Summary
Evidence from multiple avenues of pathogen recognition is accumulating that the mitochondria form an integral platform from which innate signaling takes place. Recent studies revealed that the mitochondria are shaping the innate response to intracellular pathogens, and mitochondrial function is modulating and being modulated by innate immune signaling. Further, cell biological analyses have uncovered the dynamic relocalization of key components involved in cytosolic viral recognition and signaling to the mitochondria, and the mobilization of mitochondria to the sites of viral replication. In this review, we provide an integrated view of how cellular stress and signals following cytosolic viral recognition are intimately linked and coordinated at the mitochondria. We incorporate recent findings into our current understanding of the role of mitochondrial function in antiviral immunity and suggest the existence of a ‘mitoxosome’, a mitochondrial oxidative signalosome where multiple pathways of viral recognition and cellular stress signals converge on the surface of the mitochondria to facilitate a coordinated antiviral response.
Keywords: viral, signaling proteins, Toll-like receptors/pattern recognition receptors, apoptosis/autophagy, inflammation
Introduction
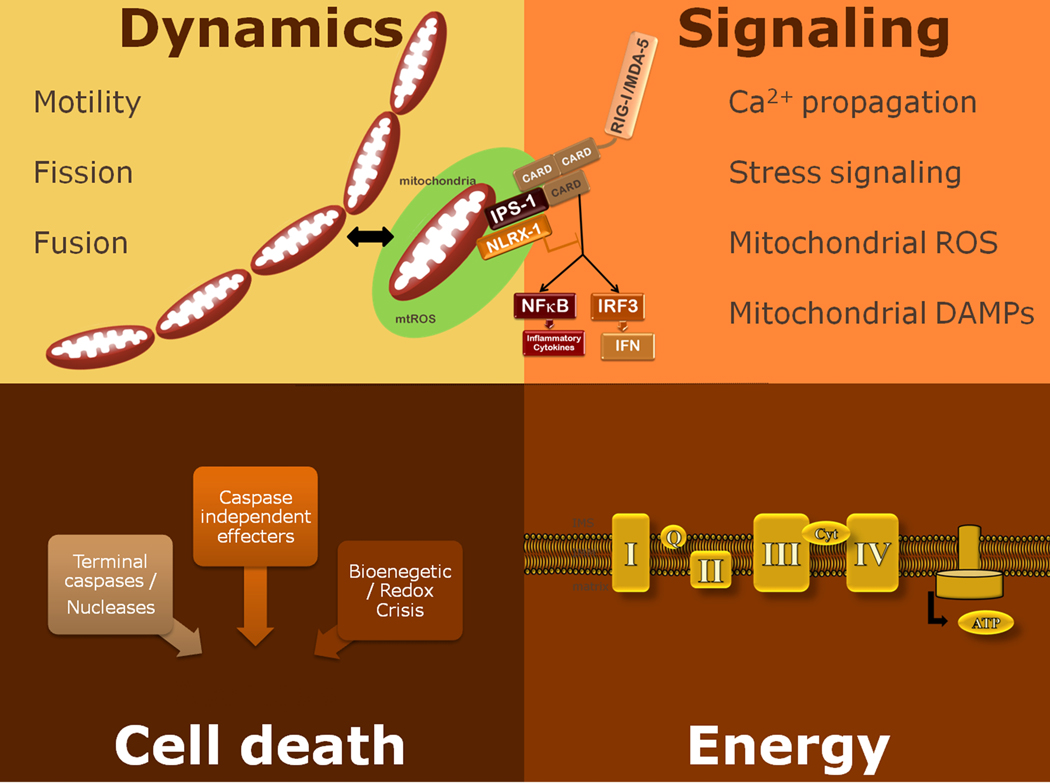
It is becoming rapidly apparent that the mitochondria serve as a crossroads between viral recognition and signaling, apoptosis, and a dysregulated state that can lead to various human diseases. This review will examine our current understanding of how viral sensing is affected by cellular stress with an emphasis on how the mitochondria may be integrating these signals to impact multiple steps within the viral recognition cascade (Fig. 1). Research in the 20th century has revealed that in the process of generating adenosine triphosphate (ATP), mitochondria produce reactive oxygen species (ROS) as a byproduct. ROS function to potentiate cell signaling, possess microbicidal functions and under pathological conditions cause internal damage to the host cells. Mitochondria have the capacity to sense the energy status of the cell as well as a variety of cellular stresses and can respond to dire situations with the initiation of cell death. Interest in the role of mitochondria in antiviral defense was sparked by identification of viral proteins that targeted the mitochondria, and towards the end of the century rapid advancements were being made on the interplay between viruses and apoptosis. It was also known that viruses could both induce ROS and be affected by ROS, and that there are high energy demands of viral replication. As we entered into the 21st century, identification of the mechanisms of viral recognition within an infected cell has uncovered molecular links between virus recognition and mitochondrial functions. In this review, we examine recent evidence that reveal the mitochondria as an integral platform whereby antiviral defense initiation pathways are coordinated with sensing of cellular stress.
Fig. 1. The mitochondria as a crossroads.
Mitochondria are dynamic organelles that have the capacity to sense the energy status of the cell as well as a variety of cellular stresses and the power to respond to dire situations with the initiation of cell death. Mitochondria power the cell processes by producing energy in the form of ATP through oxidative phosphorylation, and the reactive oxygen species generated as a byproduct of mitochondrial respiration are potent signaling molecules that can modulate protein functions.
Innate viral recognition modules
Cells utilize innate immune sensors to constantly monitor for indications of foreign or damage-causing agents. Distinct sets of pattern recognition receptors (PRRs), which detect conserved microbial components termed pathogen-associated molecular patterns (PAMPs), are used by most cells of the body to recognize virus infection. There are broadly two types of recognition mechanisms for viruses. First, specialized viral sentinel cells such as the plasmacytoid dendritic cells (pDCs) recognize viral presence without needing to be infected themselves through endosomal Toll-like receptors (TLRs). The endosomal TLRs recognize viral nucleic acids as signatures of infection (1). Second, cell-intrinsic recognition occurs in virally infected cells, whereby cytosolic viral PRRs that detect either the presence of viral nucleic acids or viral-inflicted damage are engaged. Once stimulated, viral PRRs induce the expression of potent antiviral factors, type I interferons (IFNs), which are capable of limiting the replication and spread of viruses, as well as inducing the proinflammatory cytokines and chemokines required for the initiation of innate and adaptive antiviral immunity (2). Recent evidence indicates that certain elements within these recognition pathways feed into one another. Here, we focus on the cell-intrinsic viral recognition modules and highlight intersections between these pathways, with an emphasis on the mitochondria as the convergence point.
Cytosolic RNA viral recognition
Within infected cells, signatures of viral nucleic acids activate the retinoic-acid-inducible gene I (RIG-I)-like receptor (RLR) family to mediate signaling that induces type I IFNs and cytokines. The RLR family consists of RIG-I [also known as DEAD box polypeptide 58 (DDX58)], which recognizes 5’ triphosphate RNA (3,4), present only in the viral genome of certain single-stranded RNA (ssRNA) viruses, and melanoma differentiation-associated gene 5 (MDA5), which recognizes the viral RNA of picornaviruses and synthetic dsRNA such as polyinosine-polycytidic acid [poly (I:C)] within the cytosol (5,6). A recent study indicates that MDA5 likely recognizes large dsRNA structures lacking 2’-O-methylation generated in the cytosol during virus infection (7). Activated RLRs bind to the adapter protein IFN-β promoter stimulator 1 (IPS-1) (8), also known as MAVS (9), Cardif (10), or VISA (11,12), through the caspase recruitment domain (CARD). IPS-1 activates two major signaling pathways: (i) activation of the IKK kinase complex and NF-κB leading to the induction of proinflammatory cytokines, and (ii) activation of IKK-related kinases TBK1 and IKKε, which activate IRF3 to induce expression of type I IFNs. Expression of RLRs in turn is highly enhanced by type I IFNs, thus ensuring efficient positive feedback regulation of RLR signaling. Another DEAD box helicase, DDX3, was shown to interact with IPS-1. However unlike the IFN-inducible RIG-I and MDA-5 PRRs, DDX3 is constitutively expressed in cells and has been suggested play a role in early ligand recognition when RIG-I and MDA-5 protein levels are very low (13, 14).
Tumor necrosis factor receptor-associated death domain (TRADD) was shown to interact with IPS-1 and induce NFκB and IRF3 activation (15). While recognition of viral nucleic acids by RIG-I and MDA-5 occurs in the cytosol, these molecules need to interact with the membrane-bound IPS-1 adapter protein found on either the mitochondria or the peroxisome to propagate antiviral signaling. The specific organelle localization of IPS-1 and its relevance in signaling is discussed in detail below. A protein named stimulator of interferon genes (STING) (16) was simultaneously identified by two additional groups that named it MITA (17) and MPYS (18), and both of these groups found this protein to be localized to the mitochondria. An additional group identified the protein and named it ERIS and found it to be localized to the endoplasmic reticulum (ER) and dimerize upon activation (19). STING was shown to be involved in RNA virus recognition as STING-deficient MEFs had a reduced type I IFN response to vesicular stomatitis virus (VSV) or Sendai virus. However, STING-deficient MEFs respond normally to poly (IC) and encephalomyocarditis virus (EMCV), suggesting that STING is downstream of RIG-I but not MDA5 (16). One of the oldest known ISGs, ISG56, has also been reported to interrupt IPS-1 and STING interaction, potentially as a mechanism of negative regulation (20).
Cytosolic DNA recognition
There are likely multiple sensors that recognize distinct types of DNA in the cytosol. Such DNAs include DNA from viruses, bacteria, apoptotic host cells, and synthetic B-form DNAs [particularly poly (dA:dT)] and interferon stimulatory DNA (ISD). ISDs are double-stranded DNA containing > 25 base pair oligonucleotides, which in a sequence independent manner, trigger stimulation of type I IFNs through a pathway involving TBK1, IRF3 but not IPS-1 (21). Interestingly, ISD does not engage NF-κB or MAPK pathways, thus activating only IRF3-induced pathways. ISD recognition pathway exists only in primary cells but is lost from transformed cells. The sensor for ISD still remains to be identified.
In contrast, B-form DNA, or poly(dA-dT)·poly(dT-dA) dsDNA, triggers type I IFNs via IRF3 and NF-κB, suggesting that this form of DNA triggers a separate sensor from ISD (22). Several sensor(s) for this type of DNA have been identified. First, a molecule called DAI (also known as DLM-1/ZBP1) was identified as an intracellular DNA sensing molecule (23). IFN induction following poly(dA-dT)·poly(dT-dA) DNA treatment was abrogated by a specific interfering RNA directed against DAI, suggesting that DAI is a critical cytoplasmic DNA sensor. However, DAI is not the sole sensor of DNA in the cytosol, as DAI-knockout mice still responded to B-DNA or plasmid DNA (24). Second, poly(dA-dT)·poly(dT-dA) DNA was identified to be recognized by RIG-I upon transcription by RNA polymerase III (Pol III) (25,26). Pol III transcribes dsRNA containing 5’ triphosphate using dsDNA as a template, generating a RIG-I agonist in the cytosol. Therefore, RIG-I is the sensor for cytosolic poly(dA-dT)·poly(dT-dA) dsDNA upon transcription by Pol III. Recently, the E3 protein encoded by vaccinia virus was found to inhibit the Pol III recognition pathway, indicating the importance of vaccinia virus recognition through Pol III/RIG-I pathway (27). Therefore the Pol III pathway links DNA recognition to IPS-1, which was originally thought to be involved specifically in RNA viral recognition. Third, in addition to the indirect sensing of poly(dA-dT)·poly(dT-dA) by RIG-I, IFI16, a PYHIN protein, can bind to poly(dA-dT)·poly(dT-dA), and knockdown of IFI16 expression led to reduction in IFN-β production (28).
It has been proposed that in addition to its role in RNA innate signaling pathways, STING is also an adapter protein downstream of cytosolic DNA recognition (16). While there remains controversy as to whether STING functions downstream or in parallel to IPS-1 in response to specific stimuli and whether STING can mitigate signaling through both IRF-3 and NF-κB, it is clear that STING requires TBK-1 to activate the IRF-3-dependent induction of type I IFNs. It has also been shown that an autophagy protein, ATG9a, but not other autophagy proteins, negatively regulates STING activation which requires translocation from the ER to the Golgi apparatus and finally to cytoplasmic punctuate structures to assemble with TBK-1 (29). Therefore, STING seems to plays a key role downstream of both RNA and DNA viral recognition through its dynamic movement within the cell.
NOD-like receptors
NOD-like receptors (NLRs) comprise a large family of intracellular PRRs that regulates innate immunity in response to recognition of various PAMPs and stress signals (30). Although the functions of many of the NLRPs are largely unknown, several NLRPs play a key role in the activation of caspase-1 by forming a multi-protein complex known as the ‘inflammasome’ (31). Caspase-1 is an essential mediator of inflammatory response through its capacity to cleave and generate active forms of IL-1β and IL-18, which are potent proinflammatory cytokines (32). In vitro studies have indicated that the formation and secretion of mature IL-1β and IL-18 require a two-step activation mechanism: first, transcriptional and translational upregulation of the pro-forms of these cytokines are induced by TLR signaling, and a second signal that leads to the proteolytic activation of caspase-1. The latter process is mediated by the inflammasomes. Inflammasome complexes that are important in antiviral defense are described below.
NLRP3-ASC inflammasome
NLRP3, also known as NALP3/Cryopyrin/CIAS1/PYPAF1 (33), forms an inflammasome with an adapter, ASC. NLRP3 inflammasomes require two signals for activation: signal 1 mediated by TLR and RLR stimulation leading to the expression of NLRP3 mRNA, and signal 2 mediated by damage signals such as membrane perturbation. The signal 2 of NLRP3 inflammasome can be activated by a variety of stimuli including endogenous signals from dying cells (uric acid), crystals (asbestos, silica, alum) as well as microbial signals such as whole bacteria, bacterial RNA, extracellular ATP, pore-forming toxins, or viral infections (30). It is unclear whether microbial ligands directly activate the NLRP3 inflammasome. Instead, the NLRP3 inflammasomes sense cellular stress such as disruption in membrane integrity and extracellular ATP released from stressed or damaged cells (34). Virus infection also results in the activation of inflammasomes. Sendai and influenza viruses activated the NLRP3 inflammasome in macrophages pulsed transiently with ATP (signal 2) in vitro (35). DNA viruses such as adenovirus stimulate the NLRP3-ASC-caspase-1 inflammasomes in vivo (36). However, inflammasomes are not activated by transfection of RNA, poly(I:C), or infection with reovirus (dsRNA virus) or VSV (-ssRNA virus)(37) indicating that RNA PAMPs are insufficient to trigger inflammasome activation. However, under some circumstance, RNA PAMPs are sufficient to trigger RIG-I-inflammasomes (see discussion below). Influenza virus induces both signal 1 and signal 2 to activate the NLRP3 inflammasome. Viral nucleic acid induces TLR7 activation (signal 1), while infection induces NRLP3 inflammasome through detection of ionic perturbation in the trans-Golgi network by M2 channel activity (signal 2) (38).
AIM2-ASC inflammasome
Not all inflammasomes are activated by NLRPs. Recent studies showed that AIM2 couples dsDNA recognition to ASC-dependent inflammasome (39–42). AIM2 is a HIN200 family of protein that contains a dsDNA binding domain (HIN domain) and the PYD domain, which promotes interaction with the PYD domain of ASC. AIM2 recognizes dsDNA in the cytosol and induces oligomerization of ASC and caspase-1, leading to activation of caspase-1 and cleavage of pro-forms of cytokines. Vaccinia virus, which contains a dsDNA genome, was shown to require AIM2 for recognition and activation of caspase1 inflammasomes (40). It is interesting to note that different classes of dsDNA viruses are recognized by NLRP3 (adenovirus) or AIM2 (vaccinia virus) for inflammasome activation. This likely depends on either the intracellular location within which the viral genome is accessible to the innate receptors or on the nature of PAMP being recognized.
NOD2 as a viral sensor
NOD2, originally characterized as a bacterial sensor, also has the capacity to facilitate recognition of ssRNA through IPS-1. It was found that upon ssRNA viral infection, viral RNA was found in NOD2 complexes, and NOD2 bound to IPS-1 to activate type I IFN production (43). NOD2 has also been implicated in enhancing the interferon-stimulated gene (ISG), RNAse-L activity in response to transfected poly(I:C), through direct interaction with a different ISG, oligoadenylate synthetases (OAS)(44). RNAse L not only has direct antiviral activity but also has the capacity to amplify RLR signaling. ATP cleavage by OAS activates RNAse L to cleave self RNA into small RNA fragments that can be recognized by RIG-I or MDA-5 to trigger IPS-1 activation and subsequent IFN production and ISG upregulation (45,46). Thus, NOD2 sensing of viruses leads not only to IPS-1-dependent type I IFN production but also likely to the amplification of RLR signaling. However, the relative importance of RLR vs. NOD2-dependent IPS-1 activation after a given virus infection is unclear.
Additional players in cytosolic viral PRR signaling
NLR family member X1 (NLRX1) localizes to the outer mitochondrial membrane and has been recently identified to bind to IPS-1 and negatively regulate its activity (47) as well as induce ROS through its mitochondrial localization (48). Both NLRX1 and NOD2 use their NBD (and LRR for NOD2) domains to facilitate their interaction with IPS-1(43, 47).
CARD9 is found to be involved in both RLR and NLR signaling. The protein CARD9 was shown to function as an adapter in RIG-I mediated proinflammatory cytokine responses. While CARD9 was dispensable for RIG-I mediated induction of type I IFNs through IPS-1, CARD9 was shown to be a component of RIG-I dependent IPS-1-mediated induction of proinflammatory cytokine production (49). Furthermore, RIG-I was shown to bind to ASC and form an inflammasome activating caspase-1, a process independent of CARD9 or IPS-1 (49). Both NOD2 and CARD9 have been reported to have a role in ROS upregulation in bacterial defense (50, 51). Interestingly, NOD2 and CARD9 have been shown to directly interact in detection of intracellular bacteria (52). NOD2 was found to mediate IPS-1 dependent IFN production in response to ssRNA viral ligands, while CARD9 mediates inflammatory cytokine induction by IPS-1, but not affect IFN induction. Therefore, it will be interesting to determine whether the activities of NOD2 and CARD9 are somehow interlinked to provide optimal signaling from intracellular viral PRRs.
Impact of mitochondrial oxidative stress on RLR signaling
Mitochondria turnover
There are two main sources of ROS within a cell: the mitochondrial electron transport chain (ETC), and the NADPH oxidases (NOXs). In non-phagocytic cells, mitochondrial respiration is thought to be the most significant source of ROS (53). These ROS are potent modifiers that can reversibly oxidize reactive cysteine residues within a protein to result in activation, inactivation, or multimerization of that protein. Oxidative modification can also induce dissociation of a regulatory protein resulting in the subsequent activation of its partner protein, as in the case with oxidized thioredoxin (Trx) and apoptosis signal-regulating kinase 1 (ASK-1)(54). Proteins susceptible to modification by ROS are termed redox regulated proteins. ROS also have a dark side as a destructive force within the cell, through their ability to oxidize nucleic acids, proteins, lipids, and carbohydrates, leading to the accumulation of damaged cellular constituents. However, the cell is equipped with a powerful network of antioxidants and scavenger proteins that can neutralize ROS. The mitochondrial manganese-superoxide dismutase (mnSOD) catalyzes the reduction of the superoxide anion O2− to H2O2 which is the longest lived form of ROS. H2O2 is further reduced to water through glutathione peroxidases or catalase (55). As mitochondria are constantly generating and are exposed to ROS, they are at an increased risk of succumbing to some of its damaging effects. Mitochondrial DNA is not protected by histone as is nuclear DNA, and mitochondrial DNA more rapidly fall prey to the mutagenic effect of ROS. Also, the mitochondrial DNA repair mechanisms are less advanced and less capable of repairing accumulating damage. The mitochondria form a dynamic interconnecting network that both affects and is affected by cellular stresses and can mediate the protective exchange of DNA between mitochondria (56). Mitochondria elongate and network together via a process known as mitochondrial fusion, and fragmentation of mitochondria is mediated by a process called mitochondrial fission. Mitochondria can also sustain damage to the critical components of the ETC that need to be rapidly degraded and replaced. This results in a critical need to maintain mitochondrial turnover within the cell and specific mechanisms for clearing damaged and potentially harmful mitochondria that will inherently accumulate over time.
Autophagy is a highly conserved process that maintains the integrity of long-lived intracellular components through the engulfment of cytosolic constituents to facilitate their subsequent degradation in the lysosomes. This process can be upregulated during starvation to provide additional nutrients and energy, and both starvation-induced and constitutive autophagy is considered to be nonspecific (57). However, there is also evidence for specific autophagy of damaged organelles, as seen for the autophagy of damaged mitochondria, coined ‘mitophagy’, and autophagy is the only major pathway for mitochondrial degradation. At least 30 autophagy-related gene (ATG) proteins have been identified as having an essential role in autophagy, including ATG5, ATG7, ATG12, and ATG16L1 (58). Deletion of these genes results in death of the neonate within 24 h of birth, and therefore mouse models such as conditional knockouts, tissue specific deletion, mouse embryonic fibroblast, or fetal derived liver cells are utilized in the investigation of autophagy. Autophagy has been shown to play an important role in immunity through involvement in antigen presentation, pathogen degradation, and delivery of cytosolic viral ligands to the endosomal compartment for stimulation of TLRs in plasmacytoid dendritic cells and modulation of IFN and proinflammatory signaling (reviewed in 59). However, lack of mitophagy leads to excess accumulation of ROS within the cell (60). Mitophagy of damaged mitochondria has been reported for depolarized mitochondria, which are rapidly sequestered into autophagic vesicles and degraded upon fusion with lysosomes (61). Recent studies have demonstrated that mitochondrial dynamics are important in facilitating selective mitophagy and maintaining the integrity of the mitochondrial components within the cell. Further, the mitochondria undergo fission, followed with a selective fusion of damaged components that specifically targeted the damaged mitochondria for autophagy (62). Considering the fact that cells undergo dramatic changes in redox status following viral infections, versatile antioxidant and ROS scavenging pathways, as well as quality control pathways such as mitophagy are critical for cellular homeostasis and innate antiviral signaling.
The inhibitor’s matchbox – common misconceptions about rotenone and DPI
It has long been known that ROS impacts NFκB signaling, and that NFκB can be redox regulated, but the specifics have been clouded in controversy. NOX enzymes play a critical role in bacterial killing by phagocytes, while mitochondrial ROS as a byproduct of mitochondrial respiration is the major source of ROS in most cell types. Therefore, much research has been aimed at understanding the source of ROS that is impacting different aspects of pathogen recognition. It is important to clarify some misconceptions as to the exact function of two common inhibitors that are utilized to manipulate ROS levels. Misuse of these inhibitors have resulted in underestimation of the role of mitochondrial ROS in specific on innate immune signaling. Rotenone is an inhibitor of complex 1 of the respiratory chain and has a dose dependent effect on ROS production in mitochondria. At very high doses, rotenone can almost completely inhibit complex 1 electron transfer and therefore block mitochondrial respiration. Such doses quickly become toxic and result in mitochondrial membrane permeabilization and cell death. However, at lower doses, rotenone only partially interferes with complex 1 resulting in increased mitochondrial superoxide production. The other inhibitor that has caused confusion is the use of diphenyleneiodonium (DPI), which inhibits flavin-containing enzymes and was first identified in the 1970s as an inhibitor of mitochondrial respiration but was then found to be more potent in inhibiting NOX enzymes and nitric oxide synthases (63). A careful examination of DPI function in regards to NOX mediated vs. mitochondrial ROS production was performed by Balua et al. (63) after they show in macrophages that LPS-dependent inflammatory cytokine production is not dependent on NOX1, 2, 3, or 4 by using macrophages containing a mutation in the p22phox subunit. They then utilized the differential potency of DPI to show that high dose DPI treatment blocks mitochondrial oxygen consumption as effectively as high dose rotenone. A dose titration showed a tight correlation between DPI’s reduction of mitochondrial ROS levels with inhibitory effect on inflammatory cytokine production. Low dose DPI, capable of inhibiting NOX enzymes but not mitochondrial ROS, did not significantly reduce inflammatory cytokine production (63). Therefore, it is imperative that titration of rotenone and DPI be performed to predetermine and use precisely the dose required to affect mitochondrial ROS levels in a given experimental system.
Hyperstimulation of RLR signaling in the absence of mitochondrial turnover
To understand the consequence of lack of mitochondrial turnover on the RLR signaling, we employed VSV infection (RIG-I stimulation) or poly (I:C) transfection (MDA-5 stimulation) in autophagy deficient cells. We observed increased mRNA and protein levels of IFN-α, IFN-β, and IL-6 in both ATG5-deficient mouse embryonic fibroblasts (MEFs) and macrophages in response to RLR stimulation. Consequently, ATG5-deficient MEFs were less susceptible to VSV infection than wildtype (WT) MEFs (64). Given that the key adapter protein of RLRs, IPS-1, is on the mitochondrial membrane, we hypothesized that autophagy could regulate RLR signaling either by maintenance of overall mitochondrial mass, mitochondrial integrity, or both. Indeed, we observed increased levels of mitochondrial mass (as measured by protein and mitochondrial DNA) in the absence of autophagy. Further, by staining the mitochondria with both a fluorescent stain that measures mitochondrial lipid mass (Mitotracker Green) and a stain indicative of mitochondrial membrane potential (Mitotracker Red), we detected many cells that have accumulated damaged mitochondria in the autophagy-deficient MEFs that were cleared in WT MEFs.
We then investigated two distinct possibilities that may explain how this accumulation of damaged mitochondria in the absence of autophagy could lead to enhanced RLR signaling. First, IPS-1 might accumulate along with the mitochondria, leading to amplification of both IFN and inflammatory cytokine signaling. Secondly, as mitochondria are the major producers of ROS as discussed above, the lack of clearance of damaged and potentially leaky mitochondria could provide a source of ROS within the cell, which could in turn affect both IFN and inflammatory cytokine signaling. We found evidence to support both of these possibilities. First, IPS-1 levels in ATG5-deficient MEFs and macrophages were elevated compared to WT cells due to mitochondrial accumulation. Overexpression of IPS-1 increased IFN production similarly in both WT and ATG5-deficient cells and failed to resolve the inherent difference in IFN production resulting from the absence of autophagy. Second, ROS levels were increased in ATG5-deficient cells in comparison to WT cells. While it was known that ROS plays a key role in NFκB-dependent inflammatory cytokine production (65), it was unknown whether ROS could affect IFN production. We demonstrated that using rotenone at very low levels, it was possible to increase mitochondria-associated ROS levels in WT cells, which resulted in increased IFN production that matched untreated ATG5 knockout (KO) levels. Further, rotenone-treated ATG5 KO cells produced even greater levels of IFN. Antioxidant treatment subsequently reduced IFN signaling in ATG5-deficient cells, demonstrating that increased ROS levels resulted in increased IFN induction, while reduced ROS levels decreased IFN induction (64).
A role for ROS produced by NOX2 was reported subsequently for the RIG-I mediated activation of IRF3. The authors utilize the epithelial cell line A549 stimulated with Sendai virus (SeV) or sheared poly (I:C) to examine the role of NOX2 in RIG-I-mediated signaling. NOX2 was required for basal and SeV-induced expression of IKKε and for TBK-1 activity as well as for IRF3 phosphorylation and dimerization (66). The authors proposed a role for NOX2 and ROS in stabilizing IPS-1 mRNA levels (66). As discussed above, NOD2 can directly interact with IPS-1 upon ssRNA stimulation (43). NOD2 has a NOX2-dependent mechanism of inducing ROS production upon bacterial stimulation (51), and therefore, these data indicate a possible link between NOD2-IPS-1-NOX2.
IPS-1 signaling platforms: beyond the mitochondria
In 2005 when IPS-1 was first characterized simultaneously by four independent groups, it was well accepted that IPS-1 requires localization to the mitochondria for its function. This requirement was illustrated through relocalization studies exchanging the IPS-1 transmembrane domain with that of proteins that localize to the plasma membrane (PM), the mitochondria and ER, or the ER exclusively. Targeting to the PM or ER exclusively resulted in decreased IPS-1-mediated IFNβ induction (9). It was also shown that viral suppression of RLR signaling inhibited antiviral signaling via cleavage of IPS-1 off of the mitochondria (67). The first indication that the transmembrane (TM) domain is required for an additional purpose came from a study that showed that IPS-1 functions as a dimer and that the TM domain is required for facilitating this IPS-1 dimerization (68). In the past two years, however, a new paradigm is arising in which IPS-1 is not only a mitochondrial protein, but that its signaling pathway is greatly affected by interactions with other cellular organelles.
IPS-1 is not exclusively localized to the mitochondria as previously thought but indeed has a second home on the peroxisome (69). The disparate function of IPS-1 on these two organelles was elucidated by utilizing distinct targeting motifs to localize IPS-1 to mitochondria exclusively, peroxisomes exclusively, both mitochondria and the peroxisomes, or to the cytosol. These experiments demonstrated that IPS-1 on the peroxisomes serves to provide the infected cell with immediate antiviral effector function through upregulation of interferon stimulated genes (ISGs) in an IFN-independent manner (69). This rapid process could provide the infected cell with the precious time that it takes for the mitochondrial IPS-1 to induce IFNs that can feedback on the cell to elicit ISG production and also warn neighboring cells of the invading virus and trigger an antiviral state. Might this subcompartmentalization of IPS-1 exploit differences in the kinetics of peroxisomal vs. mitochondrial motility in order to mediate an immediate effector response from the former? It is interesting to speculate whether IPS-1 might cycle between these organelles in a stimulus dependent manner similar to STING, which has been shown to translocate from the ER to the Golgi apparatus to punctate cytoplasmic structures upon stimulation (29).
Mitochondrial dynamics shape IPS-1 signaling
IPS-1 located on the mitochondrial membrane mediates the induction of proinflammatory cytokines via activation of the IKK kinase complex and NFκB, while it also mediates the induction of IFNs through the IKK-related kinases TBK1 and IKKε which activate IRF3. An intriguing story is currently unfolding in regards to how intertwined the process of cytosolic antiviral signaling is with mitochondrial dynamics. A first glimpse of this came with a study of how influenza A virus infection impacted the proteomics of the cytosolic and mitochondrial components of infected cells (70). This study found that actin and microtubule cytoskeleton components as well as heat shock proteins (HSP) such as HSP 90 become upregulated upon flu infection (70). Subsequently, it has been suggested that HSP 90 along with translocases of outer membrane 70 (TOM 70) can bridge IPS-1 with the downstream signaling molecules TBK1 and IRF3 (71). HSP90 has also been shown to play a role in NLR activation, where it was found to interact with NLRP3 NBD and LRR domains and mediate both NLRP3 and NOD2 dependent cytokine production (72). We speculate the existence of a redox regulated super-molecular complex on the mitochondria, or ‘mitoxosome’, where multiple pathways of viral recognition and cellular stress signals converge on the surface of the mitochondria to facilitate a coordinated antiviral response. The study by Ohman et al. (70) focused in particular on how RIG-I moves to the mitochondria upon viral stimulation, but did not explore the additional possibility suggested by their microscopy that it may be that the mitochondria are on the move. Indeed, their findings in regards to a role for actin dynamics in viral infection are particularly interesting in light of the fact that both the microtubule and actin cytoskeletons are important for mitochondrial motility and distribution throughout the cell. While it seems that microtubules are responsible for the bulk of mitochondrial movement, the actin cytoskeleton supports short distance movement of mitochondria and anchoring at intracellular sites (e.g. sites of high ATP utilization)(73). Interestingly, fluctuations in actin dynamics are involved in mitochondrial production of ROS, Ca2+ signaling, mitochondrial membrane potential, induction of the stress response, and apoptosis (74).
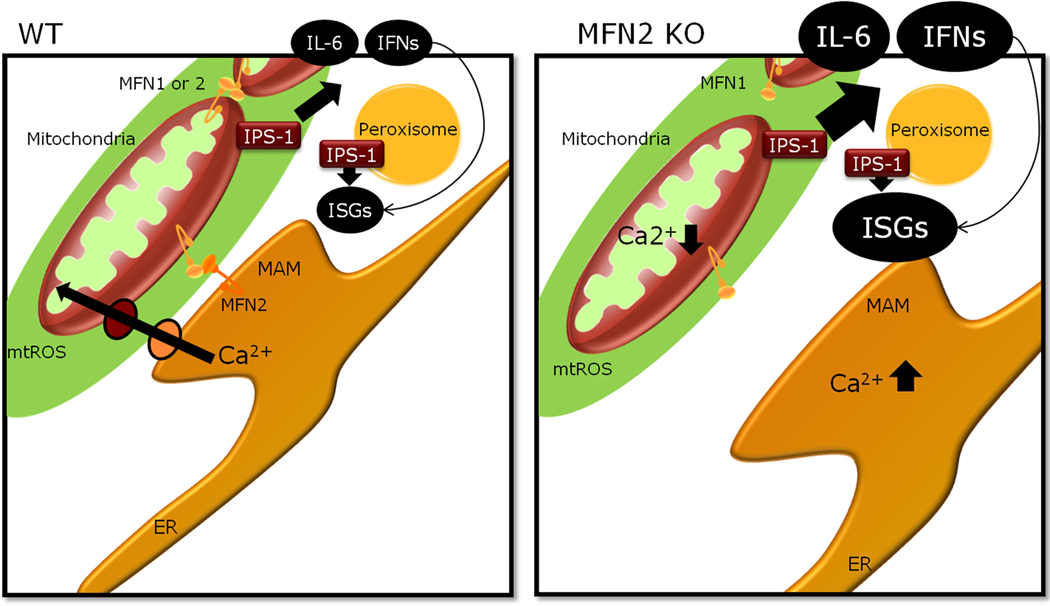
While mitochondria are often thought of as singular organelles, they continuously undergo cycles of fission and fusion, which impact mitochondrial bioenergetics, ATP production, apoptosis, and mitotophagy. At the level of individual mitochondria, the fission and fusion events facilitate the critical exchange of the metabolic, genetic, and proteomic contents (reviewed in 75). Examples of specific proteins that can mediate fission are dynamin-related protein 1 (Drp1) GTPase and fission 1 (Fis1), and examples of fusion mediating proteins are the transmembrane GTPases mitofusin 1 (MFN1) and mitofusin 2 (MFN2) as well as the optic atrophy 1 (Opa1) GTPase. One report demonstrated that IPS-1 stimulation is enhanced by mitochondrial elongation promoted by mitochondrial fusion proteins (Mfn1 and Opa1), while mitochondrial fragmentation mediated by mitochondrial fission proteins (Drp1 and Fis1) reduced IPS-1 signaling (76). Considering our current understanding that IPS-1 is localized to the mitochondria as well as to the peroxisome, it is important to mention that Drp1 and Fis1 have both been shown to mediate peroxisomal fission in addition to mitochondrial fission (77, 78). Castanier et al. (76) also reported that viral stimuli promote elongation of mitochondria and increased association between IPS-1 and STING at mitochondria-ER tethering sites. This seemed incongruous with the simultaneous report that the mitochondrial fusion protein MFN2 inhibits RLR signaling through direct interaction with IPS-1 (79). However, MFN1 and MFN2 do not have entirely redundant functions. There is ~60% homology between the MFN1 and MFN2 fusion proteins, with one of their defining differences being that MFN2 is required for mitochondria-ER tethering at the mitochondria-associated membrane (MAM) where the ER membrane is in close contact with mitochondria (80). This leads us to question the relative importance of mitochondria-ER tethering vs. mitochondrial fusion in the regulation of IPS-1 signaling. In the absence of MFN2, abnormal ER morphology is observed along with an increase in ER Ca2+ levels. While the reduction in ER-mitochondria proximity in the absence of MFN2 results in decreased mitochondrial uptake of Ca2+, the increased Ca2+ release upon discharge of the ER Ca2+ stores also sparks an increased mitochondrial Ca2+ peak (80). Whether Ca2+ levels in the ER or in the mitochondria regulate IPS-1 signaling is unknown. Additionally, it is important to assess the state of ER stress in the absence of MFN2. If the unfolded protein response (UPR) is initiated, that has the potential to modulate both IFN and inflammatory signaling (as discussed below). Also, ER stress can induce autophagy either through increased cytosolic Ca2+ levels and inhibition of mTOR through AMPK, or through phosphorylation of eIF2 which is a shared autophagy induction pathway between starvation, viral infection and the UPR (81).
Further evidence for a crucial role for MFN1 in propagating IPS-1 signaling came from a study by Onoguchi et al. (82), where the localization of IPS-1 after a series of different viral infections was examined. There was a striking change in IPS-1 localization pattern after virus infection. Because the images examined the pattern of IPS-1 at 9 or 12 hrs post infection, it is possible that the change in the IPS-1 localization reflects either the redistribution of IPS-1 or the degradation pattern of IPS-1. Most strikingly, electron microscopy analysis showed that IPS-1 proteins surrounded sites of Newcastle disease virus replication 9 h post infection, and this relocalization was shown to be both RIG-I and MFN1 dependent (82). The authors proposed a model whereby fragmentation, reorganization, and elongation of mitochondria allows for the redistribution of IPS-1 to sites of viral replication (82). This model is in line with the study by Castanier et al. (76), in promoting the idea that mitochondrial fusion enhances IPS-1 signaling, possibly by enabling redistribution towards virus replication centers within the cell.
Mitochondrial dynamics are clearly important for RLR signaling, yet discrepancies between the studies leave us asking if mitochondria elongate or fragment upon RLR signaling. Under normal physiological conditions, mitochondria are constantly cycling between fission and fusion events. Therefore, it may be that both are true, and the outcome is simply a matter of difference in assay time points. Additionally, since deletion of either MFN-1 or MFN-2 results in mitochondrial fragmentation and yet they have opposite phenotypes with respect to RLR signaling, mitochondrial fragmentation may not be detrimental to signaling. However, a recent study that examined the phenotype of MFN2-deficient MEFs in comparison to the MEFs doubly deficient for both MFN1 and MFN2 found that while MFN2 depletion increases RLR signaling, the double mutant has completely ablated RLR signaling in response to SeV infection (83). As the absence of MFN2 cannot increase RLR signaling in the additional absence of MFN1, this indicates that MFN1 or its function is required for RLR signaling. This also leads us to wonder if aside from fragmentation, if there is a differential affect of MFN1 vs. MFN2 on mitochondrial dysfunction (Fig. 2). For instance, as opposed to cells deficient in either MFN1 or MFN2, mitochondria from cells deficient for both MFN1 and MFN2 undergo severe mitochondrial DNA depletion (84). Mitochondria function and dynamics have a profound effect not only on virally induced RLR signaling but also on NLRP3 inflammasome signaling (as discussed below). The dynamic mitochondrial networks within the cell may create microenvironments that facilitate the necessary proximity to important signaling mediators found on or complexed to the mitochondria and to induce post-transcriptional modifications on such mediators through oxidation or multimerization. Evidence discussed in next section indicates that in addition, mitochondrial membrane potential is required to optimize such cross talk between the mitochondria and the immune signaling complexes.
Fig. 2. MFN2 negatively regulates IPS-1 signaling.
Mitochondrial dynamics affect IPS-1-dependent RLR-mediated production of IFNs and pro-inflammatory cytokines. Two proteins required for mitochondrial fusion, MFN1 and MFN2, have different roles in IPS-1 signaling. MFN2 is required for mitochondria-ER tethering and efficient mitochondrial uptake of Ca2+ from the ER. In the absence of MFN2, but not MFN1/MFN2 double ablation, IPS-1 dependent cytokine production is increased. We speculate that altered Ca2+ levels may play a role in IPS-1 signaling.
IPS-1 needs the mitochondrial potential to signal
It is becoming increasingly clear that there is a functional purpose for IPS-1 localization to the mitochondria to closely tie IPS-1 signaling to mitochondrial function. Using drugs that either depolarize or hyperpolarize the mitochondrial membrane, Koshiba et al. (83) show that there is a requirement for mitochondrial membrane potential in IPS-1 generated signaling but not downstream mediators of RLR signaling. This finding could prove to be of even broader importance in scope, considering that mitochondrial membrane potential is defined by the electrochemical gradient that forms along the mitochondrial membrane and is maintained by calcium uptake. The membrane potential is the driving force behind mitochondrial respiration and therefore is inherently integrated with both calcium uptake and mtROS production. Furthermore, overexpression of the mitochondrial protein, uncoupling protein-2 (UCP-2) to depolarize the membrane, greatly reduced IPS-1 dependent IFN production (83). This is intriguing, as UCP-2 has been shown to negatively regulate mitochondrial ROS production independent of NOX function, and UCP-2-deficient macrophages have increased proinflammatory cytokine production in response to Listeria infection (85).
Mitochondrial regulation of NLRP3 signaling
It is commonly believed that ROS is required for the activation of NLRP3 inflammasome (reviewed in 86, 87). However, the source of ROS mediating NLRP3 inflammasome as well as the mechanism by which ROS influence NLRP3 activation remains controversial. This is in part due to the fact that ROS scavengers, inhibitors, and inducers used in many studies are not specific and can also inhibit NF-κB activation as discussed above. Recent studied indicate that mitochondrial ROS plays an important role in supporting NLRP3 inflammasome activation. This has been accomplished on both fronts, with evidence that NADPH oxidases are not involved in producing the ROS required for the NLRP3 inflammasome, as well as reports that mitochondrial ROS is key in activating the NLRP3 inflammasome (88–90).
Two recent reports examined how the mitochondria activate the NLRP3 inflammasome, by examining increased levels of mitochondrial ROS such as seen during defective autophagy and mitochondrial DNA (mtDNA) release (89, 90). Absence of autophagy results in increased levels of mitochondrial ROS through cellular inability to clear damaged mitochondria (64). Absence of autophagy also results in strikingly increased levels of caspase 1 activation and IL-1β and IL-18 production in response to LPS, which was further propagated by IFN or ROS (91). An additional mechanism by which autophagy decreases IL-1β secretion was proposed by a recent study using inhibitors of autophagy showing colocalization of IL-1β and LC3 labeled autophagosomes by 24 h of LPS stimulation (92). This selective degradation of IL-1β has yet to be shown in a genetic model of autophagy. An additional level of complexity is that mitochondrial ROS is also involved in IL-1β cytokine induction upstream of the inflammasome (63, 93). At the transcriptional level, it has been shown that IL-1β mRNA levels are increased by mitochondrial ROS (93) but unaffected by autophagy deficiency through ATG16L1 deletion (in response to LPS) (91). ROS may also be involved in a negative feedback loop that later inactivates inflammasome-mediated cytokine secretion through redox modulation of caspase-1 (94).
Both studies confirm that the absence of autophagy results in the accumulation of damaged mitochondria and mitochondrial ROS and go on to show the impact of this on NLRP3 activation. Zhou et al. (90) demonstrate that in the absence of stimulation, NLRP3 localizes to the ER. Upon stimulation, both NLRP3 and its adapter protein ASC show a change in their localization pattern where they move to the MAM (sites of close interaction between ER and mitochondria). This finding is further providing evidence for a mitoxosome, where mitochondrial localization of cytosolic components may be crucial for signaling. NOD2 also localizes to the mitochondria upon stimulation with ssRNA and binding to IPS-1 is facilitated through the NBD and LRR domains (43). As NLRX1 utilizes its NBD domain to interact with IPS-1, it would be intriguing to know if the NBD and/or LRR domains are important for NLRP3 localization to the proximity of the mitochondria. As discussed above, HSP90 interacts with the NBD and LRR domains of NLRP3, and HSP90 has been implicated in mediating interactions between IPS-1 and cytosolic proteins as well as in cytokine production through NOD2. While there is no evidence to suggest any direct interaction between NLRP3 and IPS-1, there is evidence for cross-talk between these molecules, and it would be interesting if the NLR NBD and LRR domains are important in recruitment to the mitoxosome (Fig. 5).
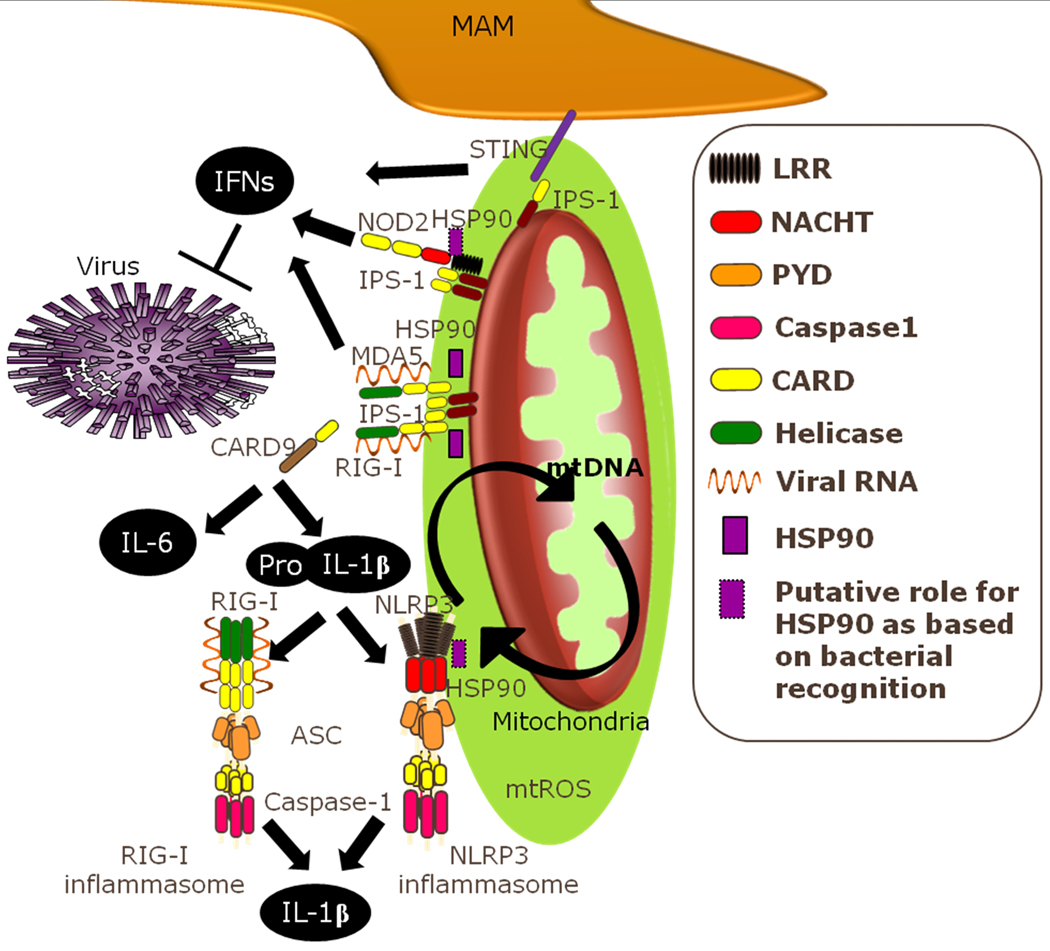
Fig. 5. The mitoxosome.
Recognition of viral ligands by RIG-I or MDA5 initiates their binding to mitochondrial IPS-1 and subsequent induction of IFNs and proinflammatory cytokines, a process augmented by mtROS. NOD2 and STING have both also been shown to contribute to IPS-1 production of IFNs, though not through direct binding of viral ligands. NFκB-dependent production of proinflammatory cytokines such as IL-6 involves the adapter protein CARD9. The inactive pro-inflammatory cytokine IL-1β can be induced through either TLR or RLR viral recognition pathways and is then subject to inflammasome dependent processing to release the active form of IL-1β. The NLRP3 inflammasome plays an important role in viral recognition and upon activation relocalizes to the MAM. HSP90 facilitates cytokine production induced by IPS-1 activation in response to viruses, as well as for NOD2 and NLRP3 in response to bacteria. Localization of these antiviral signaling pathways proximal to the mitochondria helps to centralize cellular stress signaling to an organelle that can integrate these stimuli and facilitate a coordinated response through redox modulation of IPS-1 and NLRP3 signaling. This could also allow the specific combinations of stimuli to affect the exact nature of the response as well as providing a centralized location from which such signals can be readily terminated.
Activation of NLRP3 appears to require close proximity to the mitochondria for sensing of mitochondrial ROS (90). CCCP treatment at levels that breakdown the mitochondrial membrane potential also inhibited mitochondrial respiration and the resultant ROS production (as has been previously shown with similar treatments and uncoupling proteins) and failed to induce NLRP3 activation. With a very low dose treatment of CCCP, Zhou et al. (90) report a partial disturbance of membrane potential with a dramatic increase in mitochondrial ROS comparable to rotenone treatment and also similarly increased levels of IL-1β production. To inhibit ROS generation, the authors inhibit voltage-dependent anion channel (VDAC) isoforms either though targeted shRNA knockdown or though overexpression of Bcl-2 which results in partial closure of VDAC (90). The interpretation of these experiments is complicated by the findings that VDAC has been implicated in multiple roles including flux of mitochondrial metabolites, ATP/ADP exchange, mitochondrial dynamics, intermembrane communication between the ER and mitochondria, ROS modulation, and a controversial role in mitochondrial initiation of apoptosis (95–99). Both VDAC1 and VDAC2 play a partially redundant but fundamental role in cell survival upon ROS insult, while VDAC3 is not involved (98, 100). The fact that IL-1β production was significantly reduced with VDAC1 and VDAC2 knockdown but not VDAC3 knockdown suggests that mitochondrial ROS are playing a key role (90). Zhou et al. (90) examined a prolonged high dose of rotenone treatment (6 h with 10µM) when inducing mitochondrial ROS and examined the effect that this has on NLRP3 activation in the absence of a PAMP or second signal. It would be interesting to examine the role of mitochondrial ROS as generated naturally during virus infection on NLRP3-inflammasome activation. In addition, it still remains a mystery as to what molecule(s) are targeted by ROS to enhance NLRP3 inflammasome activation. While a candidate protein, thioredoxin (TRX)-interacting protein (TXNIP), was reported to link oxidative stress to inflammasome activation (101), whether this and other molecules are involved remains to be established.
Mitochondrial DNA triggers NLRP3 inflammasome
Mitochondria have retained certain similarities to the saprophytic bacteria that they evolved from, with genomes that are rich in non-methylated CpG DNA repeats and that encode formylated peptides. Both of these signatures are PAMPs. It has been shown that mtDNA released into the circulation upon cellular injury can be recognized by neutrophils to induce overwhelming inflammation (102). Even mammalian genomic DNA, when transfected into the cytosol can elicit ASC-dependent inflammasome activation (37). In the study by Nakahira et al. (91), both mitochondrial ROS and mitochondrial DNA release were examined for their impact on NLRP3 activation. In this way, the investigators were able to both confirm but also further define the mechanism that is responsible for increased LPS-stimulated IL-1β production in the absence of autophagy (91). When either of two proteins involved in autophagy, LC3B or beclin 1, was depleted, increased levels of mitochondrial ROS and failed clearance of dysfunctional mitochondria were observed (89). The number of cells with increased levels of dysfunctional mitochondria (as measured by cells with low staining for a probe of mitochondrial membrane potential) increased strikingly upon stimulation with LPS and ATP (89). Upon LPS stimulation, there is an obvious shift in these populations with increased levels of cells with dysfunctional mitochondria. In the absence of functional autophagy, the cells have a dramatic reduction in mitochondrial membrane potential staining. Together, these data indicated that defective mitochondria accumulate in the absence of autophagy and that autophagy is required to maintain healthy mitochondrial function upon immune stimulation (64, 89). The study went on to mechanistically delineate the importance of mitochondrial ROS on IL-1β production and caspase-1 activation. The mitochondrial DNA encodes for 13 critical mitochondrial genes necessary for oxidative phosphorylation and mitochondrial function. Using chronic ethidium bromide treatment, the authors generated ρ0 cells that lack mitochondrial DNA. These cells allowed the examination of the dependency on mitochondrial DNA on NLRP3 signaling. The authors found that ρ0 cells had reduced caspase-1 activation and IL-1β secretion, despite intact pro-IL-1β expression, indicating that mtDNA is required for inflammasome activation (89). To examine how increased mitochondrial ROS affects NLRP3, rotenone was used to increase mitochondrial ROS which resulted in increased IL-1β secretion and caspase 1 activation, an effect that was ablated in the ρ0 cells as well as NLRP3 deficient cells (89). Using a mitochondrial antioxidant, mito-TEMPO, they show that both IL-1β and IL-18 decrease in a dose-dependent manner, while TNF secretion is unaffected. Furthermore, the increased cleavage of caspase 1 and increased secretion of IL-1β caused by defective autophagy is greatly reduced upon mito-TEMPO treatment, strongly suggesting that the aberrant increase in mitochondrial ROS caused by defective autophagy is causing the increase in IL-1β production (89). Interestingly, the mechanism of action for ROS-mediated NLRP3 activation is unprecedented. High levels of mitochondrial ROS activate the NLRP3 inflammasome as well as induce a transient mitochondrial membrane permeability transition that results in release of mitochondrial components into the cytosol including mtDNA (89). Such mtDNA released into the cytosol activates NLRP3, but NLRP3 is also required for mtDNA release (Fig. 3). The amount of mitochondrial ROS produced in NLRP3-deficient cells is comparable to WT and LPS with ATP stimulation results in a similar increase in ROS in both the WT and NLRP3-deficient cells. However, the amount of mtDNA present in the cytosol after LPS stimulation is significantly reduced in both NLRP3 and ASC-deficient cells, as is the amount of cytochrome C released and the percentage of cells displaying reduced mitochondrial membrane potential (89).
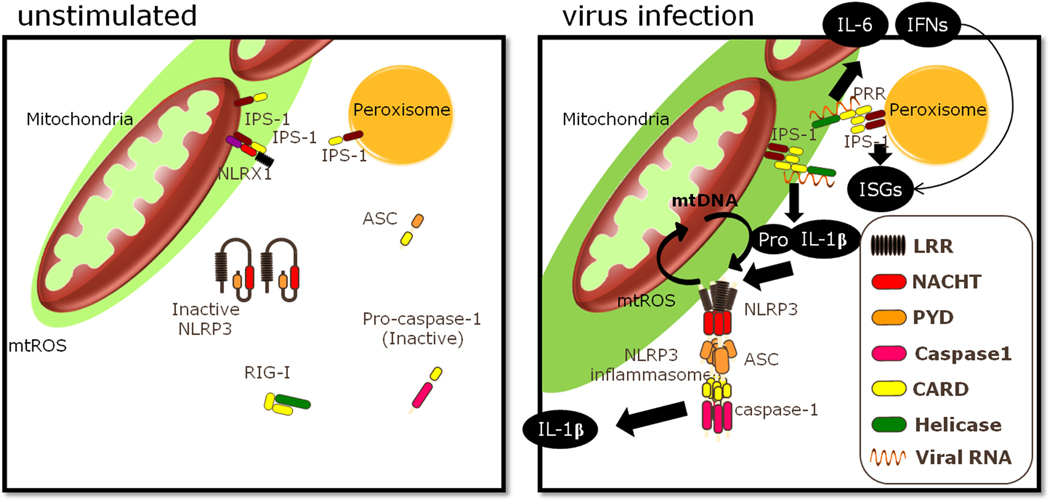
Fig. 3. Impact of mtROS on RLR and NLRP3.
Mitochondrial ROS can increase IPS-1 signaling, resulting in expression of pro-IL-1β. Additionally, mitochondrial ROS enhances the activation of NLRP3 inflammasome elicited by LPS + ATP, which leads to the release of mtDNA that feeds forward to activate further NLRP3 inflammasomes.
It was known that mtDNA can be released into the extracellular space as a weapon to capture bacteria in a sticky net of DNA, such as seen with neutrophils, or it can be released into the extracellular space upon traumatic injury and cell rupture (102, 103). Mitochondrial DNA is constantly being exchanged among mitochondria within a cell through fission and fusion events between the mitochondria (104). Mitochondrial DNA can even be exchanged between species, such has been shown in host and parasite plants (105). Now we can add to that that transient mitochondrial membrane permeabilization following LPS stimulation can release mitochondrial DNA into the cytosol to activate NLRP3, which is responsible for causing a greater release of mtDNA (89).
The common finding by both Nakahira et al. (89) and Zhou et al. (90) is that mitochondrial ROS is activating the NLRP3 inflammasome. However, one particular point of contention between the studies is how exclusive this phenomenon is the NLRP3 inflammasome as opposed to other inflammasomes. Nakahira et al. (89) find a partial role for the AIM2 inflammasome in mtDNA recognition, though it is unknown if AIM2 inflammasome activation had any effect on mtDNA release. Zhou et al. (90) showed that IL-1β production following stimulation with Salmonella or Poly (dA-dT) to induce NLRC4- or AIM-2 inflammasome activation respectively, was unaffected by shRNA knockdown of VDAC1. This suggests that the feature of VDAC knockdown (presumably decreased mitochondrial ROS production) that is decreasing inflammasome activation is specific to NLRP3. While it still remains to be shown whether mitochondrial ROS is important in NLRP3 activation upon viral recognition as well, though it certainly stands to reason that this will be an important element in NLRP3 inflammasome mediated viral recognition as well.
It is critical that inflammatory responses are carefully coordinated so as to provide effective defense against viral infections while inflicting minimal damage to the host. Hyperactivation of RLR or NLRP3 signaling could lead to detrimental levels of inflammation or have other immunopathological consequences that are harmful to the host. An intriguing idea is localization of these antiviral signaling pathways proximal to the mitochondria could centralize cellular stress signaling to an organelle that can facilitate a coordinated response. Integration of these stimuli could generate a heightened antiviral response, at the same time providing a centralized location from which such signals can be readily terminated.
A role for ER stress in RLR signaling
ER stress elicits a carefully orchestrated stress response called the unfolded protein response (UPR) which functions to (i) restore normal function of the cell by halting protein translation and (ii) activate the signaling pathways for the production of molecular chaperones involved in protein folding. The most highly conserved axis of the UPR is between the ER stress sensor kinase inositol requiring enzyme 1 (IRE1) and the transcription factor X-box binding protein 1 (XBP-1), which is a critical mediator of the UPR. It has been shown in invertebrates that XBP-1 can recruit DAF-16, a FOXO transcription factor to enhance ER stress resistance and promote longevity (106). XBP-1 plays a fundamental immune modulatory role that is critical for C. elegans to survive the innate immune response following bacterial infection (107). In mammalian cells, ER stress that elicits initiation of the UPR results in increased IFNβ transcript production even in the absence of immune stimuli. When cells were treated with thapsigargin (Tpg), which inhibits the ER calcium pumps and elicits the UPR, there was a log-fold increase in IFNβ mRNA and protein levels upon LPS, Poly (I:C), transfected Poly (I:C), but not CpG, stimulation. IFNβ induction in response to Poly (I:C) was reduced in the absence of XBP-1, and Tpg treatment together with Poly (I:C) was only able to increase IFN induction upon re-introduction of XBP-1 into the XBP-1 deficient cells (108). This suggests that ER stress in combination with TLR4, TLR3, or MDA5, but not TLR9, signaling results in amplified IFNβ production. TLR4 and TLR2 were shown to activate ER stress sensor IRE1α as well as the transcription factor XBP-1. Additionally, TLR-activated XBP1 increased IFNβ and proinflammatory cytokine production in macrophages even without chemically pre-inducing ER stress, and ER stress further increased XBP-1 mediated cytokine production. A subsequent study showed that LPS stimulation in conjunction with ER stress led to the binding of XBP-1 and recruitment of IRF3 and CREB binding protein/p300 to the ifnb1 cis-acting enhancer (109). Intriguingly, HSP90 is an important modulator of the UPR (110). This may provide an additional connection between ER stress and cytosolic pathogen recognition, as HSP90 has been shown to play a role in cytokine production through IPS-1, NOD2, and NLRP3, as discussed above.
As viruses induce various cellular stresses (including both oxidative and ER stress), it is intriguing to postulate an integrated viral sensing stress response. A cell at a critical crossroads, upon that first recognition of viral presence through PRR, would benefit from incorporating any other non-PRR-based indication of cellular stress that might accompany a viral infection to initiate a more robust response that could potentially lead to a more rapid viral clearance. This could explain how two very different forms of cellular stress signals, namely oxidative stress and ER stress, greatly amplify antiviral signaling pathways. Possibly, integration of these signals allows the distinct outcome between combating imminent danger (by turning on antiviral genes) and irreversible damage that marks a point of no return for the cell (by inducing apoptosis).
Cytosolic viral sensors and human diseases
There are a great number of pathogenic conditions that are associated with mitochondria both morphologically and functionally including chronic infection, hypoxia, lupus, sepsis, type II diabetes in addition to the inevitable effects of age exemplified in late onset diseases such as neurodegenerative diseases, cardiovascular disease, and cancer. Therefore, further research aimed at understanding cytosolic antiviral immunity within its physiological context, localization, and its interactions with other intracellular pathways also charged with restoring homeostasis is critical. It is now clear that mitochondrial ROS greatly affects both RLR and NLRP3-mediated signaling and that autophagy is critical for the regulation of signaling emanating from mitochondria. The body is in constant contact with the microbial world, and the immune system is charged with vigilant surveillance to assess the nature of these contacts and ensure that appropriate measures are taken to protect the host. Control of these relations by the immune system can range from symbiotic coexistence with commensal microbiota to a full-blown attack on a detected pathogen, which can potentially wreak havoc on host tissues. As it is very important for inflammatory responses to be carefully regulated, it is possible that an excessive IFN and proinflammatory signaling in response to viral infection could be detrimental to the host, resulting in severe tissue damage and possibly increasing the risk of developing certain types of autoimmunity.
Autophagy and RLR/NLR dysregulation in Crohn’s disease
Crohn’s disease (CD) is a multigenic disease that manifests with ulceration and chronic inflammation of the gastrointestinal tract. CD is reported to have a roughly 50% concordance rate in monozygotic twins, suggesting a strong environmental risk factor. In CD, there is strong evidence for microbial triggers, as a characteristic feature of the disease is an aberrant immune response to commensal bacteria in the intestine (111). Acute flares as well as new-onset of CD has been associated with viral infections, and virally induced aphthous ulcers have been documented proximal to diseased intestinal segments responsible for clinical symptoms (112). Reports of viral infection as a trigger in the development of CD had been controversial in light of the disease being mediated by excessive inflammation and inappropriate immune response to commensal bacteria (113). Targeted research following the identification of multiple genetic risk factors in the development of CD, including NOD2, ATG16L1, and IRGM demonstrated the potential involvement of autophagy and an aberrant response to viral infection in this autoimmune disorder and are discussed herein. The development of autoimmune disease is often multifactoral, with a combination of both genetic predisposition and environmental conditions that come together in determining ultimate risk of developing disease. As viral infections can alter the immune response to subsequent bacterial infections and deficiencies in autophagy can result in excessive inflammation upon viral sensing, it is becoming increasingly evident that we must reassess our current understanding of the mechanisms that increase risk of developing CD and the role that viral infections may play in this regard.
The CD variant of ATG16L1 has been reported to result in a less stable protein (114), and two different models of ATG16L1 deficiency have been examined to better understand the role of this risk variant and autophagy in the development of CD. One mouse model is lacking the ATG16L1 gene altogether and is phenotypically identical to mice lacking other essential autophagy genes, such as either the ATG5 or ATG7 genes. Cells lacking ATG16L1 produced higher levels of IL-1β and IL-18 in response to LPS, further propagated by IFN or ROS (91). As mice lacking functional autophagy die in their first day of life, mice lacking ATG16L1 in the hematopoietic compartment were studied. Such mice were prone to dextran sulphate sodium (DSS)-induced acute colitis, which was reversed by neutralizing the elevated IL-1β and IL-18 levels (91). Another model to examine the role of a less stable variant of ATG16L1 examined the effect of reduced levels of ATG16L1 by generating a viable ATG16L1 hypomorph (ATG16L1HM)(115). Interestingly, these ATG16L1HM show Paneth cell morphological abnormalities of aberrant, disorganized granules that resemble the Paneth cell morphology in CD patients with the ATG16 risk allele. Additionally, degenerating mitochondria were visible only in the ATG16L1HM Paneth cells and not in WT cells, though it was not reported if these are present in the CD patients, as would be suggested by the other reported similarities. There were no significant defects in basal or starvation-induced autophagy in the ATG16L1 risk variant or in the ATG16L1HM. This bares similarity to the viable ATG5 heterozygotes, where it has been well established that they undergo normal basal and starvation-induced autophagy and in these instances there have been no indications for haploinsuffinciency. However, we see a haploinsufficient mitophagy-specific defect in cells from ATG5 heterozygotes, where these cells display an intermediate phenotype of mitochondrial accumulation, ROS levels, and increased IFN and inflammatory cytokines upon RLR signaling in ATG5 heterozygotes that becomes particularly pronounced with advanced age or multiple passages of cells (Fig. 4). ATG16L1, ATG5, and ATG7 have been reported to have a common role (through the process of autophagy) in CD (116). It is interesting why other Atg genes have not been found to associate with CD risk.
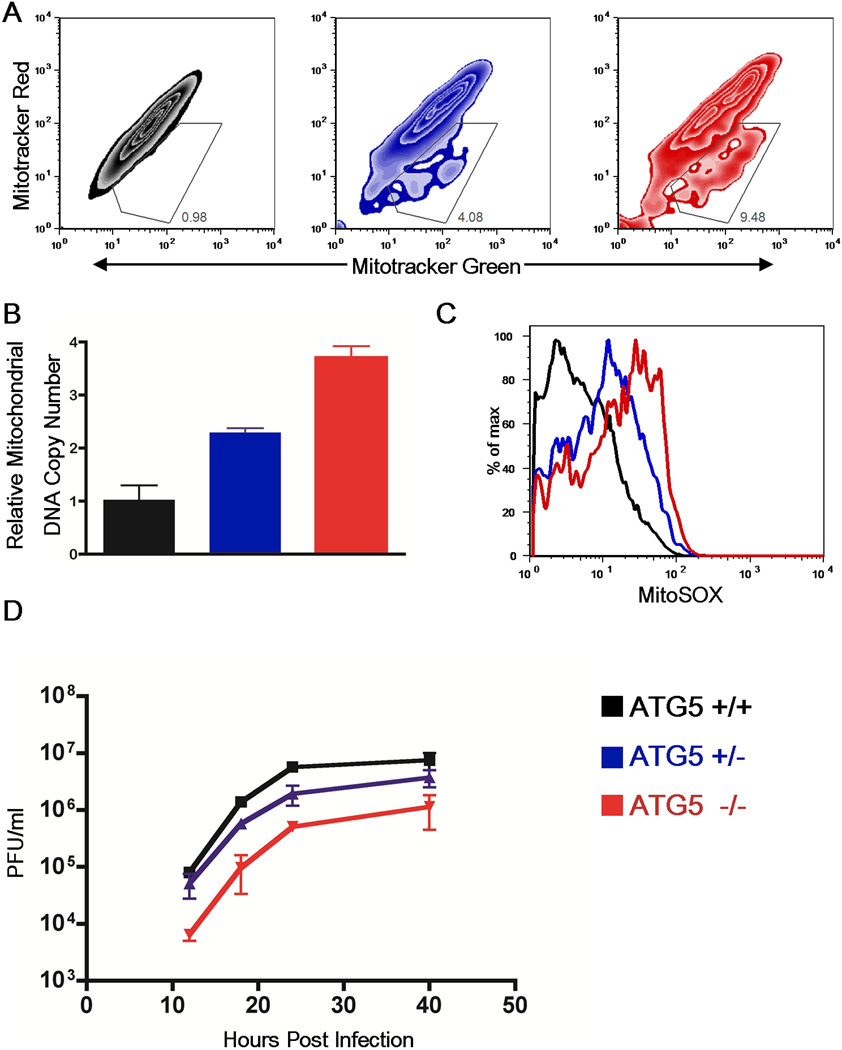
Fig. 4. Gene dose-dependent regulation of mitophagy and IPS-1 signaling by Atg5.
(A) Atg5+/+, Atg5+/− and Atg5−/− MEFs were stained with 100nM Mitotracker Green (which stains the lipid membrane of the mitochondria) and 100nM Mitotracker Red (which is an indicator of membrane potential). Contour plots of FACS analysis are depicted. (B) Mitochondrial DNA copy number was measured by qPCR and normalized to nuclear DNA levels in a ratio of mtDNA COI over 18s. Relative mitochondrial DNA copy numbers are depicted. These data are representative of three similar experiments. (C) ROS levels within WT, Atg5+/−, and ATG5−/− MEFs were examined by staining cells with MitoSOX (which labels mitochondria-associated ROS) and analyzed by FACS. (D) WT, Atg5+/−, and Atg5−/− MEFs were infected with 1 MOI of VSV-GFP and the levels of infection were determined by measuring GFP viral titers in the supernatants by plaque assay at the indicated time points. These results are representative of three separate experiments.
A landmark study revealed an unappreciated role for viral infection in ATG16L1HM intestinal pathology. This study provides a proof of concept that viral infection can spark the powder keg of specific genetic predispositions that impact the host immune response to ignite autoimmune disease pathology (117). Murine norovirus (MNV) strains, recognized by MDA-5 (118), are endemic to many mouse facilities, and Cadwell et al. (117) therefore rederived the ATG16L1HM in an enhanced-barrier facility, which provides protection against viral and bacterial infections. The abnormal Paneth cell pathology that had been observed in the ATG16L1HM derived in a pathogen-free facility was absent until the mice were infected with a strain of MNV that establishes persistent infection. Additionally, the model of DSS-induced colitis also only showed an aberrant response to tissue injury in MNV infected ATG16L1HM mice (117). This is an extremely important element for us to consider as we are all chronically infected with multiple viruses, and for better or worse, they could be shaping our immune response in more ways than we imagine.
Additional evidence for defects in selective forms of autophagy in the absence of a defect in bulk autophagy comes from a study examining the recruitment of autophagosomes in host defense against bacteria. Travassos et al. (119) report a role for NOD1 and NOD2 proteins in targeting autophagosome formation around invasive bacteria through NOD interaction with ATG16L1, independent of NFκB signaling through the NOD proteins. Importantly, the CD associated polymorphism in ATG16L1 was examined along with one of the three known NOD2 polymorphisms associated with CD known to cause a frameshift mutation in Nod2 (119). Cooney et al. (120) examined dendritic cells from patients with the CD risk alleles for either NOD2 or ATG16L1 and demonstrated that this selective autophagy of invasive bacteria is defective. Interestingly, even NOD2 overexpression in the absence of invasive bacteria has a dose dependent affect on the induction of autophagy, and Paneth cells express NOD2 at higher levels than surrounding epithelial cells (121).
An additional example of cell stress linking to the development of CD is seen in a study by Kaser et al.(122). They verify the association for variants of XBP-1 in CD and ulcerative colitis (UC), providing a link between ER stress and genetic susceptibility to inflammatory bowel disease. Using mouse models with a deletion of XBP1 in the intestinal epithelium, they see spontaneous enteritis and increased DSS-induced colitis(122).
Human immunity-related GTPase family, M (IRGM) is a GTPase that has been shown to be involved in autophagy (123). An IRGM variant associated with CD was originally thought to be non-causative, as it neither altered protein sequence or splice sites. However, a striking finding by Brest et al. (124) demonstrates that this IRGM variant results in loss of a microRNA (miRNA) regulation site for miR-196. In inflamed mucosa from patients with CD it was found that miR-196 is overexpressed. While miR-196 downregulated the IRGM protective variant, it was unable to downregulate the risk-associated allele (124). IRGM has recently been reported to translocate to mitochondria where it can modulate mitochondrial dynamics and membrane potential (125). It is intriguing to speculate that IRGM manipulation of mitochondrial function could have inflammatory implications for cytosolic viral signaling and that loss of regulation of IRGM could therefore increase autoimmune susceptibility to CD upon viral insult (124, 125).
An old but still poorly understood concept in IBD is the presence of ROS and lipid peroxidation, not only in the intestinal tract, but also in other tissues as well. Plasma from patients with mild UC was shown to contain a decreased antioxidant profile as well as markers of lipid peroxidation indicative of oxidative stress (126). In another study, mucosal resection specimens were examined from patients with CD or UC. Expression levels of the SOD isoforms were examined in paired inflamed and non-inflamed mucosa in comparison to normal control mucosa. Increased levels of the mitochondrial Mn-SOD protein levels in both inflamed and non-inflamed mucosa were found as well as increased percentage of epithelial cells expressing Mn-SOD from IBD patients (127). An intriguing study has recently examined the ROS status of immune cells from patients with active CD vs. controls or patients with CD in remission. They find that Mn-SOD is overexpressed in patients with active CD along with damaged mitochondria as measured by membrane potential and that H2O2 levels are elevated during active disease (128). These markers more closely resembled controls in patients with CD in remission. Lipid peroxidation and DNA oxidative damage were increased in both active CD patients and patients in remission in comparison to controls. Together, this evidence strongly implicates mitochondrial ROS in the oxidative damage seen in inflammatory bowel disease. Increased levels of damaged mitochondria correlate with inflammatory markers in active CD (128). A comprehensive microarray study that compared mRNA signatures of three different murine colitis models provides additional evidence that oxidative stress is playing a role in the initiation and perpetuation of the disease (129). ROS not only impact inflammatory cytokine production, but they have also been shown to impact mucosal secretions that provide the first line of defense against luminal encounters with the microbial world. One study demonstrated that while low levels of ROS in the colonic lumen increased the protection provided by the mucus barrier in vivo, high levels of ROS significantly reduced this protection and the thickness of the mucus barrier was reduced in half. While colonic mucus gels were resistant to ROS, mucins showed protein peroxidation and loss of terminal sugars(130). It is interesting to investigate if damaged mitochondria and increased mitochondrial ROS specifically contribute to the pathology of IBD. This could suggest a role for CD associated risk alleles such as the ATG16L1 risk allele in selective clearance of damaged mitochondria.
Certain viruses can inhibit autophagy and could therefore contribute to the accumulation of damaged mitochondria and increased mitochondrial ROS. Virus infections often lead to ROS production. Increased levels of ROS then lead to a hyper-proinflammatory response, which can result in immune-mediated pathology that causes damage to the host mucosa. The dynamics of this interaction could determine the difference between clearance of the pathogen and further susceptibility to disease.
Concluding remarks
It is clear that cytosolic viral recognition is far more integrated than the simplistic categorization of RLR, DNA recognition, and NLR pathways. There is orchestrated sensing of viral ligands and of the stress induced by viral infections, which can be integrated at multiple points throughout the antiviral signal transduction cascade. Both RLR and NLR-mediated cytokine production are modulated by ROS, and increasing evidence is pointing towards a central role for mitochondrial ROS specifically. Indeed, multiple aspects of mitochondrial function, other than mtROS, such as mtDNA release, mitochondrial membrane potential, and mitochondrial dynamics, have all been shown to affect and/or be affected by these signaling pathways. Now, as we are just beginning to understand how the mitochondria and pathogen recognition systems communicate, we realize that it is a bigger conversation then had been previously appreciated. So far we have identified components of the RLR and NLR signaling pathways in this dialogue, but certainly as we dig deeper there will be more to come. IPS-1 localization to the mitochondria has made it clear that RLR signaling is occurring at the mitochondrial face. The recent report that the NLRP3 inflammasome relocalizes to the MAM where it is modulated by the mitochondria, suggests the possibility of a mitoxosome, a mitochondrial oxidative signalosome that dictates the inflammatory and antiviral cytokine response to pathogen recognition (Fig. 5). NLRP3 is activated by both mtROS and mtDNA and then induces the further release of mtDNA. Intriguingly, the stress response protein HSP90, which is involved in the response to both oxidative and ER stress has been shown to have a role in facilitating the cytokine production induced by IPS-1 activation in response to virus, as well as by NOD2 and NLRP3 in response to bacteria. One exciting possibility for the localization of these antiviral signaling pathways proximal to the mitochondria is to centralize cellular stress signaling to an organelle that can integrate these stimuli and facilitate a coordinated response. This could also allow the specific combinations of stimuli to affect the exact nature of the response, as well as providing a centralized location from which such signals can be readily terminated.
While the mitochondria provide the main source of ROS in most cell types, mitochondrial ROS have been regarded simply as an unregulated byproduct of oxidative respiration. We now find ourselves at a turning point in our understanding of mitochondrial ROS production and their intricate involvement in host defense. This has become clear for innate immune signaling, with the recent body of evidence discussed above that mitochondrial ROS is fueling innate antiviral signaling. A study currently in press reports that signaling through TLR1, TLR2, and TLR4 can both increase mitochondrial ROS production, and also elicit the mitochondria to surround phagosomes containing PAMP-coated latex beads. This response was mediated through TRAF6 translocating to the mitochondria and engaging a mitochondrial respiratory chain protein, ECSIT, which was able to facilitate increased mitochondrial and cellular ROS production. Decreasing mitochondrial ROS resulted in defective bacterial killing (Phillip West and Sankar Ghosh, personal communication). It is therefore quite apparent that the mitochondria are playing a much larger role in innate defense then we had previously imagined.
Mitochondria are playing an essential role in host defense and are modulating and being modulated by innate signaling. Therefore, future research exploring the nature of the interactions between these pathways and the localization dynamics will allow us to reach a true understanding of how these components all fit together. Is there a mitoxosome, a super-molecular complex that brings all of these components together at the face of the organelle capable of facilitating their action?
As the mitochondrial ship sets sail, off to search and destroy any that would dare to invade, we enter into a new level of understanding of the role of the mitochondria in innate defense. Future research will hopefully reveal how the mitochondria use stress to ramp up host defense, and how this impacts our response to viruses. As we will never rid the world of viruses, hopefully new findings will uncover how these responses increase susceptibility to other diseases and steer us towards understanding how to tip the balance towards a tactical viral defeat followed by a return to health.
Acknowledgements
This work was supported by National Institutes of Health (NIH) (AI054359, AI062428, AI064705 and AI083242 to A.I.). A.I. holds an Investigators in Pathogenesis of Infectious Disease Award from the Burroughs Wellcome Fund. M.T. was supported by the NIH Ruth L. Kirschstein National Research Service Awards for Individual Predoctoral Fellows (F31 AG039163). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Footnotes
Authors declare no conflicts of interest.
References
- 1.Pichlmair A, Reis e Sousa C. Innate Recognition of Viruses. Immunity. 2007;27:370–383. doi: 10.1016/j.immuni.2007.08.012. [DOI] [PubMed] [Google Scholar]
- 2.Takeuchi O, Akira S. Recognition of viruses by innate immunity. Immunol Rev. 2007;220:214–224. doi: 10.1111/j.1600-065X.2007.00562.x. [DOI] [PubMed] [Google Scholar]
- 3.Pichlmair A, et al. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5'-phosphates. Science. 2006;314:997–1001. doi: 10.1126/science.1132998. [DOI] [PubMed] [Google Scholar]
- 4.Hornung V, Ellegast J, Kim S, Brzozka K, Jung A, Kato H, et al. 5'-Triphosphate RNA is the ligand for RIG-I. Science. 2006;314:994–997. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- 5.Kato H, et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- 6.Gitlin L, et al. Essential role of mda-5 in type I IFN responses to polyriboinosinic:polyribocytidylic acid and encephalomyocarditis picornavirus. Proc Natl Acad Sci USA. 2006;103:8459–8464. doi: 10.1073/pnas.0603082103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Züst R, et al. Ribose 2'-O-methylation provides a molecular signature for the distinction of self and non-self mRNA dependent on the RNA sensor Mda5. Nat Immunol. 2011;12:137–143. doi: 10.1038/ni.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kawai T, et al. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat Immunol. 2005;6:981–988. doi: 10.1038/ni1243. [DOI] [PubMed] [Google Scholar]
- 9.Seth RB, Sun L, Ea CK, Chen ZJ. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell. 2005;122:669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 10.Meylan E, et al. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature. 2005;437:1167–1172. doi: 10.1038/nature04193. [DOI] [PubMed] [Google Scholar]
- 11.Xu Y, Jagannath C, Liu X-, Sharafkhaneh A, Kolodziejska KE, Eissa NT. Toll-like Receptor 4 Is a Sensor for Autophagy Associated with Innate Immunity. Immunity. 2007;27:135–144. doi: 10.1016/j.immuni.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu LG, Wang YY, Han KJ, Li LY, Zhai Z, Shu HB. VISA is an adapter protein required for virus-triggered IFN-beta signaling. Mol Cell. 2005;19:727–740. doi: 10.1016/j.molcel.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 13.Oshiumi H, Sakai K, Matsumoto M, Seya T. DEAD/H BOX 3 (DDX3) helicase binds the RIG-I adaptor IPS-1 to up-regulate IFN-β-inducing potential. Eur J Immunol. 2010;40:940–948. doi: 10.1002/eji.200940203. [DOI] [PubMed] [Google Scholar]
- 14.Schröder M, Baran M, Bowie AG. Viral targeting of DEAD box protein 3 reveals its role in TBK1/IKKε-mediated IRF activation. EMBO J. 2008;27:2147–2157. doi: 10.1038/emboj.2008.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Michallet M, et al. TRADD protein is an essential component of the RIG-like helicase antiviral pathway. Immunity. 2008;28:651–661. doi: 10.1016/j.immuni.2008.03.013. [DOI] [PubMed] [Google Scholar]
- 16.Ishikawa H, Barber GN. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature. 2008;455:674–678. doi: 10.1038/nature07317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhong B, et al. The Adaptor Protein MITA Links Virus-Sensing Receptors to IRF3 Transcription Factor Activation. Immunity. 2008;29:538–550. doi: 10.1016/j.immuni.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 18.Jin L, Waterman PM, Jonscher KR, Short CM, Reisdorph NA, Cambier JC. MPYS, a novel membrane tetraspanner, is associated with major histocompatibility complex class II and mediates transduction of apoptotic signals. Mol Cell Biol. 2008;28:5014–5026. doi: 10.1128/MCB.00640-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sun W, et al. ERIS, an endoplasmic reticulum IFN stimulator, activates innate immune signaling through dimerization. Proc Natl Acad Sci USA. 2009;106:8653–8658. doi: 10.1073/pnas.0900850106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Y, et al. ISG56 is a negative-feedback regulator of virus-triggered signaling and cellular antiviral response. Proc Natl Acad Sci USA. 2009;106:7945–7950. doi: 10.1073/pnas.0900818106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stetson DB, Medzhitov R. Recognition of cytosolic DNA activates an IRF3-dependent innate immune response. Immunity. 2006;24:93–103. doi: 10.1016/j.immuni.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 22.Ishii KJ, et al. A toll-like receptor-independent antiviral response induced by double-stranded B-form DNA. Nat Immunol. 2006;7:40–48. doi: 10.1038/ni1282. [DOI] [PubMed] [Google Scholar]
- 23.Takaoka A, et al. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature. 2007;448:501–505. doi: 10.1038/nature06013. [DOI] [PubMed] [Google Scholar]
- 24.Ishii KJ, et al. TANK-binding kinase-1 delineates innate and adaptive immune responses to DNA vaccines. Nature. 2008;451:725–729. doi: 10.1038/nature06537. [DOI] [PubMed] [Google Scholar]
- 25.Chiu Y-, MacMillan JB, Chen ZJ. RNA Polymerase III Detects Cytosolic DNA and Induces Type I Interferons through the RIG-I Pathway. Cell. 2009;138:576–591. doi: 10.1016/j.cell.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ablasser A, Bauernfeind F, Hartmann G, Latz E, Fitzgerald KA, Hornung V. RIG-I-dependent sensing of poly(dA:dT) through the induction of an RNA polymerase III-transcribed RNA intermediate. Nat Immunol. 2009;10:1065–1072. doi: 10.1038/ni.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Valentine R, Smith GL. Inhibition of the RNA polymerase III-mediated dsDNA-sensing pathway of innate immunity by vaccinia virus protein E3. J Gen Virol. 2010;91:2221–2229. doi: 10.1099/vir.0.021998-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Unterholzner L, et al. IFI16 is an innate immune sensor for intracellular DNA. Nat Immunol. 2010;11:997–1004. doi: 10.1038/ni.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saitoh T, et al. Atg9a controls dsDNA-driven dynamic translocation of STING and the innate immune response. Proc Natl Acad Sci USA. 2009;106:20842–20846. doi: 10.1073/pnas.0911267106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martinon F, Mayor A, Tschopp J. The inflammasomes: guardians of the body. Annu Rev Immunol. 2009;27:229–265. doi: 10.1146/annurev.immunol.021908.132715. [DOI] [PubMed] [Google Scholar]
- 31.Martinon F, Burns K, Tschopp J. The Inflammasome: A molecular platform triggering activation of inflammatory caspases and processing of proIL-β. Mol Cell. 2002;10:417–426. doi: 10.1016/s1097-2765(02)00599-3. [DOI] [PubMed] [Google Scholar]
- 32.Dinarello CA. The IL-1 family and inflammatory diseases. Clin Exp Rheumatol. 2002;20 Suppl.:S1–S13. [PubMed] [Google Scholar]
- 33.Ting JP, et al. The NLR gene family: a standard nomenclature. Immunity. 2008;28:285–287. doi: 10.1016/j.immuni.2008.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen GY, Nunez G. Sterile inflammation: sensing and reacting to damage. Nat Rev Immunol. 2010;10:826–837. doi: 10.1038/nri2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kanneganti T, et al. Critical role for Cryopyrin/Nalp3 in activation of caspase-1 in response to viral infection and double-stranded RNA. J Biol Chem. 2006;281:36560–36568. doi: 10.1074/jbc.M607594200. [DOI] [PubMed] [Google Scholar]
- 36.Muruve DA, et al. The inflammasome recognizes cytosolic microbial and host DNA and triggers an innate immune response. Nature. 2008;452:103–107. doi: 10.1038/nature06664. [DOI] [PubMed] [Google Scholar]
- 37.Muruve DA, et al. The inflammasome recognizes cytosolic microbial and host DNA and triggers an innate immune response. Nature. 2008;452:103–107. doi: 10.1038/nature06664. [DOI] [PubMed] [Google Scholar]
- 38.Ichinohe T, Pang IK, Iwasaki A. Influenza virus activates inflammasomes via its intracellular M2 ion channel. Nat Immunol. 2010;11:404–410. doi: 10.1038/ni.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bürckstümmer T, Baumann C, Blüml S, Dixit E, Dürnberger G, Jahn H, et al. An orthogonal proteomic-genomic screen identifies AIM2 as a cytoplasmic DNA sensor for the inflammasome. Nat Immunol. 2009;10:266–272. doi: 10.1038/ni.1702. [DOI] [PubMed] [Google Scholar]
- 40.Hornung V, et al. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature. 2009;458:514–518. doi: 10.1038/nature07725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fernandes-Alnemri T, Yu J, Datta P, Wu J, Alnemri ES. AIM2 activates the inflammasome and cell death in response to cytoplasmic DNA. Nature. 2009;458:509–513. doi: 10.1038/nature07710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roberts TL, et al. HIN-200 proteins regulate caspase activation in response to foreign cytoplasmic DNA. Science. 2009;323:1057–1060. doi: 10.1126/science.1169841. [DOI] [PubMed] [Google Scholar]
- 43.Sabbah A, et al. Activation of innate immune antiviral responses by Nod2. Nat Immunol. 2009;10:1073–1080. doi: 10.1038/ni.1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dugan JW, et al. Nucleotide oligomerization domain-2 interacts with 2′-5′-oligoadenylate synthetase type 2 and enhances RNase-L function in THP-1 cells. Mol Immunol. 2009;47:560–566. doi: 10.1016/j.molimm.2009.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Malathi K, Dong B, Gale M, Jr., Silverman RH. Small self-RNA generated by RNase L amplifies antiviral innate immunity. Nature. 2007;448:816–819. doi: 10.1038/nature06042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Luthra P, Sun D, Silverman RH, He B. Activation of IFN-β expression by a viral mRNA through RNase L and MDA5. Proc Natl Acad Sci USA. 2011;108:2118–2123. doi: 10.1073/pnas.1012409108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moore CB, et al. NLRX1 is a regulator of mitochondrial antiviral immunity. Nature. 2008;451:573–577. doi: 10.1038/nature06501. [DOI] [PubMed] [Google Scholar]
- 48.Tattoli I, et al. NLRX1 is a mitochondrial NOD-like receptor that amplifies NF-κB and JNK pathways by inducing reactive oxygen species production. EMBO Rep. 2008;9:293–300. doi: 10.1038/sj.embor.7401161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Poeck H, et al. Recognition of RNA virus by RIG-I results in activation of CARD9 and inflammasome signaling for interleukin 1B production. Nat Immunol. 2010;11:63–69. doi: 10.1038/ni.1824. [DOI] [PubMed] [Google Scholar]
- 50.Wu W, Hsu YM, Bi L, Songyang Z, Lin X. CARD9 facilitates microbe-elicited production of reactive oxygen species by regulating the LyGDI-Rac1 complex. Nat Immunol. 2009;10:1208–1214. doi: 10.1038/ni.1788. [DOI] [PubMed] [Google Scholar]
- 51.Lipinski S, et al. DUOX2-derived reactive oxygen species are effectors of NOD2-mediated antibacterial responses. J Cell Sci. 2009;122:3522–3530. doi: 10.1242/jcs.050690. [DOI] [PubMed] [Google Scholar]
- 52.Hsu Y-S, et al. The adaptor protein CARD9 is required for innate immune responses to intracellular pathogens. Nat Immunol. 2007;8:198–205. doi: 10.1038/ni1426. [DOI] [PubMed] [Google Scholar]
- 53.Balaban RS, Nemoto S, Finkel T. Mitochondria, oxidants, and aging. Cell. 2005;120:483–495. doi: 10.1016/j.cell.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 54.Janssen-Heininger YMW, et al. Redox-based regulation of signal transduction: Principles, pitfalls, and promises. Free Radic Biol Med. 2008;45:1–17. doi: 10.1016/j.freeradbiomed.2008.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Finkel T, Holbrook NJ. Oxidants, oxidative stress and the biology of ageing. Nature. 2000;408:239–247. doi: 10.1038/35041687. [DOI] [PubMed] [Google Scholar]
- 56.Chen H, Vermulst M, Wang YE, Chomyn A, Prolla TA, McCaffery JM, et al. Mitochondrial fusion is required for mtdna stability in skeletal muscle and tolerance of mtDNA mutations. Cell. 2010;141:280–289. doi: 10.1016/j.cell.2010.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mizushima N, Yamamoto A, Matsui M, Yoshimori T, Ohsumi Y. In Vivo Analysis of Autophagy in Response to Nutrient Starvation Using Transgenic Mice Expressing a Fluorescent Autophagosome Marker. Mol Biol Cell. 2004;15:1101–1111. doi: 10.1091/mbc.E03-09-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ohsumi Y. Molecular dissection of autophagy: two ubiquitin-like systems. Nat Rev Mol Cell Biol. 2001;2:211–216. doi: 10.1038/35056522. [DOI] [PubMed] [Google Scholar]
- 59.Virgin HW, Levine B. Autophagy genes in immunity. Nat Immunol. 2009;10:461–470. doi: 10.1038/ni.1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tal MC, Iwasaki A. Autophagy and innate recognition systems. Curr Top Microbiol Immunol. 2009;335:107–121. doi: 10.1007/978-3-642-00302-8_5. [DOI] [PubMed] [Google Scholar]
- 61.Rodriguez-Enriquez S, Kim I, Currin RT, Lemasters JJ. Tracker dyes to probe mitochondrial autophagy (mitophagy) in rat hepatocytes. Autophagy. 2006;2:39–46. doi: 10.4161/auto.2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Twig G, et al. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J. 2008;27:433–446. doi: 10.1038/sj.emboj.7601963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bulua AC, et al. Mitochondrial reactive oxygen species promote production of proinflammatory cytokines and are elevated in TNFR1-associated periodic syndrome (TRAPS) J Exp Med. 2011;208:519–533. doi: 10.1084/jem.20102049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tal MC, Sasai M, Lee HK, Yordy B, Shadel GS, Iwasaki A. Absence of autophagy results in reactive oxygen species-dependent amplification of RLR signaling. Proc Natl Acad Sci USA. 2009;106:2774–2775. doi: 10.1073/pnas.0807694106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gloire G, Legrand-Poels S, Piette J. NF-κB activation by reactive oxygen species: Fifteen years later. Biochem Pharmacol. 2006;72:1493–1505. doi: 10.1016/j.bcp.2006.04.011. [DOI] [PubMed] [Google Scholar]
- 66.Soucy-Faulkner A, Mukawera E, Fink K, Martel A, Jouan L, Nzengue Y, et al. Requirement of NOX2 and reactive oxygen species for efficient RIG-I-mediated antiviral response through regulation of MAVS expression. PLoS Pathog. 2010;6(6) doi: 10.1371/journal.ppat.1000930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lin R, et al. Dissociation of a MAVS/IPS-1/VISA/Cardif-IKKε molecular complex from the mitochondrial outer membrane by hepatitis C virus NS3-4A proteolytic cleavage. J Virol. 2006;80:6072–6083. doi: 10.1128/JVI.02495-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tang ED, Wang C. MAVS self-association mediates antiviral innate immune signaling. J Virol. 2009;83:3420–3428. doi: 10.1128/JVI.02623-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dixit E, et al. Peroxisomes Are Signaling Platforms for Antiviral Innate Immunity. Cell. 2010;141:668–681. doi: 10.1016/j.cell.2010.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ohman T, Rintahaka J, Kalkkinen N, Matikainen S, Nyman TA. Actin and RIG-I/MAVS Signaling Components Translocate to Mitochondria upon Influenza A Virus Infection of Human Primary Macrophages. J Immunol. 2009;182:5682–5692. doi: 10.4049/jimmunol.0803093. [DOI] [PubMed] [Google Scholar]
- 71.Liu XY, Wei B, Shi HX, Shan YF, Wang C. Tom70 mediates activation of interferon regulatory factor 3 on mitochondria. Cell Res. 2010;20:994–1011. doi: 10.1038/cr.2010.103. [DOI] [PubMed] [Google Scholar]
- 72.Mayor A, Martinon F, De Smedt T, Pétrilli V, Tschopp J. A crucial function of SGT1 and HSP90 in inflammasome activity links mammalian and plant innate immune responses. Nat Immunol. 2007;8:497–503. doi: 10.1038/ni1459. [DOI] [PubMed] [Google Scholar]
- 73.Boldogh IR, Pon LA, Sheetz MP, De Vos KJ. Cell Free Assays for Mitochondria–Cytoskeleton Interactions. In: Pon LA, Schon EA, editors. Methods in Cell Biology. New York: Academic Press; 2007. pp. 683–706. [DOI] [PubMed] [Google Scholar]
- 74.Gourlay CW, Ayscough KR. The actin cytoskeleton: a key regulator of apoptosis and ageing? Nat Rev Mol Cell Biol. 2005;6:583–589. doi: 10.1038/nrm1682. [DOI] [PubMed] [Google Scholar]
- 75.Hyde BB, Twig G, Shirihai OS. Organellar vs cellular control of mitochondrial dynamics. Semin Cell Dev Biol. 2010;21:575–581. doi: 10.1016/j.semcdb.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Castanier C, Garcin D, Vazquez A, Arnoult D. Mitochondrial dynamics regulate the RIG-I-like receptor antiviral pathway. EMBO Rep. 2010;11:133–138. doi: 10.1038/embor.2009.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Koch A, Yoon Y, Bonekamp NA, McNiven MA, Schrader M. A role for Fis1 in both mitochondrial and peroxisomal fission in mammalian cells. Mol Biol Cell. 2005;16:5077–5086. doi: 10.1091/mbc.E05-02-0159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Koch A, Thiemann M, Grabenbauer M, Yoon Y, McNiven MA, Schrader M. Dynamin-like protein 1 is involved in peroxisomal fission. J Biol Chem. 2003;278:8597–8605. doi: 10.1074/jbc.M211761200. [DOI] [PubMed] [Google Scholar]
- 79.Yasukawa K, et al. Mitofusin 2 inhibits mitochondrial antiviral signaling. Sci Signal. 2009;2:ra47. doi: 10.1126/scisignal.2000287. [DOI] [PubMed] [Google Scholar]
- 80.De Brito OM, Scorrano L. Mitofusin 2 tethers endoplasmic reticulum to mitochondria. Nature. 2008;456:605–610. doi: 10.1038/nature07534. [DOI] [PubMed] [Google Scholar]
- 81.Høyer-Hansen M, Jäättelä M. Connecting endoplasmic reticulum stress to autophagy by unfolded protein response and calcium. Cell Death Differ. 2007;14:1576–1582. doi: 10.1038/sj.cdd.4402200. [DOI] [PubMed] [Google Scholar]
- 82.Onoguchi K, et al. Virus-infection or 5'ppp-RNA activates antiviral signal through redistribution of IPS-1 mediated by MFN1. PLoS Pathog. 2010;6:e1001012. doi: 10.1371/journal.ppat.1001012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Koshiba T, Yasukawa K, Yanagi Y, Kawabata S. Mitochondrial membrane potential is required for MAVS-mediated antiviral signaling. Sci Signal. 2011;4:ra7. doi: 10.1126/scisignal.2001147. [DOI] [PubMed] [Google Scholar]
- 84.Chen H, et al. Mitochondrial fusion is required for mtdna stability in skeletal muscle and tolerance of mtDNA mutations. Cell. 2010;141:280–289. doi: 10.1016/j.cell.2010.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Emre Y, Nübel T. Uncoupling protein UCP2: When mitochondrial activity meets immunity. FEBS Lett. 2010;584:1437–1442. doi: 10.1016/j.febslet.2010.03.014. [DOI] [PubMed] [Google Scholar]
- 86.Martinon F. Signaling by ROS drives inflammasome activation. Eur J Immunol. 2010;40:616–619. doi: 10.1002/eji.200940168. [DOI] [PubMed] [Google Scholar]
- 87.Tschopp J, Schroder K. NLRP3 inflammasome activation: The convergence of multiple signalling pathways on ROS production? Nat Rev Immunol. 2010;10:210–215. doi: 10.1038/nri2725. [DOI] [PubMed] [Google Scholar]
- 88.Van Bruggen R, et al. Human NLRP3 inflammasome activation is Nox1–4 independent. Blood. 2010;115:5398–5400. doi: 10.1182/blood-2009-10-250803. [DOI] [PubMed] [Google Scholar]
- 89.Nakahira K, et al. Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. Nat Immunol. 2011;12:222–230. doi: 10.1038/ni.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhou R, Yazdi AS, Menu P, Tschopp J. A role for mitochondria in NLRP3 inflammasome activation. Nature. 2011;469:221–226. doi: 10.1038/nature09663. [DOI] [PubMed] [Google Scholar]
- 91.Saitoh T, et al. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1beta production. Nature. 2008;456:264–268. doi: 10.1038/nature07383. [DOI] [PubMed] [Google Scholar]
- 92.Harris J, et al. Autophagy Controls IL-1β Secretion by Targeting Pro-IL-1β for Degradation. J Biol Chem. 2011;286:9587–9597. doi: 10.1074/jbc.M110.202911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Van De Veerdonk FL, et al. Reactive oxygen species-independent activation of the IL-1β inflammasome in cells from patients with chronic granulomatous disease. Proc Natl Acad Sci USA. 2010;107:3030–3033. doi: 10.1073/pnas.0914795107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Meissner F, Molawi K, Zychlinsky A. Superoxide dismutase 1 regulates caspase-1 and endotoxic shock. Nat Immunol. 2008;9:866–872. doi: 10.1038/ni.1633. [DOI] [PubMed] [Google Scholar]
- 95.Park J, et al. Drosophila porin/VDAC affects mitochondrial morphology. PLoS ONE. 2010;5:e13151. doi: 10.1371/journal.pone.0013151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Shoshan-Barmatz V, De Pinto V, Zweckstetter M, Raviv Z, Keinan N, Arbel N. VDAC, a multi-functional mitochondrial protein regulating cell life and death. Mol Asp Med. 2010;31:227–285. doi: 10.1016/j.mam.2010.03.002. [DOI] [PubMed] [Google Scholar]
- 97.Rostovtseva TK, Bezrukov SM. VDAC regulation: role of cytosolic proteins and mitochondrial lipids. J Bioenerg Biomembr. 2008;40:163–170. doi: 10.1007/s10863-008-9145-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Galluzzi L, Kroemer G. Mitochondrial apoptosis without VDAC. Nature Cell Biol. 2007;9:487–489. doi: 10.1038/ncb0507-487. [DOI] [PubMed] [Google Scholar]
- 99.Shoshan-Barmatz V, Zalk R, Gincel D, Vardi N. Subcellular localization of VDAC in mitochondria and ER in the cerebellum. Biochim Biophys Acta Bioenerg. 2004;1657:105–114. doi: 10.1016/j.bbabio.2004.02.009. [DOI] [PubMed] [Google Scholar]
- 100.De Pinto V, Guarino F, Guarnera A, Messina A, Reina S, Tomasello FM, et al. Characterization of human VDAC isoforms: A peculiar function for VDAC3? Biochimica et Biophysica Acta - Bioenergetics. 2010;1797:1268–1275. doi: 10.1016/j.bbabio.2010.01.031. [DOI] [PubMed] [Google Scholar]
- 101.Zhou R, Tardivel A, Thorens B, Choi I, Tschopp J. Thioredoxin-interacting protein links oxidative stress to inflammasome activation. Nat Immunol. 2010;11:136–140. doi: 10.1038/ni.1831. [DOI] [PubMed] [Google Scholar]
- 102.Zhang Q, et al. Circulating mitochondrial DAMPs cause inflammatory responses to injury. Nature. 2010;464:104–107. doi: 10.1038/nature08780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Yousefi S, et al. Catapult-like release of mitochondrial DNA by eosinophils contributes to antibacterial defense. Nat Med. 2008;14:949–953. doi: 10.1038/nm.1855. [DOI] [PubMed] [Google Scholar]
- 104.Sato A, Nakada K, Hayashi J. Mitochondrial complementation preventing respiratory dysfunction caused by mutant mtDNA. Biofactors. 2009;35:130–137. doi: 10.1002/biof.14. [DOI] [PubMed] [Google Scholar]
- 105.Archibald JM, Richards TA. Gene transfer: anything goes in plant mitochondria. BMC Biol. 2010;8:147. doi: 10.1186/1741-7007-8-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Henis-Korenblit S, et al. Insulin/IGF-1 signaling mutants reprogram ER stress response regulators to promote longevity. Proc Natl Acad Sci USA. 2010;107:9730–9735. doi: 10.1073/pnas.1002575107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Richardson CE, Kooistra T, Kim DH. An essential role for XBP-1 in host protection against immune activation in C. elegans. Nature. 2010;463:1092–1095. doi: 10.1038/nature08762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Smith JA, Turner MJ, Delay ML, Klenk EI, Sowders DP, Colbert RA. Endoplasmic reticulum stress and the unfolded protein response are linked to synergistic IFN-β induction via X-box binding protein 1. Eur J Immunol. 2008;38:1194–1203. doi: 10.1002/eji.200737882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zeng L, Liu Y-, Sha H, Chen H, Qi L, Smith JA. XBP-1 couples endoplasmic reticulum stress to augmented IFN-β induction via a cis-acting enhancer in macrophages. J Immunol. 2010;185:2324–2330. doi: 10.4049/jimmunol.0903052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Marcu MG, Doyle M, Bertolotti A, Ron D, Hendershot L, Neckers L. Heat shock protein 90 modulates the unfolded protein response by stabilizing IRE1α. Mol Cell Biol. 2002;22:8506–8513. doi: 10.1128/MCB.22.24.8506-8513.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Knösel T, Schewe C, Petersen N, Dietel M, Petersen I. Prevalence of infectious pathogens in Crohn's disease. Pathol Res Pract. 2009;205:223–230. doi: 10.1016/j.prp.2008.04.018. [DOI] [PubMed] [Google Scholar]
- 112.Van Kruiningen HJ, et al. Search for evidence of recurring or persistent viruses in Crohn's disease. APMIS. 2007;115:962–968. doi: 10.1111/j.1600-0463.2007.apm_564.x. [DOI] [PubMed] [Google Scholar]
- 113.Foxman EF, Iwasaki A. Genome-virome interactions: examining the role of common viral infections in complex disease. Nat Rev Microbiol. 2011;9:254–264. doi: 10.1038/nrmicro2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kuballa P, Huett A, Rioux JD, Daly MJ, Xavier RJ. Impaired autophagy of an intracellular pathogen induced by a Crohn's disease associated ATG16L1 variant. PLoS ONE. 2008;3:e3391. doi: 10.1371/journal.pone.0003391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Cadwell K, et al. A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature. 2008;456:259–263. doi: 10.1038/nature07416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Cadwell K, Patel KK, Komatsu M, Virgin HW, 4th, Stappenbeck TS. A common role for Atg16L1, Atg5 and Atg7 in small intestinal Paneth cells and Crohn disease. Autophagy. 2009;5:250–252. doi: 10.4161/auto.5.2.7560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Cadwell K, et al. Virus-Plus-Susceptibility Gene Interaction Determines Crohn's Disease Gene Atg16L1 Phenotypes in Intestine. Cell. 2010;141:1135–1145. doi: 10.1016/j.cell.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.McCartney SA, Thackray LB, Gitlin L, Gilfillan S, Virgin HW, Colonna M. MDA-5 recognition of a murine norovirus. PLoS Pathog. 2008;4:e1000108. doi: 10.1371/journal.ppat.1000108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Travassos LH, et al. Nod1 and Nod2 direct autophagy by recruiting ATG16L1 to the plasma membrane at the site of bacterial entry. Nat Immunol. 2010;11:55–62. doi: 10.1038/ni.1823. [DOI] [PubMed] [Google Scholar]
- 120.Cooney R, et al. NOD2 stimulation induces autophagy in dendritic cells influencing bacterial handling and antigen presentation. Nat Med. 2010;16:90–97. doi: 10.1038/nm.2069. [DOI] [PubMed] [Google Scholar]
- 121.Homer CR, Richmond AL, Rebert NA, Achkar J, McDonald C. ATG16L1 and NOD2 interact in an autophagy-dependent antibacterial pathway implicated in crohn's disease pathogenesis. Gastroenterology. 2010;139:1630–1641. doi: 10.1053/j.gastro.2010.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Kaser A, et al. XBP1 Links ER Stress to Intestinal Inflammation and Confers Genetic Risk for Human Inflammatory Bowel Disease. Cell. 2008;134:743–756. doi: 10.1016/j.cell.2008.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Singh SB, Davis AS, Taylor GA, Deretic V. Human IRGM induces autophagy to eliminate intracellular mycobacteria. Science. 2006;313:1438–1441. doi: 10.1126/science.1129577. [DOI] [PubMed] [Google Scholar]
- 124.Brest P, et al. A synonymous variant in IRGM alters a binding site for miR-196 and causes deregulation of IRGM-dependent xenophagy in Crohn's disease. Nat Genet. 2011;43:242–245. doi: 10.1038/ng.762. [DOI] [PubMed] [Google Scholar]
- 125.Singh SB, Ornatowski W, Vergne I, Naylor J, Delgado M, Roberts E, et al. Human IRGM regulates autophagy and cell-autonomous immunity functions through mitochondria. Nature Cell Biol. 2010;12:1154–1165. doi: 10.1038/ncb2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Barbosa DS, Cecchini R, El Kadri MZ, Rodríguez MAM, Burini RC, Dichi I. Decreased oxidative stress in patients with ulcerative colitis supplemented with fish oil ω-3 fatty acids. Nutrition. 2003;19:837–842. doi: 10.1016/s0899-9007(03)00162-x. [DOI] [PubMed] [Google Scholar]
- 127.Kruidenier L, et al. Differential mucosal expression of three superoxide dismutase isoforms in inflammatory bowel disease. J Pathol. 2003;201:7–16. doi: 10.1002/path.1407. [DOI] [PubMed] [Google Scholar]
- 128.Beltŕan B, et al. Mitochondrial dysfunction, persistent oxidative damage, and catalase inhibition in immune cells of naïve and treated Crohn's disease. Inflammatory Bowel Dis. 2010;16:76–86. doi: 10.1002/ibd.21027. [DOI] [PubMed] [Google Scholar]
- 129.Te Velde AA, Pronk I, De Kort F, Stokkers PCF. Glutathione peroxidase 2 and aquaporin 8 as new markers for colonic inflammation in experimental colitis and inflammatory bowel diseases: An important role for H2O2? Eur J Gastroenterol Hepatol. 2008;20:555–560. doi: 10.1097/MEG.0b013e3282f45751. [DOI] [PubMed] [Google Scholar]
- 130.Brownlee IA, Knight J, Dettmar PW, Pearson JP. Action of reactive oxygen species on colonic mucus secretions. Free Radic Biol Med. 2007;43:800–808. doi: 10.1016/j.freeradbiomed.2007.05.023. [DOI] [PubMed] [Google Scholar]