Abstract
We show that the Saccharomyces cerevisiae recombination protein Rad52 and the single-strand DNA-binding protein RPA assemble into cytologically detectable subnuclear complexes (foci) during meiotic recombination. Immunostaining shows extensive colocalization of Rad52 and RPA and more limited colocalization of Rad52 with the strand exchange protein Rad51. Rad52 and RPA foci are distinct from those formed by Rad51, and its meiosis-specific relative Dmc1, in that they are also detected in meiosis during replication. In addition, RPA foci are observed during mitotic S phase. Double-strand breaks (DSBs) promote formation of RPA, Rad52, and Rad51 foci. Mutants that lack Spo11, a protein required for DSB formation, are defective in focus formation, and this defect is suppressed by ionizing radiation in a dose-dependent manner. DSBs are not sufficient for the appearance of Rad51 foci; Rad52, Rad55, and Rad57 are also required supporting a model in which these three proteins promote meiotic recombination by promoting the assembly of strand exchange complexes.
Keywords: RAD52, RAD51, RPA, DMC1, recombination, meiosis
Double-strand breaks (DSBs) in DNA can be caused by chemical damaging agents, ionizing radiation, and replication at sites of single-strand nicks (for review, see Friedberg et al. 1995; Kogoma 1997). Accurate repair of DSBs can occur by a recombinagenic mechanism involving homologous joint molecule intermediates. RecA is the central protein required for this process in Escherichia coli (for review, see Kowalczykowski et al. 1994). The eukaryotic Rad51 protein is structurally and functionally similar to RecA and can promote the formation of homologous joints by strand exchange in vitro (for review, see Ogawa et al. 1993; see also Sung 1994; Baumann et al. 1996; Maeshima et al. 1996). RAD51 is required along with other members of the RAD52 epistasis group for recombinagenic repair of damage-induced DSBs in budding yeast (for review, see Game 1993; Friedberg et al. 1995; Shinohara and Ogawa 1995).
DSBs are also induced specifically during meiosis and are intermediates in meiotic recombination (Fig. 1) (for comprehensive reviews, see Kleckner 1996; Kupiec et al. 1997; Roeder 1997). The Spo11 protein appears to play a direct role in formation of meiotic DSBs by way of a transesterase cleavage activity (Keeney et al. 1997). Several other yeast genes, including MER2, RAD50, and MRE11, are also required for the formation of meiotic DSBs, although less is known about their mechanism of action. DNA ends at meiotic DSBs are resected to yield 3′ single-stranded tails, and this resection requires RAD50; breaks form but remain unresected in certain non-null mutants termed rad50S. The mechanism that promotes resolution of meiotic DSBs is closely related to that which leads to recombinational repair of DSBs in mitosis (for review, see Game 1993; Shinohara and Ogawa 1995). Resected DSBs are converted to homologous joint molecules by invasion of a homologous duplex by single-strand DNA (ssDNA). Normal formation of homologous joint molecules and recombination products during meiosis requires several mitotic repair genes including RAD52 (Borts et al. 1986; Ogawa et al. 1993; N. Kleckner, pers. comm.), RAD51, RAD55, and RAD57 (Shinohara et al. 1992; Schwacha and Kleckner 1997). The conversion of DSBs to homologous joint molecules during meiosis involves additional functions that are not required during mitotic repair of DSBs, including the meiosis-specific recA homolog DMC1 (Bishop et al. 1992; Schwacha and Kleckner 1997). Consistent with an in vivo role in homologous joint molecule formation, strand exchange activity has been detected recently for human Dmc1 in vitro (Li et al. 1997).
Figure 1.
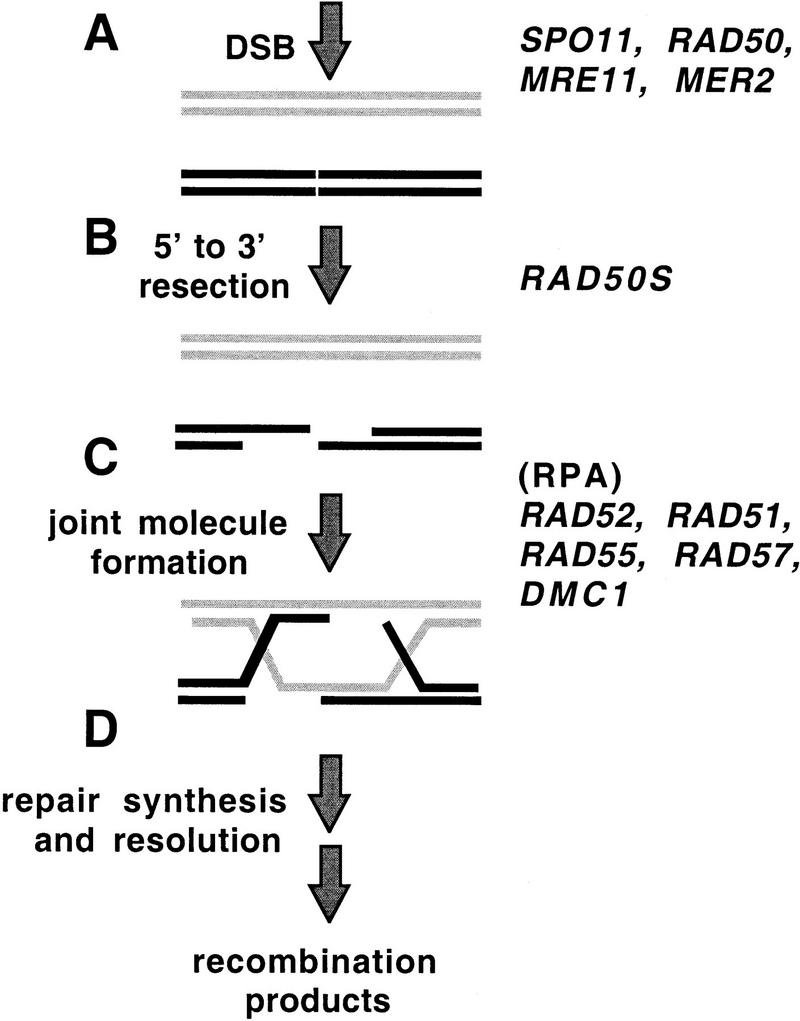
DNA intermediates in DSB-mediated meiotic recombination. Many other genes are also required for one or more of the steps shown (Shinohara and Ogawa 1995; Kupiec et al. 1997); however, only those genes relevant to the work in this paper are listed. (A) Meiosis-specific DSBs are generated by the product of the SPO11 gene. RAD50, MRE11, and MER2 are also required. (B) The ends at the sites of DSB are processed resulting in resection of the 5′ single strands. RAD50S refers to the activity that is absent in rad50S mutants. (C) ssDNA ends at DSBs invade homologous target duplex. Invasion leads to strand exchange with the “like” strand of a target duplex yielding a heteroduplex joint. All of the genes listed have been shown by mutant analysis to be required for optimal efficiency of this process. Mutant analysis has not shown directly that the genes that encode RPA are required for this step; however, several observations, including some presented in this paper, make it likely that RPA is involved at this step. (D) DNA repair synthesis generates two complete duplexes joined by a double Holliday junction, which are then resolved into recombination products.
Immunostaining of spread meiotic nuclei of Saccharomyces cerevisiae has shown that Rad51 and Dmc1 protein complexes assemble at multiple discrete sites (foci) on chromatin (Bishop 1994; Dresser et al. 1997). Rad51 and Dmc1 foci show extensive colocalization suggesting both proteins assemble at common sites during recombination. These foci are likely to be associated with recombination as they are detected specifically when DNA intermediates are present and as mutations that prevent the appearance and disappearance of meiotic DSBs have corresponding effects on foci (Bishop 1994; Shinohara et al. 1997). Foci are likely to represent higher order structures of Rad51 and Dmc1 proteins involved in RecA-like strand exchange because this process requires assembly of higher order multimers on ssDNA (for review, see Kowalczykowski et al. 1994; Stasiak and Egelman 1994). Homologs of yeast Rad51 and Dmc1 have been shown to assemble into foci during meiosis in a wide variety of eukaryotes (for review, see Roeder 1997), and immuno-electron microscopic studies indicate that antibodies raised against Rad51 localize to recombination-associated “zygotene nodules” in lily and mouse spermatocytes (Anderson et al. 1997; Moens et al. 1997).
Subnuclear foci of Rad51 and RPA have been detected in mitotic mammalian cells. Rad51 foci can be induced by treatment with DNA-damaging agents but are also found in untreated cells during S-phase (Haaf et al. 1995; Tashiro et al. 1996; Maser et al. 1997; Scully et al. 1997; Bishop et al. 1998). RPA also forms subnuclear foci, which are components of “replication centers” in vertebrate nuclei (for review, see Newport and Yan 1996). These observations are consistent with the role of RPA in replication (for review, see Wold 1997). Recently, immunostaining of spread mouse spermatocyte nuclei localized RPA to sites of homolog pairing and to Rad51 foci consistent with a role for RPA during pairing and recombination (Plug et al. 1997, 1998).
Here we show that two important recombination proteins, Rad52 and RPA, are present in multiple subnuclear complexes during meiosis in budding yeast. These proteins codistribute with each other and with Rad51 supporting the idea that they are all components of multiprotein recombination complexes. Meiotic DSB formation is shown to be associated with assembly of RPA, Rad52, and Rad51. Mutants defective in DSB formation are also defective in complex formation of all three proteins during midprophase, and this defect can be rescued with ionizing radiation, a treatment that induces DSBs. Although DSBs may be necessary for assembly of Rad51 complexes, they are not sufficient. Rad52, Rad55, and Rad57 are also required for Rad51 focus formation. We also report detection of a second type of RPA complex in both meiotic and mitotic cells that is likely to represent a form of the protein engaged in DNA replication.
Results
Rad52 and RPA are present in multiple subnuclear complexes
The properties of the Rad52 and RPA proteins strongly suggested that both Rad52 and RPA function in recombination as oligomeric complexes (Mortensen et al. 1996; Wold 1997; Shinohara et al. 1998) and would therefore be detectable during meiotic prophase by immunostaining of spread meiotic nuclei similar to Rad51 and Dmc1. As predicted, multiple subnuclear foci were detected by immunostaining wild-type nuclei with polyclonal antibodies directed against each of the proteins (Figs. 2A and 3). The frequency of positive staining nuclei and the number of Rad52 and RPA foci per nucleus peaked in meiosis when the majority of cells were in midprophase (∼3–4 hr after induction of meiosis and sporulation). At 3 hr after induction the average number of foci per nucleus was 27 ± 11 s.d. for Rad52 and 55 ± 18 s.d. for RPA with maximums of 98 and 142, respectively. Unlike Rad51 foci, which were not detected until 3 hr after induction of meiosis (Fig. 3A), Rad52 and RPA foci were detected in a large fraction of nuclei starting immediately after transfer to sporulation medium. The Rad52 and RPA foci detected between 0 and 2 hr tended to be fewer in number (averages of 15 ± 10 s.d. and 24 ± 13 s.d. foci per nucleus, respectively) and to stain less intensely than those detected at 3 and 4 hr.
Figure 2.
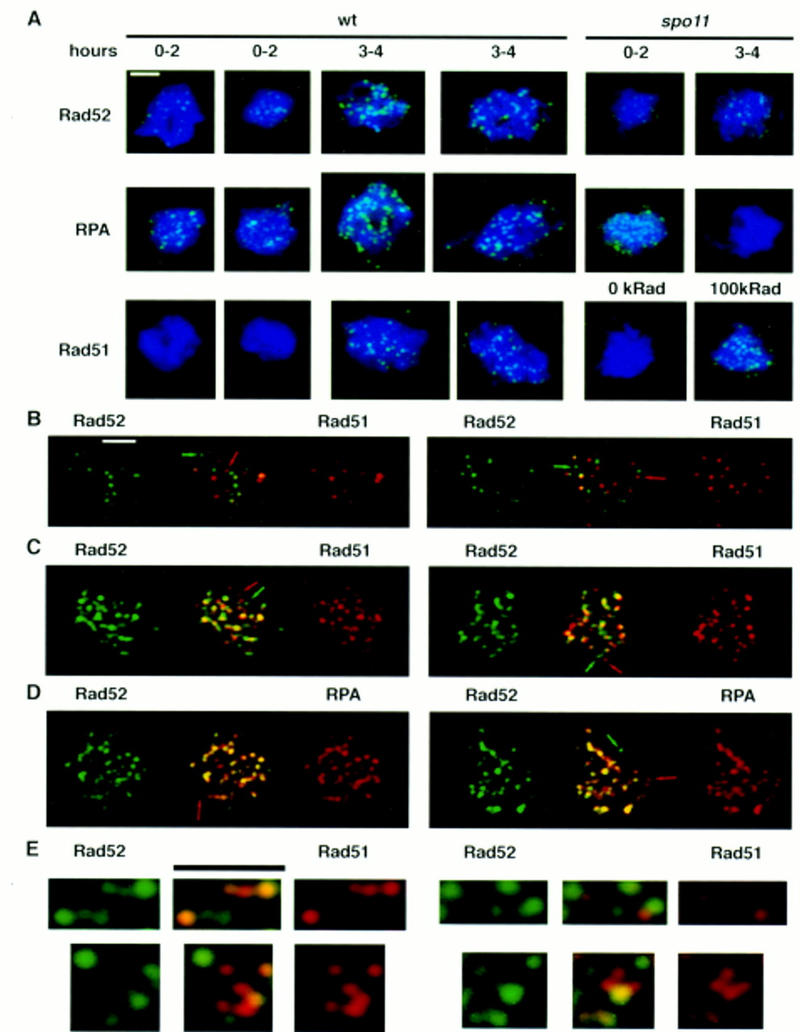
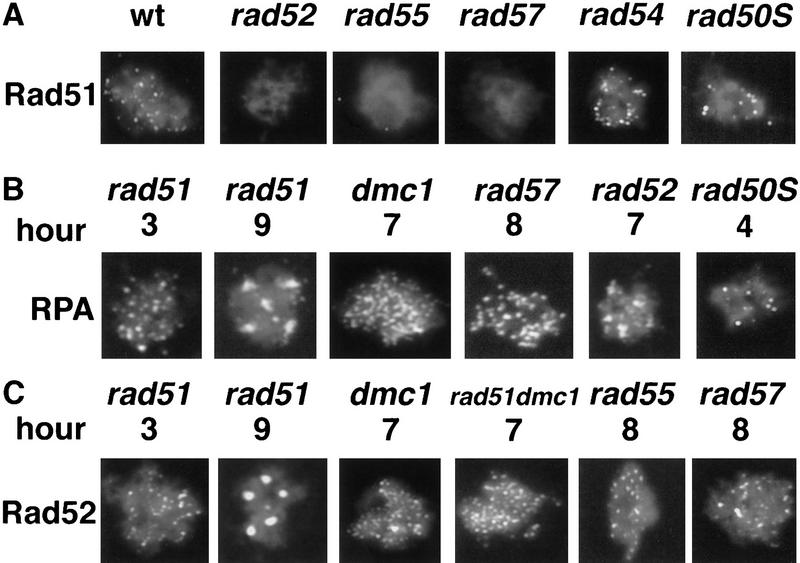
Rad52, RPA, and Rad51 localize to discrete complexes in spread meiotic nuclei. Cells were induced to undergo sporulation and spread meiotic nuclei were prepared and immunostained with antibodies that bind Rad52, RPA, or Rad51, (see Materials and Methods). All nuclei were stained with DAPI to detect chromatin. Pictures shown are pseudocolored composites of monochrome images. Immunostaining patterns are shown in green and DAPI staining patterns in blue. (A) Anti-Rad52, anti-RPA, and anti-Rad51 immunostaining of wild-type and spo11 meiotic nuclei. Nuclei shown are typical of those seen at the times indicated. In the Rad51-staining set one of the spo11 mutant nuclei shown was treated with a dose of 100 krads of ionizing radiation 1 hr before harvesting (cells were irradiated at 3 hr after meiotic induction). (B,C) Colocalization of Rad52 with Rad51. Double immunostaining with a combination of Rad52 and Rad51 antibodies in wild type (B, 3 hr) and dmc1 (C, 7 hr). Antibodies used were rabbit anti-Rad52 and guinea pig anti-Rad51. Single-staining images are pseudocolored from the original black and white images and the merged image is a two-channel combination of the original black and white images. Yellow indicates combination of the two signals. Arrows indicate single-staining foci. (D) Rat anti-Rad52 and rabbit anti-RPA were used for detection in dmc1. The nuclei shown are from the same preparations as in C. (E) Close ups of Rad52–Rad51 double-stained nuclei (using directly conjugated antibodies) showing single-staining foci and double-staining foci with different relative contributions of Rad52- and Rad51-specific signals. Bars, 2 μm.
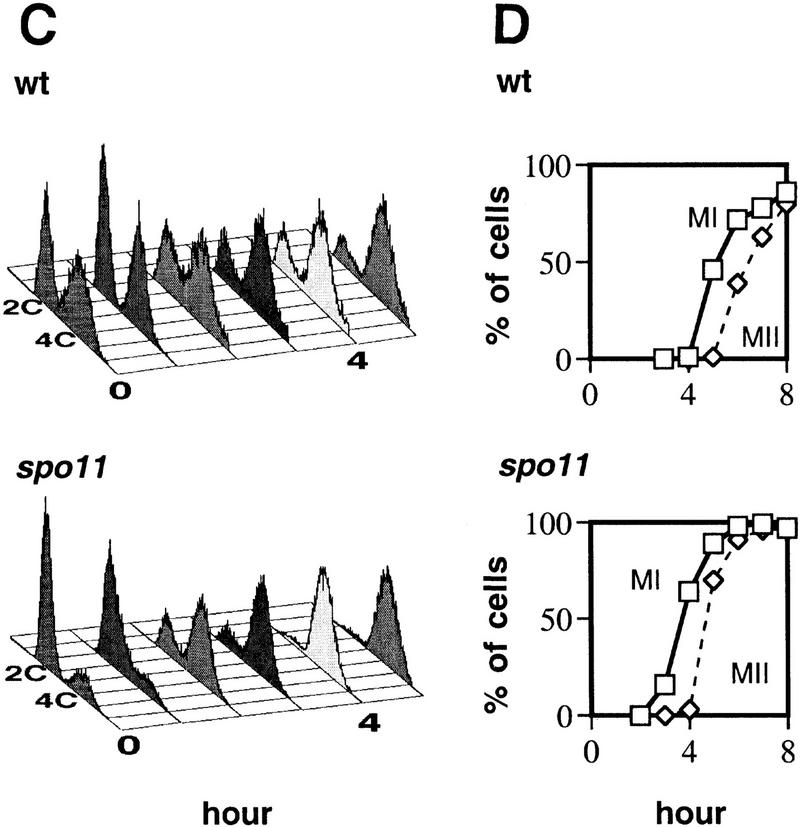
Figure 3.
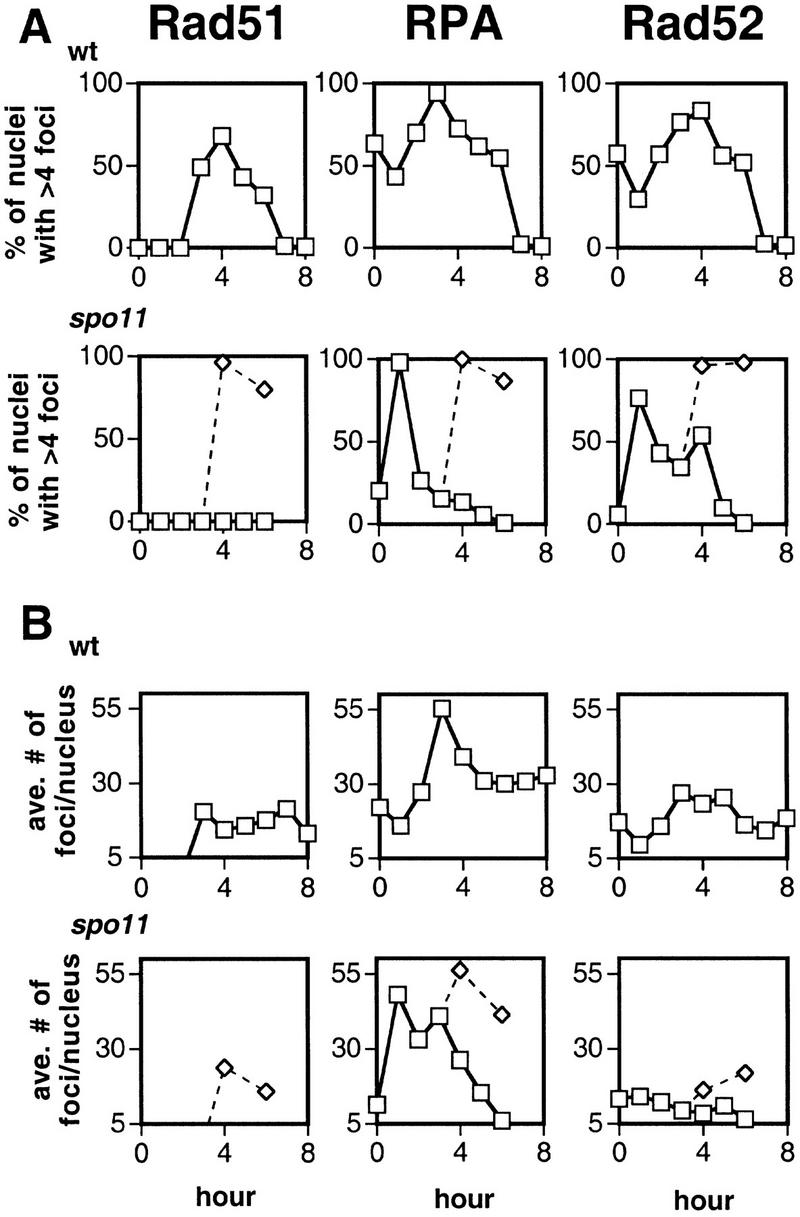
Time course analysis of Rad51, RPA, and Rad52 staining in wild-type and spo11 nuclei. The spo11 culture was split at 3 hr and 50% was irradiated with a dose of 100 krad (⋄). (A) Percent of nuclei that contain subnuclear foci. Counts are derived from images of 50–55 random nuclei from each time point. Nuclei were scored as staining positive if they contained at least five foci. Spores present on the slide were included in the calculation of total nuclei. (B) The average number of foci among staining positive nuclei in wild type and spo11. (C) Analysis of DNA content by FACS for wild type and spo11. Because preparation of spread nuclei requires an additional incubation of unfixed cells for at least 25 min at 30°C, the samples in this analysis are expected not to have progressed as far in the cycle as those used to prepare spread nuclei. (D) Analysis of meiotic divisions by fluorescent microscopy of DAPI-stained cells from wild type and spo11.
Colocalization of Rad52 with Rad51 and RPA
To determine whether Rad52 and RPA foci colocalize with each other and with Rad51 foci, double-immunostaining experiments were carried out (Fig. 2B–E; Table 1). In wild type ∼26 ± 0.5% s.e. of foci detected were Rad52–Rad51 double-staining foci (i.e., foci that contained a detectable level of both Rad51- and Rad52-specific signal). We also examined a dmc1 mutant in which Rad51 and Rad52 foci accumulate and stain more intensely than those in wild-type cells (Bishop 1994; discussed below). Although the frequency of double-staining foci in the dmc1 mutant was significantly higher than that in wild type (42 ± 2.9% s.e., P = 0.0011, <0.0001), the majority of foci detected in the mutant were single-staining foci.
Table 1.
Analysis of Rad52, Rad51, and RPA distributions
| Probe 1a
|
Probe 2
|
Strainb
|
Method
|
Percentages
|
No. of foci/nucleusc
|
||||
|---|---|---|---|---|---|---|---|---|---|
| both
|
1 only
|
2 only
|
both
|
1 only
|
2 only
|
||||
| A. Colocalization frequencies | |||||||||
| α-Rad52 (rabbit) | α-Rad51 (guinea pig) | wt | merged | 25 | 42 | 33 | 7.7 (2.5) | 14 (4.9) | 11 (6.6) |
| α-Rad52TR (rabbit) | α-Rad51F (rabbit) | wt | overlay | 26 | 37 | 37 | 14 (5.0) | 20 (6.0) | 20 (7.8) |
| α-Rad52 (rabbit) | α-Rad51 (guinea pig) | dmc1Δ | merged | 45 | 40 | 15 | 28 (9.0) | 25 (7.3) | 8.8 (3.9) |
| α-Rad52 (rat) | α-Rad51 (rabbit) | dmc1Δ | merged | 38 | 28 | 34 | 17.8 (5.6) | 13 (2.7) | 16 (5.4) |
| α-Rad52TR (rabbit) | α-Rad51F (rabbit) | dmc1Δ | overlay | 42 | 27 | 31 | 29 (6.6) | 19 (5.5) | 21 (7.1) |
| α-Rad52TR (rabbit)d | α-RPA (rabbit) | dmc1Δ | merged | 91 | 1.1 | 8.1 | 70 (12.6) | 0.8 (1.4) | 6.2 (4.7) |
| α-Rad52 (rat) | α-RPA (rabbit) | dmc1Δ | merged | 81 | 6.2 | 13 | 39 (6.5) | 3.0 (1.7) | 6.5 (5.1) |
| α-Rad52TR (rabbit)d | α-RPA p34 (rabbit) | dmc1Δ | merged | 78 | 3.0 | 19 | 56 (9.3) | 2.2 (1.7) | 14 (8.6) |
| α-RPA (rabbit) | α-Rad51 (guinea pig) | wt | merged | 45 | 27 | 28 | 23 (7.2) | 13 (4.5) | 15 (7.2) |
| α-RPA (rabbit) | α-Rad51 (guinea pig) | dmc1Δ | merged | 55 | 20 | 25 | 38 (5.8) | 14 (4.9) | 18 (6.3) |
| B. Estimates of fortuitous Rad52–Rad51 colocalization | |||||||||
| Overlay sourcee | |||||||||
| α-Rad52TR (rabbit) | α-Rad51F (rabbit) | unflipped | 31 | 35 | 34 | 8.4 (3.1) | 10 (5.4) | 10 (4.5) | |
| α-Rad52TR (rabbit) | α-Rad51F (rabbit) | flipped | 6.1 | 48 | 46 | 2.4 (2.1) | 16 (5.0) | 16 (5.4) | |
| “Rad52”f | “Rad51”f | simulated | 3.2 | 49 | 48 | 1.3 (0.95) | 18 (5.9) | 17 (6.2) | |
(TR) Antibody is directly conjugated to Texas Red-X; (F) antibody is directly conjugated to fluorescein.
Strains used are wild type (wt), NKY1314, or NKY1551 at 3 hr; dmc1Δ, ASY103, at 7 hr.
Numbers shown are averages of foci per nucleus, considering only nuclei with five or more foci; numbers in parentheses indicate standard deviations.
Staining used direct conjugate after blocking goat anti-rabbit secondary with preimmune serum.
Overlay sources are described in Materials and Methods. (Unflipped) Rad51 and Rad52 overlays from circular regions of wild-type nuclei. These data serve as a reference for comparison with flipped and simulated overlays. (Flipped) Rad52 overlays from unflipped analysis misoriented in six ways. (Simulated) The average of 600,000 focal patterns generated with Dotstat.
“Rad52” and “Rad51” refer to the source of input data for generation of simulated focus patterns (see Materials and Methods for details).
The intensity of both Rad52 foci and Rad51 foci was variable within a single nucleus. This range in staining intensity was seen for both single- and double-staining foci and in both wild-type and dmc1 mutant nuclei. Furthermore, the relative intensity of the two signals varied from one double-staining focus to the next (Fig. 2E).
Because the frequency of Rad51–Rad52 colocalization was fairly low, it was of interest to estimate the fraction of colocalization accounted for by fortuitous superposition of complexes. Two methods were used to estimate random colocalization frequency and these yielded similar estimates (3% and 6%; Table 1; see Materials and Methods for a discussion of details and limitations of these methods). These analyses indicate that the observed level of Rad52 and Rad51 codistribution is not the result of random distribution of foci on the spreading surface.
Although the amount of Rad52–Rad51 colocalization is significant, Rad52 foci show more extensive colocalization with RPA. Rad52–RPA double immunostaining of nuclei from a dmc1 mutant at 7 hr in meiosis showed that 86 ± 5.0% s.e. of foci were double stained. The limited sensitivity of our Rad52 probes prevented analysis of relative Rad52–RPA codistribution in wild-type cells (see Materials and Methods).
Association of DSBs with Rad51, RPA, and Rad52 foci
Four genes required for DSBs are required for Rad51 foci
To determine whether the initiation of recombination is required for the formation of Rad51 foci, we examined strains carrying null mutations in one of four genes required for DSB formation: SPO11, RAD50, MRE11, and MER2. We did not detect Rad51 foci in any of the four mutants indicating that execution of an early stage in meiotic recombination is required for the formation of Rad51 foci (Figs. 2A and 3; Table 2).
Table 2.
Genetic requirements for focus formation I
| Straina
|
Probeb
|
||
|---|---|---|---|
| α-Rad51
|
α-Rad52
|
α-RPA
|
|
| wt | + | + | + |
| spo11 | − | +/−c | +/−d |
| rad50 | − | N.D. | N.D. |
| mer2 | − | N.D. | N.D. |
| mre11 | − | N.D. | N.D. |
Strains used are wild type (wt), NKY1314 and NKY1551; spo11Δ, NKY676 and DKB490; rad50Δ, NKY1352 mer2Δ, TNT464; mre11Δ, OSY17.
Antibodies as per Materials and Methods. (N.D.) Not determined.
+/− indicates less intense staining than wild type at 3–4 hr.
Foci only prevalent at early times (see text).
SPO11 is required for some but not all RPA and Rad52 foci
Given that most DSBs are detected starting ∼2 hr after induction of meiosis, the detection of RPA and Rad52 foci at earlier times suggests that recombination is not required for appearance of all Rad52 and RPA foci. To determine whether later Rad52 and RPA foci are associated with ongoing recombination, the effects of a spo11 mutation were examined. A spo11 mutation did not alter dramatically the frequency of RPA foci during premeiotic S phase (Fig. 3A). However, spo11 did reduce the frequency of RPA staining to <15% at times after the bulk of the culture completed premeiotic S phase (after 2 hr in spo11 compared to after 3 hr in wild type in the experiment shown). Thus, although S-phase RPA foci were SPO11 independent, the majority of post S-phase foci were SPO11 dependent.
The effect of a spo11 mutation on the frequency of Rad52-positive nuclei was significant, but not as pronounced as it was for RPA-positive nuclei; ∼50% of nuclei retained Rad52 foci up to 3 hr after S phase was complete, although the intensity of the foci remained as low as that of early prophase foci (Figs. 2A and 3A). The number of Rad52 foci per nucleus was also significantly lower at similar times after S phase in spo11 compared to wild type (Fig. 3B; P = 0.001, <0.0001).
Radiation induces formation of Rad51, Rad52, and RPA foci in a
spo11 mutant The observation that SPO11 is required for normal appearance of foci suggested that focus formation was promoted by DSBs. This suggestion predicted that DSBs induced artificially by ionizing radiation would promote formation of recombination protein foci in the spo11 mutant. This approach was also suggested by a previous study showing that ionizing radiation partially suppresses the spore inviability phenotype of spo11 mutants (Thorne and Byers 1993). spo11 cells were induced to initiate meiosis and allowed to complete the bulk of premeiotic DNA synthesis. The cells were then exposed to 100 krads of γ-rays (Friedberg et al. 1995) and returned to sporulation conditions. As predicted, irradiation induced the appearance of Rad51, RPA, and Rad52 foci (Figs. 2A and 3). These results suggest that DSBs promote recombination protein focus formation.
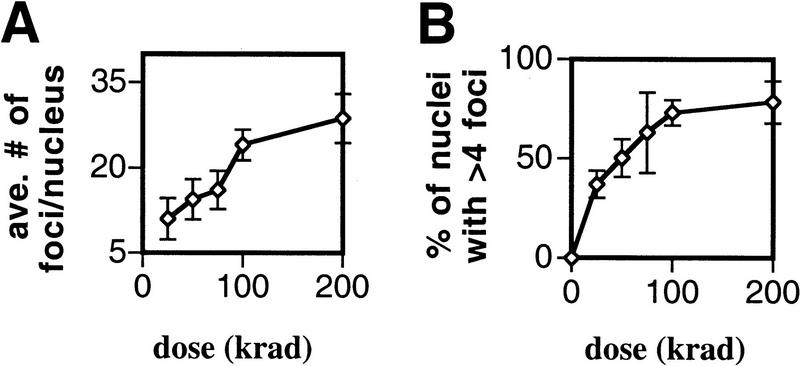
To determine whether the number of Rad51 foci induced by radiation was related to the amount of radiation-induced damage, a meiotic culture of spo11 cells was split into several aliquots and these aliquots were treated with different doses of radiation. Two dose-dependent effects on Rad51 foci were observed. First, considering only the fraction of the population that responded to radiation (i.e., that contained more than the background level of foci), the average number of foci per nucleus increased with increasing dose in a roughly linear fashion up to 100 krads (Fig. 4A). The number of foci induced in this responding fraction was ∼0.25 foci per krad. Second, the fraction of cells that contain foci 3 hr after cells were irradiated increased with dose (Fig. 4B). This increase in the responding fraction is not predicted if a random distribution of foci among a single population of nuclei is assumed (see legend of Fig. 4 for details). This result suggests that the size of the radiation dose can determine whether some cells contain foci 3 hr after irradiation.
Figure 4.
Ionizing radiation induces a dose-dependent response for Rad51 focus formation. A spo11 mutant (NKY676) was induced to undergo sporulation and exposed to γ rays after 3 hr in meiotic medium. Nuclei were prepared for immunostaining 3 hr after irradiation. (A) The average number of foci per positive staining nucleus (i.e., per nucleus with more than four foci). Error bars represent the s.e.m. obtained from three experiments. (B) The percent of positive staining nuclei. The fraction of nuclei with less than five foci is larger than predicted if a normal distribution of foci among nuclei is assumed. Normal distributions based on the mean and variance of the focus counts in the total population were generated for each dose and used to determine the expected frequency of nuclei with less than five foci, assuming a single population. This method yielded predicted frequencies for nuclei with less than five foci of 0.52, 0.40, 0.26, 0.16, and 0.12 for 25, 50, 75, 100, and 200 krads, respectively. The corresponding observed frequencies were 0.63, 0.50, 0.37, 0.27, and 0.22. All of these values are significantly higher than the predicted frequencies using the binomial probability test (P < 0.0058). This analysis indicates that the distribution of foci in nuclei is not accounted for by a random distribution of foci in a single population. Thus, the population of cells irradiated displays heterogeneity with respect to the ability to form or retain foci.
RPA foci are detected during mitotic S phase
A potential function of early RPA and Rad52 foci in premeiotic replication is suggested by detection of SPO11-independent foci at early times and by previous results in metazoan cells (see Discussion). To determine whether RPA forms visible complexes during mitosis in yeast, 100 spread nuclei from a log phase culture were examined; 43% of nuclei showed a similar punctate pattern to that seen at early times after induction of meiosis (data not shown). To determine whether mitotic nuclei that contain RPA foci represent one or more specific stages of the cell cycle, haploid MATa cells were arrested in G1 with the mating pheromone α-factor, washed free of the pheromone, and returned to normal growth medium. This treatment allows examination of at least one round of synchronous mitosis. Aliquots of synchronized cells were removed from the culture at 15-min intervals and assayed for the formation of RPA foci and DNA content (Fig. 5). RPA foci are rarely detected in α-factor-arrested cells, but appear in >90% of nuclei after the cells are released from arrest (around the time of the G1/S transition). The frequency of nuclei containing RPA foci drops dramatically when the majority of cells have completed S phase. Thus, when cells are released from α-factor arrest, RPA foci are found predominantly within an interval that includes late G1 and S phase. Although Rad52 foci were detected in a subset of mitotic nuclei, these foci stained very faintly. We did not observe significant changes in the frequency of Rad52 focus-containing nuclei as the culture progressed through the mitotic cell cycle (data not shown).
Figure 5.
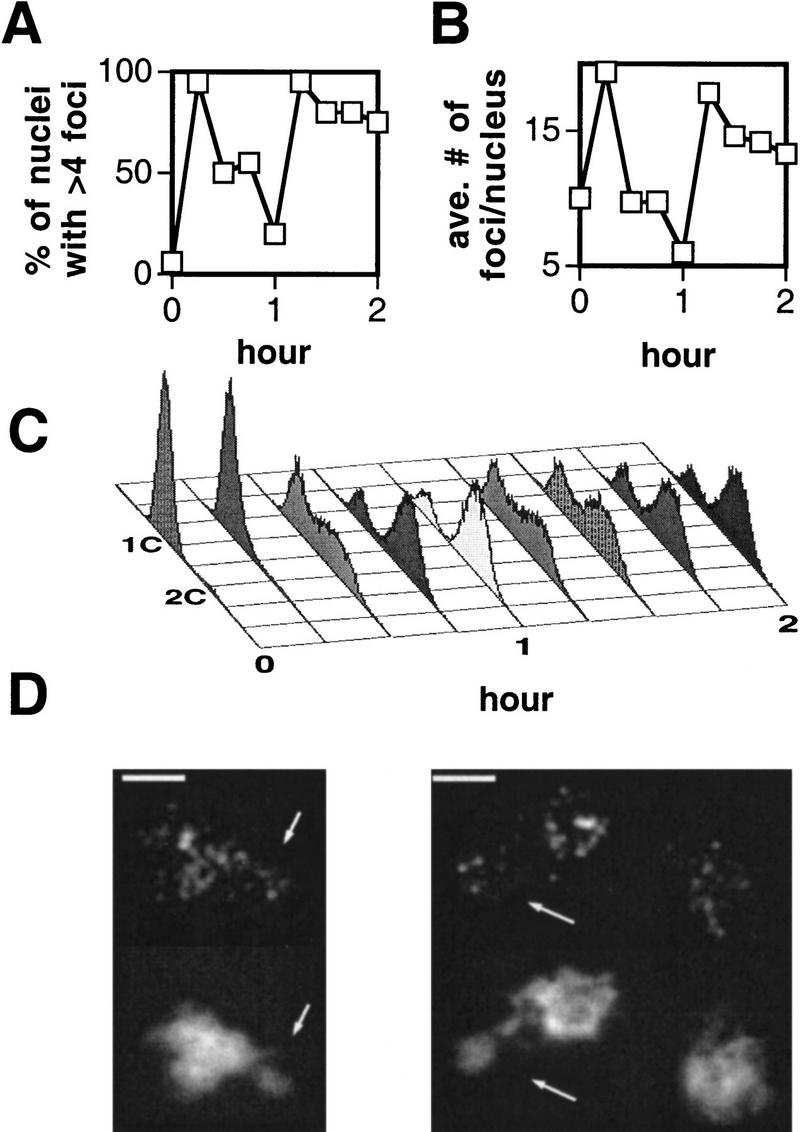
Time course of RPA staining in wild-type haploid nuclei after release from α-factor arrest. Haploid cells were released from α-factor arrest, spread, and probed with anti-RPA antibody. (A) Percent of unselected staining-positive nuclei. (B) The average number of foci per focus-positive nucleus seen in a sample of 20 nuclei (with the exception of the 0 hr time point, sample size = 14). The nucleoli of these spread nuclei tended to separate from the rest of the DAPI-stained material. Foci were detected in the nucleolus in a fraction of nuclei in all time points, but these were excluded from the analyses above. (C) FACS profiles of the same time course. As explained in the legend of Fig. 3, the cells used in this analysis are not expected to have progressed as far in the cycle as those used to prepare spread nuclei. (D) Representative nuclei from the 15-min time point. Arrows indicate nucleoli. Bars, 2 μm.
Genetic requirements for appearance and disappearance of foci after DSB formation
Normal resection at DSBs is not required for focus formation
To determine whether normal resection of DSBs to ssDNA is necessary for the formation of Rad51 or RPA foci, a rad50S mutant was assayed. Foci of each type formed in the rad50S mutant (Table 3; Fig. 6) and were retained at later times. At 8 hours after induction of meiosis, 98% of rad50S nuclei contained RPA foci; whereas 26% contained Rad51 foci. The average numbers of Rad51 and RPA foci detected in the rad50S mutant were less than the numbers seen in wild type: 9.6 ± 4.5 s.d. and 13 ± 3.0 s.d. at 4 hr in the mutant compared to 29 ± 14 s.d. and 47 ± 11 s.d. in wild type at 4 hr for Rad51 and RPA, respectively. These results indicate that a single-stranded binding protein and a strand exchange protein can assemble into oligomeric complexes in the absence of normal single-strand resection of duplex ends.
Table 3.
Genetic requirements for focus formation II
| Straina
|
Probeb
|
||
|---|---|---|---|
| α-Rad51
|
α-Rad52
|
α-RPA
|
|
| wt | + | + | + |
| dmc1 | +++c | ++ | ++ |
| rad50S | + | N.D. | ++ |
| rad51 | (−)d | ++ | ++ |
| rad51dmc1 | (−) | ++ | ++ |
| rad52 | − | (−) | ++ |
| rad55 | − | ++ | ++ |
| rad57 | − | ++ | ++ |
| rad54 | + | N.D. | N.D. |
Strains used are wild-type (wt), NKY1314 and NKY1551; dmc1Δ, ASY103; rad50S, NKY1392; rad51Δ, ASY102; rad52Δ, DKB988xDKB992; rad54Δ, MSY241 or DKB1322; rad55Δ, D012; rad57Δ, D0018 or D006.
Antibodies as per Materials and Methods. (N.D.) Not determined.
(++) Brighter foci and/or accumulate at late hours in meiosis (see text).
[−] Negative control.
Figure 6.
(A) Anti-Rad51 immunostaining of mutant nuclei. Nuclei representative of those seen at 3–4 hr are shown. (B–C) Anti-RPA and anti-Rad52 immunostaining of rad51, rad51dmc1, rad52, rad55, rad57, and rad50S mutants at the times indicated. Sample nuclei with large numbers of intensely staining foci or aggregates typical of those seen at later hours are shown. The RPA staining example in late hour rad52 is a typical intermediate phenotype. Bar, 2 μm.
RAD52,
RAD55, and RAD57 are required for formation of Rad51 foci RAD52, RAD55, and RAD57 are required for the transition from DSB to homologous joint molecule, and the effect of null mutations in these genes on the formation of Rad51 foci was determined. Mutations in RAD52, RAD55, and RAD57 prevented the appearance of Rad51 foci (Table 3; Fig. 6). Western analysis and whole cell immunostaining were carried out in each of the three single mutants to show that failure of Rad51 focus formation was not attributable to failure to express Rad51 nor to a failure to transport it to the nucleus (data not shown). We also did not detect Rad51 foci after irradiation of meiotic rad52 cells. This result is as expected because the failure of rad52 cells to form Rad51 foci is not the result of a defect in DSB formation. Unlike the other mutants examined, a rad54 mutant did not have a strong effect on the appearance or disappearance of Rad51 foci in meiosis (Table 3; Fig. 6). This result is consistent with previous data indicating that rad54 single mutants have limited effects on meiotic recombination and spore viability owing to partial redundancy of RAD54 function (for review, see Game 1993; see also Klein 1997; Shinohara et al. 1997).
Rad52 and RPA accumulate in mutants that block recombination at later stages
To determine whether assembly of Rad52 and RPA foci depends on other proteins that contribute to the conversion of DSBs to homologous joint molecules, the effects of rad51, rad55, rad57, dmc1, and rad51dmc1 mutations were examined. We also examined the effect of a rad52 mutation on RPA foci. Brightly staining foci were detected in all experiments indicating that the appearance of RPA and Rad52 foci is independent of RAD51, RAD55, RAD57, and DMC1 (Table 3 and typical examples of staining patterns shown in Fig. 6). However, the disappearance of RPA and Rad52 foci does depend on all four genes; foci did not disappear at the normal time in any of the mutants. The fraction of nuclei containing foci increased above the wild-type level to >80% of nuclei after 7 hr in sporulation medium as the mutants arrested in prophase. In addition, the number of foci detected per nucleus was 2- and 1.5-fold higher for Rad52 and RPA compared to wild type at 3 hr and the focus intensity was greater on average than that seen in wild type at 3 hr.
Starting at 7 hr after induction of meiosis, a dramatic change in RPA and Rad52 staining patterns was seen in the rad51, rad55, and rad57 mutants. Rather than continuing to display a fine punctate pattern, spread nuclei displayed four to six structures that appeared to be roughly spherical or ellipsoidal with diameters of 0.7 ± 0.1 s.d. (in μm). By 9 hr 95% of nuclei showed this pattern. These larger structures were not found in spreads of dmc1 mutant nuclei where the punctate pattern persisted.
Discussion
DSBs promote assembly of recombination complexes
Assembly of recombination complexes comprising Rad51, Rad52, and RPA is closely associated with the formation of meiotic DSBs. The assembly of such complexes is observed whether DSBs are formed during normal initiation of meiotic recombination or by ionizing radiation (IR). In irradiated spo11 nuclei the efficiency of focus induction is roughly 0.25 foci per kilorad in the subpopulation of cells that responds to radiation. This number is remarkably similar to estimates of the efficiency of DSB induction in response to IR in diploids (0.33–0.65 DSBs per krad) (Resnick and Martin 1976; Frankenberg et al. 1980). The impact of mutations that block the formation of DSBs, as well as dose dependence after IR, make it likely that assembly of recombination proteins during normal meiosis depends on formation of DSBs. An alternative possibility, which we think less likely, is that SPO11 might have a second function that promotes assembly independent of its direct role in DSB formation. If this were the case, assembly could be closely coordinated with break formation but not mechanistically dependent on it. For a discussion of DSB-independent interactions between homologs in meiosis see Kleckner (1996).
In addition to observing a dose-dependent increase in the number of IR-induced Rad51 foci, we also observed that the fraction of nuclei that contained foci was greater after high doses of radiation than after low doses. This observation suggests that not all the cells given low doses of radiation are equally competent to induce or retain Rad51 foci. This heterogeneity requires further study.
Normal degradation of DNA at DSB sites is not required for subnuclear assembly of Rad51 or RPA
The formation of foci in a rad50S mutant indicates that normal resection at DSBs is not required for assembly of RPA and Rad51. In vitro studies on Rad51 indicate that its strand exchange activity depends on the ability of the protein to assemble on ssDNA (Sung and Robberson 1995). Single strand tails form at the sites of DSBs in vivo and these tailed species accumulate in rad51 single mutants suggesting that they are normal substrates for Rad51 assembly (Shinohara et al. 1992). Given these observations, the ability of Rad51 foci to form in a rad50S mutant was not expected because a rad50S mutation dramatically reduces single-strand resection at DSBs (Alani et al. 1990). The ability of Rad51 and RPA to form foci in rad50S mutants can be explained in one of two ways. First, Rad51 assembly may occur by a mechanism that does not depend on a ssDNA substrate. Although it is unlikely that Rad51 and RPA can assemble directly on the duplex DNA ends that accumulate in rad50S, the Spo11 protein is attached covalently to the 5′ ends of unresected DSBs in a rad50S mutant (Keeney et al. 1997), and other proteins may be associated with ends as well. It is possible, therefore, that assembly of Rad51 and RPA is promoted by interaction with one or more proteins that are associated with DSB ends in the rad50S mutant. A second type of explanation for Rad51 and RPA foci in the rad50S mutant is that the ends at DSBs, while not resected normally, might contain single-stranded regions that go undetected in DSB assays. A helicase might unwind DNA at the sites of breaks and produce “split ends” in a manner roughly analogous to the action of the E. coli exonuclease V (RecBCD proteins) in the absence of RecD subunit (for review, see Myers and Stahl 1994). Alternatively, limited resection of ends might occur in the rad50S mutant.
Disassembly of recombination protein complexes
Disassembly of Rad52 and RPA complexes depends on Rad51, Rad55, Rad57, and Dmc1. Previously it was shown that Rad51 foci do not disappear at the normal time in a dmc1 mutant (Bishop 1994). Rad52 and RPA foci behave in a similar manner in dmc1 mutants as well as in rad51, rad55, rad57, and rad51dmc1 mutants. RPA foci also accumulate in a rad52 mutant. The increase in the fraction of cells containing foci and in the number of foci detected per nucleus at later times in these mutants is likely to be related to the fact that recombination events are blocked between the DSB and the homologous joint molecule stages in these mutants. The mutational blocks that prevent disappearance of foci also cause an apparent increase in the number of molecules in each focus, indicated by an increase in staining intensity in these mutants. This increase may be a result of the increase in the average size of ssDNA tracts caused by these mutations (Bishop et al. 1992; Shinohara et al. 1992; A. Shimohara, unpubl.).
Rad52, Rad55, and Rad57 promote assembly of Rad51 during meiotic recombination
Although they are likely necessary, DSBs are not sufficient for the appearance of Rad51-containing recombination complexes. At least one of the functions of Rad52, Rad55, and Rad57 during meiotic recombination is either to recruit Rad51 into oligomeric complexes or to stabilize the complexes once they form; we use the phrase “promotes assembly” to refer to both possibilities. The involvement of Rad52, Rad55, and Rad57 in assembly of Rad51 is fully consistent with the observation that all three of these proteins play a role in the normal conversion of DSBs to homologous joint molecules in vivo (Schwacha and Kleckner 1997; N. Kleckner, pers. comm.). Given that RPA interacts with Rad52 in vivo and in vitro (as discussed below), RPA may also be required for Rad51 assembly, but we have yet to test this possibility because the experiment is complicated by the fact that all the RPA-encoding genes are essential (Heyer et al. 1990; Brill and Stillman 1991).
Rad52 may promote assembly by direct contact with RPA and Rad51. Rad52 has been shown to interact directly with both Rad51 (Shinohara et al. 1992; Milne and Weaver 1993; Donovan et al. 1994; Shen et al. 1996; Sung 1997a) and RPA (Park et al. 1996; Shinohara et al. 1998). Our results show that Rad52 colocalizes with the same two proteins providing support for the view that these interactions are biologically relevant as have genetic suppressor studies (Milne and Weaver 1993; Firmenich et al. 1995; Smith and Rothstein 1995). The protein–protein interactions of Rad52 are likely to promote assembly of complexes by forming a bridge between Rad51 monomers and the RPA–ssDNA complex. Recruitment by protein–protein interaction may be essential for the recombination function of Rad51 because, unlike RecA, Rad51 does not bind ssDNA more efficiently than dsDNA (Shinohara et al. 1992; Benson et al. 1994; Sung and Robberson 1995; Baumann et al. 1996).
In vitro observations mirror the assembly function of Rad52, Rad55, and Rad57 in vivo. RPA has been shown to stimulate both yeast and human Rad51 strand exchange (Sung 1994; Baumann et al. 1996; Baumann and West 1997) at least in part by removing ssDNA secondary structure (for review, see Kowalczykowski et al. 1994). However, RPA inhibits the reaction when added before Rad51 (Sugiyama et al. 1997; Sung 1997b). The Rad55/Rad57 heterodimer (Sung 1997b) and Rad52 protein (Sung 1997a; New et al. 1998; Shinohara and Ogawa 1998) stimulate Rad51-mediated strand exchange in vitro by overcoming RPA-mediated inhibition. This stimulation of strand exchange proteins is thought to result at least in part from the ability of the accessory factors to allow initiation of assembly of Rad51 on RPA-coated ssDNA. Consistent with the idea that stimulation occurs by nucleation of cooperative Rad51 assembly is the low cellular abundance of Rad52 and the Rad55/Rad57 heterodimer compared to Rad51 (Sung 1997a,b).
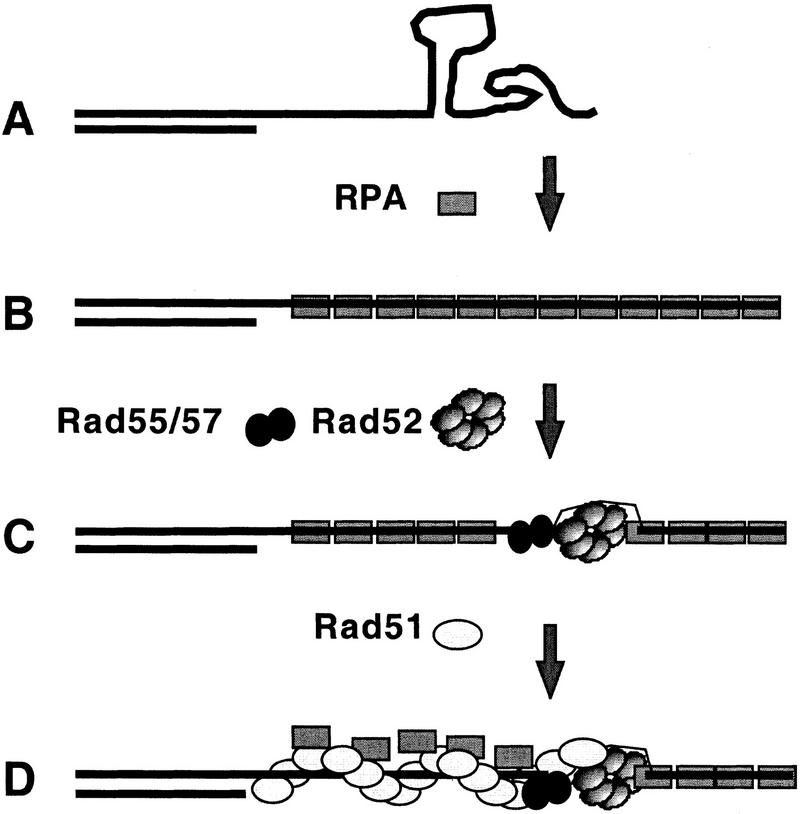
The results of many experiments, including those mentioned above and studies of functionally related proteins encoded by E. coli and phage T4 (Kowalczykowski et al. 1994; Salinas and Kodadek 1995 and references therein), have provided the basis for a model of Rad51 assembly as shown in Figure 7. Not included in the model is assembly of the meiosis-specific RecA homolog Dmc1, a process partially dependent on Rad51 (Bishop et al. 1992; Shinohara et al. 1997), because less is known about the nature of the interaction between Rad51 and Dmc1. Our results provide in vivo evidence supporting key features of the model. In addition, the version of the model shown reflects our observation that Rad52, Rad55, and Rad57 are not redundant with respect to their influence on Rad51 assembly during meiosis; all three single mutants fail to form Rad51 complexes in vivo. This model predicts that an appropriate combination of Rad52 and the Rad55/Rad57 heterodimer in vitro should yield more efficient stimulation of Rad51 strand exchange than either protein alone.
Figure 7.
Model for the assembly of RPA, Rad52, and Rad51. Several features of the model are borrowed from previous work (as indicated in the text). (A) Resection of DSB produces 3′ single-stranded tails. (B) RPA assembles onto resected DSBs removing secondary structures. (C) Rad52 and the Rad55/57 heterodimer bind the ssDNA–RPA filament generating a nucleation site for Rad51 filament assembly. A novel feature of this model is that both Rad52 and Rad55/Rad57 are required for generating the nucleation site. (D) Rad51 filaments are assembled in a cooperative manner displacing RPA from binding sites on ssDNA.
RPA and Rad52 foci are present during meiotic S phase
Meiotic and mitotic time course analysis indicates that there are two types of RPA foci. Several observations make it likely that early RPA protein complexes are engaged in replication including the detection of SPO11-independent RPA foci at early times in meiosis and during mitotic S phase, the known role of RPA in DNA replication, and previous experiments with metazoans showing that RPA localizes to subnuclear sites of DNA synthesis during mitosis (for review, see Newport and Yan 1996; Wold 1997). Thus, we think it is likely that early SPO11-independent RPA foci are functionally distinct from later SPO11-dependent foci in that they are engaged in replication. Although we have not been able to resolve temporally two rounds of RPA focus appearance and disappearance in wild-type nuclei, this result is expected if the different nuclei or even different chromosomes within each nucleus make the transition from replication to recombination in an asynchronous manner. Consistent with this possibility, the commitment to recombination occurs at different times on different chromosomes (Esposito and Klapholz 1981). It is also possible that completion of replication and initiation of recombination occur in distinct stages and that early RPA complexes do not disassemble when wild-type cells make the transition from premeiotic S phase to later recombination-associated phases. Replication-associated RPA complexes could evolve into recombination complexes at some point after the bulk of DNA synthesis is complete, but if this scenario is true then the persistence of RPA complexes, rather than their reassembly, depends on SPO11.
Like RPA foci, Rad52 foci are detected during meiotic S phase and these foci are SPO11 independent. It is possible that SPO11-independent Rad52 foci, like early RPA foci, represent a form of the protein that plays a functional role during premeiotic S phase. Consistent with a role for Rad52 during premeiotic S phase are observations linking Rad52 function to mitotic replication (Prakash 1981; Malkova et al. 1996; Zou and Rothstein 1997). However, a large fraction of nuclei containing Rad52 foci were found at times after the bulk of S phase was complete in the spo11 mutant (i.e., in nuclei where no DNA intermediates of replication or recombination were expected to be present). Thus, although brightly staining SPO11-dependent Rad52 foci are likely to be functioning recombination complexes, we cannot eliminate the possibility that SPO11-independent or mitotic Rad52 foci are nonfunctional aggregates that form in vivo or as an artifact of the spreading procedure.
Recombination complexes do not appear to have a consistent stoichiometry
Although significant colocalization is seen between Rad51, Rad52, and RPA the relative distributions of Rad51 and Rad52 suggest that recombination complexes vary in composition. The relative contribution of the two fluorescent probes to the signal at double-staining foci varies in wild-type nuclei for Rad51 and Rad52. If a double-staining focus represents a single protein complex, variation in double-staining signals from one focus to the next is likely to indicate that complexes do not have a set stoichiometry. In addition, single-staining foci are detected. Of particular note are the Rad51 single-staining foci detected under conditions where detection of Rad51 foci is entirely dependent on Rad52 function. The complexity of the staining pattern can be explained in three ways. First, more than one distinct recombination pathway may operate in the wild-type cell (Dresser et al. 1997). Second, the composition of complexes could evolve from one type to another (Plug et al. 1998 and references therein), that is, Rad52 might dissociate (or become inaccessible to the antibody) after promoting Rad51 assembly. Third, the assembly pathways might be substantially insensitive to the differences in composition revealed by the double-staining method. For example, Rad51 assembly might be promoted by assemblies of Rad52 that vary dramatically in size or epitope accessibility with some of the assemblies escaping detection. Although our results do not allow us to eliminate completely any of these explanations, the second explanation does not easily account for the mixture of focus types observed when disappearance of Rad51 and Rad52 foci is blocked by a dmc1 mutation. An alternative explanation for the increase in Rad51–Rad52 double-staining foci caused by a dmc1 mutation is that there are different relative amounts of the two proteins at each focus. Stalling progression of recombination with a dmc1 mutation results in an accumulation of both types of molecules, which in turn results in detection of both pecies at a larger number of sites.
The results of this study and previous studies indicate that DSBs promote the assembly of recombination complexes containing RPA, Rad52, Rad51, and Dmc1, which in turn promote the formation of homologous joint molecules. Immunostaining patterns suggest that these recombination complexes vary significantly in composition and this variation could reflect functional differences between complexes. Most important, the results provide the first in vivo evidence supporting a model in which the accessory factors Rad52, Rad55, and Rad57 promote strand exchange by promoting assembly of Rad51 into functional oligomeric complexes.
Materials and methods
Yeast strains
All yeast strains for meiotic cytology are isogenic SK-1 derivatives. All strains used share the following markers: MATa/MATα, lys2/lys2, ho::LYS2/ho::LYS2, ura3/ura3, leu2::hisG/leu2::hisG. Additional relevant markers and strains are wild type, NKY1314, and NKY1551; dmc1Δ::LEU2/dmc1Δ::LEU2, ASY103; rad50Δ::hisG/rad50Δ::hisG, NKY1352; rad50KI81:: URA3/rad50KI81::URA3, NKY1392; rad51Δ::hisG–URA3–hisG/rad51Δ::hisG–URA3–hisG, ASY102; rad51Δ::hisG–URA3–hisG dmc1Δ::LEU2/rad51Δ::hisG–URA3–hisG dmc1Δ:: LEU2, ASY106; rad54Δ::hisG–URA3–hisG/rad54Δ::hisG–URA3–hisG, MSY241; rad54Δ::hisG/rad54Δ::hisG, DKB1322; rad55Δ::hisG–URA3–hisG/rad55Δ::hisG–URA3–hisG, D012; rad57Δ::hisG–URA3–hisG/rad57Δ::hisG–URA3–hisG, D0018 or D006; spo11::hisG–URA3–hisG/spo11::hisG–URA3–hisG, NKY676 or DKB490; mer2Δ::hisG/mer2Δ::hisG, TNT464; mre11Δ::hisG/mre11Δ::hisG, OSY17. All rad52Δ strains are derived from crosses with NKY2502, a gift of A. Schwacha and N. Kleckner (Harvard University, Cambridge, MA). NKY2502 has all but first six amino acids of the RAD52 coding region deleted. Diploid strains that carry homozygous rad52Δ mutations have a tendency to lose the ability to undergo meiosis when grown vegetatively. Therefore, fresh diploids were generated by mating colony-purified haploid parents for each experiment (DKB988, MATα, lys2, ho::LYS2, ura3, leu2::hisG, his4X, trp1::hisG, rad52Δ::URA3 with DKB994, MATa, lys2, ho::LYS2, ura3, leu2::hisG, his4B, ade2::LK, rad52Δ::URA3). RM157 (MATa, ade2-101, can1-100, his3-11,15; leu2-3,112; trp1-1, ura3-1; W303 strain background) was used for mitotic cell cycle analysis.
Cell culture and irradiation
Conditions for sporulation of yeast cultures were described previously (Alani et al. 1990; Bishop 1994). For mitotic synchrony experiments, a log phase culture of RM157 at a concentration of 2 × 106 to 4 × 106 cells/ml was arrested with 7.5 μg/ml α-factor (Sigma, St. Louis, MO) for 2 hr. Cells were then washed twice and returned to YPD. At various times, 15-ml samples taken for cytology and for FACS analysis. A GammaCell 60Co source (Atomic Energy of Canada Ltd., Kanata, Ontario) was used for irradiation. Cells were transferred to 50-ml polyurethane tubes and irradiated on ice.
FACS analysis
Cells were prepared for FACS analysis by a published protocol (Longhese et al. 1994) after fixation in 70% ethanol. Proteinase K (Boehringer Mannheim, Indianapolis, IN) was used in place of pepsin at 2 mg/ml at pH 7.5. Becton Dickenson Immuno Cytometry Systems (San Jose, CA) was used to analyze DNA content.
Protein analysis
In some cases, aliquots of resuspended spheroplasted cells were taken for parallel preparation of whole-cell protein extracts by adding 0.5 ml of spheroplasted cells to 0.5 ml of boiling 2× loading buffer with protease inhibitors [8% SDS, 100 mm Tris pH 7.5, 20 mm EDTA, 10 mm β-mercaptoethanol, 0.2 μg/ml leupeptin, 4 μg/ml aprotinin, 10 μg/ml antipain, 0.10 mm PMSF, and 1% bromophenol blue] and boiled for 1 additional min. Western analysis was performed by a standard method described in Shinohara et al. (1992) using Immobilon-P (Millipore, Bedford, MA), alkaline phosphatase-conjugated secondary antibodies (Bio-Rad, Hercules, CA), and enhanced chemiluminescense detection reagents (NEN Life Sciences Products, Boston, MA). A dilution series was run in parallel to demonstrate that the detection method produced a linear response at the relevant concentrations of protein.
Preparation and immunostaining of spread nucleoids, scoring of divisions, and immunofluorescence microscopy
Meiotic and mitotic cells were spheroplasted and spread as described previously (Klein et al. 1992; Bishop 1994). Methods for immunostaining and microscopy were described previously (Bishop 1994; Shinohara et al. 1997) with antibody concentrations described below.
Meiotic cultures were examined to determine the fraction of cells that had carried out the first and second divisions of meiosis (MI and MII). Aliquots (0.5 ml) were fixed by addition of 0.5 ml of 95% ethanol. Ethanol-fixed cells were stained with 0.1 μg/ml DAPI. Cells were considered to have completed MI if they contained two to four DAPI staining bodies (nuclei) and MII if they contained three to four staining bodies.
Preparation of IgG and direct conjugate antibodies
Rabbit anti-sera raised against Rad52 and Rad51 were purified on protein A columns using a standard protocol (Harlow and Lane 1988). For use in some experiments, anti-RPA was affinity purified on a 1-ml NHS-activated Sepharose column (Pharmacia, Upsala, Sweden) conjugated with 1 mg of purified RPA. Antibodies were conjugated directly to fluorescein and Texas Red-X using the amide reactive compounds fluorescein-5-isothiocyanate and Texas Red–sulfonyl chloride following the instructions provided by the manufacturer (Molecular Probes, Eugene, OR). Rabbit polyclonal IgG was used for detection of Rad52 and Rad51 proteins unless otherwise noted in figure legends. Antibodies were diluted 1:200 for anti-Rad52 and 1:1000 for anti-Rad51 from 2 mg/ml stocks. Directly conjugated anti-Rad52-Texas Red-X and anti-Rad51-fluorescein were used at concentrations of 2 and 6 μg/ml, respectively. Specificity of antibody probes was demonstrated by staining spread nuclei from rad52Δ and rad51Δ mutant strains. Nonspecific nucleolar staining with anti-Rad52 IgG was eliminated by preabsorption with protein extract from meiotic rad52Δ cells. Evidence for specificity of the anti-RPA antibody, a gift from A. Sugino (Osaka University, Toyonaka, Osaka, Japan), was obtained by titration of staining by addition of purified RPA protein to anti-serum. A 1:100 dilution of anti-RPA antibody was incubated overnight at 4°C with 35 ng/ml of purified RPA, a gift of R. Kolodner (UCSD, La Jolla). This solution was then diluted a further 1:10 for staining wild-type meiotic spreads (a final dilution of 1:1000 for the antibody). No staining was visible in blocked antibody slides, whereas controls were positive for RPA foci. All experiments were carried out both with and without previous affinity purification of antibody with equivalent results.
Indirect immunostaining with rabbit anti-Rad52 was the most sensitive method available to us and was used in single-staining experiments. This antibody or two direct conjugates were also used for Rad51–Rad52 double-staining experiments. We were not able to use this method for Rad52–RPA double-staining experiments because our anti-RPA antibody was also raised in a rabbit. Instead, direct immunostaining with fluorochrome-conjugated anti-Rad52 rabbit antibody or indirect immunostaining with rat anti-Rad52 antibody was used. This method was successful at detection of Rad52 foci in dmc1 nuclei where foci stain particularly brightly but not in wild-type nuclei.
Conjugated secondary antibodies were obtained from Molecular Probes with the exception of fluorescein-conjugated goat anti-rat (ICN Pharmaceuticals, Costa Mesa, CA) and Texas Red-conjugated goat anti-rat (Vector, Burlingame, CA). Fluorescein-conjugated anti-rabbit was used at 1 μg/ml. Texas Red-conjugated anti-rabbit was used at 2 μg/ml. Fluorescein-conjugated anti-rat for Rad52 detection was used at 5 μg/ml. Texas Red anti-guinea pig for Rad51 detection was used at 5 μg/ml.
Focus scoring and statistical analyses
In double-staining experiments, monochrome images from each filter set were combined to produce a color composite image as described previously (Bishop 1994), except IP Lab Spectrum from Scanalytics (Fairfax, VA) was used. Colocalization was scored by examination of the composite images. A second method of scoring colocalization was also used in some experiments. In these cases the foci in the single pass images were marked by drawing artificial circles on top of the center of the foci using an overlay. These circles were of constant diameter which was similar to the foci in the nucleus. One set of artificial circles from the Texas Red filter set image was superimposed on an overlay prepared in a similar fashion from the fluorescein image. Greater than 50% overlap between the circles was considered colocalization. This analysis was done to facilitate the comparison of wild-type colocalization to the two methods for estimating fortuitous colocalization described below.
Two methods were used to estimate the frequency of fortuitous colocalization of staining foci in double-staining experiments. In both methods only the portion of the each nucleus defined by the largest circle that could be drawn within a uniform DAPI staining region was evaluated. The first method to generate random foci used the program Dotstat (Gotta et al. 1996). Dotstat generates two random sets of foci on a two-dimensional circular surface and then determines the frequency of colocalization of the two types of foci. The program uses the following parameters: number of foci of type 1, number of foci of type 2, diameter of surface, and diameter of foci. Input for all of these parameters is obtained from the experimental images. The diameter of the foci used was the diameter of the artificial circles used for scoring colocalization in the same samples. Input from 10 nuclei were used, and 600,000 focus patterns were generated and averaged.
A second method involved misorienting an overlay corresponding to the signal from one of the two probes. Copies of the overlay from the Texas Red image were misoriented in one of six different ways and superimposed on the unaltered overlay from the fluorescein image. New orientations used were: 90° rotation, 180° rotation, 270° rotation, horizontal flip, vertical flip, and a 270° rotation combined with a horizontal flip. Colocalization frequencies were scored for all six misoriented composites. Data were collected using images from a total of 10 nuclei and the results were averaged.
Both of the methods for estimating fortuitous colocalization ignore the possibility that the foci might only be capable of residing in a subnuclear domain. It is possible, for example, that focus assembly is limited to chromosome axes. We chose nuclei with a uniform diffuse DAPI staining pattern for analysis to minimize this possibility, but it cannot be excluded. If foci can only form in a nuclear subdomain, our estimates of fortuitous colocalization are low. Another shortcoming that applies only to the Dotstat analysis is that it assumes the pattern generated by spreading a three dimensional nucleus containing randomly distributed foci on a surface will be the same as patterns generated by random placement of foci on a two-dimensional surface.
Wilcoxon’s rank sum test was used to determine whether spo11 effected the frequency of foci in the focus-containing nuclei (i.e., in nuclei with more than four foci/cell). This test was used because the threshold used produces an asymmetric sample distribution. Time points were compared that represented equivalent times with respect to completion of the bulk of DNA synthesis. For example, in the experiment of Figure 3, the 4-hr wild-type time point was compared to the 3-hr spo11 time point. Wilcoxon’s rank sum test was also used to compare the frequency of Rad51–Rad52 double staining foci in wild-type and dmc1 nuclei. P values for Rad52–Rad51 colocalization in wild type versus dmc1 reflect comparisons within experiments in which the same antibody detection method, slide preparation, and scoring method were used (see Table 1).
Acknowledgments
We thank Anamitra Bhattacharyya, Uy Ear, Jeremy Grushcow, Howard Halpern, Nancy Kleckner, Eurie Lee, Bob McCarroll, John Petrini, Frank Stahl, and Phil Shumm for helpful comments and suggestions. We are also grateful to Hideyuki Ogawa and Tony Schwacha for strains; to Mike Snyder, Steve Brill, and Akio Sugino for antibodies; and Richard Kolodner for RPA protein. This work was supported by National Institute of General Medical Sciences grant GM50936 to D.K.B.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL dbishop@midway.uchicago.edu; FAX (773) 702-1968.
References
- Alani E, Padmore R, Kleckner N. Analysis of wild-type and rad50 mutants of yeast suggests an intimate relationship between meiotic chromosome synapsis and recombination. Cell. 1990;61:419–436. doi: 10.1016/0092-8674(90)90524-i. [DOI] [PubMed] [Google Scholar]
- Anderson LK, Offenberg HH, Verkuijlen WMHC, Heyting C. RecA-like proteins are components of early meiotic nodules in lily. Proc Natl Acad Sci. 1997;94:6868–6873. doi: 10.1073/pnas.94.13.6868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann P, West SC. The human Rad51 protein: Polarity of strand transfer and stimulation by hRP-A. EMBO J. 1997;16:5198–5206. doi: 10.1093/emboj/16.17.5198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann P, Benson FE, West SC. Human Rad51 protein promotes ATP-dependent homologous pairing and strand transfer reactions in vitro. Cell. 1996;87:757–766. doi: 10.1016/s0092-8674(00)81394-x. [DOI] [PubMed] [Google Scholar]
- Benson FE, Stasiak A, West SC. Purification and characterization of the human Rad51 protein, an analogue of E. coli RecA. EMBO J. 1994;13:5764–5771. doi: 10.1002/j.1460-2075.1994.tb06914.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop DK. RecA homologs Dmc1 and Rad51 interact to form multiple nuclear complexes prior to meiotic chromosome synapsis. Cell. 1994;79:1081–1092. doi: 10.1016/0092-8674(94)90038-8. [DOI] [PubMed] [Google Scholar]
- Bishop DK, Park D, Xu L, Kleckner N. DMC1: A meiosis-specific yeast homolog of E. coli recA required for recombination, synaptonemal complex formation, and cell cycle progression. Cell. 1992;69:439–456. doi: 10.1016/0092-8674(92)90446-j. [DOI] [PubMed] [Google Scholar]
- Bishop, D.K., U. Ear, A. Bhattacharyya, C. Calderone, M. Beckett, R.R. Weichselbaum, and A. Shinohara. 1998. Xrcc3 is required for assembly of Rad51-complexes in vivo. J. Biol. Chem. (in press). [DOI] [PubMed]
- Borts RH, Lichten M, Haber JE. Analysis of meiosis-defective mutations in yeast by physical monitoring of recombination. Genetics. 1986;113:551–567. doi: 10.1093/genetics/113.3.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brill SJ, Stillman B. Replication factor-A from Saccharomyces cerevisiae is encoded by three essential genes coordinately expressed at S phase. Genes & Dev. 1991;5:1589–1600. doi: 10.1101/gad.5.9.1589. [DOI] [PubMed] [Google Scholar]
- Donovan JW, Milne GT, Weaver DT. Homotypic and heterotypic protein associations control Rad51 function in double-strand break repair. Genes & Dev. 1994;8:2552–2562. doi: 10.1101/gad.8.21.2552. [DOI] [PubMed] [Google Scholar]
- Dresser ME, Ewing DJ, Conrad MN, Dominguez AM, Barstead R, Jiang H, Kodadek T. DMC1 functions in a Saccharomyces cerevisiae meiotic pathway that is largely independent of the RAD51 protein. Genetics. 1997;147:533–544. doi: 10.1093/genetics/147.2.533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esposito RE, Klapholz S. Meiosis and ascospore development. In: Strathern J, Jones E, Broach J, editors. The molecular biology of the yeast Saccharomyces. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1981. pp. 181–209. [Google Scholar]
- Firmenich AA, Elias AM, Berg P. A novel allele of Saccharomyces cerevisiae RFA1 that is deficient in recombination and repair and suppressible by RAD52. Mol Cell Biol. 1995;15:1620–1631. doi: 10.1128/mcb.15.3.1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankenberg SM, Frankenberg D, Blocher D, Adamczyk C. The linear relationship between DNA double-strand breaks and radiation dose (30 MeV electrons) is converted into a quadratic function by cellular repair. Int J Rad Biol. 1980;37:207–212. doi: 10.1080/09553008014550251. [DOI] [PubMed] [Google Scholar]
- Friedberg EC, Walker GC, Siede W. DNA repair and mutagenesis. Washington, D.C.: ASM Press; 1995. [Google Scholar]
- Game JC. DNA double-strand breaks and the RAD50–RAD57 genes in Saccharomyces. Semin Can Biol. 1993;4:73–83. [PubMed] [Google Scholar]
- Gotta M, Laroche T, Formenton A, Maillet L, Scherthan H, Gasser SM. The clustering of telomeres and colocalization with Rap1, Sir3, and Sir4 proteins in wild-type Saccharomyces cerevisiae. J Cell Biol. 1996;134:1349–1363. doi: 10.1083/jcb.134.6.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haaf T, Golub EI, Reddy G, Radding CM, Ward DC. Nuclear foci of mammalian Rad51 recombination protein in somatic cells after DNA damage and its localization in synaptonemal complexes. Proc Natl Acad Sci. 1995;92:2298–2302. doi: 10.1073/pnas.92.6.2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlow E, Lane D. Antibodies: A laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- Heyer WD, Rao MR, Erdile LF, Kelly TJ, Kolodner RD. An essential Saccharomyces cerevisiae single-stranded DNA binding protein is homologous to the large subunit of human RP-A. EMBO J. 1990;9:2321–2329. doi: 10.1002/j.1460-2075.1990.tb07404.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeney S, Giroux CN, Kleckner N. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell. 1997;88:375–384. doi: 10.1016/s0092-8674(00)81876-0. [DOI] [PubMed] [Google Scholar]
- Kleckner N. Meiosis: How could it work? Proc Natl Acad Sci. 1996;93:8167–8174. doi: 10.1073/pnas.93.16.8167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein F, Laroche T, Cardenas ME, Hofmann JF, Schweizer D, Gasser SM. Localization of RAP1 and topoisomerase II in nuclei and meiotic chromosomes of yeast. J Cell Biol. 1992;117:935–948. doi: 10.1083/jcb.117.5.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein HL. RDH54, a RAD54 homologue in Saccharomyces cerevisiae, is required for mitotic diploid-specific recombination and repair and for meiosis. Genetics. 1997;147:1533–1543. doi: 10.1093/genetics/147.4.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kogoma T. Stable DNA replication: Interplay between DNA replication, homologous recombination, and transcription. Microbiol Mol Biol Rev. 1997;61:212–238. doi: 10.1128/mmbr.61.2.212-238.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalczykowski SC, Dixon DA, Eggleston AK, Lauder SD, Rehrauer WM. Biochemistry of homologous recombination in E. coli. Microbiol Rev. 1994;58:401–465. doi: 10.1128/mr.58.3.401-465.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kupiec M, Byers B, Esposito RE, Mitchell AP. Meiosis and sporulation in Saccharomyces cerevisiae. In: Pringle JR, Broach JR, Jones EW, editors. The molecular and cellular biology of the yeast Saccharomyces. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1997. pp. 889–1036. [Google Scholar]
- Li Z, Golub EI, Gupta R, Radding CM. Recombination activities of HsDmc1 protein, the meiotic human homolog of RecA protein. Proc Natl Acad Sci. 1997;94:11221–11226. doi: 10.1073/pnas.94.21.11221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longhese MP, Plevani P, Lucchini G. Replication factor A is required in vivo for DNA replication, repair, and recombination. Mol Cell Biol. 1994;14:7884–7890. doi: 10.1128/mcb.14.12.7884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeshima K, Morimatsu K, Horii T. Purification and characterization of XRad51.1 protein, Xenopus RAD51 homologue: Recombinant XRad51.1 promotes strand exchange reaction. Genes Cells. 1996;1:1057–1068. doi: 10.1046/j.1365-2443.1996.d01-224.x. [DOI] [PubMed] [Google Scholar]
- Malkova A, Ivanov EL, Haber JE. Double-strand break repair in the absence of RAD51 in yeast: A possible role for break-induced DNA replication. Proc Natl Acad Sci. 1996;93:7131–7136. doi: 10.1073/pnas.93.14.7131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maser RS, Monsen KJ, Nelms BE, Petrini JH. hMre11 and hRad50 nuclear foci are induced during the normal cellular response to DNA double-strand breaks. Mol Cell Biol. 1997;17:6087–6096. doi: 10.1128/mcb.17.10.6087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milne GT, Weaver DT. Dominant negative alleles of RAD52 reveal a DNA repair/recombination complex including Rad51 and Rad52. Genes & Dev. 1993;7:1755–1765. doi: 10.1101/gad.7.9.1755. [DOI] [PubMed] [Google Scholar]
- Moens PB, Chen DJ, Shen Z, Kolas N, Tarsounas M, Heng H, Spyropoulos B. Rad51 immunocytology in rat and mouse spermatocytes and oocytes. Chromosoma. 1997;106:207–215. doi: 10.1007/s004120050241. [DOI] [PubMed] [Google Scholar]
- Mortensen UH, Bendixen C, Sunjevaric I, Rothstein R. DNA strand annealing is promoted by the yeast Rad52 protein. Proc Natl Acad Sci. 1996;93:10729–10734. doi: 10.1073/pnas.93.20.10729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers RS, Stahl FW. Chi and the RecBC D enzyme of Escherichia coli. Annu Rev Genet. 1994;28:49–70. doi: 10.1146/annurev.ge.28.120194.000405. [DOI] [PubMed] [Google Scholar]
- New JH, Sugiyama T, Zaitseva E, Kowalczykowski SC. Rad52 protein stimulates DNA strand exchange by Rad51 and replication protein A. Nature. 1998;39:407–410. doi: 10.1038/34950. [DOI] [PubMed] [Google Scholar]
- Newport J, Yan H. Organization of DNA into foci during replication. Curr Opin Cell Biol. 1996;8:365–368. doi: 10.1016/s0955-0674(96)80011-1. [DOI] [PubMed] [Google Scholar]
- Ogawa T, Shinohara A, Nabetani A, Ikeya T, Yu X, Egelman EH, Ogawa H. RecA-like recombination proteins in eukaryotes: Functions and structures of RAD51 genes. Cold Spring Harbor Symp Quant Biol. 1993;58:567–576. doi: 10.1101/sqb.1993.058.01.063. [DOI] [PubMed] [Google Scholar]
- Park MS, Ludwig DL, Stigger E, Lee SH. Physical interaction between human RAD52 and RPA is required for homologous recombination in mammalian cells. J Biol Chem. 1996;271:18996–19000. doi: 10.1074/jbc.271.31.18996. [DOI] [PubMed] [Google Scholar]
- Plug AW, Peters AHFM, Xu Y, Keegan KS, Hoekstra MF, Baltimore D, de Boer P, Ashley T. ATM and RPA in meiotic chromosome synapsis and recombination. Nature Genet. 1997;17:457–461. doi: 10.1038/ng1297-457. [DOI] [PubMed] [Google Scholar]
- Plug AW, Peters AHFM, Keegan KS, Hoekstra MF, de Boer P, Ashley T. Changes in protein composition of meiotic nodules during mammalian meiosis. J Cell Science. 1998;111:413–423. doi: 10.1242/jcs.111.4.413. [DOI] [PubMed] [Google Scholar]
- Prakash L. Characterization of postreplication repair in Saccharomyces cerevisiae and effects of rad6, rad18, rev3 and rad52 mutations. Mol & Gen Genet. 1981;184:471–478. doi: 10.1007/BF00352525. [DOI] [PubMed] [Google Scholar]
- Resnick MA, Martin P. The repair of double-strand breaks in the nuclear DNA of Saccharomyces cerevisiae and its genetic control. Mol & Gen Genet. 1976;143:119–129. doi: 10.1007/BF00266917. [DOI] [PubMed] [Google Scholar]
- Roeder GS. Meiotic chomosomes: It takes two to tango. Genes & Dev. 1997;11:2600–2621. doi: 10.1101/gad.11.20.2600. [DOI] [PubMed] [Google Scholar]
- Salinas F, Kodadek T. Phage T4 homologous strand exchange: A DNA helicase, not the strand transferase, drives polar branch migration. Cell. 1995;82:111–119. doi: 10.1016/0092-8674(95)90057-8. [DOI] [PubMed] [Google Scholar]
- Schwacha A, Kleckner N. Interhomolog bias during meiotic recombination: Meiotic functions promote a highly differentiated interhomolog-only pathway. Cell. 1997;90:1123–1135. doi: 10.1016/s0092-8674(00)80378-5. [DOI] [PubMed] [Google Scholar]
- Scully R, Chen JJ, Plug A, Xiao YH, Weaver D, Feunteun J, Ashley T, Livingston DM. Association of BRCA1 with Rad51 in mitotic and meiotic cells. Cell. 1997;88:265–275. doi: 10.1016/s0092-8674(00)81847-4. [DOI] [PubMed] [Google Scholar]
- Shen Z, Cloud KG, Chen DJ, Park MS. Specific interactions between the human RAD51 and RAD52 proteins. J Biol Chem. 1996;271:148–152. doi: 10.1074/jbc.271.1.148. [DOI] [PubMed] [Google Scholar]
- Shinohara A, Ogawa T. Homologous recombination and the roles of double-strand breaks. Trends Biochem Sci. 1995;20:387–391. doi: 10.1016/s0968-0004(00)89085-4. [DOI] [PubMed] [Google Scholar]
- ————— Stimulation by Rad52 of yeast Rad51-mediated recombination. Nature. 1998;39:404–407. doi: 10.1038/34943. [DOI] [PubMed] [Google Scholar]
- Shinohara A, Ogawa H, Ogawa T. Rad51 protein involved in repair and recombination in S. cerevisiae is a RecA-like protein. Cell. 1992;69:457–470. doi: 10.1016/0092-8674(92)90447-k. [DOI] [PubMed] [Google Scholar]
- Shinohara A, Gasior S, Ogawa T, Kleckner N, Bishop DK. Saccharomyces cerevisiae recA homologues RAD51 and DMC1 have both distinct and overlapping roles in meiotic recombination. Genes Cells. 1997;2:615–629. doi: 10.1046/j.1365-2443.1997.1480347.x. [DOI] [PubMed] [Google Scholar]
- Shinohara A, Shinohara M, Ohta T, Matsuda S, Ogawa T. Rad52 forms ring structures and co-operates with RPA in single-strand DNA annealing. Genes Cells. 1998;3:145–156. doi: 10.1046/j.1365-2443.1998.00176.x. [DOI] [PubMed] [Google Scholar]
- Shinohara M, Shita-Yamaguchi E, Buerstedde J-M, Shinagawa H, Ogawa H, Shinohara A. Characterization of the roles of the Saccharomyces cerevisiae RAD54 gene and a homolog of RAD54, RDH54/TID1, in mitosis and meiosis. Genetics. 1997;147:1545–1556. doi: 10.1093/genetics/147.4.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J, Rothstein R. A mutation in the gene encoding the Saccharomyces cerevisiae single-stranded DNA-binding protein Rfa1 stimulates a RAD52-independent pathway for direct-repeat recombination. Mol Cell Biol. 1995;15:1632–1641. doi: 10.1128/mcb.15.3.1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stasiak A, Egelman EH. Structure and function of RecA-DNA complexes. Experientia. 1994;50:192–203. doi: 10.1007/BF01924002. [DOI] [PubMed] [Google Scholar]
- Sugiyama T, Zaitseva EM, Kowalczykowski SC. A single-stranded DNA-binding protein is needed for efficient presynaptic complex formation by the Saccharomyces cerevisiae Rad51 protein. J Biol Chem. 1997;272:7940–7945. doi: 10.1074/jbc.272.12.7940. [DOI] [PubMed] [Google Scholar]
- Sung P. Catalysis of ATP-dependent homologous DNA pairing and strand exchange by yeast RAD51 protein. Science. 1994;265:1241–1243. doi: 10.1126/science.8066464. [DOI] [PubMed] [Google Scholar]
- ————— Function of yeast Rad52 protein as a mediator between replication protein A and the Rad51 recombinase. J Biol Chem. 1997a;272:28194–28197. doi: 10.1074/jbc.272.45.28194. [DOI] [PubMed] [Google Scholar]
- ————— Yeast Rad55 and Rad57 proteins form a heterodimer that functions with replication protein A to promote DNA strand exchange by Rad51 recombinase. Genes & Dev. 1997b;11:1111–1121. doi: 10.1101/gad.11.9.1111. [DOI] [PubMed] [Google Scholar]
- Sung P, Robberson DL. DNA strand exchange mediated by a RAD51–ssDNA nucleoprotein filament with polarity opposite to that of RecA. Cell. 1995;82:453–461. doi: 10.1016/0092-8674(95)90434-4. [DOI] [PubMed] [Google Scholar]
- Tashiro S, Kotomura N, Shinohara A, Tanaka K, Ueda K, Kamada N. S phase specific formation of the human Rad51 protein nuclear foci in lymphocytes. Oncogene. 1996;12:2165–2170. [PubMed] [Google Scholar]
- Thorne LW, Byers B. Stage-specific effects of X-irradiation on yeast meiosis. Genetics. 1993;134:29–42. doi: 10.1093/genetics/134.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wold MS. Replication protein A: A heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu Rev Biochem. 1997;66:61–92. doi: 10.1146/annurev.biochem.66.1.61. [DOI] [PubMed] [Google Scholar]
- Zou H, Rothstein R. Holliday junctions accumulate in replication mutants via a RecA homolog-independent mechanism. Cell. 1997;90:87–96. doi: 10.1016/s0092-8674(00)80316-5. [DOI] [PubMed] [Google Scholar]