Abstract
Structural water molecules are found in many protein–ligand complexes. They are known to be vital in mediating hydrogen-bonding interactions and, in some cases, key for facilitating tight binding. It is thus very important to consider water molecules when attempting to model protein–ligand interactions for cognate ligand identification, virtual screening and drug design. While the rigid treatment of water molecules present in structures is feasible, the more relevant task of treating all possible positions and orientations of water molecules with each possible ligand pose is computationally daunting. Current methods in molecular docking provide partial treatment for such water molecules, with modest success. Here we describe a new method employing dead-end elimination to place water molecules within a binding site, bridging interactions between protein and ligand. Dead-end elimination permits a thorough, though still incomplete, treatment of water placement. The results show that this method is able to place water molecules correctly within known complexes and to create physically reasonable hydrogen bonds. The approach has also been incorporated within an inverse molecular design approach, to model a variety of compounds in the process of de novo ligand design. The inclusion of structural water molecules, combined with ranking based on the electrostatic contribution to binding affinity, improves a number of otherwise poor energetic predictions.
Keywords: dead-end elimination, inverse molecular design, protein–ligand interaction, scoring function, structural water molecules
Introduction
A number of challenges remain for the modeling, design and evaluation of protein–ligand interactions. Rigorous treatment of structural water molecules is one of the challenges that has received relatively little attention, in comparison with modeling ligand and protein flexibility and developing accurate scoring functions. Water has long been known to play an important role in protein–ligand binding interactions (Poornima and Dean, 1995a; Poornima and Dean, 1995b; Poornima and Dean, 1995c; Mancera, 2002; Li and Lazaridis, 2007). Analysis of complexes from the PDBbind database (Wang et al., 2004) shows that small-molecule ligands in complex with proteins are bound to an average of 4.6 water molecules (Lu et al., 2007). The importance of structural water can be seen in complexes with both cognate ligands and with designed inhibitors. Many natural complexes include key water molecules that either are functionally important within the protein or stabilize the required conformation of the ligand. Enzyme families of aspartyl proteases, β–lactamases and alcohol dehydrogenases utilize such water molecules to effect catalysis. Modeling structural water molecules can be vital in determining the cognate ligand that binds to an active site or a modulatory site in cases where this is not known. Water can also be important in modeling proteins where no ligand is present, bridging interactions between surface residues or across domains.
The positions of the oxygen atoms of highly ordered water molecules can often be identified by structural elucidation with techniques such as X-ray diffraction. Structural data are now available for many apo and holo proteins, and this provides useful information about structural water molecules. However, the positions of the hydrogen atoms are not routinely determined by X-ray diffraction and are thus undefined. Furthermore, serious difficulties occur when considering potential drug molecules. Some water molecules from the apo or a holo state may remain in place within some ligand complexes, but they often shift subtly, and many are simply displaced by portions of a ligand. A partial solution when modeling a new ligand is to consider a combinatorial set of cases, where structural water molecules from other known apo or holo structures of the same target can be present or absent. However, this technique is cumbersome, vastly incomplete and cannot account for the formation of new water-mediated interactions that may occur between a protein and a ligand. A more complete solution to this problem is the focus of this paper.
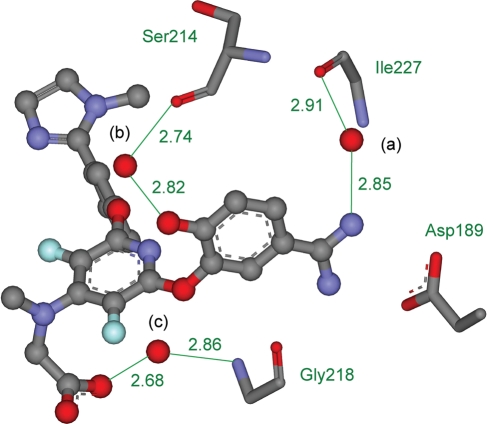
An example illustrating the importance of water molecules is the complex of Factor Xa with the inhibitor ZK-807834 from PDB ID 1FJS (Fig. 1) (Adler et al., 2000). There are three structural water molecules making important interactions at the interface. Water a makes a hydrogen bond with the protein backbone of residue Ile227 and with the amidino group of the inhibitor and is an important structural element in many such binding sites. Water b satisfies the hydrogen-bonding requirements of the phenolic group of the inhibitor while making a hydrogen bond with the protein backbone of Ser214. The final water c is the only binding site element that interacts with the carboxylate group of the inhibitor and is likely to be very important in stabilizing this complex. It bridges to the protein backbone through hydrogen bonds with Gly218. This example demonstrates how structural water molecules can participate in hydrogen bonding and satisfy otherwise uncompensated or undercompensated polar or charged groups. They can also have more subtle structural and electrostatic effects. Neglect or misplacement of structural water molecules often leads to improper scoring of the correct ligand conformational pose (the conformation, translation, and rotation of the ligand in the active site) and may result in selection of an incorrect pose (Huang and Shoichet, 2008). This can produce false positives and false negatives, leading to poor enrichment. False negatives are less of a problem for molecular design than other errors, but they represent lost opportunities and can be problematic when negative design is necessary for specificity.
Fig. 1.
The crystal structure of Factor Xa bound with the inhibitor ZK-807834 from PDB ID 1FJS. The inhibitor is displayed as atom-colored balls and sticks and the three water molecules are displayed as red balls and labeled (a), (b) and (c) in black. Important interactions are marked in green, distances are given in Å, and protein residues Asp189, Ser214, Gly218 and Ile227 are displayed as atom colored sticks and named in green. Some atoms are not shown, for clarity.
A number of methods have been devised to mark key water molecules within a crystal structure (Raymer et al., 1997; Amadasi et al., 2006) and also to calculate the probability that certain water molecules will be displaced on ligand binding (Garcia-Sosa et al., 2003; Barillari et al., 2007). This can provide very useful information, but deciding how to use the predictions wisely in a design context is not always clear. Within protein–ligand docking, algorithms such as GOLD have been modified to include this aspect by allowing each water molecule within the target structure to switch on and off and to rotationally reorient (Verdonk et al., 2005). Other methods have also been developed using this concept (Schnecke and Kuhn, 2000; Lippow et al., 2007). The approach shows some success, but it can only deal with water molecules whose positions are known ahead of time, and one cannot anticipate placement of unexpected water molecules. The method also suffers when water molecules shift subtly with different inhibitors. AutoDock includes the potential for considering structural water molecules by considering multiple target structures within one calculation, some with water molecules and some without (Osterberg et al., 2002). This again takes advantage of water molecules of known position within the site but does not account for new or shifted water molecules. FlexX has been modified using the concept of phantom particles (Rarey et al., 1999; Claussen et al., 2001). Water molecules are treated as spheres and an ensemble of favorably placed spheres is docked into the binding site. These spheres can then be switched on or off during the docking. Glide works similarly by docking explicit water molecules into each energetically competitive ligand pose and then rescores the complex (Friesner et al., 2004). These two techniques can account for unexpected water molecules, and we focus on such a technique in this paper. The improvements that the new method offers are two-fold. First, water molecules are grown from the ligand, and thus hydrogen-bonding groups are automatically aligned to make strongly favorable hydrogen-bonding interactions with at least one of the binding partners. This is a good determinant of affinity and very important in drug design. Second, we use the A* algorithm to generate an energetically ranked list of ligand poses and retain solutions that may be initially relatively poor. These can then be ranked much more highly when water molecules are added. The method also represents the first use of de novo water placement in a molecular design framework.
Displacement of water molecules from an active site has been noted in a number of drug development projects (Lam et al., 1996; Chen et al., 1998). The importance of water molecules is highlighted by the design of a set of novel inhibitors of tRNA-guanine transglycosylase (TGT) (Gradler et al., 2001). One of the main sources of interest in the crystal structures was the presence of two interstitial water molecules mediating the interactions between the ligands and the protein. One of these water molecules had not been observed at that site in any other complex of TGT or in the apo structure (Brenk et al., 2006). Structural water molecules also play an important role in the recognition of ligands by the heat-shock protein HSP90 (Yan et al., 2008). Four water molecules bridge the interaction between HSP90 and ADP (Obermann et al., 1998) and are found at similar locations in the complexes of HSP90 with the inhibitor geldanamycin (Stebbins et al., 1997) and PU3 (Wright et al., 2004). However, the inhibitor radicolol displaces one of these water molecules and noticeably shifts two of the others (Roe et al., 1999). The possibility of displaced or unexpected water molecules is clearly a very important concern in rational inhibitor design. However, current methods do not deal adequately with this problem. Unexpected structural water molecules have been incorporated into rational protein design. Baker and co-workers modeled a number of protein–protein complexes, with water molecules mediating interactions by extending side-chain rotamers to include optional water molecules appended to polar groups (Jiang et al., 2005). Selection of a solvated rotamer that was energetically favorable in the complex corresponded frequently to interfacial structural solvent. This method allowed placement of a number of structural water molecules at the barnase–barstar interface. However, the problem of solving this task is more difficult in the case of protein–ligand interfaces due to the greater diversity in molecular structure. There are only 20 commonly used amino acids, and there are significant data on each with respect to the preferred geometry of forming hydrogen bonds with water. In contrast, there is an enormous variety in the functional groups present in small molecules, and each has its peculiar set of preferred geometries. One possibility for modeling unexpected water molecules is to introduce structural water molecules only from the protein side of the interface using extended rotamers. Another is to introduce an extended approach to append water molecules to small-molecule ligands. A third is to combine both approaches. Here we have focused on the second approach, in the context of an inverse design method under development in this laboratory employing dead-end elimination (DEE) and A* using a physical potential function (Altman et al., 2008). This approach has the advantage that it models water molecules near the ligand, where they are most relevant to the design. More distal water molecules are known to affect binding, but do not make direct bridging hydrogen bonds between the protein and the ligand.
One of the main difficulties in placing structural water molecules in protein–ligand complexes is the deviation from ideal hydrogen-bond geometry involving water molecules. This makes systematic placement problematic and requires extensive sampling of the possible geometries for each potential interaction. Analysis of ten complexes from the PDB highlights this variability. Some of the exact distances and angles cannot be measured exactly due to the lack of hydrogen atoms in the structure. However, we estimated their values by using CHARMM's HBUILD function (Brooks et al., 1983; Brünger and Karplus, 1988) to build protein hydrogen atom positions, assuming standard bond lengths and angles and searching for favorable interactions. This allowed all distances and angles to be measured. The data for distances and angles between ligands and water molecules can be seen in Table I. The hydrogen-bond lengths span a large range for water both as donor and acceptor. The average hydrogen-bonding distance with water molecules within protein structures is ∼1.9 Å, as noted previously in the context of protein design (Jiang et al., 2005). However, there is significant variation, with the majority of the distribution lying in the range 1.7–2.1 Å. This variation must be considered in design. The range of bond angles is even more diverse and varies depending on the nature of the hydrogen bond. It is interesting to note that in cases where the hydrogen bond acceptor is an sp2 center, the angle about the acceptor atom ranges from ∼120° (predicted by orbital considerations) to ∼180° (predicted by simple Coulombic and steric considerations). The mean value of 150° lies exactly between these two extremes. The distances used by Baker and co-workers when performing protein design match these values (Jiang et al., 2005). The distances they used vary between 1.70 and 2.20 Å and the angles vary between 120° and 180°. This large range of geometries requires a method that can evaluate a variety of poses in the face of the associated combinatorial complexity. Here we describe such a method and detail the process we use to evaluate its performance.
Table I.
The range of hydrogen-bonding distances and angles observed between ligands and structural waters in 10 complexes from the PDB, showing donor (D) and acceptor (A) atomsa
| Bond parameter | Minimum observed value | Maximum observed value | Mean value |
|---|---|---|---|
| A–-H–OH distance | 1.72 Å | 2.26 Å | 1.87 Å |
| D–-OH2 distance | 1.70 Å | 2.04 Å | 1.87 Å |
| A1–-A2–-H–OH angle (sp2) | 113.5° | 174.9° | 149.4° |
| A1–-A2–-H–OH angle (sp3) | 107.5° | 124.6° | 116.0° |
| D1–D2––OH2 angle | 140.4° | 168.9° | 154.5° |
| A––H–-OH angle (sp2) | 93.2° | 175.0° | 140.9° |
| A–-H–-OH angle (sp3) | 99.3° | 160.0° | 127.7° |
| D–-O(H)–-H angle | 82.2° | 171.1° | 115.5° |
aThe hydrogen bond distances being measured are shown as dashed lines between the two atoms that form the bond. The hydrogen bond angles being measured are shown as dashed lines between the three atoms that form the angle. The atoms at the vertices of the angles are underlined.
Materials and methods
The method developed in this study, for incorporating structural water molecules within molecular design, builds on previous work from this laboratory. The existing inverse-design approach has been applied to the rational design of HIV-1 protease inhibitors, which have shown subnanomolar binding in experimental validation (Altman et al., 2008). Here we describe the process used to prepare the molecular structures, create an ensemble of scaffold poses, combine these with a set of side groups and then place energetically favorable bridging water molecules at the protein–ligand interface.
Design methodology
The existing design approach considers a library of potential–ligands created combinatorially from a set of scaffolds and side groups (Altman et al., 2008; Huggins et al., 2009). Each chosen scaffold is first placed systematically in the binding site in many different acceptable conformations and orientations. All scaffolds contain a number of attachable positions where side groups can be substituted, generally at hydrogen-atom positions. Each attachable position is associated with a library of side groups, each member of which exists as a set of discrete rotameric conformations. This problem framing is analogous to that used for inverse protein design (Drexler, 1981; Pabo, 1983; Ponder and Richards, 1987; Desmet et al., 1992; Dahiyat and Mayo, 1997; Hellinga, 1997; Leach and Lemon, 1998; Kuhlman et al., 2003; Lippow et al., 2007). We use a pairwise-decomposable physical-potential function with the DEE (Desmet et al., 1992; Pierce et al., 2000) and A* (Leach and Lemon, 1998) algorithms to prune poor scoring compound poses to produce an energetically ranked list of the best-computed binders. This ranked list is then cut down, such that only solutions within a cutoff of the global minimum energy conformation (GMEC) are retained. This set is then re-evaluated using more sophisticated energy functions, which need not be pairwise additive, yielding a set of compounds predicted to bind tightly. The method is described in detail in a study performed in an engineered binding site (Huggins et al., 2009). In the remainder of this section, we discuss the methodological improvements developed to incorporate de novo placement of structural water molecules, describe the process employed for evaluation, outline the design process used and detail differences from our prior work.
Evaluation of ligand and water placement
We chose to evaluate the water placement method in two ways. The first evaluation involved the placement of structural water molecules in known complexes using an ensemble of ligand placements. This omitted the ligand selection stage and focused solely on the ability of the algorithm to place each known ligand and its associated water molecules correctly. This was a useful test as it evaluated the process of ligand and water placement, disentangling it from the evaluation of the process of side group selection and placement. The second evaluation involved using the inverse design method to grow a set of ligands from a scaffold and a number of side groups, including water molecules, to construct each complex. This assessed the ability of the algorithm to incorporate water molecules within the design methodology.
For the first stage of the evaluation, we selected five complexes from the protein data bank (Berman et al., 2000) for testing. Multiple factors were considered when making the choice of which complexes to test and which particular structure to use. Each was from a high-resolution crystal structure to yield an accurate model of the complex. Each complex contained between three and six water molecules in close association with the ligand and, more importantly, the set of water molecules across all the complexes provided a variety of geometries to test the design protocol rigorously. We were also interested in evaluating the ability of the methodology within a drug-design framework, and thus all five complexes are linked with drug discovery. The five complexes are detailed in Table II. Each complex was prepared in the manner detailed below and subjected to three different analyses. In the first analysis, all the water molecules from the crystal structure were retained and many ligand conformations and placements were generated within the site. Each of these was analyzed and scored. This is termed as the explicit water case. In the second analysis, all the water molecules from the crystal structure were deleted prior to pose generation. This is termed as the omitted water case. In the third analysis, all the water molecules were deleted prior to pose generation, but the water placement design was performed on the ensemble of ligand poses. This is termed as the designed water case. A comparison of these three cases permitted us to explore the effect of ignoring bridging water molecules and the ability of the method to compensate for this by replacing them.
Table II.
The five test cases from the PDB that were used in the first stage of the evaluation
| PDB IDa | 1VZQ | 1KI8 | 1NNC | 1JIO | 1DF7 |
|---|---|---|---|---|---|
| Protein | Thrombin | Thymidine Kinase | Neuraminidase | Cytochrome P450 | Dihydrofolate Reductase |
| Ligand | SHY | 5-Bromovinyl Deoxyuridine | Relenza | 6-Deoxy Erythronolide B | Methotrexate |
| Therapeutic relevance | Blood Clotting | Herpes | Influenza | Drug Metabolism | Tuberculosis |
| Crystal water molecules | 6 | 3 | 4 | 3 | 4 |
| Resolution (Å) | 1.54 | 2.20 | 1.80 | 2.10 | 1.70 |
| Prior usesb | FlexX, AutoDock, GOLD | FlexX, AutoDock, GOLD |
aThe references for the test cases are 1VZQ (Scharer et al., 2004), 1KI8 (Champness et al., 1998), 1NNC (Vonitzstein et al., 1993), 1JIO (Cupp–Vickery et al., 2001) and 1DF7 (Li et al., 2000).
bThe reference for the prior uses for FlexX, AutoDock and GOLD is (de Graaf et al., 2005).
Ligand and water placement in design
The second evaluation of this method involved using the water placement scheme within a molecular-design framework. The design was accomplished by performing two rounds of DEE and A*. In the first round, a variety of side groups was grown on a particular scaffold and; in the second round, the resulting molecules were then subjected to solvation by growing water molecules. Growable water positions were placed on both the scaffold and the side groups during design, such that the entire resulting molecule was able to make interactions with water. For the purposes of validation, we selected the test case of the oligopeptide binding protein (OppA), which has been crystallized in complex with all the tripeptides KXK, where X represents a naturally occurring amino acid (Sleigh et al., 1999). Water is known to be very important in allowing many different tripeptides to bind (Tame et al., 1996). We treated the backbone of the tripeptide as the scaffold and used side-chain libraries containing only lysine at the terminal positions and a library containing the 20 common amino acids, except for proline, at the central position. Histidine was modeled in all three of the protonation states that occur near physiological pH. This yielded a total of 21 substitutable side chains. The scaffold and each of the side-chains were decorated with growable water positions before beginning.
To assess the ability of the algorithm to place the ligand in the correct orientation, each of the crystal structures of the tripeptides bound with OppA was rotated onto the crystal structure of OppA in complex with the tripeptide KNK from PDB ID 1B5I (Sleigh et al., 1999). Fitting was performed using the McLachlan algorithm (McLachlan, 1982) as implemented in the program ProFit (Martin, A.C.R., http://www.bioinf.org.uk/software/profit/). Only backbone atoms were considered. The structures are all very similar, with a maximum root mean square deviation (RMSD) value of 0.394 Å between structures. Rotation into the same coordinate frame facilitated an estimate of the accuracy of prediction. For comparison, we also completed a design with only one round of DEE, omitting the water molecule placement stage. This allowed us to evaluate the effect of the water placement scheme on the results of the design.
Ligand preparation
The small-molecule ligand structures from all test cases were prepared with an identical process. The ligand coordinates were taken from the PDB files along with any hetero-groups and co-factors. Hydrogen atoms were added with GaussView (Frisch et al., 2003) and the molecules were then geometry optimized with Gaussian03 (Frisch et al., 2004) using the restricted Hartree–Fock model employing the 3-21G basis set. The resulting structures were then re-evaluated with a single-point Gaussian03 energy calculation using the restricted Hartree–Fock model with the 6-31G* basis set. The partial atomic charges were then assigned using a RESP fit (Bayly et al., 1993; Cornell et al., 1993). Previous work has shown that this scheme performs well compared with performing both the minimization and single-point calculation with the 6-31G* basis set (Green and Tidor, 2003). For the purposes of calculating total molecular charges, all guanidine and amidine groups were considered protonated and all carboxylate groups were considered deprotonated. The pteridine ring of methotrexate from PDB ID 1DF7 was considered protonated, as indicated in previous studies (Bennett et al., 2005). The SHY inhibitor from PDB ID 1VZQ contains a tertiary amine and such groups are generally protonated at neutral pH. However, the molecule possesses another site that is positively charged, which could depress the amine pKa. A pKa calculation performed using Gaussian03 gave a pKa of 6.4 and this amine was not protonated for this work.
For the five test cases omitting the ligand design stage, the scaffold used was simply the entire molecule. In the OppA test case, the scaffold was created by trimming the side-chains of the KNK tripeptide from PDB ID 1B5I to yield just the peptide backbone (Sleigh et al., 1999). No side groups are necessary for the five test cases that omitted the ligand design stage. In the OppA test case, side-chain libraries were created from the standard CHARMM22 amino acid parameters for each residue. The residues were modified to remove all backbone atoms and then treated in the same manner as the scaffolds. This yielded geometry-optimized structures with defined partial atomic charges for each amino acid side-chain. A set of conformations was then created systematically for each scaffold and for each side-chain by rotating each rotatable torsion angle in increments of 30°. Van der Waals radii were scaled by 0.75 and each conformation where the scaled radii of any two non-bonded atoms overlapped was discarded. For the entire work with small molecules, CHARMm22 torsional parameters and van der Waals parameters were used throughout (Momany and Rone, 1992). This yielded an ensemble of conformations without steric clashes for the scaffold and the side groups.
Protein preparation
The protein structures from all test cases were prepared with an identical process. Coordinates for the protein and all the water atoms were taken from the PDB and the ligand was removed from the bound state. The protein and water hydrogen-atom positions were built using the HBUILD facility (Brünger and Karplus, 1988) of the CHARMM program package (Brooks et al., 1983) with the CHARMM22 energy function (MacKerell et al., 1998). All asparagine and glutamine residues were then checked manually for potential hydrogen bonding and analyzed by NQ-Flipper (Weichenberger and Sippl, 2006). For 1DF7, Gln28 was altered and for 1JIO, Asn89 was altered by swapping the coordinates of the nitrogen and oxygen atoms to improve the hydrogen-bonding patterns. Histidine residues were examined for orientation and protonation state in the same manner. For 1KI8, His58 and His213 were chosen as epsilon protonated. For 1NNC, His184 and His274 were chosen as epsilon protonated. For 1VZQ, His91, His119 and His230 were chosen as epsilon protonated. For 1JIO, His13 and His330 were chosen as epsilon protonated. All other histidines were assigned as delta protonated. The residues lysine, arginine, aspartate, glutamate, cysteine and tyrosine were also analyzed in this manner to examine their protonation state. There was no evidence of any unusual protonation states, and thus all lysine and arginine residues were assigned as positively charged, all aspartate and glutamate residues were assigned as negatively charged and all cysteine and tyrosine residues were assigned as neutral. From the revised structures, all the hydrogen-atom positions were then rebuilt using HBUILD (Brooks et al., 1983). The final structures were then rotated to provide advantageous grid resolution for finite-difference continuum electrostatic calculations and the protein atoms were assigned PARSE charges (Sitkoff et al., 1994) for use with Delphi (Gilson and Honig, 1988; Sharp and Honig, 1990a; Sharp and Honig, 1990b). The active site was defined as in previous studies to create a shape inside which the design was performed (Huggins et al., 2009). Precomputations for a grid-based estimation of van der Waals and continuum electrostatic interactions were then carried out with Delphi. The methodology for creating these grid-based estimates was as described previously (Huggins et al., 2009). In this case, we used a van der Waals grid spacing of 0.2 Å and an electrostatic grid spacing of 0.75 Å. For all Delphi calculations, we used a salt concentration of 0.145 M, a solvent dielectric of 80 and an internal dielectric constant of 4. For each test case, two sets of active site shapes and energy grids were created. One set with water present was used for the explicit water case and another set with no water present was used for the designed water case and for the omitted water case.
Scaffold pose generation
The first stage of scaffold pose generation was the creation of a large number of low-energy poses within the binding site for further analysis. Each conformation of every scaffold pose within the ensemble was placed into the site. For the current work, we focused the search by limiting the initially placed scaffold to the neighborhood around the crystal structure position. We placed the centroid of the search at the centroid of the scaffold crystal structure (using heavy atoms only) and searched within a Cartesian grid of side 4.0 Å. We considered this resource conserving approach to be acceptable as the goal was to examine the quality of water molecule placement and ligand scoring. More distant searches would be necessary to ascertain that other structures do not score well as false positives, although this is unlikely. In any new application of this approach, crystal structures of analogs may well be available and core placements can be based on these. Each conformation of every scaffold was then subject to systematic placement in the defined grid with a translational enumeration of 0.2 Å and rotational enumeration such that the maximum arc length of atoms from the centroid swept out a distance of 1.0 Å between orientations. Scaffold poses were discarded if their calculated van der Waals binding energy was >15.0 kcal/mol or the 0.75-scaled radii of any two non-bonded atoms overlapped. For the five test cases which omitted the ligand design stage, the scaffold used was simply the entire molecule. For the OppA test case, the scaffold used was the peptide backbone. For the explicit water case and the omitted water case, the results of this stage were passed straight to the binding free energy calculation. For the designed water case and the OppA test case, water molecules were placed in the site prior to the binding free energy calculation.
Water placement
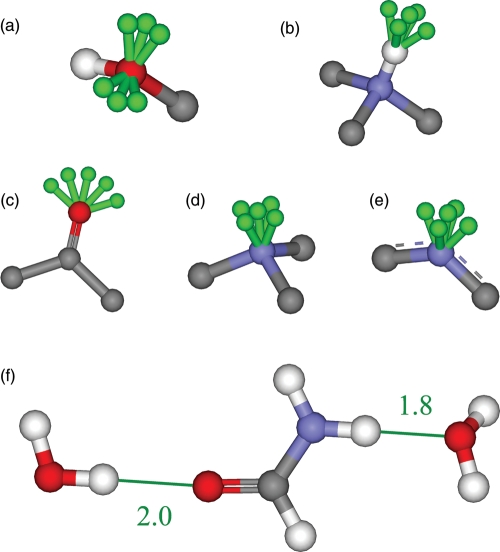
The next step was analyzing each ligand and flagging atoms with the potential to form hydrogen bonds with water. These include all oxygen atoms, all polar-hydrogen atoms, all free sp2-hybridized nitrogen atoms and all unprotonated amine nitrogen atoms in both the scaffold and the added side groups. It is very important to consider a variety of different hydrogen-bonding geometries as these can vary significantly. The angles and distances that were used for each atom type were based on statistics from known structures. Water atoms were constructed using discrete allowed geometries with pseudo-bond lengths and angles. We allowed two hydrogen-to-acceptor pseudo-bond lengths of 1.8 and 2.0 Å. The angles were as defined in Table III and the resulting geometries are illustrated in Fig. 2a. Ligand acceptor atoms were elaborated by placing the first hydrogen atom of the water molecule at the end of the pseudo-bond and the water oxygen atom diametrically opposite to the acceptor. The second hydrogen atom of the water molecule was placed by sampling at 30° around the pseudo-bond. The donor hydrogen atoms were elaborated by placing the water oxygen atom at the end of the pseudo-bond. The first hydrogen atom of the water molecule was placed using an angle of 109.5° between the donor hydrogen atom, the water oxygen atom and the water hydrogen atom, sampling at 30° around the pseudo-bond. The second hydrogen atom of the water molecule was also placed using an angle of 109.5°, to create an approximately tetrahedral geometry around the oxygen. Both of these angles vary in observation, as shown in Table I, but were not varied in this initial study. All water molecules had equal bond lengths of 0.96 Å and bond angles of 104.5° (Csaszar et al., 2005). The two types of water molecule placements are illustrated in Fig. 2b.
Table III.
The five atom types considered when making hydrogen bonds and the geometries around these atoms where growable positions are placed, as illustrated in Fig. 2
| Defined | One | Two | Three | Four | Five | |
|---|---|---|---|---|---|---|
| Hydrogen | In/out-of-plane angles | 210°/180° | 150°/180° | 180°/180° | 180°/210° | 180°/150° |
| sp3 oxygen(±)a | In-plane angle | 89.5 | 109.5 | 129.5 | NA | NA |
| sp2 oxygen | In-plane angle | 120° | 150° | 180° | 210° | 240° |
| Amine nitrogen | In/out of plane angles | 200°/180° | 160°/180° | 180°/180° | 180°/200° | 180°/160° |
| sp2 nitrogen | In/out-of-plane angles | 210°/180° | 150°/180° | 180°/180° | 180°/210° | 180°/150° |
aThe sign in brackets indicates that the bond vector can point in two opposite directions away from the plane, yielding six growable positions from an sp3-hybridized oxygen atom.
Fig. 2.
A diagram of the spatial placement of water molecules. The five atom types considered when making hydrogen bonds: sp3-hybridized oxygen atom (a), polar hydrogen atom (b), sp2-hybridized oxygen atom including carbonyl oxygen atoms but excluding aromatic oxygen atoms (c), unprotonated amine nitrogen atom (d) and sp2-hybridized nitrogen atom including aromatic nitrogen atoms (e). Hydrogen atoms are colored white, oxygen atoms are colored red, nitrogen atoms are colored blue, carbon atoms are colored gray and the growable positions for each case are colored green. Each green growable position represents a direction vector and each water molecule was placed at a discrete position along these vectors. Extra atoms are shown for clarity. Two water molecules grown from the molecule formamide (f). One water molecule is grown from the carbonyl oxygen atom and the other is grown from the amide hydrogen atom. Distances in Å are displayed in green.
Optimization
For the five test cases omitting the ligand design stage, only the water optimization stage was necessary. For the OppA test case, each pose created in the scaffold pose generation stage was treated as rigid and was first subjected to an optimization of the side groups. The DEE/A* algorithm was run, placing all side groups in all conformations and combinations on each scaffold pose. The bonds joining the scaffold to the side groups are all sp3–sp3 linkages and thus the dihedral angle of the join was enumerated at energetically favored angles; 50°, 60°, 70°, 170°, 180°, 190°, 290°, 300° and 310°. The binding free energy was computed for all the species created with the low-resolution energy function detailed in previous work (Huggins et al., 2009). Enumeration yielded an energy-ranked list of molecules with the guarantee that no solutions were missed. For every scaffold pose, only the lowest-energy conformation of each particular designed molecule was retained. All of the solutions from this design were then used as the scaffold molecules for the water optimization stage. This was performed by placing water molecule using the DEE/A* routine with the low-resolution energy function. All combinations of water molecules were considered, where a water molecule could be absent or could be present at any combination of allowed lengths and angles around a potential hydrogen bonding site. The interaction energy was calculated between every allowed water molecule and the protein. In many cases there were nearly 100 growable water positions, making the A* algorithm computationally unmanageable. Thus, only the GMEC was calculated using DEE. This step yielded a set of complexes of the protein with the ligand and a set of water molecules. The DEE optimization of the energy ensured that the water made strong interactions with the protein and the use of near-optimal geometries ensured that the water made strong interactions with the ligand. Once this set of complexes had been created, the next stage was to determine the actual interaction energy we wished to calculate, between the ligand and the water/protein complex.
Binding free energy calculation
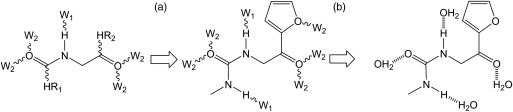
For the OppA test case, we limited the number of molecules carried from the side group optimization stage to the water molecule placement stage. We initially chose to retain solutions within 15 kcal/mol of the GMEC. However, though the algorithm was able to design all 21 side groups within the binding site of the KNK-bound structure, there were no instances of the tripeptide KWK within 15 kcal/mol of the GMEC. This was due to the steric constraints of fitting the large tryptophan side-chain within the site. To avoid this problem, the A* algorithm was used to collect the 500 lowest-energy instances of each designed tripeptide, which were retained for the water placement stage. This permitted us to perform the second level of design with each tripeptide. These same 500 lowest-energy instances of each designed tripeptide were also sent directly for high-level analysis without water placement for comparison. For the five test cases omitting the side group placement stage, each test case created a different number of results with a different spread of energies. For the water placement stage, only a subset of the results was retained for high-level analysis, due to computational cost. We retained the subset of solutions within 5 kcal/mol of the GMEC for every design. We then increased the size of each subset by incrementally raising the energy cut off above the GMEC by 5 kcal/mol, until the size of the subset was >50 000 molecules. We then used a more sophisticated energy function to re-score the interaction. This involved using CHARMM energy minimization on the ligand and water coordinates, a separate desolvation calculation using Delphi, a buried surface area term and a calculation of the internal energy deformation penalty. This scoring function proved useful in the design of a set of high-affinity HIV-1 protease inhibitors (Altman et al., 2008) and was found to perform well in an evaluation using a standard test case (Huggins et al., 2009). The entire design process thus occurred in two stages. The first stage was an inverse design using DEE/A* to create a set of protein–ligand complexes with favorable intermolecular interactions. This set of complexes was then further arrayed by placing water molecules in favorable placements to make interactions with both the protein and the ligand. This two-stage process is illustrated in Fig. 3. The final results were evaluated by calculating the binding free energy to yield a set of protein–ligand complexes with the incorporation of bridging water molecules not in the original complex. For the final binding free energy calculations, the designed water molecules are considered to be part of the protein.
Fig. 3.
Schematic illustrating the process of design incorporating solvation of the ligand. The symbols HR1 and HR2 represent functional group positions. The symbol W1 represents a growable water position from a hydrogen-bond donor and the symbol W2 represents a growable water position from a hydrogen-bond acceptor. Wavy lines indicate a potential bond and dashed lines indicate a hydrogen bond. The first stage of design involves growing functional groups (a) and the second involves growing water molecules (b).
Results
Ligand placement results
The aim of this work was to create and test a method for placing water molecules within a binding site, as part of an inverse molecular design algorithm. We first evaluated the ability of the algorithm to place and score ligands within their native binding site, performing three separate calculations. One was performed with pre-existing water molecules, one without water molecules and one allowing water molecules to be placed by the method. Table IV shows the scores of the lowest-energy conformations for each design and the heavy-atom RMSD of the computed ligand conformation with the crystal structure. The results show that the designed water scheme compares well with using explicit water molecules in design in terms of ligand placement. The RMSD of the ligand using the designed water scheme and the omitted water scheme is comparable with that when using the explicit water scheme in all cases. However, in each case, the energy of the designed water scheme is similar to the explicit water scheme and substantially lower than the omitted water scheme. This is significant because accurate scoring of hits and decoys is very important in molecular design methods. Any errors in scoring will be translated into poorer enrichment. In some cases the designed water scheme predicts a lower energy than is found for the explicit water case. This occurs because more water molecules are placed by the design than are seen in the crystal structure (see Table V). In reality, there is an energetic cost to fixing a water molecule that is not considered in the scoring, and thus the design may place too many water molecules.
Table IV.
The scores of the lowest-energy conformation and the heavy-atom RMSD against the crystal structure, showing results of the three water treatments for the five test cases
| Complex | Water treatmenta | Lowest energy (kcal/mol) | RMSD versus crystal (Å) |
|---|---|---|---|
| Designed | −65.88 | 0.54 | |
| 1DF7 | Explicit | −64.65 | 0.41 |
| Omitted | −57.23 | 0.46 | |
| Designed | −52.11 | 0.71 | |
| 1JIO | Explicit | −51.44 | 0.46 |
| Omitted | −44.87 | 0.75 | |
| Designed | −33.22 | 0.46 | |
| 1KI8 | Explicit | −37.71 | 0.33 |
| Omitted | −30.58 | 0.45 | |
| Designed | −41.92 | 0.42 | |
| 1NNC | Explicit | −38.36 | 0.55 |
| Omitted | −31.32 | 0.92 | |
| Designed | −54.41 | 0.25 | |
| 1VZQ | Explicit | −59.41 | 0.31 |
| Omitted | −43.92 | 0.27 |
aIn the explicit case, all the water molecules from the crystal structure were retained. In the omitted case, all the water molecules from the crystal structure were deleted. In the designed case, all the water molecules were deleted, but the water placement design was performed on the ensemble of ligand poses.
Table V.
For each test case, the table shows the number of water molecules predicted, the fraction of total water molecules correctly predicted (RMSD < 2.0 Å), the total RMSD of the water molecules predicted, and the individual RMSD values for each water molecule
| Complex | Waters Predicted | Fraction Correct | RMSD (Å)a |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Total | 1 | 2 | 3 | 4 | 5 | 6 | |||
| 1DF7 | 10 | 3/4 | 1.29 | 0.50 | 1.69 | 1.37 | NA | NA | NA |
| 1NNC | 6 | 1/3 | 0.99 | 0.99 | NA | NA | NA | NA | NA |
| 1JIO | 4 | 3/3 | 0.94 | 1.48 | 0.62 | 0.32 | NA | NA | NA |
| 1KI8 | 1 | 1/3 | 0.32 | 0.32 | NA | NA | NA | NA | NA |
| 1VZQ | 8 | 5/6 | 0.74 | 0.32 | 0.36 | 1.47 | 0.42 | 0.44 | NA |
aWaters not predicted or not present are marked as not applicable (NA).
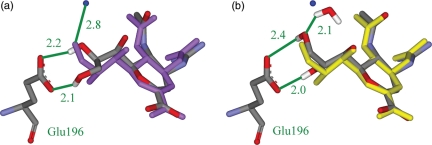
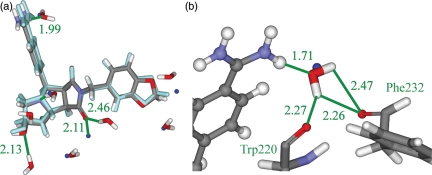
It is interesting to examine the case of neuraminidase bound with the inhibitor Relenza from PDB ID 1NNC. In this case, the designed water scheme does a better job of predicting the crystal-structure conformation (RMSD 0.42 Å) than the omitted water scheme (RMSD 0.92 Å). This can be seen in Fig. 4, which highlights the problem of performing docking or molecular design without water molecules. The ligand placement using the omitted water scheme is shown in Fig. 4a. The portion of the molecule on the left-hand side is predicted incorrectly, as one of the hydroxyl groups cannot interact with the water molecule that should be there. Instead, it makes an interaction with Glu196, which is less favorable than the interaction with water (see Table IV). The designed water scheme is shown in Fig. 4b. The algorithm places a water molecule in the binding site close to the correct location, allowing the ligand to make the correct interaction. This yields a smaller RMSD of prediction and a lower interaction energy. This is precisely the issue that this designed water scheme attempts to address.
Fig. 4.
The crystal structure of neuraminidase bound with the inhibitor Relenza from PDB ID 1NNC. The crystal-structure pose, overlaid with the predicted structure using the omitted water scheme (a). The true pose is displayed as purple balls and sticks and the designed pose is displayed as atom-colored balls and sticks. One true water molecule is displayed as a blue ball. The crystal-structure pose, overlaid with the predicted structure using the designed water scheme (b). The true pose is displayed as yellow balls and sticks and the designed pose is displayed as atom-colored balls and sticks. One true water molecule is displayed as a blue ball. One designed water molecule and the protein residue Glu196 are displayed as atom-colored sticks. Important interactions are marked, distances are given in Å, and the protein residue is named in green. Some hydrogen atoms are omitted for clarity.
Water placement results
The calculations performed on the five test cases were also analyzed to test the ability of the algorithm to place water molecules correctly within the site. A water molecule was considered to be placed correctly if the predicted water oxygen atom was within 2.0 Å of a crystal-structure position. This generates a sphere of volume 33.5 Å3 which may be too large to truly identify that the correct water molecule has been placed. However, a cut off of 2.0 Å for heavy atom RMSD is commonly used to measure ligand similarity (Jones et al., 1997; Morris et al., 1998) and in this case provides a good estimate of whether the correct hydrogen-bonding bridging interaction is being made. We also placed a restriction that a predicted water molecule could only satisfy one of the crystal-structure water molecules. The results are given in Table V. The designed water scheme placed water molecules correctly and scored them well in the great majority of cases. This means that the water molecules interact favorably with the protein during the initial DEE design and then favorably with the ligand in the higher-level scoring functions. One concern is that too many water molecules will be predicted and that this will be detrimental to the design. Certainly, more water molecules are generally predicted than are actually found, but this does not appear to affect the results adversely, as seen in Table IV. Analysis of these extra water molecules shows that they are commonly placed away from the protein, out into the solvent, and do not make significant interactions with the protein. They contribute a small amount to the calculated binding energy, but they are not likely to contribute to the true binding energy as, even if they are fixed to some degree, the interactions can occur in the solvent as well as in the complex. It is interesting to note that many of the water molecules were placed correctly, despite the small range of bonds and angles used. In particular, the angle about the hydrogen-bonding atom of the water shows a huge variation, as shown in the last two columns of Table I, but was not varied in this model. Further work exploring more water placements may increase performance and allow more bridging interactions to be identified correctly.
Analysis of case 1VZQ
It is interesting to analyze two of the cases from this design in terms of water placement. The design from PDB ID 1VZQ yielded very good results, placing five of the six water molecules from the crystal structure correctly, four of them with a distance of below 0.5 Å (Fig. 5a). The five correctly predicted water molecules are placed well and the remaining three predicted water molecules make sensible hydrogen-bonding interactions with the ligand. Analysis of the exact nature of the interactions shows that the designed water scheme is able to generate good hydrogen-bonding interactions with both the ligand and the protein (Fig. 5b). The one true water molecule that is not predicted correctly in this case is not predicted in any of the designs at any level of analysis. When the crystal structure is analyzed, it appears that this water molecule makes no hydrogen bonds with the protein but may interact with the π orbital of Tyr47. Further development of this method should consider the possibility of growing water molecules from π orbitals, as this type of interaction is not uncommon. The water oxygen is also very close to the carbonyl oxygen of the ligand (2.11 Å), suggesting a highly non-linear hydrogen bond between them. This is unlikely to be predicted by our methodology, which begins with linear hydrogen bonds in the sense of the hydrogen atom lying between the donor and the acceptor. However, accurate predictions of weak interactions such as this are less important in terms of binding free energy than strong interactions and, within molecular design, it is important to design strong interactions, such as those that are the focus of the current implementation of this method.
Fig. 5.
The crystal structure of thrombin bound with the inhibitor SHY from PDB ID 1VZQ. The crystal structure pose, overlaid with the predicted pose from the designed water scheme (a). The true pose is displayed as cyan balls and sticks and the designed pose is displayed as atom-colored balls and sticks. Six true water molecules are displayed as blue balls and eight designed water molecules are displayed as atom-colored sticks. A close-up of the interaction between a predicted water molecule and the amidine group of the inhibitor (b). The predicted ligand pose is displayed as atom-colored balls and sticks. The predicted water molecule and the protein residues Trp220 and Phe232 are displayed as atom-colored sticks. The true water molecule from the crystal structure is displayed as a blue ball. Important interactions are marked, distances are given in Å, and protein residues are named in green.
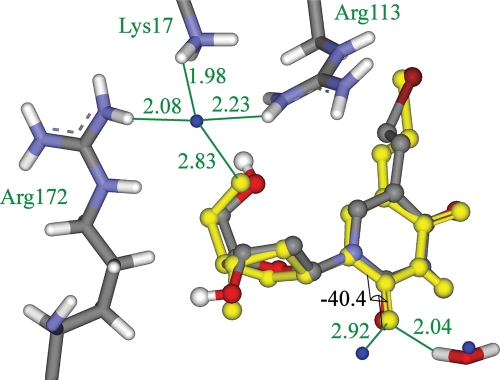
Analysis of case 1KI8
The design from PDB ID 1KI8 yielded relatively poor results, placing only one of the three water molecules from the crystal structure correctly. This test case was uniformly poorly predicted. None of the top five solutions predicted more than one more water molecule correctly. The binding site is shown in Fig. 6. The first water molecule has an interesting problem. It is not found in the conformational search because the interactions it makes with the protein are too tight. It forms very close interactions with residues Lys17, Arg113 and Arg172 and there is only an extremely small channel in which the water molecule can be placed favorably. In this case, the rotamer search is not fine enough to locate this channel and thus no water molecule is placed here. This water molecule is not predicted in previous studies on this test case (de Graaf et al., 2005). This is a case where an explicit water molecule modeled in the site would perform better. The second water molecule has a different issue. In this case, the problem is the out-of-plane angle of 40.4° between the water molecule and the carbonyl groups of the ligand (see Fig. 6). The current implementation of the algorithm only searches in-plane interactions for carbonyl groups and thus misses this interaction. This water is also not predicted in previous studies on this test case (de Graaf et al., 2005). It is certainly computationally feasible to include more growable positions within the designs, and this is an issue that will be addressed in further development. Despite some deficiencies, the results from the first stage of validation show that overall the method can place water molecules in physically realistic positions and make good interactions with both the ligand and the protein.
Fig. 6.
The crystal structure of thymidine kinase bound with the inhibitor 5-bromovinyl deoxyuridine from PDB ID 1VZQ. The crystal structure of the ligand pose, overlaid with the predicted pose from the designed water scheme. The true pose is displayed as yellow balls and sticks and the designed pose is displayed as atom-colored balls and sticks. Three true water molecules are displayed as blue balls and one designed water molecule is displayed as atom-colored sticks. Protein residues Lys17, Arg113 and Arg172 are displayed as atom-colored sticks. Important interactions are marked, distances are given in Å, and protein residues are named in green. A dihedral angle from the true pose is marked and the value is displayed in black. Some hydrogen atoms are omitted for clarity.
Water placement in design
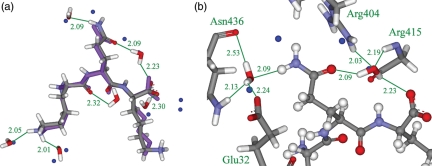
The results of the tripeptide design, with respect to water placement, can be seen in Table VI. The algorithm predicts approximately half of the crystallographic water molecules correctly in each case, without predicting an unreasonable total number of water molecules. The majority of the predicted water molecules interact with the side-chains of the tripeptide, as only three water molecules form hydrogen bonds with scaffold atoms. The distances between the predicted and true water molecule positions are also very good, with many predictions <0.5 Å and many averages below 1.0 Å. This is illustrated by the predicted and actual structures of the tripeptide KQK shown in Fig. 7. The method places six of the water molecules correctly, including two of the water molecules interacting with the glutamine side-chain. All water molecules make good hydrogen bonds with both the ligand and the protein. The results of this validation show that this methodology is able to place structural water molecules sensibly and correctly.
Table VII.
The heavy atom RMSD and the scores of the lowest-energy conformation for all 21 tripeptides KXK
| Amino acid X (KXK)a | RMSD (Å) | Energy (kcal/mol) |
|---|---|---|
| ALA | 0.42 | −66.73 |
| ARG | 0.74 | −68.59 |
| ASN | 0.98 | −70.49 |
| ASP | 1.04 | −66.07 |
| CYS | 0.67 | −68.88 |
| GLN | 0.63 | −75.01 |
| GLU | 1.28 | −67.75 |
| GLY | 0.50 | −64.18 |
| HIS(D) | 0.83 | −72.97 |
| HIS(E) | 0.64 | −73.40 |
| HIS(P) | 0.82 | −69.30 |
| ILE | 0.39 | −70.29 |
| LEU | 0.85 | −51.07 |
| LYS | 0.69 | −77.02 |
| MET | 0.58 | −71.51 |
| PHE | 0.78 | −73.04 |
| SER | 0.68 | −64.60 |
| THR | 0.78 | −71.21 |
| TRP | 1.66 | −52.79 |
| TYR | 0.81 | −72.20 |
| VAL | 0.58 | −71.42 |
aHistidine was modeled in its three physiologically relevant states, delta protonated (D), epsilon protonated (E) and doubly protonated (P).
Fig. 7.
The crystal structure of OppA bound with the tripeptide KQK from PDB ID 1B5I. The crystal structure pose, overlaid with the predicted pose from the designed water scheme (a). The true pose is displayed as purple balls and sticks and the designed pose is displayed as atom-colored balls and sticks. Ten true water molecules are displayed as blue balls and eight designed water molecules are displayed as atom-colored sticks. A close-up of the interaction between the protein and the predicted ligand pose (b). The predicted ligand pose is displayed as atom-colored balls and sticks. Protein residues Glu32, Arg404, Gly415 and Asn436 and eight designed water molecules are displayed as atom-colored sticks. Four true water molecules are displayed as blue balls. Important interactions are marked, distances are given in Å, and protein residues are named in green.
Ligand placement in design
The results of the complete design for ligand placement can be seen in Table VII. The designed water scheme performed very well in terms of ligand placement, predicting the position of every ligand within an RMSD of 2.0 Å. It also performed well in terms of scoring, predicting a favorable binding free energy for each tripeptide. The energetic predictions were reasonable, with the known weaker binders KDK, KGK, KRK and KLK being predicted to bind weakly (> −69.0 kcal/mol) and the known stronger binders KQK, KVK, KFK and KMK being predicted to bind well (< −71.0 kcal/mol). One issue is that the predicted energy for KKK (−77.02 kcal/mol) is considerably stronger than might be expected, as experiments mark it as one of the weaker binders (Sleigh et al., 1999). This is likely to be due to inaccuracies computing solvation energetics of charged amines. It is known that atomic charges for amines calculated by commonly employed quantum mechanical methods (Ding et al., 1995; Morgantini and Kollman, 1995) can lead to estimations for hydration free energies that vary by as much as 5.0 kcal/mol from experiment (Green and Tidor, 2003) or by as much as 8.4 kcal/mol for charged amines (Gallicchio et al., 2002). Previous work using this method has highlighted a problem with correctly ranking charged amines (Huggins et al., 2009). It is also possible that the error stems from KKK changing protonation state upon binding (lysine was modeled as charged in all cases).
Table VI.
For each of the twenty-one peptides KXK, the table shows the number of water molecules predicted, the fraction of total water molecules predicted correctly (RMSD < 2.0 Å), and the total RMSD of the water molecules predicted
| Amino acid X (KXK)a | Waters predicted | Fraction correct | RMSD total (Å) |
|---|---|---|---|
| ALA | 3 | 3/7 | 0.54 |
| ARG | 4 | 3/7 | 0.32 |
| ASN | 5 | 2/8 | 0.57 |
| ASP | 6 | 6/10 | 1.06 |
| CYS | 4 | 2/6 | 0.45 |
| GLN | 6 | 5/10 | 0.72 |
| GLU | 6 | 5/12 | 1.40 |
| GLY | 4 | 2/5 | 0.93 |
| HIS(D) | 6 | 4/11 | 0.91 |
| HIS(E) | 5 | 4/11 | 1.24 |
| HIS(P) | 5 | 4/11 | 0.90 |
| ILE | 4 | 4/6 | 1.09 |
| LEU | 3 | 2/7 | 1.22 |
| LYS | 7 | 5/8 | 0.97 |
| MET | 3 | 2/8 | 1.60 |
| PHE | 4 | 2/3 | 0.92 |
| SER | 4 | 3/8 | 0.27 |
| THR | 5 | 2/5 | 1.11 |
| TRP | 3 | 3/9 | 0.34 |
| TYR | 5 | 5/9 | 1.08 |
| VAL | 5 | 4/7 | 1.02 |
aHistidine was modeled in its three physiologically relevant states, delta protonated (D), epsilon protonated (E) and doubly protonated (P).
Enrichment analysis
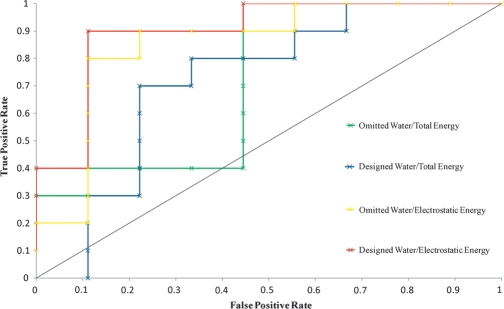
Molecular design is used within drug discovery to generate many compounds with the aim of correctly selecting those that bind strongly to a chosen target. It is thus very useful to examine the effect of the water placement scheme on the selection of strongly binding versus weakly binding compounds. The affinities for all tripeptides have been measured with isothermal titration calorimetry (Sleigh et al., 1999) and we used this data to partition them into two sets of 10; the set of stronger binders and the set of weaker binders. The affinities are uniformly distributed, making this a reasonable approach. We then looked at the ability of the algorithm to select the strong binders before the weak binders in the process of enrichment. KPK was not included in this study and thus only nine of the 10 weaker binders were modeled. Recent work suggests that a good way to examine enrichment is with a receiver operating characteristic (ROC) curve plot (Jain and Nicholls, 2008). An ROC curve is calculated by first ranking all the predicted compounds in the order of increasing binding free energy. Compounds are then selected in order from this list and the fraction of true positives is plotted against the fraction of false positives. The predicted binding of the KHK tripeptide was taken as the lowest-energy prediction among the three protonation states tested. This was uniformly the epsilon protonated form. The crystal structure of the KHK-bound structure from PDBID 1B3F (Sleigh et al., 1999) supports this prediction. The epsilon nitrogen atom of the histidine is only 2.97 Å from a side-chain oxygen atom of glutamate 32, suggesting a hydrogen bond between them. Figure 8 shows ROC curves for the designed water scheme in blue and the omitted water scheme in green. In both cases, the enrichments are better than random, but could not be considered acceptable in a drug development project.
Fig. 8.
Receiver operating characteristic curve plots of the false-positive rate against the true positive rate for a set of KXK tripeptides partitioned into 10 strong binders and nine weak binders. For the omitted water scheme, scoring using the total energy is plotted in green and scoring using the electrostatic energy is plotted in yellow. For the designed water scheme, scoring using the total energy is plotted in blue and scoring using the electrostatic energy is plotted in red. The diagonal dotted black line represents a random selection.
Previous computational protein design work employing a similar set of energy functions found that the predicted electrostatic contribution to binding affinity correlated better with the experimental binding free energies than did the predicted total interaction (Lippow et al., 2007). The van der Waals contribution was actually misleading, enriching for bulkier groups at the expense of smaller but more electrostatically optimal groups. We thus re-ranked the 500 lowest-energy instances of each designed tripeptide using an electrostatic scoring method by excluding the van der Waals contribution to binding. The lowest electrostatic score from each set of 500 tripeptides was used as the prediction for that tripeptide. Figure 8 shows ROC curves for the designed water scheme in red and the omitted water scheme in yellow. The re-ranking radically improves the enrichment for both the designed water scheme and the omitted water scheme, improving the ranking of the smaller tripeptides such as KSK and KAK, which are known to bind strongly, and lowering the ranking of the larger tripeptides such as KKK and KYK, which are known to bind weakly. A comparison of the results for the designed water scheme and the omitted water scheme suggests that including the water molecules does slightly improve the enrichment. However, the enrichment is already excellent and it is not surprising that a marked effect is not observed. A more expansive evaluation would be necessary to rigorously examine the effect of including water molecules on enrichment. This would be performed on numerous test cases, using the lessons learned in this study.
Discussion
Consideration of structural water molecules is vital within both molecular docking and molecular design. Numerous biological complexes include such water molecules and, in the future, the most successful drug design efforts must incorporate them. The ability to build these waters from the ligand and/or the protein helps to avoid the subtle problem of water molecules shifting when considering different ligands and the more radical problem that can occur when new water molecules appear in the complex. We chose a two-stage process to place ligand side groups and waters independently. This differs from the solvated rotamers approach used previously and was chosen as the A* algorithm proved computationally unmanageable when considering the huge number of possible solvated rotamers. The two stage process allows many energetically favorable complexes to be passed on to the solvation stage, and thus to the more sophisticated energy function. It is possible that the GMEC of the solvated complex would not be found with this approach, but this is unlikely, as long as enough unsolvated complexes were kept after the first stage. We have shown that inverse design approaches are able to place water molecules in physically reasonable positions that make sensible hydrogen-bonding interactions with both the ligand and the protein. The water placement scheme could be implemented with any search method and this would likely allow finer sampling, but in this case it was implemented with DEE/A* to yield a guaranteed optima in a discrete space. This has been shown to be functional within both molecular docking and molecular design, generally making a great improvement in ligand scoring and a slight improvement in results in terms of ligand placement. Such a method has already been employed within protein design, with some success. However, protein–protein complexes, such as the one used for evaluation of this method, are the result of evolution. The interactions within the complex, including the water molecules, are thus the result of selective pressure. Functionally or structurally important water molecules needing to be strongly bound in the correct orientation are more likely to be nearer perfect geometry. The water molecules that mediate interactions between proteins and drug molecules are not the result of evolution and could be expected to show more variation from ideal geometry. This would require increased sampling to predict their positions. We chose OppA as the test case for the molecular design algorithm and this provides a useful evaluation of the design methodology due to the highly flexible ligands. However, we would expect a similar performance on more rigid and more hydrophobic drug-like molecules.
Despite the successes of this method, the validation procedure suggests a number of improvements to be implemented. Structural water molecules interact with ligands and proteins in a variety of geometries. Inclusion of extra geometries within the design is easily implementable and could be computationally tractable. This should improve the predictions in cases where the current method was not able to place water molecule in less common hydrogen-bonding geometries as they were not sampled. Analysis of a wide range of protein–ligand complexes will allow the most energetically favorable geometries to be selected. The increased sampling will also increase the likelihood of selecting very tightly bound water molecules such as the one in Fig. 6. This problem can also be addressed by placing growable positions on both the ligand and the protein. Relevant protein residues can be selected prior to run time and single and pair energies can be calculated for DEE/A*. The best set of water molecules from both ligand and protein growable positions can then be found. The ability to grow water molecules from the protein will also be useful in solvating the unbound states for protein design calculations. The current implementation of the method looks only at structural water molecules in the complex. In reality, the protein and ligand will both interact closely with structural water molecules in the unbound state. This is a very important consideration for protein design calculations. The predicted binding energies can be updated by considering the difference in energy between the solvated complex and the two solvated unbound states. This should improve the quality of the prediction and is also a computationally feasible task. In terms of scoring the interactions, there should be an entropic bonus for freeing structural water molecules from the bound state to enter the solvent continuum and an entropic penalty for fixing water molecules from the solvent continuum to the bound state. This bonus and penalty has been estimated in the past (Rarey et al., 1999; Verdonk et al., 2005; Friesner et al., 2006) and incorporating them should improve scoring and enrichment. It may also improve predictions in cases where the current method placed additional water molecules that were not seen in the crystal structure and more accurately predict cases where ligands are able to displace observed water molecules. The combination of DEE and A* may prove particularly useful when considering entropic contributions as it provides an ordered list of all available energy levels of the system at the chosen level of discretization.
A number of methods have been presented for including structural water molecules within protein design and within molecular docking. To date, no work has been published on including them within small-molecule design. Furthermore, many of the existing treatments of structural water molecules suffer from problems when considering new or slightly altered ligand–water interactions and thus are inapplicable to design methods where many different ligands must be considered. Neglect or misplacement of even one single water molecule can skew predictions of binding free energies and negatively impact on enrichment. We have shown that optimizing water molecules by growing from the ligand during the design process can place water molecules correctly and that this improves the modeling and the scoring. This technique is thus a valuable addition to the analytical tools available for considering ligand–protein interactions and will be very useful in future design work.
Funding
The work was partially supported by the National Institutes of Health (GM065418, GM066524).
Acknowledgments
We thank Michael Altman for helpful discussions.
References
- Adler M., Davey D.D., Phillips G.B., Kim S.H., Jancarik J., Rumennik G., Light D.R., Whitlow M. Biochemistry-US. 2000;39:12534–12542. doi: 10.1021/bi001477q. [DOI] [PubMed] [Google Scholar]
- Altman M.D., Ali A., Reddy G.S., Nalam M.N., Anjum S.G., Cao H., Chellappan S., Kairys V., Fernandes M.X., Gilson M.K., et al. J. Am. Chem. Soc. 2008;130:6099–6113. doi: 10.1021/ja076558p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amadasi A., Spyrakis F., Cozzini P., Abraham D.J., Kellogg G.E., Mozzarelli A. J.f Mol. Biol. 2006;358:289–309. doi: 10.1016/j.jmb.2006.01.053. [DOI] [PubMed] [Google Scholar]
- Barillari C., Taylor J., Viner R., Essex J.W. J. Am. Chem. Soc. 2007;129:2577–2587. doi: 10.1021/ja066980q. [DOI] [PubMed] [Google Scholar]
- Bayly C.I., Cieplak P., Cornell W.D., Kollman P.A. J. Phys. Chem. 1993;97:10269–10280. [Google Scholar]
- Bennett B.C., Meilleur F., Myles D.A.A., Howell E.E., Dealwis C.G. Acta. Crystallogr. D Biol. Crystallogr. 2005;61:574–579. doi: 10.1107/S0907444905004804. [DOI] [PubMed] [Google Scholar]
- Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenk R., Vetter S.W., Boyce S.E., Goodin D.B., Shoichet B.K. J. Mol. Biol. 2006;357:1449–1470. doi: 10.1016/j.jmb.2006.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks B.R., Bruccoleri R.E., Olafson B.D., States D.J., Swaminathan S., Karplus M. J. Comput. Chem. 1983;4:187–217. [Google Scholar]
- Brünger A.T., Karplus M. Proteins. 1988;4:148–156. doi: 10.1002/prot.340040208. [DOI] [PubMed] [Google Scholar]
- Champness J.N., Bennett M.S., Wien F., Visse R., Summers W.C., Herdewijn P., de Clercq E., Ostrowski T., Jarvest R.L., Sanderson M.R. Proteins Struct. Funct. Genet. 1998;32:350–361. doi: 10.1002/(sici)1097-0134(19980815)32:3<350::aid-prot10>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- Chen J.M., Xu S.L., Wawrzak Z., Basarab G.S., Jordan D.B. Biochemistry-US. 1998;37:17735–17744. doi: 10.1021/bi981848r. [DOI] [PubMed] [Google Scholar]
- Claussen H., Buning C., Rarey M., Lengauer T. J. Mol. Biol. 2001;308:377–395. doi: 10.1006/jmbi.2001.4551. [DOI] [PubMed] [Google Scholar]
- Cornell W.D., Cieplak P., Bayly C.I., Kollman P.A. J. Am. Chem. Soc. 1993;115:9620–9631. [Google Scholar]
- Csaszar A.G., Czako G., Furtenbacher T., Tennyson J., Szalay V., Shirin S.V., Zobov N.F., Polyansky O.L. J. Chem. Phys. 2005;122:214305. doi: 10.1063/1.1924506. [DOI] [PubMed] [Google Scholar]
- Cupp-Vickery J.R., Garcia C., Hofacre A., McGee-Estrada K. J. Mol. Biol. 2001;311:101–110. doi: 10.1006/jmbi.2001.4803. [DOI] [PubMed] [Google Scholar]
- Dahiyat B.I., Mayo S.L. Science. 1997;278:82–87. doi: 10.1126/science.278.5335.82. [DOI] [PubMed] [Google Scholar]
- de Graaf C., Pospisil P., Pos W., Folkers G., Vermeulen N.P.E. J. Med. Chem. 2005;48:2308–2318. doi: 10.1021/jm049650u. [DOI] [PubMed] [Google Scholar]
- Desmet J., Demaeyer M., Hazes B., Lasters I. Nature. 1992;356:539–542. doi: 10.1038/356539a0. [DOI] [PubMed] [Google Scholar]
- Ding Y.B., Bernardo D.N., Kroghjespersen K., Levy R.M. J. Phys. Chem. 1995;99:11575–11583. [Google Scholar]
- Drexler K.E. Proc. Natl Acad. Sci. USA. 1981;78:5275–5278. doi: 10.1073/pnas.78.9.5275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friesner R.A., Banks J.L., Murphy R.B, et al. J. Med. Chem. 2004;47:1739–1749. doi: 10.1021/jm0306430. [DOI] [PubMed] [Google Scholar]
- Friesner R.A., Murphy R.B., Repasky M.P., Frye L.L., Greenwood J.R., Halgren T.A., Sanschagrin P.C., Mainz D.T. J. Med. Chem. 2006;49:6177–6196. doi: 10.1021/jm051256o. [DOI] [PubMed] [Google Scholar]
- Frisch A., Dennington R., Keith T., Nielsen A., Holder A. GaussView. Pittsburg: Gaussian Inc.; 2003. [Google Scholar]
- Frisch M.J., Trucks G.W., Schlegel H.B., et al. Gaussian03. Pittsburg: Gaussian Inc.; 2004. [Google Scholar]
- Gallicchio E., Zhang L.Y., Levy R.M. J. Comput. Chem. 2002;23:517–529. doi: 10.1002/jcc.10045. [DOI] [PubMed] [Google Scholar]
- Garcia-Sosa A.T., Mancera R.L., Dean P.M. J. Mol. Model. 2003;9:172–182. doi: 10.1007/s00894-003-0129-x. [DOI] [PubMed] [Google Scholar]
- Gilson M.K., Honig B. Proteins Structure Function Genet. 1988;4:7–18. doi: 10.1002/prot.340040104. [DOI] [PubMed] [Google Scholar]
- Gradler U., Gerber H.D., Goodenough-Lashua D.M., Garcia G.A., Ficner R., Reuter K., Stubbs M.T., Klebe G. J. Mol. Biol. 2001;306:455–467. doi: 10.1006/jmbi.2000.4256. [DOI] [PubMed] [Google Scholar]
- Green D.F., Tidor B. J. Phys. Chem. B. 2003;107:10261–10273. [Google Scholar]
- Hellinga H.W. Proc.Natl Acad. Sci. USA. 1997;94:10015–10017. doi: 10.1073/pnas.94.19.10015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang N., Shoichet B.K. J. Med. Chem. 2008;51:4862–4865. doi: 10.1021/jm8006239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huggins D.J., Altman M.D., Tidor B. Proteins. 2009;75:168–186. doi: 10.1002/prot.22226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain A.N., Nicholls A. J. Comput. Aided Mol. Des. 2008;22:133–139. doi: 10.1007/s10822-008-9196-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L., Kuhlman B., Kortemme T.A., Baker D. Proteins Structure Function Bioinform. 2005;58:893–904. doi: 10.1002/prot.20347. [DOI] [PubMed] [Google Scholar]
- Jones G., Willett P., Glen R.C., Leach A.R., Taylor R. J. Mol. Biol. 1997;267:727–748. doi: 10.1006/jmbi.1996.0897. [DOI] [PubMed] [Google Scholar]
- Kuhlman B., Dantas G., Ireton G.C., Varani G., Stoddard B.L., Baker D. Science. 2003;302:1364–1368. doi: 10.1126/science.1089427. [DOI] [PubMed] [Google Scholar]
- Lam P.Y.S., Ru Y., Jadhav P.K., et al. J. Med. Chem. 1996;39:3514–3525. doi: 10.1021/jm9602571. [DOI] [PubMed] [Google Scholar]
- Leach A.R., Lemon A.P. Proteins Structure Function Genet. 1998;33:227–239. [Google Scholar]
- Li R.B., Sirawaraporn R., Chitnumsub P., Sirawaraporn W., Wooden J., Athappilly F., Turley S., Hol W.G.J. J. Mol. Biol. 2000;295:307–323. doi: 10.1006/jmbi.1999.3328. [DOI] [PubMed] [Google Scholar]
- Li Z., Lazaridis T. Phys. Chem. Chem. Phys. 2007;9:573–581. doi: 10.1039/b612449f. [DOI] [PubMed] [Google Scholar]
- Lippow S.M., Wittrup K.D., Tidor B. Nat. Biotechnol. 2007;25:1171–1176. doi: 10.1038/nbt1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y.P., Wang R.X., Yang C.Y., Wang S.M. J. Chem. Inform. Model. 2007;47:668–675. doi: 10.1021/ci6003527. [DOI] [PubMed] [Google Scholar]
- MacKerell A.D., Bashford D., Bellott M., et al. J. Phys. Chem. B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- Mancera R.L. J. Comput. Aided Mol. Des. 2002;16:479–499. doi: 10.1023/a:1021273501447. [DOI] [PubMed] [Google Scholar]
- McLachlan A.D. Acta. Crystallogr. D Biol. Crystallogr. 1982;38:871–873. [Google Scholar]
- Momany F.A., Rone R. J. Comput. Chem. 1992;13:888–900. [Google Scholar]
- Morgantini P.Y., Kollman P.A. J. Am. Chem. Soc. 1995;117:6057–6063. [Google Scholar]
- Morris G.M., Goodsell D.S., Halliday R.S., Huey R., Hart W.E., Belew R.K., Olson A.J. J. Comput. Chem. 1998;19:1639–1662. [Google Scholar]
- Obermann W.M.J., Sondermann H., Russo A.A., Pavletich N.P., Hartl F.U. J. Cell Biol. 1998;143:901–910. doi: 10.1083/jcb.143.4.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterberg F., Morris G.M., Sanner M.F., Olson A.J., Goodsell D.S. Proteins Struct. Function Genet. 2002;46:34–40. doi: 10.1002/prot.10028. [DOI] [PubMed] [Google Scholar]
- Pabo C. Nature. 1983;301:200. doi: 10.1038/301200a0. [DOI] [PubMed] [Google Scholar]
- Pierce N.A., Spriet J.A., Desmet J., Mayo S.L. J. Comput. Chem. 2000;21:999–1009. [Google Scholar]
- Ponder J.W., Richards F.M. J. Mol. Biol. 1987;193:775–791. doi: 10.1016/0022-2836(87)90358-5. [DOI] [PubMed] [Google Scholar]
- Poornima C.S., Dean P.M. J. Comput. Aided Mol. Des. 1995a;9:500–512. doi: 10.1007/BF00124321. [DOI] [PubMed] [Google Scholar]
- Poornima C.S., Dean P.M. J. Comput. Aided Mole. Des. 1995b;9:513–520. doi: 10.1007/BF00124322. [DOI] [PubMed] [Google Scholar]
- Poornima C.S., Dean P.M. J. Computer Aided Mol. Des. 1995c;9:521–531. doi: 10.1007/BF00124323. [DOI] [PubMed] [Google Scholar]
- Rarey M., Kramer B., Lengauer T. Proteins Struct. Function Genet. 1999;34:17–28. [PubMed] [Google Scholar]
- Raymer M.L., Sanschagrin P.C., Punch W.F., Venkataraman S., Goodman E.D., Kuhn L.A. J. Mol. Biol. 1997;265:445–464. doi: 10.1006/jmbi.1996.0746. [DOI] [PubMed] [Google Scholar]
- Roe S.M., Prodromou C., O'Brien R., Ladbury J.E., Piper P.W., Pearl L.H. J. Med. Chem. 1999;42:260–266. doi: 10.1021/jm980403y. [DOI] [PubMed] [Google Scholar]
- Scharer K., Morgenthaler M., Seiler P., Diederich F., Banner D.W., Tschopp T., Obst-Sander U. Helvetica Chim. Acta. 2004;87:2517–2538. [Google Scholar]
- Schnecke V., Kuhn L.A. Perspect. Drug Discov. Des. 2000;20:171–190. [Google Scholar]
- Sharp K.A., Honig B. J. Phys. Chem. 1990a;94:7684–7692. [Google Scholar]
- Sharp K.A., Honig B. Ann. Rev. Biophys. Biophys. Chem. 1990b;19:301–332. doi: 10.1146/annurev.bb.19.060190.001505. [DOI] [PubMed] [Google Scholar]
- Sitkoff D., Sharp K.A., Honig B. J. Phys. Chem. 1994;98:1978–1988. [Google Scholar]
- Sleigh S.H., Seavers P.R., Wilkinson A.J., Ladbury J.E., Tame J.R.H. J. Mol. Biol. 1999;291:393–415. doi: 10.1006/jmbi.1999.2929. [DOI] [PubMed] [Google Scholar]
- Stebbins C.E., Russo A.A., Schneider C., Rosen N., Hartl F.U., Pavletich N.P. Cell. 1997;89:239–250. doi: 10.1016/s0092-8674(00)80203-2. [DOI] [PubMed] [Google Scholar]
- Tame J.R.H., Sleigh S.H., Wilkinson A.J., Ladbury J.E. Nature Struct. Biol. 1996;3:998–1001. doi: 10.1038/nsb1296-998. [DOI] [PubMed] [Google Scholar]
- Verdonk M.L., Chessari G., Cole J.C., Hartshorn M.J., Murray C.W., Nissink J.W.M., Taylor R.D., Taylor R. J. Med. Chem. 2005;48:6504–6515. doi: 10.1021/jm050543p. [DOI] [PubMed] [Google Scholar]
- Vonitzstein M., Wu W.Y., Kok G.B., Pegg M.S., Dyason J.C., Jin B., Phan T.V., Smythe M.L., White H.F., Oliver S.W., et al. Nature. 1993;363:418–423. doi: 10.1038/363418a0. [DOI] [PubMed] [Google Scholar]
- Wang R.X., Fang X.L., Lu Y.P., Wang S.M. J. Med. Chem. 2004;47:2977–2980. doi: 10.1021/jm030580l. [DOI] [PubMed] [Google Scholar]
- Weichenberger C.X., Sippl M.J. Bioinformatics. 2006;22:1397–1398. doi: 10.1093/bioinformatics/btl128. [DOI] [PubMed] [Google Scholar]
- Wright L., Barril X., Dymock B., et al. Chem. Biol. 2004;11:775–785. doi: 10.1016/j.chembiol.2004.03.033. [DOI] [PubMed] [Google Scholar]
- Yan A., Grant G.H., Richards W.G. J. R Soc. Interface. 2008;5(Suppl 3):S199–S205. doi: 10.1098/rsif.2008.0331.focus. [DOI] [PMC free article] [PubMed] [Google Scholar]