peptides bind natural p53-response elements and bend the DNA. Proc Natl Acad Sci. 1995;92:8591–8595. doi: 10.1073/pnas.92.19.8591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brugarolas J, Chandrasekaran C, Gordon JI, Beach D, Jacks T, Hannon GJ. Radiation-induced cell cycle arrest compromised by p21 deficiency. Nature. 1995;377:552–557. doi: 10.1038/377552a0. [DOI] [PubMed] [Google Scholar]
- Buckbinder L, Talbott R, Velasco-Miguel S, Takenaka I, Faha B, Seizinger BR, Kley N. Induction of the growth inhibitor IGF-binding protein 3 by p53. Nature. 1995;377:646–649. doi: 10.1038/377646a0. [DOI] [PubMed] [Google Scholar]
- Cross SM, Sanchez CA, Morgan CA, Schimke MK, Ramel S, Izerda RL, Rashkind WH, Reid BJ. A p53-dependent mouse spindle checkpoint. Science. 1995;267:1353–1355. doi: 10.1126/science.7871434. [DOI] [PubMed] [Google Scholar]
- Deng C, Zhang P, Harper JW, Elledge SJ, Leder P. Mice lacking p21CIP1/WAF1 undergo normal development, but are defective in G1 checkpoint control. Cell. 1995;82:675–684. doi: 10.1016/0092-8674(95)90039-x. [DOI] [PubMed] [Google Scholar]
- El-Deiry WS, Kern SE, Pietenpol JA, Kinzler KW, Vogelstein B. Definition of a consensus binding site for p53. Nature Genet. 1992;1:45–49. doi: 10.1038/ng0492-45. [DOI] [PubMed] [Google Scholar]
- El-Deiry WS, Tokino T, Velculescu VE, Levy DB, Parsons R, Trent JM, Lin D, Mercer WE, Kinzler K, Vogelstein B. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- El-Deiry WS, Tokino T, Waldman T, Oliner JD, Velculescu VE, Burrell M, Hill DE, Healy E, Rees JL, Hamilton SR, Kinzler KW, Vogelstein B. Topological control of p21WAF1/CIP1 expression in normal and neoplastic tissues. Cancer Res. 1995;55:2910–2919. [PubMed] [Google Scholar]
- Friedlander P, Haupt Y, Prives C, Oren M. A mutant p53 that discriminates between p53-responsive genes cannot induce apoptosis. Mol Cell Biol. 1996;16:4961–4971. doi: 10.1128/mcb.16.9.4961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman PN, Chen X, Bargonetti J, Prives C. The p53 protein is an unusually shaped tetramer that binds directly to DNA. Proc Natl Acad Sci. 1993;90:3319–3323. doi: 10.1073/pnas.90.8.3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funk WD, Pak DT, Karas RH, Wright WE, Shay JW. A transcriptionally active DNA-binding site for human p53 protein complexes. Mol Cell Biol. 1992;12:2866–2871. doi: 10.1128/mcb.12.6.2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottlieb TM, Oren M. p53 in growth control and neoplasia. Biochim Biophys Acta. 1996;1287:77–102. doi: 10.1016/0304-419x(95)00019-c. [DOI] [PubMed] [Google Scholar]
- Gu W, Roeder RG. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- Halazonetis TD, Davis LJ, Kandil AN. Wild-type p53 adopts a “mutant”-like conformation when bound to DNA. EMBO J. 1993;12:1021–1028. doi: 10.1002/j.1460-2075.1993.tb05743.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hupp TR, Meek DW, Midgely CA, Lane DP. Regulation of the specific DNA binding function of p53. Cell. 1992;71:875–886. doi: 10.1016/0092-8674(92)90562-q. [DOI] [PubMed] [Google Scholar]
- Hupp TR, Sparks A, Lane DP. Small peptides activate the latent sequence-specific DNA binding function of p53. Cell. 1995;83:237–245. doi: 10.1016/0092-8674(95)90165-5. [DOI] [PubMed] [Google Scholar]
- Ko LJ, Prives C. p53: Puzzle and paradigm. Genes & Dev. 1996;10:1054–1072. doi: 10.1101/gad.10.9.1054. [DOI] [PubMed] [Google Scholar]
- Levine AJ. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- Ludwig RL, Bates S, Vousden KH. Differential activation of target cellular promoters by p53 mutants with impaired apoptotic function. Mol Cell Biol. 1996;16:4952–4960. doi: 10.1128/mcb.16.9.4952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macleod KF, Sherry N, Hannon G, Beach D, Tokino T, Kinzler K, Vogelstein B, Jacks T. p53-dependent and independent expression of p21 during cell growth, differentiation, and DNA damage. Genes & Dev. 1995;9:935–944. doi: 10.1101/gad.9.8.935. [DOI] [PubMed] [Google Scholar]
- Meek DW, Simon S, Kikkawa U, Eckhart W. The p53 tumour suppressor protein is phosphorylated at serine 389 by casein kinase II. EMBO J. 1990;9:3253–3260. doi: 10.1002/j.1460-2075.1990.tb07524.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyashita T, Reed JC. Tumor suppressor p53 is a direct transcriptional activator of the human bax gene. Cell. 1995;80:293–299. doi: 10.1016/0092-8674(95)90412-3. [DOI] [PubMed] [Google Scholar]
- Mundt M, Hupp T, Fritsche M, Merkle C, Hansen S, Lane D, Groner B. Protein interactions at the carboxyl terminus of p53 result in the induction of its in vitro transactivation potential. Oncogene. 1997;15:237–244. doi: 10.1038/sj.onc.1201174. [DOI] [PubMed] [Google Scholar]
- Nagaich AK, Appella E, Harrington RE. DNA bending is essential for the site-specific recognition of DNA response elements by the DNA binding domain of the tumor suppressor protein p53. J Biol Chem. 1997a;272:14842–14849. doi: 10.1074/jbc.272.23.14842. [DOI] [PubMed] [Google Scholar]
- Nagaich AK, Zhurkin VB, Sakamoto H, Gorin AA, Clore GM, Gronenborn AM, Appella E, Harrington RE. Architectural accommodation in the complex of four p53 DNA binding domain peptides with the p21/waf1/cip1 DNA response element. J Biol Chem. 1997b;272:14830–14841. doi: 10.1074/jbc.272.23.14830. [DOI] [PubMed] [Google Scholar]
- Shaulsky G, Goldfinger N, Peled A, Rotter V. Involvement of wild-type p53 in pre-B-cell differentiation in vitro. Proc Natl Acad Sci. 1991;88:8982–8986. doi: 10.1073/pnas.88.20.8982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soddu S, Blandino G, Citro G, Scardigli R, Piaggio G, Ferber A, Calabretta B, Sacchi A. Wild-type p53 gene expression induces granulocytic differentiation of HL-60 cells. Blood. 1994;83:2230–2237. [PubMed] [Google Scholar]
- Sugrue MM, Shin DY, Lee SW, Aaronson SA. Wild-type p53 triggers a rapid senescence program in human tumor cells lacking functional p53. Proc Natl Acad Sci. 1997;94:9648–9653. doi: 10.1073/pnas.94.18.9648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takenaka I, Morin F, Seizinger BR, Kley N. Regulation of the sequence-specific DNA binding function of p53 by protein kinase C and protein phosphatases. J Biol Chem. 1995;270:5405–5411. doi: 10.1074/jbc.270.10.5405. [DOI] [PubMed] [Google Scholar]
- Wang Y, Prives C. Increased and altered DNA binding of human p53 by S and G2/M but not G1 cyclin-dependent kinases. Nature. 1995;376:88–91. doi: 10.1038/376088a0. [DOI] [PubMed] [Google Scholar]