Summary
Porphyromonas gingivalis, Treponema denticola, and Tannerella forsythia are consistently associated with adult periodontitis. This study sought to document the host transcriptome to a P. gingivalis, T. denticola, and T. forsythia challenge as a polymicrobial infection using a murine calvarial model of acute inflammation and bone resorption. Mice were infected with P. gingivalis, T. denticola, and T. forsythia over the calvaria, after which the soft tissues and calvarial bones were excised. A Murine GeneChip® array analysis of transcript profiles showed that 6997 genes were differentially expressed in calvarial bones (P < 0.05) and 1544 genes were differentially transcribed in the inflamed tissues after the polymicrobial infection. Of these genes, 4476 and 1035 genes in the infected bone and tissues were differentially expressed by upregulation. Biological pathways significantly impacted by the polymicrobial infection in calvarial bone included leukocyte transendothelial migration (LTM), cell adhesion molecules, adherens junction, major histocompatibility complex antigen, extracellular matrix-receptor interaction (ECM), and antigen processing and presentation resulting in inflammatory/cytokine/chemokine transcripts stimulation in bone and soft tissue. Intense inflammation and increased activated osteoclasts was observed in calvarias compared to sham-infected controls. Quantitative real-time RT-PCR analysis confirmed mRNA level of selected genes corresponded with the microarray expression. The polymicrobial infection regulated several LTM and extracellular membrane (ECM) pathway genes in a manner distinct from monoinfection with P. gingivalis, T. denticola, or T. forsythia. To our knowledge, this is the first definition of the polymicrobial induced transcriptome in calvarial bone and soft tissue in response to periodontal pathogens.
Keywords: P. gingivalis, T. denticola, T. forsythia, polymicrobial infection, gene expression, calvarial bone, tissue, microarray
Introduction
Polymicrobial infections induce distinct pathogenic characteristics compared to monoinfection with a single pathogen (Brogden, 2002; Brogden et al., 2005). The predominant polymicrobial infection of humans is expressed clinically as periodontal disease (PD), a complex, immunoinflammatory condition, with multiple bacterial species responsible for triggering the destructive host responses. A predominant consortia identified in a majority of adult periodontitis patients consists of Porphyromonas gingivalis (Pg), Treponema denticola (Td) and Tannerella forsythia (Tf) (red complex pathogens) (Socransky et al., 1998; 2005). While with modern metagenomic microbiome technologies, it is clear that periodontopathic biofilms are comprised of a wide array of bacteria, many uncultivable and thus, minimally studied (Chen et al., 2010). Nevertheless, this consortium is strongly associated with disease lesions and is nearly always co-isolated or identified in biofilm samples from adult periodontitis lesions. Additionally, these pathogens elaborate numerous virulence determinants that may mediate adherence to mucosal surfaces (Lamont & Jenkinson, 1998; Holt et al., 1999), enable penetration into gingival epithelial cells (Lamont et al., 1995; Tribble & Lamont, 2010), inhibit host defense mechanisms (Holt & Ebersole, 2005), create a more anaerobic environment, and elicit synergistic gingival/periodontal tissue destruction, and alveolar bone resorption (Kesavalu et al., 2007; Hajishengallis 2009). We have reported that P. gingivalis, T. denticola, and T. forsythia not only exists as a pathogenic consortium in periodontitis lesions, but also exhibit synergistic virulence resulting in immunoinflammatory alveolar bone resorption in rodent models (Kesavalu et al., 2007). Our recent mouse microarray studies have determined the in vivo transcriptional profiles of host soft tissue and calvarial bone following acute infection with P. gingivalis (Meka et al., 2010), T. denticola (Bakthavatchalu et al., 2010a), and T. forsythia (Bakthavatchalu et al., 2010b) in a calvarial model of inflammation and bone resorption. A recent gingival transcriptome study in human patients demonstrated that the microbial content of the periodontal pocket is a determinant of gene expression in the adjacent gingival tissues, and provided new insights into the differential ability of periodontal species to elicit a local host response (Papapanou et al., 2009). In addition, a gene ontology analysis of healthy and diseased gingival tissues from patients with advanced periodontitis identified 61 differentially expressed pathways of genes, including those regulating apoptosis, antimicrobial immune response, and antigen presentation (Demmer et al., 2008). However, global host gene expression in response to defined polymicrobial infection in vivo remains to be defined.
This investigation is to determine the transcriptome profiles to P. gingivalis, T. denticola, and T. forsythia as a polymicrobial infection. To better understand the distinct local gene expression profiles, we performed a genome-wide transcriptional analysis of the calvarial bone and overlying soft tissues. The analysis focused on altered biological gene pathways which were significantly changed during the polybacterial infection, which were compared to those found after monoinfection with P. gingivalis, T. denticola or T. forsythia, yielding distinct results.
Methods
Mice
Female BALB/c mice 8-10 weeks of age (Harlan, Indianapolis, IN, USA) were routinely acclimatized before polymicrobial infection. BALB/c mice were infected with bacteria as described below following isoflurane inhalation anesthesia. Mice polymicrobial infection and sham-infection procedures were performed in accordance with the approved guidelines of the Institutional Animal Care and Use Committee at the University of Kentucky (Lexington, KY, USA).
Bacterial Strains and growth conditions
P. gingivalis FDC 381, T. denticola ATCC 35404, and T. forsythia ATCC 43037 were grown under anaerobic conditions at 37°C as previously described (Meka et al., 2010; Bakthavatchalu et al., 2010a; Bakthavatchalu et al., 2010b).
Polymicrobial inoculums and mouse infection
We prepared the polymicrobial inoculum based on our previously published report (Kesavalu et al., 2007), and consistent with the detection of these species in subgingival biofilm in adult periodontitis, synergism in biofilm formation between P. gingivalis and T. denticola (Kuramitsu et al., 2005), and specific molecular interactions between T. denticola and T. forsythia (Nishihara & Koseki, 2004). For polymicrobial infection, members of the P. gingivalis+T. denticola+T. forsythia consortium were individually prepared as described previously (Meka et al., 2010; Bakthavatchalu et al., 2010a; Bakthavatchalu et al., 2010b). Briefly, P. gingivalis was gently mixed in a vortex for 1-2 min with an equal volume of T. denticola, allowed to interact for 5 min, subsequently T. forsythia was added to the P. gingivalis+T. denticola tubes, mixed gently for 1-2 min, and allowed to interact for an additional 5 min to allow interactions among these species. Bacterial culture, growth phase, viability, enumeration, interaction times, suspension medium, and infection procedures were all standardized as described (Kesavalu et al., 2007). P. gingivalis, T. denticola, and T. forsythia suspended in reduced transport fluid (RTF) were injected at 1.5 × 109 into the soft tissues overlying the calvariae of the mice (n = 10) mice (Meka et al., 2010; Bakthavatchalu et al., 2010a; Bakthavatchalu et al., 2010b; Zubery et al., 1998). Polymicrobial cultures (5 × 108cells of each) were injected (suspended in 30 μl of RTF into the soft tissues overlying the calvariae of the mice (Meka et al., 2010; Bakthavatchalu et al., 2010a; Bakthavatchalu et al., 2010b). The sham-infected control group (n = 10 mice) was injected with 30 μl of RTF once daily for 3 days. Bacterial infection, mice euthanasia, collection and preparation of calvarial bone and soft tissue for histology were performed as previously described (Meka et al., 2010; Bakthavatchalu et al., 2010a; Bakthavatchalu et al., 2010b).
Total RNA isolation and mouse GeneChip hybridization
RNA was isolated from the calvarial soft tissue and calvaria bone from each mouse [polymicrobial (P. gingivalis/T. denticola/T. forsythia) infected and control mice, N=5 in each group] with Trizol reagent (Invitrogen, Carlsbad, CA) (Meka et al., 2010). Quantification of RNA yield was done by spectrophotometric analysis and the absorbance at 260 and 280 nm was checked to determine the RNA purity and concentration. For GeneChip analyses, individual RNA samples were further purified with Qiagen RNeasy columns (Qiagen, Valencia, CA). All calvarial tissue and bone samples were processed individually and RNA samples were not pooled. The biotin-labelled complementary RNA was synthesized by in vitro transcription, fragmented and hybridized on a mouse GeneChip MG- MOE430A (Affymetrix), following the protocol described in the GeneChip Expression Analysis Technical manual (Affymetrix, Santa Clara, CA). After hybridization, the GeneChip arrays were stained and scanned in Affymetrix GCS 3000 7G Scanner as previously described (Meka et al., 2010; Bakthavatchalu et al., 2010a).
Murine Microarray data analysis
The polymicrobial murine microarray raw data were normalized, evaluation of the dataset by both unsupervised and supervised analyses, hierarchal clustering analysis, following which differences between the various treatment tissue classes, and determination of fold-change of significantly impacted genes were determined as described previously (Draghici et al., 2003; Feezor et al., 2003; Meka et al., 2010; Bakthavatchalu et al., 2010a). Pathway express was used to compile a list of impacted pathways and to populate KEGG pathways according to the gene regulation uncovered by microarray. Pathways were prioritized and further investigated (http://bortex.cs.wayne.edu/fag.htm).
Real-time reverse transcription-polymerase chain reaction analysis
Expression of selected genes, that showed significant differential expression to infection compared to controls in the microarray analyses, was confirmed by qRT-PCR analysis (LightCycler FastStart DNA Master SYBR Green I, Roche, Indianapolis, IN) as described previously (Meka et al., 2010; Bakthavatchalu et al., 2010a). Six representative upregulated genes based on a broad overview of the different functional categories such as extracellular matrix, cell adhesion, cell proliferation, immune and defense responses, transport, and other categories from the both polymicrobial challenged soft tissues and calvarial bone were evaluated. These genes were: defensin B (Defb3), small proline rich (Sprr2d), matrix metalloproteinases 13 (Mmp13) from calvarial tissue and chemokine (C-X-C Motif) ligand 7 (Cxcl7), matrix metalloproteinases 9 (Mmp9), and peptidoglycan recognition protein 1 (Pglyrp1) from calvarial bone.
Calvarial bone histology
The mouse polymicrobial infected and sham-infected calvariae were fixed in 10% neutral phosphate-buffered formalin, decalcified, embedded, sectioned, stained, and analyzed for osteoclasts as described previously for mono infection with P. gingivalis (Meka et al., 2010), T. denticola (Bakathavatchalu et al., 2010a), and T. forsythia (Bakathavatchalu et al., 2010b).
Statistical analysis
Polymicrobial transcriptome murine microarray data were analyzed as described above (Meka et al., 2010; Bakathavatchalu et al., 2010a; Bakathavatchalu et al., 2010b). P values of 0.05 or less were considered significant. The qRT-PCR data from two independent experiments were combined and results were presented as means ± SD.
Microarray data accession numbers
The array results have been deposited in the GEO repository (http://www.ncbi.nlm.nih.gov/projects/geo/) under accession numbers GSE17110, GSE 29670.
Results
Ontology of gene expression changes in murine calvarial bone and soft tissue
To analyze host global gene transcription following the polymicrobial infection, the mouse gene chip MOE430A containing 22,690 probe sets, with 17, 809 and 17,908 probe sets provided positive readable signals in soft tissue and bone, respectively to polymicrobial infection in comparison to sham-infected control. Significant differences were observed in mean gene expression levels of 6997 and 1544 probes sets in bone and soft tissue in response to polymicrobial infection (P < 0.05), respectively in comparison to sham-infected control. Of the significantly regulated genes, 4476 probe sets were upregulated and 2521 genes were downregulated in calvarial bone. In contrast, 1035 genes were upregulated and 459 were downregulated in calvarial soft tissue samples following the polymicrobial infection compared to control tissues. The results of this gene expression analysis indicate that polymicrobial infection stimulated greater changes in the transcriptome of up- and downregulated genes in calvarial bone compared to the overlying inflamed soft tissues. The majority of genes with altered expression in calvarial bone to polymicrobial infection were primarily associated with basic cellular functions [transcription, cell proliferation, cell cycle, cell adhesion, extracellular matrix (ECM), transport, apoptosis] for maintaining tissue integrity.
The significantly altered probe sets following a polybacterial challenge were analyzed by the PATHWAY EXPRESS tool as previously described (Draghici et al., 2007, Khatri et al., 2007). Probe sets significant at P ≤ 0.05 to differentiate between the polymicrobial infection and sham-infected groups were confirmed by LOOCV analysis (Supporting Information, Table S1 and S2). Pathways significantly impacted by P. gingivalis, T. denticola, and T. forsythia at the P ≤ 0.05 level in calvarial bone and soft tissue types included: Leukocyte Transendothelial Migration (LTM) (Figs. 1 and 2), Cell Adhesion Molecules (CAM) (Supporting information, Fig. S1), Adherens Junction (AJ) (Supporting information, Fig. S2), major histocompatibility complex (MHC) Antigen I and II Processing and Presentation (APP) (Supporting information, Fig. S3), and ECM-receptor interaction (Fig. 3). Table 1 shows calvarial bone and soft-tissue pathways generated from this analysis that were predominantly affected in order of their impact factors. P. gingivalis, T. denticola, and T. forsythia significantly impacted 23 pathways in calvarial bone and eight in soft tissue with an impact factor >5. The high impact factors associated with these pathways predicts that the effects of P. gingivalis, T. denticola, and T. forsythia-induced gene expression changes in the bone or tissue should have a significant biological effect downstream. There are more than 5 pathways including Leukocyte Transendothelial Migration, Cell Adhesion Molecules, Adherens Junction, Phosphatidylinositol Signaling system and ribosome that were significantly impacted and overlapped in both the calvarial bone and soft tissue (Table 1).
Figure 1.
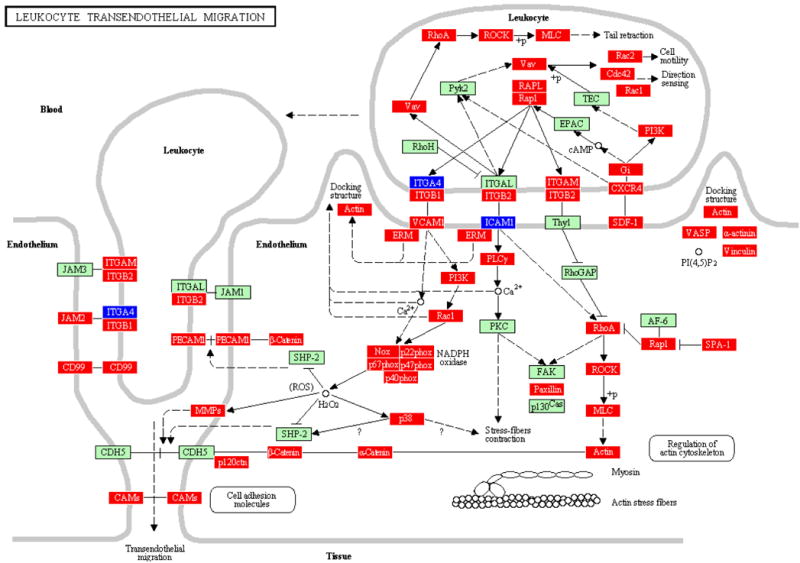
LTM pathway containing genes differentially regulated by polymicrobial infection with P. gingivalis+T. denticola+T. forsythia in calvarial bone compared to sham-infected controls at P ≤ 0.05, adapted from PATHWAY EXPRESS and using the Kyoto Encyclopedia of Genes and Genomes nomenclature. Genes shown in red are upregulated, genes shown in blue are downregulated, and green indicates no change in gene expression at the P < 0.05 significance level. +p = phosphorylation event; -p dephosphorylation event; ? = receptors that are yet to be identified; and O = other molecule. An arrow indicates a molecular interaction resulting in transendothelial migration, leukocyte activation, regulation of actin cytoskeleton and a line without an arrowhead indicates a molecular interaction resulting in inhibition.
Figure 2.
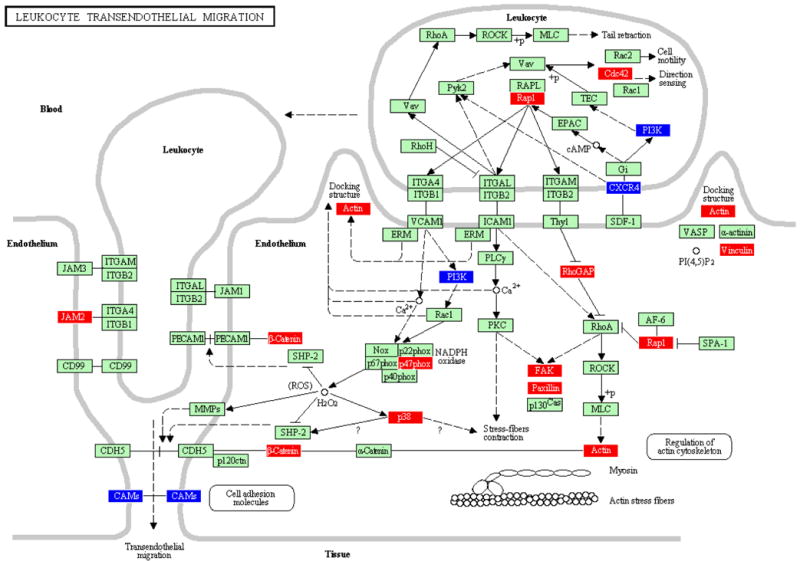
LTM pathway containing genes differentially regulated by polymicrobial infection with P. gingivalis+T. denticola+T. forsythia in calvarial soft tissue compared to sham-infected controls at P ≤ 0.05. An arrow indicates a molecular interaction resulting in transendothelial migration, leukocyte activation, regulation of actin cytoskeleton and a line without an arrowhead indicates a molecular interaction resulting in inhibition.
Figure 3.
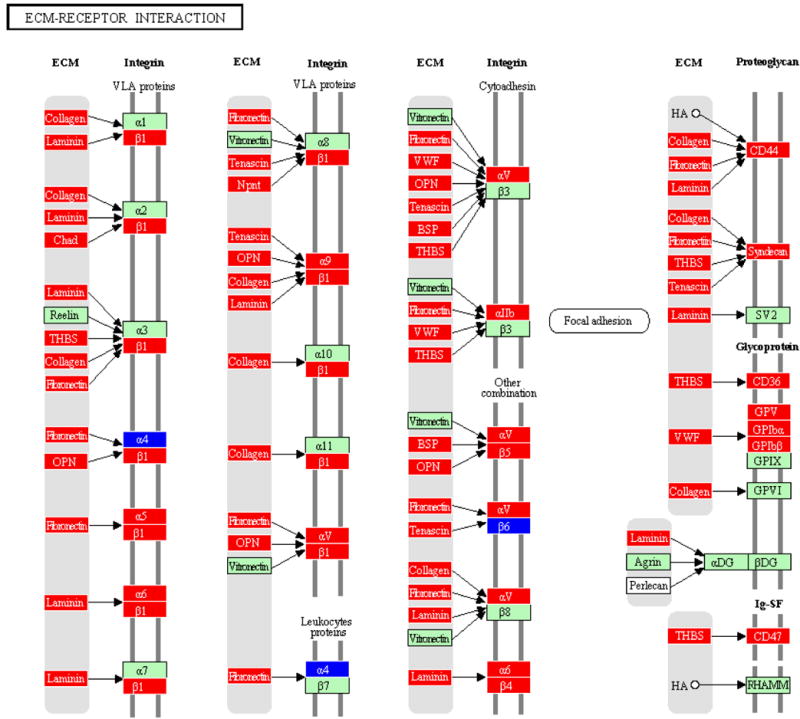
Extracelular Membrane receptor interaction pathway containing genes differentially regulated by polymicrobial infection with P. gingivalis+T. denticola+T. forsythia in calvarial bone compared to sham-infected controls at P ≤ 0.05. An arrow indicates a molecular interaction resulting in ECM-receptor activation, regulation of integrin (VLA proteins, leukoproteins, cytoadhesin, focal adhesion, proteoglycan, glycoprotein).
Table 1.
Ontology analysis of calvarial bone and soft tissue pathways impacted by polymicrobial infection with P. gingivalis+T. denticola+T. forsythia1
| Impacted pathway2 | Impact Factor3 | Input genes/no. of pathway genes4 | P Value |
|---|---|---|---|
| Calvarial bone | |||
| Leukocyte transendothelial migration | 55.967 | 49/119 | 0.000E0 |
| Cell adhesion molecules | 45.448 | 49/159 | 0.000E0 |
| Adherens junction | 31.999 | 37/77 | 3.510E-13 |
| Antigen processing and presentation | 17.26 | 37/100 | 5.829E-7 |
| Phosphatidylinositol signaling system | 15.717 | 28/75 | 2.497E-6 |
| Ribosome | 13.463 | 55/115 | 2.058E-5 |
| DNA replication | 12.825 | 26/36 | 3.723E-5 |
| ECM-receptor interaction | 11.145 | 45/81 | 1.755E-4 |
| Focal adhesion | 9.608 | 94/199 | 7.127E-4 |
| B cell receptor signaling pathway | 7.748 | 35/71 | 3.776E-3 |
| Apoptosis | 6.122 | 41/93 | 1.563E-2 |
| Natural killer cell mediated cytotoxicity | 5.945 | 47/109 | 1.819E-2 |
| Cell cycle | 5.81 | 54/124 | 2.041E-2 |
| Toll-like receptor signaling pathway | 5.591 | 48/101 | 2.459E-2 |
| VEGF signaling pathway | 5.518 | 36/76 | 2.616E-2 |
| Calvarial tissue | |||
| Leukocyte transendothelial migration | 79.756 | 14/119 | 0.000E0 |
| Cell adhesion molecules | 71.416 | 13/159 | 0.000E0 |
| Adherens junction | 22.027 | 13/77 | 6.282E-9 |
| Phosphatidylinositol signaling system | 14.206 | 7/75 | 1.029E-5 |
| Ribosome | 10.313 | 21/115 | 3.756E-4 |
| PPAR signaling pathway | 7.475 | 14/76 | 4.806E-3 |
| mTOR signaling pathway | 7.117 | 11/55 | 6.584E-3 |
| Insulin signaling pathway | 5.621 | 20/140 | 2.397E-2 |
ECM = extracellular matrix; VEGF = vascular endothelial growth factor; PPAR = peroxisome proliferator-activated receptor; mTOR = mammalian target of rapamycin.
The calvarial bone and tissue gene pathways were determined by Pathway Express (Draghici et al. 2007; Khatri et al. 2007).
Kyoto Encyclopedia of genes and genome pathways (http://www.genome.jp/kegg/).
The impact factor measures the pathways most affected by changes in gene expression in calvarial bone and soft tissue by considering the proportion of differentially regulated genes, the perturbation factors of all pathway genes, and the propagation of these perturbations throughout the pathway. Only pathways with an impact factor greater than 5 are included in this table.
Number of regulated genes in a pathway/total number of genes currently mapped to this pathway. First three pathways that are impacted both in bone and tissue are indicated in bold.
We have selected LTM and ECM-interaction pathways to examine how P. gingivalis (Meka et al., 2010) or T. denticola (Bakthavatchalu et al., 2010a) or T. forsythia mono-infection (Bakthavatchalu et al., 2010b) impacted regulation of pathway genes compared to a polybacterial infection in bone and soft tissues. T. denticola monoinfection impacted (impact factor: 306) the LTM pathway to a greater extent than monoinfection with T. forsythia or polymicrobial infection in calvarial bone and soft tissue. Polymicrobial infection regulated more LTM pathway genes (49/119) in bone than monoinfection with P. gingivalis or T. denticola or T. forsythia. P. gingivalis regulated more of the LTM pathway (39/115) genes in soft tissue than the polymicrobial infection-induced genes (Table 2). Three LTM pathway genes (claudin 1, chemokine (C-X-C motif) ligand 12, Mmp 2) were regulated following both mono- and the polymicrobial infection in bone and a few LTM pathway genes (CD99 antigen, integrin beta 1, junctional adhesion molecule 2, vav 1 oncogene) were uniquely regulated only following the polymicrobial infection in bone (Table 3) and not in tissue (Supporting information, Table S4).
Table 2.
Ontology analysis of leukocyte transendothelial migration (LTM) pathway impacted by mono- and polymicrobial infection1.
| Impacted LTM pathway2 | Impact Factor3 | Input genes/no. of pathway genes4 | P value |
|---|---|---|---|
| Calvarial bone | |||
| P. gingivalis | 2.912 | 18/115 | 2.127E-1 |
| T. denticola | 306.541 | 14/119 | 0.000E0 |
| T. forsythia | 85.099 | 37/119 | 0.000E0 |
| P. gingivalis/T. denticola/T. forsythia | 55.967 | 49/119 | 0.000E0 |
| Calvarial tissue | |||
| P. gingivalis | 18.487 | 39/115 | 1.194E-2 |
| T. denticola | 220.561 | 5/119 | 0.000E0 |
| T. forsythia | 148.247 | 35/119 | 0.000E0 |
| P. gingivalis/T. denticola/T. forsythia | 79.756 | 14/119 | 0.000E0 |
The calvarial bone and tissue gene pathways were determined by Pathway Express (Draghici et al. 2007; Khatri et al. 2007).
Kyoto Encyclopedia of genes and genome pathways (http://www.genome.jp/kegg/).
The impact factor measures the pathways most affected by changes in gene expression in calvarial bone and soft tissue to monoinfection (P. gingivalis or T. denticola or T. forsythia) and polymicrobial infection with P. gingivalis+T. denticola+T. forsythia by considering the proportion of differentially regulated genes, the perturbation factors of all pathway genes, and the propagation of these perturbations throughout the pathway.
Number of regulated genes in a pathway/total number of genes currently mapped to this pathway.
Table 3.
Leukocyte transendothelial migration (LTM) pathway genes impacted by polymicrobial and mono-infection in calvarial bone1.
| NO | Gene | LTM pathway genes2 | Pg+Td+Tf | P. gingivalis | T. denticola | T. forsythia |
|---|---|---|---|---|---|---|
| 1 | Actb | actin, beta | 2.033 | 0 | -1.69 | 0 |
| 2 | Actg1 | actin, gamma, cytoplasmic 1 | 1.63 | 0 | 0 | 1.57 |
| 3 | Actn1 | actinin, alpha 1 | 2.36 | 0 | 0 | 2.04 |
| 4 | Actn3 | actinin alpha 3 | 0 | 0 | 0 | -1.50 |
| 5 | Actn4 | 1.49 | 0 | 0 | 0 | |
| 6 | Arhgap5 | Rho GTPase activating protein 5 | 0 | -1.54 | 0 | 0 |
| 7 | Cd99 | CD99 antigen | 1.53 | 0 | 0 | 0 |
| 8 | Cdc42 | 1.69 | 1.46 | 0 | 1.42 | |
| 9 | Cdh5 | cadherin 5 | 0 | 0 | 0 | 1.37 |
| 10 | Cldn1 | claudin 1 | 4.62 | 9.36 | 1.93 | 3.39 |
| 11 | Cldn13 | claudin 13 | 0 | 0 | 1.78 | 0 |
| 12 | Cldn14 | claudin 14 | 0 | 0 | -1.40 | 0 |
| 13 | Cldn19 | claudin 19 | 0 | 0 | 2.63 | 0 |
| 14 | Cldn3 | claudin 3 | 0 | 0 | 0 | 2.69 |
| 15 | Cldn5 | claudin 5 | 0 | -2.37 | 0 | 0 |
| 16 | Cldn6 | claudin 6 | 0 | 2.12 | 0 | 0 |
| 17 | Cldn7 | claudin 7 | 0 | 0 | 1.82 | 0 |
| 18 | Ctnna1 | catenin (cadherin associated protein), alpha 1 | 1.62 | 2.24 | 0 | 1.48 |
| 19 | Ctnnb1 | catenin (cadherin associated protein), beta 1 | 2.65 | 1.42 | 0 | 0 |
| 20 | Ctnnd1 | catenin (cadherin associated protein), delta 1 | 1.28 | 1.66 | 0 | 0 |
| 21 | Cxcl12 | chemokine (C-X-C motif) ligand 12 | 3.35 | 1.65 | 2.01 | 2.24 |
| 22 | Cxcr4 | chemokine (C-X-C motif) receptor 4 | 1.81 | 0 | 0 | 2.42 |
| 23 | Cyba | cytochrome b-245, alpha polypeptide | 5.15 | 0 | 0 | 3.63 |
| 24 | Cybb | cytochrome b-245, beta polypeptide | 9.68 | 0 | 0 | 7.53 |
| 25 | Esam | endothelial cell-specific adhesion molecule | 0 | -1.46 | -1.27 | 0 |
| 26 | Ezr | ezrin | 3.16 | 0 | 0 | 3.78 |
| 27 | Gnai2 | guanine nucleotide binding protein (G protein), alpha inhibiting 2 | 2.65 | 0 | 0 | 2.31 |
| 28 | Gnai3 | guanine nucleotide binding protein (G protein), alpha inhibiting 3 | 2.53 | 1.98 | 0 | 1.80 |
| 29 | Icam1 | intercellular adhesion molecule 1 | -1.93 | 0 | 0 | -1.40 |
| 30 | Itga4 | integrin alpha 4 | -1.58 | -1.57 | 0 | -1.86 |
| 31 | Itgam | integrin alpha M | 4.92 | 0 | 0 | 0 |
| 32 | Itgb1 | integrin beta 1 (fibronectin receptor beta) | 2.00 | 0 | 0 | 0 |
| 33 | Itgb2 | integrin beta 2 | 4.41 | 0 | 0 | 3.44 |
| 34 | Itgb2l | integrin beta 2-like | -1.65 | 0 | 0 | 0 |
| 35 | Jam2 | junction adhesion molecule 2 | 1.41 | 0 | 0 | 0 |
| 36 | Mapk11 | mitogen-activated protein kinase 11 (EC:2.7.11.24) | 0 | 0 | 0 | -2.98 |
| 37 | Mapk14 | mitogen-activated protein kinase 14 | 2.55 | -1.97 | 0 | 0 |
| 38 | Mmp2 | matrix metallopeptidase 2 | 2.84 | 3.33 | 2.47 | 1.84 |
| 39 | Mmp9 | matrix metallopeptidase 9 | 16.29 | 0 | 4.71 | 10.58 |
| 40 | Msn | moesin | 5.53 | 0 | 0 | 2.94 |
| 41 | Myl9 | myosin, light polypeptide 9, regulatory | 2.63 | 0 | 1.62 | 2.29 |
| 42 | Ncf1 | neutrophil cytosolic factor 1 | 8.08 | 0 | 0 | 4.17 |
| 43 | Ncf2 | neutrophil cytosolic factor 2 | 1.94 | 0 | 0 | 1.71 |
| 44 | Ncf4 | neutrophil cytosolic factor 4 | 4.05 | 0 | 0 | 3.89 |
| 45 | Pecam1 | platelet/endothelial cell adhesion molecule 1 | 2.09 | 0 | 0 | 1.82 |
| 46 | Pik3cd | phosphatidylinositol 3-kinase catalytic delta polypeptide | 1.38 | -1.45 | -1.63 | 1.68 |
| 47 | Pik3cg | 2.34 | 0 | 0 | 2.28 | |
| 48 | Plcg2 | phospholipase C, gamma 2 | 2.52 | 0 | 0 | 2.49 |
| 49 | Pxn | paxillin | 1.70 | -1.43 | -1.38 | 0 |
| 50 | Rac1 | RAS-related C3 botulinum substrate 1 | 1.79 | 0 | 0 | 0 |
| 51 | Rac2 | RAS-related C3 botulinum substrate 2 | 3.44 | 0 | 0 | 2.73 |
| 52 | Rap1b | 2.05 | 0 | 0 | 0 | |
| 53 | Rassf5 | Ras association (RalGDS/AF-6) domain family member 5 | 2.58 | 0 | 0 | 2.93 |
| 54 | Rhoa | ras homolog gene family, member A | 1.41 | 0 | 0 | 0 |
| 55 | Rock1 | Rho-associated coiled-coil containing protein kinase 1 | 1.65 | -1.87 | 0 | 1.67 |
| 56 | Rock2 | Rho-associated coiled-coil containing protein kinase 2 (EC:2.7.11.1) | 0 | 0 | 0 | -1.34 |
| 57 | Sipa1 | signal-induced proliferation associated gene 1 | 2.56 | 0 | 0 | 0 |
| 58 | Vasp | vasodilator-stimulated phosphoprotein | 4.63 | 0 | 0 | 4.30 |
| 59 | Vav1 | vav 1 oncogene | 3.92 | 0 | 0 | 0 |
| 60 | Vav3 | vav 3 oncogene | 3.37 | 0 | 2.07 | 3.45 |
| 61 | Vcam1 | vascular cell adhesion molecule 1 | 2.17 | -1.37 | 0 | 0 |
| 62 | Vcl | vinculin | 4.68 | 0 | 1.65 | 2.81 |
The calvarial bone and tissue gene pathways were determined by Pathway Express (Draghici et al. 2007; Khatri et al. 2007).
Kyoto Encyclopedia of genes and genome pathways (http://www.genome.jp/kegg/).
Fold change describes the ratio of mean expression in infected calvarial bone over the mean expression in uninfected control calvarial bone. Genes that are impacted to mono- and polymicrobial infection are indicated in bold.
Similarly, the T. forsythia monoinfection impacted ECM receptor-interaction pathway (impact factor: 23) higher than T. denticola and the polymicrobial infection in bone, whereas the polymicrobial infection regulated more ECM receptor-interaction pathway genes (45/81) in bone than a T. denticola monoinfection and the P. gingivalis-infection did not significantly impact ECM genes in calvarial bone. In contrast, neither the mono- nor polymicrobial infection significantly impacted ECM receptor-interaction pathway genes in soft tissue (Table 4). Five ECM pathway genes (collagen type XI alpha 1, integrin alpha V, syndecan 1, thrombospondin 1, tenascin C) were robustly regulated following both mono- and polymicrobial infection in bone and a few ECM pathway genes (CD36 antigen, integrin beta 1, integrin beta 5, laminin gamma 1, syndecan 4) were uniquely regulated only after the polymicrobial infection in bone (Table 5) and not in tissue (Supporting information, Table S5). The significant ECM protein gene transcription of Mmp9 (16-fold), laminin B1 subunit 1 (Lamb1-1; 4-fold), Mmp13 (5-fold), Mmp14 (4-fold), Mmp8 (3-fold), Mmp3 (5-fold), Mmp2 (3-fold), Mmp23 (2-fold), Ctgf (connective tissue growth factor: 4-fold), Fmod (fibromodulin: 4-fold), Fn1 (fibronectin 1: 5-fold), Matn2 (matrilin 2: 2-fold), Ecm1 (4-fold), Timp1, (7-fold) Timp2, (2-fold) and Timp3 (2-fold) were altered in calvaria.
Table 4.
Ontology analysis of ECM-receptor interaction pathway impacted by infection with mono- and polymicrobial infection1.
| Impacted ECM-receptor interaction pathway2 | Impact Factor3 | Input genes/no. of pathway genes4 | P value |
|---|---|---|---|
| Calvarial bone | |||
| P. gingivalis | NSI | NSI | NSI |
| T. denticola | 17.812 | 32/81 | 3.458E-7 |
| T. forsythia | 23.467 | 35/81 | 1.574E-9 |
| P. gingivalis, T. denticola, T. forsythia | 11.145 | 45/81 | 1.775E-4 |
| Calvarial tissue | |||
| P. gingivalis | NSI5 | NSI | NSI |
| T. denticola | NSI | NSI | NSI |
| T. forsythia | NSI | NSI | NSI |
| P. gingivalis, T. denticola, T. forsythia | NSI | NSI | NSI |
The calvarial bone and tissue gene pathways were determined by Pathway Express (Draghici et al. 2007; Khatri et al. 2007).
Kyoto Encyclopedia of genes and genome pathways (http://www.genome.jp/kegg/).
The impact factor measures the pathways most affected by changes in gene expression in calvarial bone and soft tissue to monoinfection (P. gingivalis or T. denticola or T. forsythia) and polymicrobial infection with P. gingivalis, T. denticola, and T. forsythia by considering the proportion of differentially regulated genes, the perturbation factors of all pathway genes, and the propagation of these perturbations throughout the ECM pathway.
Number of regulated genes in ECM receptor-interaction pathway/total number of genes currently mapped to ECM pathway.
NSI stands for not significantly impacted pathway.
Table 5.
ECM-receptor interaction pathway genes impacted by polymicrobial and mono-infection in calvarial bone1.
| NO | Gene | ECM-receptor pathway genes2 | Pg+Td+Tf | P. gingivalis | T. denticola | T. forsythia |
|---|---|---|---|---|---|---|
| 1 | Cd36 | CD36 antigen | 1.873 | 0 | 0 | 0 |
| 2 | Cd44 | CD44 antigen | 6.71 | 0 | 1.93 | 3.72 |
| 3 | Cd47 | CD47 antigen (Rh-related antigen, integrin-associated signal | 2.56 | 0 | 0 | 1.86 |
| 4 | Chad | chondroadherin | 2.83 | 0 | 2.32 | 1.80 |
| 5 | Col11a1 | collagen, type XI, alpha 1 | 2.82 | 0 | 3.58 | 2.25 |
| 6 | Col11a2 | collagen, type XI, alpha 2 | 0 | 0 | 1.37 | 1.51 |
| 7 | Col1a1 | collagen, type I, alpha 1 | 4.44 | 0 | 4.55 | 3.34 |
| 8 | Col1a2 | collagen, type I, alpha 2 | 5.06 | 0 | 6.24 | 3.76 |
| 9 | Col3a1 | collagen, type III, alpha 1 | 5.66 | 6.27 | 7.57 | 4.36 |
| 10 | Col4a1 | collagen, type IV, alpha 1 | 1.52 | 0 | 0 | 0 |
| 11 | Col5a1 | collagen, type V, alpha 1 | 5.47 | 0 | 4.68 | 3.28 |
| 12 | Col5a2 | collagen, type V, alpha 2 | 4.44 | 0 | 4.51 | 2.95 |
| 13 | Col6a1 | collagen, type VI, alpha 1 | 1.72 | 0 | 1.87 | 0 |
| 14 | Col6a2 | collagen, type VI, alpha 2 | 2.66 | 0 | 2.83 | 1.70 |
| 15 | Comp | cartilage oligomeric matrix protein | 3.16 | 0 | 2.27 | 3.35 |
| 16 | Dag1 | dystroglycan 1 | 0 | -1.44 | -1.22 | -1.56 |
| 17 | Fn1 | fibronectin 1 | 5.12 | 0 | 4.82 | 4.35 |
| 18 | Gp1ba | glycoprotein 1b, alpha polypeptide | 1.46 | 0 | 0 | 1.93 |
| 19 | Gp1bb | glycoprotein Ib, beta polypeptide | 4.95 | 0 | 0 | 4.36 |
| 20 | Gp5 | glycoprotein 5 (platelet) | 2.36 | 0 | 1.37 | 2.27 |
| 21 | Ibsp | integrin binding sialoprotein | 21.94 | 0 | 11.91 | 7.13 |
| 22 | Itga2b | integrin alpha 2b | 2.34 | 0 | 0 | 2.58 |
| 23 | Itga4 | integrin alpha 4 | -1.58 | -1.57 | 0 | -1.86 |
| 24 | Itga5 | integrin alpha 5 (fibronectin receptor alpha) | 3.07 | 0 | 0 | 2.09 |
| 25 | Itga6 | integrin alpha 6 | 1.49 | 0 | 0 | 2.32 |
| 26 | Itga7 | integrin alpha 7 | 0 | 0 | -1.38 | 0 |
| 27 | Itga9 | integrin alpha 9 | 2.30 | 1.84 | -1.38 | 0 |
| 28 | Itgav | integrin alpha V | 3.43 | 1.62 | 2.11 | 1.87 |
| 29 | Itgb1 | integrin beta 1 (fibronectin receptor beta) | 2.00 | 0 | 0 | 0 |
| 30 | Itgb3 | integrin beta 3 | 0 | 0 | 0 | 1.52 |
| 31 | Itgb4 | integrin beta 4 | 2.18 | 0 | 0 | 2.29 |
| 32 | Itgb5 | integrin beta 5 | 1.61 | 0 | 0 | 0 |
| 33 | Itgb6 | integrin beta 6 | -2.07 | -1.99 | 0 | 0 |
| 34 | Lama3 | laminin, alpha 3 | 0 | 0 | 1.33 | 1.75 |
| 35 | Lama4 | laminin, alpha 4 | 2.19 | 1.84 | 1.63 | 0 |
| 36 | Lama5 | laminin, alpha 5 | -1.50 | 0 | 0 | 0 |
| 37 | Lamb1-1 | laminin B1 subunit 1 | 3.82 | 1.82 | 2.08 | 0 |
| 38 | Lamc1 | laminin, gamma 1 | 2.85 | 0 | 0 | 0 |
| 39 | Lamc2 | laminin, gamma 2 | -1.62 | 0 | 0 | -1.77 |
| 40 | Npnt | 2.15 | 4.95 | 0 | 0 | |
| 41 | Sdc1 | syndecan 1 | 9.68 | 5.99 | 3.68 | 5.88 |
| 42 | Sdc2 | syndecan 2 | 2.01 | 0 | 1.95 | 0 |
| 43 | Sdc3 | syndecan 3 | 0 | -1.50 | 0 | 1.66 |
| 44 | Sdc4 | syndecan 4 | 1.55 | 0 | 0 | 0 |
| 45 | Spp1 | secreted phosphoprotein 1 | 5.64 | 0 | 4.01 | 4.14 |
| 46 | Thbs1 | thrombospondin 1 | 6.38 | 5.79 | 1.88 | 3.76 |
| 47 | Thbs2 | thrombospondin 2 | 2.18 | 7.03 | 2.66 | 0 |
| 48 | Thbs3 | thrombospondin 3 | 0 | 2.39 | 1.92 | 0 |
| 49 | Thbs4 | thrombospondin 4 | 5.90 | 3.99 | 3.24 | 4.94 |
| 50 | Tnc | tenascin C | 5.14 | 4.27 | 6.08 | 4.02 |
| 51 | Tnxb | tenascin XB | -1.23 | 0 | 0 | 0 |
| 52 | Vtn | vitronectin | 0 | 0 | -1.33 | -1.93 |
| 53 | Vwf | Von Willebrand factor homolog | 4.06 | 0 | 1.48 | 3.12 |
The calvarial bone and tissue gene pathways were determined by Pathway Express (Draghici et al. 2007; Khatri et al. 2007).
Kyoto Encyclopedia of genes and genome pathways (http://www.genome.jp/kegg/).
Fold change describes the ratio of mean expression in infected calvarial bone over the mean expression in uninfected control calvarial bone. Genes that are impacted to mono- and polymicrobial infection are indicated in bold.
Inflammatory and immune response gene expression profiles
Studies of infection with periodontal pathogens have examined acute and chronic inflammatory responses to microbial challenge leading to localized tissue destruction. Thus, we also focused on examination of gene profiles related to inflammatory and immune responses to the polymicrobial infection. Several inflammation/cytokine/chemokine transcripts including Cxcl7 (12-fold), Cxcl12 (4-fold), Ccl4 (3-fold), Ccl9, Ccl12, Ccl17, Ccl20, IL-1 receptor (Ilr2: 5-fold), TNF receptor, lymphotoxin B, chitinase 3-Like 3, and neutrophil cytosolic factor were significantly altered in calvarial bone. The significantly upregulated immune and defense response genes included Orm1 (orosomucoid 1: 7-fold), Hp (Haptoglobulin: 8-fold), C1qtnf1 (C1q And Tumor Necrosis Factor Related Protein 1: 6-fold), Bst1 (Bone Marrow Stromal Cell Antigen 1: 7-fold), Saa1 (Serum Amyloid A 1: 2-fold), Ifi30 (Interferon Gamma Inducible Protein 30: 2-fold), and Ifitm3 (Interferon Induced Transmembrane Protein 3: 8-fold). Other altered genes that were increased expression were associated with cell cycle & proliferation [Ribonucleotide Reductase M2 (Rrm2), 8-fold; cyclin B (Ccnb2), 11-fold; Transforming Growth Factor, Beta 1 (Tgfb1), 3-fold), transport [Lactotransferrin (Ltf), 12-fold; Solute Carrier Family 6 (Slc6) 5-fold; Ceruloplasmin (Cp), 7-fold], cell adhesion [CEA-Related Cell Adhesion Molecule (Cecam1), 10-fold; CD44 Antigen (Cd44) 8-fold; Laminin B1 Subunit 1 (Lamb1-1) 4-fold), apoptosis [Peptidoglycan Recognition Protein 1 (Pglyrp1) 14-fold], bone remodeling & ossification [Integrin Binding Sialoprotein (Ibsp) 22-fold; Secreted Phosphoprotein 1 (Spp1) 6-fold], and other category genes [acid phosphatase 5 tartrate resistant (Acp5) 11-fold); immunoglobulin superfamily member 6 (Igsf6) 8-fold].
The majority of genes upregulated during a polymicrobial infection in soft tissue were associated with inflammatory, immune & defense response, transport, ECM, cell cycle & differentiation, and cell adhesion and ‘other’ genes. Inflammatory, immune & defense response transcripts Cxcl7, Saa1, Ncf1, Selp, and Orm1 were modestly induced in soft tissue, while defensin beta 3 (Defb3) was induced significantly at 58-fold. The ECM protein genes Mmp12 and Mmp13 transcription were again modestly induced in soft tissue. A potentially interesting observation was, that the top 5 genes upregulated in response to polymicrobial infection in soft tissue, Defb3, Sprr2d, Sprr2i, Sprr2f, and Sprr2h were not enhanced at all in calvarial bone suggesting tissue specific unique gene upregulation. These comparative analyses of transcription clearly demonstrate that the polymicrobial infection induced transcript alterations distinctively different in soft tissue and bone.
Real-Time RT-PCR confirmations
The polymicrobial infection-induced mRNA expression defined by the microarray studies were confirmed by qRT-PCR, including Defb3, Sprr2d, and Mmp13 in soft tissue and Cxcl7, Mmp9, and Pglyrp1 in calvarial bone using aliquots of the pooled RNA samples that were evaluated in the microarrays (Supporting Information, Table S3). The expression levels of Mmp13 and Mmp9 by qRT-PCR were higher than microarray expression levels and the expression levels of Defb3, Sprr2d, Cxcl7, and Pglyrp1 by qRT-PCR were lower than microarray expression levels. Temporal qRT-PCR data confirmed the direction of change of mRNA levels of expression of selected genes in calvarial soft tissue and bone indicating our microarray data reliability as well as the high sensitivity of the mouse DNA microarray technology. However, the magnitude of the fold-change in levels differed between the two analyses, a common phenomenon largely due to differences in dynamic range and threshold detection between microarray and qRT-PCR technologies in agreement with other studies (Cobb et al., 2002; Higgins et al., 2003).
Calvarial histology
The uninfected control mice showed a lack of edema and minimal inflammation in the soft tissue over the calvaria at the site of injection (Fig. 4A). Soft tissue swelling occurred at the injection site and increased in size following 48 and 72 h in almost all of the mice injected with P. gingivalis, T. denticola, and T. forsythia, but not with control mice. None of the mice infected with P. gingivalis, T. denticola, and T. forsythia had soft tissue abscesses, ulceration of the overlying skin, or any evidence of spread of infection to neighboring sites. Microscopic examination of the tissue sections showed prominent edema and a mixed inflammatory cell infiltrate consisting of aggregates of lymphocytes, polymorphonuclear leukocytes, and macrophages (Fig. 4B). Several multi-nucleated and activated osteoclasts were seen at the suture area on the inner aspect of the calvaria compared to controls. Activated osteoclasts were noted along the entire suture area corresponding to the areas of bone resorption (Fig. 4C and 4D).
Figure 4.
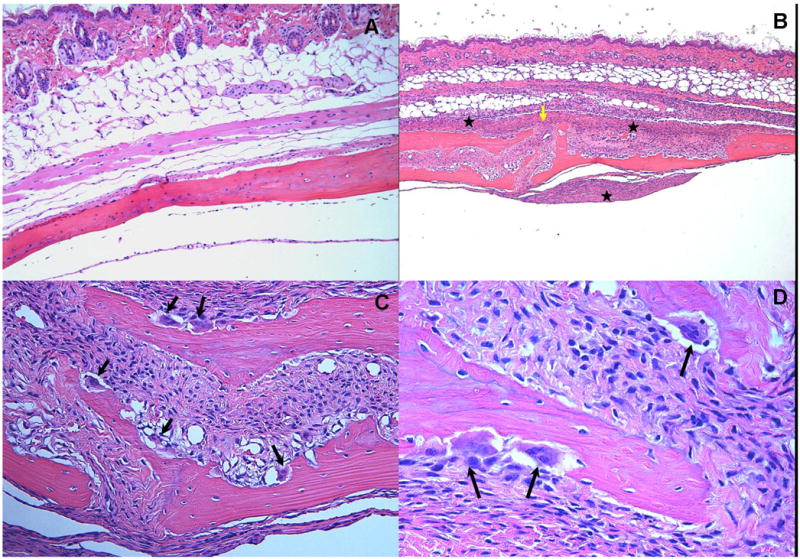
Effects of polymicrobial infections with P. gingivalis/T. denticola/T. forsythia on mouse calvaria. Live P. gingivalis+T. denticola+T. forsythia (5 × 108 cells/each) bacteria as polymicrobial consortium were injected once daily for 3 days into the s.c. tissues overlying the calvaria of mice. All photomicrographs are of slides stained with hematoxylin and eosin. A. Lack of edema and inflammation in the calvarial soft tissue of the sham-infected control mouse (magnification × 10). B. A section from a polymicrobial infected animal demonstrates significant disruption of the suture area (yellow arrow) with an intense mixed inflammatory infiltrate consisting primarily of neutrophils, lymphocytes and macrophages along with areas of edema and increased vascularity (magnification × 5). Inflammation is noted on both the dermal and supraosteal areas (asterix).C. Numerous osteoclasts (black arrows) are seen throughout the inner aspects of the calvarial bone mainly in the suture area. (magnification × 20). D. Activated osteoclasts within resorption lacunae at higher magnification (× 40).
Discussion
This is the first study that has evaluated the global gene transcriptional profiles during a polymicrobial infection with P. gingivalis, T. denticola, and T. forsythia in infected soft tissues and underlying bone. This type of seminal information is required to understand the complex synergistic interactions that can take place in situ affecting host cells, tissues, organs and systems, as well as provide unique insights into characteristics of the regulation of these interactions. While the report of this transcriptome study does not include a total catalogue of specific differentially impacted biological pathways in calvarial bone and soft tissue, it targets a few pathways that were found to be highly significantly altered in expression. Recently, three studies have reported the use of microarrays to determine the polymicrobial sepsis “transcriptome” in vivo and measured the broad-scale gene expression profiles for septic liver and spleen and compared these responses with controls using a well-accepted model of murine polymicrobial abdominal sepsis (Cobb et al., 2002; Nemeth et al., 2006; Weighardt et al., 2006). Furthermore, a recent review summarized several studies that monitored the changes in gene expression that take place in host cells such as macrophages, neutrophils, epithelial cells, fibroblasts, dendritic cells after contact with a specific pathogen in vitro using publicly available datasets (Jenner & Young, 2005). Using microarray analysis, we now demonstrate calvarial soft tissue and calvarial bone transcriptional signature profiling to specific periodontal pathogens P. gingivalis, T. denticola, and T. forsythia as a distinct polymicrobial infection to compare with gene expression patterns that have been reported in human periodontitis processes (Demmer et al., 2008; Papapanou et al., 2009).
In the present study, approximately 4476 and 1035 genes were differentially expressed in the bone and soft tissue in response to the polymicrobial infection, indicating the polymicrobial infection-induced a more robust activation of host gene expression in calvarial bone than in overlying soft tissue. Our findings indicate that the gene expression profiles induced by polymicrobial infection in these two tissues are distinct classes of gene with very little overlap. The greatest number of these altered genes was identified as being related to biologic pathways of transport, cell proliferation, cell cycle, defense & immune response, transcription, apoptosis, and inflammatory response, suggesting that the polymicrobial infection was able to induce a multitude of specific gene expression changes during infection. To our knowledge, several highly upregulated genes in bone (Ltb4r1, Ceacam1, Evi2b, Cxcl7, Mmp9; Orm1, Hp, Bst1) and soft tissue (Defb3, Sprr2d, Sprr2i, Sprr2f, Sprr2h) in response to the polymicrobial infection in mice have not been reported as being elicited in chronic periodontitis and aggressive periodontitis diseased gingival tissues (Demmer et al., 2008; Papapanou et al., 2009) and during the induction and resolution of experimental gingivitis in humans (Offenbacher et al., 2009; Jonsson et al., 2011) and their functional significance will need to be further evaluated to determine the biological events underlying polymicrobial periodontitis pathogenesis.
We anticipated that the combined P. gingivalis, T. denticola, and T. forsythia infection would induce a strong inflammatory response that would be reflected by a coordinated and controlled array of cytokines, chemokines, and oxidative burst effectors (Cohen et al., 2000; Eskra et al., 2003). However, we observed that the major proinflammatory cytokines IL-1, IL-6, TNF that have been predicted to be crucial in the induction of chronic inflammation and bone resorption were only modestly upregulated in bone following the polymicrobial infection. However, mRNAs for defensin beta 3, chemokine ligand 7, neutrophil cytosolic factor 1, lymphotoxin B, chitinase 3, chemokine ligand 9, chemokine ligand 12, and bone morphogenetic protein1 were modestly induced. Beta-defensins are believed to contribute to the host defense system by eradicating pathogens at the mucosal surface. Beta-defensins are expressed predominantly at epithelial surfaces, suggesting that these molecules are an important component of the innate immune system and their expression is inducible in response to inflammatory stimuli including both Gram-positive and Gram-negative bacteria (Maxwell et al., 2003). The significantly highly upregulated level of Defb3 gene (58-fold) expression during polymicrobial infection clearly demonstrates innate immune defense antimicrobial peptides are a portion of the gene repertoire induced to overcome this Gram-negative bacterial infection. The small proline-rich (Sprr) proteins are the primary constituents of the cornified layer of the epidermis, which is the major barrier against the environment, and are expressed in all squamous epithelium of the skin, scalp, and most of the epithelial lining of the digestive tract including oral epithelium (Tesfaigzi & Carlson, 1999). High levels of Sprr genes are detected in various diseases (inflammatory dermatoses), cancers of the skin, human papilloma virus infection, as well as being upregulated under stress (De Heller-Milev et al., 2000; Lehr et al., 2004). A recent study identified SPRR1A as a novel stress-inducible downstream mediator of gp130 cytokines in cardiomyocytes and documented its cardioprotective effect against ischemic stress (Pradervand et al., 2004). The current finding of upregulation of expression of Sprr2d, Sprr2i, Sprr2f, and Sprr2h genes (6 to 24-fold) in soft tissue clearly suggested a novel view of the response to this polymicrobial infection and these genes may function in vivo as a major barrier against the complex bacterial challenge.
A major difference between the calvarial bone and soft tissue sample responses was the impact of polymicrobial infection on the LTM pathway in bone resulting in the activation of actin (regulation of cytoskeleton) and α-cadherin. Leukocytes have a number of functions, including activation of endothelial cell signals, production of reactive oxygen species (ROS) with subsequent activation of ITGB2 (integrin beta 2), platelet endothelial cell adhesion molecule-1 (PECAM1), MMPs, CDH5 (cadherin 5), and CAMs. Many of the LTM pathway genes such as integrins, focal adhesion molecules, and cadherins have been reported to be upregulated during induction of experimental gingivitis in humans which is consistent with the activation of the LTM pathway (Offenbacher et al., 2009). PECAM-1 is one of the most abundant proteins on the endothelial cell surfaces, is expressed on the surface of platelets and leukocytes, and its expression increases significantly with increasing size of inflammatory infiltrates in the lesions of gingivitis and periodontitis (Gemmell et al., 1994). A similar robust impact on LTM pathway genes was observed after T. forsythia mono infection (Bakthavatchalu et al., 2010b) and to a lesser extent following a T. denticola infection (Bakthavatchalu et al., 2010a). In contrast, this pathway was unaffected in bone and soft tissue after P. gingivalis infection (Meka et al., 2010). P. gingivalis induced greater differential regulation of genes in a mixed infection compared to a monoinfection, consistent with its characterization as a stealth pathogen (Hajishengallis, 2009). In addition, the phenotypic properties of P. gingivalis change dramatically when the organism is in a community with other oral bacteria (Kuboniwa et al., 2009). These findings suggested that the complex infection with P. gingivalis, T. denticola, and T. forsythia impacted the LTM path way genes in bone and to a lesser extent in soft tissues with the host recognizing the challenge as a summation of the 3 individual bacteria.
A robust transcriptional changes was also observed with ECM proteins (N=14) in calvarial bone. Surprisingly, almost all the components of the ECM-receptor interaction pathway in calvarial bone (but not in soft tissue) including collagen, laminin, chondroadherein, fibronectin, osteopontin, tenascin, bone sialoprotein, VWF, and thrombospondin were upregulated following the polymicrobial infection. However, these ECM-receptor interaction pathway components were not altered with P. gingivalis infection (Meka et al., 2010), modestly affected following T. forsythia challenge (Bakthavatchalu et al., 2010b) and significantly increased with T. denticola monoinfection (Bakthavatchalu et al., 2010a). Collectively, collagen, fibronectin, osteopontin, and bone sialoprotein are the major constituents of periodontal tissues. They are critical for their regeneration after injury and seem to have a unique distribution within the periodontium and accumulate predominantly at the hard tissue interfaces. MMPs 2, 3, 9, 12, 13, 14, and 23 were upregulated in both bone and soft tissue after the polymicrobial challenge. Probe sets representing TIMPs 1, 2 and 3 also demonstrated a modest upregulation during the polymicrobial infection. This murine infection model outcomes for the MMPs and TIMPs are similar to recent microarray data showing several MMPs (1, 2, 3, 7, 9, 13, 14, 28) and TIMPs (2, 3) were significantly upregulated in human periodontitis gingival tissues (Demmer et al., 2008; Kubota et al., 2008). Our results with ECM proteins regulation are also in agreement with Escherichia coli lipopolysaccharide stimulation of canine transcriptional changes (Higgins et al., 2003). As P. gingivalis, T. denticola, and T. forsythia levels are quantitatively higher in subgingival plaque samples from deep periodontal pockets of patients with adult periodontitis (Socransky et al., 1998) they may synergistically induce expression of several MMP's and cathepsins which collectively can degrade ECM proteins in the periodontium and may contribute to bone resorption and ligament attachment loss observed in PD (Demmer et al., 2008; Kubota et al., 2008). These observations are consistent with the “red complex” microorganisms a functionally critical members of the pathogenic biofilms eliciting hard tissue destruction observed in periodontitis.
Many acute-phase proteins were upregulated in soft tissue and bone during the polymicrobial infection including serum amyloid A1, orosomucoid 1, aquaporin 7, aquaporin 9, haptoglobin, and complement protein 1q and tumor necrosis factor related protein 1. During bacterial infection innate and adaptive immunity co-regulate through soluble factors, such as cytokines, complement proteins and by their specific receptors expressed on various cells. The complement protein 1q and tumor necrosis factor related protein 1 (C1qtnf1) gene were overexpressed in calvarial bone following infection. This member of the C1q and TNF superfamily represents a group of proteins involved in host defense, inflammation, apoptosis, autoimmunity, cell differentiation, and insulin-resistant obesity (C1qtnf1 is an adiponectin paralog in mice). C1q triggers the production of IL-8, IL-6 and MCP-1 by endothelial cells may also contribute to the acute-phase response. These data clearly indicate that during the polymicrobial infection the host initiates a strong acute-phase response by expressing C1qtnf1, Saa1, and Orm1 in calvarial bone and soft tissue (Higgins et al., 2003).
The leukotriene B4 receptor 1 (Ltb4r1; 18-fold) was highly expressed in calvarial bone following the polymicrobial infection. Leukotriene B4, a product of 5 lipoxygenase, is a potent mediator of inflammation expressed in several inflammatory diseases including periodontitis, bronchial asthma, and atherogenesis. Additionally, they play a role in the regulation of innate and adaptive immunity, since the LTB4-BLT1 (leukotriene B4 receptor 1) axis is required for the development of a Th2-type immune response in bronchial asthma (Terawaki et al., 2005). Moreover, LTB4 was reported to be significantly elevated in gingival tissue and gingival crevicular fluid from patients with chronic periodontitis and generalized aggressive periodontitis (Emingil et al., 2001). The example of Ltb4r1 gene expression suggested that the periodontal pathogens induced potent lipid inflammatory mediators that could contribute to the role in the pathophysiology of periodontitis.
The characteristic inflammation and osteoclastic full thickness calvarial bone resorption defects observed in response to polymicrobial infection were similar to the effects that we observed in the calvariae of mice following infection with P. gingivalis (Meka et al., 2010), T. denticola (Bakthavatchalu et al., 2010a), T. forsythia (Bakthavatchalu et al., 2010b), C. rectus and F. nucleatum (Zubery et al., 1998), suggesting that there is not a synergistic action of these micro-organisms to induce genes that regulate osteoclastogenesis or that the response to individual organisms is maximal.
In conclusion, the present study provided a comprehensive gene expression profile of mouse calvarial soft tissue and calvarial bone that accompanied a localized, acute infection with a polybacterial challenge with P. gingivalis, T. denticola, and T. forsythia. In addition, this study demonstrated that while the polymicrobial infection resulted in a multitude of specific gene expression changes, there appeared to be some targeted biological pathways that were more selectively altered and could provide some guidance for further understanding the polymicrobial challenge leading to the pathogenesis of periodontitis. Lastly, by depositing our raw data in GEO repository (http://www.ncbi.nlm.nih.gov/projects/geo/), we enable prospective researchers to conduct more targeted analyses focusing on specific pathways and genes of their interest related to the pathogenesis, diagnosis, and/or therapeutic targets in periodontal disease.
Supplementary Material
Figure S1. Cell adhesion molecules (immune system) pathway.
Figure S2. Tight adherens junction in calvarial bone.
Figure S3. Antigen processing and presentation in calvarial bone.
Table S1. Leave-one-out cross validation (calvarial bone).
Table S2. Leave-one-out cross validation (calvarial soft tissue).
Table S3. Comparison of expression of selected genes in calvarial tissue and calvariae by microarray and real-time quantitative polymerase chain reaction.
Table S4. Leukocyte transendothelial migration (LTM) pathway genes impacted by polymicrobial and mono-infection in calvarial tissue.
Table S5. Extracellular matrix (ECM) receptor-interaction pathway genes impacted by polymicrobial and mono-infection in calvarial tissue.
Acknowledgments
We thank Drs Arnold Stromberg, Cidambaram Srinivasan, Christopher Saunders, and Matt Hersh (Department of Statistics, University of Kentucky, Lexington, USA) for initial data management and microarray data analysis. We also thank Dr Quey-Chen and Ms Donna Wall (University of Kentucky, Lexington, Microarray Core Facility) for microarray technical assistance. This study was supported by National Institute of Dental and Craniofacial Research DE015720 to L.K., DE11111 to R.J.L., United States Public Health Services research grant U24 DE016509 (Robert Burne), and by a National Institute of Arthritis, Musculoskeletal and Skin Diseases grant (AR43510) to B.F.B.
Footnotes
Disclosures The authors have no financial conflict of interest.
References
- Bakthavatchalu V, Meka A, Sathishkumar S, et al. Molecular characterization of Treponema denticola infection-induced bone and tissue transcriptional profiles. Mol Oral Microbiol. 2010a;25:1–15. doi: 10.1111/j.2041-1014.2010.00575.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakthavatchalu V, Meka A, Sathishkumar S, et al. Tannerella forsythia infection-induced bone and tissue Transcriptional profiles. Mol Oral Microbiol. 2010b;25:317–330. doi: 10.1111/j.2041-1014.2010.00583.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brogden KA. Polymicrobial diseases of animals and humans. In: Brogden K, Guthmiller J, editors. Polymicrobial Diseases. ASM Press; Washington, DC: 2002. pp. 3–20. [PubMed] [Google Scholar]
- Brogden KA, Guthmiller JM, Taylor CE. Human polymicrobial infections. Lancet. 2005;365:253–255. doi: 10.1016/S0140-6736(05)17745-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T, Wen-Han Y, Izard J, et al. The human oral microbiome database : a web accessible resource for investigating oral microbe taxonomic and genomic information. Database. 2010;6:1–10. doi: 10.1093/database/baq013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobb JP, Laramie JM, Stormo GD, Morrissey JJ, et al. Sepsis gene expression profiling: murine splenic compared with hepatic responses determined by using complementary DNA microarrays. Crit Care Med. 2002;30:2711–2721. doi: 10.1097/00003246-200212000-00016. [DOI] [PubMed] [Google Scholar]
- Cohen P, Bouaboula M, Bellis M, et al. Monitoring cellular responses to Listeria monocytogenes with oligonucleotide arrays. J Biol Chem. 2000;275:11181–11190. doi: 10.1074/jbc.275.15.11181. [DOI] [PubMed] [Google Scholar]
- Dennis G, Jr, Sherman BT, Hosack DA, et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- De Heller-Milev M, Huber M. Expression of small proline rich proteins in neoplastic and inflammatory skin diseases. Br J Dermatol. 2000;143:733–740. doi: 10.1046/j.1365-2133.2000.03768.x. [DOI] [PubMed] [Google Scholar]
- Demmer RT, Behle JH, Wolf DL, et al. Transcriptomes in healthy and diseased gingival tissues. J Periodontol. 2008;79:2112–2124. doi: 10.1902/jop.2008.080139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draghici S, Khatri P, Martins RP, Ostermeier GC, Krawetz SA. Global functional profiling of gene expression. Genomics. 2003;81:98–104. doi: 10.1016/s0888-7543(02)00021-6. [DOI] [PubMed] [Google Scholar]
- Dudoit S, Gentleman RC, Quackenbush J. Open source software for the analysis of microarray data. Biotechniques. 2003;(Suppl):45–51. [PubMed] [Google Scholar]
- Emingil G, Cinarcik S, Baylas H, Coker I, Huseyinov A. Levels of leukotriene B4 in gingival crevicular fluid and gingival tissue in specific periodontal diseases. J Periodontol. 2001;72:1025–31. doi: 10.1902/jop.2001.72.8.1025. [DOI] [PubMed] [Google Scholar]
- Eskra L, Mathison A, Splitter G. Microarray analysis of mRNA levels from RAW264.7 macrophages infected with Brucella abortus. Infect Immun. 2003;71:1125–1133. doi: 10.1128/IAI.71.3.1125-1133.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feezor RJ, Oberholzer C, Baker HV, et al. Molecular characterization of the acute inflammatory response to infections with gram-negative versus gram-positive bacteria. Infect Immun. 2003;71:5803–13. doi: 10.1128/IAI.71.10.5803-5813.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gemmell E, Walsh LJ, Savage NW, Seymour GJ. Adhesion molecule expression in chronic inflammatory periodontal disease tissue. J Periodont Res. 1994;29:46–53. doi: 10.1111/j.1600-0765.1994.tb01090.x. [DOI] [PubMed] [Google Scholar]
- Hajishengallis G. Porphyromonas gingivalis-host interactions: open war or intelligent guerilla tactics? Microbes Infect. 2009;11:637–645. doi: 10.1016/j.micinf.2009.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins MA, Berridge BR, Mills BJ, et al. Gene expression analysis of the acute phase response using a canine microarray. Toxicol Sci. 2003;74:470–484. doi: 10.1093/toxsci/kfg142. [DOI] [PubMed] [Google Scholar]
- Holt SC, Ebersole JL. Porphyromonas gingivalis, Treponema denticola, and Tannerella forsythia: the “red complex”, a prototype polybacterial pathogenic consortium in periodontitis. Periodontol 2000. 2005;38:72–122. doi: 10.1111/j.1600-0757.2005.00113.x. [DOI] [PubMed] [Google Scholar]
- Holt SC, Kesavalu L, Walker S, Genco CA. Virulence factors of Porphyromonas gingivalis. Periodontol 2000. 1999;20:168–238. doi: 10.1111/j.1600-0757.1999.tb00162.x. [DOI] [PubMed] [Google Scholar]
- Hosack DA, Dennis G, Sherman BT, Lane HC, Lempicki RA. Identifying biological themes within lists of genes with EASE. Genome Biol. 2003;4:R70. doi: 10.1186/gb-2003-4-10-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenner RG, Young RA. Insights into host responses against pathogens from transcriptional profiling. Nat Rev Microbiol. 2005;3:281–294. doi: 10.1038/nrmicro1126. [DOI] [PubMed] [Google Scholar]
- Jonsson D, Ramberg P, Demmer RT, Klebschull M, Dahlen G, Papapanou PN. Gingival tissue transcriptomes in experimental gingivitis. J Clin Periodontol. 2011;10 doi: 10.1111/j.1600-051X.2011.01719.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kesavalu L, Chandrasekar B, Ebersole JL. Induction of proinflammatory cytokine expression in vivo mediated by Porphyromonas gingivalis and Actinobacillus actinomycetemcomitans. Oral Microbiol Immunol. 2002;17:177–180. doi: 10.1034/j.1399-302x.2002.170307.x. [DOI] [PubMed] [Google Scholar]
- Kesavalu L, Holt SC, Ebersole JL. Virulence of a polymicrobic complex, Treponema denticola and Porphyromonas gingivalis, in a murine model. Oral Microbiol Immunol. 1998;13:373–377. doi: 10.1111/j.1399-302x.1998.tb00694.x. [DOI] [PubMed] [Google Scholar]
- Kesavalu L, Sathishkumar S, Vasudevan B, et al. Rat model of polymicrobial infection, immunity, and alveolar bone resorption in Periodontal disease. Infect Immun. 2007;75:1704–1712. doi: 10.1128/IAI.00733-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuboniwa M, Hendrickson EL, Xia Q, Wang T, Xie H, Hackett M, Lamont RJ. Proteomics of Porphyromonas gingivalis within a model oral microbial community. BMC Microbiol. 2009;9:98. doi: 10.1186/1471-2180-9-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota T, Itagaki M, Hoshino C, et al. Altered gene expression levels of matrix metalloproteinases and their inhibitors in periodontitis-affected gingival tissue. J Periodontol. 2008;79:166–173. doi: 10.1902/jop.2008.070159. [DOI] [PubMed] [Google Scholar]
- Kuramitsu HK, Chen W, Ikegami A. Biofilm formation by the periodontopathic bacteria Treponema denticola and Porphyromonas gingivalis. J Periodontol. 2005;76:2047–51. doi: 10.1902/jop.2005.76.11-S.2047. [DOI] [PubMed] [Google Scholar]
- Lamont RJ, Chan A, Belton CM, Izutsu KT, Vasel D, et al. Porphyromonas gingivalis invasion of gingival epithelial cells. Infect Immun. 1995;63:3878–3885. doi: 10.1128/iai.63.10.3878-3885.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamont RJ, Jenkinson HF. Life below the gum line: pathogenic mechanisms of Porphyromonas gingivalis. Microbiol Mol Biol Rev. 1998;62:1244–1263. doi: 10.1128/mmbr.62.4.1244-1263.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehr E, Hohl D, Huber M, Brown D. Infection with human papillomavirus alters expression of the small proline proteins 2 and 3. J Med Virol. 2004;72:478–483. doi: 10.1002/jmv.20011. [DOI] [PubMed] [Google Scholar]
- Maxwell AI, Morrison GM, Dorin JR. Rapid sequence divergence in mammalian beta-defensins by adaptive evolution. Mol Immunol. 2003;40:413–421. doi: 10.1016/s0161-5890(03)00160-3. [DOI] [PubMed] [Google Scholar]
- Meka A, Bakthavatchalu V, Sathishkumar S, et al. Porphyromonas gingivalis infection-induced tissue and bone transcriptional profiles. Mol Oral Microbiol. 2010;25:61–74. doi: 10.1111/j.2041-1014.2009.00555.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemeth ZH, Csoka B, Wilmanski J, et al. Adenosine A2A receptor inactivation increases survival in polymicrobial sepsis. J Immunol. 2006;176:5616–5626. doi: 10.4049/jimmunol.176.9.5616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishihara T, Koseki T. Microbial etiology of periodontitis. Periodontol 2000. 2004;36:14–26. doi: 10.1111/j.1600-0757.2004.03671.x. [DOI] [PubMed] [Google Scholar]
- Offenbacher S, Barros SP, Paquette DW, et al. Gingival transcriptome patterns during induction and resolution of experimental gingivitis in humans. J Periodontol. 2009;80:1963–1982. doi: 10.1902/jop.2009.080645. [DOI] [PubMed] [Google Scholar]
- Papapanou PN, Behle JM, Kebschull M, et al. Subgingival bacterial colonization profiles correlate with gingival tissue gene expression. BMC Microbiology. 2009;9:221. doi: 10.1186/1471-2180-9-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradervand S, Yasukawa H, Muller OG, et al. Small proline-rich protein 1A is a gp130 pathway- and stress-indicible cardioprotective action. Embo J. 2004;23:4517–25. doi: 10.1038/sj.emboj.7600454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rachman H, Strong M, Ulrichs T, et al. Unique transcriptome signature of Mycobacterium tuberculosis in pulmonary tuberculosis. Infect Immun. 2006;74:1233–42. doi: 10.1128/IAI.74.2.1233-1242.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Socransky SS, Haffajee AD. Periodontal microbial ecology. Periodontol 2000. 2005;38:135–187. doi: 10.1111/j.1600-0757.2005.00107.x. [DOI] [PubMed] [Google Scholar]
- Socransky SS, Haffajee AD, Cugini MA, Smith C, Kent RL. Microbial complexes in subgingival plaque. J Clin Periodontol. 1998;25:134–144. doi: 10.1111/j.1600-051x.1998.tb02419.x. [DOI] [PubMed] [Google Scholar]
- Terawaki K, Yokomizo T, Nagase T, et al. Absence of leukotriene B4 receptor 1 confers resistance to airway hyperresponsiveness and Th2-type immune responses. J Immunol. 2005;175:4217–25. doi: 10.4049/jimmunol.175.7.4217. [DOI] [PubMed] [Google Scholar]
- Tesfaigzi J, Carlson DM. Expression, regulation, and function of the SPR family of proteins. A review. Cell Biochem Biophys. 1999;30:243–65. doi: 10.1007/BF02738069. [DOI] [PubMed] [Google Scholar]
- Treister NS, Richards SM, Lombardi MJ, et al. Sex-related differences in gene expression in salivary glands of BALB/c mice. J Dent Res. 2005;84:160–5. doi: 10.1177/154405910508400210. [DOI] [PubMed] [Google Scholar]
- Tribble GD, Lamont RJ. Bacterial invasion of epithelial cells and spreading in periodontal tissue. Periodontol 2000. 2010;52:68–83. doi: 10.1111/j.1600-0757.2009.00323.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weighardt H, Mages J, Jusek G, et al. Organ-specific role of MyD88 for gene regulation during polymicrobial peritonitis. Infect Immun. 2006;74:3618–3632. doi: 10.1128/IAI.01681-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneda M, Hirofuji T, Anan H, et al. Mixed infection of Porphyromonas gingivalis and Bacteroides forsythus in a murine abscess model: involvement of gingipains in a synergistic effect. J Periodontal Res. 2001;36:237–243. doi: 10.1034/j.1600-0765.2001.036004237.x. [DOI] [PubMed] [Google Scholar]
- Zubery Y, Dunstan CR, Story B, et al. Bone resorption caused by three periodontal pathogens In vivo in mice is mediated in part by prostaglandin. Infect Immun. 1998;66:4158–4162. doi: 10.1128/iai.66.9.4158-4162.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Cell adhesion molecules (immune system) pathway.
Figure S2. Tight adherens junction in calvarial bone.
Figure S3. Antigen processing and presentation in calvarial bone.
Table S1. Leave-one-out cross validation (calvarial bone).
Table S2. Leave-one-out cross validation (calvarial soft tissue).
Table S3. Comparison of expression of selected genes in calvarial tissue and calvariae by microarray and real-time quantitative polymerase chain reaction.
Table S4. Leukocyte transendothelial migration (LTM) pathway genes impacted by polymicrobial and mono-infection in calvarial tissue.
Table S5. Extracellular matrix (ECM) receptor-interaction pathway genes impacted by polymicrobial and mono-infection in calvarial tissue.


