Abstract
Successful immunity depends upon the activity of multiple cell types. Therefore, commitment of pluripotent precursor cells to specific lineages, such as T or B cells, is obviously fundamental. However, it is also becoming clear that continued differentiation and specialization of lymphoid cells is equally important for immune system integrity. Several members of the BTB-ZF family have emerged as critical factors that control development of specific lineages and also of specific effector subsets within these lineages. For example, BTB-ZF genes have been shown to control T cell versus B cell commitment and CD4 versus CD8 lineage commitment. Others, such as PLZF for NKT cells and Bcl6 for T follicular helpers cells, are necessary for the acquisition of effector functions. Here we summarize current findings concerning the BTB-ZF family members with reported role in the immune system.
BTB-ZF Proteins Are Transcriptional Repressors that Recruit Co-Repressors and Chromatin Remodeling Factors to Target Genes
BTB-ZF [Broad complex, Tramtrack, Bric a`brac and Zinc Finger] proteins are an evolutionary conserved family of transcriptional regulators. Members of this group, of which there are >45 in human and mice, are characterized as having one or more C-terminal C2H2 Krüppel-type zinc finger DNA binding domains in combination with an N-terminal BTB domain that mediates protein-protein interactions. Transcriptional regulation, most often repression, is achieved by sequence-specific binding by the ZF domain to regulatory regions in target genes, coupled with the recruitment of co-factors involved in chromatin remodeling and transcriptional silencing/activation.
Co-factor complex formation is largely mediated by the BTB domain, which has been shown to directly interact with corepressors and histone modification enzymes, including SMRT, ETO, N-Cor, B-Cor, CtBP, Sin3A, DRAL/FHL2, and HDAC-1, -2, -4, -5, and -7 (1-11). Though most of these interactions were described in non-hematopoietic cells or transformed cell lines, BTB-ZF proteins likely regulate gene expression in primary lymphocytes via a similar mechanism. For example, the BTB-ZF protein, PLZP, has been shown to associate with HDAC-2 in Th2-skewed CD4+ and CD8+ T-cells, and these two proteins colocalize at regulatory elements in the IL-13 gene where they likely act in concert to modulate transcription (12).
In addition to co-repressor recruitment, the BTB domain and, in some cases, the ZF domain, also facilitate hetero- and homo-dimerization among the different gene family members. For example, the BTB-ZF protein, Bcl6, can exist as a homodimer (13) but may also form heterodimers with other BTB-ZF proteins, including NAC-1, PLZF, LRF, BAZF, and Miz-1 (14-18). Similarly, overexpression and co-transfection systems have demonstrated an interaction between PLZF and PLZP, though PLZF can also exist as a homodimer (1, 19, 20). Like the co-factor studies, nearly all of this work has been done in non-hematopoietic cells or cell lines; nevertheless, the finding that Miz-1 and Bcl-6 physically interact in primary germinal center B-cells (18) suggests that heterodimerization may be physiological relevant to BTB-ZF protein function in primary cells of the immune system and further studies are needed to shed light on this topic.
BTB-ZF Proteins Control Lineage Commitment, Development, and Function in Lymphocytes
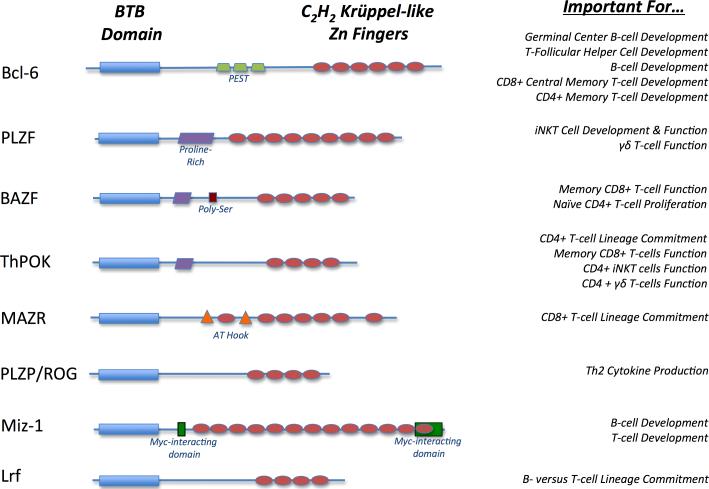
As powerful regulators of gene expression, BTB-ZF proteins are critical players in a wide variety of biological processes, including developmental events such as gastrulation and limb formation, control of DNA damage and cell cycle progression in normal and oncogenic tissues, maintenance of the stem cell pool, and gamete formation (21). Moreover, recent studies have highlighted a fundamental and non-redundant role for many BTB-ZF factors in the development and function of cells in the immune system. This review will summarize current findings on the eight family members with known roles in orchestrating lymphocyte development: Bcl-6, PLZF, ThPOK, PLZP, MAZR, BAZF, LRF, and MIZ-1 (Figure 1).
Fig. 1.
The key structural features of the eight BTB-ZF transcription factors discussed in this review are highlighted and the reported functions of each transcription factor are listed.
Bcl6
The BTB-ZF protein Bcl6 was first identified as an oncogene in diffuse large B cell lymphoma (DLBCL), the most common form of Non-Hodgkin's Lymphoma. The transformative properties of Bcl6 stem largely from its ability to repress transcription of tumor suppressor and cell cycle arrest genes, including p53, ATR, CHEK1 and CDKN1A/p21 (18, 22-26). Bcl6 is normally expressed at high levels in germinal center (GC) B-cells. Early studies showed that mice lacking Bcl6 were unable to form germinal centers following immunization with T-cell-dependent antigens. Moreover, antigen-specific B-cells in these mice were impaired for affinity maturation and class switch recombination (CSR) to IgG subtypes (27, 28). Reconstitution of Rag1-KO mice with Bcl6-deficient bone marrow showed that Bcl-6 was required in the hematopoietic compartment for germinal center formation and somatic hypermutation (SHM), but not for primary IgG responses (29, 30). Bcl6 also represses expression of Blimp-1, a transcription factor that promotes plasma cell differentiation (31, 32). Given that Blimp1 represses Bcl6, the reciprocal antagonism of these two genes has been proposed to serve as a bimodal “switch”, by which B-cell fate, as either a germinal center B-cell or an antibody-secreting plasma cell, is established and maintained (33). Beyond promoting germinal center, and suppressing plasma cell gene programs, Bcl6 inhibits cell cycle arrest and apoptosis in GC B-cells, allowing DNA damage, a natural byproduct of CSR and SHM, to occur in the absence of cell cycle checkpoint activation. To this end, it represses many of the same genes dysregulated during Bcl6-mediated transformation, including p53, ATR, CHEK1 and CDKN1A/p21 (18, 24-26).
In addition to peripheral lineage commitment, Bcl-6 is important for B-cell development in the bone marrow (34). High Bcl-6 expression is induced by pre-BCR signaling during the pro- to pre-B-cell transition. Bcl-6 in turn suppresses DNA damage response genes, including CDKN1A/p21, CDKN1B/p27, and CDKN2A/Arf, during Ig light chain rearrangement, which would otherwise activate apoptosis and cellular senescence in response to RAG-induced DNA lesions. Consequently, Bcl6-deficient mice have an immature B cell pool that is reduced in both size and clonal diversity (34).
Hints at a role for Bcl6 in T-cell function originated from early studies in which Bcl6-deficient mice were found to develop spontaneous Th2 inflammation, characterized by enhanced IgE production and severe eosinophilia (27, 28). Upon stimulation in vitro, T-cells from Bcl6-deficient mice produce elevated levels of IL-4, IL-5, and IL-13, a phenotype that has been linked to direct binding by Bcl-6 in regulatory regions of genes for IL-5, Ig epsilon, and IL-4 (27, 35, 36).
More recently Bcl6 was shown to be necessary and sufficient for the development of CD4+ T follicular helper (Tfh) cells, which provide critical help to germinal center B-cells undergoing SHM and CSR (37-39). Constitutive Bcl-6 expression in CD4+ T-cells in vivo drives nearly complete commitment to the Tfh lineage and these helper cells are highly effective inducers of germinal center formation and antibody production by B-cells. Conversely, Bcl-6 deficiency abrogates Tfh differentiation and CD4+ T-cells from these mice fail to mediate germinal center formation (38, 39). At a mechanistic level, Bcl-6 is induced in CD4+ T-cells by IL-6 and IL-21, though neither cytokine is independently required for Tfh differentiation (40). Bcl6 drives expression of molecules involved in Tfh homing and function, including CXCR5, CXCR4, IL-21R, IL-6R, PD-1, and IL-21, and is required to maintain Tfh identity by suppressing the expression of factors associated with other helper T-cell lineages, including IFNγ, IL-17, T-bet, IL-4, GATA-3, and Blimp-1 (37-39). As in B-cells, Blimp-1 and Bcl-6 counter-repress each other and play antagonistic roles in CD4+ T-cell differentiation, with Blimp-1 suppressing and Bcl-6 promoting Tfh lineage commitment (39).
Bcl-6 is also expressed at high levels in memory CD8+ T-cells. In mice lacking Bcl6, CD8+ T-cells proliferate poorly and fail to develop into central memory cells, highlighting a critical role for Bcl6 in memory T-cell formation. Overexpression of Bcl-6 leads to increased numbers of central memory CD8+ T cells following immunization and enhances CD8+ T-cell proliferation after secondary stimulation (41). Similarly, Bcl6-deficient CD4+ T-cells exhibit increased apoptosis at the effector cell stage and fail to persist as long-lived memory cells (42). Beyond its role in traditional T-cell subsets, Bcl6 is also important for the function of human CD8+ T suppressor cells, which may play a role in immunological tolerance in transplantation settings (43).
PLZF
Like Bcl6, the BTB-ZF protein Promyelocytic Leukemia Zinc Finger (PLZF) was first identified in the context of hematopoietic cancer. In acute promyelocytic leukemia (APL), chromosomal translocation fuses the genes for PLZF and the nuclear retonic acid receptor alpha (RARα), ultimately leading to oncogenic transformation (44). In keeping with a putative function as a tumor suppressor gene in APL, PLZF has been associated with cellular quiescence and growth suppression in non-transformed cells of hematopoietic and non-hematopoietic origin (45-51).
Recent studies on PLZF have elucidated its function as a critical regulator of innate T-cell lineages. PLZF is highly expressed in immature CD1d-restricted invariant NKT (iNKT) cells. In PLZF-deficient mice, iNKT cells fail to undergo thymic expansion and are substantially reduced in the thymus, liver, and spleen. The iNKT that do develop in these mice behave more like conventional naïve T-cells: these cells preferentially traffic to the lymph nodes and fail to express characteristic activation markers, e.g. CD44 and CD69. Moreover, PLZF-deficient iNKT cells show a marked reduction in cytokine secretion upon primary stimulation - most notable is their inability to simultaneously produce both IL-4 and IFNγ - and instead require secondary activation to fully elaborate effector responses (52, 53). In contrast, ectopic PLZF expression in T cells results in the spontaneous acquisition of memory/effector phenotypes and functions. PLZF-transgenic T-cells exhibit an activated phenotype (e.g. CD44hi and CD62Llo), migrate to non-lymphoid tissues, and show enhanced cytokine production upon primary activation even in the absence of costimulation (52, 54, 55).
In addition to iNKT cells, PLZF expression was recently shown in a subset of mature γδ T-cells expressing the Vγ1.1+Vδ6.3+ TCR (56, 57). Vγ1.1+Vδ6.3+ T-cells share many features with αβ TCR-expressing iNKT cells, including constitutive expression of activation markers, rapid and simultaneous production of IFNγ and IL-4, requiring the SLAM-associated Adaptor Protein, SAP, for their development. Although PLZF-deficient mice harbored normal numbers of these cells, their function is dramatically impaired in the absence of PLZF. Indeed, in contrast to WT cells, PLZF-deficient Vγ1.1+Vδ6.3+ T-cells produce almost undetectable levels of IFNγ and IL-4 in response to TCR stimulation (57, 58). Interestingly, transgenic mice expressing the Vγ1.1+Vδ6.4+ TCR have large numbers of PLZF+ T-cells and develop spontaneous dermatitis, perhaps underscoring a pro-inflammatory and sometimes pathogenic role for these cells (56).
Several mouse strains have been described with increased numbers of innate CD8+ T-cells, including mice lacking the Tec kinase Itk (59), the coactivator CBP (60), and the transcription factor KLF2 (61). Recently, this CD8+ T-cell expansion was shown to be a cell extrinsic consequence of elevated IL-4 produced by an expanded PLZF+ T cell population in these mice (61).
In addition to αβ and γδ NKT cell subsets, PLZF is expressed in human MR1-specific MAIT cells and in fetal MHC class II-restricted T cells that develop as a result of positive selection on other T-cells (52, 62). PLZF may also impact NK cell function either directly or indirectly; PLZF-deficiency impairs protection against MCMV infection and this susceptibility was associated with reduced interferon-induced NK cell cytotoxicity (63). Given the innate lymphocyte features of all of these cells, future studies will be useful for understanding the role of PLZF in their development and/or function.
ThPOK
ZBTB7b, also known as ThPOK or cKrox, encodes a three zinc finger BTB domain protein originally identified as a regulator of development and function in cells from connective tissues. Within the hematopoietic compartment, ThPOK is upregulated in CD4+, but not CD8+, T-cells upon differentiation from double- to single-positive thymocytes, and its expression is stably maintained in CD4+ T-cells (64, 65). A spontaneous mutation in one of its zinc finger domains resulted in the loss of helper T cells in HD (helper deficient) mice (65). MHC class II-restricted T-cells from HD mice, as well as T-cells with submaximal ThPok expression, exhibit features of transdifferentation to the CD8+ T-cell lineage, highlighting a critical role for ThPOK in driving the CD4 vs CD8 lineage fate in naïve T-cells (65, 66). Indeed, constitutive ThPOK expression in developing thymocytes induces redirection to the CD4+ lineage, even among MHC class I-restricted cells, with many of the features of helper T-cells (65).
In a temporal and developmental sense, ThPOK functions downstream of Gata-3, an early CD4 lineage transcription factor, and ThPOK upregulation in conventional thymocytes appears to depend on effective TCR signaling at the CD4+CD8lo stage (67-69). MHC class II-signaled CD4+CD8- thymocytes with impaired signaling through the TCR-associated kinases, Zap70 or Itk, fail to upregulate ThPOK and instead express cytotoxic T-cell markers, including the transcription factor Runx3 and its targets, Eomesodermin and Perforin (70-72). Analogous to the antagonistic relationship of Blimp-1 and Bcl6 in B-cell differentiation, ThPOK and the CD8+ T-cell determinant, Runx3, counter-repress expression of each other. Thus, Runx-deficient MHC class-I restricted thymocytes, which lack functionality in both Runx3 and Runx1 or are genetically deficient for the obligatory Runx binding protein, Cbfβ, retain ThPOK expression and exhibit features of the helper T-cell lineage, showing that Runx-mediated silencing of ThPOK is required to maintain CD8+ lineage commitment (73). Surprisingly, in animals lacking both Cbfβ and ThPOK, MHC class II-restricted thymocytes maintain helper T cell characteristics, suggesting that in settings where commitment to the CD8+ T-cell lineage is intrinsically limited (i.e. as a result of Runx deficiency), ThPOK is more of importance for the maintenance, rather than the induction, of the CD4+ T-cell fate (74). Nevertheless, in mice capable of CD8+ vs. CD4+ fate decisions, ThPOK plays a critical and active role in repressing CD8+ T-cell lineage commitment.
Recent studies have uncovered critical roles for ThPOK in the development and function of other T-cells subsets. In mature CD8+ T-cells, ThPOK expression is upregulated as a result of TCR activation and is required for effective expansion during the primary and secondary responses to acute viral infection (75). In mice lacking functional ThPOK, memory CD8+ T-cells are impaired for IL-2 secretion and granzyme B expression following antigen rechallenge (75). In innate T-cell lineages, ThPOK controls the development and functional maturation of PLZF+ NKT cells of both the γδ and αβ TCR lineages (57, 76-78). ThPOK is highly expressed in Vγ1.1+Vδ6.3+ γδ T-cells and CD4+ iNKTs. Although ThPOK-deficiency does not significantly alter the frequency of either population in mice, both subsets show dramatically impaired CD4 co-receptor expression, much like conventional T-cells (57, 76, 78). In addition, a separate study reported a reduction in the total pool of mature CD24- γδ T-cells, particularly those expressing NK1.1, in ThPOK-deficient mice, suggestive of a broader role for ThPOK in the development of some, though not all, γδ T-cell subsets (77). More striking, however, is the role of ThPOK in the effector function of iNKT and Vγ1.1+Vδ6.3+ γδ T-cells. ThPOK-deficient iNKTs show impaired expression of markers associated with NKT function, including granzyme B, CD69, and, in one study, NK1.1, and produce less IL-4, IFNγ and TNFα, in response to αGal-Cer stimulation (76, 78). As in conventional T-cells, ThPOK expression in iNKTs depends on Gata-3 (76). Similarly, Vγ1.1+Vδ6.3+ γδ T-cells produce less IL-4, but more IFNγ, in response to stimulation and this impairment correlated with reduced PLZF expression (57).
PLZP
The BTB protein PLZF-Like Zinc Finger Protein (PLZP), also known as FAZF, TZFP, and ROG, shares many features with PLZF, including a high level of sequence similarity, recognition of the same target DNA sequences, and declining expression in hematopoietic progenitors cells as a function of lineage-specific differentiation (19, 79, 80). Among lymphocytes, PLZP is upregulated in primary CD4+ and CD8+ T-cells following TCR activation in vitro (12, 80, 81). T-cells from PLZP-deficient mice produce more IL-2 and are hyper-proliferative in response to TCR stimulation as compared to WT cells, suggesting an anti-proliferative role for PLZP in activated T-cells (80, 82).
In addition to controlling proliferation, several studies have highlighted a role for PLZP in controlling lymphocyte cytokine responses. When overexpressed in T-cells lines, PLZP was shown to directly interact with the Th2-promoting transcription factor, GATA-3, and could antagonize GATA-3 binding to target genes, such as IL-5 (81). Similarly, PLZP repressed GATA-3-induced IL-4 production when both transcription factors were co-transfected into primary CD4+ T-cells (12). Moreover, overexpression in either Th1- or Th2-skewed T-cells impaired cytokine production on a more global level, leading to reduced IL-4, IL-5, IL-10, IFNγ, and IL-17 (12, 81). In Tc2-skewed CD8+ T-cells, endogenous PLZP was shown to directly bind a regulatory region in the IL-13 gene, hinting at a similar role for PLZP in cytokine production by CD8+ T-cells (12).
Consistent with the overexpression studies, CD4+ T-cells from PLZP-deficient mice express higher levels of IL-4, IL-5, and IL-13 when stimulated in vitro under Th2-promoting culture conditions, and PLZP-deficient CD8+ T-cells produced more IFNγ upon stimulation under neutral conditions (80). Nevertheless, PLZP-deficient CD4+ T-cells are fully capable of differentiating into Th1 or Th2 cells in vitro and PLZP-deficient mice mount normal Th1 and Th2 responses to MOG-induced EAE and KLH immunization, respectively (82). In contrast, allergic responses in airway hypersensitivity models are enhanced in PLZP-deficient mice, reflecting increased Th2 differentiation and Th2-driven inflammation in the affected airways (83). Similarly, these mice exhibit heightened Th2 inflammation in the context of a hapten-induced model of contact hypersensitivity, leading to exacerbated edema and mast cell degranulation at reaction sites and increased levels of IgE and hapten-specific IgG antibodies in the circulation (84). These defects are directly linked to a cell intrinsic requirement for PLZP in T-cells, as transfer of PLZP-deficient or PLZP-overexpressing T-cells exacerbated or ameliorated, respectively, disease progression in wild-type animals. Thus, additional studies will be needed to clarify the exact role of PLZP in lymphocyte function in vivo.
Beyond regulating cytokine production in T-cells, PLZP may also play a role in lineage commitment during T-cell development. In one study, overexpression of PLZP in developing double-positive thymocytes, in the context of a fetal thymic organ culture model, led to a preferential accumulation of single positive CD8+ T-cells. Similar to its function in mature T-cells, this accumulation was linked to the ability of PLZP to inhibit GATA-3 function (85).
BAZF
BAZF, also known as Bcl6b, is a five zinc finger BTB protein with high similarity to Bcl6. In addition to sharing significant sequence homology, both can recognize the same target DNA sequences (17). Within in the lymphocyte compartment, BAZF mRNA is detectable in CD4+ and CD8+ naïve T-cells (86). Interestingly, both Bcl6- and BAZF-deficient animals exhibit aberrant hematopoietic progenitor cell proliferation, a defect that was linked to a cell-intrinsic requirement for BAZF in CD8+ T-cells (87). In addition to naïve T-cells, BAZF is expressed at high levels in some antigen-specific, memory CD8+ T-cells and, although BAZF-deficient mice show normal CD8+ T-cell activation in response to primary viral infection, recall responses by memory CD8+ T-cells are greatly impaired in vitro and in vivo (88).
In contrast to its role in memory CD8+ T-cells, naïve CD4+ T cells require BAZF expression for maximal TCR-induced proliferation in vitro, whereas memory CD4+ T-cells do not (86). Moreover, while naïve CD4+ T-cells from mice that ectopically express BAZF are hyper-proliferative in response to TCR stimulation, memory CD4+ T-cells from these mice behave normally.
MAZR
Compared to the BTB-ZF proteins detailed above, relatively little is known about the role of MAZR in lymphocyte and development. MAZR is expressed in thymocytes and B-cells and, in the latter, activated c-Myc in overexpression studies (89). In double negative thymocytes, MAZR has been postulated to function as a transcriptional repressor of the CD8 locus and its downregulation is required, in part, for CD8 expression at the transition to the double positive stage (90). In double positive thymocytes, ectopically expressed MAZR could bind enhancer elements in the CD8 gene and suppress CD8 expression (90). Recent studies, however, show that MAZR-deficiency is insufficient to allow CD8 expression in double negative thymocytes. Instead, development of single positive CD8+ T-cells was impaired in these animals and MHC class I-restricted thymocytes were redirected into the CD4+ lineage, leading to an increased CD4+ to CD8+ T-cell ratio among mature thymocytes and peripheral T-cells. This phenotype was linked to MAZR-mediated repression of the Th-POK gene in double positive thymocytes (91).
LRF
Leukemia/lymphoma Related Factor (LRF), also known as OCZF, Zbtb7a, FBI-1 and Pokemon, is a BTB-ZF transcriptional repressor and oncogene associated with malignancy in lymphomas and solid epithelial tumors, including breast cancer, and non-small cell lung carcinoma (92-94). LRF is expressed in a broad range of myeloid and lymphoid lineages, including most subsets of developing and peripheral B-cells (95). Conditional deletion of the LRF gene in HSCs in vivo led to a profound reduction in peripheral B-cells, consistent with a cell intrinsic requirement for LRF for progression past the prepro-B cell stage during bone marrow development (93, 95). Unexpectedly, these mice exhibited extrathymic T-cell development in the bone marrow. Additional experiments revealed that LRF was required in lymphoid progenitor cells in the bone marrow to suppress Notch-dependent T-cell lineage commitment and allow B-cell development to progress. In the absence of LRF, Notch genes are aberrantly upregulated in hematopoietic progenitors, abrogating B-cell development and driving spontaneous T-cell development outside of the thymus (95). It remains to be determined whether Notch is a direct target of LRF or whether aberrant Notch signaling in these mice is an indirect consequence of impaired LRF-dependent signals.
MIZ-1
A critical role for Myc-interacting Zinc Finger Protein-1 (Miz-1) in lymphocyte development has recently been appreciated. In the past year, two separate studies using Miz1-defective mice revealed an essential function for Miz-1 in the development of both T- and B-lymphocytes (96, 97). Animals lacking the BTB domain of Miz-1 exhibit a profound reduction in the number of thymic early T-cell progenitor cells (ETPs). Moreover, T-cell development in these animals is blocked at the double negative to double positive transition causing a severe reduction in thymic cellularity, which was mirrored by specific reductions in the number of αβ and γδ T-cells. Transfer of Miz1-deficient hematopoietic progenitors into wild-type mice confirmed that the requirement for Miz-1 in T-cell development was cell intrinsic. Consistently, pro-T cells from these mice failed to differentiate in vitro as a result of increased apoptosis (96). In addition to the T-cell defect, Miz-1 deficiency leads to a complete loss of follicular B-cells, underscoring a parallel role for Miz-1 in the development of certain B-cell subsets (97). In both lymphocyte populations, Miz-1 was shown to be required downstream of IL-7R signaling in lymphocyte progenitor populations to promote STAT5 activation and expression of the anti-apoptotic protein, Bcl-2 (96, 97). In developing T-cells, these effects were mediated in part by MIZ-1-dependent repression of the STAT inhibitor, SOCS-1; consistent with this, binding of MIZ-1 to the SOCS-1 promoter could be detected in primary double negative thymocytes. A similar relationship between SOCS-1 and Miz-1 was observed in developing B-cells, though loss of Miz-1 also abrogated expression of two transcription factors, Tcf3 and Ebf1, critical for the survival and function of early B-cell progenitors (97).
Conclusions and Future Directions
New studies are required to further delineate the exact molecular mechanisms by which BTB-ZF proteins regulate gene expression in lymphocytes. This will require a more thorough investigation of the co-factors recruited by these proteins for the purpose of regulating target gene transcription, as well as the identity of the gene targets themselves. In addition, relatively little is known about the upstream signals that regulate the spatial/temporal function of these transcription factors in distinct lymphocyte lineages. In this vein, several studies have highlighted post-translational modification of BTB-ZF proteins as an important regulatory signal controlling their expression and function. For example, acetylation and sumoylation of PLZF may promote DNA binding and transcriptional repression (98-100), and acetylation of ThPok blocks ubiquitin-mediated degradation, thereby stabilizing its expression in CD4+ T-cells (101). In contrast, phosphorylation of PLZF and Bcl6, by CDK2 and ATM, respectively, has been shown to trigger ubiquitylation and degradation via proteasome-dependent pathways (102, 103).
From development to effector function, recent studies have highlighted a central and indispensible role for BTB-ZF transcriptions factors in controlling nearly every aspect of lymphocyte biology. Moreover, the fact that fewer than ten of the more than 45 factors in this protein family have been evaluated in this context suggests that future work is likely to uncover additional BTB-ZF proteins with roles in lymphocyte function, fate, and phenotype.
Acknowledgements
The authors are grateful for the engaging and challenging intellectual environment created by the members of the Sant'Angelo, Denzin and Chaudhuri laboratories.
Work related to this review was supported by NIAID R01s AI083988 and AI059739. A.M.B. is supported by T32 CA009149
References
- 1.Melnick A, Carlile G, Ahmad KF, Kiang CL, Corcoran C, Bardwell V, Prive GG, Licht JD. Critical residues within the BTB domain of PLZF and Bcl-6 modulate interaction with corepressors. Mol Cell Biol. 2002;22:1804–1818. doi: 10.1128/MCB.22.6.1804-1818.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chauchereau A, Mathieu M, de Saintignon J, Ferreira R, Pritchard LL, Mishal Z, Dejean A, Harel-Bellan A. HDAC4 mediates transcriptional repression by the acute promyelocytic leukaemia-associated protein PLZF. Oncogene. 2004;23:8777–8784. doi: 10.1038/sj.onc.1208128. [DOI] [PubMed] [Google Scholar]
- 3.Lemercier C, Brocard MP, Puvion-Dutilleul F, Kao HY, Albagli O, Khochbin S. Class II histone deacetylases are directly recruited by BCL6 transcriptional repressor. J Biol Chem. 2002;277:22045–22052. doi: 10.1074/jbc.M201736200. [DOI] [PubMed] [Google Scholar]
- 4.Tao RH, Kawate H, Wu Y, Ohnaka K, Ishizuka M, Inoue A, Hagiwara H, Takayanagi R. Testicular zinc finger protein recruits histone deacetylase 2 and suppresses the transactivation function and intranuclear foci formation of agonist-bound androgen receptor competitively with TIF2. Mol Cell Endocrinol. 2006;247:150–165. doi: 10.1016/j.mce.2005.12.052. [DOI] [PubMed] [Google Scholar]
- 5.Brenner C, Deplus R, Didelot C, Loriot A, Vire E, De Smet C, Gutierrez A, Danovi D, Bernard D, Boon T, Pelicci PG, Amati B, Kouzarides T, de Launoit Y, Di Croce L, Fuks F. Myc represses transcription through recruitment of DNA methyltransferase corepressor. EMBO J. 2005;24:336–346. doi: 10.1038/sj.emboj.7600509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barna M, Merghoub T, Costoya JA, Ruggero D, Branford M, Bergia A, Samori B, Pandolfi PP. Plzf mediates transcriptional repression of HoxD gene expression through chromatin remodeling. Dev Cell. 2002;3:499–510. doi: 10.1016/s1534-5807(02)00289-7. [DOI] [PubMed] [Google Scholar]
- 7.Jeon BN, Yoo JY, Choi WI, Lee CE, Yoon HG, Hur MW. Proto-oncogene FBI-1 (Pokemon/ZBTB7A) represses transcription of the tumor suppressor Rb gene via binding competition with Sp1 and recruitment of co-repressors. J Biol Chem. 2008;283:33199–33210. doi: 10.1074/jbc.M802935200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McLoughlin P, Ehler E, Carlile G, Licht JD, Schafer BW. The LIM-only protein DRAL/FHL2 interacts with and is a corepressor for the promyelocytic leukemia zinc finger protein. J Biol Chem. 2002;277:37045–37053. doi: 10.1074/jbc.M203336200. [DOI] [PubMed] [Google Scholar]
- 9.Laudes M, Bilkovski R, Oberhauser F, Droste A, Gomolka M, Leeser U, Udelhoven M, Krone W. Transcription factor FBI-1 acts as a dual regulator in adipogenesis by coordinated regulation of cyclin-A and E2F-4. J Mol Med. 2008;86:597–608. doi: 10.1007/s00109-008-0326-2. [DOI] [PubMed] [Google Scholar]
- 10.Costoya JA. Functional analysis of the role of POK transcriptional repressors. Brief Funct Genomic Proteomic. 2007;6:8–18. doi: 10.1093/bfgp/elm002. [DOI] [PubMed] [Google Scholar]
- 11.Ci W, Polo JM, Melnick A. B-cell lymphoma 6 and the molecular pathogenesis of diffuse large B-cell lymphoma. Curr Opin Hematol. 2008;15:381–390. doi: 10.1097/MOH.0b013e328302c7df. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Omori M, Yamashita M, Inami M, Ukai-Tadenuma M, Kimura M, Nigo Y, Hosokawa H, Hasegawa A, Taniguchi M, Nakayama T. CD8 T cell-specific downregulation of histone hyperacetylation and gene activation of the IL-4 gene locus by ROG, repressor of GATA. Immunity. 2003;19:281–294. doi: 10.1016/s1074-7613(03)00210-3. [DOI] [PubMed] [Google Scholar]
- 13.Dhordain P, Albagli O, Ansieau S, Koken MH, Deweindt C, Quief S, Lantoine D, Leutz A, Kerckaert JP, Leprince D. The BTB/POZ domain targets the LAZ3/BCL6 oncoprotein to nuclear dots and mediates homomerisation in vivo. Oncogene. 1995;11:2689–2697. [PubMed] [Google Scholar]
- 14.Davies JM, Hawe N, Kabarowski J, Huang QH, Zhu J, Brand NJ, Leprince D, Dhordain P, Cook M, Morriss-Kay G, Zelent A. Novel BTB/POZ domain zinc-finger protein, LRF, is a potential target of the LAZ-3/BCL-6 oncogene. Oncogene. 1999;18:365–375. doi: 10.1038/sj.onc.1202332. [DOI] [PubMed] [Google Scholar]
- 15.Korutla L, Wang P, Jackson TG, Mackler SA. NAC1, a POZ/BTB protein that functions as a corepressor. Neurochem Int. 2009;54:245–252. doi: 10.1016/j.neuint.2008.12.008. [DOI] [PubMed] [Google Scholar]
- 16.Dhordain P, Albagli O, Honore N, Guidez F, Lantoine D, Schmid M, The HD, Zelent A, Koken MH. Colocalization and heteromerization between the two human oncogene POZ/zinc finger proteins, LAZ3 (BCL6) and PLZF. Oncogene. 2000;19:6240–6250. doi: 10.1038/sj.onc.1203976. [DOI] [PubMed] [Google Scholar]
- 17.Okabe S, Fukuda T, Ishibashi K, Kojima S, Okada S, Hatano M, Ebara M, Saisho H, Tokuhisa T. BAZF, a novel Bcl6 homolog, functions as a transcriptional repressor. Mol Cell Biol. 1998;18:4235–4244. doi: 10.1128/mcb.18.7.4235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Phan RT, Saito M, Basso K, Niu H, Dalla-Favera R. BCL6 interacts with the transcription factor Miz-1 to suppress the cyclin-dependent kinase inhibitor p21 and cell cycle arrest in germinal center B cells. Nat Immunol. 2005;6:1054–1060. doi: 10.1038/ni1245. [DOI] [PubMed] [Google Scholar]
- 19.Hoatlin ME, Zhi Y, Ball H, Silvey K, Melnick A, Stone S, Arai S, Hawe N, Owen G, Zelent A, Licht JD. A novel BTB/POZ transcriptional repressor protein interacts with the Fanconi anemia group C protein and PLZF. Blood. 1999;94:3737–3747. [PubMed] [Google Scholar]
- 20.Melnick A, Ahmad KF, Arai S, Polinger A, Ball H, Borden KL, Carlile GW, Prive GG, Licht JD. In-depth mutational analysis of the promyelocytic leukemia zinc finger BTB/POZ domain reveals motifs and residues required for biological and transcriptional functions. Mol Cell Biol. 2000;20:6550–6567. doi: 10.1128/mcb.20.17.6550-6567.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kelly KF, Daniel JM. POZ for effect--POZ-ZF transcription factors in cancer and development. Trends Cell Biol. 2006;16:578–587. doi: 10.1016/j.tcb.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 22.Kawamata N, Miki T, Fukuda T, Hirosawa S, Aoki N. The organization of the BCL6 gene. Leukemia. 1994;8:1327–1330. [PubMed] [Google Scholar]
- 23.Cattoretti G, Pasqualucci L, Ballon G, Tam W, Nandula SV, Shen Q, Mo T, Murty VV, Dalla-Favera R. Deregulated BCL6 expression recapitulates the pathogenesis of human diffuse large B cell lymphomas in mice. Cancer Cell. 2005;7:445–455. doi: 10.1016/j.ccr.2005.03.037. [DOI] [PubMed] [Google Scholar]
- 24.Ranuncolo SM, Polo JM, Melnick A. BCL6 represses CHEK1 and suppresses DNA damage pathways in normal and malignant B-cells. Blood Cells Mol Dis. 2008;41:95–99. doi: 10.1016/j.bcmd.2008.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ranuncolo SM, Wang L, Polo JM, Dell'Oso T, Dierov J, Gaymes TJ, Rassool F, Carroll M, Melnick A. BCL6-mediated attenuation of DNA damage sensing triggers growth arrest and senescence through a p53-dependent pathway in a cell context-dependent manner. J Biol Chem. 2008;283:22565–22572. doi: 10.1074/jbc.M803490200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ranuncolo SM, Polo JM, Dierov J, Singer M, Kuo T, Greally J, Green R, Carroll M, Melnick A. Bcl-6 mediates the germinal center B cell phenotype and lymphomagenesis through transcriptional repression of the DNA-damage sensor ATR. Nat Immunol. 2007;8:705–714. doi: 10.1038/ni1478. [DOI] [PubMed] [Google Scholar]
- 27.Dent AL, Shaffer AL, Yu X, Allman D, Staudt LM. Control of inflammation, cytokine expression, and germinal center formation by BCL-6. Science. 1997;276:589–592. doi: 10.1126/science.276.5312.589. [DOI] [PubMed] [Google Scholar]
- 28.Ye BH, Cattoretti G, Shen Q, Zhang J, Hawe N, de Waard R, Leung C, Nouri-Shirazi M, Orazi A, Chaganti RS, Rothman P, Stall AM, Pandolfi PP, Dalla-Favera R. The BCL-6 proto-oncogene controls germinal-centre formation and Th2-type inflammation. Nat Genet. 1997;16:161–170. doi: 10.1038/ng0697-161. [DOI] [PubMed] [Google Scholar]
- 29.Fukuda T, Yoshida T, Okada S, Hatano M, Miki T, Ishibashi K, Okabe S, Koseki H, Hirosawa S, Taniguchi M, Miyasaka N, Tokuhisa T. Disruption of the Bcl6 gene results in an impaired germinal center formation. J Exp Med. 1997;186:439–448. doi: 10.1084/jem.186.3.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Toyama H, Okada S, Hatano M, Takahashi Y, Takeda N, Ichii H, Takemori T, Kuroda Y, Tokuhisa T. Memory B cells without somatic hypermutation are generated from Bcl6-deficient B cells. Immunity. 2002;17:329–339. doi: 10.1016/s1074-7613(02)00387-4. [DOI] [PubMed] [Google Scholar]
- 31.Tunyaplin C, Shaffer AL, Angelin-Duclos CD, Yu X, Staudt LM, Calame KL. Direct repression of prdm1 by Bcl-6 inhibits plasmacytic differentiation. J Immunol. 2004;173:1158–1165. doi: 10.4049/jimmunol.173.2.1158. [DOI] [PubMed] [Google Scholar]
- 32.Shaffer AL, Yu X, He Y, Boldrick J, Chan EP, Staudt LM. BCL-6 represses genes that function in lymphocyte differentiation, inflammation, and cell cycle control. Immunity. 2000;13:199–212. doi: 10.1016/s1074-7613(00)00020-0. [DOI] [PubMed] [Google Scholar]
- 33.Crotty S, Johnston RJ, Schoenberger SP. Effectors and memories: Bcl-6 and Blimp-1 in T and B lymphocyte differentiation. Nat Immunol. 2010;11:114–120. doi: 10.1038/ni.1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Duy C, Yu JJ, Nahar R, Swaminathan S, Kweon SM, Polo JM, Valls E, Klemm L, Shojaee S, Cerchietti L, Schuh W, Jack HM, Hurtz C, Ramezani-Rad P, Herzog S, Jumaa H, Koeffler HP, de Alboran IM, Melnick AM, Ye BH, Muschen M. BCL6 is critical for the development of a diverse primary B cell repertoire. J Exp Med. 2010;207:1209–1221. doi: 10.1084/jem.20091299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hartatik T, Okada S, Okabe S, Arima M, Hatano M, Tokuhisa T. Binding of BAZF and Bc16 to STAT6-binding DNA sequences. Biochem Biophys Res Commun. 2001;284:26–32. doi: 10.1006/bbrc.2001.4931. [DOI] [PubMed] [Google Scholar]
- 36.Arima M, Toyama H, Ichii H, Kojima S, Okada S, Hatano M, Cheng G, Kubo M, Fukuda T, Tokuhisa T. A putative silencer element in the IL-5 gene recognized by Bcl6. J Immunol. 2002;169:829–836. doi: 10.4049/jimmunol.169.2.829. [DOI] [PubMed] [Google Scholar]
- 37.Yu D, Rao S, Tsai LM, Lee SK, He Y, Sutcliffe EL, Srivastava M, Linterman M, Zheng L, Simpson N, Ellyard JI, Parish IA, Ma CS, Li QJ, Parish CR, Mackay CR, Vinuesa CG. The transcriptional repressor Bcl-6 directs T follicular helper cell lineage commitment. Immunity. 2009;31:457–468. doi: 10.1016/j.immuni.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 38.Nurieva RI, Chung Y, Martinez GJ, Yang XO, Tanaka S, Matskevitch TD, Wang YH, Dong C. Bcl6 mediates the development of T follicular helper cells. Science. 2009;325:1001–1005. doi: 10.1126/science.1176676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Johnston RJ, Poholek AC, DiToro D, Yusuf I, Eto D, Barnett B, Dent AL, Craft J, Crotty S. Bcl6 and Blimp-1 are reciprocal and antagonistic regulators of T follicular helper cell differentiation. Science. 2009;325:1006–1010. doi: 10.1126/science.1175870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Poholek AC, Hansen K, Hernandez SG, Eto D, Chandele A, Weinstein JS, Dong X, Odegard JM, Kaech SM, Dent AL, Crotty S, Craft J. In vivo regulation of Bcl6 and T follicular helper cell development. J Immunol. 2010;185:313–326. doi: 10.4049/jimmunol.0904023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ichii H, Sakamoto A, Kuroda Y, Tokuhisa T. Bcl6 acts as an amplifier for the generation and proliferative capacity of central memory CD8+ T cells. J Immunol. 2004;173:883–891. doi: 10.4049/jimmunol.173.2.883. [DOI] [PubMed] [Google Scholar]
- 42.Ichii H, Sakamoto A, Arima M, Hatano M, Kuroda Y, Tokuhisa T. Bcl6 is essential for the generation of long-term memory CD4+ T cells. Int Immunol. 2007;19:427–433. doi: 10.1093/intimm/dxm007. [DOI] [PubMed] [Google Scholar]
- 43.Chang CC, Vlad G, D'Agati VD, Liu Z, Zhang QY, Witkowski P, Torkamani AA, Stokes MB, Ho EK, Cortesini R, Suciu-Foca N. BCL6 is required for differentiation of Ig-like transcript 3-Fc-induced CD8+ T suppressor cells. J Immunol. 2010;185:5714–5722. doi: 10.4049/jimmunol.1001732. [DOI] [PubMed] [Google Scholar]
- 44.Zelent A, Guidez F, Melnick A, Waxman S, Licht JD. Translocations of the RARalpha gene in acute promyelocytic leukemia. Oncogene. 2001;20:7186–7203. doi: 10.1038/sj.onc.1204766. [DOI] [PubMed] [Google Scholar]
- 45.Good KL, Tangye SG. Decreased expression of Kruppel-like factors in memory B cells induces the rapid response typical of secondary antibody responses. Proc Natl Acad Sci U S A. 2007;104:13420–13425. doi: 10.1073/pnas.0703872104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Joko T, Nanba D, Shiba F, Miyata K, Shiraishi A, Ohashi Y, Higashiyama S. Effects of promyelocytic leukemia zinc finger protein on the proliferation of cultured human corneal endothelial cells. Mol Vis. 2007;13:649–658. [PMC free article] [PubMed] [Google Scholar]
- 47.Shiraishi A, Joko T, Higashiyama S, Ohashi Y. Role of promyelocytic leukemia zinc finger protein in proliferation of cultured human corneal endothelial cells. Cornea. 2007;26:S55–58. doi: 10.1097/ICO.0b013e31812f6b67. [DOI] [PubMed] [Google Scholar]
- 48.Kikugawa T, Kinugasa Y, Shiraishi K, Nanba D, Nakashiro K, Tanji N, Yokoyama M, Higashiyama S. PLZF regulates Pbx1 transcription and Pbx1-HoxC8 complex leads to androgen-independent prostate cancer proliferation. Prostate. 2006;66:1092–1099. doi: 10.1002/pros.20443. [DOI] [PubMed] [Google Scholar]
- 49.Felicetti F, Errico MC, Bottero L, Segnalini P, Stoppacciaro A, Biffoni M, Felli N, Mattia G, Petrini M, Colombo MP, Peschle C, Care A. The promyelocytic leukemia zinc finger-microRNA-221/-222 pathway controls melanoma progression through multiple oncogenic mechanisms. Cancer Res. 2008;68:2745–2754. doi: 10.1158/0008-5472.CAN-07-2538. [DOI] [PubMed] [Google Scholar]
- 50.Shiraishi K, Yamasaki K, Nanba D, Inoue H, Hanakawa Y, Shirakata Y, Hashimoto K, Higashiyama S. Pre-B-cell leukemia transcription factor 1 is a major target of promyelocytic leukemia zinc-finger-mediated melanoma cell growth suppression. Oncogene. 2007;26:339–348. doi: 10.1038/sj.onc.1209800. [DOI] [PubMed] [Google Scholar]
- 51.Bernardo MV, Yelo E, Gimeno L, Campillo JA, Parrado A. Identification of apoptosis-related PLZF target genes. Biochem Biophys Res Commun. 2007;359:317–322. doi: 10.1016/j.bbrc.2007.05.085. [DOI] [PubMed] [Google Scholar]
- 52.Savage AK, Constantinides MG, Han J, Picard D, Martin E, Li B, Lantz O, Bendelac A. The transcription factor PLZF directs the effector program of the NKT cell lineage. Immunity. 2008;29:391–403. doi: 10.1016/j.immuni.2008.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kovalovsky D, Uche OU, Eladad S, Hobbs RM, Yi W, Alonzo E, Chua K, Eidson M, Kim HJ, Im JS, Pandolfi PP, Sant'Angelo DB. The BTB-zinc finger transcriptional regulator PLZF controls the development of invariant natural killer T cell effector functions. Nat Immunol. 2008;9:1055–1064. doi: 10.1038/ni.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Raberger J, Schebesta A, Sakaguchi S, Boucheron N, Blomberg KE, Berglof A, Kolbe T, Smith CI, Rulicke T, Ellmeier W. The transcriptional regulator PLZF induces the development of CD44 high memory phenotype T cells. Proc Natl Acad Sci U S A. 2008;105:17919–17924. doi: 10.1073/pnas.0805733105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kovalovsky D, Alonzo ES, Uche OU, Eidson M, Nichols KE, Sant'Angelo DB. PLZF induces the spontaneous acquisition of memory/effector functions in T cells independently of NKT cell-related signals. J Immunol. 2010;184:6746–6755. doi: 10.4049/jimmunol.1000776. [DOI] [PubMed] [Google Scholar]
- 56.Kreslavsky T, Savage AK, Hobbs R, Gounari F, Bronson R, Pereira P, Pandolfi PP, Bendelac A, von Boehmer H. TCR-inducible PLZF transcription factor required for innate phenotype of a subset of gammadelta T cells with restricted TCR diversity. Proc Natl Acad Sci U S A. 2009;106:12453–12458. doi: 10.1073/pnas.0903895106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Alonzo ES, Gottschalk RA, Das J, Egawa T, Hobbs RM, Pandolfi PP, Pereira P, Nichols KE, Koretzky GA, Jordan MS, Sant'Angelo DB. Development of promyelocytic zinc finger and ThPOK-expressing innate gamma delta T cells is controlled by strength of TCR signaling and Id3. J Immunol. 2010;184:1268–1279. doi: 10.4049/jimmunol.0903218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Verykokakis M, Boos MD, Bendelac A, Adams EJ, Pereira P, Kee BL. Inhibitor of DNA binding 3 limits development of murine slam-associated adaptor protein-dependent “innate” gammadelta T cells. PLoS One. 2010;5:e9303. doi: 10.1371/journal.pone.0009303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Felices M, Yin CC, Kosaka Y, Kang J, Berg LJ. Tec kinase Itk in gammadeltaT cells is pivotal for controlling IgE production in vivo. Proc Natl Acad Sci U S A. 2009;106:8308–8313. doi: 10.1073/pnas.0808459106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fukuyama T, Kasper LH, Boussouar F, Jeevan T, van Deursen J, Brindle PK. Histone acetyltransferase CBP is vital to demarcate conventional and innate CD8+ T-cell development. Mol Cell Biol. 2009;29:3894–3904. doi: 10.1128/MCB.01598-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Weinreich MA, Odumade OA, Jameson SC, Hogquist KA. T cells expressing the transcription factor PLZF regulate the development of memory-like CD8+ T cells. Nat Immunol. 2010;11:709–716. doi: 10.1038/ni.1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lee YJ, Jeon YK, Kang BH, Chung DH, Park CG, Shin HY, Jung KC, Park SH. Generation of PLZF+ CD4+ T cells via MHC class II- dependent thymocyte-thymocyte interaction is a physiological process in humans. J Exp Med. 2010;207:237–246. doi: 10.1084/jem.20091519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xu D, Holko M, Sadler AJ, Scott B, Higashiyama S, Berkofsky-Fessler W, McConnell MJ, Pandolfi PP, Licht JD, Williams BR. Promyelocytic leukemia zinc finger protein regulates interferon-mediated innate immunity. Immunity. 2009;30:802–816. doi: 10.1016/j.immuni.2009.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sun G, Liu X, Mercado P, Jenkinson SR, Kypriotou M, Feigenbaum L, Galera P, Bosselut R. The zinc finger protein cKrox directs CD4 lineage differentiation during intrathymic T cell positive selection. Nat Immunol. 2005;6:373–381. doi: 10.1038/ni1183. [DOI] [PubMed] [Google Scholar]
- 65.He X, Dave VP, Zhang Y, Hua X, Nicolas E, Xu W, Roe BA, Kappes DJ. The zinc finger transcription factor Th-POK regulates CD4 versus CD8 T-cell lineage commitment. Nature. 2005;433:826–833. doi: 10.1038/nature03338. [DOI] [PubMed] [Google Scholar]
- 66.Muroi S, Naoe Y, Miyamoto C, Akiyama K, Ikawa T, Masuda K, Kawamoto H, Taniuchi I. Cascading suppression of transcriptional silencers by ThPOK seals helper T cell fate. Nat Immunol. 2008;9:1113–1121. doi: 10.1038/ni.1650. [DOI] [PubMed] [Google Scholar]
- 67.Wang L, Wildt KF, Zhu J, Zhang X, Feigenbaum L, Tessarollo L, Paul WE, Fowlkes BJ, Bosselut R. Distinct functions for the transcription factors GATA-3 and ThPOK during intrathymic differentiation of CD4(+) T cells. Nat Immunol. 2008;9:1122–1130. doi: 10.1038/ni.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.van Hamburg JP, de Bruijn MJ, Ribeiro de Almeida C, Dingjan GM, Hendriks RW. Gene expression profiling in mice with enforced Gata3 expression reveals putative targets of Gata3 in double positive thymocytes. Mol Immunol. 2009;46:3251–3260. doi: 10.1016/j.molimm.2009.08.004. [DOI] [PubMed] [Google Scholar]
- 69.He X, Park K, Wang H, Zhang Y, Hua X, Li Y, Kappes DJ. CD4-CD8 lineage commitment is regulated by a silencer element at the ThPOK transcription-factor locus. Immunity. 2008;28:346–358. doi: 10.1016/j.immuni.2008.02.006. [DOI] [PubMed] [Google Scholar]
- 70.Cruz-Guilloty F, Pipkin ME, Djuretic IM, Levanon D, Lotem J, Lichtenheld MG, Groner Y, Rao A. Runx3 and T-box proteins cooperate to establish the transcriptional program of effector CTLs. J Exp Med. 2009;206:51–59. doi: 10.1084/jem.20081242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hu J, Qi Q, August A. Itk derived signals regulate the expression of Th-POK and controls the development of CD4 T cells. PLoS One. 2010;5:e8891. doi: 10.1371/journal.pone.0008891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Liu X, Taylor BJ, Sun G, Bosselut R. Analyzing expression of perforin, Runx3, and Thpok genes during positive selection reveals activation of CD8-differentiation programs by MHC II-signaled thymocytes. J Immunol. 2005;175:4465–4474. doi: 10.4049/jimmunol.175.7.4465. [DOI] [PubMed] [Google Scholar]
- 73.Setoguchi R, Tachibana M, Naoe Y, Muroi S, Akiyama K, Tezuka C, Okuda T, Taniuchi I. Repression of the transcription factor Th-POK by Runx complexes in cytotoxic T cell development. Science. 2008;319:822–825. doi: 10.1126/science.1151844. [DOI] [PubMed] [Google Scholar]
- 74.Egawa T, Littman DR. ThPOK acts late in specification of the helper T cell lineage and suppresses Runx-mediated commitment to the cytotoxic T cell lineage. Nat Immunol. 2008;9:1131–1139. doi: 10.1038/ni.1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Setoguchi R, Taniuchi I, Bevan MJ. ThPOK derepression is required for robust CD8 T cell responses to viral infection. J Immunol. 2009;183:4467–4474. doi: 10.4049/jimmunol.0901428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang L, Carr T, Xiong Y, Wildt KF, Zhu J, Feigenbaum L, Bendelac A, Bosselut R. The sequential activity of Gata3 and Thpok is required for the differentiation of CD1d-restricted CD4+ NKT cells. Eur J Immunol. 2010;40:2385–2390. doi: 10.1002/eji.201040534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Park K, He X, Lee HO, Hua X, Li Y, Wiest D, Kappes DJ. TCR-mediated ThPOK induction promotes development of mature (CD24-) gammadelta thymocytes. EMBO J. 2010;29:2329–2341. doi: 10.1038/emboj.2010.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Engel I, Hammond K, Sullivan BA, He X, Taniuchi I, Kappes D, Kronenberg M. Co-receptor choice by V alpha14i NKT cells is driven by Th-POK expression rather than avoidance of CD8-mediated negative selection. J Exp Med. 2010;207:1015–1029. doi: 10.1084/jem.20090557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Dai MS, Chevallier N, Stone S, Heinrich MC, McConnell M, Reuter T, Broxmeyer HE, Licht JD, Lu L, Hoatlin ME. The effects of the Fanconi anemia zinc finger (FAZF) on cell cycle, apoptosis, and proliferation are differentiation stage-specific. J Biol Chem. 2002;277:26327–26334. doi: 10.1074/jbc.M201834200. [DOI] [PubMed] [Google Scholar]
- 80.Piazza F, Costoya JA, Merghoub T, Hobbs RM, Pandolfi PP. Disruption of PLZP in mice leads to increased T-lymphocyte proliferation, cytokine production, and altered hematopoietic stem cell homeostasis. Mol Cell Biol. 2004;24:10456–10469. doi: 10.1128/MCB.24.23.10456-10469.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Miaw SC, Choi A, Yu E, Kishikawa H, Ho IC. ROG, repressor of GATA, regulates the expression of cytokine genes. Immunity. 2000;12:323–333. doi: 10.1016/s1074-7613(00)80185-5. [DOI] [PubMed] [Google Scholar]
- 82.Kang BY, Miaw SC, Ho IC. ROG negatively regulates T-cell activation but is dispensable for Th-cell differentiation. Mol Cell Biol. 2005;25:554–562. doi: 10.1128/MCB.25.2.554-562.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hirahara K, Yamashita M, Iwamura C, Shinoda K, Hasegawa A, Yoshizawa H, Koseki H, Gejyo F, Nakayama T. Repressor of GATA regulates TH2-driven allergic airway inflammation and airway hyperresponsiveness. J Allergy Clin Immunol. 2008;122:512–520. e511. doi: 10.1016/j.jaci.2008.06.004. [DOI] [PubMed] [Google Scholar]
- 84.Hirasaki Y, Iwamura C, Yamashita M, Ito T, Kitajima M, Shinoda K, Namiki T, Terasawa K, Nakayama T. Repressor of GATA negatively regulates murine contact hypersensitivity through the inhibition of type-2 allergic responses. Clin Immunol. 2011 doi: 10.1016/j.clim.2011.02.009. [DOI] [PubMed] [Google Scholar]
- 85.Hernandez-Hoyos G, Anderson MK, Wang C, Rothenberg EV, Alberola-Ila J. GATA-3 expression is controlled by TCR signals and regulates CD4/CD8 differentiation. Immunity. 2003;19:83–94. doi: 10.1016/s1074-7613(03)00176-6. [DOI] [PubMed] [Google Scholar]
- 86.Takamori M, Hatano M, Arima M, Sakamoto A, Fujimura L, Hartatik T, Kuriyama T, Tokuhisa T. BAZF is required for activation of naive CD4 T cells by TCR triggering. Int Immunol. 2004;16:1439–1449. doi: 10.1093/intimm/dxh144. [DOI] [PubMed] [Google Scholar]
- 87.Broxmeyer HE, Sehra S, Cooper S, Toney LM, Kusam S, Aloor JJ, Marchal CC, Dinauer MC, Dent AL. Aberrant regulation of hematopoiesis by T cells in BAZF-deficient mice. Mol Cell Biol. 2007;27:5275–5285. doi: 10.1128/MCB.01967-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Manders PM, Hunter PJ, Telaranta AI, Carr JM, Marshall JL, Carrasco M, Murakami Y, Palmowski MJ, Cerundolo V, Kaech SM, Ahmed R, Fearon DT. BCL6b mediates the enhanced magnitude of the secondary response of memory CD8+ T lymphocytes. Proc Natl Acad Sci U S A. 2005;102:7418–7425. doi: 10.1073/pnas.0501585102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kobayashi A, Yamagiwa H, Hoshino H, Muto A, Sato K, Morita M, Hayashi N, Yamamoto M, Igarashi K. A combinatorial code for gene expression generated by transcription factor Bach2 and MAZR (MAZ-related factor) through the BTB/POZ domain. Mol Cell Biol. 2000;20:1733–1746. doi: 10.1128/mcb.20.5.1733-1746.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bilic I, Koesters C, Unger B, Sekimata M, Hertweck A, Maschek R, Wilson CB, Ellmeier W. Negative regulation of CD8 expression via Cd8 enhancer-mediated recruitment of the zinc finger protein MAZR. Nat Immunol. 2006;7:392–400. doi: 10.1038/ni1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sakaguchi S, Hombauer M, Bilic I, Naoe Y, Schebesta A, Taniuchi I, Ellmeier W. The zinc-finger protein MAZR is part of the transcription factor network that controls the CD4 versus CD8 lineage fate of double-positive thymocytes. Nat Immunol. 2010;11:442–448. doi: 10.1038/ni.1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhao ZH, Wang SF, Yu L, Wang J, Chang H, Yan WL, Zhang J, Fu K. Overexpression of Pokemon in non-small cell lung cancer and foreshowing tumor biological behavior as well as clinical results. Lung Cancer. 2008;62:113–119. doi: 10.1016/j.lungcan.2008.02.014. [DOI] [PubMed] [Google Scholar]
- 93.Maeda T, Hobbs RM, Merghoub T, Guernah I, Zelent A, Cordon-Cardo C, Teruya-Feldstein J, Pandolfi PP. Role of the proto-oncogene Pokemon in cellular transformation and ARF repression. Nature. 2005;433:278–285. doi: 10.1038/nature03203. [DOI] [PubMed] [Google Scholar]
- 94.Zu X, Ma J, Liu H, Liu F, Tan C, Yu L, Wang J, Xie Z, Cao D, Jiang Y. Pro-oncogene Pokemon promotes breast cancer progression by upregulating survivin expression. Breast Cancer Res. 2011;13:R26. doi: 10.1186/bcr2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Maeda T, Merghoub T, Hobbs RM, Dong L, Maeda M, Zakrzewski J, van den Brink MR, Zelent A, Shigematsu H, Akashi K, Teruya-Feldstein J, Cattoretti G, Pandolfi PP. Regulation of B versus T lymphoid lineage fate decision by the proto-oncogene LRF. Science. 2007;316:860–866. doi: 10.1126/science.1140881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Saba I, Kosan C, Vassen L, Moroy T. IL-7R-dependent survival and differentiation of early T-lineage progenitors is regulated by the BTB/POZ domain transcription factor Miz-1. Blood. 2011 doi: 10.1182/blood-2010-09-310680. [DOI] [PubMed] [Google Scholar]
- 97.Kosan C, Saba I, Godmann M, Herold S, Herkert B, Eilers M, Moroy T. Transcription factor miz-1 is required to regulate interleukin-7 receptor signaling at early commitment stages of B cell differentiation. Immunity. 2010;33:917–928. doi: 10.1016/j.immuni.2010.11.028. [DOI] [PubMed] [Google Scholar]
- 98.Guidez F, Howell L, Isalan M, Cebrat M, Alani RM, Ivins S, Hormaeche I, McConnell MJ, Pierce S, Cole PA, Licht J, Zelent A. Histone acetyltransferase activity of p300 is required for transcriptional repression by the promyelocytic leukemia zinc finger protein. Mol Cell Biol. 2005;25:5552–5566. doi: 10.1128/MCB.25.13.5552-5566.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kang SI, Choi HW, Kim IY. Redox-mediated modification of PLZF by SUMO-1 and ubiquitin. Biochem Biophys Res Commun. 2008;369:1209–1214. doi: 10.1016/j.bbrc.2008.03.037. [DOI] [PubMed] [Google Scholar]
- 100.Kang SI, Chang WJ, Cho SG, Kim IY. Modification of promyelocytic leukemia zinc finger protein (PLZF) by SUMO-1 conjugation regulates its transcriptional repressor activity. J Biol Chem. 2003;278:51479–51483. doi: 10.1074/jbc.M309237200. [DOI] [PubMed] [Google Scholar]
- 101.Zhang M, Zhang J, Rui J, Liu X. p300-mediated acetylation stabilizes the Th-inducing POK factor. J Immunol. 2010;185:3960–3969. doi: 10.4049/jimmunol.1001462. [DOI] [PubMed] [Google Scholar]
- 102.Costoya JA, Hobbs RM, Pandolfi PP. Cyclin-dependent kinase antagonizes promyelocytic leukemia zinc-finger through phosphorylation. Oncogene. 2008;27:3789–3796. doi: 10.1038/onc.2008.7. [DOI] [PubMed] [Google Scholar]
- 103.Phan RT, Saito M, Kitagawa Y, Means AR, Dalla-Favera R. Genotoxic stress regulates expression of the proto-oncogene Bcl6 in germinal center B cells. Nat Immunol. 2007;8:1132–1139. doi: 10.1038/ni1508. [DOI] [PubMed] [Google Scholar]