Abstract
Class D β-lactamases, a major source of bacterial resistance to β-lactam antibiotic therapies, represent a distinct subset of the β-lactamase superfamily. They share a serine hydrolase mechanism with Classes A/C vs. Class B. Further understanding of their sequence–structure–function relationships would benefit efforts to design a new generation of antibiotics as well as to predict evolutionary mechanisms in response to such therapies. Here we describe analyses based on our high-resolution multiple sequence alignment and phylogenetic tree of ∼80 Class D β-lactamases that leverage several 3D structures of these enzymes. We observe several sequence clusters on the phylogenetic tree, some that are species specific while others include several species from α-, β- and γ-proteobacteria. Residues characteristic of a specific cluster were identified and shown to be located just outside the active site, possibly modulating the function of the catalytic residues to facilitate reactions with specific types of β-lactams. Most significant was the discovery of a likely disulfide bond in a large group composed of α-, β- and γ-proteobacteria that would contribute to enzyme stability and hence bacterial viability under antibiotic assault. A network of co-evolving residues was identified which suggested the importance of maintaining a surface for binding a highly conserved Phe69.
Keywords: Class D β-lactamase, disulfide bond, group entropy, protein evolution, structure–function relationship
Introduction
β-Lactamases are a major source of bacterial resistance to β-lactam antibiotic therapies. Other, less common, mechanisms include lowering of β-lactam affinity to penicillin binding proteins, down-regulation of porins and up-regulation of efflux pumps (Wilke et al., 2005). β-Lactamases have been assigned to four classes: A, B, C and D. Classes A, C and D share a common serine hydrolase mechanism as shown in Fig. 1 while Class B are metalloenzymes. The serine hydrolase β-lactamases exhibit a wide range of hydrolytic profiles and have been the subject of numerous reviews (Fisher et al., 2005; Bush and Jacoby, 2010; Drawz and Bonomo, 2010). Of particular clinical concern is the emerging and growing incidence of carbapenemase activity among bacterial species (Livermore and Woodford, 2000; Queenan and Bush, 2007). Carbapenems remain resistant to hydrolysis by a majority of commonly encountered β-lactamases (including extended spectrum β-lactamases) (Drawz and Bonomo, 2010) and therefore are crucial in the current arsenal of antibiotics as the last-line treatment of bacterial infections caused by pathogens resistant to other β-lactams. Currently, the majority of β-lactamases able to hydrolyze carbapenems belong to Class B (IMiPenem resistant and Verona Integron-encoded Metallo type β-lactamases) (Walsh et al., 2005; Bush, 2010) though several Class A enzymes have also been reported to hydrolyze carbapenems (Queenan and Bush, 2007; Drawz and Bonomo, 2010). In addition, weak carbapenemase activity is emerging worldwide in Class D serine β-lactamases (for example OXA 24/40 and 48 enzymes) (Afzal-Shah et al., 2001; Walther-Rasmussen and Høiby, 2006; Queenan and Bush, 2007).
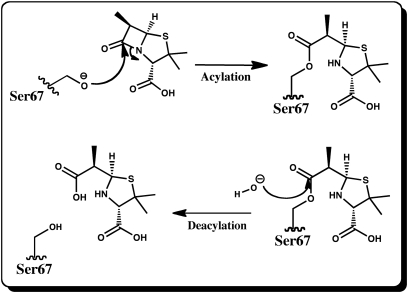
Fig. 1.
Mechanism for the acylation and deacylation reactions of Class D β-lactamases. In the first half-reaction nucleophilic Ser residue attacks the carbonyl carbon of the β-lactam ring leading to cleavage of the ring and formation of a stable doripenem-enzyme acylate. In the second step, a water molecule attacks the same carbon, the Ser-doripenem bond breaks and the inactivated ligand is released from the active site.
Class D (oxacillinases, OXAs) (Poirel et al., 2010) has not been studied nearly as extensively as Classes A and C though this may change with the recent increase in the number of clinical studies reporting emergence of pathogens utilizing OXA-mediated mechanisms of resistance (Euro Surveill., 2010). Probably the most characteristic feature of Class D is the presence of a conserved carboxylated lysine residue in the active site (Golemi et al., 2001; Fisher et al., 2005). In addition, at least some OXAs have been confirmed to form dimers (Danel et al., 2001; Vercheval et al., 2010) and dimer–monomer equilibrium is implicated in kinetic behavior of these enzymes (Danel et al., 2001).
Given the clinical and pharmacological implications of Class D resistance to various antibiotics, there is an urgent need to understand not only their detailed enzymatic mechanisms of hydrolysis but also the mechanisms through which these enzymes evolve new functional profiles, information that indeed may enable prediction of the course of Class D genetic evolution in response to antibiotics (Weinreich et al., 2006; Novais et al., 2010). In part, this understanding can be gained through sequence-based bioinformatics methods where for many protein families there is a high correspondence between phylogenetic and functional classification (Sjölander, 2010). Yet, due to horizontal gene transfer in bacteria that takes place often when genes exist on plasmids and due to very recent evolutionary adaptations to man-made antibiotics, the structure–function relationships may be more subtle than those found in other protein families. Class D enzymes have been assigned to three different groups in a functional classification (Bush and Jacoby, 2010) which has provided some insight into the diversity of this particular class. Other phylogenetic analyses of Class D have been reported (Barlow and Hall, 2002; Hall and Barlow, 2004; Poirel et al., 2010) though there has been no subsequent analysis of any distinguishing sequence or structural features between clusters of Class D sequences. Finally, beyond the role of the nucleophilic serine and the carboxylated lysine to catalysis (Fisher et al., 2005; Schneider et al., 2009), the role of other active site residues remains unknown.
Here, we report our results employing (i) sequence-based bioinformatics that identify evolutionary conserved residues, intermolecular interactions and surfaces among the entire Class D family and among various sub-groups that likely contribute to their respective functional characteristics and (ii) molecular modeling using hybrid quantum chemical/molecular mechanical (QC/MM) methods that provide insight into the function of residues in the active site that are less understood. Of course, other residues, appropriately mutated, could also play large roles in determining function but our results offer insight into the mechanisms by which evolution has acted to tailor these functional characteristics and hence how it may design mutations to respond to further environmental pressures. Finally, our findings are critical for placing the results of site-directed mutagenesis experiments in context of the entire Class D family and for making decisions on the type and locations of residues to mutate that would yield the most insight.
Materials and methods
Sequences were gathered from the iProClass database (Wu et al., 2004) and from literature searches. A subset of 84 representative sequences was retained after deleting some that were 100% identical due to a recent horizontal transfer event. The initial multiple sequence alignment (MSA) was constructed using MUSCLE (Edgar, 2004). The MEME program (Bailey and Elkan, 1994) was employed using the zoops model (zero or one motif per sequences) to search for the 30 most conserved patterns or motifs ranging in length between 6 and 50 residues over the entire list of diverse Class D sequences. We have found (Perozich et al., 1999) that these parameters to MEME generally return motifs that are immensely useful for efficiently refining MSAs with the Genedoc editor (sb.nrbsc.org) as well as assisting in assigning sequences to groups (described in more detail below). The information content of a motif is determined by the conservation of residues along the motif, the length of the motif and the distribution of the motif residues in the submitted data set (Bailey and Elkan, 1994). Only the top five motifs that span all the Class D sequences are presented in the Results section. In addition, a structural alignment of five OXA structures [protein data bank (PDB) entries 3ISG, 1K38, 1K57, 3FV7 and 3HBR] was performed using STAMP within MultiSeq module (Roberts et al., 2006) contained within the VMD program (Humphrey et al., 1996). The MSA was then manually adjusted using the motifs and structural information as a guide within the GeneDoc editor to create a high-resolution MSA. The final MSA and information on all the sequences in the alignment is provided in the Supplementary material.
The quality of a phylogenetic tree is highly dependent on the quality of the MSA and the regions included for tree construction (Sjölander, 2004, 2010). The final MSA was manually trimmed to 245 characters by deleting sections where the alignment was equivocal; primarily at the N- and C-termini. All active site residues were included. The trimmed MSA file was used to create a distance-based phylogenetic tree using the PHYLIP suite software (Felsenstein, 1989). A total of 1000 data sets were generated through bootstrapping analysis using SEQBOOT. A distance matrix was computed for each of these data sets with the program PROTDIST using the Jones–Taylor–Thornton substitution matrix (Jones et al., 1992). The resulting distance matrix data were used with NEIGHBOR to generate a tree for each of the 1000 data sets, all of which were run through CONSENSE to produce a consensus tree. The phylogenetic tree was visualized and a figure created with FigTree version 1.2.2. A file that can be used to visualize all sequence labels on the phylogenetic tree has been included in the Supplementary material. The retrieval of functional characteristics for all sequences, ordering of the MSA file based on the phylogenetic tree and other information gathering tasks were facilitated by our program HarvestSeq (sb.nrbsc.org).
The phylogenetic tree was used as a guide to partition the sequences into separate groups, typically those that cluster together with high bootstrap support. The MSA and a partitioning of sequences into defined groups serves as input to the GEnt program (Hempel et al., 2003) with the PAM160 substitution matrix and a pseudocount multiplier of 1.0. This program detects amino acid residues characteristic of sub-groups within a set of orthologous proteins or characteristic of individual protein families within a collection of paralogous proteins. Often these group-specific residues are most responsible for changes in biochemical properties like substrate selectivity (Lichtarge et al., 1996; Hempel et al., 2003). These characteristic residues are identified as having (i) low overall family relative entropy defined as
where for each of the 20 amino acids pi is the fraction of residue type i at that alignment position and qi is the fraction of residue type i expected in a random sequence. qi is usually taken from an appropriate non-redundant database and (ii) high group cross-entropy computed as
where pi is the fraction of residue type i at a particular position in the alignment for sequences in the predefined group while qi is the fraction of residue type i at that position for sequences not in the predefined group. Other details relating to this method can be found in the literature (Hempel et al., 2003).
We employed a mutual information program (Gloor et al., 2005) to detect covarying (or coevolving) residues. In the study by Gloor et al., they used sets of sequences that were orthologous implying that residues that were coevolving did so in order to maintain some characteristic of biochemical function. Since it is quite difficult to conclusively establish whether all Class D sequences are orthologous and because their active sites are so well conserved, we decided to analyze them as an orthologous set. In this report, we will primarily use the term covarying since some residue pairs that appear to be coevolving could be false positives, the result of phylogenetic linkage (Ashkenazy and Kliger, 2010).
Molecular modeling of OXA-1 using hybrid QC/MM methods
The crystal structure of OXA-1 was retrieved from the PDB (Rose et al., 2011) (entry 3ISG). A model of this enzyme was constructed in CHARMM version 34b2 (Brooks et al., 2009). This construction included adding hydrogen atoms to all the protein atoms found in the deposition using HBUILD. All protein residues were set to their canonical protonation state. All oxygen atoms identified as water molecules in the crystal structure were retained and hydrogen atoms added to these as for the protein structure. A molecular mechanical force field was developed for the doripenem substrate molecule and the carboxylated lysine through analogy to similar structures already present in the CHARMM force fields (MacKerell et al., 1998). The entire ternary complex was solvated with TIP3P water molecules (Jorgensen et al., 1983). This structure was energy minimized using pDynamo (Field, 2008) where the entire doripenem molecule and the Ser67, Thr68, carboxylated Lys70, Ser115, Lys212 side chains were represented with the PM6 (Stewart, 2007) semiempirical molecular orbital method and the surroundings were represented by the CHARMM force field. The polarization of the QC region by the surroundings takes place through the SCF procedure (Field, 2008). This minimization facilitated an accurate atomistic model of the Michaelis complex and the acylated intermediate used in our structural analysis. Further simulation details and results will be reported in the near future.
Results and discussion
Unless otherwise noted, sequence numbering refers to the numbering found in the OXA-1 structure (PDB entry 3ISG). Numbering of secondary structures elements is consistent with PDB entry 3ISG (authors' assignment).
Structure–function analysis of conserved residues in Class D
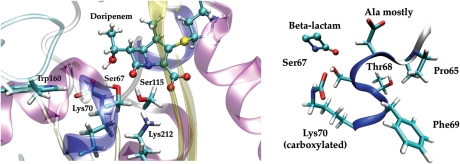
The highly conserved residues (>95%) of the 84 Class D sequences are listed in Table I. The strictly conserved residues that participate in the acylation and deacylation enzymatic reactions include the Ser67 nucleophile (Frère et al., 1999; Paetzel et al., 2000), the carboxylated Lys70 that likely serves as a general base activating Ser67 (although not essential for acylation, it is essential for the deacylation reaction (Schneider et al., 2009)), and Lys212 that likely plays a role in protonation of the antibiotic substrate in concert with the acylation reaction (see Fig. 2). In our modeling of the OXA-1 structure with hybrid QC/MM methods, Lys212 is shown to interact strongly with the lactam nitrogen atom of the doripenem substrate either directly or through Ser115. In the serine hydrolase type β-lactamases, this lysine is often mentioned in the context of the conserved KTG triad (Paetzel et al., 2000) but in Class D the Thr is substituted several times by Ser. Trp160, part of the Ω-loop, is in a partial stacking arrangement with Phe69 and also hydrogen bonds with the carboxylated Lys70 through its Nε proton. The interaction between Trp160 and Lys70 promotes carboxylation of Lys70 and an optimal orientation of the Lys70 side chain with respect to the nucleophilic Ser67, and has an impact on antibiotic specificity (Danel et al., 1999; Paetzel et al., 2000; Golemi et al., 2001; Baurin et al., 2009). In addition, the Ω-loop becomes significantly more flexible when Trp160 is substituted (Baurin et al., 2009). Other absolutely conserved residues include Trp228, Phe243 and three glycines, two of which are located outside the active site. In addition, a contiguous stretch of residues, Pro65-Lys70, are highly conserved except for residue 66 which is nevertheless mostly Ala. This conserved helical region and its relation to a β-lactam bound in the active site of a QC/MM model of the OXA-1 Michaelis complex is shown in Fig. 2.
Table I.
Ninety-five percent conserved residues among the 84 Class D sequences
| 3ISG sequence #s | 3ISG PDB #s | Function/comment |
|---|---|---|
| Pro44 | Pro65 | |
| Ser46a | Ser67b | Nucleophile that attacks β-lactam |
| Thr47 | Thr68b | |
| Phe48 | Phe69b | |
| Lys49a | Lys70b | General base |
| Ser93a | Ser115c | |
| Gly106 | Gly128 | |
| Tyr119 (some Phe) | Tyr141d | Ω-loop Class D motif |
| Gly120a | Gly142d | Ω-loop Class D motif |
| Asn121 | Asn143d | Ω-loop Class D motif |
| Trp138a | Trp160e | Interacts with general base |
| Ile145 | Ile167 | Val makes up the remainder |
| Leu162 | Leu184 | Val, Phe make up the remainder |
| Lys190a | Lys212f | Likely proton donor |
| Gly192a | Gly214f | Mainchain amide contributes to oxyanion hole |
| Trp205a | Trp228 | Part of hydrophobic core |
| Gly208a | Gly231 | |
| Phe220a | Phe243 |
aStrictly conserved.
bSTFK (Paetzel et al., 2000).
cSXV (Paetzel et al., 2000).
dYGN (Paetzel et al., 2000).eBaurin et al. (2009).
fKTG family signatures (Paetzel et al., 2000).
Fig. 2.
(left) One hundred percent conserved residues in the active site shown on a model of OXA-1—doripenem Michealis complex. The complex was constructed from the OXA-1 structure crystallized in the acylated form (pdb 3ISG). (right) Highly conserved helix (α3) and its relation to the β-lactam ring in our model structure.
Most of the highly conserved residues, not surprisingly, cluster around the β-lactam ring, primarily on the side that the carbonyl dipole of the β-lactam ring points, extending back to the base of the Ω-loop. These residues come primarily from helix 3–4 (3–10/α) and β6, which are in close proximity to highly conserved Trp228 and Gly231 on β5. Ser115 is the only conserved residue behind the β-lactam carbonyl dipole. Trp228 is positioned like a platform that interacts with the highly conserved residues of the Ω-loop region. Tyr141 and Ile167, at separate ends of the Ω-loop, form a hydrophobic stacking interaction. The Tyr141Phe substitution has been shown to confer the extended spectrum beta-lactamase phenotype (Fisher et al., 2005). Of these 18 highly conserved residues, 10 are located in loop regions between secondary structure elements.
Conserved motifs in Class D
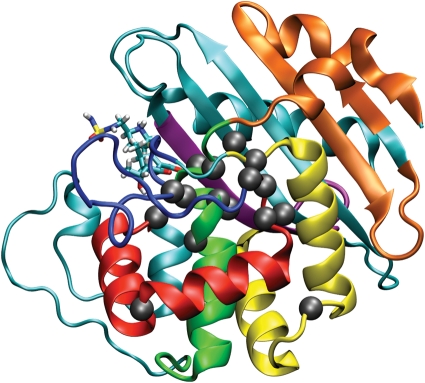
Figure 3 shows the top five motifs, determined statistically through the MEME (Bailey and Elkan, 1994), annotated on the OXA-1 structure model that are present in nearly all sequences along with two others that are present only outside a group of related sequences composed of α-, β- and γ-proteobacteria (see Fig. 3 and Table II). These motifs demonstrate that considerable information exists besides the highly conserved residues enabling the refinement of the MSA and to demarcate those larger segments that are most conserved from those less conserved. Excluding motif 2, the top five motifs come together in space to make up the substrate-binding site and to preserve the catalytic function of Class D enzymes. Motif 1 runs from Ser115 to Asn143 and is located just after the long-loop region (that spans from Glu81 to Thr107) making up helices α7 and α8—up to the Ω-loop. The four conserved residues in this motif are located in loop regions: Ser115 between α6 and α7, Gly128 between α7 and α8, and Gly142–Asn143 on the Ω-loop. Motif 2 runs from Ile167 to Asp202 and is located just past the end of the Ω-loop comprising helices α10 and α11, and leading into β7. Again the conserved residues in this motif are located in the loop regions connecting the secondary structure elements. Interestingly, this motif does not contain any residues that make up the substrate-binding site. Motif 3 runs from Met63 through Gln86, primarily comprising helices 3 (310) and 4 (α4), which contain many of the important catalytic residues in Class D, including the STFK signature of Class D β-lactamases. Given the high conservation of the slightly downstream Pro65, a change in reference to the Class D signature PASTFK may be warranted. Motif 4 runs from Leu203 to Gly216 and comprises mostly β6 containing two strictly conserved residues, namely Lys212 and Gly214. Motif 5 is primarily the Ω-loop and runs from Gln144 to Ser164 and is not present in all the sequences possibly because their sequences lie just outside the threshold for motif inclusion. Only one conserved residue, Trp160, is present in this motif, although four others are found in flanking motifs just outside of it.
Fig. 3.
The five most conserved motifs in Class D β-lactamases detected with MEME (Bailey and Elkan, 1994) colored from first to fifth in information content (red, yellow, green, purple, blue) on the OXA-1 structure model. The last two motifs colored in orange are only present outside a large evolutionary related group of α-, β- and γ-proteobacteria sequences. Highly conserved residues are shown with a gray sphere centered on the Cα atom. The doripenem molecule from our OXA-1 model is shown in stick form.
Table II.
Five most conserved motifs identified by MEME (Bailey and Elkan, 1994) within Class D with two motifs (6 and 7) contained in sequences outside the large α-, β- and γ-proteobacteria group
| Motif number | Length | Information content (bits) | Motif | Indices (3ISG) |
|---|---|---|---|---|
| 1 | 29 | 71.7 | S[AV]V[WP][VY][YS]QE[LI][AT]RR[IL]GxE[RK][MF]QKY[LV]K[KRA][FL]DYGN | 115–143 |
| 2 | 36 | 70.8 | IS[AP]x[EQ]Q[VI]xFLR[KR]L[YA]R[NG]KLP[FV]xSA[ER][AS]Q[DR][LM][VT]KDI[ML][IL]VE | 167–202 |
| 3 | 23 | 59.9 | [SL]PAxSTFKI[PA][NL][SAT]L[IM][GA]L[DE][AT]G[AV][VL]KD | 63–86 |
| 4 | 15 | 30.2 | [AE]GxG[GD][WY][KI][LIV][RYH][GA]KTGWG | 203–216 |
| 5 | 21 | 32.2 | ADI[SG]GDxGxx[GN]G[LIV][DT]x[FA]WL[DE][GS][SP] | 144–164 |
| 6 | 29 | 45.7 | E[RN]x[DS]x[WL]AKLFS[DE]AG[AVT]KG[TV][FI]V[LI]YDERxxxx | 19–46 |
| 7 | 11 | 23.0 | [VT][NH][ND]L[AE]R[AS]x[QTK]R[YF] | 49–59 |
Highly conserved residues (>95%) are shown in bold font.
Interestingly, two additional motifs that begin at the N-terminus and go almost continuously to residue 59 are not present in the largest group of Class D sequences made up of α-, β- and γ-proteobacteria (see Fig. 3). There are a few hydrogen bonding interactions between these two motifs and motif 2. This section of the protein likely plays a role in the proper folding of the rest of the protein and in maintaining the hydrophobic core. The absence of these motifs from a particular group of Class D sequences is discussed below.
Structural conservation of Class D
Although the remaining parts of the Class D enzyme sequence are not conserved, they display a high degree of structural conservation (see Supplementary material). Five Class D β-lactamases sharing between 19 and 44% sequence identity clearly demonstrate an overall shared structure with the major differences in loops and at the end of β-strands. Two very long-loop regions are also structurally well conserved, the Ω-loop comprising residues 140–168 and a region we designate the P-loop comprising residues 81–107. In Class A, Ω-loop is important for orienting a glutamic acid that serves as the general base (Sampson et al., 2011 and references therein). In all the aligned structures, a hydrophobic stacking interaction between Phe90 and Val119 as well as a salt bridge between Asp93 and Lys97 in the P-loop is found. These interactions help position Met99, Trp102, Ser115 and Val117 that form part of the binding pocket, with Ser115 possibly playing a role in catalysis as well. Although we have aligned Class D structures from different sections of the phylogenetic tree (described below), there is still some uncertainty as to whether other OXA sequences would adopt the same P-loop structure.
Phylogenetic analysis of Class D
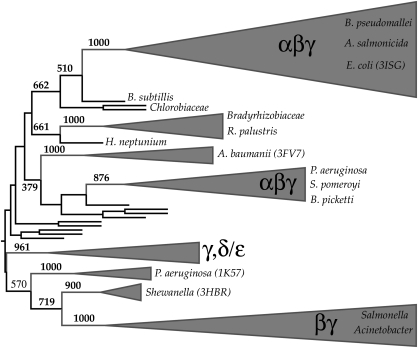
The phylogenetic tree and high bootstrap values were used to assign sequences into groups (see Fig. 4). Interestingly, there is high bootstrap support for assigning a large fraction of Class D sequences into two large groups that contain sequences that are not all species related, though within these two groups, sequences from the same species can be distinguished as different minor branches on the phylogenetic tree. All of the smaller groups each contain at most nine sequences from the same bacterial species and have high bootstrap support for such an assignment. The largest group, containing the OXA-1 sequence of known structure (PDB entry 3ISG), consists of α-, β- and γ-proteobacteria. The other group, containing the OXA-2 sequence of known structure (PDB entry 1K38), consists of β- and γ-proteobacteria.
Fig. 4.
Phylogenetic tree of Class D β-lactamases. The triangulated regions are sequences that could be confidently assigned to a group for further analysis. Groups that contain several different species are labeled by type of proteobacteria. Selected PDB sequences are shown in parentheses next to the species origination. Full details on all the sequences and a complete phylogenetic tree with UniProt identifiers is given in the Supplementary material.
The sequences within each of these two large groups are related possibly due to past horizontal gene transfer or speciation (sequences are orthologous). Ideally, sequences that are grouped together will be orthologous sharing similar structures and especially biochemical function enabling an even more in-depth profiling of their specific properties (see below). Yet, since there appeared no strict, consistent way of experimentally characterizing all Class D sequences (Fisher et al., 2005; Bush and Jacoby, 2010), we were unable to confirm that indeed all sequences in a group share similar biochemical characteristics. For millions of years, this enzyme family has evolved in response to environmental pressures other than man-made antibiotics (Barlow and Hall, 2002) such that similar sequences will have similar structures and biochemical properties. The recent onslaught of man-made antibiotics has required evolutionary mechanisms to counteract this pressure. Some of these bacterial responses to human efforts to eradicate them have required only a trivial substitution such as seen in the single residue mutation at OXA-10's Ω-loop, to give OXA-11, extending its ability to hydrolyze ceftazidime (Poirel et al., 2010).
Group-specific residues
Residues conserved within a group but largely absent from other family sequences are often possible determinants of specificity. The group-specific residues determined by the GEnt program are given in Table III. The group-specific residues are concentrated in the α-helical regions (helices 4, 6–7) (see Fig. 5) that are adjacent to the active site surface with a few exceptions. We primarily restrict our remarks to just the two largest groups which given their size gives the most robust conclusions.
Table III.
Group-specific residues determined through Gent (Hempel et al., 2003; shannon.psc.edu/harvest)
| Alignment index | Family entropy | Group entropy | Residue | AA out |
|---|---|---|---|---|
| α-, β- and γ-proteobacteria (3ISG) 22–93% | ||||
| 182 | 3.36 | 24.23 | Trp111 | Ala |
| 191 | 3.04 | 19.79 | Ser120 | Phe/Tyr |
| 179 | 2.26 | 17.94 | Pro108 | Leu |
| 121 | 2.78 | 18.38 | Cys59 | Ala/Ser |
| 135 | 2.33 | 17.12 | Leu73 | Asn/His |
| 98 | 2.60 | 11.69 | Cys37 | Thr/Val |
| Pseudomonas aeruginosa (1K57) 76–94% | ||||
| 311 | 3.34 | 14.12 | Ala220 | Gly |
| 102 | 0.87 | 12.48 | Cys44 | Varies |
| β- and γ-proteobacteria (1K38) 49–90% | ||||
| 319 | 1.48 | 16.59 | Trp221 | Lys,Arg |
| 138 | 2.82 | 14.96 | Phe76 | Ile, Met |
| 203 | 2.46 | 13.90 | Ala132 | Varies |
| Shewanellaceae (3HBR) 46–92% | ||||
| 180 | 1.21 | 11.74 | Ile112 | Arg, Lys |
| 102 | 0.87 | 11.30 | Trp47 | Varies |
| Acinetobacter (3FV7) 44–99% | ||||
| 202 | 2.32 | 15.45 | Leu144 | Arg, Lys |
| 206 | 2.94 | 14.64 | Glu148 | Tyr |
Highest common taxonomy classifier, a PDB structure chosen as a representative of the group and the sequence identity range are given for each group. Second to last column gives the identity and position of the group-specific residue as it relates to the representative PDB. The final column gives the most common amino acid outside of the group in that alignment position.
Fig. 5.
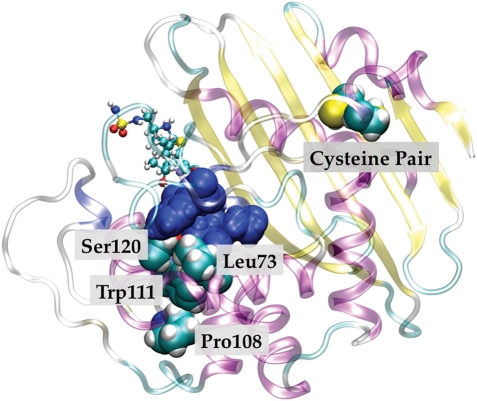
Group-specific residues for the α-, β- and γ-proteobacteria group annotated on the OXA-1 structure. Group-specific residues for the α-, β- and γ-proteobacteria group annotated on the OXA-1 structure. The group-specific residue pairs Pro108/Trp111 and Leu173/Ser120 (cyan) both make contact with the surface of the active site (blue).
In the largest group made up of α-, β- and γ-proteobacteria ranging in pairwise sequence identity from 22 to 93% and containing the OXA-1 sequence, there are three pairs of two group-specific residues in contact with one another. As in other parts of this report, the numbering corresponds to the PDB entry 3ISG. Pro108/Trp111 makes contact with one another with Trp111 making contact with the active site surface. Leu73/Ser120 is also in contact with one another and both make contact with the active site surface. Thus within this particular class, evolutionary constraints exist beyond the active site with these residues modulating the active site surface possibly enabling the detoxification of specific β-lactams.
Finally, there are two group-specific cysteine residues (Cys37 and Cys59) that we discovered through refinement of the sequence alignment. Examination of OXA-1 crystal structures reveals that these two residues reside on β3 and helix-2, and have their sulfur atoms 3.75 Å (PDB entry 3ISG) and 2.8 Å (PDB entry 1M6K) apart from each other. These atoms are clearly defined in the electron density maps (Sun et al., 2003; Schneider et al., 2009). The solution used to grow these crystals was pH 7.5. Yet, given both their proximity and their conservation within the largest Class D sub-group raises an important question. Does the Cys37–Cys59 pair form a disulfide bridge in an oxidizing environment? Certainly, no residue is in a position to sterically hinder such a minor movement of these residues to bring them into a reactive position. The absence of a disulfide bond in the crystal is likely due to the reducing cytoplasmic environment for the vector expression vs. the enzyme's physiological environment, i.e. more oxidizing periplasmic space (D.A.Leonard, private communication). A disulfide bond would have a huge impact on the stability of the protein (Trivedi et al., 2009) and hence immensely assist the survival of the species under assault from antibiotics. If such a mechanism were true, it may also partially explain why motifs 6/7 are not observed in this sub-group, since sequences of this group have other mechanisms by which to stabilize the hydrophobic core reducing the evolutionary constraint on this part of the enzyme in comparison to the other Class D enzymes. Regardless of any confirmation of these predictions, this finding also provides compelling evidence that these sequences constitute a unique group within the Class D family. Interestingly, the group consisting of sequences from Pseudomonas aeruginosa also contains a disulfide bond specific to this group. This disulfide is not in the exact location as the one for the group of α-, β- and γ-proteobacteria, residing instead on β3 and β2 (Cys44 and Cys51 in OXA-10 PDB entry 1K57; authors' assignment) and demonstrating presumably that additional stabilization of this region confers an advantage. This stabilization may also allow them to more readily adopt new functions (Tokuriki et al., 2008). A disulfide bond located within the β-lactam binding site of Class A SME is critical for activity (Majiduddin and Palzkill, 2003).
Finally, three Bradyrhizobium sequences have two conserved cysteines located 16 residues apart near the C-terminus (Cys262 and Cys278 in the sequence labeled L1_BRAJA, see Supplementary material). Using the MSA and other known structures, we predict that these cysteines may be located in relative proximity on the adjacent β4 and α12. However, the high sequence divergence in this area of the Class D MSA and lack of a structural template belonging to the same phylogenetic group tempers confidence in predicting the existence of a further disulfide bond in this extended sheet.
The other large group made up of β- and γ-proteobacteria contains the OXA-2 sequence of known structure (PDB entry 1K38); the residue numbering in the following discussion of this group will correlate with the numbering in PDB entry 1K38. As with the other large group, two group-specific residues are in contact. Phe76 and Ala132 are located on adjacent helices that make contact with the Ω-loop. Again, these residues may modulate the active site allowing it to respond more effectively to certain types of antibiotics. The other group-specific residue, Trp221, is located on the surface of the enzyme and thus may play an important role in the monomer–dimer equilibria that has been established in OXA-10 (Paetzel et al., 2000).
As is typical for sequences clustering into different groups within a phylogenetic tree, OXA-1 and OXA-2 show differences in their functional profiles. OXA-1 is active against amino- and ureido-penicillins as well as, to a smaller extent, narrow and broad spectrum cephalosporins (Poirel et al., 2010). And while none of the known OXA-1 like enzymes can hydrolyze ceftazidime (a third generation cephalosporin), OXA-2 like enzymes can (Poirel et al., 2010). Interestingly, two group-specific residues within the group containing the OXA-2 sequence are located near the Ω-loop at locations where substitutions are known to increase the hydrolytic profile both in Class D (Poirel et al., 2010) and Class A enzymes (Sampson et al., 2011).
Covarying Class D residues
A network of covarying residues was discovered through the use of a mutual information program (Gloor et al., 2005) using the MSA as input (see Fig. 6). As in previous sections, the residue identification and numbering follows that for the 3ISG PDB entry unless otherwise noted (but obviously, since these residues covary, there will be differences when visualizing other OXA structures).
Fig. 6.
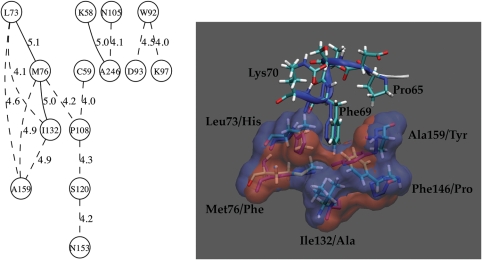
Covarying residues of Class D β-lactamases. (left) The network of covarying residues determined from the Mutual Information program (Gloor et al., 2005). Circled numbers correspond to numbering in the OXA-1 structure with the strength of connection shown along connecting line. (right) Structure comparison of OXA-1 QC/MM model and OXA-2 structures that provide a surface for binding Phe69. Surface is colored red for OXA-1 and blue for OXA-2. Residues are numbered according to OXA-1 structure with the OXA-2 residue shown after the slash.
The connections between Trp92 and Asp93/Lys97 occur in the P-loop and in the majority of sequences are Trp92–Asp93–Arg97. In the OXA-1 structure, residue 97 is not Arg but the similar amino acid, Lys that forms an ion pair with Asp93. In the MSA, when Trp92 changes to Tyr, then residues 93/97 change to Gly/Gln which likely do not interact. Therefore, we predict that these sequences may have different P-loop structures and hence a different shape to the substrate-binding pocket that forms opposite the concentration of highly conserved residues.
Despite being distant in sequence, the covarying Lys58 and Ala246 are in contact with each other while Asn105 is quite distant. It is possible that residue 105 is a false positive caused by the size of our data set. Regardless, residues 58 and 246 are in the same region as the Cys37–Cys59 pair. For many Class D sequences, this 58/246 pair is either Arg/Asn or Asp/His. In the OXA-2 structure (PDB entry 1K38), the Nε atom of Arg58 forms a hydrogen bond with the Oδ atom of Asn231 (equivalent to positions 58/246 in 3ISG). Given the distance this pair is from the active site and its proximity to a possible disulfide bond in the α-, β- and γ-proteobacteria OXA sub-group, we would predict that these residues are important for the stability of the protein or that it facilitates folding.
Examination of the connections between Leu73, Met76, Ile132 and Ala159 reveals that they are in close proximity with one another, and taken together present a surface for which to bind Phe69 of the active site helix, part of the PASTFK signature of Class D (see Fig. 6). Proper positioning of the Phe69 side chain is likely a crucial anchor for the entire segment that carries both nucleophilic residue (Ser67) and general base (Lys70) which have to be in their optimal position for catalysis. The compensations seem to be related to changes in either size, polarity or branched vs. unbranched side chains. Alignment of OXA-1 (3ISG) with OXA-2 (1K38) shows that in both structures these residues have largely overlapping surfaces. In most Class D sequences, when residue 132 switches from an Ala to a Phe then residue 159 switches from Tyr to Ala; a nearly symmetric swap. Yet in OXA-1, residue 132 is an Ile instead of Phe so it does not totally compensate for the big change in residue 159 from a Tyr to a Phe. But a change from Pro146 in OXA-2 to Phe146 in OXA-1 enables Phe146 to occupy a similar space as Tyr153 (equivalent to Ala159 in OXA-1) does in OXA-2. Thus, despite the fact that the OXA-1 structure is not a good representative to demonstrate these compensations in a majority of the sequences, the structure nevertheless preserves the surface for Phe69 through another substitution.
The network of covarying residues that connects to the Leu73–Met76–Ile132–Ala159 network through Pro108 all reside adjacent to other residues that covary, almost like another layer of covariation extending out from the active site. None of these residues are in close contact with each other although Pro108 is within 11 Å of Ser120 which may explain the covariation between the two.
In conclusion, the high-resolution MSA of 80 Class D β-lactamases and the analyses based on this effort (phylogenetics, group entropy, mutual information) has resulted in a extensive view of the evolutionary constraints in the Class D β-lactamase family. Class D enzymes have a highly conserved network of residues that extend from the active site to the ends of the Ω-loop; the Cα of the highly conserved Gly142 is 17 Å from the carbonyl oxygen atom of the β-lactam ring in our Michaelis complex model. All but two of the highly conserved residues are part of motifs that make up the active site and the adjacent secondary structure elements. The top five conserved motifs can tolerate mutations in the secondary structure elements outside the catalytic site as long as they preserve the secondary structure. Across the phylogenetic tree are sequences that have had their 3D structures solved by crystallography. These structures are well conserved including two long-loop regions, the Ω-loop and what we have designated the P-loop for its shape.
Phylogenetic analysis has enabled the separation of Class D sequences into distinct groups. Further experimental data are needed before it can be established if sequences within these groups share similar biochemical properties (Bush and Jacoby, 2010). Pairwise sequence identities within these groups can be very small (as low as 22% in the largest group), yet there are distinct signatures or group-specific residues that demarcate the groups. These group-specific residues are mainly located adjacent to the active site and likely modulate the properties of the conserved active site catalytic residues (Majiduddin and Palzkill, 2005; Phillips, 2008) and thus may correlate with their spectrum of activity. The largest sub-group of Class D sequences made up of α-, β- and γ-proteobacteria contains a group-specific Cys pair that is positioned to form a disulfide bond while a group of P. aeruginosa also contain a group-specific Cys pair in a disulfide bond but located on the opposite end of a β-sheet. Patterns of evolutionary constraint seen in these analyses support the prediction that formation of this disulfide bond may be important for survival in an oxidizing environment by maintaining the hydrophobic core of the enzyme. From the experimental standpoint, our findings are critical for helping to make decisions on the type and locations of residues to mutate that would yield the most insight into the structure–function and evolution of Class D β-lactamases.
Supplementary data
Funding
This work was supported by the National Center for Research Resources at the National Institutes of Health (grant number RR06009).
Supplementary Material
Acknowledgments
We would like to thank Dr John Hempel for editorial assistance.
References
- Afzal-Shah M., Woodford N., Livermore D.M. Antimicrob. Agents Chemother. 2001;45:583–588. doi: 10.1128/AAC.45.2.583-588.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashkenazy H., Kliger Y. Protein Eng. Des. Sel. 2010;23:321–326. doi: 10.1093/protein/gzp078. [DOI] [PubMed] [Google Scholar]
- Bailey T.L., Elkan C. Proceedgs of the Second International Conference on Intelligent Systems for Molecular Biology; 1994. pp. 28–36. [PubMed] [Google Scholar]
- Barlow M., Hall B.G. J. Mol. Evol. 2002;55:314–321. doi: 10.1007/s00239-002-2328-y. [DOI] [PubMed] [Google Scholar]
- Baurin S., Vercheval L., Bouillenne F., et al. Biochemistry. 2009;48:11252–11263.. doi: 10.1021/bi901548c. [DOI] [PubMed] [Google Scholar]
- Brooks B.R., Brooks C.L., III, Mackerell A.D., et al. J. Comp. Chem. 2009;30:1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush K. Curr. Opin. Microbiol. 2010;13:558–564. doi: 10.1016/j.mib.2010.09.006. [DOI] [PubMed] [Google Scholar]
- Bush K., Jacoby G.A. Antimicrob. Agents Chemother. 2010;54:969–976. doi: 10.1128/AAC.01009-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danel F., Hall L.M.C., Livermore D.M. J. Antimicrob. Chemother. 1999;43:339–344. doi: 10.1093/jac/43.3.339. [DOI] [PubMed] [Google Scholar]
- Danel F., Paetzel M., Strynadka N.C.J., Page M.G.P. Biochemistry. 2001;40:9412–9420. doi: 10.1021/bi0025969. [DOI] [PubMed] [Google Scholar]
- Drawz S.M., Bonomo R.A. Clin. Microbiol. Rev. 2010;23:160–201. doi: 10.1128/CMR.00037-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R.C. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Euro Surveill. 2010;15:pii=19719. ECDC Antimicrobial Resistance and Healthcare-Associated Infections Programme. Antimicrobial resistance 2010: global attention on carbapenemase-producing bacteria. [Google Scholar]
- Felsenstein J. Cladistics. 1989;5:164–166. [Google Scholar]
- Field M.J. J. Chem. Theory Comp. 2008;4:1151–1161. doi: 10.1021/ct800092p. [DOI] [PubMed] [Google Scholar]
- Fisher J.F., Meroueh S.O., Mobashery S. Chem. Rev. 2005;105:395–424. doi: 10.1021/cr030102i. [DOI] [PubMed] [Google Scholar]
- Frère J.M., Dubus A., Galleni M., Matagne A., Amicosante G. Biochem. Soc. Trans. 1999;27:58–63. doi: 10.1042/bst0270058. [DOI] [PubMed] [Google Scholar]
- Gloor G.B., Martin L.C., Wahl L.M., Dunn S.D. Biochemistry. 2005;44:7156–7165. doi: 10.1021/bi050293e. [DOI] [PubMed] [Google Scholar]
- Golemi D., Maveyraud L., Vakulenko S., Samama J.-P., Mobashery S. Proc. Natl Acad. Sci. 2001;98:14280–14285. doi: 10.1073/pnas.241442898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall B.G., Barlow M. Drug Resist. Updates. 2004;7:111–123. doi: 10.1016/j.drup.2004.02.003. [DOI] [PubMed] [Google Scholar]
- Hempel J., Perozich J., Wymore T., Nicholas H.B. Chem. Biol. Int. 2003;143–144:23–28. doi: 10.1016/s0009-2797(02)00165-5. [DOI] [PubMed] [Google Scholar]
- Humphrey W., Dalke A., Schulten K. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- Jones D.T., Taylor W.R., Thornton J.M. Comput. Appl. Biosci. 1992;8:275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- Jorgensen W.L., Chandrasekhar J., Madura J.D., Impey R.W., Klein M.L. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- Lichtarge O., Bourne H.R., Cohen F.E. J. Mol. Biol. 1996;257:342–358. doi: 10.1006/jmbi.1996.0167. [DOI] [PubMed] [Google Scholar]
- Livermore D.M., Woodford N. Curr. Opin. Microbiol. 2000;3:489–495. doi: 10.1016/s1369-5274(00)00128-4. [DOI] [PubMed] [Google Scholar]
- MacKerell A.D., Jr, Bashford D., Bellott M., et al. J. Phys. Chem. B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- Majiduddin F.K., Palzkill T. Antimicrob. Agents Chemother. 2003;47:1062–1067. doi: 10.1128/AAC.47.3.1062-1067.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majiduddin F.K., Palzkill T. Antimicrob. Agents Chemother. 2005;49:3421–3427. doi: 10.1128/AAC.49.8.3421-3427.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novais A., Comas I., Baquero B., Cantón R., Coque T.M., Moya A., González-Candelas F., Galán J.-C. PLoS Pathog. 2010;6:e10000735. doi: 10.1371/journal.ppat.1000735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paetzel M., Danel F., de Castro L., Mosimann S.C., Page M.G.P., Strynadka N.C.J. Nat. Struct. Biol. 2000;7:918–924. doi: 10.1038/79688. [DOI] [PubMed] [Google Scholar]
- Perozich J., Nicholas H., Wang B.C., Lindahl R., Hempel J. Protein Sci. 1999;8:137–146. doi: 10.1110/ps.8.1.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips P.C. Nat. Rev. Genet. 2008;9:855–867. doi: 10.1038/nrg2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirel L., Naas T., Nordmann P. Antimicrob. Agents Chemother. 2010;54:24–38. doi: 10.1128/AAC.01512-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queenan A.M., Bush K. Clin. Microbiol. Rev. 2007;20:440–458. doi: 10.1128/CMR.00001-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts E., Eargle J., Wright D., Luthey-Schulten Z. BMC Bioinformatics. 2006;7:382–392. doi: 10.1186/1471-2105-7-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose P.W., Beran B., Bi C., et al. Nucleic Acids Res. 2011;39:D392–D401. doi: 10.1093/nar/gkq1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampson J.M., Ke W., Bethel C.R., Pagadala S.R.R., Nottingham M.D., Bonomo R.A., Buynak J.D., van den Akker F. Antimicrob. Agents Chemother. 2011;55:2303–2309. doi: 10.1128/AAC.01360-10. doi:10.1128/AAC.01360–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider K.D., Bethel C.R., Distler A.M., Hujer A.M., Bonomo R.A., Leonard D.A. Biochmistry. 2009;48:6136–6145. doi: 10.1021/bi900448u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjölander K. Bioinformatics. 2004;20:170–179. doi: 10.1093/bioinformatics/bth021. [DOI] [PubMed] [Google Scholar]
- Sjölander K. PLoS Comp. Biol. 2010;6:1–3. doi: 10.1371/journal.pcbi.1000621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart J.J. J. Mol. Model. 2007;13:1173–1213. doi: 10.1007/s00894-007-0233-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun T., Nukaga M., Mayama K., Braswell E.H., Knox J.R. Prot. Sci. 2003;12:82–91. doi: 10.1110/ps.0224303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokuriki N., Stricher F., Serrano L., Tawfik D. PLoS Comp. Biol. 2008;4:e1000002. doi: 10.1371/journal.pcbi.1000002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trivedi M., Laurence J.S., Siahaan T.J. Curr. Protein Peptide Sci. 2009;10:614–625. doi: 10.2174/138920309789630534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vercheval L., Bauvois C., Di Paolo A., et al. Biochem. J. 2010;432:495–504. doi: 10.1042/BJ20101122. [DOI] [PubMed] [Google Scholar]
- Walsh T.R., Toleman M.A., Poirel L., Nordmann P. Clin. Microbiol. Rev. 2005;18:306–325. doi: 10.1128/CMR.18.2.306-325.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther-Rasmussen J., Høiby N. J. Antimicrob. Chemother. 2006;57:373–383. doi: 10.1093/jac/dki482. [DOI] [PubMed] [Google Scholar]
- Weinreich D.M., Delaney N.F., Depristo M.A., Hartl D.L. Science. 2006;312:111–114. doi: 10.1126/science.1123539. [DOI] [PubMed] [Google Scholar]
- Wilke M.S., Lovering A.L., Strynadka N.C.J. Curr. Opin. Microbiol. 2005;8:525–533. doi: 10.1016/j.mib.2005.08.016. [DOI] [PubMed] [Google Scholar]
- Wu C.H., Huang H., Nikolskaya A., Hu Z., Barker W.C. Comp. Biol. Chem. 2004;28:87–96. doi: 10.1016/j.compbiolchem.2003.10.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.