Summary
The innate immune system plays a crucial role in the rapid recognition and elimination of invading microbes. Detection of microbes relies on germ-line encoded pattern recognition receptors (PRRs) that recognize essential bacterial molecules, so-called pathogen-associated molecular patterns (PAMPs). A subset of PRRs, belonging to the NOD-like receptor (NLR) and the PYHIN protein families, detects viral and bacterial pathogens in the cytosol of host cells and induces the assembly of a multi-protein signaling platform called the inflammasome. The inflammasome serves as an activation platform for the mammalian cysteine protease caspase-1, a central mediator of innate immunity. Active caspase-1 promotes the maturation and release of interleukin-1β (IL-1β) and IL-18 as well as protein involved in cytoprotection and tissue repair. In addition, caspase-1 initiates a novel form of cell death called pyroptosis. Here we discuss latest advances and our insights on inflammasome stimulation by two model intracellular pathogens, Francisella tularensis and Salmonella typhimurium. Recent studies on these pathogens have significantly shaped our understanding of the molecular mechanisms of inflammasome activation and how microbes can evade or manipulate inflammasome activity. In addition, we review the role of the inflammasome adapter ASC in the caspase-1 autoproteolysis and new insights into the structure of the inflammasome complex.
Keywords: inflammasome, Caspase-1, NLRs, ASC, Francisella tularensis, Salmonella typhimurium
Introduction
The host innate immune system plays an important role in the rapid recognition and elimination of invading microbes through phagocytosis and the induction of inflammation. Recognition of microbes is based on germ-line encoded pattern-recognition receptors (PRRs), which recognize conserved microbial molecules called pathogen-associated molecular patterns (PAMPs) (1). In addition to PAMPs, PRRs also respond to endogenous, host-derived danger signals, termed DAMPs (danger-associated molecular patterns) or alarmins, which are released in response to tissue injury, stress or necrotic cell death (2). Such damage-induced responses could also play a role in the response to pathogen, since microbes as well as the host immune responses can lead to the release of DAMPs from infected cells.
The first group of PRRs to be characterized were the Toll-like receptors (TLRs), membrane-bound sensors that recognize PAMPs in the extracellular compartment and in endosomes (3). Following ligand binding, TLRs dimerize and induce gene expression of cytokines and pro-inflammatory molecules through the nuclear factor κB (NFκB) signaling pathway. Similarly membrane bound C-type lectin receptors (CLRs) initiate inflammatory responses to extracellular PAMPs like β-glucans and mannan (4). Most of the recent progress however has been focused on understanding how the innate immune system detects the presence of microbial pathogens in the cytosol. This focus has led to the discovery of an array of distinct cytoplasmic PRRs, which belong to the NOD-like receptor (NLR), retinoic-acid inducible gene-I (RIG-I)-like helicase, and the PYHIN protein families. These proteins constitute a structural and functional heterologous group, but they can be roughly divided into two categories based on the downstream signaling events that follow their activation. The first group of receptors promotes transcriptional activation of cytokine expression trough the NFκB or IRF3-transcriptional activator pathways, while the second group of receptors initiates the assembly of a cytoplasmic signaling complex known as the inflammasome. The inflammasome serves as an activation platform for the mammalian cysteine protease caspase-1, a key mediator of innate immune defenses (5). Active caspase-1 promotes the proteolytic maturation and secretion of pro-inflammatory cytokines IL-1β and IL-18 and the release of proteins involved in tissue repair and cytoprotection. In addition, caspase-1 induces a pro-inflammatory form of cell death called pyroptosis, which is thought to remove the replicative niche of intracellular pathogens and re-exposes the pathogen to extracellular immune responses. Assembly of the inflammasome complex is tightly regulated and linked to other signaling pathways. In many cases, assembly of the inflammasome requires a preceding priming signal, often referred to as ‘signal 1’, via TLRs or intracellular receptors, which is required to upregulate the expression of certain inflammasome receptors and the substrates IL-1β and IL-18, before ‘signal 2’ can initiate complex formation (5). In this review, we focus on recent advances in pathogen recognition by the cytosolic inflammasome receptors (i.e. receptors that initiate inflammasome assembly), and discuss other receptors mainly in context of inflammasome priming.
Caspase-1, a founding father
Caspase-1 is a founding member of the caspase family of aspartate-specific cysteine proteases, peptidases that use a cysteine residue as the catalytic nucleophile and that share an exquisite specificity for cleaving target proteins at recognition sites next to aspartic acid residues. All caspases are synthesized as inactive zymogens consisting of an N-terminal prodomain of variable length, followed by a large and small subunit (6). Caspases can be subdivided according to the length of their pro-domain into initiator and effector caspases. Initiator caspases have a large pro-domain containing a homotypic protein-protein interaction motif belonging to the death domain (DD) superfamily, specifically either a CARD (caspase activation and recruitment domain) (caspase-1, caspase-2, caspase-4, caspase-5, caspase-11, and caspase-12) or a DED (death effector domain) (caspase-8 and caspase-10) (6). These domains are used to recruit the initiator caspase into multiprotein complexes, such as the apoptosome or the inflammasome. Within these complexes, the caspase is supposed to be activated by proximity-induced dimerization and autoproteolytic processing (7). Activated initiator caspases proteolytically activate downstream effector caspases (caspase-3, caspase-6, and caspase-7), thus amplifying the signal. Caspases have been shown to have essential roles in apoptosis, cell survival, proliferation, differentiation, and inflammation. Caspase-1 is the best-characterized inflammatory caspases and plays a prominent role in innate immune responses. Originally termed interleukin converting enzyme (ICE), caspase-1 was identified based on its ability to cleave the pro-form of IL-1β to mature, bioactive IL-1β (8). Caspase-1 also directs a novel cell death pathway called pyroptosis, which has characteristics of both apoptosis and necrosis (9). Inappropriate activation of caspase-1 and excess IL-1β secretion has been linked to several autoimmune inflammatory disorders in humans illustrating the importance of tight regulation of caspase-1 activity (10). Clearly inflammation can be a double-edged sword but it is essential to combat infection and restore tissue homeostasis after infection (11).
Inflammasome receptors detect cytoplasmic intracellular pathogens and danger signals
Caspase-1 is activated in multi-protein signaling complexes termed inflammasomes (5). Assembly of an inflammasome is initiated upon recognition of specific cytosolic signals by members of the NLR and PYHIN protein families. In its simplest form, such a complex consists of an activated inflammasome receptor, the adapter protein ASC (apoptosis-associated speck-like protein containing a CARD), and pro-caspase-1. Different types of inflammasomes can be formed and are named after the receptor that promotes its assembly.
The human genome encodes 23 NLR family members, while over 34 NLRs are known in mice (5). NLR family members possess a characteristic domain architecture, consisting of an N-terminal effector domain, a central nucleotide binding/oligomerization domain (NBD), and C-terminal leucine-rich repeats (LRRs). Analogous to TLRs, the LRRs are thought to function in ligand-binding sensing and autorepression, while the effector domains mediate homotypic protein-protein interaction for downstream signaling (5). However, in many cases, a direct interaction between an NLR and a specific PAMP or DAMP has not been demonstrated. NLR subclasses can be distinguished based on their distinct effector domains, which can be a baculovirus IAP repeat (BIRC), a Pyrin-like domain (PYD), or a CARD. The founding members of this family, NOD1 and NOD2, activate NFκB and mitogen-activated protein kinase (MAPK)-dependent gene expression via a CARD-dependent association with the signaling kinase RIP2. The NLR Class II transactivator (CIITA) has been shown to regulate the transcription of major histocompatibility complex (MHC) class II by associating with the MHC enhanceosome (12). Another NLR, NLRC5, has recently been suggested to act as a MHC class I transactivator (13); however, the role of NLRC5 remains controversial, since it has also been implicated in regulating NFκB and type I interferon signaling pathways as well as inflammasome activation (14–16). Thus far, only three members of the NLR family, NLRP1, NLRP3, and NLRC4, have definitely been shown to initiate inflammasome assembly (5). Others, such as NAIP5, NLRP6, NLRP12, and NLRC5, have auxiliary or regulatory function in inflammasome assembly but cannot by themselves initiate the assembly of the complex. Many of the remaining NLR family members are poorly characterized at present, but emerging reports suggest roles in inflammasome regulation and NFκB signaling.
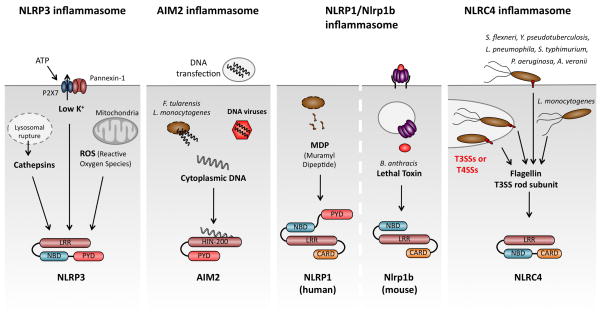
The different NLRs allow the host to detect a vast variety of distinct intracellular pathogens and danger signals. The first inflammasome to be described was the NLRP1 inflammasome (17); however, it still remains the least studied and understood (Fig. 1). This might be in part due to differences between human NLRP1 and its three murine homologs Nlrp1a, Nlrp1b, and Nlrp1c, which are highly polymorphic between different inbred mouse strains. Strain variation in the mouse Nlrp1b locus appears to underlie susceptibility to Bacillus anthracis lethal toxin (LeTx), as macrophages from susceptible, but not resistant, mouse strains activate caspase-1 after LeTx exposure (18). In addition, human NLRP1 was shown to recognize MDP in vitro; however, this association has not been reported for its murine homologs (19). The exact mechanisms of NLRP1 activation and how it can sense these two very distinct ligands remain obscure. Human and murine NLRP1 also differ in their domain architecture. Human NLRP1 possesses a CARD and PYD, while its murine homologs lack the PYD. In addition to caspase-1, NLRP1 interacts with caspase-5, which may contribute to IL-1β processing in human cells (17).
Fig. 1. Inflammasome receptors recognize a variety of microbial pathogens and danger signals.
NLRP3 responds to numerous stimuli. Common terminal signals appear to involve lysosomal rupture and the release of Cathepsins, potassium release and the production of reactive oxygen species (ROS). AIM2 functions as a cytosolic DNA sensor, detecting DNA introduced by transfection, infection with the cytosolic bacterial pathogens F. tularensis or L. monocytogenes or DNA viruses. Human NLRP1 responds to muramyl dipeptide, while Anthrax lethal toxin triggers murine Nlrp1b. NLRC4 detects flagellin or the T3SS rod subunit in the cytosol. LRR, leucine-rich repeats; NBD, nucleotide binding and oligomerization domain; PYD, Pyrin-like domain; CARD, caspase activation and recruitment domain.
By far the most widely studied receptor is NLRP3, which has been shown to respond to a variety of structurally and chemically diverse molecules (Fig. 1), originating from viruses (Sendai, influenza, and adenoviral strains) (20, 21), fungi (Candida albicans and Saccharomyces cerevisiae) (22, 23), and bacteria (Staphylococcus aureus, Neisseria gonorrhoeae, Legionella monocytogenes and Salmonella typhimurium) (24–27). NLRP3 is activated by danger signals such as elevated concentrations of ATP (24), ultraviolet B (UVB) irradiation, and particulate matter such as crystalline forms of monosodium urate (MSU) (28), asbestos, and silica (29–31), and amyloid β aggregates (32). Since it is unclear how a single receptor can respond to such a wide array of stimuli, the current hypothesis is that NLRP3 senses a common terminal signal. Release of Cathepsins from damaged lysosomes, ROS production, and/or the release of potassium have been implicated as the terminal signal, but each has caveats. Therefore, the precise molecular mechanism of NLRP3 activation remains a mystery.
In contrast to NLRP3, NLRC4 seems to be a sensor that is dedicated to the detection of bacterial pathogens and has been shown to respond to bacterial flagellin and the rod component of type 3 secretion systems (T3SS) (33–35)(Fig. 1). These molecules are believed to be accidentally secreted into the host cell cytoplasm by active bacterial type 3 and type 4 secretion systems. A variety of flagellated and unflagellated bacteria are known to induce NLRC4 activation during infection, such as S. typhimurium (36), Pseudomonas aeruginosa (37), Aeromonas veronii (38), Yersinia pseudotuberculosis (39), L. monocytogenes (25), and Legionella pneumophila (40). In the case of L. pneumophila, infections NLRC4 requires NAIP5 (Birc1e), another member of the NLR family, to respond to flagellin.
Studies of NLRP1, NLRP3, and NLRC4 have expanded our understanding of the role of the NLR family and inflammasomes in the host response to pathogens. However, it had been known that transfection of synthetic, bacterial, viral, or mammalian dsDNA triggers caspase-1 activation in a manner that required the adapter ASC but not any of the known NLRs (20). Recently, absent in melanoma 2 (AIM2), a member of the family of hemopoietic IFN-inducible nuclear proteins with a 200-amino acid motif (HIN-200), was shown to activate the inflammasome in response to cytosolic double stranded DNA (dsDNA) (41–44) (Fig. 1). In addition to AIM2, this family also includes IFIX (Pyhin1), MNDA, and IFI-16 in humans, and Ifi202a, Ifi202b, Ifi203, Ifi204, Ifi205 in mice. A characteristic feature of these proteins is one or several C-terminal HIN200 domains. AIM2 contains a HIN domain that facilitates dsDNA binding and a PYD domain that recruits ASC, allowing for caspase-1 activation. While AIM2 induces the assembly of an inflammasome, p202 has been suggested to have a regulatory role on DNA-mediated inflammasome activation (44). Most recently, IFI16 (murine p204) was shown to act as a cytoplasmic DNA sensor and to induce IFN-β trough the signaling adapter STING (45). This was the first example of a non-NLR family protein triggering inflammasome assembly. Moreover, AIM2 is interferon-inducible, establishing a link between two cytosolic innate immune responses.
How microbial pathogens shape our understanding of the inflammasome function
Although some inflammasome receptors (e.g. NLRP3) also detect danger signals, most of the receptors that have been characterized thus far are dedicated to the recognition of intracellular viral or bacterial pathogens. Microbial infections have become an important tool in inflammasome study, since they represent relevant model systems for inflammasome activation, which cannot be achieved using artificial stimuli, such as transfection of DNA. In addition, most pathogens activate several pattern recognition receptors during the course of the infection, thus providing the possibility to study the complex network of innate immune signaling pathways. Finally, in vivo infection models have been established for most pathogens allowing for a validation of in vitro findings. In the following, we discuss recent advances in our insight of inflammasome stimulation by two model intracellular pathogens, Francisella tularensis and Salmonella typhimurium. We describe how these studies have shaped our understanding of the molecular mechanisms of inflammasome activation.
F. tularensis activates the AIM2 inflammasome
F. tularensis is a non-flagellated, Gram-negative bacterium that has evolved the capacity to successfully colonize eukaryotic hosts, sometimes causing disease, even in the face of a robust immune response. Crucial for Francisella virulence is it ability to survive and replicate within the macrophage, its primary niche in vivo. After uptake, F. tularensis initially resides in a membrane-bound vacuole termed the Francisella-containing phagosome (FCP). The FCP acquires the early endosomal antigen 1 (EEA1) within 5 min of uptake, but this marker rapidly dissociates from the phagosome followed by the acquisition of late endosomal markers Lamp1, Lamp2, and the Rab7 GTPase within 15–30 min (46–48). The FCP does not significantly fuse with lysosomes, and though not absolutely required, acidification of the phagosome acts as a cue for F. tularensis to escape this compartment and enter into the host cell cytosol (49, 50). Phagosomal escape is rapid, occurring within 60 minutes post macrophage infection. Once in the cytosol, F. tularensis replicates to high numbers. Both phagosomal escape and intracellular replication are mediated by a locus of F. tularensis genes known as the Francisella pathogenicity island (FPI) (51). FPI mutants remain in the initial phagosome, which progresses to lysosomes (52). FPI mutants are also avirulent in vivo (53, 54).
After F. tularensis escapes the phagosome, it is subject to cytosolic innate immune recognition (Fig. 2). In murine macrophages, F. tularensis subsp. novicida and the live vaccine strain (LVS) induced inflammasome activation did not require Nlrp1b, NLRC4, and NLRP3, but required the adapter protein ASC (24, 55). In addition, macrophages deficient in the type I IFN receptor failed to activate the inflammasome in response to F. novicida (56). Similary, L. monocytogenes-mediated activation of the inflammasome was deficient in Ifnar−/− macrophages (56), suggesting that type I IFN signaling is a specifically required during infection with bacteria that escape into the cytosol. The recent discovery of the interferon-inducible inflammasome receptor AIM2 and its role in viral infections led us and others to investigate whether this receptor mediates recognition of intracellular bacterial pathogens such as F. novicida and L. monocytogenes. Indeed, infection of primary bone-marrow derived macrophages from AIM2-deficient mice demonstrated that AIM2 was required for caspase-1 processing, release of mature IL-1β, and host cell death in response to F. novicida (57–59). Interestingly, confocal microscopy showed that a subpopulation of cytosolic F. novicida seems to lyse during infection and that these bacteria release DNA into the macrophage cytosol (57, 58). Such DNA foci also colocalized with AIM2, consistent with previous data showing a direct interaction of dsDNA with AIM2 (41). The DNA/AIM2 complexes seemed to serve as a nucleation point for the assembly of an ASC/caspase-1 focus, which was generally found adjacent to a DNA/AIM2 complex but not directly co-localizing (58). Intriguingly, if several lysing bacteria were present within a single cell, DNA/AIM2 complexes formed in close proximity to all of these bacteria; however, only one of these DNA/AIM2 complexes served as a nucleation point for ASC/caspase-1 focus formation. This observation represented the first visualization of an endogenous inflammasome component complexed with its ligand. Similarly, L. monocytogenes was later shown to release DNA through lysis during infection and activate the AIM2 inflammasome (59–62). Inflammasome activation was also shown to be critical to host defense against Francisella in vivo as mice lacking the inflammasome components AIM2, caspase-1, or ASC have increased bacterial burden in systemic organs like liver, lung, and spleen, reduced serum IL-18 levels, and succumb to infection much faster than wildtype mice (57–59).
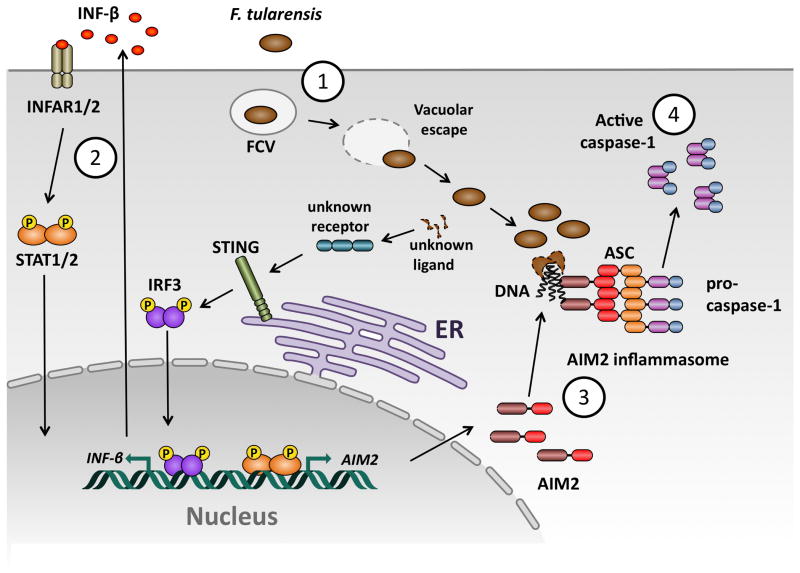
Fig. 2. Model of innate immune recognition of F. tularensis by macrophages.
(1) F. tularensis is phagocytosed by macrophages and quickly escapes from the FCV (Francisella containing vacuole) into the cytosol. In the cytosol, F. tularensis releases an unknown ligand, which is recognized by an unknown cytoplasmic receptor, leading to STING- and IRF3-dependent production of type I interferons. (2) Autocrine and paracrine signaling through the type-I-interferon receptor (IFNAR) leads to STAT1/2 dependent expression of interferon inducible genes, among them AIM2. (3) A subset of cytocolic F. tularensis lyse, releasing bacterial DNA, which is recognized by AIM2. DNA-AIM2 complexes serve as a nucleation point for rapid ASC oligomerization, which leads to the formation of an ASC focus. (4) Pro-caspase-1 is recruited and activated in the ASC focus and promotes cell death and cytokine maturation.
The role of type I interferon for the AIM2 inflammasome
In addition to the inflammasome, F. novicida activates a cytosolic surveillance pathway that leads to the production of type I interferons (56, 63). The exact nature of the host cytosolic sensor responsible for type I interferon production remains unknown. However, we have recently shown that the adapter protein stimulator of interferon genes (STING) is required for type I interferon production in response to F. novicida in murine macrophages (58). STING is a signaling adapter protein known to be required for TBK1-dependent IFN-β responses to viruses and DNA (64). Recently, two new cytosolic DNA sensors have been identified, LRRFIP1 (65) and IFI16 (45), a PYHIN protein that contains a PYD and a HIN domain, similar to AIM2. LRRFIP1 was shown to mediate type-I IFN production in macrophages in response to L. monocytogenes and vesicular stomatitus virus (65). IFI16 mediates type I interferon production in response to dsDNA (synthetic or from viral and bacterial sources) and herpes simplex virus-1 (HSV-1) and is able to associate with the signaling adapter STING (45). Yet, it remains to be determined whether LRRFIP1 and/or IFI16 are inducing the type I interferon response to intracellular Francisella and if the ligand is indeed DNA.
Autocrine and paracrine signaling through IFNAR leads to an increase in AIM2 protein expression in the macrophage (57, 58), which effectively primes the cell for recognition of cytosolic dsDNA. However, it is unclear how important that priming step is for the activity of the AIM2 inflammasome. Even though basal levels of AIM2 are expressed even without type I interferon priming (57, 58), we and others have observed that macrophages deficient for components of the type I interferon signaling pathway (STING, IRF-3, and IFNAR) do not process caspase-1, release proinflammatory cytokines, or die in response to F. novicida (56–58). Consistent with type I interferon signaling being upstream of inflammasome stimulation, priming of macrophages with IFN-β overcomes IRF-3 and STING deficiency, but not the lack of IFNAR. These findings indicate an important role of type I interferon in F. novicida-mediated AIM2 activation. In contrast, transfection of macrophages with dsDNA induces comparable levels of AIM2 inflammasome activation in WT and Irf3−/− macrophages (57). Possible explanations for this discrepancy might be that the DNA transfection results in a strong activation of the existing basal levels of AIM2, while bacterial infections release less DNA and thus need preceding type I interferon priming and the resulting upregulation of receptor expression to be detected. Alternatively, it has been proposed that type I interferon signaling enhances killing and lysis of intracellular F. novicida (57).
A positive feedback loop between type I interferon signaling and inflammasome activation in macrophages exists, but the link in vivo seems to be less clear. Previous reports show that mice deficient in inflammasome components are more susceptible to infection with F. novicida (55, 57, 66). In contrast, mice deficient in type I interferon signaling were shown to be more resistant to infection (56). Similar results were also obtained for L. monocytogenes infections (56). This apparent discrepancy between the in vitro and the in vivo results is partially explained by the control of type I interferon signaling on IL-17 production (67). Mice deficient in type I interferon signaling produce more IL-17, which leads to a greater influx of neutrophils that can better control bacterial infections. Another possible explanation for the in vivo phenotype is that type I interferon has been shown to promote apoptosis of immune cells such as macrophages, neutrophils, and lymphocytes during Listeria and Francisella infections (68–70). Additionally, IFN-γ can restore inflammasome activation in vivo in Ifnar−/− mice by signaling through the IFN-γ receptor, which would result in increased expression of AIM2 and subsequent inflammasome activation (authors’ unpublished results). Thus, an interesting paradox exists, where type I interferon is beneficial to the host in vitro and detrimental in vivo during bacterial infections.
Inflammasome/caspase-1 activation by Salmonella typhimurium
Salmonella enterica is a Gram-negative, flagellated bacterium and a leading cause of enteric disease today (71). Various Salmonella serovars can cause a range of diseases in humans. For example, S. enterica serovar Typhi causes systemic disease (Typhoid), whereas S. enterica serovar Typhimurium (S. typhimurium) causes a self-limiting gastroenteritis in humans. S. typhimurium causes a Typhoid-like systemic disease in mice and is commonly used to study Salmonella interactions with a mammalian host (71). In the gut, S. typhimurium actively induces intestinal inflammation to establish its niche and to better compete with the intestinal microbiota (72). Host-adapted serovars are able to disseminate from the gastrointestinal tract and cause a systemic infection. Salmonella virulence relies heavily on two pathogenicity island called SPI-1 and SPI-2, which encode type 3 secretion systems (T3SS). Salmonella uses the SPI-1 T3SS to invade epithelial cells and to cross the intestinal barrier to the lamina propria, where cells of the immune system like macrophages and dendritic cells take up the bacteria (71). Once inside the host cell, Salmonellae express the SPI-2 T3SS to prevent phagosome maturation and establish its intracellular niche within a vacuolar compartment called the Salmonella containing vacuole (SCV). S. typhimurium is one of the best characterized intracellular pathogens and one of the first bacteria to be studied in context of caspase-1 activation. However, only recently the full picture of S. typhimurium induced caspase-1 activation has emerged.
Early studies showed that S. typhimurium activates caspase-1 in J774 and peritoneal macrophages and suggested that SipB, a component of the T3SS translocation pore is involved (73). Later studies with bone-marrow derived macrophages showed that caspase-1 activation required a functional SPI-1 T3SS and the host inflammasome components ASC and NLRC4 (36). Activation of the NLRC4-inflammasome was also dependent on Salmonella flagellin, leading to the hypothesis that flagellin is translocated through the SPI-1 T3SS into the macrophage cytoplasm, most likely due to evolutionary similarities between the T3SS and the flagellar biosynthesis machinery (34, 35). However, since non-flagellated Salmonella strains still induced significant SPI-1-dependent caspase-1 activation, it was hypothesized that an additional ligand is injected into the host cell cytosol. Indeed, it was recently shown that PrgJ, which forms the periplasmic rod of the T3SS needle complex, is also recognized by NLRC4 (33). PrgJ and flagellin share a common sequence motif that triggers NLRC4 activation. Interestingly, PrgI, the SPI-1 T3SS needle component, and SsaI, the SPI-2 T3SS rod component, lack this motif and are not sensed by NLRC4. Homologous rod proteins in related T3SS from E. coli, S. flexneri, and P. aeruginosa were also shown to induce NLRC4 activation, providing an explanation for why non-flagellated, T3SS-endowed bacteria are capable of activating the NLRC4 inflammasome.
In addition, caspase-1 is important for innate immune defenses during systemic S. typhimurium infections in mice, which is consistent with the in vitro findings in macrophages. C57/BL6 mice deficient in caspase-1 are more sensitive to S. typhimurium infection, as measured by increased bacterial burden in the Peyer’s patches, mesenteric lymph nodes, spleen, and liver as well as reduced serum IL-18 levels after oral infections (27, 74). In addition, a significant reduction in survival was observed with caspase-1 deficient mice, which was much more evident with Nramp1+/+ animals such as 129Sv or Nramp1+/+ C57/BL6 (74, 75). Consistent with these findings, mice deficient for IL-1β, IL-18, or the IL-1receptor (IL-1R) had attenuated immune responses comparable to caspase-1-deficient animals. In contrast to the robust NLRC4-dependent caspase-1 activation observed in vitro, mice deficient for NLRC4 were phenotypically comparable to wildtype animals, as were NLRP3- and ASC-deficient mice (74). It was therefore hypothesized that S. typhimurium activates several redundant pathways of caspase-1 activation in vivo or that a yet unknown receptor/s was involved.
We recently provided an explanation for this apparent discrepancy between the in vitro and in vivo studies of Salmonella infection (27). Previous in vitro studies had used Salmonella grown to logarithmic phase, which results in the expression of the SPI-1 T3SS (Fig. 3A). However, in vivo SPI-1 expression is downregulated during the systemic phase of the infection and the second T3SS encoded on SPI-2 is expressed. To investigate if Salmonella can activate caspase-1 independently of the SPI-1 T3SS, we infected macrophages with S. typhimurium grown to stationary phase, which results in downregulation of SPI-1 expression and expression of the SPI-2 T3SS. Surprisingly, we observed that under these conditions, S. typhimurium activated two inflammasome receptors, NLRP3 and NLRC4 (27) (Fig. 3B). Activation of NLRC4 was dependent on a functional SPI-2 T3SS and on bacterial flagellin. The SPI-2 rod subunit SsaI did not contribute to caspase-1 activation, as expected from the results of Miao and colleagues. The exact signal that activates NLRP3 during Salmonella infections remains unknown, but interestingly, NLRP3 detects Salmonella SPI-2 mutants, which are not replicating intracellularly, suggesting that NLRP3 has the ability to detect persisting bacteria. NLRP3-induced inflammasome activation relied completely on the adaptor protein ASC, while ASC enhanced NLRC4-dependent inflammasome activation.
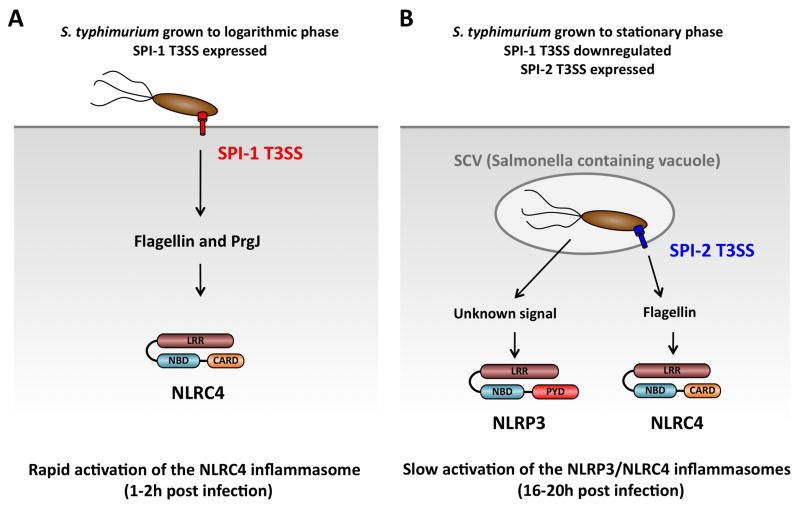
Fig. 3. Model of S. typhimurium-induced inflammasome activation in cultured macrophages.
Distinct growth conditions of S. typhimurium results in different innate immune responses in cultured macrophages. (A). Growth of S. typhimurium to logarithmic phase results in the expression of the SPI-1 T3SS (red). Infection of bone-marrow derived macrophages under this condition leads to a rapid recognition of flagellin and PrgJ by NLRC-4 resulting in inflammasome activation within 1-2 hours post-infection. (B). Growth of S. typhimurium to stationary phase leads to the downregulation of SPI-1 expression and the up-regulation of the SPI-2 T3SS (blue). Infection under these condition results in a slow activation of the NLRC4 and NLRP3 inflammasomes starting approximately 10 hours and a peak at 16-20 hours post-infection. NLRC4 responds to SPI-2 T3SS-dependent translocation of flagellin into the cytosol, while NLRP3 responds to a T3SS-independent signal. LRR, leucine-rich repeats; NBD, nucleotide binding and oligomerization domain; PYD, Pyrin-like domain; CARD, caspasea activation and recruitment domain.
We showed that mice deficient in caspase-1 or NLRP3/NLRC4 had higher bacterial loads in mesenteric lymph nodes, spleen, and liver, and reduced serum levels of IL-18 in comparison to wildtype animals after oral infection (27). These results demonstrated that redundant pathways of caspase-1 activation can be engaged in response to microbial pathogens. S. typhimurium is known to downregulate flagellin expression during systemic infections. Thus, it will be interesting to determine in which organs these receptors actually play a role in detecting S. typhimurium (76). We also observed that ASC-deficient animals had slightly reduced levels of serum IL-18 but comparable bacterial loads as wildtype animals (27). A recent study demonstrated that Salmonellae that constitutively express flagellin induced NLRC4-inflammasome activation that resulted in macrophage pyroptosis, bacterial release, and neutrophil-mediated killing (77). NLRC4-dependent bacterial clearance was independent of ASC, NALP3, IL-18, and IL-1R, providing a possible explanation for the phenotype of ASC-deficient animals. In line with these results we have recently demonstrated that ASC is not necessary for NLRC4 to induce pyroptosis, but it is crucial for cytokine maturation (78). Nevertheless, the relative importance of caspase-1-mediated cytokine maturation and pyroptosis during animal infections with wildtype S. typhimurium still remains to be investigated.
In addition to NLRC4 and NLRP3, recent reports suggest that S. typhimurium activates caspase-1 directly trough T3SS effector proteins. Hardt and collegues (79) reported that SopE, a guanine exchange factor (GEF) of RhoGTPases, leads to caspase-1 activation, in a manner dependent on its GEF activity and the RhoGTPases Rac1 and Cdc42. In vivo, SopE triggered mucosal inflammation in wildtype but not caspase-1−/−, IL-1R−/−, or IL-18−/−mice. Bone marrow chimeras indicated that caspase-1 was more important in stromal cells, most likely enterocytes, than in bone marrow-derived cells. However, the mechanism of SopE-induced caspase-1 activation and its dependence on known inflammasome receptors remains unclear. It is intriguing to speculate that Salmonella could specifically exploit caspase-1 activation in gut epithelial cells to trigger intestinal inflammation, which is beneficial for the pathogen, while avoiding inflammasome activation in other organs.
Manipulation of inflammasome signaling by microbial pathogens
During infections, microbial pathogens need to establish a replicative niche in the presence of a robust host innate immune system. The inflammasome is crucial to mount effective innate immune responses against a variety of viral and bacterial pathogens, including F. novicida and S. typhimurium. Successful pathogens have been shown to manipulate host cell signaling pathways in many different ways that ultimately lead to successful evasion of host defense mechanisms (80). Similarly, it has been proposed that microbes must have developed ways to inhibit or avoid inflammasome activation. Indeed, viral pathogens have been shown to use a panoply of different strategies to block inflammasome assembly and caspase-1 activation. For example, myxoma virus and Shope fibroma virus, both members of the poxvirus family, express the viral PYD-only proteins (vPOPs) m013 and gp013L (81, 82). These proteins are homologs of cellular PYD-only proteins (cPOPs), which have been shown to regulate the inflammasome (83). Consistently, vPOPs co-localize with ASC and reduce caspase-1 activation and IL-1β processing in response to NLRP3 activation when overexpressed in an inflammasome reconstitution system. As another mode of action, these proteins also repress NF-κB-dependent transcription of pro-IL-1β and other cytokines (82, 84). Another viral strategy of inflammasome inhibition is to sequester or block the activity of inflammasome receptors. For example, Kaposi’s sarcoma-associated herpesvirus (KSHV) encodes a protein called Orf3, which is a homolog of NLRP1, but lacks the CARD and PYD domains (85). This protein interacts with NLRP1, NLRP3, and NOD2 and was necessary to repress IL-1β production during KSHV infections. An additional mechanism of inhibiting inflammasome activity is to target caspase-1 directly. Viral serpins (serine protease inhibitors), such as cowpox crmA, vaccinia virus B13R, myxoma virus Serp2, and ectromelia virus SPI-2, have been shown to either inhibit caspase-1 proteolytic activity or to bind directly to caspase-1 (86–89). In conclusion, these results highlight the surprising variety of viral strategies to inhibit the assembly of the inflammasome complex itself, blocking caspase-1 activity or limiting expression of caspase-1 substrates.
There is no evidence yet for a direct inflammasome inhibition by bacterial pathogens, and no bacterial effectors with homology to NLRs or cPOPs have been identified thus far. Nevertheless, many bacterial pathogens avoid detection by the inflammasome and other innate immune receptors by downregulating the expression of PAMPs (80). For example, Salmonella is known to downregulate the expression of flagellin and the SPI-1 T3SS during systemic infections. Thus, Salmonellae avoid detection of flagellin by TLR5 as well as NLRC4-mediated detection of flagellin and of the SPI-1 rod subunit PrgJ. The rod-subunit of the SPI-2 T3SS, which is expressed during systemic infections, does not activate NLRC4. Early studies with Y. enterocolitica demonstrated that formation of the T3SS translocation pore formation can also trigger caspase-1 activation in cultured cell lines and that this activation was inhibited by bacterial T3SS effector proteins (90). Indeed, a recent study highlighted the importance of maintaining the integrity of the T3SS translocation pore, since Yersinia pseudotuberculosis mutants lacking YopK, a component of the T3SS translocon, induce higher levels of caspase-1 activation and IL-1β release (39). It was subsequently proposed, that YopK masks the T3SS apparatus and prevents its recognition by the innate immune system. However, as YopK was previously proposed to modulate the size of the translocation pore (91), it is also possible that YopK acts as a gatekeeper for the T3SS translocon and that in its absence proteins like flagellin or small molecule PAMPs might leak into the host cell cytoplasm.
Several other bacterial pathogens have been reported to possibly possess effector proteins that inhibit inflammasome activation. Infections with Mycobacterium tuberculosis are typically not associated with high levels of IL-1β, but mycobacteria lacking the Zn2+ metalloprotease Zmp1 induced increased levels of caspase-1 activation and IL-1β maturation (92). Zmp1 also appeared to inhibit lysosomal maturation of mycobacterium-containing phagosomes and prevented mycobacterial clearance by macrophages. Intriguingly, infection with Zmp1+ mycobacteria prevented the activation of caspase-1 triggered by the simultaneous administration of NLRP3 agonists, suggesting an active repression of inflammasome activation. Further research will be necessary to define the exact mode of action of Zmp1 during mycobacterial infections. Similarly, P. aeruginosa normally induces activation of the NLRC4-inflammasome; however, the PA103 strain does not induce caspase-1 activation and IL-1β maturation (93). This effect was attributed to the T3SS effector ExoU and was dependent on its phospholipase activity, yet it remains unclear how this activity relates mechanistically to its inhibitory action on caspase-1. In addition, ExoS was shown to negatively regulate caspase-1 mediated IL-1β processing; however, this effect is likely to be indirect, since ExoS deficiency switched the mode of macrophage death from apoptosis to pyroptosis (94). An indirect effect on caspase-1 activation could also be responsible for the elevated levels of caspase-1 activation and IL-1β release observed during the infection of cultured macrophages with Y. enterocolitica deficient in T3SS effectors YopT and YopE (90). Since both of these effectors are known to rapidly disrupt the actin cytoskeleton and prevent phagocytosis, the increased inflammasome activation observed with these mutants could be due to indirect effects.
Francisella genes that were identified in various genetic screens were speculated to suppress inflammasome activation, because bacterial mutants lacking these genes stimulated elevated levels of pro-inflammatory secretion and macrophage death (54, 66, 95, 96). For example, a microarray-based negative selection screen for bacterial genes necessary for survival in mice identified FTT0584, a gene of unknown function (54). The ΔFTT0584 deletion mutant induced increased levels of IL-1β release and was hypercytotoxic in cultured macrophages compared to wildtype bacteria (54). These phenotypes were dependent on caspase-1 and ASC (54). Similarly, F. tularensis live vaccine strain (LVS) mutants that lack mviN, which encodes a putative lipid II flippase, or ripA, which encodes a cytosolic membrane protein of unknown function, have been reported to trigger increased levels of pyroptosis (66, 95). In addition, Francisella tularensis subsp. tularensis mutant strains that lack one or more of the genes that play a role in O-antigen and capsule biosynthesis, FTT1236, FTT1237, and FTT1238, induce higher levels of macrophage cytotoxicity compared to wildtype bacteria (96). Based on these results, we and others had initially speculated that these Francisella genes might encode virulence factors that suppress inflammasome activation. However, in light of our increased knowledge about the mechanisms of inflammasome activation during Francisella infections, we revisited this hypothesis.
It has been shown recently that a small number of wildtype Francisella lyse in the macrophage cytosol and that the released bacterial DNA is recognized by AIM2, which results in activation of the inflammasome. Since many of the previously described hypercytotoxic Francisella mutants are deficient for genes that encode proteins with diverse biological functions and yet exhibit a similar phenotype in macrophages, we postulated that the mechanism for inducing elevated macrophage cytotoxicity involves increased bacterial lysis and therefore increased release of bacterial DNA as well as other PAMPs into the intracellular milieu. Therefore, we have recently examined F. novicida hypercytotoxic mutants that are deficient for membrane-associated proteins (ΔFTT0584, ΔmviN, ΔripA, ΔfopA and ΔFTN1217) or for O-antigen or LPS biosynthesis genes (ΔwbtA and ΔlpxH) to determine if increased lysis could account for their hypercytotoxic phenotype in macrophages. All of the F. novicida mutants tested stimulated increased levels of AIM2 inflammasome activation compared to wildtype bacteria (authors’ unpublished data). In addition, the hypercytotoxic F. novicida mutants released higher levels of bacterial DNA into the host cell cytosol compared to wildtype bacteria, as measured by delivery of a luciferase reporter plasmid (97), indicating that the mutants lyse more than wildtype bacteria within the host cell (authors’ unpublished data). Consistent with this finding, the hypercytotoxic mutants induced significantly more AIM2-DNA complexes in macrophages compared to wildtype bacteria. Finally, these bacterial mutants hyperstimulated additional innate immune recognition pathways, as indicated by increased levels of TNF-α and IL-6 production (authors’ unpublished data), suggesting that these mutants are either releasing more PAMPs or have additional PAMPs exposed on their surface. Our results indicate that F. novicida does not actively suppress or modulate the inflammasome. Importantly, our results have implications for other bacterial pathogens by demonstrating that mutations in membrane-associated proteins seem to affect stability or integrity of the bacterial membrane and increase susceptibility of the pathogen to lysis and subsequent release of PAMPs.
Structure of the inflammasome complex
Even though components of the inflammasome have been known for over a decade, structural data on the inflammasome complex remains scarce. Functionally, the inflammasome is related to the multi-protein complexes that activate apoptotic initiator caspases such as the apoptosome, the DISC complex, or the PIDDosome. The best characterized of these is the apoptosome that serves as an activation platform for Caspase-9. Biochemical and structural analysis of the in vitro reconstituted human apoptosome revealed a heptameric, wheel-shaped complex consisting of Apaf-1, cytochrome c and pro-caspase-9, with an apparent molecular weight of 700–1400 kDa. Similarly, the CED-4/3 apoptosome of Caenorhabditis elegans forms a ring-like structure (7). Consistently, inflammasome components can also form oligomers between 500 and 700 kDa in size (98, 99) under certain lysis conditions. In addition, in vitro studies with purified inflammasome components suggested that NLRP1 oligomerizes to form a donut-shaped platform following incubation with its ligand MDP (19). Although the structures of single domains (CARD, PYD) as well as full-length and processed caspase-1 have been determined (100), high-resolution structures of the multi-protein inflammasome complex still remain an exciting task for crystallographers to pursue.
In addition to the above-mentioned oligomers, components of the inflammasome have also been shown to assemble a much larger, macro-molecular structure, called a ‘speck’, ‘ASC focus’, or ‘pyroptosome’. As the field has not agreed on a particular name for these structures, we will refer to them as ASC foci, since ASC is a structural component of the focus (27). Early studies of the inflammasome adapter ASC reported its peculiar tendency to aggregate and form an ASC focus when overexpressed in HL-60 cells or in HeLa cells (101, 102). In addition, these ASC foci could recruit other proteins, such as Pyrin, through homotypic PYD-PYD interactions (103). Alnemri and collegues (104) provided the first detailed study of ASC foci that were formed when THP-1 cells overexpressing ASC-GFP were stimulated with LPS and ATP. Crude purification of these structures revealed that they consisted of ASC-dimers and had a star-shaped crystal-like structure. In contrast to this report, ASC, NLRP3, and caspase-1 formed a large donut-shaped perinuclear structure when overexpressed in HEK293 cells (105). However, since these ASC aggregates were observed in overexpression studies their biological relevance remained unclear.
We recently investigated the subcellular localization of endogenous inflammasome components in murine bone marrow-derived macrophages before and after infection with S. typhimurium (27). We observed that ASC, which was mostly nuclear in uninfected macrophages, re-localized to the cytoplasm following infection, where it formed a dense speck of 1–2 micron in diameter, which we named an ASC focus. Formation of the ASC focus was observed with Salmonella expressing the SPI-1 T3SS (activate NLRC4) or Salmonella repressing SPI-1 expression (activate NLRC4 and NLRP3). Interestingly, even two receptors can be activated by Salmonella, only one ASC focus forms per cell. A recent study showed that aggregation of YFP-tagged ASC in HeLa cells was a rapid all or nothing reaction, making it highly unlikely that a second focus would form in the same cell (106), but the relevance of these observations to endogenous ASC in immune cells is unclear. ASC foci also formed in close proximity of DNA/AIM2 complexes during the infection of murine macrophages with F. tularensis (27, 58). The ASC focus did not co-localize with the DNA/AIM2 complexes formed during Francisella infection but formed right next to one of these structures. This observation suggested that the receptor AIM2 is not directly part of the structure but most likely provides nucleation sites on which the focus forms through homotypic PYD-PYD and CARD-CARD interaction between ASC molecules. If NLRC4 and NLRP3 are part of the ASC focus during Salmonella infection remains to be determined.
Since the formation of the ASC foci strongly correlated with the release of mature cytokines but not with cell death, we investigated whether the ASC foci might represent sites of IL-1β maturation (27). Immunofluorescence microscopy confirmed that pro-caspase-1 is recruited to the ASC focus, even though the presence of caspase-1 is not necessary for focus formation. Staining with FLICA, a fluorescent caspase-1 activity probe, also revealed that catalytically active caspase-1 is present in the focus (27). Similarly, foci of active caspase-1 have been reported to form during the stimulation of murine macrophages with Anthrax lethal toxin, suggesting that Nlrp1b can also induce ASC focus formation (107). Interestingly, in macrophages treated with a caspase-1 inhibitor or expressing a catalytically dead mutant caspase-1, we observed that pro-caspase-1 accumulated at the site of the speck (27, 78). These results suggest that caspase-1 is only recruited temporally to the ASC focus trough CARD-CARD interactions and released again once cleavage between the p20/p10 complex and the CARD domain has occurred. Similarly, pro-IL-1β accumulated in the ASC focus, if caspase-1 activity was inhibited (27). Since ASC is necessary to induce pro-caspase-1 autoproteolysis, a prerequisite for efficient cytokine processing (78), we suggest that the macromolecular ASC focus could represent the major subcellular site of caspase-1 activation and cytokine processing. However, it cannot be excluded that smaller inflammasome complexes, such as the ASC-independent death-complexes that are formed by CARD-containing receptors (see next section), co-exist in the same cell with the large ASC focus during infection.
The exact function of the ASC focus remains to be determined. The relocation of inflammasome components into one focus could serve to integrate signals originating from different NLRs and/or enhance inflammasome-mediated cytokine processing. Alternatively, since ASC is important for cytokine processing, the focus could link the inflammasome to organelles mediating the unconventional secretion pathway, which releases mature IL-1β and IL-18. Future studies addressing the exact composition and molecular architecture of the ASC focus will hopefully lead to a greater understanding of the function of this large macromolecular structure.
ASC: more than just an inflammasome adapter
Thus far, the inflammasome adapter protein ASC has received less attention than inflammasome receptors, most likely since it is only considered to be an adapter protein of the inflammasome complex. However, recent results have expanded our knowledge of ASC and suggest additional roles for ASC beyond its function as adapter protein. The protein was originally identified as a small 22 kDa protein that formed a perinuclear speck in apoptotic HL-60 cells (101); hence it was called ASC for ‘apoptosis-associated speck-like protein containing a CARD’. It is also known as Pycard, since it consists of an N-terminal PYRIN-like domain (PYD) followed by a C-terminal CARD. Interestingly, the PYD of ASC forms filamentous structures when overexpressed, while the full-length protein formed the above-mentioned specks (103). At the time of its discovery, the role of ASC was elusive; depending on the context, it could act as an activator or repressor of NF-κB signaling or was involved in apoptotic signaling (108, 109). However, a role for ASC in caspase-1 activation had also been observed (110), which became more evident, when the group of J. Tschopp (17) identified a caspase-1-activating complex called the inflammasome in the human THP-1 monocyte cell line consisting of caspase-1, caspase-5, NLRP1, and ASC. They concluded that ASC acts as an adapter protein, interacting with NLRP1 through its PYD and recruiting pro-caspase-1 by homotypic CARD-CARD interactions.
Although ASC is generally referred to as an essential part of the inflammasome, it remained unclear if all types of inflammasome complexes require the recruitment ASC. Based on their distinct effector domain, inflammasome receptors can be divided into two subclasses, the PYD-containing receptors (NLRP3 and AIM2) and the CARD-containing receptors (NLRC4 and murine Nlrp1b) (Fig. 1). The generation of ASC-deficient mice established that ASC was a crucial component of inflammasome complexes formed by the PYD-containing sensors NLRP3 and AIM2 (24, 57–59), which cannot recruit pro-caspase-1 in the absence of ASC. However the role of ASC for CARD-containing receptors, like NLRC4 or Nlrp1b remained controversial, since for example NLRC4 was shown to directly interact with pro-caspase-1 via its CARD domain (111). Nevertheless, ASC seemed to be required for the full activity of the NLRC4-inflammasome during microbial infections (27, 36, 112).
We and others reported earlier that ASC-deficient macrophages infected with bacteria that activate NLRC4 are severely impaired in their ability to mature cytokines; however, they still undergo pyroptotic cell death to levels comparable to wildtype macrophages. This phenotype of Asc−/−macrophages was observed with S. typhimurium, S. flexneri, and L. pneumophila, and in the following, we refer to it as ASC-independent cell death (36, 112, 113). Interestingly, ASC-independent cell death seems to progress with slower kinetics during S. typhimurium infection (36), while the kinetics of ASC-independent cell death were undistinguishable from cell death in wildtype macrophages during S. flexneri infections and L. pneumophila infections (112, 113). The most remarkable feature of ASC-independent cell death, however, is the absence of any detectable pro-caspase-1 autoproteolytic processing. This feature was observed during infections with all pathogens activating NLRC4, such as S. typhimurium, L. pneumophila, P. aeruginosa, and Y. pseudotuberculosis (39), and it remained mysterious since pro-caspase-1 autoproteolysis was regarded as the hallmark of caspase-1 activation. It was therefore hypothesized that other caspases, such as caspase-11 which is absent in caspase-1 knockout mice, could be responsible for cell death in ASC-deficient macrophages or even that the catalytic activity of caspase-1 was not required for cell death. Alternatively, since ASC was known to also mediate NF-κB signaling, it was proposed that ASC might promote prosurvival signals in the absence of caspase-1 and NLRC4, and that these cues would be missing in ASC-deficient macrophages (112).
We investigated the molecular basis of ASC-independent cell death (78). We observed that stimulation of the CARD-containing receptors NLRC4 with S. typhimurium, L. pneumophila, and P. aeruginosa or Nlrp1b with Anthrax lethal toxin induced pyroptosis in the absence of ASC. Since NLRC4 and Nlrp1b contain CARDs, these results led to the hypothesis that a CARD domain was sufficient to promote ASC-independent pyroptosis in the absence of pro-caspase-1 autoproteolysis. To prove this hypothesis, we generated a chimeric CARD-AIM2 fusion receptor and activated the receptor by DNA transfection or F. novicida infections in wildtype or Asc−/− cells. Indeed, the CARD-AIM2 fusion receptor induced pyroptosis in the absence of ASC and independently of pro-caspase-1 autoproteolysis (78). Importantly, cell death in ASC-deficient macrophages was still dependent on the catalytic activity of caspase-1 and was independent of caspase-11. These results suggested that even uncleaved pro-caspase-1 can have catalytic activity and that proximity induced dimerization in the inflammasome complex is the actual activation step of the caspase. To prove this hypothesis, we generated an uncleavable pro-caspase-1 by mutating the autoproteolysis cleavage sites, and expressed wildtype caspase-1 and the uncleavable mutant protein in caspase-1-deficient bone marrow-derived macrophages. Surprisingly, uncleavable pro-caspase-1 was capable of initiating pyroptosis to the same levels as wildtype caspase-1. However, only wildtype caspase-1 promoted efficient cytokine processing and autoproteolysis. These results suggest that in addition to acting as an adapter protein for PYD-containing sensors (NLRP3, AIM2), the recruitment of ASC to inflammasomes assembled by CARD-containing receptors was necessary to induce pro-caspase-1 autoproteolysis in these complexes, which is a prerequisite for efficient cytokine processing (78). However, CARD-containing receptors can still initiate the formation of a subtype of inflammasome complex in the absence of ASC. These complexes, which are only able to efficiently promote pyroptotic cell death, are referred to as ‘death complexes’, to distinguish them from ASC-containing complexes (Fig. 4).
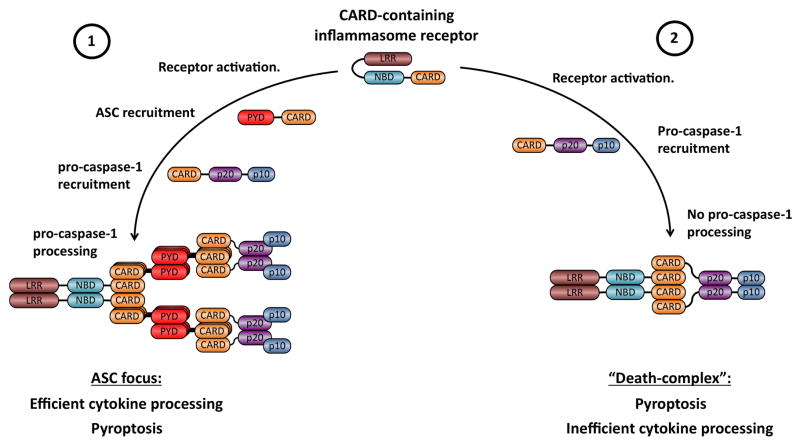
Fig. 4. Model for the formation of functionally distinct inflammasomes by CARD-containing receptors.
(1) Recruitment of ASC to activated receptors is followed by rapid oligomerization of ASC and the formation of the macro-molecular ASC focus through homotypic CARD-CARD and PYD-PYD interactions. This complex recruits and activates pro-caspase-1 through proximity-induced dimerization. In the ASC focus, conformational changes in the pro-caspase-1 dimer allow for auto-proteolytic processing into the p20 and p10 subunits. Only fully processed Caspase-1 efficiently promotes cytokine maturation. (2) Recruitment of pro-caspase-1 to activated receptors leads to the formation of a “Death-complex”. Pro-caspase-1 is not autoproteolytically processed in this type of a complex. Nevertheless, pro-caspase-1 is activated in this complex and promotes pyroptosis and inefficient cytokine processing. LRR, leucine-rich repeats; NBD, nucleotide binding and oligomerization domain; PYD, Pyrin-like domain; CARD, caspase activation and recruitment domain.
We speculate that the death complex could represent a functional analog of the apoptosome complexes that activates apoptotic caspases. The caspase-9 apoptosome, for example, has an apparent molecular weight of 700–1400 kDa, and no macromolecular assemblies similar to the ASC focus have been reported for the caspase-9 apoptosome. Similarly, NLRC4-death complexes do not form any macromolecular structure in the cytosol; caspase-1 can be found distributed in a diffuse, speckled pattern in Asc−/− macrophages (78). Another striking similarity between caspase-1 and caspase-9 is that the activation of caspase-9 does not seems to require autoproteolytic processing either (114). Apoptotic initiator caspases (caspase-9, caspase-8, and caspase-2, Dronc) are thought to be activated by proximity-driven dimerization in the corresponding multiprotein activation platforms (115); however, the importance of the autoproteolytic processing that occurs simultaneously with dimerization is unclear. Processing seems to be important to stabilize dimers of caspase-2, caspase-8, and Dronc, since autocleaved forms of these caspases preferentially form dimers and only these dimers have sufficient catalytic activity to induce cell death (7). In contrast, noncleavable caspase-9 retains the ability to cleave substrates and promote cell death (114, 116). Recently, Malladi et al. (116) proposed that autoproteolysis of caspase-9 within the apoptosome could serve as a ‘molecular timer’ that limits the proteolytic activity of the complex through displacement of bound caspase-9 molecules. In an elegant set of experiments, the investigators showed that autoproteolytic processing of pro-caspase-9 reduced the affinity of the caspase for the complex and consequently processed caspase-9 was quickly displaced from the apoptosome by unprocessed pro-caspase-9. Importantly, caspase-9 is only active during the short time it is part of the apoptosome and is inactive once released. Since uncleavable pro-caspase-9 does not have reduced affinity for the complex, it was able to override the molecular timer and consistently hyper-induced cell death (116). According to this model, the intracellular concentration of pro-caspase-9 sets the duration of the timer (i.e. duration of the apoptotic signal). Pro-caspase-9 autoprocessing activates the timer, and the rate at which the processed caspase-9 dissociates from the complex (and thus loses its capacity to activate effector caspases) dictates how fast the timer ‘ticks’ over. Thus, cells with low concentrations of caspase-9 may escape full-blown caspase activation and, consequently, death, in circumstances in which few apoptosomes become assembled within a particular window of time. It remains to be determined whether caspase-1 auto-proteolysis might also reduce its affinity for the complex and whether uncleavable caspase-1 is hyperactive in inducing cell death. In addition, it is unclear why caspase-1 auto-proteolysis is required for efficient cytokine processing. It might be possible that auto-proteolysis simply enhances caspase-1 activity or that processed caspase-1 has different substrate specificity. Additional studies will be required to fully understand the molecular mechanisms of caspase-1 activation and the down-stream signaling pathways.
Conclusions and future directions
Innate immunity and inflammation are crucial for the host defense against invading pathogens as well as for tissue repair. In particular, the inflammasome has emerged as a central signaling complex driving the maturation of pro-inflammatory cytokines and regulating pyroptotic cell death. In addition to being important for pathogen recognition, the high incidence of hereditary and acquired diseases, in which inflammasome activity is deregulated, highlights the importance of this signaling platform for human health.
There has been rapid advance in recent years in the characterization of pathogens and ligands that lead to inflammasome assembly and in the identification of receptors that recognize these ligands. However, despite these advances, many aspects of the specific signaling events that lead to ligand recognition, inflammasome assembly, and finally to IL-1β/IL-18 release and cell death remain uncharacterized. For example, even though NLRs are referred to as receptors, it is not clear how these proteins recognize their ligands; a direct interaction of ligand and receptor has only been shown for AIM2. In particular, the wide variety of signals detected by NLRP3 suggests that additional upstream signaling pathways are involved. It is possible that additional proteins serve as receptors or that adapters are needed for the recognition, similar to the function of NAIP5 in flagellin recognition. In addition, very little is known about the assembly of the complex itself and its structure following receptor activation. Considering the similarities of the inflammasome to the apoptosome complexes, defining the structure of the complex and its components will provide exciting opportunities for structural biologists to pursue. In addition, the formation of the macromolecular ASC focus and its significance for inflammasome signaling remains mysterious.
The downstream signaling pathways promoted by active caspase-1 remain largely unstudied. Active caspase-1 appears to direct an unconventional endoplasmic reticulum/Golgi-independent secretion pathway that is responsible for the secretion of a large number of proteins involved in tissue repair and cytoprotection (117), in addition to the mature IL-1β and IL-18 cytokines. The majority of these proteins do not appear to be cleavage substrates of caspase-1, but nonetheless depend on the activity of caspase-1 for their secretion (117). Several possible pathways of IL-1β secretion have been proposed such as secretion via exocytosis of secretory lysosomes, shedding of plasma membrane microvesicles, release of exosomes or a specialized membrane transporter, but the exact mechanism of unconventional secretion remains an area of further exploration (118). In addition, the nature of the novel form of caspase-1-dependent cell death, called pyroptosis, is still uncharacterized (9). Pyroptosis has many similarities to apoptosis, like nuclear condensation and PARP1 cleavage; however, it is also accompanied by swelling and rupture of the cell. Activation of caspase-3 and caspase-7 has been observed to occur during pyroptosis (119, authors’ unpublished results), but it is unclear if these effector caspases contribute to caspase-1 induced cell death. Undoubtedly, characterization of the downstream signaling pathways of the inflammasome will be a major task for the years to come and will not only contribute to a better understanding of inflammasome importance but also could provide new targets for the development of therapeutic strategies.
Acknowledgments
Denise M. Monack is supported by grants AI063302 and AI065359 from the NIHNIAID. Petr Broz is supported by a long-term postdoctoral fellowship from the Human Frontiers in Science Program (HFSP).
Footnotes
The authors declare no conflicts of interest
References
- 1.Janeway CA., Jr Approaching the asymptote? Evolution and revolution in immunology. Cold Spring Harb Symp Quant Biol. 1989;54:1–13. doi: 10.1101/sqb.1989.054.01.003. [DOI] [PubMed] [Google Scholar]
- 2.Vance RE, Isberg RR, Portnoy DA. Patterns of pathogenesis: discrimination of pathogenic and nonpathogenic microbes by the innate immune system. Cell Host Microbe. 2009;6:10–21. doi: 10.1016/j.chom.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Iwasaki A, Medzhitov R. Toll-like receptor control of the adaptive immune responses. Nat Immunol. 2004;5:987–995. doi: 10.1038/ni1112. [DOI] [PubMed] [Google Scholar]
- 4.Brown GD. Dectin-1: a signalling non-TLR pattern-recognition receptor. Nat Rev Immunol. 2006;6:33–43. doi: 10.1038/nri1745. [DOI] [PubMed] [Google Scholar]
- 5.Schroder K, Tschopp J. The inflammasomes. Cell. 2010;140:821–832. doi: 10.1016/j.cell.2010.01.040. [DOI] [PubMed] [Google Scholar]
- 6.Taylor RC, Cullen SP, Martin SJ. Apoptosis: controlled demolition at the cellular level. Nat Rev Mol Cell Biol. 2008;9:231–241. doi: 10.1038/nrm2312. [DOI] [PubMed] [Google Scholar]
- 7.Bao Q, Shi Y. Apoptosome: a platform for the activation of initiator caspases. Cell Death Differ. 2007;14:56–65. doi: 10.1038/sj.cdd.4402028. [DOI] [PubMed] [Google Scholar]
- 8.Thornberry NA, Lazebnik Y. Caspases: enemies within. Science. 1998;281:1312–1316. doi: 10.1126/science.281.5381.1312. [DOI] [PubMed] [Google Scholar]
- 9.Bergsbaken T, Fink SL, Cookson BT. Pyroptosis: host cell death and inflammation. Nat Rev Microbiol. 2009;7:99–109. doi: 10.1038/nrmicro2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martinon F, Mayor A, Tschopp J. The inflammasomes: guardians of the body. Annu Rev Immunol. 2009;27:229–265. doi: 10.1146/annurev.immunol.021908.132715. [DOI] [PubMed] [Google Scholar]
- 11.Medzhitov R. Inflammation 2010: new adventures of an old flame. Cell. 2010;140:771–776. doi: 10.1016/j.cell.2010.03.006. [DOI] [PubMed] [Google Scholar]
- 12.Steimle V, Otten LA, Zufferey M, Mach B. Complementation cloning of an MHC class II transactivator mutated in hereditary MHC class II deficiency (or bare lymphocyte syndrome) Cell. 1993;75:135–146. [PubMed] [Google Scholar]
- 13.Meissner TB, et al. NLR family member NLRC5 is a transcriptional regulator of MHC class I genes. Proc Natl Acad Sci USA. 2010;107:13794–13799. doi: 10.1073/pnas.1008684107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cui J, et al. NLRC5 negatively regulates the NF-kappaB and type I interferon signaling pathways. Cell. 2010;141:483–496. doi: 10.1016/j.cell.2010.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kumar H, et al. NLRC5 deficiency does not influence cytokine induction by virus and bacteria infections. J Immunol. 2011;186:994–1000. doi: 10.4049/jimmunol.1002094. [DOI] [PubMed] [Google Scholar]
- 16.Davis BK, et al. Cutting edge: NLRC5-dependent activation of the inflammasome. J Immunol. 2011;186:1333–1337. doi: 10.4049/jimmunol.1003111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martinon F, Burns K, Tschopp J. The inflammasome: a molecular platform triggering activation of inflammatory caspases and processing of proIL-beta. Mol Cell. 2002;10:417–426. doi: 10.1016/s1097-2765(02)00599-3. [DOI] [PubMed] [Google Scholar]
- 18.Boyden ED, Dietrich WF. Nalp1b controls mouse macrophage susceptibility to anthrax lethal toxin. Nat Genet. 2006;38:240–244. doi: 10.1038/ng1724. [DOI] [PubMed] [Google Scholar]
- 19.Faustin B, et al. Reconstituted NALP1 inflammasome reveals two-step mechanism of caspase-1 activation. Mol Cell. 2007;25:713–724. doi: 10.1016/j.molcel.2007.01.032. [DOI] [PubMed] [Google Scholar]
- 20.Muruve DA, et al. The inflammasome recognizes cytosolic microbial and host DNA and triggers an innate immune response. Nature. 2008;452:103–107. doi: 10.1038/nature06664. [DOI] [PubMed] [Google Scholar]
- 21.Kanneganti TD, et al. Critical role for Cryopyrin/Nalp3 in activation of caspase-1 in response to viral infection and double-stranded RNA. J Biol Chem. 2006;281:36560–36568. doi: 10.1074/jbc.M607594200. [DOI] [PubMed] [Google Scholar]
- 22.Hise AG, Tomalka J, Ganesan S, Patel K, Hall BA, Brown GD, et al. An essential role for the NLRP3 inflammasome in host defense against the human fungal pathogen Candida albicans. Cell Host Microbe. 2009;5:487–497. doi: 10.1016/j.chom.2009.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gross O, et al. Syk kinase signalling couples to the Nlrp3 inflammasome for anti-fungal host defence. Nature. 2009 May 21;459:433–436. doi: 10.1038/nature07965. [DOI] [PubMed] [Google Scholar]
- 24.Mariathasan S, et al. Cryopyrin activates the inflammasome in response to toxins and ATP. Nature. 2006;440:228–232. doi: 10.1038/nature04515. [DOI] [PubMed] [Google Scholar]
- 25.Warren SE, Mao DP, Rodriguez AE, Miao EA, Aderem A. Multiple Nod-like receptors activate caspase 1 during Listeria monocytogenes infection. J Immunol. 2008;180:7558–7564. doi: 10.4049/jimmunol.180.11.7558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Duncan JA, et al. Neisseria gonorrhoeae activates the proteinase cathepsin B to mediate the signaling activities of the NLRP3 and ASC-containing inflammasome. J Immunol. 2009;182:6460–6469. doi: 10.4049/jimmunol.0802696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Broz P, Newton K, Lamkanfi M, Mariathasan S, Dixit VM, Monack DM. Redundant roles for inflammasome receptors NLRP3 and NLRC4 in host defense against Salmonella. J Exp Med. 2010;207:1745–1755. doi: 10.1084/jem.20100257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martinon F, Petrilli V, Mayor A, Tardivel A, Tschopp J. Gout-associated uric acid crystals activate the NALP3 inflammasome. Nature. 2006;440:237–241. doi: 10.1038/nature04516. [DOI] [PubMed] [Google Scholar]
- 29.Dostert C, Petrilli V, Van Bruggen R, Steele C, Mossman BT, Tschopp J. Innate immune activation through Nalp3 inflammasome sensing of asbestos and silica. Science. 2008;320:674–677. doi: 10.1126/science.1156995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hornung V, et al. Silica crystals and aluminum salts activate the NALP3 inflammasome through phagosomal destabilization. Nat Immunol. 2008;9:847–856. doi: 10.1038/ni.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cassel SL, et al. The Nalp3 inflammasome is essential for the development of silicosis. Proc Natl Acad Sci USA. 2008;105:9035–9040. doi: 10.1073/pnas.0803933105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Halle A, et al. The NALP3 inflammasome is involved in the innate immune response to amyloid-beta. Nat Immunol. 2008;9:857–865. doi: 10.1038/ni.1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miao EA, et al. Innate immune detection of the type III secretion apparatus through the NLRC4 inflammasome. Proc Natl Acad Sci USA. 2010;107:3076–3080. doi: 10.1073/pnas.0913087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miao EA, et al. Cytoplasmic flagellin activates caspase-1 and secretion of interleukin 1beta via Ipaf. Nat Immunol. 2006;7:569–575. doi: 10.1038/ni1344. [DOI] [PubMed] [Google Scholar]
- 35.Franchi L, et al. Cytosolic flagellin requires Ipaf for activation of caspase-1 and interleukin 1beta in salmonella-infected macrophages. Nat Immunol. 2006;7:576–582. doi: 10.1038/ni1346. [DOI] [PubMed] [Google Scholar]
- 36.Mariathasan S, et al. Differential activation of the inflammasome by caspase-1 adaptors ASC and Ipaf. Nature. 2004;430:213–218. doi: 10.1038/nature02664. [DOI] [PubMed] [Google Scholar]
- 37.Miao EA, Ernst RK, Dors M, Mao DP, Aderem A. Pseudomonas aeruginosa activates caspase 1 through Ipaf. Proc Natl Acad Sci USA. 2008;105:2562–2567. doi: 10.1073/pnas.0712183105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McCoy AJ, et al. Cytotoxins of the human pathogen Aeromonas hydrophila trigger, via the NLRP3 inflammasome, caspase-1 activation in macrophages. Eur J Immunol. 2010;40:2797–2803. doi: 10.1002/eji.201040490. [DOI] [PubMed] [Google Scholar]
- 39.Brodsky IE, et al. A Yersinia effector protein promotes virulence by preventing inflammasome recognition of the type III secretion system. Cell Host Microbe. 2010;7:376–387. doi: 10.1016/j.chom.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Amer A, et al. Regulation of Legionella phagosome maturation and infection through flagellin and host Ipaf. J Biol Chem. 2006;281:35217–35223. doi: 10.1074/jbc.M604933200. [DOI] [PubMed] [Google Scholar]
- 41.Hornung V, et al. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature. 2009;458:514–518. doi: 10.1038/nature07725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fernandes-Alnemri T, Yu JW, Datta P, Wu J, Alnemri ES. AIM2 activates the inflammasome and cell death in response to cytoplasmic DNA. Nature. 2009;458:509–513. doi: 10.1038/nature07710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Burckstummer T, et al. An orthogonal proteomic-genomic screen identifies AIM2 as a cytoplasmic DNA sensor for the inflammasome. Nat Immunol. 2009;10:266–272. doi: 10.1038/ni.1702. [DOI] [PubMed] [Google Scholar]
- 44.Roberts TL, et al. HIN-200 proteins regulate caspase activation in response to foreign cytoplasmic DNA. Science. 2009;323:1057–1060. doi: 10.1126/science.1169841. [DOI] [PubMed] [Google Scholar]
- 45.Unterholzner L, et al. IFI16 is an innate immune sensor for intracellular DNA. Nat Immunol. 2010;11:997–1004. doi: 10.1038/ni.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Checroun C, Wehrly TD, Fischer ER, Hayes SF, Celli J. Autophagy-mediated reentry of Francisella tularensis into the endocytic compartment after cytoplasmic replication. Proc Natl Acad Sci USA. 2006;103:14578–14583. doi: 10.1073/pnas.0601838103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Clemens DL, Lee BY, Horwitz MA. Virulent and avirulent strains of Francisella tularensis prevent acidification and maturation of their phagosomes and escape into the cytoplasm in human macrophages. Infect Immun. 2004;72:3204–3217. doi: 10.1128/IAI.72.6.3204-3217.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Santic M, Molmeret M, Klose KE, Jones S, Kwaik YA. The Francisella tularensis pathogenicity island protein IglC and its regulator MglA are essential for modulating phagosome biogenesis and subsequent bacterial escape into the cytoplasm. Cell Microbiol. 2005;7:969–979. doi: 10.1111/j.1462-5822.2005.00526.x. [DOI] [PubMed] [Google Scholar]
- 49.Santic M, Asare R, Skrobonja I, Jones S, Abu Kwaik Y. Acquisition of the vacuolar ATPase proton pump and phagosome acidification are essential for escape of Francisella tularensis into the macrophage cytosol. Infect Immun. 2008;76:2671–2677. doi: 10.1128/IAI.00185-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chong A, et al. The early phagosomal stage of Francisella tularensis determines optimal phagosomal escape and Francisella pathogenicity island protein expression. Infect Immun. 2008;76:5488–5499. doi: 10.1128/IAI.00682-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nano FE, et al. A Francisella tularensis pathogenicity island required for intramacrophage growth. J Bacteriol. 2004;186:6430–6436. doi: 10.1128/JB.186.19.6430-6436.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bonquist L, Lindgren H, Golovliov I, Guina T, Sjostedt A. MglA and Igl proteins contribute to the modulation of Francisella tularensis live vaccine strain-containing phagosomes in murine macrophages. Infect Immun. 2008;76:3502–10. doi: 10.1128/IAI.00226-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Brotcke A, et al. Identification of MglA-regulated genes reveals novel virulence factors in Francisella tularensis. Infect Immun. 2006;74:6642–6655. doi: 10.1128/IAI.01250-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Weiss DS, Brotcke A, Henry T, Margolis JJ, Chan K, Monack DM. In vivo negative selection screen identifies genes required for Francisella virulence. Proc Natl Acad Sci USA. 2007;104:6037–6042. doi: 10.1073/pnas.0609675104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mariathasan S, Weiss DS, Dixit VM, Monack DM. Innate immunity against Francisella tularensis is dependent on the ASC/caspase-1 axis. J Exp Med. 2005;202:1043–1049. doi: 10.1084/jem.20050977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Henry T, Brotcke A, Weiss DS, Thompson LJ, Monack DM. Type I interferon signaling is required for activation of the inflammasome during Francisella infection. J Exp Med. 2007;204:987–994. doi: 10.1084/jem.20062665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fernandes-Alnemri T, et al. The AIM2 inflammasome is critical for innate immunity to Francisella tularensis. Nat Immunol. 2010;11:385–393. doi: 10.1038/ni.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jones JW, et al. Absent in melanoma 2 is required for innate immune recognition of Francisella tularensis. Proc Natl Acad Sci USA. 2010;107:9771–9776. doi: 10.1073/pnas.1003738107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rathinam VA, et al. The AIM2 inflammasome is essential for host defense against cytosolic bacteria and DNA viruses. Nat Immunol. 2010;11:395–402. doi: 10.1038/ni.1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Warren SE, et al. Cutting edge: cytosolic bacterial DNA activates the inflammasome via Aim2. J Immunol. 2010;185:818–821. doi: 10.4049/jimmunol.1000724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wu J, Fernandes-Alnemri T, Alnemri ES. Involvement of the AIM2, NLRC4, and NLRP3 inflammasomes in caspase-1 activation by Listeria monocytogenes. J Clin Immunol. 2010;30:693–702. doi: 10.1007/s10875-010-9425-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tsuchiya K, et al. Involvement of absent in melanoma 2 in inflammasome activation in macrophages infected with Listeria monocytogenes. J Immunol. 2010;185:1186–1195. doi: 10.4049/jimmunol.1001058. [DOI] [PubMed] [Google Scholar]
- 63.Cole LE, et al. Macrophage proinflammatory response to Francisella tularensis live vaccine strain requires coordination of multiple signaling pathways. J Immunol. 2008;180:6885–6891. doi: 10.4049/jimmunol.180.10.6885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ishikawa H, Ma Z, Barber GN. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature. 2009;461:788–792. doi: 10.1038/nature08476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yang P, et al. The cytosolic nucleic acid sensor LRRFIP1 mediates the production of type I interferon via a beta-catenin-dependent pathway. Nat Immunol. 2010;11:487–494. doi: 10.1038/ni.1876. [DOI] [PubMed] [Google Scholar]
- 66.Ulland TK, et al. Cutting edge: mutation of Francisella tularensis mviN leads to increased macrophage absent in melanoma 2 inflammasome activation and a loss of virulence. J Immunol. 2010;185:2670–2674. doi: 10.4049/jimmunol.1001610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Henry T, et al. Type I IFN signaling constrains IL-17A/F secretion by gammadelta T cells during bacterial infections. J Immunol. 2010;184:3755–3767. doi: 10.4049/jimmunol.0902065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.O’Connell RM, et al. Type I interferon production enhances susceptibility to Listeria monocytogenes infection. J Exp Med. 2004;200:437–445. doi: 10.1084/jem.20040712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Navarini AA, et al. Increased susceptibility to bacterial superinfection as a consequence of innate antiviral responses. Proc Natl Acad Sci USA. 2006;103:15535–15539. doi: 10.1073/pnas.0607325103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Carrero JA, Calderon B, Unanue ER. Lymphocytes are detrimental during the early innate immune response against Listeria monocytogenes. J Exp Med. 2006;203:933–940. doi: 10.1084/jem.20060045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Monack DM, Mueller A, Falkow S. Persistent bacterial infections: the interface of the pathogen and the host immune system. Nat Rev Microbiol. 2004;2:747–765. doi: 10.1038/nrmicro955. [DOI] [PubMed] [Google Scholar]
- 72.Winter SE, et al. Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature. 2010;467:426–429. doi: 10.1038/nature09415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hersh D, Monack DM, Smith MR, Ghori N, Falkow S, Zychlinsky A. The Salmonella invasin SipB induces macrophage apoptosis by binding to caspase-1. Proc Natl Acad Sci USA. 1999;96:2396–23401. doi: 10.1073/pnas.96.5.2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lara-Tejero M, et al. Role of the caspase-1 inflammasome in Salmonella typhimurium pathogenesis. J Exp Med. 2006;203:1407–1412. doi: 10.1084/jem.20060206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Raupach B, Peuschel SK, Monack DM, Zychlinsky A. Caspase-1-mediated activation of interleukin-1beta (IL-1beta) and IL-18 contributes to innate immune defenses against Salmonella enterica serovar Typhimurium infection. Infect Immun. 2006;74:4922–4926. doi: 10.1128/IAI.00417-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cummings LA, Wilkerson WD, Bergsbaken T, Cookson BT. In vivo, fliC expression by Salmonella enterica serovar Typhimurium is heterogeneous, regulated by ClpX, and anatomically restricted. Mol Microbiol. 2006;61:795–809. doi: 10.1111/j.1365-2958.2006.05271.x. [DOI] [PubMed] [Google Scholar]
- 77.Miao EA, et al. Caspase-1-induced pyroptosis is an innate immune effector mechanism against intracellular bacteria. Nat Immunol. 2010;11:1136–1142. doi: 10.1038/ni.1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Broz P, von Moltke J, Jones JW, Vance RE, Monack DM. Differential requirement for Caspase-1 autoproteolysis in pathogen-induced cell death and cytokine processing. Cell Host Microbe. 2010;8:471–483. doi: 10.1016/j.chom.2010.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Muller AJ, et al. The S. Typhimurium effector SopE induces caspase-1 activation in stromal cells to initiate gut inflammation. Cell Host Microbe. 2009;6:125–136. doi: 10.1016/j.chom.2009.07.007. [DOI] [PubMed] [Google Scholar]
- 80.Finlay BB, McFadden G. Anti-immunology: evasion of the host immune system by bacterial and viral pathogens. Cell. 2006;124:767–782. doi: 10.1016/j.cell.2006.01.034. [DOI] [PubMed] [Google Scholar]
- 81.Johnston JB, et al. A poxvirus-encoded pyrin domain protein interacts with ASC-1 to inhibit host inflammatory and apoptotic responses to infection. Immunity. 2005;23:587–598. doi: 10.1016/j.immuni.2005.10.003. [DOI] [PubMed] [Google Scholar]
- 82.Dorfleutner A, et al. A Shope Fibroma virus PYRIN-only protein modulates the host immune response. Virus Genes. 2007;35:685–694. doi: 10.1007/s11262-007-0141-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Stehlik C, Krajewska M, Welsh K, Krajewski S, Godzik A, Reed JC. The PAAD/PYRIN-only protein POP1/ASC2 is a modulator of ASC-mediated nuclear-factor-kappa B and pro-caspase-1 regulation. Biochem J. 2003;373:101–113. doi: 10.1042/BJ20030304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rahman MM, Mohamed MR, Kim M, Smallwood S, McFadden G. Co-regulation of NF-kappaB and inflammasome-mediated inflammatory responses by myxoma virus pyrin domain-containing protein M013. PLoS Pathog. 2009;5:e1000635. doi: 10.1371/journal.ppat.1000635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gregory SM, et al. Discovery of a viral NLR homolog that inhibits the inflammasome. Science. 2011;331:330–334. doi: 10.1126/science.1199478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ray CA, et al. Viral inhibition of inflammation: cowpox virus encodes an inhibitor of the interleukin-1 beta converting enzyme. Cell. 1992;69:597–604. doi: 10.1016/0092-8674(92)90223-y. [DOI] [PubMed] [Google Scholar]
- 87.Kettle S, Alcami A, Khanna A, Ehret R, Jassoy C, Smith GL. Vaccinia virus serpin B13R (SPI-2) inhibits interleukin-1beta-converting enzyme and protects virus-infected cells from TNF- and Fas-mediated apoptosis, but does not prevent IL-1beta-induced fever. J Gen Virol. 1997;78:677–685. doi: 10.1099/0022-1317-78-3-677. [DOI] [PubMed] [Google Scholar]
- 88.Petit F, Bertagnoli S, Gelfi J, Fassy F, Boucraut-Baralon C, Milon A. Characterization of a myxoma virus-encoded serpin-like protein with activity against interleukin-1 beta-converting enzyme. J Virol. 1996;70:5860–5866. doi: 10.1128/jvi.70.9.5860-5866.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Turner SJ, Silke J, Kenshole B, Ruby J. Characterization of the ectromelia virus serpin, SPI-2. J Gen Virol. 2000;81:2425–2430. doi: 10.1099/0022-1317-81-10-2425. [DOI] [PubMed] [Google Scholar]
- 90.Schotte P, Denecker G, Van Den Broeke A, Vandenabeele P, Cornelis GR, Beyaert R. Targeting Rac1 by the Yersinia effector protein YopE inhibits caspase-1-mediated maturation and release of interleukin-1beta. J Biol Chem. 2004;279:25134–25142. doi: 10.1074/jbc.M401245200. [DOI] [PubMed] [Google Scholar]
- 91.Holmstrom A, et al. YopK of Yersinia pseudotuberculosis controls translocation of Yop effectors across the eukaryotic cell membrane. Mol Microbiol. 1997;24:73–91. doi: 10.1046/j.1365-2958.1997.3211681.x. [DOI] [PubMed] [Google Scholar]
- 92.Master SS, et al. Mycobacterium tuberculosis prevents inflammasome activation. Cell Host Microbe. 2008;3:224–232. doi: 10.1016/j.chom.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sutterwala FS, Mijares LA, Li L, Ogura Y, Kazmierczak BI, Flavell RA. Immune recognition of Pseudomonas aeruginosa mediated by the IPAF/NLRC4 inflammasome. J Exp Med. 2007;204:3235–3245. doi: 10.1084/jem.20071239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Galle M, et al. The Pseudomonas aeruginosa Type III secretion system plays a dual role in the regulation of caspase-1 mediated IL-1beta maturation. J Cell Mol Med. 2008;12:1767–1776. doi: 10.1111/j.1582-4934.2007.00190.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Huang MT, et al. Deletion of ripA alleviates suppression of the inflammasome and MAPK by Francisella tularensis. J Immunol. 2010;185:5476–5485. doi: 10.4049/jimmunol.1002154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lindemann SR, et al. Francisella tularensis Schu S4 O-antigen and capsule biosynthesis gene mutants induce early cell death in human macrophages. Infect Immun. 2011;79:581–594. doi: 10.1128/IAI.00863-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sauer JD, Witte CE, Zemansky J, Hanson B, Lauer P, Portnoy DA. Listeria monocytogenes triggers AIM2-mediated pyroptosis upon infrequent bacteriolysis in the macrophage cytosol. Cell Host Microbe. 2010;7:412–419. doi: 10.1016/j.chom.2010.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Martinon F, Agostini L, Meylan E, Tschopp J. Identification of bacterial muramyl dipeptide as activator of the NALP3/cryopyrin inflammasome. Curr Biol. 2004;14:1929–1934. doi: 10.1016/j.cub.2004.10.027. [DOI] [PubMed] [Google Scholar]
- 99.Hsu LC, et al. A NOD2-NALP1 complex mediates caspase-1-dependent IL-1beta secretion in response to Bacillus anthracis infection and muramyl dipeptide. Proc Natl Acad Sci USA. 2008;105:7803–7808. doi: 10.1073/pnas.0802726105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Liepinsh E, Barbals R, Dahl E, Sharipo A, Staub E, Otting G. The death-domain fold of the ASC PYRIN domain, presenting a basis for PYRIN/PYRIN recognition. J Mol Biol. 2003;332:1155–1163. doi: 10.1016/j.jmb.2003.07.007. [DOI] [PubMed] [Google Scholar]
- 101.Masumoto J, et al. ASC, a novel 22-kDa protein, aggregates during apoptosis of human promyelocytic leukemia HL-60 cells. J Biol Chem. 1999;274:33835–33838. doi: 10.1074/jbc.274.48.33835. [DOI] [PubMed] [Google Scholar]
- 102.Richards N, et al. Interaction between pyrin and the apoptotic speck protein (ASC) modulates ASC-induced apoptosis. J Biol Chem. 2001;276:39320–39329. doi: 10.1074/jbc.M104730200. [DOI] [PubMed] [Google Scholar]
- 103.Moriya M, Taniguchi S, Wu P, Liepinsh E, Otting G, Sagara J. Role of charged and hydrophobic residues in the oligomerization of the PYRIN domain of ASC. Biochemistry. 2005;44:575–583. doi: 10.1021/bi048374i. [DOI] [PubMed] [Google Scholar]
- 104.Fernandes-Alnemri T, et al. The pyroptosome: a supramolecular assembly of ASC dimers mediating inflammatory cell death via caspase-1 activation. Cell Death Differ. 2007;14:1590–1604. doi: 10.1038/sj.cdd.4402194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Bryan NB, Dorfleutner A, Rojanasakul Y, Stehlik C. Activation of inflammasomes requires intracellular redistribution of the apoptotic speck-like protein containing a caspase recruitment domain. J Immunol. 2009;182:3173–3182. doi: 10.4049/jimmunol.0802367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Cheng J, et al. Kinetic properties of ASC protein aggregation in epithelial cells. J Cell Physiol. 2009;222:738–747. doi: 10.1002/jcp.22005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Fink SL, Bergsbaken T, Cookson BT. Anthrax lethal toxin and Salmonella elicit the common cell death pathway of caspase-1-dependent pyroptosis via distinct mechanisms. Proc Natl Acad Sci USA. 2008;105:4312–4317. doi: 10.1073/pnas.0707370105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ohtsuka T, et al. ASC is a Bax adaptor and regulates the p53-Bax mitochondrial apoptosis pathway. Nat Cell Biol. 2004;6:121–128. doi: 10.1038/ncb1087. [DOI] [PubMed] [Google Scholar]
- 109.Stehlik C, et al. The PAAD/PYRIN-family protein ASC is a dual regulator of a conserved step in nuclear factor kappaB activation pathways. J Exp Med. 2002;196:1605–1615. doi: 10.1084/jem.20021552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Stehlik C, Lee SH, Dorfleutner A, Stassinopoulos A, Sagara J, Reed JC. Apoptosis-associated speck-like protein containing a caspase recruitment domain is a regulator of procaspase-1 activation. J Immunol. 2003;171:6154–6163. doi: 10.4049/jimmunol.171.11.6154. [DOI] [PubMed] [Google Scholar]
- 111.Poyet JL, Srinivasula SM, Tnani M, Razmara M, Fernandes-Alnemri T, Alnemri ES. Identification of Ipaf, a human caspase-1-activating protein related to Apaf-1. J Biol Chem. 2001;276:28309–28313. doi: 10.1074/jbc.C100250200. [DOI] [PubMed] [Google Scholar]
- 112.Suzuki T, Franchi L, Toma C, Ashida H, Ogawa M, Yoshikawa Y, et al. Differential regulation of caspase-1 activation, pyroptosis, and autophagy via Ipaf and ASC in Shigella-infected macrophages. PLoS Pathog. 2007;3:e111. doi: 10.1371/journal.ppat.0030111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Case CL, Shin S, Roy CR. Asc and Ipaf Inflammasomes direct distinct pathways for caspase-1 activation in response to Legionella pneumophila. Infect Immun. 2009;77:1981–1991. doi: 10.1128/IAI.01382-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Stennicke HR, Deveraux QL, Humke EW, Reed JC, Dixit VM, Salvesen GS. Caspase-9 can be activated without proteolytic processing. J Biol Chem. 1999;274:8359–8362. doi: 10.1074/jbc.274.13.8359. [DOI] [PubMed] [Google Scholar]
- 115.Boatright KM, et al. A unified model for apical caspase activation. Mol Cell. 2003;11:529–541. doi: 10.1016/s1097-2765(03)00051-0. [DOI] [PubMed] [Google Scholar]
- 116.Malladi S, Challa-Malladi M, Fearnhead HO, Bratton SB. The Apaf-1*procaspase-9 apoptosome complex functions as a proteolytic-based molecular timer. EMBO J. 2009;28:1916–1925. doi: 10.1038/emboj.2009.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Keller M, Ruegg A, Werner S, Beer HD. Active caspase-1 is a regulator of unconventional protein secretion. Cell. 2008;132:818–831. doi: 10.1016/j.cell.2007.12.040. [DOI] [PubMed] [Google Scholar]
- 118.Eder C. Mechanisms of interleukin-1beta release. Immunobiology. 2009;214:543–553. doi: 10.1016/j.imbio.2008.11.007. [DOI] [PubMed] [Google Scholar]
- 119.Lamkanfi M, et al. Targeted peptidecentric proteomics reveals caspase-7 as a substrate of the caspase-1 inflammasomes. Mol Cell Proteomics. 2008;7:2350–2363. doi: 10.1074/mcp.M800132-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]