Abstract
Aims
To test whether the TCF7L2 gene was associated with gestational diabetes, whether the association between TCF7L2 and gestational diabetes was independent of HLA-DQB1*0602 and islet cell autoantibodies, as well as maternal age, number of pregnancies, family history of diabetes and the HLA-DQB1 genotypes, and to test whether the distribution of HLA-DQB1 alleles was affected by country of birth.
Methods
We genotyped the rs7903146, rs12255372 and rs7901695 single nucleotide polymorphisms of the TCF7L2 gene in 826 mothers with gestational diabetes and in 1185 healthy control subjects in the Diabetes Prediction in Skåne Study. The mothers were also typed for HLA-DQB1 genotypes and tested for islet cell autoantibodies against GAD65, insulinoma-associated antigen-2 and insulin.
Results
The heterozygous genotypes CT, GT and TC of the rs7903146 (T is risk for Type 2 diabetes), rs12255372 (T is risk for Type 2 diabetes) and rs7901695 (C is risk for Type 2 diabetes), respectively, as well as the homozygous genotypes TT, TT and CC of the rs7903146, rs12255372 and rs7901695, respectively, were strongly associated with gestational diabetes (P < 0.0001). These associations remained statistically significant after adjusting for maternal age, number of pregnancies, family history of diabetes and HLA-DQ genotypes and were independent of the presence of islet cell autoantibodies. No interaction was observed between TCF7L2 and HLA-DQB1*0602, which was shown to be negatively associated with gestational diabetes in mothers born in Sweden (P = 0.010).
Conclusions
The TCF7L2 was associated with susceptibility for gestational diabetes independently of the presence of HLA-DQB1*0602 and islet cell autoantibodies and other factors such as maternal age, number of pregnancies, family history of diabetes and other HLA-DQ genotypes. The HLA-DQB1*0602 was negatively associated with gestational diabetes in mothers born in Sweden.
Keywords: gestational diabetes mellitus, HLA-DQ, islet cell autoantibodies, transcription factor 7-like 2
Introduction
Gestational diabetes mellitus is considered to be a heterogeneous metabolic disorder with mixed genetic aetiology and phenotypes [1]. Approximately 10% of women with gestational diabetes test positive for islet cell autoantibodies (GAD, IA-2, IA), which in previous studies have been shown to be risk factors for Type 1 diabetes post-partum [2–6]. Islet autoimmunity in gestational diabetes was reported to be related to higher frequencies of HLA-DR3 and -DR4 alleles, which are associated with Type 1 diabetes [7]. Previous studies have shown that there was a positive association between HLA-DR3 and HLA-DR4 and gestational diabetes in women with positive islet cell autoantibodies in some [8–10], but not all studies [11, 12]. In a previous report from our study, we observed that there was a negative association between gestational diabetes and HLA-DQB1*0602, which is negatively associated with Type 1 diabetes [13].
However, gestational diabetes in most cases expresses the same clinical features as Type 2 diabetes, including inadequate insulin secretion in relation to increased insulin resistance, and it is widely known that the majority of women with gestational diabetes develop Type 2 diabetes post-partum [14, 15]. Gestational diabetes has been associated with a number of genetic factors that have been shown to confer risk for Type 2 diabetes [16–20]. Among those genes, the transcription factor 7-like 2 (TCF7L2) gene was reported to be associated with the highest risk for Type 2 diabetes [21, 22] and even with risk for gestational diabetes [23, 24]. The TCF7L2 rs7903146 single nucleotide polymorphism (SNP) was associated with an increased risk of gestational diabetes in Scandinavian women [23], while the rs12255372 single nucleotide polymorphism was associated with susceptibility for gestational diabetes and affected the insulin response to oral glucose through interaction with the percentage of body fat in probands with gestational diabetes [24]. The three TCF7L2 single nucleotide polymorphisms most studied and shown to confer the highest risk to Type 2 diabetes and gestational diabetes are rs7903146 C>T, rs12255372 G>T and rs7901695 T>C [25], which are located within a well-defined linkage disequilibrium block [21, 26, 27]. These single nucleotide polymorphisms of the TCF7L2 gene were also reported to be associated with impaired insulin secretion in response to oral and intravenous glucose as well as arginine in subjects with Type 2 diabetes, suggesting that the TCF7L2 gene predisposes to the development of Type 2 diabetes through impaired insulin secretion, possibly attributable to an underlying mechanism in the β-cells [28].
The aims of the present study were to test (1) whether rs7903146 C>T, rs12255372 G>T and rs7901695 T>C single nucleotide polymorphisms of the TCF7L2 gene were associated with gestational diabetes, (2) whether the association between TCF7L2 and gestational diabetes was affected by the presence of either HLA-DQB1*0602, which was previously shown to be negatively associated with gestational diabetes, islet cell autoantibodies or both and (3) whether the association between TCF7L2 and gestational diabetes was affected by other confounding factors, such as maternal age, number of pregnancies, family history of diabetes and the HLA-DQB1 genotypes. We also studied the distribution of HLA-DQB1 alleles in our mothers with gestational diabetes, stratifying by country of birth.
Patients and methods
Patients
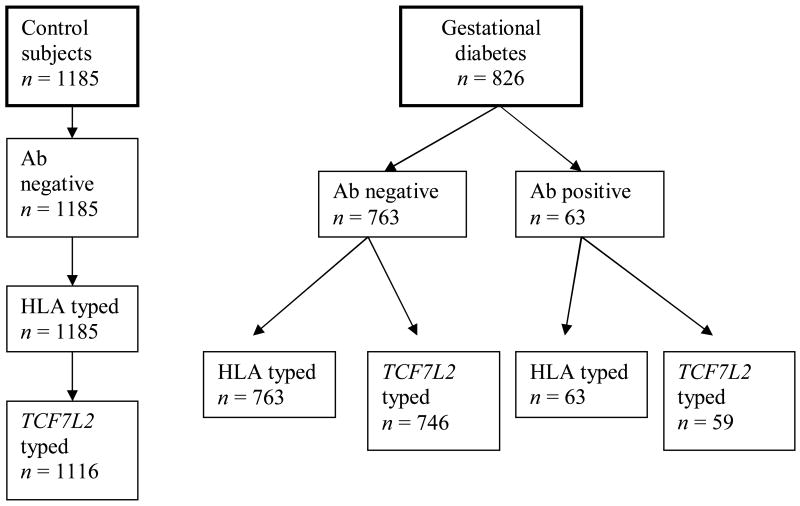
Mothers in this study were recruited from the Diabetes Prediction in Skåne (DiPiS) study, which is a population-based study conducted in the Skåne County in Southern Sweden and examining genetic and environmental risk factors that could contribute to development of Type 1 diabetes in children [13, 29–31]. All pregnant women were informed about DiPiS study at all maternity healthcare clinics in Skåne. From September 2000 until August 2004, cord and venous blood from approximately 35 000 mothers were obtained as dried blood spots (DBS) on filter papers (Schleicher & Schuell, Stockholm, Sweden) at the time of delivery. Out of those, 902 mothers were diagnosed with gestational diabetes at least once during the 4-year period, a number which is compatible with the 2–3% prevalence of gestational diabetes in Skåne, and we were able to find the stored blood samples for a total of 826 mothers with gestational diabetes. As 83% of all pregnancies in 2000–2004 were included, our study can be considered as population based, which gives us the opportunity to study the genetic background of mothers with gestational diabetes. If the mother had had more than one pregnancy complicated with gestational diabetes, the first pregnancy with gestational diabetes was chosen. Additionally, we randomly selected 1185 healthy mothers with normal glucose tolerance during pregnancy as control subjects (Fig. 1). Out of 826 mothers with gestational diabetes, 63 (7.6%) were found positive for at least one autoantibody against GAD65, insulinoma-associated antigen-2 (IA-2) and insulin (Fig. 1). All of the 1185 control subjects were selected because they were negative for all three islet cell autoantibodies. A total of 805 mothers with gestational diabetes and 1116 control subjects were genotyped for TCF7L2, while all 826 mothers with gestational diabetes and all 1185 control subjects were genotyped for HLA-DQB1 alleles.
Figure 1.
Percentage of HLA and TCF7L2 genotyped control subjects and mothers with gestational diabetes in the study
Gestational diabetes was diagnosed with a 75-g oral glucose tolerance test at gestational week 25–week 28 or at gestational week 12 if the pregnant woman had had a previous history of gestational diabetes or positive family history of diabetes. Gestational diabetes was defined as a 2-h capillary blood glucose ≥ 9.0 mmol/l according to the criteria recommended by the International Association of Diabetes and Pregnancy Study Groups (IADPSG) of the European Association for the Study of Diabetes [32]. If glucose concentrations were between 7.8 and 8.9 mmol/l, the women were tested again with a second 75-g oral glucose tolerance test within 1 week and, if the second value was normal (< 9.0 mmol/l), no more measures were required. The midwife recorded the health status of the mother on the filter for the dried blood spots.
Ethical approval was given for the DiPiS study by the Southern Sweden Ethical Review Board for the collection of cord and venous blood, together with genetic studies and biochemical analyses of these blood samples.
DNA extraction
Venous blood samples were prepared as dried blood spots on filter papers. DNA extraction from dried blood spots was carried out as reported previously [33]. Dried blood spots 3 mm in diameter were punched in a 96-well PCR plate (Abgene House, Epsom, UK) with the help of a DBS-puncher (1296-071 Delfia Dried Blood Spot punch, Wallac DBS puncher; PerkinElmer, Turku, Finland). In each of the 96 wells, 100 μl washing buffer [150 mmol/l sodium chloride (NaCl), 20 mmol/l Tris, 15% Tween 20, 0.1% bovine serum albumin (BSA), pH 7.4] was added and they were then sealed with Mylar plate-sealers (Thermo Labsystems, Franklin, MA, USA). The plates were left to shake (1296-003 Delfia plateshake; Wallac, Turku, Finland) overnight at 3–8 °C and were then centrifuged. Washing buffer was removed from the wells and 50 μl 10 mmol/l sodium hydroxide (NaOH) was added. The plates were then centrifuged and boiled in 100 °C for 10 min (PTC-100 Peltier thermal cycler; MJ Research, NV, USA). After boiling, 50 μl 100 mmol/l Tris-hydrogen chloride (HCl) was added and the samples were centrifuged again.
In order to identify the DNA concentration in the samples before PCR, we used the PicoGreen (Picogreen® dsDNA Quantitation Kit; Molecular Probes, Eugene, OR, USA). Approximately 10 μl solution from each well in the 96-well plates was taken and added in the wells of a 384-well plate. Sixteen wells were left empty, where a diluted series of λ DNA with concentrations: 0; 15 625; 31, 25; 62.5; 125; 250; 500; and 1000 ng/μl was added in the samples. Picogreen was diluted by 1:200 in TE buffer (10 mmol/l Tris, 1 mmol/l EDTA, pH 7.5) and 10 μl were added in each well. The plates were left to shake for 10 min and then the DNA concentration was measured by the use of ABI Prism 7900HT SDS (Applied Biosystems, Foster City, CA, USA). The DNA concentration varied between 1 and 372 ng/ μl for the different samples and the mean DNA concentration was 132 ng/ μl.
TCF7L2 genotyping
TCF7L2 genotyping was carried out in a 384-well plate (Eppendorf Mastercykler ep 384; Eppendorf North America, New York, NY, USA) using TaqMan allelic discrimination assay with 3 μl of a reaction solution which contained 2.5 μl/well TaqMan Universal PCR Mastermix (No AmpErase UNG), 0.125 μl/well TCF7L2 assay mix (TaqMan Pre-Designed SNP Genotyping Assays; Applied Biosystems) and 0.375 μl/well sterile water. In addition, 2 μl of extracted DNA was added to the 384-well plate and was analysed with the use of ABI Prism 7900HT SDS Thermal Cycle Conditions (10 min at 95° C, 15 s at 92 °C and 1 min at 60 °C (Applied Biosystems). For every plate, there were three samples—a –control sample and two samples of non-template controls). The results for these controls were the same for every analysis.
HLA
The HLA-DQB1 genotypes were analysed as previously described [29, 30, 31, 34]. HLA-DQB1 and DQA1 genotypes were typed by sequence-specific oligonucleotide probes using a Delfia hybridization assay (Perkin Elmer, Boston, MA, USA). The first set of probes defined the presence of HLA-DQB1*02, 0302, 0301, 0602, 0603 and 0604. The second set of probes defined the presence of additional DQB1 alleles. HLA-DQA1 probes defined the DQA1*0201, 03 and 05 alleles.
Islet cell autoantibodies
Islet cell autoantibodies against GAD65, IA-2 and insulin were measured in the cord blood of newborns with the use of standard radioligand binding assays as previously described [13, 31]. Autoantibodies were determined in a first combined screen in eluates from dried blood spots. The eluates were incubated overnight in duplicate with labelled antigen. The antibody-bound antigen was precipitated with Protein A Sepharose and the unbound antigen was removed by washing. The radioactivity of antibody-bound GAD65 and/or IA-2 was counted in a β-counter. The combined screen compared a positive reference with two negative reference samples in order to determine high levels. The diagnostic sensitivity for GAD65 autoantibodies using a cut-off at 31 RU/ml was 68% (1317/1950 patiens with Type 1 diabetes diagnosed during May 2005–September 2008 in Sweden). In the IA-2 autoantibody assay using a cut-off of 5 RU/ml the diagnostic sensitivity was 75% (1465/1950 patients). The GAD65 and IA-2 autoantibody assays showed mean interassay coefficients of variation of 14% and intra-assays of 8%. Samples greater than the 99th percentile of the combined screen were individually analysed for each antibody. In children born after 2001 with high individual GAD65 or IA-2 autoantibodies above the 99th percentile, the dried blood spots sample from the mother was also analysed for the two autoantibodies. The 99th percentile was defined for the entire population.
Insulin autoantibodies were analysed in cord blood serum samples incubated in duplicate wells in 96-well plates. Antibody-bound and free-labelled insulin were separated with 40% Protein A Sepharose and the radioactivity counted in a β-counter. Results were expressed in arbitrary units and samples above the 99th percentile were reanalysed in duplicate wells with 8 U/ml cold insulin in order to identify serum samples with non-specific binding. The data were expressed in relative units based on the degree of blocking 125I-insulin to the highest positive reference standard by cold insulin and the inter- and intra-assays coefficient of variations were 6.8–7.8% and 5.2–7.8%, respectively. Samples above the 99th percentile were considered to have high levels.
Statistical analysis
The Hardy–Weinberg equilibrium for the three single nucleotide polymorphisms, both among control subjects and mothers with gestational diabetes, were tested by a permutation based χ2- test that was available in the ‘genetics’ library of the statistical package R (version 2.10.1; XXXXXX, XXXXX). Among Swedish-born and non-Swedish-born mothers with gestational diabetes, linkage disequilibrium between each pair of single nucleotide polymorphisms was estimated by direct correlation (r2) and by the scaled linkage disequilibrium estimate (D′). Marker independence (r2, D′ = 0) was tested by the χ2 statistic.
The association of each TCF7L2 single nucleotide polymorphism genotype with mothers with gestational diabetes was first examined using a simple logistic regression model. The genotype containing only non-susceptible alleles was chosen as the reference group. All mothers with gestational diabetes were initially included as cases and compared with control subjects. However, as none of the mothers in the control group had islet cell autoantibodies, analysis was repeated with and without autoimmune mothers with gestational diabetes as case subjects to examine if autoimmune gestational diabetes explained the association. Other possible confounding factors such as HLA-DQB1, maternal age, number of pregnancies, country of birth and family history were later included in the model to test for the independent association of the single nucleotide polymorphism with gestational diabetes status. We included a HLA-DQB*TCF7L2 single nucleotide polymorphism interaction term to test for statistical (multiplicative) interaction of the HLA-DQB1*0602 allele with the susceptible TCF7L2 single nucleotide polymorphism allele. Additionally, we examined for biological interaction using an additive model. A logistic regression model was fitted that included a created variable of four mutually exclusive groups coded as: 0 = subject had DQB1*0602 and non-susceptible TCF7L2 allele; 1 = subject had DQB1*0602 and susceptible TCF7L2 allele; 2 = subject had non-DQB1*0602 and non-susceptible TCF7L2 allele; and 3 = subject had non-DQB1*0602 and susceptible TCF7L2 allele. Departure from the following—odds ratio (OR)3 = OR1 + OR2 – 1—indicated a biological (additive) interaction. Chi-square tests or, when appropriate, Fisher's exact tests, tested for difference in categorical variables between mothers with gestational diabetes and control subjects. The logistic regression modelling was performed using SPSS 18.0 (www.spss.com). It is estimated, with 80% power and with a susceptible allele frequency of 35% among control subjects, that a total of 350 mothers with gestational diabetes will be needed to estimate a true odds ratio of 1.50 given a 1 to 1.3 ratio of case subjects to control subjects. P-values less than 0.05 were considered statistically significant.
Results
Out of 1185 randomly selected control subjects, 1185 were genotyped for HLA and 1116 were genotyped for TCF7L2. All the control subjects were negative for islet cell autoantibodies (Fig. 1). Out of 826 mothers with gestational diabetes who gave birth during 2000–2004, 763 were found negative for all three islet cell autoantibodies, while 63 (7.6%) were found positive for at least one of the three islet cell autoantibodies. The prevalence of GAD, IA-2 and IA antibodies in mothers with gestational diabetes was 4.3, 1.3 and 3.2%, respectively. Out of 763 antibody-negative mothers with gestational diabetes, 763 were genotyped for HLA and 746 were genotyped for TCF7L2. Out of 63 autoantibody-positive mothers with gestational diabetes, 63 were genotyped for HLA and 59 were genotyped for TCF7L2 (Fig. 1). The data in Table 1 demonstrate that the distribution of HLA-DQ genotypes was different between control mothers and those with gestational diabetes (P = 0.001).
Table 1.
Characteristics of control subjects and mothers with gestational diabetes in the study
| Characteristics | Control subjects n = 1185 (%) |
Mothers with gestational diabetes n = 826 (%) |
P-value | |
|---|---|---|---|---|
| Age in 2000 | < 25 years | 182 (15.4%) | 123 (14.9%) | |
| 26–34 years | 843 (71.1%) | 520 (63.0%) | ||
| ≥ 35 years | 160 (13.5%) | 183 (22.2%) | < 0.0001 | |
| No. of pregnancies | 0 | 934 (78.8%) | 708 (85.7%) | |
| ≥ 1 | 251 (21.2%) | 118 (14.3%) | < 0.0001 | |
| Hereditary | ||||
| Mother with diabetes | Yes | 28 (2.4%) | 97 (11.7%) | |
| No | 736 (62.1%) | 471 (57.0%) | ||
| Unknown† | 421 (35.5%) | 258 (31.2%) | < 0.0001 | |
| Mother with gestational diabetes | Yes | 15 (1.3%) | 21 (2.5%) | |
| No | 592 (50.0%) | 320 (38.7%) | ||
| Unknown† | 578 (48.8%) | 485 (58.7%) | < 0.0001 | |
| Father with diabetes | Yes | 46 (3.9%) | 76 (9.2%) | |
| No | 699 (59.0%) | 460 (55.7%) | ||
| Unknown† | 440 (37.1%) | 290 (35.1%) | < 0.0001 | |
| Country of origin | ||||
| Sweden | 741 (62.5) | 563 (57.2) | ||
| Outside Sweden | 45 (3.8) | 140 (17.3) | ||
| Not available | 399 (33.7) | 207 (25.6) | < 0.0001 | |
| HLA-DQ genotype | ||||
| DQB1*0302/*02¶ | 42 (3.5) | 37 (4.5) | ||
| DQB1*0302/‡ | 117 (9.9) | 92 (11.1) | ||
| DQB1*02¶/‡ | 76 (6.4) | 70 (8.5) | ||
| DQB1*0602/§ | 300 (25.3) | 145 (17.6) | ||
| Other (non-T1D risk) | 650 (54.9) | 482 (58.4) | 0.001 |
‘Unknown’ is missing data because of lack of filled-out questionnaires to parents when the children participating in the DiPiS study [40] were 2 months of age.
Does not included DQB1*0301, 0603 or 0602.
All other haplotypes.
DQA1*0501-DQB1*0201.
Characteristics of control subjects and mothers with gestational diabetes in the study
Mothers with gestational diabetes were older (P < 0.0001), had had fewer pregnancies (P < 0.0001), had a positive family history of diabetes in the family (P < 0.0001) and were more often born outside Sweden than control subjects (P < 0.0001) (Table 1).
TCF7L2 in mothers with gestational diabetes and control subjects
We genotyped 1116 control subjects and 805 mothers with gestational diabetes for the three TCF7L2 single nucleotide polymorphisms rs7903146 (C is a non-risk and T is a risk allele), rs12255372 (G is a non-risk and T is a risk allele) and rs7901695 (T is a non-risk and C is a risk allele).
Compared with the wild-type CC genotype, carriers of the heterozygous CT genotype and the homozygous TT genotype of rs7903146 had a 1.6-fold increased risk [OR (95% CI) 1.63 (1.34–1.97), P < 0.0001] and a 1.9-fold increased risk [OR (95% CI) 1.90 (1.37–2.64), P < 0.0001) for gestational diabetes, respectively. The heterozygous GT and the homozygous TT genotype carriers of rs12255372 had also an increased risk for gestational diabetes [OR (95% CI) 1.42 (1.17–1.72), P < 0.0001] and 1.58 [OR (95% CI) 1.58 (1.13–2.19), P < 0.001], respectively, compared with the wild-type GG genotype carriers. Strong associations were also shown between the rs7901695 variant and gestational diabetes in the heterozygous TC [OR (95% CI) 1.56 (1.28–1.89), P < 0.0001] as well as the homozygous CC genotype carriers [OR (95% CI) 1.87 (1.36–2.57), P < 0.0001] (Table 2).
Table 2.
TCF7L2 single nucleotide polymorphisms rs7903146, rs12255372 and rs7901695 genotypes in control subjects and mothers with gestational diabetes
| TCF7L2 | Control subjects 1185 n (%) |
Mothers with gestational diabetes 826 n (%) |
P-value† | OR (95% CI) |
|---|---|---|---|---|
| rs7903146 | ||||
| CC | 644 (54.3) | 363 (43.9) | 1.00 | |
| CT | 384 (32.4) | 352 (42.6) | 1.63 (1.34–1.97) | |
| TT | 82 (6.9) | 88 (10.7) | < 0.0001 | 1.90 (1.37–2.64) |
| Unknown‡ | 75 (6.3) | 23(2.8) | ||
| rs12255372 | ||||
| GG | 633 (53.4) | 387 (46.9) | 1.00 | |
| GT | 385 (32.5) | 333 (40.3) | 1.42 (1.17–1.72) | |
| TT | 84 (7.1) | 81 (9.8) | < 0.0001 | 1.58 (1.13–2.19) |
| Unknown‡ | 83 (7.0) | 25 (3.0) | ||
| rs7901695 | ||||
| TT | 607 (51.2) | 343 (41.5) | 1.00 | |
| TC | 405 (34.2) | 356 (43.1) | 1.56 (1.28–1.89) | |
| CC | 90 (7.6) | 95 (11.5) | < 0.0001 | 1.87 (1.36–2.57) |
| Unknown‡ | 83 (7.0) | 32 (3.9) |
P-value is test of overall difference between controls and mothers with gestational diabetes among subjects typed for TCFL2 genotype.
Seventy-five control subjects and 21 mothers with gestational diabetes were not typed for any of the three TCF7L2 single nucleotide polymorphisms. The missing remainder were undetermined for each specific single nucleotide polymorphism.
TCF7L2 single nucleotide polymorphisms rs7903146, rs12255372 and rs7901695 in autoimmune and non-autoimmune gestational diabetes and in control subjects and testing for interaction with HLA-DQB1*0602
When analysing the distribution of HLA-DQB1 alleles in control subjects and mothers with gestational diabetes, stratifying by country of birth, we observed that the HLA-DQB1*0602 allele, which is considered to be protective for Type 1 diabetes, is negatively associated with gestational diabetes only in mothers born in Sweden [OR (95% CI) 0.69 (0.52–0.91), P = 0.010] (see also Supporting Information, Table S1].
The presence of risk T, T and C alleles for the TCF7L2 gene polymorphisms rs7903146, rs12255372 and rs7901695, respectively, was associated with gestational diabetes both in the autoimmune and in the non-autoimmune gestational diabetes group of mothers. No interaction was shown between the risk alleles T, T and C for the TCF7L2 gene polymorphisms rs7903146, rs12255372 and rs790165, respectively, and the HLA DQB1*0602 (Table 3).
Table 3.
Test for possible interaction between HLA-DQ6 and TCF7L2 single nucleotide polymorphisms rs7903146, rs12255372 and rs7901695, respectively, in autoimmune and non-autoimmune gestational diabetes: (a) the risk (T) allele of the TCF7L2 gene variant of rs7903146; (b) the risk (T) allele of the TCF7L2 gene variant of rs12255372 and (c) the risk (C) allele of the TCF7L2 gene variant of rs7901695
| (a) | ||||
|---|---|---|---|---|
| DQ*0602 | T allele | All gestational diabetes No. of controls/cases OR (95% CI) P-value |
Non-autoimmune gestational diabetes No. of controls/cases OR (95% CI) P-value |
Autoimmune gestational diabetes No. of controls/cases OR (95% CI) P-value |
| + | − | 154/60 | 154/58 | 154/2 |
| 1.00† | 1.00† | 1.00† | ||
| + | + | 126/85 | 126/78 | 126/7 |
| 1.73 (1.15–2.60) | 1.64 (1.09–2.49) | 4.28 (0.87–20.9) | ||
| 0.008 | 0.018 | 0.073 | ||
| − | − | 490/303 | 490/280 | 490/23 |
| 1.59 (1.14–2.21) | 1.52 (1.09–2.12) | 3.61 (0.84–15.5) | ||
| 0.006 | 0.015 | 0.084 | ||
| − | + | 340/355 | 340/329 | 340/26 |
| 2.68 (1.92–3.74) | 2.57 (1.83–3.60) | 5.89 (1.38–25.1) | ||
| < 0.0001 | < 0.0001 | 0.017 | ||
| (b) | ||||
|---|---|---|---|---|
| DQ*0602 | T allele | All gestational diabetes No. of controls/cases OR (95% CI) P-value |
Non-autoimmune gestational diabetes No. of controls/cases OR (95% CI) P-value |
Autoimmune gestational diabetes No. of controls/cases OR (95% CI) P-value |
| + | − | 152/63 | 152/61 | 152/2 |
| 1.00† | 1.00† | 1.00† | ||
| + | + | 123/80 | 123/73 | 123/7 |
| 1.57 (1.05–2.36) | 1.48 (0.98–2.24) | 4.33 (0.88–21.2) | ||
| 0.030 | 0.065 | 0.071 | ||
| − | − | 481/324 | 481/299 | 481/25 |
| 1.63 (1.17–2.25) | 1.55 (1.11–2.15) | 3.95 (0.93–16.9) | ||
| 0.003 | 0.009 | 0.064 | ||
| − | + | 346/334 | 346/309 | 346/25 |
| 2.34 (1.68–3.24) | 2.23 (1.59–3.11) | 5.49 (1.29–23.5) | ||
| < 0.0001 | < 0.0001 | 0.022 | ||
| (c) | ||||
|---|---|---|---|---|
| DQ*0602 | C allele | All gestational diabetes No. of controls/cases OR (95% CI) P-value |
Non-autoimmune gestational diabetes No. of controls/cases OR (95% CI) P-value |
Autoimmune gestational diabetes No. of controls/cases OR (95% CI) P-value |
| + | − | 148/55 | 148/54 | 148/1 |
| 1.00† | 1.00† | 1.00† | ||
| + | + | 130/87 | 130/79 | 130/8 |
| 1.80 (1.19–2.72) | 1.67 (1.10–2.53) | 9.11 (1.12–73.8) | ||
| 0.005 | 0.017 | 0.038 | ||
| − | − | 459/288 | 459/266 | 459/22 |
| 1.69 (1.20–2.38) | 1.59 (1.12–2.25) | 7.09 (0.93–53.1) | ||
| 0.003 | 0.009 | 0.056 | ||
| − | + | 365/364 | 365/337 | 365/27 |
| 2.68 (1.91–3.78) | 2.53 (1.79–3.57) | 11.0 (1.28–81.3) | ||
| < 0.0001 | < 0.0001 | 0.019 | ||
Represents the reference group.
The association between TCF7L2 and gestational diabetes remained statistically significant also after adjusting for maternal age, number of pregnancies, family history of diabetes and HLA-DQ genotype (Table 4).
Table 4.
The TCF7L2 single nucleotide polymorphisms rs7903146, rs12255372 and rs7901695 genotypes in control subjects and mothers with gestational diabetes after adjusting for age of mother, number of pregnancies, family history of diabetes and HLA-DQ genotype
| TCF7L2 | OR (95% CI) | P-value† |
|---|---|---|
| rs7903146 | ||
| CC | 1.00 | |
| CT | 1.66 (1.34–2.03) | |
| TT | 1.76 (1.24–2.49) | < 0.0001 |
|
| ||
| rs12255372 | ||
| GG | 1.00 | |
| GT | 1.46 (1.18–1.79) | |
| TT | 1.56 (1.10–2.21) | < 0.0001 |
|
| ||
| rs7901695 | ||
| TT | 1.00 | |
| TC | 1.56 (1.27–1.91) | |
| CC | 1.78 (1.27–2.48) | < 0.0001 |
Unknown data on family history of diabetes was included as a separate category.
Linkage disequilibrium between rs7903146, rs7901695 and rs1225372
As expected, a very significant pairwise linkage disequilibrium was observed between the three alleles, both among mothers born in Sweden and non-Swedish-born mothers with gestational diabetes. However, among the Swedish-born mothers, the degree of linkage disequilibrium of rs1225372 with either rs7903146 or rs7901695 appeared higher than among non-Swedish born mothers (see also Supporting Information, Table S2]
Combined genotype frequencies for rs7903146 and rs1225372
We next calculated the genotype frequencies for combined rs7903146 and rs1225372 to evaluate whether there were specific genotype combinations more often observed in mothers with gestational diabetes with and without islet cell autoantibodies. Because of the almost complete linkage disequilibrium between rs7903146 and rs7901695, genotype combinations were made only for rs7903146 and rs1225372. The frequencies of the combined genotypes for the single nucleotide polymorphisms rs7903146 and rs1225372 were similar in the autoimmune and non-autoimmune mothers with gestational diabetes (see also Supporting Information, Table S3).
Discussion
We report that the TCF7L2 single nucleotide polymorphisms rs7903146, rs12255372 and rs7901695 were associated with gestational diabetes and that the risk alleles are the same as those previously shown to be associated with high risk for Type 2 diabetes [25]. These associations were independent of the presence of HLA-DQB1*0602, which was negatively associated with gestational diabetes in mothers born in Sweden and even independent of the presence of islet cell autoantibodies against GAD65, IA-2 and insulin. Mothers with gestational diabetes were found to be older, had fewer pregnancies and more often had a positive family history of diabetes compared with the control subjects. The association between gestational diabetes and TCF7L2 remained, even after adjusting for other confounding factors, such as maternal age, number of pregnancies, family history of diabetes and the HLA-DQ genotypes.
In a previous study from DiPiS, we tested if there was any association between gestational diabetes and HLA-DQB1 genotypes and found that the DQB1*0602 allele, which is thought to confer protection for Type 1 diabetes [35], was negatively associated with gestational diabetes [13]. In our present study, we therefore tested for interaction between HLA-DQB1*0602 and TCF7L2 and found that these two genetic factors are associated with protection and risk, respectively, independent of each other. In a previous study, it was tested whether there was an interaction between Type 1 and Type 2 diabetes-associated genes [36]. No convincing evidence was found that HLA class II genotypes interacted with Type 2 diabetes-associated gene regions. These findings strengthen our present conclusion that there does not seem to be an interaction between TCF7L2 that confers the strongest susceptibility to Type 2 diabetes and HLA-DQB1*0602, which is considered to have a negative association with Type 1 diabetes. This outcome may be considered to be as expected, as the two genes are associated with different types of diabetes, most likely representing two different mechanisms of action. However, HLA-DQB1 genes have been shown to interact with single nucleotide polymorphisms in genes associated with risk for Type 1 diabetes [37, 38].
It is well known that the rs7903146, rs12255372 and rs7901695 single nucleotide polymorphisms are located within a well-defined linkage disequilibrium block [25, 39]. We therefore tested for linkage disequilibrium between those three single nucleotide polymorphisms in our mothers with gestational diabetes, stratifying by country of birth. We observed an almost complete linkage disequilibrium between the rs7903146 and the rs7901695. Because of the high linkage disequilibrium between the two single nucleotide polymorphisms and the lack of statistical power in our study, we were unfortunately not able to test separately the effect of each single nucleotide polymorphism in mothers with gestational diabetess who had only the one of the two risk alleles. Larger cohort studies will be needed in order to find out if these two single nucleotide polymorphisms represent independent associations and to find out which single nucleotide polymorphism is driving the association.
We then continued to study the observed combined genotype frequencies for the rs7903146 and the rs12255372, which did not differ in the mothers with gestational diabetes with and without islet cell autoantibodies.
The minor allele frequency of the three different single nucleotide polymorphisms rs7903146, rs12255372 and rs7901695 did not deviate from previously reported frequencies among North Europeans [25].
In our study, we lack complete information regarding the genetic background of our mothers as we have information only about the country of birth and not on the precise origin of their families. In addition, there is a relatively large number of mothers (n = 223) for whom the country of birth was unknown, as approximately only 22 000 out of 35 000 mothers participating in the DiPiS study returned the questionnaire that was sent to the parents 2 months after delivery [40]. For these reasons, it was not possible to precisely define groups according to the mothers' ethnic background based only on the current available information on country of birth. However, when studying the HLA-DQB1 allele distribution and the linkage disequilibrium between the three TCF7L2 single nucleotide polymorphisms and stratifying by country of birth, we still had an adequate number of cases in each group in order to draw conclusions whether the data differed by country of birth. However, we were not able to draw any conclusions from that information as to whether the ethnic background of the mothers involved in the study affected the analysis.
In a previous report [10], it was suggested that gestational diabetes could be further classified as autoimmune and non-autoimmune based on the presence of islet cell autoantibodies as well as the HLA distribution. The term ‘autoimmune’ applied to mothers who were at risk of developing Type 1 diabetes and the term ‘non-autoimmune’ to those who had high risk of developing Type 2 diabetes post-partum. In the present study, we showed that the TCF7L2 gene, which has been shown to be associated with increased risk for Type 2 diabetes [21, 22], is strongly associated with gestational diabetes in mothers in the DiPiS study, suggesting that in most cases gestational diabetes shares the same genetic background and clinical features with Type 2 diabetes. The fact that a high percentage of women with previously diagnosed gestational diabetes develop Type 2 diabetes post-partum [15] strengthens this hypothesis. However, in a subgroup of women with gestational diabetes, this metabolic disorder relates more to Type 1 diabetes, as proved by the 7% of islet autoantibody-positive mothers with gestational diabetes, as well as the negative association with the HLA-DQB1*0602 allele.
The current study supports the fact that TCF7L2 is a highly susceptible gene for gestational diabetes, even in those women positive for islet cell autoantibodies, and that the HLA-DQB1*0602 allele is a protective genetic factor for gestational diabetes in women born in Sweden. These associations are independent of each other. Further analysis on the biological mechanism of these associations, as well as the association between gestational diabetes and other genes [41], is needed in order to better understand the underlying pathogenesis and also if ethnic background may be of importance.
We conclude that the TCF7L2 gene was associated with risk for gestational diabetes in mothers both negative and positive for islet cell autoantibodies against GAD65, IA-2 and insulin and this association was independent of maternal age, number of pregnancies, family history of diabetes and HLA-DQ genotypes. There was no interaction between TCF7L2 and HLA-DQB1*0602, which was found to be negatively associated with gestational diabetes in mothers born in Sweden.
Supplementary Material
Table S1. The distribution of HLA-DQB1 genes in non-autoimmune mothers with gestational diabetes and control subjects stratified by country of birth.
Table S2. Pairwise linkage disequilibrium between TCF7L2 single nucleotide polymorphisms rs7903146, rs12255372 and rs7901695 both among Swedish and non-Swedish born mothers with gestational diabetes.
Table S3. Combined genotype frequencies for TCF7L2 rs7903146 and rs12255372 in control subjects and in mothers with gestational diabetes with and without islet cell autoantibodies.
Acknowledgments
The members of the DiPiS Study Group included: A. Carlsson, E. Cederwall, C. M. Cilio, B. Jönsson, H. Larsson, K. Larsson, B. Lernmark, B. Lindberg, J. Neiderud, S. Resic-Lindehammer and S. Sjöblad. We thank B. Buveris-Svendburg, I. Hansson, B. Gustavsson, J. Gerardsson, G. Hansson and H. Rastkhani for expert technical assistance. We also thank all the participating parents and their children. The study was supported by the Swedish Research Council (14064), the National Institutes of Health (DK26190), the Skåne County Council Funds for Research and Development, the Swedish Diabetes Association, the Childhood Diabetes Fund, UMAS Research Funds, Knut and Alice Wallenberg Foundation, and Lund University.
Abbreviations
- DiPiS
Diabetes Prediction in Skåne
- TCF7L2
transcription factor 7-like 2
Footnotes
Competing interests: Nothing to declare.
Supporting Information: Additional Supporting Information may be found in the online version of this article:
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than for missing material) should be directed to the corresponding author for the article.
References
- 1.Lapolla A, Dalfrà MG, Fedele D. Diabetes related autoimmunity in gestational diabetes mellitus: is it important? Nutr Metab Cardiovasc Dis. 2009;19:674–682. doi: 10.1016/j.numecd.2009.04.004. [DOI] [PubMed] [Google Scholar]
- 2.Damm P, Kühl C, Buschard K, Jakobsen BK, Svejgaard A, Sodoyez-Goffaux F, et al. Prevalence and predictive value of islet cell antibodies and insulin autoantibodies in women with gestational diabetes. Diabet Med. 1994;11:558–563. doi: 10.1111/j.1464-5491.1994.tb02035.x. [DOI] [PubMed] [Google Scholar]
- 3.Järvelä IY, Juutinen J, Koskela P, Hartikainen AL, Kulmala P, Knip M, et al. Gestational diabetes identifies women at risk for permanent type 1 and type 2 diabetes in fertile age: predictive role of autoantibodies. Diabetes Care. 2006;29:607–612. doi: 10.2337/diacare.29.03.06.dc05-1118. [DOI] [PubMed] [Google Scholar]
- 4.Füchtenbusch M, Ferber K, Standl E, Ziegler AG. Prediction of type 1 diabetes postpartum in patients with gestational diabetes mellitus by combined islet cell autoantibody screening: a prospective multicenter study. Diabetes. 1997;46:1459–1467. doi: 10.2337/diab.46.9.1459. [DOI] [PubMed] [Google Scholar]
- 5.Petersen JS, Dyrberg T, Damm P, Kühl C, Mølsted-Pedersen L, Buschard K. GAD65 autoantibodies in women with gestational or insulin dependent diabetes mellitus diagnosed during pregnancy. Diabetologia. 1996;39:1329–1333. doi: 10.1007/s001250050578. [DOI] [PubMed] [Google Scholar]
- 6.Nilsson C, Ursing D, Törn C, Aberg A, Landin-Olsson M. Presence of GAD antibodies during gestational diabetes mellitus predicts type 1 diabetes. Diabetes Care. 2007;30:1968–1971. doi: 10.2337/dc07-0157. [DOI] [PubMed] [Google Scholar]
- 7.Wolf E, Spencer KM, Cudworth AG. The genetic susceptibility to type 1 (insulin-dependent) diabetes: analysis of the HLA-DR association. Diabetologia. 1983;24:224–230. doi: 10.1007/BF00282704. [DOI] [PubMed] [Google Scholar]
- 8.Rubinstein P, Walker M, Krassner J, Carrier C, Carpenter C, Dobersen MJ, et al. HLA antigens and islet cell antibodies in gestational diabetes. Hum Immunol. 1981;3:271–275. doi: 10.1016/0198-8859(81)90023-9. [DOI] [PubMed] [Google Scholar]
- 9.Shaat N, Ekelund M, Lernmark A, Ivarsson S, Nilsson A, Perfekt R, et al. Genotypic and phenotypic differences between Arabian and Scandinavian women with gestational diabetes mellitus. Diabetologia. 2004;47:878–884. doi: 10.1007/s00125-004-1388-5. [DOI] [PubMed] [Google Scholar]
- 10.Törn C, Gupta M, Sanjeevi CB, Åberg A, Frid A, Landin-Olsson M. Different HLA-DR-DQ and MHC class I chain-related gene A (MICA) genotypes in autoimmune and non-autoimmune gestational diabetes in a Swedish population. Hum Immunol. 2004;65:1443–1450. doi: 10.1016/j.humimm.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 11.Stangenberg M, Agarwal N, Rahman F, Sheth K, al Sedeiry S, De Vol E. Frequency of HLA genes and islet cell antibodies (ICA) and result of postpartum oral glucose tolerance tests (OGTT) in Saudi Arabian women with abnormal OGTT during pregnancy. Diabetes Res. 1990;14:9–13. [PubMed] [Google Scholar]
- 12.Vambergue A, Fajardy I, Bianchi F, Cazaubiel M, Verier-Mine O, Goeusse P, et al. Gestational diabetes mellitus and HLA class II (-DQ, -DR) association: The Digest Study. Eur J Immunogenet. 1997;24:385–394. doi: 10.1046/j.1365-2370.1997.d01-114.x. [DOI] [PubMed] [Google Scholar]
- 13.Papadopoulou A, Lynch KF, Shaat N, Nilsson A, Lernmark B, Berntorp K, et al. The Type 1 diabetes protective HLA DQB1*0602 allele is less frequent in gestational diabetes mellitus. Diabetologia. 2009;52:1339–1342. doi: 10.1007/s00125-009-1351-6. [DOI] [PubMed] [Google Scholar]
- 14.Bellamy L, Casas JP, Hingorani AD, Williams D. Type 2 diabetes mellitus after gestational diabetes: a systematic review and meta-analysis. Lancet. 2009;373:1773–1779. doi: 10.1016/S0140-6736(09)60731-5. [DOI] [PubMed] [Google Scholar]
- 15.Kim C, Newton KM, Knopp RH. Gestational diabetes and the incidence of type 2 diabetes: a systematic review. Diabetes Care. 2002;25:1862–1868. doi: 10.2337/diacare.25.10.1862. [DOI] [PubMed] [Google Scholar]
- 16.Lauenborg J, Grarup N, Damm P, Borch-Johnsen K, Jørgensen T, Pedersen O, et al. Common type 2 diabetes risk gene variants associate with gestational diabetes. J Clin Endocrinol Metab. 2009;94:145–150. doi: 10.1210/jc.2008-1336. [DOI] [PubMed] [Google Scholar]
- 17.Cho YM, Kim TH, Lim S, Choi SH, Shin HD, Lee HK, et al. Type 2 diabetes-associated genetic variants discovered in the recent genome-wide association studies are related to gestational diabetes mellitus in the Korean population. Diabetologia. 2009;52:253–261. doi: 10.1007/s00125-008-1196-4. [DOI] [PubMed] [Google Scholar]
- 18.Freathy RM, Hayes MG, Urbanek M, Lowe LP, Lee H, Ackerman C, et al. HAPO Study Cooperative Research Group. Hyperglycemia and Adverse Pregnancy Outcome (HAPO) study: common genetic variants in GCK and TCF7L2 are associated with fasting and postchallenge glucose levels in pregnancy and with the new consensus definition of gestational diabetes mellitus from the International Association of Diabetes and Pregnancy Study Groups. Diabetes. 2010;59:2682–2689. doi: 10.2337/db10-0177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shaat N, Ekelund M, Lernmark A, Ivarsson S, Almgren P, Berntorp K, et al. Association of the E23K polymorphism in the KCNJ11 gene with gestational diabetes mellitus. Diabetologia. 2005;48:2544–2551. doi: 10.1007/s00125-005-0035-0. [DOI] [PubMed] [Google Scholar]
- 20.Zhou Q, Zhang K, Li W, Liu JT, Hong J, Qin SW, et al. Association of KCNQ1 gene polymorphism with gestational diabetes mellitus in a Chinese population. Diabetologia. 2009;52:2466–2468. doi: 10.1007/s00125-009-1500-y. [DOI] [PubMed] [Google Scholar]
- 21.Grant SF, Thorleifsson G, Reynisdottir I, Benediktsson R, Manolescu A, Sainz J, et al. Variant of transcription factor 7-like 2 (TCF7L2) gene confers risk of type 2 diabetes. Nat Genet. 2006;38:320–323. doi: 10.1038/ng1732. [DOI] [PubMed] [Google Scholar]
- 22.Cauchi S, Meyre D, Dina C, Choquet H, Samson C, Gallina S, et al. Transcription factor TCF7L2 genetic study in the French population: expression in human beta-cells and adipose tissue and strong association with type 2 diabetes. Diabetes. 2006;55:2903–2908. doi: 10.2337/db06-0474. [DOI] [PubMed] [Google Scholar]
- 23.Shaat N, Lernmark Å, Karlsson E, Ivarsson S, Parikh H, Berntorp K, et al. A variant in the transcription factor 7-like 2 (TCF7L2) gene is associated with an increased risk of gestational diabetes mellitus. Diabetologia. 2007;50:972–979. doi: 10.1007/s00125-007-0623-2. [DOI] [PubMed] [Google Scholar]
- 24.Watanabe RM, Allayee H, Xiang AH, Trigo E, Hartiala J, Lawrence JM, et al. Transcription factor 7-like 2 (TCF7L2) is associated with gestational diabetes mellitus and interacts with adiposity to alter insulin secretion in Mexican Americans. Diabetes. 2007;56:1481–1485. doi: 10.2337/db06-1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tong Y, Lin Y, Zhang Y, Yang J, Zhang Y, Liu H, et al. Association between TCF7L2 gene polymorphisms and susceptibility to type 2 diabetes mellitus: a large Human Genome Epidemiology (HuGE) review and meta-analysis. BMC Med Genet. 2009;19:15–39. doi: 10.1186/1471-2350-10-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Helgason A, Pálsson S, Thorleifsson G, Grant SF, Emilsson V, Gunnarsdottir S, et al. Refining the impact of TCF7L2 gene variants on type 2 diabetes and adaptive evolution. Nat Genet. 2007;39:218–225. doi: 10.1038/ng1960. [DOI] [PubMed] [Google Scholar]
- 27.Scott LJ, Bonnycastle LL, Willer CJ, Sprau AG, Jackson AU, Narisu N, et al. Association of transcription factor 7-like 2 (TCF7L2) variants with type 2 diabetes in a Finnish sample. Diabetes. 2006;55:2649–2653. doi: 10.2337/db06-0341. [DOI] [PubMed] [Google Scholar]
- 28.Lyssenko V, Lupi R, Marchetti P, Del Guerra S, Orho-Melander M, Almgren P, et al. Mechanisms by which common variants in the TCF7L2 gene increase risk of type 2 diabetes. J Clin Invest. 2007;117:2155–2163. doi: 10.1172/JCI30706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Larsson HE, Lynch K, Lernmark B, Nilsson A, Hansson G, Almgren P, et al. Diabetes-associated HLA genotypes affect birthweight in the general population. Diabetologia. 2005;48:1484–1491. doi: 10.1007/s00125-005-1813-4. [DOI] [PubMed] [Google Scholar]
- 30.Larsson HE, Lynch K, Lernmark B, Hansson G, Lernmark Å, Ivarsson SA. Relationship between increased relative birthweight and infections during pregnancy in children with a high-risk diabetes HLA genotype. Diabetologia. 2007;50:1161–1169. doi: 10.1007/s00125-007-0648-6. [DOI] [PubMed] [Google Scholar]
- 31.Lynch KF, Lernmark B, Merlo J, Cilio CM, Ivarsson SA, Lernmark Å. Cord blood islet autoantibodies and seasonal association with the type 1 diabetes high-risk genotype. J Perinatol. 2008;28:211–217. doi: 10.1038/sj.jp.7211912. [DOI] [PubMed] [Google Scholar]
- 32.Lind T, Phillips PR. Influence of pregnancy on the 75-g OGTT. A prospective multicenter study. The Diabetic Pregnancy Study Group of the European Association for the Study of Diabetes. Diabetes. 1991;40:S8–13. doi: 10.2337/diab.40.2.s8. [DOI] [PubMed] [Google Scholar]
- 33.Kiviniemi M, Nurmi J, Lövgren T, Ilonen J. Locked nucleic acid (LNA) probes in high-throughput genetic analysis: application to an assay for type 1 diabetes-related HLA-DQB1 alleles. Clin Biochem. 2005;38:1015–1022. doi: 10.1016/j.clinbiochem.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 34.Sjöroos M, Iitiä A, Ilonen J, Reijonen H, Lövgren T. Triple-label hybridization assay for type-1 diabetes-related HLA alleles. Biotechniques. 1995;18:870–877. [PubMed] [Google Scholar]
- 35.Graham J, Kockum I, Sanjeevi CB, Landin-Olsson M, Nyström L, Sundkvist G, et al. Negative association between type 1 diabetes and HLA DQB1*0602-DQA1*0102 is attenuated with age at onset. Swedish Childhood Diabetes Study Group. Eur J Immunogenet. 1999;26:117–127. [PubMed] [Google Scholar]
- 36.Raj SM, Howson JM, Walker NM, Cooper JD, Smyth DJ, Field SF, et al. No association of multiple type 2 diabetes loci with type 1 diabetes. Diabetologia. 2009;52:2109–2116. doi: 10.1007/s00125-009-1391-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aminkeng F, Van Autreve JE, Weets I, Quartier E, Van Schravendijk C, Gorus FK, et al. IFIH1 gene polymorphisms in type 1 diabetes: genetic association analysis and genotype-phenotype correlation in the Belgian population. Hum Immunol. 2009;70:706–710. doi: 10.1016/j.humimm.2009.06.013. [DOI] [PubMed] [Google Scholar]
- 38.Maziarz M, Janer M, Roach JC, Hagopian W, Palmer JP, Deutsch K, et al. The association between the PTPN22 1858C>T variant and type 1 diabetes depends on HLA risk and GAD65 autoantibodies. Genes Immun. 2010;11:406–415. doi: 10.1038/gene.2010.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bonetti S, Trombetta M, Malerba G, Boselli L, Trabetti E, Muggeo M, et al. Variants and haplotypes of TCF7L2 are associated with β-cell function in patients with newly diagnosed Type 2 diabetes: The Verona Newly Diagnosed Type 2 Diabetes Study (VNDS) 1. J Clin Endocrinol Metab. 2010 doi: 10.1210/jc.2010-1677. XX: XX–XX. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 40.Lernmark B, Elding-Larsson H, Hansson G, Lindberg B, Lynch K, Sjöblad S. Parent responses to participation in genetic screening for diabetes risk. Pediatr Diabetes. 2004;5:174–181. doi: 10.1111/j.1399-543X.2004.00070.x. [DOI] [PubMed] [Google Scholar]
- 41.Shaat N, Groop L. Genetics of gestational diabetes mellitus. Curr Med Chem. 2007;14:569–583. doi: 10.2174/092986707780059643. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. The distribution of HLA-DQB1 genes in non-autoimmune mothers with gestational diabetes and control subjects stratified by country of birth.
Table S2. Pairwise linkage disequilibrium between TCF7L2 single nucleotide polymorphisms rs7903146, rs12255372 and rs7901695 both among Swedish and non-Swedish born mothers with gestational diabetes.
Table S3. Combined genotype frequencies for TCF7L2 rs7903146 and rs12255372 in control subjects and in mothers with gestational diabetes with and without islet cell autoantibodies.