Abstract
Background
For the Solanaceae-type self-incompatibility, also possessed by Rosaceae and Plantaginaceae, the specificity of self/non-self interactions between pollen and pistil is controlled by two polymorphic genes at the S-locus: the S-locus F-box gene (SLF or SFB) controls pollen specificity and the S-RNase gene controls pistil specificity.
Scope
This review focuses on the work from the authors' laboratory using Petunia inflata (Solanaceae) as a model. Here, recent results on the identification and functional studies of S-RNase and SLF are summarized and a protein-degradation model is proposed to explain the biochemical mechanism for specific rejection of self-pollen tubes by the pistil.
Conclusions
The protein-degradation model invokes specific degradation of non-self S-RNases in the pollen tube mediated by an SLF, and can explain compatible versus incompatible pollination and the phenomenon of competitive interaction, where SI breaks down in pollen carrying two different S-alleles. In Solanaceae, Plantaginaceae and subfamily Maloideae of Rosaceae, there also exist multiple S-locus-linked SLF/SFB-like genes that potentially function as the pollen S-gene. To date, only three such genes, all in P. inflata, have been examined, and they do not function as the pollen S-gene in the S-genotype backgrounds tested. Interestingly, subfamily Prunoideae of Rosaceae appears to possess only a single SLF/SFB gene, and competitive interaction, observed in Solanaceae, Plantaginaceae and subfamily Maloideae, has not been observed. Thus, although the cytotoxic function of S-RNase is an integral part of SI in Solanaceae, Plantaginaceae and Rosaceae, the function of SLF/SFB may have diverged. This highlights the complexity of the S-RNase-based SI mechanism. The review concludes by discussing some key experiments that will further advance our understanding of this self/non-self discrimination mechanism.
Keywords: Competitive interaction, Petunia inflata, Plantaginaceae, protein degradation, Rosaceae, self-incompatibility, S-locus F-box protein, Solanaceae, S-RNase, ubiquitination
INTRODUCTION
Flowering plants (angiosperms) represent the most diverse group of land plants, and most of them have perfect flowers, with the male reproductive organ (anther) and the female reproductive organ (pistil) located in close proximity. As self-fertilization results in inbreeding, the outcome of which is deleterious to the long-term survival of any population, flowering plants have adopted various strategies to circumvent the strong tendency to self-fertilize so as to promote out-crossing and generate genetic variability within a species. Self-incompatibility (SI) is one such strategy widely adopted by flowering plants (de Nettancourt, 2001) and it has long been considered responsible for their explosive success (Whitehouse, 1950). SI allows the pistil to distinguish between genetically related (self) and genetically unrelated (non-self) pollen; self pollen is rejected, either on the stigmatic surface or during its tube growth in the style, whereas non-self pollen is accepted for fertilization. In the simplest cases, self/non-self recognition between pollen and the pistil is controlled by one highly polymorphic locus, named the S-locus, with variants of the locus referred to as ‘haplotypes’ and designated S1, S2, S3, etc.
To date, extensive molecular studies have been carried out on five of the families that possess SI, all of which have a single-locus SI system. The results have revealed that (a) flowering plants have adopted different SI mechanisms to accomplish the same goal, and (b) for each mechanism, two separate genes at the S-locus control pollen and pistil functions in SI. Based on the chemical nature of the pollen and pistil S-genes, three of these five families, Solanaceae, Rosaceae and Plantaginaceae, are thought to employ a similar SI mechanism, whereas Brassicaceae and Papaveraceae each employ different mechanisms (Takayama and Isogai, 2005). This paper focuses on the Solanaceae mechanism, and particularly the results obtained in the authors' laboratory using Petunia inflata as a model. Recent reviews of this mechanism can be found in Kao and Tsukamoto (2004), Sims (2007), Franklin-Tong (2008), Hua et al. (2008), McClure (2009) and Chen et al. (2010).
For the Solanaceae-type SI, pollen behaviour is determined by its own S-genotype (i.e. pollen is recognized as self-pollen and rejected by the pistil only if its S-haplotype is identical to one of the two S-haplotypes carried by the pistil). Two tightly linked genes at the S-locus have been identified and shown to be involved in SI, the S-RNase gene controlling pistil specificity (Lee et al., 1994; Murfett et al., 1994) and the S-locus F-box (SLF or SFB) gene controlling pollen specificity (Entani et al., 2003; Yamane et al., 2003; Qiao et al., 2004b; Sijacic et al., 2004; Ushijima et al., 2004; Sonneveld et al., 2005; Sassa et al., 2007). In this type of SI, the rejection of self-pollen occurs during the growth of self-pollen tubes in the style, around the time when growth shifts from the autotrophic phase to the heterotrophic phase (Herrero and Hormaza, 1996).
S-RNASE: THE PISTIL SPECIFICITY DETERMINANT
The S-RNase gene was first identified in Nicotiana alata by Anderson et al. (1986) after its allelic products had been identified based on their showing S-haplotype-specific differences in molecular mass and isoelectric point. S-RNase exhibits properties expected of the protein functioning as the pistil specificity determinant for the following reasons. S-RNase is a pistil-specific protein, and it is initially synthesized in the transmitting cells of the style and then secreted into the extracellular space of the transmitting tract. S-RNase is most abundant in the upper third segment of the style, a location coinciding with the site of growth arrest of self-pollen tubes after incompatible pollination (Ai et al., 1990). S-RNase is present at very low levels in immature pistils, which do not exhibit SI, but is abundantly present in mature pistils, accounting for up to 10 % of total pistil protein (Roalson and McCubbin, 2003), when SI is fully functional. S-RNase shows a high degree of allelic sequence diversity, with the most divergent pair sharing only approx. 38 % amino acid sequence identity (Tsai et al., 1992), as expected of a protein functioning in self/non-self recognition. The function of the S-RNase gene in SI was definitively established from gain-of-function and loss-of-function experiments conducted in transgenic plants (Lee et al., 1994; Murfett et al., 1994), which showed that S-RNase is necessary and sufficient for the pistil to recognize and reject self-pollen. Specifically, the S3-RNase gene of P. inflata was introduced into plants of S1S2 genotype and it was found that the transgenic plants that produced S3-RNase from the transgene acquired the ability to reject S3 pollen. When an antisense S3-RNase gene was introduced into plants of S2S3 genotype to suppress the production of S3-RNase, the transgenic plants lost the ability to reject S3 pollen, but still retained the ability to reject S2 pollen (Lee et al., 1994). Thus, the S-RNase gene alone determines pistil specificity in SI.
S-RNase was so named because it has ribonuclease activity (McClure et al., 1989; Broothaerts et al., 1991; Singh et al., 1991), which was shown to be essential for rejection of self-pollen (Huang et al., 1994). Thus, the biochemical mechanism of rejection of self-pollen tubes by the pistil most likely involves degradation of pollen RNA in incompatible pollen tubes. S-RNases are glycoproteins with various numbers of N-linked glycan chains; however, the carbohydrate moiety is not required for their RNase activity or recognition function (Karunanandaa et al., 1994). Thus, the allelic specificity determinant of S-RNase resides in its protein backbone. Comparison of the sequences of S-RNases from different solanaceous species revealed five conserved regions, named C1 to C5, and two hypervariable regions, named HVa and HVb (Ioerger et al., 1991). The crystal structure of an S-RNase of N. alata revealed that the HVa and HVb regions are located on the outside surface of the molecule, suggesting that they are likely to be involved in interactions with allelic products of SLF (Ida et al., 2001). However, domain-swapping experiments showed that these hypervariable regions are necessary but not sufficient for the allelic specificity of S-RNases of P. inflata (Kao and McCubbin, 1996) and of N. alata (Zurek et al., 1997). Thus, it remains unknown which amino acid residues of S-RNase are involved in S-allele specificity.
S-LOCUS F-BOX PROTEIN: THE POLLEN SPECIFICITY DETERMINANT
Identification of the gene encoding the pollen specificity determinant turned out to be much more difficult than the identification of the S-RNase gene. Attempts to identify pollen/pollen tube proteins that showed S-haplotype-specific differences in molecular mass and/or isoelectric point did not result in any potential candidate, nor did attempts to identify pollen/pollen tube proteins that interact with S-RNase (Dowd et al., 2000). RNA differential display was used to identify pollen-specific genes that are tightly linked to the S-locus (McCubbin et al., 2000a); however, none of the genes identified were deemed as good candidates because they all showed low degrees of allelic sequence diversity at the amino acid sequence level (Wang et al., 2003). Interestingly, two of the genes found through RNA differential display encode F-box proteins and were later identified as SLF-like proteins (Hua et al., 2007). Ultimately, it was through genomic sequencing of the S-locus that SLF was identified, first in Antirrhinum (Plantaginaceae) (Lai et al., 2002) and then in two rosaceous species (Entani et al., 2003; Ushijima et al., 2003) and P. inflata (Wang et al., 2004). In P. inflata, a BAC library of S2S2 genotype was constructed (McCubbin et al., 2000b), and screening of the library using the S2-RNase gene as a probe, followed by chromosome walking, resulted in an 881-kb contig. Sequencing of a 328-kb region of this contig that included the S2-RNase gene revealed that it was rich in repetitive sequences and that the majority of the predicted genes with sequence similarities to known genes in the database encoded retrotransposons or polyproteins, with the only exception being PiSLF (P. inflata SLF), located 161 kb downstream from the S2-RNase gene (Wang et al., 2004). PiSLF exhibited several characteristics expected of the pollen S-gene, including S-haplotype-specific RFLP, developing pollen- and mature pollen/pollen tube-specific expression, and the deduced amino acid sequences of the S1, S2 and S3 alleles showing approx. 10 % pair-wise sequence diversity.
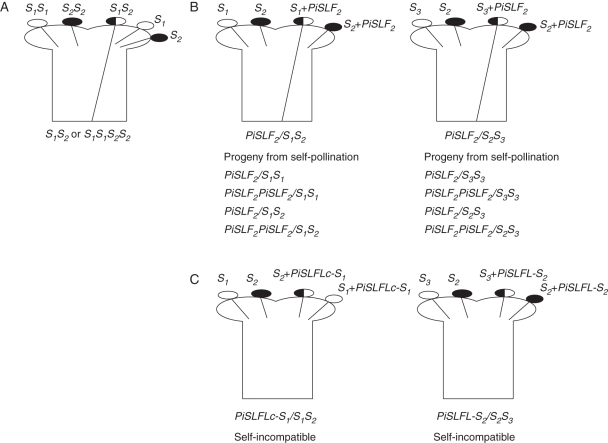
As stated earlier, expression of an allele of S-RNase different from the two alleles of S-RNase present in the pistil of transgenic plants confers on the pistil the S-allele specificity of the introduced S-RNase (Lee et al., 1994). Similarly, one might expect that, if PiSLF is the pollen S-gene, expression of an additional allele of PiSLF different from the allele of PiSLF present in pollen grains of transgenic plants would lead to gain of the S-allele specificity of the introduced PiSLF in the transgenic pollen. The competitive interaction phenomenon (Fig. 1A), discovered from earlier genetic studies in solanaceous species including P. inflata (Brewbaker and Natarajan, 1960; Entani et al., 1999; de Nettancourt, 2001), provided the basis for predicting the outcome of such an experiment. It was found that diploid pollen carrying two different pollen S-alleles (heteroallelic pollen, e.g. resulting from duplication of the entire set of chromosomes in a diploid plant with two different S-haplotypes) failed to function in SI, whereas diploid pollen carrying two copies of the same pollen S-allele (homoallellic pollen) functioned normally in SI. Although the biochemical basis of the phenomenon was unknown, taking advantage of competitive interaction, the function of PiSLF in SI was ascertained (Sijacic et al., 2004). The prediction was that if PiSLF was the pollen S-gene, then the expression of two different alleles of PiSLF in a pollen grain should cause breakdown of SI. The S2-allele of PiSLF (PiSLF2) was introduced into S1S1, S1S2 and S2S3 plants, and the presence of PiSLF2 in S1 and S3 pollen grains, but not in S2 pollen grains, was found to cause the breakdown of SI, as predicted by competitive interaction and confirmed through progeny analysis. For example, self-pollination of a PiSLF2/S2S3 transgenic plant resulted in S2S3 and S3S3 progeny plants, but no S2S2 progeny plants, and all progeny plants carried the PiSLF2 transgene (Fig. 1B). A biochemical model for competitive interaction will be presented in the next section.
Fig. 1.
Competitive interaction and its use in establishing the function of PiSLF and assessing potential function of PiSLF-like genes in SI. (A) Competitive interaction. For a diploid self-incompatible plant of S1S2 genotype, S1 and S2 pollen are rejected by the S1S2 pistil due to matching of S-haplotypes. A tetraploid plant of S1S1S2S2 genotype produces three S-genotypes of pollen and, whereas S1S1 and S2S2 pollen are rejected by S1S2 and S1S1S2S2 pistils, S1S2 (heteroallelic) pollen is not. (B) Breakdown of SI in S1S2 and S2S3 transgenic plants caused by expression of PiSLF2 in pollen. The left panel shows self-pollination of a transgenic plant PiSLF2/S1S2. The plant produces four different genotypes of pollen and, based on inheritance of the PiSLF2 transgene and the S-genotypes in the progeny, only S1 pollen carrying the PiSLF2 transgene was accepted by the pistil. The right panel shows self-pollination of a transgenic plant PiSLF2/S2S3. Progeny analysis suggests that the PiSLF2 transgene caused breakdown of SI in S3 pollen, but not in S2 pollen. That PiSLF2 caused breakdown of SI in S1 and S3 pollen, but not in S2 pollen, is consistent with the prediction made by competitive interaction, that pollen carrying two different S-alleles fails to function in SI. (C) Testing potential function of PiSLF-like genes. The left panel shows self-pollination of an S1S2 transgenic plant carrying the S1-allele of a PiSLF-like gene, PiSLFLc. The transgenic plant remained self-incompatible, suggesting that PiSLFLc-S1 did not cause breakdown of SI in S2 pollen. The right panel shows self-pollination of an S2S3 transgenic plant carrying the S2-allele of either PiSLFLb or PiSLFLd (labelled as PiSLFL-S2). The resulting transgenic plants remained self-incompatible, suggesting that neither PiSLFLb-S2 nor PiSLFLd-S2 caused breakdown of SI in S3 pollen.
BIOCHEMICAL BASIS FOR SPECIFIC REJECTION OF SELF-POLLEN
With the identification of the S-RNase and SLF genes, a model, based on the biochemical properties of the protein products of these genes, their locations within the pollen tube and the interactions between different allelic products of these two proteins, has been proposed to explain S-haplotype-specific rejection of self-pollen tube growth by the pistil in P. inflata. A model, which is likely to be applicable to other solanaceous species, Antirrhinum (Plantaginaceae) and the Maloideae subfamily of Rosaceae, will be proposed, followed by a discussion of the possible differences in the function of SLF/SFB in the Prunoideae subfamily of Rosaceae.
An SLF selectively detoxifies its non-self S-RNases by mediating their degradation inside pollen tubes
Luu et al. (2000) and Goldraij et al. (2006) showed that uptake of S-RNase by growing pollen tubes is not S-haplotype-specific, because both self and non-self S-RNases were found inside pollen tubes after pollination (e.g. both S1-RNase and S2-RNase are taken up by S1 pollen tubes growing in an S1S2 style). This is an important finding, as it rules out any model that invokes specific uptake of self S-RNase by a pollen tube as being responsible for rejection of self-pollen tubes (Kao and McCubbin, 1996). However, Luu et al. (2000) observed that all S-RNases were localized in the cytoplasm of the pollen tube, whereas Goldraij et al. (2006) observed that most, if not all, S-RNases were initially sequestered in a vacuole-like compartment, which was later specifically disrupted in incompatible pollen tubes, releasing the sequestered S-RNases into the cytoplasm. Regardless of whether S-RNases are taken up directly into the cytoplasm or into a vacuolar compartment, they must eventually enter the cytoplasm in order to exert their cytotoxic activity, as it has been shown that the RNase activity of S-RNase is required for its function (Huang et al., 1994) and that pollen rRNA is degraded after incompatible pollination, but not after compatible pollination (McClure et al., 1990). Interestingly, when S2-RNase and S3-RNase of P. inflata were expressed as green fluorescent protein (GFP) fusion proteins in their respective self and non-self pollen, all the ectopically expressed S-RNases were compartmentalized in both self and non-self pollen tubes, and direct expression of S-RNase in pollen had no effect on its viability or SI behaviour (Meng et al., 2009). Whether the S-RNase-containing compartments observed by the authors are the same as those observed by Goldraij et al. (2006) remains to be determined, but the results lend additional support to the proposal that the cytoplasm of the pollen tube is the site of the cytotoxic action of S-RNase. This is also consistent with the cytoplasmic localization of SLF (Hua et al., 2007; Meng et al., 2009).
Although both self and non-self S-RNases are taken up by a pollen tube, only self S-RNase exerts its cytotoxic activity to inhibit the growth of the pollen tube. The biochemical properties of PiSLF have allowed the authors to begin to elucidate the molecular basis for this observation. Most F-box proteins are components of SCF (Skp1-Cullin-F-box) complexes, which are a type of E3 ubiquitin ligase complex. This protein-degradation system uses E1 (ubiquitin activating enzyme), E2 (ubiquitin conjugating enzyme) and E3 (ubiquitin ligase) to transfer polyubiquitin chains to target proteins for degradation by the 26S proteasome (Hershko and Ciechanover, 1998; Moon et al., 2004; Smalle and Vierstra, 2004). The F-box protein component of an SCF complex recognizes and interacts with a specific set of proteins to result in their specific degradation (Tyers and Jorgensen, 2000). Since SLF contains an F-box domain at its N-terminus, it is reasonable to hypothesize that SLF is a component of an SCF complex, and that SLF mediates specific degradation of all its non-self S-RNases inside a pollen tube. This can explain why only self S-RNase functions inside a pollen tube, and results consistent with this model are described below.
First, SLF is a component of either a canonical SCF complex or a novel E3 ubiquitin ligase complex. A conventional SCF complex consists of four proteins, Skp1, Cullin, a RING-HC finger protein (Rbx1) and an F-box protein (Tyers and Jorgensen, 2000; Moon et al., 2004). Cullin serves as a bridge, binding Rbx1 through its C-terminal domain and Skp1 through its N-terminal domain. The F-box protein interacts with Skp1 through its F-box domain, and interacts with specific substrates with a separate protein–protein interaction domain, which may contain WD40-repeats, leucine-rich repeats, etc. (Cenciarelli et al., 1999). However, no recognizable protein–protein interaction domain has been identified within the C-terminal part of SLF. In Antirrhinum, the SLF-containing complex is thought to be a conventional SCF complex, which contains a novel Skp1-like protein, SSK1 (SLF-interacting SKP1-like1) (Huang et al., 2006; Zhao et al., 2010). SSK1 is specifically expressed in pollen, but is encoded by a monomorphic gene. Interestingly, SSK1 has a unique seven to nine amino-acid tail consisting of a disordered coil at the C-terminus, located downstream from the conventional C-terminal residues ‘WAFE’ found in most plant Skp1 homologues (Gagne et al., 2002; Risseeuw et al., 2003). Since no other Skp1-like proteins interact with AhSLF2, the unique C-terminal tail of SSK1 may be important for the interaction. In P. inflata, PiSLF is thought to be a component of a novel E3 ubiquitin ligase complex that contains PiCUL1-G and PiSBP1 (P. inflata S-RNase Binding Protein1, a RING-HC protein), but does not contain Skp1 or Rbx1 (Hua and Kao, 2006). In this complex, PiSBP1 rather than Skp1 is thought to organize PiCUL1-G and PiSLF since, like Skp1, PiSBP1 interacts with the Cullin component (PiCUL1-G) and the F-box component (PiSLF) of the putative complex, and like Rbx1, interacts with an E2. The fact that PiSBP1 is three times the size of PiRBX1 (Hua and Kao, 2006) lends support for this dual role. However, since PiSBP1 is expressed in all tissues examined (Hua and Kao, 2006), it may have a more general role than just in pollination. For example, yeast two-hybrid and in vitro pull-down assays showed that NaSBP1, SBP1 of N. alata, interacted with the C-terminal domain of two arabinogalactan proteins in the extracellular matrix of the pistil, suggesting that SBP1 might have a role in trafficking of endocytic cargo (Lee et al., 2008).
Second, Liu et al. (2009) developed a style-by-style method for quantification of S-RNase levels in Solanum chacoense (Solanaceae), and they reported that the S-RNase levels measured in unpollinated styles were approximately equal to those measured in incompatibly pollinated styles, but the S-RNase levels measured in compatibly pollinated styles were lower by up to 30 %. These results provide in vivo support for the notion that non-self S-RNases are degraded in compatible pollen tubes, whereas self S-RNase is not degraded in incompatible tubes. Moreover, in vitro, cell-free protein ubiquitination and degradation assays have been developed for P. inflata, and it has been shown that S-RNases were ubiquitinated and degraded in pollen tube extracts (Hua and Kao, 2006). The observed degradation and ubiquitination of S-RNase was not S-haplotype-specific, but this could be because the in vitro systems did not completely mimic in vivo conditions. For Antirrhinum, the physical interaction between AhSLF-S2 and S-RNase was demonstrated by pull-down, yeast two-hybrid, and co-immunoprecipitation, although allelic specificity was not shown. Upon compatible pollination, the extent of ubiquitination increased and the growth of pollen tubes was inhibited after treatment with a proteasomal inhibitor, suggesting that non-self S-RNases may be degraded by the ubiquitin-26S proteasome pathway (Qiao et al., 2004a, b). Since lysine residues are normally the attachment sites for polyubiquitin chains, each of the 20 lysine residues of S3-RNase of P. inflata was mutated to arginine, either one at a time or several at a time, and the rate of degradation of all the resulting lysine-to-arginine mutant proteins assayed in the in vitro system. It was found that changing six lysine residues near the C-terminus to arginines had the most significant effect on the extent of ubiquitination and rate of degradation of the mutant S-RNases (Hua and Kao, 2008). Interestingly, an alignment of the amino acid sequences of 32 S-RNases from several solanaceous species showed that two of these six lysines are among the most highly conserved lysines of S-RNases (Hua and Kao, 2008).
Third, in vitro protein-binding assays showed that a PiSLF interacts with its non-self S-RNases more strongly than with its self S-RNase (Hua and Kao, 2006). For example, when equal amounts of GST-tagged S1-RNase and S3-RNase were separately assayed for their interactions with His-tagged PiSLF1, GST:S3-RNase pulled down more His:PiSLF1 than did GST:S1-RNase, i.e. PiSLF1 interacted more strongly with its non-self S-RNase, S3-RNase, than with its self S-RNase, S1-RNase.
Biochemical basis for preferential non-self interaction between SLF and S-RNase
It may seem counterintuitive that non-self interactions between S-RNase and SLF are favoured over self-interactions, as self-interactions are usually viewed as between a key and a matching lock. However, SLF and S-RNase could have evolved in such a way that the overall strength of interaction is reduced when they are derived from the same S-haplotype. Thus, any S-RNase that interacts strongly with an SLF would suffer the fate of degradation, and only when an S-RNase has acquired a specificity domain that matches the specificity domain of SLF would it escape degradation.
To understand the biochemical basis for the differential interactions between an SLF and its self and non-self S-RNases, first six PiSLF-like genes of P. inflata and three alleles of PiSLF were compared (McCubbin et al., 2000a; Wang et al., 2003; Hua et al., 2007). The PiSLF-like genes share a number of properties with PiSLF. They are tightly linked to the S-locus, show S-haplotype-specific RFLP, are specifically expressed in pollen, and their deduced amino acid sequences are 50–54 % identical with PiSLF. However, when three of these PiSLF-like genes (one from S1-haplotype and two from S2-haplotype) were expressed in pollen of transgenic S1S2 or S2S3 plants, none of them caused the breakdown of SI function in their respective heteroallelic pollen grains (Fig. 1C). An in vitro protein-binding assay was also used to show that these PiSLF-like proteins either did not interact with S-RNase, or interacted with S-RNase much more weakly than did PiSLF2.
Comparison of the amino acid sequences of three allelic variants of PiSLF and ten PiSLF-like proteins (some of which are allelic variants) revealed three PiSLF-specific regions. PiSLF was then divided into three functional domains, named FD1, FD2 and FD3, each of which contains one of the PiSLF-specific regions (Hua et al., 2007). In vitro protein-binding assays using various truncated forms of PiSLF2 containing one or two domains showed that FD2 interacted with S3-RNase the strongest, even more strongly than did full-length PiSLF2, and that addition of FD1 or FD3 to FD2 reduced the strength of interaction. As FD2 is relatively conserved among the three allelic variants of PiSLF, it may be the primary region for the interaction between PiSLF and S-RNase. FD1 and FD3 each contain one of the two variable regions of PiSLF, and they may negatively regulate the strong interaction between FD2 and S-RNase. Results from in vitro protein-binding assays using chimeric proteins between PiSLF1 and PiSLF2, with their FD1 and FD3 swapped, suggested that FD1 and FD3 together determine the strength of interaction with S-RNase. For example, a chimeric protein with FD1 and FD3 from PiSLF1 and FD2 from PiSLF2 interacted with S2-RNase as strongly as did full-length PiSLF1, whereas a chimeric protein with FD1 and FD3 from PiSLF2 and FD2 from PiSLF1 interacted with S2-RNase as weakly as did full-length PiSLF2. Thus, FD1 and FD3 together appear to constitute the S-allele-specificity determinant of PiSLF. Our lab proposes that during self-interactions, matching of FD1 and FD3 of a PiSLF and the specificity domain of its self S-RNase weakens the otherwise strong interaction between FD2 and S-RNase. How this may occur is not known at the biochemical level.
Biochemical model for compatible and incompatible pollinations and competitive interaction
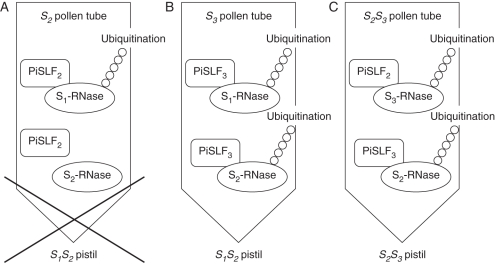
The proposed protein-degradation model can explain the outcome of compatible and incompatible pollinations and, most importantly, the phenomenon of competitive interaction (Hua et al., 2008). In the case of incompatible pollination, for example, pollination of an S1S2 style by S2 pollen (Fig. 2A), both S1-RNase and S2-RNase are taken up by the S2 pollen tube during penetration into the style, and the strong interaction between SLF2, produced in the cytoplasm of the S2 pollen tube, and S1-RNase (a non-self S-RNase for SLF2) would result in the ubiquitination and degradation of S1-RNase. However, S2-RNase (self S-RNase for SLF2) would not be ubiquitinated or degraded, and would thus degrade pollen RNA to result in inhibition of tube growth. In the case of compatible pollination, for example, pollination of an S1S2 style by S3 pollen (Fig. 2B), the strong interaction between SLF3 and S1-RNase and S2-RNase (both non-self S-RNases for SLF3) inside the S3 pollen tube would result in their ubiquitination and degradation, thus allowing the S3 pollen tube to circumvent the toxic effect of S-RNase. In the case of competitive interaction, for example, pollination of an S2S3 pistil by heteroallelic S2S3 pollen (Fig. 2C), SLF2 and SLF3 would preferentially interact with their respective non-self S-RNases (S3-RNase for SLF2 and S2-RNase for SLF3) in the cytoplasm of the S2S3 pollen tube, and as a result, both S2-RNase and S3-RNase would be ubiquitinated and degraded. Thus, heteroallelic S2S3 pollen would be compatible with the S2S3 pistil. The fact that heteroallelic pollen, regardless of the two S-haplotypes it carries, is universally accepted by pistils of any S-genotype can be explained by our model, which predicts that the two allelic products of SLF produced in heteroallelic pollen together would mediate the ubiquitination and degradation of all S-RNases.
Fig. 2.
Biochemical model for compatible and incompatible pollinations and competitive interaction. (A) Incompatible pollination. An S2 pollen tube produces PiSLF2, and as it is growing in an S1S2 pistil, it takes up S1-RNase and S2-RNase from the style. PiSLF2 preferentially interacts with S1-RNase (a non-self S-RNase) to mediate its ubiquitination and degradation. S2-RNase (self S-RNase) is not affected and can exert its cytotoxic function in degrading pollen RNA. As a result, growth of the S2 pollen tube is arrested. (B) Compatible pollination. An S3 pollen tube produces PiSLF3 and, as it is growing in an S1S2 pistil, it takes up S1-RNase and S2-RNase from the style. PiSLF3 interacts strongly with S1-RNase and S2-RNase (non-self S-RNases), and mediates their ubiquitination and degradation. Thus, the S3 pollen tube is able to grow through the style to effect fertilization. (C) Competitive interaction. An S2S3 heteroallelic pollen tube produces both PiSLF2 and PiSLF3 and, as it is growing in an S2S3 pistil, it takes up S2-RNase and S3-RNase from the style. PiSLF2 preferentially interacts with S3-RNase to mediate its ubiquitination and degradation, and PiSLF3 preferentially interacts with S2-RNase to mediate its ubiquitination and degradation. As a result, the S2S3 pollen tube is able to grow through the style to effect fertilization.
Mechanistic differences for S-RNase-based SI in subfamily Prunoideae of Rosaceae
Although Solanaceae, Rosaceae and Plantaginaceae all employ S-RNase as the pistil determinant and SLF/SFB as the pollen determinant, how SLF/SFB functions in SI interactions between pollen and pistil may not be entirely conserved (Table 1). First, the above-mentioned competitive interaction phenomenon is observed in Solanaceae, Plantaginaceae and the subfamily Maloideae of Rosaceae (Crane and Lewis, 1942; Golz et al., 1999, 2001; Adachi et al., 2009; Xue et al., 2009), but not in the subfamily Prunoideae of Rosaceae (Hauck et al., 2006b; Tsukamomo et al., 2006). [Huang et al. (2008) reported the only case of competitive interaction in a self-compatible line of tetraploid Prunus pseudocerasus (Prunoideae), but Tao and Iezzoni (2010) raised the possibility that the breakdown of SI was caused by mutations in other SI-related genes.] Second, defects in SLF of Solanaceae and Plantaginaceae most likely lead to pollen rejection by pistils of any S-genotype, as such mutants have never been found among pollen-part self-compatible mutants. In contrast, mutations in SLF/SFB of Prunoideae result in breakdown of SI (Ushijima et al., 2004; Tsukamoto et al., 2006; Yamane and Tao, 2009). These findings suggest that, in contrast to the proposed function of SLF of Solanaceae, Plantaginaceae and subfamily Maloideae of Rosaceae in mediating degradation of non-self S-RNases, the function of SLF/SFB in subfamily Prunoideae of Rosaceae may be to specifically protect self S-RNase from being degraded. Thus, loss-of-function of SLF/SFB in this subfamily results in the inability of pollen tubes to protect self S-RNase, as well as all non-self S-RNases, from being degraded.
Table 1.
Mechanistic differences for S-RNase-based SI between subfamily Prunoideae of Rosaceae and Solanaceae, Plantaginaceae and subfamily Maloideae
| Family | Subfamily | Competitive interaction | Defects in SLF/SFB | Potential number of genes for pollen specificity | Proposed function of SLF/SFB |
|---|---|---|---|---|---|
| Rosaceae | Prunoideae | No* | SC† | Single | Protect self S-RNase from being degraded |
| Rosaceae | Maloideae | Yes | Not reported | Multiple | Mediate degradation of non-self S-RNases |
| Solanaceae | Yes | Not reported | Multiple | Mediate degradation of non-self S-RNases | |
| Plantaginaceae | Yes | Not reported | Multiple | Mediate degradation of non-self S-RNases |
* Huang et al. (2008) reported the only case of competitive interaction in tetraploid Prunus pseudocerasus, but the breakdown of SI could have been caused by mutations in other SI-related genes (Tao and Iezzoni, 2010).
† SC (self-compatible) indicates breakdown of SI in pollen.
Another significant difference between Solanaceae, Plantaginaceae and subfamily Maloideae of Rosaceae, and subfamily Prunoideae of Rosaceae is the copy number of SLF/SFB (Sassa et al., 2010). In Prunoideae, although there are additional F-box genes linked to the S-locus of Prunus, they exhibit much lower allelic sequence diversity compared with SLF/SFB and are not thought to be involved in SI (Ushijima et al., 2003). On the contrary, in Maloideae, Sassa et al. (2007) identified two highly similar F-box genes (87·5 % identity in their deduced amino acid sequences) of apple, named SFBBs (S-locus F-box brothers), and proposed that both encode the pollen specificity determinant. Most recently, 20 additional SFBB-like genes (some of which may be alleles of the same gene) were isolated from screening a BAC library of an S-heterozygote of apple, suggesting that the SLF/SFB gene family in Maloideae is even larger than initially thought (Minamikawa et al., 2010). In Plantaginaceae, three AhSLF-like genes were identified and their deduced amino acid sequences are 38–54 % identical with that of AhSLF-S2 (Zhou et al., 2003). In P. inflata, as stated earlier, six PiSLF-like genes have been identified and they share similar properties with PiSLF (McCubbin et al., 2000a; Wang et al., 2003; Hua et al., 2007). In N. alata, ten genes encoding SLF-related proteins were identified, and all of them are linked to the S-locus. Seven of these ten genes are specifically expressed in pollen and at least three are located in the same chromosomal segment as the pollen S-allele (Wheeler and Newbigin, 2007). It is interesting that, in these species, there exist a number of additional F-box genes that are tightly linked to the S-locus and possess other properties expected of the pollen S-gene. To date, functional studies of only three of the SLF-like genes from these species (PiSLFLb-S2, PiSLFLc-S1 and PiSLFLd-S2 of P. inflata) have been reported (Hua et al., 2007), and none of them cause breakdown of SI in S3 pollen (in the case of PiSLFLb-S2 and PiSLFLd-S2) or S2 pollen (in the case of PiSLFLc-S1) of transgenic plants (Fig. 1C). It will be enlightening to examine the possible function of more SLF-like genes in SI by a similar transgenic approach.
CONCLUSIONS AND FUTURE PROSPECTS
The discovery in the mid-1980s that the S-RNase gene (initially simply referred to as the pistil S-gene) encoded the pistil determinant of the Solanaceae SI system (Anderson et al., 1986) marked the dawn of the molecular genetic/biochemical studies of this type of SI mechanism. Since then, there have been several major breakthroughs that led to new directions of research and enhanced our understanding of the SI mechanism. The demonstration that S-RNases have RNase activity (McClure et al., 1989) and that the RNase activity is required for the function of S-RNase (Huang et al., 1994) provided clues as to how the pistil might inhibit growth of self-pollen tubes at the biochemical level. The finding that uptake of S-RNases by the pollen tube is not S-haplotype-specific (Luu et al., 2000; Goldraij et al., 2006) suggested that specific rejection of self-pollen tubes by the pistil most likely lies in differential fates of self and non-self S-RNases inside the pollen tube. Finally, the recent identification of the SLF/SFB gene as the pollen determinant (Lai et al., 2002; Entani et al., 2003; Ushijima et al., 2003; Qiao et al., 2004a, b; Sijacic et al., 2004; Ushijima et al., 2004; Wang et al., 2004; Sonneveld et al., 2005; Tsukamoto et al., 2005, 2006; Hauck et al., 2006a; Vilanova et al., 2006; Sassa et al., 2007) allowed the formulation of new hypotheses and models for the biochemical basis of S-haplotype-specific rejection of pollen tubes.
The S-RNase gene has been very extensively studied over the past quarter century, but there are still some aspects of S-RNase that remain unknown. For example, what specific amino acid residues of each S-RNase determine its S-allele specificity? What is the role, if any, of the glycan chain(s)? What is the mechanism for the uptake of S-RNases by pollen tubes? How might S-RNases sequestered in a vacuole-like compartment in the pollen tube be specifically released into the cytoplasm of an incompatible pollen tube to exert their cytotoxic effect?
The transgenic approach used to establish the function of SLF in P. inflata (Sijacic et al., 2004) and Antirrhinum (Qiao et al., 2004b) is a robust assay for testing whether a candidate for the pollen S-gene is indeed involved in controlling pollen specificity, provided that competitive interaction has been observed in the species under study. The S-locus-linked SLF-like genes, that share similar properties with SLF but whose function has not been examined, will be good candidates for this in vivo functional assay. This assay is based on whether a particular F-box protein produced in the pollen from a transgene can cause breakdown of SI in transgenic pollen, which, based on our model, reflects whether the F-box protein can interact with the S-RNases taken up into the transgenic pollen tubes to mediate their degradation. For example, if the S1-allelic variant of SLF, when expressed in S2 pollen, causes breakdown of SI in S2 pollen, and if the S1-allelic variant of an SLF-like protein fails do so, the results would be interpreted to mean that this SLF-like protein, unlike SLF1, cannot interact with S2-RNase to mediate its degradation. As more and more SLF-like proteins of the S1-haplotype are tested, one can compare the sequences of those that do not cause breakdown of SI in S2 pollen with the sequences of those that do, if any, and SLF1 to identify any amino acid residues that might be responsible for the functional and biochemical differences between these two groups of F-box proteins.
The in vivo functional assay can also be used for structure/function studies to identify specific amino acids and domains of SLF that are involved in a particular function. For example, one can address the question of whether the F-box domain of SLF is required for its function by expressing a truncated SLF in pollen carrying a different S-allele and examining whether the truncated SLF can cause competitive interaction in transgenic pollen. Another key question about SLF is how the S-specificity is determined. FD1 and FD3 of PiSLF have been identified as the putative S-specificity determinant from in vitro protein-binding assays. One can test this model by expressing chimeric SLF proteins between two allelic variants in transgenic plants of the appropriate S-genotype, and examining the effect of the chimeric proteins on the SI behaviour of transgenic pollen. For example, based on our model, the chimeric protein containing FD1 and FD3 from PiSLF2 and FD2 from PiSLF3 would possess S2-allele specificity and would cause breakdown of SI in S3 pollen (heteroallelic), but not in S2 pollen (homoallelic), when introduced into plants of S2S3 genotype. Once a particular domain is found to possess a particular function, site-directed mutagenesis can be used to narrow down specific amino acids involved.
The tenet of the protein-degradation model proposed by our lab is that an SLF protein preferentially interacts with all its non-self S-RNases in the pollen tube to mediate their ubiquitination and ultimate degradation. This model predicts that absence of SLF in pollen would lead to the inability of the pollen to detoxify any S-RNase (self or non-self), and as a result, the pollen would be rejected by pistils of any S-genotype. One direct approach to test this prediction is to use the methodology of RNA interference (RNAi) to suppress the production of SLF in pollen, and to determine whether the transgenic pollen becomes incompatible with pistils that should normally recognize it as non-self pollen. For example, if our model is correct, then suppression of SLF1 in S1 pollen would result in the inability of the transgenic pollen to detoxify any S-RNase that it takes up, and consequently, the transgenic pollen would be rejected by pistils of any S-genotype. The same approach can be used to examine the involvement of all other genes that have been implicated in SI. For example, if PiSBP1 is indeed an essential component of the PiSLF-containing E3 ligase complex, suppression of its production in pollen would result in the inability of the E3 ligase complex to be assembled and consequently, the inability of PiSLF to mediate degradation of non-self S-RNases. The prediction is that the transgenic pollen, in which the expression of PiSBP1 is suppressed, would be rejected by pistils of any S-genotype.
In conclusion, tremendous progress has been made towards understanding the Solanaceae SI system since the cloning of the S-RNase gene was first reported many years ago, but there remain many key questions, as any new discovery invariably leads to new questions and new avenues of research. The availability of robust in vivo functional assays for S-RNase and SLF in P. inflata has made this species a good model system, and it would be of interest to determine how much of the information obtained is applicable to other taxa possessing the Solanaceae-type SI system, particularly those in subfamily Prunoideae of Rosaceae.
ACKNOWLEDGEMENTS
We thank Allison Fields and Ning Wang for comments. The work from the authors' laboratory was supported by successive grants to T.-h.K from the US National Science Foundation.
LITERATURE CITED
- Adachi Y, Komori S, Hoshikawa Y, et al. Characteristics of fruiting and pollen tube growth of apple autotetraploid cultivars showing self-compatibility. Journal of the Japanese Society for Horticultural Science. 2009;78:402–409. [Google Scholar]
- Ai Y, Singh A, Coleman CE, Ioerger TR, Kheyr-Pour A, Kao T-h. Self-incompatibility in Petunia inflata: isolation and characterization of cDNAs encoding three S-allele-associated proteins. Sexual Plant Reproduction. 1990;3:130–138. [Google Scholar]
- Anderson MA, Cornish EC, Mau S-L, et al. Cloning of cDNA for a stylar glycoprotein associated with expression of self-incompatibility in Nicotiana alata. Nature. 1986;321:38–44. [Google Scholar]
- Brewbaker JL, Natarajan AT. Centric fragments and pollen part mutation of incompatibility alleles in Petunia. Genetics. 1960;45:699–704. doi: 10.1093/genetics/45.6.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broothaerts W, Vanvinckenroye P, Decock B, Damme J, Vendrig JC. Petunia hybrida S-proteins: ribonuclease activity and the role of their glycan side chains in self-incompatibility. Sexual Plant Reproduction. 1991;4:258–266. [Google Scholar]
- Cenciarelli C, Chiaur DS, Guardavaccaro D, Parks W, Vidal M, Pagano M. Identification of a family of human F-box proteins. Current Biology. 1999;9:1177–1179. doi: 10.1016/S0960-9822(00)80020-2. [DOI] [PubMed] [Google Scholar]
- Chen G, Zhang B, Zhao Z, Sui Z, Zhang H, Xue Y. ‘A life or death decision’ for pollen tubes in S-RNase-based self-incompatibility. Journal of Experimental Botany. 2010;61:2027–2037. doi: 10.1093/jxb/erp381. [DOI] [PubMed] [Google Scholar]
- Crane MB, Lewis D. Genetical studies in pears. III. Incompatibility and sterility. Journal of Genetics. 1942;43:31–44. [Google Scholar]
- Dowd PE, McCubbin AG, Wang, et al. Use of Petunia inflata as a model for the study of solanaceous type self-incompatibility. Annals of Botany. 2000;85:87–93. [Google Scholar]
- Entani T, Takayama S, Iwano M, Shiba H, Che F-S, Isogai A. Relationship between polyploidy and pollen self-incompatibility phenotype in Petunia hybrida Vilm. Bioscience, Biotechnology and Biochemistry. 1999;63:1882–1888. doi: 10.1271/bbb.63.1882. [DOI] [PubMed] [Google Scholar]
- Entani T, Zwano M, Shiba H, Che FS, Isogai A, Takayama S. Comparative analysis of the self-incompatibility (S-) locus region of Prunus mume: identification of a pollen-expressed F-box gene with allelic diversity. Genes to Cells. 2003;8:203–213. doi: 10.1046/j.1365-2443.2003.00626.x. [DOI] [PubMed] [Google Scholar]
- Franklin-Tong VE. Self-incompatibility in flowing plants: evolution, diversity, and mechanism. Berlin: Springer-Verlag; 2008. [Google Scholar]
- Gagne JM, Downes BP, Shiu SH, Durski AM, Vierstra RD. The F-box subunit of the SCF E3 complex is encoded by a diverse superfamily of genes in Arabidopsis. Proceedings of the National Academy of Sciences of the USA. 2002;99:11519–11524. doi: 10.1073/pnas.162339999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldraij A, Kondo K, Lee CB, et al. Compartmentalization of S-RNase and HT-B degradation in self-incompatible Nicotiana. Nature. 2006;439:805–810. doi: 10.1038/nature04491. [DOI] [PubMed] [Google Scholar]
- Golz JF, Su V, Clarke AE, Newbigin E. A molecular description of mutations affecting the pollen components of the Nicotiana alata S locus. Genetics. 1999;152:1123–1135. doi: 10.1093/genetics/152.3.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golz JF, Oh H-Y, Su V, Kusaba M, Newbigin E. Genetic analysis of Nicotiana pollen-part mutants is consistent with the presence of an S-ribonuclease inhibitor at the S locus. Proceedings of the National Academy of Sciences of the USA. 2001;98:15372–15376. doi: 10.1073/pnas.261571598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauck NR, Yamane H, Tao R, Iezzoni AF. The mutated S1-haplotype in sour cherry has an altered S haplotype-specific F-box protein gene. Journal of Heredity. 2006a;97:514–520. doi: 10.1093/jhered/esl029. [DOI] [PubMed] [Google Scholar]
- Hauck NR, Yamane H, Tao R, Iezzoni AF. Accumulation of non-functional S-haplotypes results in the breakdown of gametophytic self-incompatibility in tetraploid Prunus. Genetics. 2006b;172:1191–1198. doi: 10.1534/genetics.105.049395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrero M, Hormaza JI. Pistil strategies controlling pollen tube growth. Sexual Plant Reproduction. 1996;9:343–347. [Google Scholar]
- Hershko A, Ciechanover A. The ubiquitin system. Annual Review of Biochemistry. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- Hua Z, Kao T-h. Identification and characterization of components of a putative Petunia S-locus F-box-containing E3 ligase complex involved in S-RNase-based self-incompatibility. The Plant Cell. 2006;18:2531–2553. doi: 10.1105/tpc.106.041061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua Z, Kao T-h. Identification of major lysine residues of S3-RNase of Petunia inflata involved in ubiquitin-26S proteasome-mediated degradation in vitro. The Plant Journal. 2008;54:1094–1104. doi: 10.1111/j.1365-313X.2008.03487.x. [DOI] [PubMed] [Google Scholar]
- Hua Z, Meng XY, Kao T-h. Comparison of Petunia inflata S-locus F-box protein (Pi SLF) with Pi SLF-like proteins reveals its unique function in S-RNase-based self-incompatibility. The Plant Cell. 2007;19:3593–3609. doi: 10.1105/tpc.107.055426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua Z, Fields A, Kao T-h. Biochemical models for S-RNase-based self-incompatibility. Molecular Plant. 2008;1:575–585. doi: 10.1093/mp/ssn032. [DOI] [PubMed] [Google Scholar]
- Huang J, Zhao L, Yang Q, Xue Y. AhSSK1, a novel SKP1-like protein that interacts with the S-locus F-box protein SLF. The Plant Journal. 2006;46:780–793. doi: 10.1111/j.1365-313X.2006.02735.x. [DOI] [PubMed] [Google Scholar]
- Huang S, Lee H-S, Karunanandaa B, Kao T-h. Ribonuclease activity of Petunia inflata S proteins is essential for rejection of self-pollen. The Plant Cell. 1994;6:1021–1028. doi: 10.1105/tpc.6.7.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S-X, Wu H-Q, Li Y-R, et al. Competitive interaction between two functional S-haplotypes confers self-compatibility on tetraploid Chinese cherry (Prunus pseudocerasus Lindl cv. Nanjing Chuisi) Plant Cell Reports. 2008;27:1075–1085. doi: 10.1007/s00299-008-0528-7. [DOI] [PubMed] [Google Scholar]
- Ida K, Norioka S, Yamamoto M, et al. The 1·55 Å resolution structure of Nicotiana alata SF11-RNase associated with gametophytic self-incompatibility. Journal of Molecular Biology. 2001;314:103–112. doi: 10.1006/jmbi.2001.5127. [DOI] [PubMed] [Google Scholar]
- Ioerger TR, Gohlke JR, Xu B, Kao T-h. Primary structural features of the self-incompatibility protein in Solanaceae. Sexual Plant Reproduction. 1991;4:81–87. [Google Scholar]
- Kao T-h, McCubbin AG. How flowering plants discriminate between self and non-self pollen to prevent inbreeding. Proceedings of the National Academy of Sciences of the USA. 1996;93:12059–12065. doi: 10.1073/pnas.93.22.12059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao T-h, Tsukamoto T. The molecular and genetic bases of S-RNase-based self-incompatibility. The Plant Cell. 2004;16:S72–S83. doi: 10.1105/tpc.016154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karunanandaa B, Huang S, Kao T-h. Carbohydrate moiety of Petunia inflata S3 protein is not required for self-incompatibility interactions between pollen and pistil. The Plant Cell. 1994;6:1933–1940. doi: 10.1105/tpc.6.12.1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai Z, Ma W, Han B, et al. An F-box gene linked to the self-incompatibility (S) locus of Antirrhinum is expressed specifically in pollen and tapetum. Plant Molecular Biology. 2002;50:29–42. doi: 10.1023/a:1016050018779. [DOI] [PubMed] [Google Scholar]
- Lee H-S, Huang S, Kao T-h. S proteins control rejection of incompatible pollen in Petunia inflata. Nature. 1994;367:560–563. doi: 10.1038/367560a0. [DOI] [PubMed] [Google Scholar]
- Lee CB, Swatek KN, McClure B. Pollen proteins bind to the C-terminal domain of Nicotiana alata pistil arabinogalactan proteins. Journal of Biological Chemistry. 2008;283:26965–26973. doi: 10.1074/jbc.M804410200. [DOI] [PubMed] [Google Scholar]
- Liu B, Morse D, Cappadocia M. Compatible pollinations in Solanum chacoense decrease both S-RNase and S-RNase mRNA. PLoS ONE. 2009;4:e5774. doi: 10.1371/journal.pone.0005774. doi:10.1371/journal.pone.0005774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luu D-T, Qin X, Morse D, Cappadocia M. S-RNase uptake by compatible pollen tubes in gametophytic self-incompatibility. Nature. 2000;407:649–651. doi: 10.1038/35036623. [DOI] [PubMed] [Google Scholar]
- McClure BA. Darwin's foundation for investigating self-incompatibility and the progress toward a physiological model for S-RNase-based SI. Journal of Experimental Botany. 2009;60:1069–1081. doi: 10.1093/jxb/erp024. [DOI] [PubMed] [Google Scholar]
- McClure BA, Haring V, Ebert PR, et al. Style self-incompatibility gene products of Nicotiana alata are ribonucleases. Nature. 1989;342:955–957. doi: 10.1038/342955a0. [DOI] [PubMed] [Google Scholar]
- McClure BA, Gray JE, Anderson MA, Clarke AE. Self-incompatibility in Nicotiana alata involves degradation of pollen rRNA. Nature. 1990;347:757–760. [Google Scholar]
- McCubbin AG, Wang X, Kao T-h. Identification of self-incompatibility (S-) locus linked pollen cDNA markers in Petunia inflata. Genome. 2000a;43:619–627. [PubMed] [Google Scholar]
- McCubbin AG, Zuniga C, Kao T-h. Construction of a binary artificial chromosome library of Petunia inflata and the identification of larger genomic fragments linked to the self-incompatibility (S-) locus. Genome. 2000b;43:820–826. [PubMed] [Google Scholar]
- Meng XY, Hua ZH, Wang N, Fields AM, Dowd PE, Kao T-h. Ectopic expression of S-RNase of Petunia inflata in pollen results in its sequestration and non-cytotoxic function. Sexual Plant Reproduction. 2009;22:263–275. doi: 10.1007/s00497-009-0114-3. [DOI] [PubMed] [Google Scholar]
- Minamikawa M, Kakui H, Wang S, et al. Apple S locus region represents a large cluster of related, polymorphic and pollen-specific F-box genes. Plant Molecular Biology. 2010;74:143–154. doi: 10.1007/s11103-010-9662-z. [DOI] [PubMed] [Google Scholar]
- Moon J, Parry G, Estelle M. The ubiquitin-proteasome pathway and plant development. The Plant Cell. 2004;16:3181–3195. doi: 10.1105/tpc.104.161220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murfett J, Atherton TL, Mou B, Gasser CS, McClure BA. S-RNase expressed in transgenic Nicotiana causes S-allele-specific pollen rejection. Nature. 1994;367:563–566. doi: 10.1038/367563a0. [DOI] [PubMed] [Google Scholar]
- de Nettancourt D. Incompatibility and incongruity in wild and cultivated plants. Berlin: Springer-Verlag; 2001. [Google Scholar]
- Qiao H, Wang H, Zhao L, et al. The F-box protein Ah SLF-S2 physically interacts with S-RNases that may be inhibited by the ubiquitin/26S proteasome pathway of protein degradation during compatible pollination in Antirrhinum. The Plant Cell. 2004a;16:582–595. doi: 10.1105/tpc.017673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao H, Wang F, Zhao L, et al. The F-box protein AhSLF-S2 controls the pollen function of S-RNase-based self-incompatibility. The Plant Cell. 2004b;16:2307–2322. doi: 10.1105/tpc.104.024919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risseeuw EP, Daskalchuk TE, Banks TW, et al. Protein interaction analysis of SCF ubiquitin E3 ligase subunits from Arabidopsis. The Plant Journal. 2003;34:753–767. doi: 10.1046/j.1365-313x.2003.01768.x. [DOI] [PubMed] [Google Scholar]
- Roalson EH, McCubbin AG. S-RNases and sexual incompatibility: structure, functions, and evolutionary perspectives. Molecular Phylogenetics and Evolution. 2003;29:490–506. doi: 10.1016/s1055-7903(03)00195-7. [DOI] [PubMed] [Google Scholar]
- Sassa H, Kakui H, Miyamoto M, et al. S locus F-box brothers: multiple and pollen-specific F-box genes with S haplotype-specific polymorphisms in apple and Japanese pear. Genetics. 2007;175:1869–1881. doi: 10.1534/genetics.106.068858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sassa H, Kakui H, Minamikawa M. Pollen-expresses F-box gene family and mechanism of S-RNase-based gametophytic self-incompatibility (GSI) in Rosaceae. Sexual Plant Reproduction. 2010;23:39–43. doi: 10.1007/s00497-009-0111-6. [DOI] [PubMed] [Google Scholar]
- Sijacic P, Wang X, Skirpan AL, et al. Identification of the pollen determinant of S-RNase-mediated self-incompatibility. Nature. 2004;429:302–305. doi: 10.1038/nature02523. [DOI] [PubMed] [Google Scholar]
- Sims TL. Mechanisms of S-RNase-based self-incompatibility systems. CAB Reviews. 2007;2:1–13. [Google Scholar]
- Singh A, Ai Y, Kao T-h. Characterization of ribonuclease activity of three S-allele-associated proteins of Petunia inflata. Plant Physiology. 1991;96:61–68. doi: 10.1104/pp.96.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalle J, Vierstra RD. The ubiquitin 26S proteasome proteolytic pathway. Annual Review of Plant Biology. 2004;55:555–590. doi: 10.1146/annurev.arplant.55.031903.141801. [DOI] [PubMed] [Google Scholar]
- Sonneveld T, Kenneth R, Tobutt KR, Simon P, Vaughan SP, Robbins TP. Loss of pollen-S function in two self-compatible selections of Prunus avium is associated with deletion/mutation of an S haplotype-specific F-box gene. The Plant Cell. 2005;17:37–51. doi: 10.1105/tpc.104.026963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayama S, Isogai A. Self-incompatibility in plants. Annual Review of Plant Biology. 2005;56:467–489. doi: 10.1146/annurev.arplant.56.032604.144249. [DOI] [PubMed] [Google Scholar]
- Tao R, Iezzoni AF. The S-RNase-based gametophytic self-incompatibility system in Prunus exhibits distinct genetic and molecular features. Scientia Horticulturae. 2010;124:423–433. [Google Scholar]
- Tsai D-S, Lee H-S, Post LC, Kreiling KM, Kao T-h. Sequence of an S-protein of Lycopersicon peruvianum and comparison with other solanaceous S-proteins. Sexual Plant Reproduction. 1992;5:256–263. [Google Scholar]
- Tsukamoto T, Ando T, Watanabe H, Marchesi E, Kao T-h. Duplication of the S-locus F-box gene is associated with breakdown of pollen function in an S-haplotype identified in a natural population of self-incompatible Petunia axillaris. Plant Molecular Biology. 2005;57:141–153. doi: 10.1007/s11103-004-6852-6. [DOI] [PubMed] [Google Scholar]
- Tsukamoto T, Hauck NR, Tao R, Jiang N, Iezzoni AF. Molecular characterization of three non-functional S-haplotypes in sour cherry (Prunus cerasus) Plant Molecular Biology. 2006;62:371–383. doi: 10.1007/s11103-006-9026-x. [DOI] [PubMed] [Google Scholar]
- Tyers M, Jorgensen P. Proteolysis and the cell cycle: with this RING I do thee destroy. Current Opinion in Genetics and Development. 2000;10:54–64. doi: 10.1016/s0959-437x(99)00049-0. [DOI] [PubMed] [Google Scholar]
- Ushijima K, Sassa H, Dandekar AM, Gradziel TM, Tao R, Hirano H. Structural and transcriptional analysis of the self-incompatibility locus of almond: identification of a pollen-expressed F-box gene with haplotype-specific polymorphism. The Plant Cell. 2003;15:771–781. doi: 10.1105/tpc.009290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ushijima K, Yamane H, Watari A, et al. The S haplotype-specific F-box protein gene, SFB, is defective in self-compatible haplotypes of Prunus avium and P. mume. The Plant Journal. 2004;39:573–586. doi: 10.1111/j.1365-313X.2004.02154.x. [DOI] [PubMed] [Google Scholar]
- Vilanova S, Badenes ML, Burgos L, Martinez-Calvo J, Llacer G, Romero C. Self-compatibility of two apricot selections is associated with two pollen-part mutations of different nature. Plant Physiology. 2006;142:629–641. doi: 10.1104/pp.106.083865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Wang X, McCubbin AG, Kao T-h. Genetic mapping and molecular characterization of the self-incompatibility (S) locus in Petunia inflata. Plant Molecular Biology. 2003;53:565–580. doi: 10.1023/B:PLAN.0000019068.00034.09. [DOI] [PubMed] [Google Scholar]
- Wang Y, Tsukamoto T, Yi K-w, et al. Chromosome walking in the Petunia inflata self-incompatibility (S-) locus and gene identification in an 881-kb contig containing S2-RNase. Plant Molecular Biology. 2004;54:727–742. doi: 10.1023/B:PLAN.0000040901.98982.82. [DOI] [PubMed] [Google Scholar]
- Wheeler D, Newbigin E. Expression of 10 S-class SLF-like genes in Nicotiana alata pollen and its implications for understanding the pollen factor of the S locus. Genetics. 2007;177:2171–2180. doi: 10.1534/genetics.107.076885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehouse HLK. Multiple-allelomorph incompatibility of pollen and style in the evolution of the angiosperms. Annals of Botany. 1950;14:198–216. [Google Scholar]
- Xue Y, Zhang Y, Yang Q, Li Q, Cheng Z, Dickinson HG. Genetic features of a pollen-part mutation suggest an inhibitory role for the Antirrhinum pollen self-incompatibility determinant. Plant Molecular Biology. 2009;70:499–509. doi: 10.1007/s11103-009-9487-9. [DOI] [PubMed] [Google Scholar]
- Yamane H, Tao R. Molecular basis of self-(in)compatibility and current status of S-genotyping in rosaceous fruit trees. Journal of the Japanese Society for Horticultural Science. 2009;78:137–157. [Google Scholar]
- Yamane H, Ikeda K, Ushijima K, Sassa H, Tao R. A pollen-expressed gene for a novel protein with an F-box motif that is very tightly linked to a gene for S-RNase in two species of cherry, Prunus cerasus and P. avium. Plant and Cell Physiology. 2003;44:764–769. doi: 10.1093/pcp/pcg088. [DOI] [PubMed] [Google Scholar]
- Zhao L, Huang J, Zhao Z, Li Q, Sim T, Xue Y. The Skp1-like protein SSK1 is required for cross-pollen compatibility in S-RNase-based self-incompatibility. The Plant Journal. 2010;62:52–63. doi: 10.1111/j.1365-313X.2010.04123.x. [DOI] [PubMed] [Google Scholar]
- Zhou J, Wang F, Ma W, Zhang Y, Han B, Xue Y. Structural and transcriptional analysis of S-locus F-box (SLF) genes in Antirrhinum. Sexual Plant Reproduction. 2003;16:165–177. [Google Scholar]
- Zurek DM, Mou B, Beecher B, McClure B. Exchanging sequence domains between S-RNases from Nicotiana alata disrupts pollen recognition. The Plant Journal. 1997;11:797–808. doi: 10.1046/j.1365-313x.1997.11040797.x. [DOI] [PubMed] [Google Scholar]