Abstract
Smads regulate transcription of defined genes in response to TGF-β receptor activation, although the mechanisms of Smad-mediated transcription are not well understood. We demonstrate that the TGF-β-inducible Smad3 uses the tumor suppressor Smad4/DPC4 and CBP/p300 as transcriptional coactivators, which associate with Smad3 in response to TGF-β. The association of CBP with Smad3 was localized to the carboxyl terminus of Smad3, which is required for transcriptional activation, and a defined segment in CBP. Furthermore, CBP/p300 stimulated both TGF-β- and Smad-induced transcription in a Smad4/DPC4-dependent fashion. Smad3 transactivation and TGF-β-induced transcription were inhibited by expressing E1A, which interferes with CBP functions. The coactivator functions and physical interactions of Smad4 and CBP/p300 with Smad3 allow a model for the induction of gene expression in response to TGF-β.
Keywords: TGF-β signaling, Smad, CBP/p300, transcription, PAI-1 promoter
Members of the transforming growth factor β (TGF-β) superfamily regulate cell growth and differentiation through their ability to induce or repress transcription of various genes, including cell-cycle control genes. TGF-β induces and/or stabilizes the formation of a cell-surface heteromeric receptor complex consisting of type I and II receptors, both of which are transmembrane serine/threonine kinases (Derynck and Feng 1997). In response to TGF-β binding, the constitutively active type II receptor kinase (TβRII) phosphorylates and activates the TGF-β type I receptor (TβRI) (Wrana et al. 1994; Chen and Weinberg 1995). The intracellular responses such as growth inhibition and extracellular matrix production are specified by the kinase domain of the type I receptor (Feng and Derynck 1997).
Activated type I receptors phosphorylate and thus activate the intracellular signaling mediators, named Smads, which relay TGF-β signaling into the nucleus (Derynck and Zhang 1996; Wrana and Attisano 1996; Heldin et al. 1997; Massagué et al. 1997). There are three subgroups of Smads: ligand responsive (e.g., Smad1, Smad2, Smad3, Smad5, and Smad8), shared signaling (e.g., the tumor suppressor Smad4/DPC4 in vertebrates, Medea in Drosophila, and Sma-4 in Caenorhabditis elegans), and inhibitory (e.g., Smad6 and Smad7 in vertebrates and Dad in Drosophila) (Heldin et al. 1997). Among ligand-responsive Smads, Smad1 and Smad5 respond to bone morphogenetic proteins (BMPs), whereas Smad2 and Smad3 are activated by TGF-β and activin (Heldin et al. 1997). Smad2 and Smad3 are able to associate with the TβRI/TβRII receptor complex and are carboxy-terminally phosphorylated by activated TβRI (Macías-Silva et al. 1996; Zhang et al. 1996). Activated Smad2/3 forms a heteromeric complex with Smad4 and is then translocated into the nucleus (Macías-Silva et al. 1996; Nakao et al. 1997; Zhang et al. 1997). In Xenopus, Smad2/4 complexes have been shown to associate through a DNA-binding protein with two different activin-responsive elements (Candia et al. 1997; Chen et al. 1997), whereas in Drosophila Mad binds directly to a promoter sequence (Kim et al. 1997). Although these interactions with a promoter are thought to be of critical importance, the underlying mechanism for transcriptional activation is poorly understood.
Transcription from the promoter for plasminogen activator inhibitor type I (PAI-1) is strongly induced by TGF-β and is often used as a marker for TGF-β responsiveness in mammalian cells (Keeton et al. 1991). Coexpression of the TGF-β-responsive Smad2 or Smad3 and Smad4 also induces strongly transcription from the PAI-1 promoter (Zhang et al. 1996, 1997). However, the respective roles of these two Smads in transcriptional activation are unclear and whether and how Smad3 and Smad4 interplay with the transcriptional machinery is unknown. We now show that CBP/p300 and Smad4 act as coactivators for the transcription factor Smad3 through TGF-β-inducible direct physical interactions.
Results and Discussion
The transcription activity of Smad3 is TGF-β inducible and requires its carboxy-terminal SSXS motif
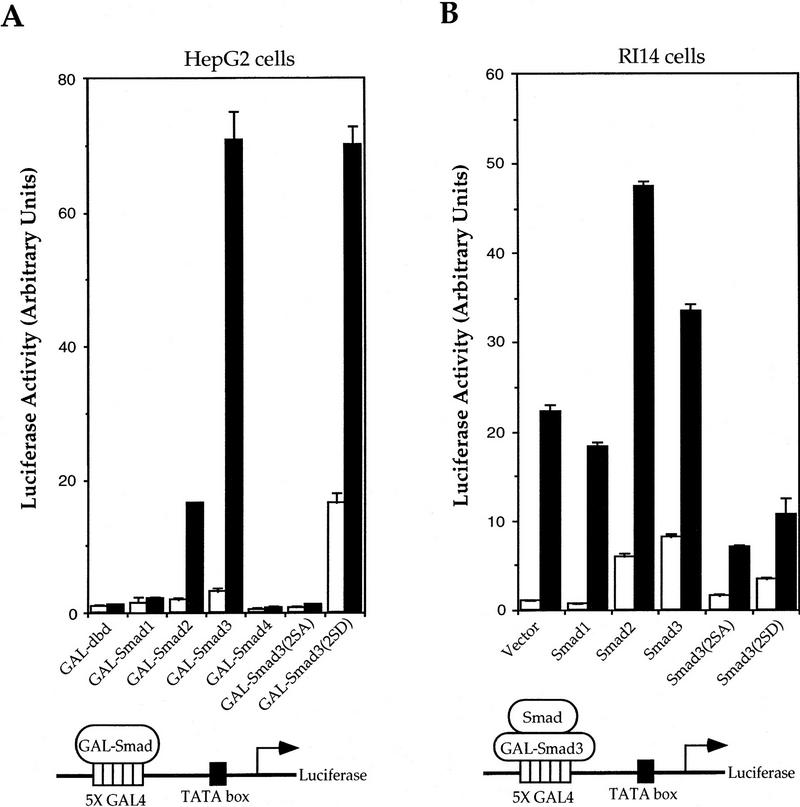
Smad3 synergizes with Smad4/DPC4 to induce a high level transcription from the PAI-1 promoter, and overexpression of carboxy-terminally truncated Smad3 or Smad4 results in dominant-negative inhibition of TGF-β-induced transcription from this promoter (Zhang et al. 1996). Because Smad3 or Smad2 associates directly with Smad4 in response to TGF-β (Nakao et al. 1997) and the heteromeric complex interacts with the promoter to induce transcription (Candia et al. 1997; Chen et al. 1997), we characterized the role of Smad3 and Smad4 in TGF-β-induced transcription. Smad3 was fused to the GAL4 DNA-binding domain, which confers nuclear localization (Silver et al. 1984), and was accordingly localized in the nucleus (data not shown). The transcriptional activity of GAL–Smad3 from a heterologous GAL4 promoter was low, but increased about 15-fold in response to TGF-β (Fig. 1A). Whereas the structurally closely related Smad2 also had a TGF-β-dependent transcriptional activity, Smad4 had only minimal activity both in the absence or presence of TGF-β. These data are consistent with the ability of Smad3 and the inability of Smad4 to activate transcription in yeast, that is, in the absence of endogenous Smads (Wu et al. 1997). The basal activity of GAL–Smad4 may be caused by functional cooperativity with endogenous Smad2 or Smad3.
Figure 1.
Transcriptional activity of Smad3 and effect of interactions with Smad2, Smad3, and Smad4 on Smad3-mediated transcription. (A) Smad3 is a TGF-β-inducible transcriptional activator. HepG2 cells were transfected with plasmids encoding the indicated GAL–Smad and the transcriptional activity from the cotransfected GAL4–luciferase reporter plasmid was measured. Assays were performed in the presence or absence of TGF-β. (B) Smad2 and Smad3 potentiate the transactivation activity of GAL–Smad3. RI14 cells were cotransfected with pFR–Luc and the plasmid encoding GAL–Smad3, and expression plasmids for the indicated Smads or mutants. (C) Smad4 is a potent transcriptional coactivator for Smad3. RI14 cells were cotransfected with pFR–Luc, pGAL–Smad3, and indicated amounts of a Smad4 expression plasmid. (D) Smad4 is not essential for transcriptional activity of Smad3. Smad4-deficient SW480.7 cells were cotransfected with pFR–Luc and pGAL–Smad3, without (open bars) or with (solid bars) TβRI (act.). The transcriptional activity was measured in the absence (open bars) or presence (black bars) of a coexpressed constitutively active TβRI. (E) Smad2 and Smad3 stimulate the low transactivation activity of pGAL–Smad4. Mv1Lu cells were cotransfected with pGAL–Smad4 and pFR–Luc, and expression plasmids for the indicated Smads or relevant mutants. (A–C,E) (Open bars) −TGF-β; (solid bars) +TGF-β. Note the lower scale of luciferase activity, when compared to A–D.
Receptor activation results in phosphorylation of the two distal serines in the carboxy-terminal SSXS sequence of Smad1, Smad2, or Smad3 (Abdollah et al. 1997; Souchelnytskyi et al. 1997). Replacement of all three serines with alanines renders these Smads biologically inactive, which may be caused by impaired release from the receptor, lack of heteromerization with Smad4, and/or inability to translocate into the nucleus (Macías-Silva et al. 1996; Kretzschmar et al. 1997). To investigate the role of ligand-induced phosphorylation in transcriptional activity of Smad3, we replaced the two distal serines in GAL–Smad3 with alanines, thus generating GAL–Smad3(2SA). This mutation abolished the TGF-β-induced transcriptional activity of Smad3 (Fig. 1A), even though GAL–Smad3(2SA) was localized in the nucleus (data not shown). In contrast, replacement of the two serines with aspartic acids in Smad3(2SD), thus providing negative charges similar to those of phosphoserines, conferred a high level of constitutive transcriptional activity, which could still be further enhanced by TGF-β. Thus, the carboxy-terminal negative charges, resulting from receptor-mediated phosphorylation, may be required for the ligand-responsive Smads to adopt a TGF-β-induced conformation for their optimal transcriptional activity. The role of the carboxy-terminal phosphorylation in transcriptional activation complements its role in heteromerization with Smad4 and subsequent nuclear translocation.
The transactivation activity of Smad3 is potentiated by Smad2 or Smad3 and Smad4/DPC4
The carboxyl domain of Smad4 forms a homotrimer (Shi et al. 1997), thus raising the possibility that Smads function as homotrimers. Homomerization of Smad3 may be required for biological activity (Hata et al. 1997; Wu et al. 1997), and Smad2 and Smad3 interact with each other (Wu et al. 1997) and synergize in their biological activity (Nakao et al. 1997). To evaluate the role of oligomerization in transcriptional activity of Smad3, we coexpressed GAL–Smad3 with Smad2 or Smad3 and measured its transcriptional activity. Smad2 and Smad3 enhanced the ligand-independent and -dependent transcriptional activity of GAL–Smad3 (Fig. 1B), without altering the extent to which GAL–Smad3 was localized in the nucleus (data not shown). In contrast, the BMP-2/4-responsive Smad1 did not affect the transcriptional activity of Smad3. Our data thus suggest that the transcriptional activity of Smad3 is enhanced by oligomerization with TGF-β-responsive Smad2 or Smad3. Coexpression of Smad3(2SA) or Smad3(2SD), however, decreased the transcriptional activity of GAL–Smad3 (Fig. 1B). Because Smad3(2SA) was transcriptionally inactive (Fig. 1A), oligomerization of wild-type Smad3 with this mutant may result in an inactive complex; alternatively, the association of Smad3(2SA) with TβRI may prevent carboxy-terminal phosphorylation of GAL–Smad3. On the other hand, Smad3(2SD) had a higher transcriptional activity than wild-type Smad3; its inhibitory effect on GAL–Smad3 is therefore less likely caused by the formation of an inactive complex, but could result from sequestration of endogenous Smad4, which interacts with and coactivates Smad3 (see below).
Because Smad4 has no transcriptional activity on its own (Fig. 1A; Wu et al. 1997), its presence in the transcription complex (Candia et al. 1997; Chen et al. 1997) raises the possibility that it may act as a transcriptional coactivator for ligand-responsive Smads. As shown in Figure 1, C and D, Smad4 strongly increased the transcriptional activity of GAL–Smad3 in the absence or presence of TGF-β. However, Smad4 was not essential for transcriptional activity of GAL–Smad3, as assessed in SW480.7 cells, which lack Smad4 (Goyette et al. 1992) (Fig. 1D). In the absence of Smad4, the transcriptional activity of GAL–Smad3 was still induced by coexpressing an activated TβRI receptor (Fig. 1D), consistent with the notion that receptor-mediated carboxy-terminal phosphorylation enhances the transcriptional activity of Smad3 (see above). Reciprocally, the minimal transcriptional activity of GAL–Smad4 was moderately enhanced by coexpressing Smad2 or Smad3, but not Smad1 (data not shown), and the carboxy-terminal serine mutations in Smad3 abolished this synergy (Fig. 1E). Taken together, Smad4 has only minimal transcriptional activity by itself and acts as a coactivator of Smad3 in the transcription complex. This coactivator role of Smad4 may complement a role in ligand-induced Smad nuclear translocation.
CBP/p300 functions as a Smad4-dependent transcriptional coactivator for Smad3
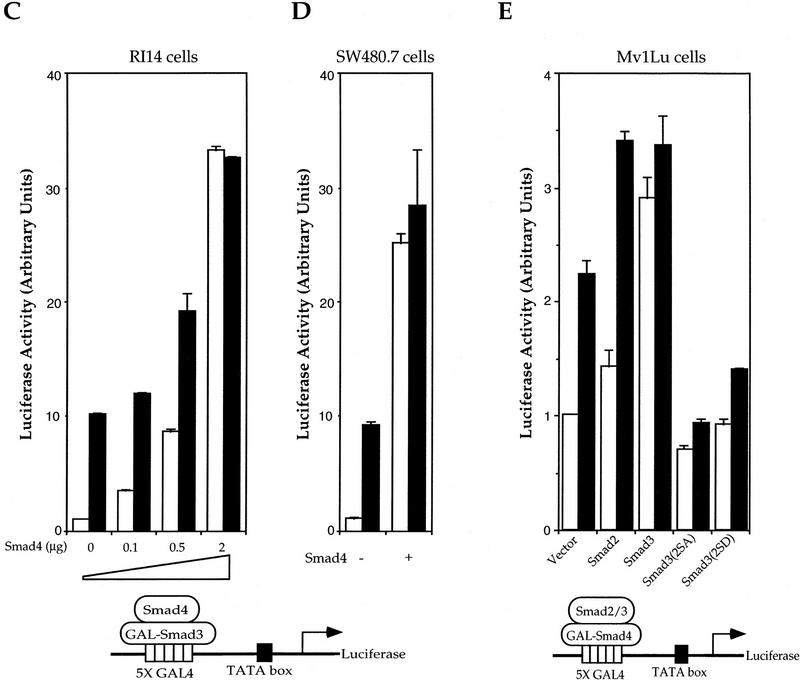
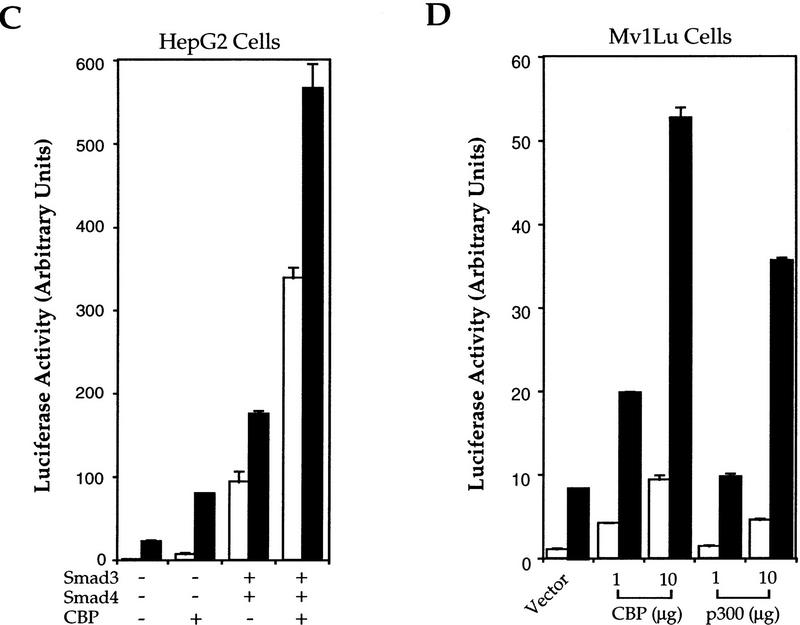
CBP (CREB-binding protein) and the structurally closely related p300 act as transcriptional coactivators for and interact with multiple transcription factors, including CREB, c-Jun, c-Fos, and basal transcription factor TFIIB (Goldman et al. 1997; Shikama et al. 1997). To evaluate whether CBP or p300 plays a role in TGF-β-induced transcription and Smad function, we tested whether p300 and CBP could regulate the transcriptional activity of Smad3. In GAL4-based transcription assays, CBP and p300 both increased the transcriptional activity of Smad3 in the presence or absence of TGF-β (Fig. 2A). This coactivation of Smad3 by CBP was not observed in Smad4-deficient SW480.7 cells (Fig. 2B) and expression of Smad4 not only increased the transcriptional activity of Smad3, consistent with its role as coactivator, but also allowed CBP to transactivate Smad3 (Fig. 2B). We then examined whether CBP/p300 could also potentiate transcription from the PAI-1 promoter in response to TGF-β (Fig. 2C,D). Consistent with the ability of CBP to transactivate Smad3, CBP expression increased transcription from the PAI-1 promoter in response to Smad3 and Smad4 (Fig. 2C), and in response to TGF-β (Fig. 2C,D).
Figure 2.


CBP/p300 functions as a transcriptional coactivator for Smad3. (A) CBP and p300 potentiate the transcriptional activity of GAL–Smad3. RI14 cells were cotransfected with pGAL–Smad3 and pFR–Luc, and indicated amounts of expression plasmids for CBP or p300. (B) Smad4 is required for efficient coactivation of Smad3 by CBP. SW480.7 cells were cotransfected with pGAL–Smad3 and pFR–Luc, and indicated amounts of an expression plasmid for Smad4, in the absence (open bar) or presence (hatched bar) of an expression plasmid for CBP. Transfected cells were treated with TGF-β and luciferase values were measured. (C) CBP stimulates Smad3/4-induced transcription from the PAI-1 promoter in the absence or presence of TGF-β. HepG2 cells were cotransfected with the PAI-1 luciferase reporter p800luc, and indicated combinations of expression plasmids for Smad3, Smad4, and CBP. (D) CBP and p300 stimulate TGF-β-induced transcription from the PAI-1 promoter. Mv1Lu cells were cotransfected with the PAI-1 luciferase reporter p800luc, and indicated amounts of expression plasmids for CBP or p300. (E) Smad3 and Smad4 stimulate the transactivation activity of CBP. Mv1Lu and SW480.7 cells were cotransfected with pGAL–CBP(1678–2441), pFR–Luc, and expression plasmids for Smad3 and/or Smad4. (F) The −732 to −635 segment of the PAI-1 promoter mediates TGF-β- and Smad3/4-inducible transcription. Mv1Lu cells were cotransfected with the PAI-1 luciferase reporter pGL5P/97, and indicated combinations of expression plasmids for Smad3 and Smad4. (G) p300, Smad3, and Smad4 participate in a complex assembled at the PAI-1 promoter. Nuclear extracts, prepared from 293 cells, were incubated with the 32P-labeled, 97-bp TGF-β- and Smad3/4-inducible segment of the PAI-1 promoter. Free DNA, DNA–protein (shift), and supershifted (SS) complexes are marked and the nuclear lysates and antibodies are also shown. (A,C–F) (Open bars) −TGF-β; (solid bars) +TGF-β.
In reciprocal experiments, we evaluated the ability of Smad3 and Smad4 to regulate the transcriptional activity of CBP. The carboxy-terminal segment (amino acids 1678–2441) mediates transcriptional activity of CBP (Kwok et al. 1994); thus GAL–CBP(1678–2441)-mediated transcription from the GAL4-promoter (Fig. 2E). In the absence of Smad4, that is, in SW480.7 cells, Smad3 increased the transcription activity of CBP, but this increase was not further enhanced by TGF-β, unless Smad4 was expressed (Fig. 2E). Thus, Smad4 provided TGF-β inducibility and enhanced the transcription activity of GAL–CBP, presumably in cooperation with endogenous Smad3. Accordingly, the transcriptional activity of CBP was enhanced by TGF-β in Mv1Lu cells and by increased Smad3 or Smad4 expression (Fig. 2E). Finally, Smad3 and Smad4 synergized to increase the ligand-independent and -dependent transcriptional activity of CBP (Fig. 2E). Taken together, our data indicate that CBP transactivates Smad3, but efficient transactivation requires Smad4, and that Smad3 and Smad4 cooperate to transactivate CBP and provide TGF-β inducibility to its transcriptional activity. The TGF-β-dependent transcriptional activity of CBP in Mv1Lu cells is thus likely regulated by endogenous Smad2/3 and Smad4.
Smad3, Smad4, and p300 interact in a nucleoprotein complex with the PAI-1 promoter
The coactivation of Smad3 by CBP/p300 prompted us to test whether Smad3 and p300/CBP interact with a 97-bp PAI-1 promoter fragment (nucleotides −732 to −635), which confers TGF-β and Smad3/4 responsiveness (Fig. 2F). Nuclear extracts from 293 cells provided a cleaner background than those from Mv1Lu or HepG2 cells in gel shift and supershift analyses using the 97-bp probe (data not shown). Whereas untransfected cells did not clearly show a gel-shifted complex (Fig. 2G, lanes 1,2), a TGF-β-dependent DNA–protein complex was detected in transfected cells expressing Smad3 (lanes 3,4), suggesting that this complex contained Smad3. This complex was specific for the 97-bp PAI-1 promoter segment, as it competed with a 25-fold excess of cold probe, but not with unrelated DNA (lanes 15,16). An anti-Smad2/3 antibody (N-19) abolished the TGF-β-inducible complex (lane 5), whereas another anti-Smad3 antibody (I-20) supershifted the complex (lane 6). Control antibodies did not abolish or supershift the complex (e.g., lane 10). The TGF-β-inducible, Smad3-dependent complex was supershifted by an antibody against endogenous p300 (lane 7), which has been validated in supershift experiments (Avantaggiati et al. 1997). Addition of anti-Smad3(N-19) antibody abolished this anti-p300-supershifted complex (lane 8), whereas anti-Smad3(I-20) antibody further decreased the electrophoretic mobility of the supershifted complex (lane 9). No anti-CBP antibodies have been reported to induce supershifts. The complex formation of Smad3 and CBP/p300 at the PAI-1 promoter was also confirmed in gel supershift analyses using tag-specific antibodies and nuclear extracts of cells transfected with tagged Smad3 and CBP/p300 (lanes 17–20). We also determined that Smad3 and Smad4 interact with the same promoter fragment. Thus, cells coexpressing Smad3 and Smad4 together (lane 10) showed a complex of the same size as the TGF-β-inducible, Smad3-dependent complex (lanes 4,17) and this complex, which reacted with anti-Smad3 (lanes 5,6,18) or anti-p300 (lanes 7,19), was supershifted using anti-Smad4 antibody (lane 11). The anti-Smad4-supershifted complex also reacted with the anti-p300 antibody (lane 13), although its intensity was decreased and only a slight supershift occurred, which is not surprising considering the large size of the complex of the DNA fragment associated with p300 and the Smad3/4 heterohexamer. The anti-Smad3(N-19) antibody interfered with the formation of the anti-Smad4-supershifted complex and the anti-Smad4/anti-p300 complex (lanes 12 vs. 11,14 vs. 13). These data thus suggest that p300 (or CBP), Smad3, and Smad4 interact in a TGF-β-dependent manner in a nucleoprotein complex associated with the TGF-β- and Smad3/4-responsive 97-bp PAI-1 promoter segment.
Smad3 and CBP associate with each other in a TGF-β-dependent manner
The functions of CBP/p300 and Smad4 as coactivators of Smad3 in TGF-β-induced transcription and their coexistence at the TGF-β- and Smad3/4-responsive PAI-1 promoter segment strongly suggested a pattern of physical interactions. As shown in Figure 3A and in previous studies (Lagna et al. 1996; Nakao et al. 1997), Smad4 associates directly with Smad3 or Smad2 in a TGF-β-dependent manner. This heteromeric association is mediated by the conserved carboxyl domains of the Smads (Hata et al. 1997; Wu et al. 1997). To investigate whether Smad3 interacts with CBP in response to TGF-β, we performed coimmunoprecipitation analyses using transfected cells. No association with CBP was observed without receptor stimulation, but TGF-β receptor activation resulted in interaction of Smad3, but not Smad4, with CBP (Fig. 3B). Smad1 did not associate with CBP, which is consistent with its responsiveness to BMP-2/4 and not to TGF-β (Hoodless et al. 1996; Liu et al. 1996; Kretzschmar et al. 1997).
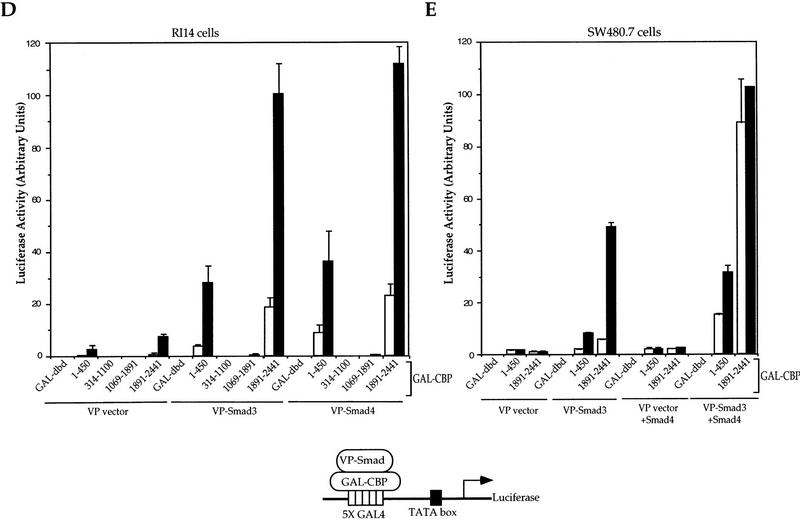
Figure 3.

TGF-β-dependent association of CBP with Smad3 and cooperation of Smad4. (A) TGF-β-dependent association of Smad3 and Smad4 . RI14 cells were transfected with Flag-tagged Smad3 and HA-tagged Smad4, and treated with (+) or without (−) TGF-β. Cell lysates were immunoprecipitated (IP) with anti-Flag antibody, followed by immunoblotting (IB) with anti-HA antibody to detect Smad3-bound Smad4, or with anti-Flag immunoblotting to demonstrate equal expression of Smad3. (B) Smad3 coimmunoprecipitates with CBP after TGF-β-receptor activation. COS-1 cells were transfected with Flag-tagged Smads as shown and HA-tagged CBP, in the absence or presence of a plasmid for an activated TβRI. Immunoprecipitation with anti-Flag antibody was followed by anti-HA immunostaining to detect Smad-bound CBP. The control panel shows the expression levels of Smad3 and Smad4. (C) TGF-β-dependent CBP–Smad3 interaction requires the carboxy-terminal SSXS site of Smad3. RI14 cells were transfected with the indicated combination of plasmids expressing GAL–CBP(1678–2441) and Flag–Smad3 or Flag–Smad3(2SA). Immunoprecipitation with anti-Flag antibody was followed by immunostaining with anti-GAL4 (BabCO) to detect Smad-bound CBP. The control panel shows the Smad3 expression levels. (D) TGF-β-dependent interaction of Smad3 and Smad4 with CBP in mammalian two-hybrid assays. Plasmids for GAL-fused CBP segments, in combination with VP16–Smad plasmids, as indicated, were transfected into RI14 cells, together with the luciferase reporter plasmid pFR–Luc. The interactions were measured by luciferase expression in the absence (−, open bars) or presence (+, solid bars) of TGF-β. GAL–DNA-binding domain is the control containing only the GAL4 DNA-binding domain, not fused to a CBP segment. (E) Smad4 enhances the association of Smad3 with CBP in mammalian two-hybrid assays. Smad4-deficient SW480.7 cells were cotransfected with plasmids for GAL-fused CBP segments and VP16–Smd3, in the absence (open bars) or presence (solid bars) of an activated TβRI plasmid, and the CBP–Smad3 association was scored in the absence or presence of coexpressed Smad4.
Furthermore, the carboxy-terminal segment (amino acids 1678–2441) of CBP associated with Smad3 in response to TGF-β stimulation, but this interaction was not seen with Smad3(2SA) (Fig. 3C). This lack of association of Smad3(2SA) could be caused by a decreased level of nuclear translocation and/or may reflect the role of the TGF-β-induced carboxy-terminal phosphorylation of Smad3 in the interaction with CBP, which is consistent with the role of phosphorylation in the transcriptional activity of Smad3 (Fig. 1A).
Mammalian two-hybrid analyses were used to evaluate the ability of defined CBP segments to interact with Smad3 or Smad4 in Mv1Lu cells. As shown in Figure 3D, the interaction of CBP with Smad3 was localized to two segments, a weakly interacting amino-terminal segment (amino acids 1–451) and the much stronger interacting carboxy-terminal segment (amino acids 1891–2441). These interactions were strongly enhanced by TGF-β, thus confirming the TGF-β-inducible coimmunoprecipitation of CBP and Smad3. Accordingly, when individual CBP domains were coexpressed with Smad3, CBP (1891–2441), but not the other segments, coimmunoprecipitated with Smad3 in response to TGF-β (Fig. 3C; data not shown). Smad4 also showed ligand-inducible interaction with the two CBP segments in two-hybrid assays in Mv1Lu cells (Fig. 3D). This is in contrast with the lack of Smad4–CBP interaction in coimmunoprecipitation (Fig. 3B) and yeast two-hybrid (see below) experiments, suggesting that this interaction is mediated through the ligand-dependent association of Smad4 with endogenous Smad3, which in turn interacts in a ligand-dependent fashion with CBP. In addition, coexpression of Smad4 in SW480.7 cells increased the interaction of Smad3 with the amino- and carboxy-terminal domains of CBP (Fig. 3E), whereas coexpression of Smad3 promoted the association of Smad4 and CBP in mammalian two-hybrid assays (data not shown). Our results thus suggest a ternary protein complex, whereby the ligand-dependent interaction of Smad3 with CBP (primarily its carboxy-terminal segment) is stabilized by Smad4. This interpretation is consistent with the participation of all three proteins in a nucleoprotein complex at the promoter (Fig. 2G; see above). The stabilization by Smad4 may be required for the ability of CBP to efficiently coactivate Smad3, as illustrated in Fig. 2B.
The direct association of Smad3 and CBP is mediated by the carboxy-terminal domains of both proteins
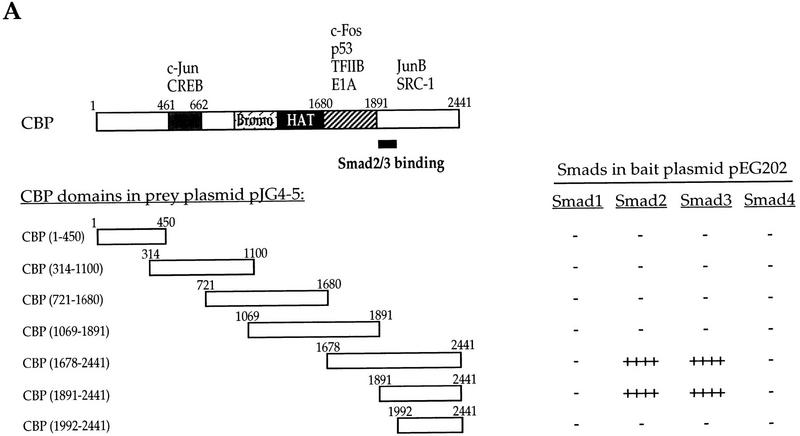
The interaction of CBP with Smad3 was also analyzed using yeast two-hybrid assays (Fig. 4A). Smad3 was only observed to interact with the carboxy-terminal segment of CBP that comprised amino acids 1891–2441, but not with amino acids 1992–2441, thus indicating a requirement of amino acids 1891–1991. The interaction of Smad3 with CBP is most likely direct, as no Smad or CBP homologs are encoded by the yeast genome, and CBP(1678–2441) interacted with GST–Smad3 in vitro (see below). Smad2, which is structurally closely related to Smad3, also interacted with the carboxy-terminal segment of CBP. In contrast, Smad4 and the BMP-2/4-responsive Smad1 did not interact with CBP in yeast two-hybrid assays. These results allow us to assign a new function to a defined segment of CBP (Fig. 4A).
Figure 4.

The association of CBP and Smad3 is mediated by carboxy-terminal domains of both proteins. (A) Yeast two-hybrid assays demonstrate the interaction of Smad2 and Smad3, but not Smad1 and Smad4, with carboxy-terminal sequences (aa 1891–2441) of CBP. Smad–CBP interactions were detected by measuring β-galactosidase activity. (−) Lack of detectable interaction; (++++) very strong interaction. The structural organization of CBP is shown with the previously characterized location of sequences required for interaction with the proteins shown. Our results now allow the localization of sequences required for association with Smad2 and Smad3. (B) Localization of the CBP-interacting sequences in Smad3 using yeast two-hybrid. Interactions were scored by measuring β-galactosidase activity from negative (−) to strongly positive (+++) and not determined (n.d.). The correlation with in vitro binding to GST-fused Smad3 or its fragments is also shown. Besides some previously defined functions shown on the schematic diagram of Smad3, the sequences that mediate interaction with CBP have now been localized to the carboxyl domain and require the carboxy-terminal sequence. (C) Direct interaction of 35S-labeled CBP(1678–2441) with GST–Smad3 and GST–Smad3C, but not GST–Smad4. Results are summarized in B.
To map the domains in Smad3 that interact with CBP, we carried out yeast two-hybrid assays that were further confirmed using GST-based in vitro binding assays. Smads have highly conserved amino and carboxyl domains, separated by a less conserved, proline-rich linker (L) region. The carboxyl domain of ligand-responsive Smads mediates transcriptional activation in GAL4 transactivation assays (Liu et al. 1996) and in transcription assays in yeast (Wu et al. 1997). As shown in Figure 4B, the interaction of Smad3 with the carboxy-terminal segment of CBP was mediated by the carboxyl domain and not by the amino domain or L segment. This interaction was most likely direct as the in vitro-synthesized 35S-labeled CBP(1678–2441) interacted with GST–Smad3 and GST–Smad3C (Fig. 4C). Consistent with the yeast two-hybrid results (Fig. 4A), the carboxy-terminal segment of CBP did not interact with GST–Smad4. Deletion of the carboxy-terminal 35 amino acids (the Δc deletion), which inactivates the transcriptional activity of Smads (Zhang et al. 1996), abolished the interaction of full-sized Smad3 or its carboxyl domain with CBP in yeast two-hybrid assays. In contrast, replacement of the carboxy-terminal phosphoacceptor serines with alanines did not abolish the interaction of Smad3 with CBP, although the affinity of this interaction may be affected. Thus, the carboxy-terminal phosphorylation is not essential for interaction in yeast with CBP, although it is required for TGF-β-induced transcriptional activity (Fig. 1A). These results suggest that binding to CBP is a new function for the carboxyl domain of Smad3, and likely Smad2 (Fig. 4B).
Taken together, our data indicate a ligand-dependent association of Smad3 with CBP, which is mediated through the carboxyl domain of Smad3 and a carboxy-terminal sequence in CBP. In vivo, this interaction requires stabilization by Smad4, even though Smad4 cannot interact directly with CBP in the absence of Smad3 (Fig. 5A). In this ternary complex, both Smad4 and CBP function as coactivators for Smad3, and Smad4 is also required for efficient coactivation of Smad3 by CBP. Because the carboxyl domain of Smad3 is the effector of its transcriptional activity and the small Δc deletion results in transcriptional inactivity, our findings correlate the transcriptional activity of Smad3 with its ability to associate with CBP, which then allows transactivation of Smad3 by CBP (and vice versa).
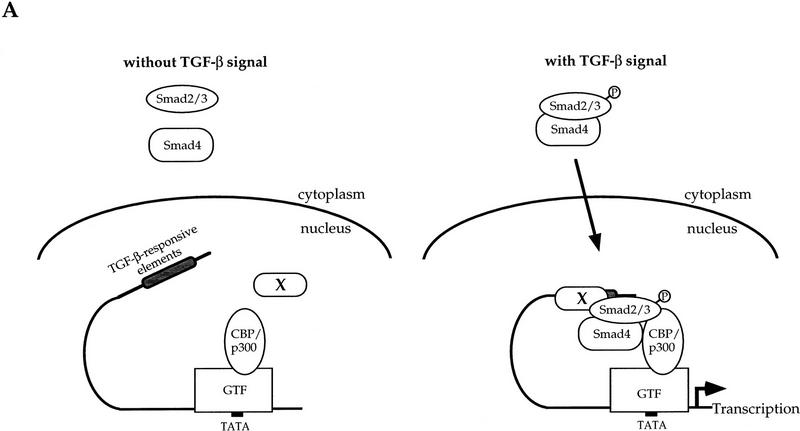
Figure 5.
Model of Smad3/Smad4/CBP interactions at the promoter and inhibition of TGF-β- and Smad3/4-induced transcription by E1A. (A) A working model for TGF-β-inducible cooperation between Smad2/3, Smad4, and CBP/p300. Nuclear factor X is a hypothetical DNA-binding protein bound to the TGF-β-responsive elements, although Smad2/3 or Smad4 may directly bind to DNA. GTF represents a group of transcription factors for general transcription associated with RNA polymerase II. (B) E1A inhibits transcription from the PAI-1 promoter, induced by Smad3 and Smad4. Smad4-deficient SW480.7 cells were cotransfected with p800luc, and indicated combinations of expression plasmids for Smad3, Smad4, and E1A. (C) E1A, but not E1AΔ2–36, which is incapable of CBP association, inhibits TGF-β-induced transcription from the PAI-1 promoter. E1A928 has a point mutation at aa 928 and does not interact with pRB but still binds to CBP. Mv1Lu cells were cotransfected with p800luc, and wild-type or mutant E1A. (D) E1A, but not E1AΔ2–36, blocks TGF-β-induced transcription activity of GAL–Smad3 and the transactivation of GAL–Smad3 by CBP. Mv1Lu cells were cotransfected with pFR-Luc, and wild-type or mutant E1A, and CBP. (B–D) (Open bars) −TGF-β; (solid bars) +TGF-β.
E1A inhibits TGF-β- and Smad3/4-induced gene transcription
The functional cooperativity between Smad3 and CBP and the inability of the transcriptionally inactive Smad3ΔC to interact with CBP strongly suggest that this physical interaction is required for TGF-β- and Smad3-induced transcriptional activation. Unfortunately, the requirement of this interaction for TGF-β-induced transcription cannot be confirmed using cells that lack both Smad2 and Smad3 or both CBP and p300. Furthermore, no CBP or p300 deletion mutants are known to act as dominant-negative inhibitors of endogenous CBP/p300 function. To this end, we used the adenoviral E1A protein to determine if CBP/p300 is necessary for TGF-β- and Smad3/4-induced transcription. E1A interacts with CBP sequences between amino acids 1680 and 1891, i.e., adjacent to the sequences that interact with Smad3, and interferes with the interaction of CBP with general transcription machinery, thereby acting as a reliable inhibitory indicator of CBP/p300-dependent transactivation (Goldman et al. 1997; Shikama et al. 1997). We therefore expected that E1A might at least partially inhibit Smad3 binding to CBP and Smad3-mediated transcription. As shown, E1A significantly inhibited transcription induced from the PAI-1 promoter by Smad3/4 (Fig. 5B) and TGF-β (Fig. 5C). In contrast, E1AΔ2–36, which has a short amino-terminal deletion and is incapable of binding CBP (Kraus et al. 1992), only poorly inhibited TGF-β-induced transcription (Fig. 5C). However, E1A928, which is defective in pRB binding but retains the ability to interact with CBP (Kraus et al. 1992), inhibited TGF-β-induced transcription as efficiently as wild-type E1A (Fig. 5C). Furthermore, E1A inhibited completely the transcriptional activity of GAL–Smad3 and the ability of CBP to transactivate GAL–Smad3 (Fig. 5D), whereas E1AΔ2–36 was ineffective. The inhibition of Smad3/4- and TGF-β-induced transcriptional activation by E1A is therefore most likely because of its ability to block the transcriptional activity of GAL–Smad3 and the ability o CBP to act as coactivator. Thus, our findings support a requirement for CBP as coactivator of Smad2/3-mediated transcription and TGF-β-induced signaling and suggest that E1A suppresses TGF-β-induced growth inhibition and induces oncogenic transformation at least in part by interfering with the Smad–CBP interaction.
Materials and methods
Expression plasmids
Coding sequences for amino-terminally HA- or Flag-tagged, or GAL4- or VP16-fused, Smad and CBP proteins or defined regions were generated by oligonucleotide- or PCR-based techniques and inserted into the ClaI–EcoRI sites of the mammalian expression plasmid pRK5 (Graycar et al. 1989) or derivatives. Detailed information on the plasmid constructions will be provided upon request. The pRK5-based expression plasmid for constitutively active TβRI has been described (Feng and Derynck 1996). Expression plasmids encoding GAL–Smad1 or GAL–Smad4 (Liu et al. 1996) were gifts from J. Massagué, whereas L. Xu and M.G. Rosenfeld provided plasmids for GAL–CBP fragments (Horvai et al. 1997). Expression plasmids for carboxy-terminally HA-tagged CBP and GAL–CBP (1678–2441) (Kwok et al. 1994) were obtained from R. Goodman, and the p300 expression plasmid (Lill et al. 1997) was provided by D. Livingston.
Cell culture, transfections, and immunoprecipitations
COS-1 cells were maintained in DME, 10% fetal bovine serum; and HepG2, Mv1Lu, and SW480.7 cells were maintained in MEM, 10% fetal bovine serum, supplemented with nonessential amino acids. RI14 cells are stably transfected R1B/L17 cells (Wrana et al. 1994), expressing TβRI (X.-H. Feng and R. Derynck, unpubl.). COS-1 and SW480.7 cells were transfected using Lipofectamine (GIBCO–BRL). Immunoprecipitations using anti-Flag antibody were carried out as described (Feng et al. 1995). For subsequent anti-HA Western blot, the immunoprecipitated proteins, separated by SDS-PAGE, were transferred onto Immobilon (Millipore), incubated with anti-HA antibody (Babco), and antibody-bound proteins were visualized by chemiluminescence (Pierce).
Transcriptional reporter assays
Plasmid p800luc containing the luciferase reporter gene under control of the PAI-1 promoter (Keeton et al. 1991) was used to measure TGF-β- and Smad-induced gene expression. pGL5P/97 contains the 97-bp upstream regulatory sequence (nucleotides −732 to −635) of PAI-1 gene to drive expression of luciferase reporter. pRKβgal, which expresses β-galactosidase under the control of the CMV promoter, was cotransfected to allow normalization of transfection efficiency. Transient transfections, TGF-β treatment, and reporter assays were done as described (Feng et al. 1995). Mv1Lu and RI14 cells were transfected using DEAE–dextran, whereas HepG2 and SW480.7 cells were transfected using Lipofectamine. The total amount of transfected DNA was always the same by adding pRK5 control DNA, as needed. All assays were done in triplicate and all values were normalized for transfection efficiency.
GAL4 transactivation assays
Plasmids encoding GAL–Smad or GAL–CBP were cotransfected with the GAL4–luciferase plasmid pFR-Luc (Stratagene) and other expression plasmids into cells, as specified in the figures (Fig. 1A–E, 2A, 2B, 2E), and transfected cells were treated for 24 hr with or without 400 pm TGF-β. The ability of the Smads or CBP to transactivate the heterologous GAL4 promoter was quantitated by measuring luciferase expression from the GAL4 promoter. Transfections, TGF-β treatment, and luciferase assays were done essentially as for the PAI-1 luciferase assays.
Mammalian two-hybrid assays
Plasmids encoding GAL–CBP, plasmids for Smads fused to the VP16 activation domain, and the luciferase reporter plasmid pFR-Luc were transfected into Mv1Lu or SW480.7 cells, which were then treated with or without TGF-β. Luciferase assays were carried out as described above.
Gel shift assays
Nuclear proteins were extracted from exponentially growing 293 cells, as described (Datto et al. 1995) and gel-shift assays were performed using a commercial kit (Promega). A 10-μl reaction, containing 4 μg of nuclear protein in 10 mm Tris-HCl at pH 7.5, 0.5 mm EDTA, 1 mm MgCl2, 50 mm NaCl, 0.5 mm DTT, 4% glycerol, and 0.05 mg/ml poly[d(I-C)] · poly[d(I-C)], was incubated at room temperature for 15–30 min with 40,000–100,000 dpm of 32P-labeled probe. The probe corresponded to the TGF-β- and Smad3/4-inducible segment of the PAI-1 promoter (nucleotide −732 to −635). Smad3 antisera (Santa Cruz Biotechnology), Smad4 antiserum (Yingling et al. 1997, gift of X.-F. Wang), and/or anti-p300 antiserum (Avantaggiati et al. 1997, obtained from M. Avantaggiati and K. Kelly) were then added, and the incubation proceeded at 4°C for 90 min. Gel-shifted and -supershifted complexes were separated in a 5% polyacrylamide gel in 0.5× TBE buffer and visualized by autoradiography.
Yeast two-hybrid assays
LexA-based yeast two-hybrid assays (Gyuris et al. 1993) were used to detect interactions between full-length Smads in bait plasmid pEG202 (Wu et al. 1997) and CBP fragments in prey plasmid pJG4-5. CBP(1–450), CBP(314–1100), CBP(1069–1891), and CBP(1891–2441) in pJG4-5 (Kamei et al. 1996) were provided by L. Xu and M.G. Rosenfeld, whereas other CBP segments were obtained by PCR and subcloned in pJG4-5. Plasmids were transformed into yeast EGY48 using Alkali Cation (Bio 101), and protein interactions were assessed by scoring β-galactosidase activity as reporter, as described previously (Wu et al. 1997).
GST fusion proteins and in vitro protein binding assays
Glutathione-S-transferase (GST) fusion proteins of Smads were described previously (Zhang et al. 1996). 35S-Labeled CBP(1678–2441), obtained by in vitro transcription/translation (Promega) from pRK5–CBP(1678–2441), was incubated with GST or GST–Smad or GST–Smad fragments bound to glutathione Sepharose beads (Pharmacia). After extensive washing in 25 mm Tris-HCl at pH 8.0, 150 mm NaCl, and 0.5% Triton X-100, associated proteins were separated by SDS-PAGE and visualized by autoradiography.
Acknowledgments
This research was supported by National Institutes of Health grant CA63101 to R.D. and postdoctoral fellowships from the American Cancer Society to X.-H. F., the American Lung Association to Y. Z., and the Leukemia Society of America to R.-Y.W. We thank X.-F. Wang, J. Massagué, R. Goodman, L. Xu, M.G. Rosenfeld, M. Avantaggiati, K. Kelly, D. Livingston, R. Roeder, Y. Shi, and D. Loskutoff for valuable reagents.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL derynck@itsa.ucsf.edu; FAX (415) 476-1499.
References
- Abdollah S, Macías-Silva M, Tsukazaki T, Hayashi H, Attisano L, Wrana JL. TβRI phosphorylation of Smad2 on Ser465 and Ser467 is required for Smad2-Smad4 complex formation and signaling. J Biol Chem. 1997;272:27678–27685. doi: 10.1074/jbc.272.44.27678. [DOI] [PubMed] [Google Scholar]
- Avantaggiati ML, Ogryzko V, Gardner K, Giordano A, Levine A, Kelly K. Recruitment of p300/CBP in p53-dependent signal pathways. Cell. 1997;89:1175–1184. doi: 10.1016/s0092-8674(00)80304-9. [DOI] [PubMed] [Google Scholar]
- Candia A, Watabe T, Hawley S, Onichtchouk D, Zhang Y, Derynck R, Niehrs C, Cho KWY. Cellular interpretation of multiple TGF-β signals: Intracellular antagonism between activin/BVg1 and BMP-2/4 signaling mediated by Smads. Development. 1997;124:4467–4480. doi: 10.1242/dev.124.22.4467. [DOI] [PubMed] [Google Scholar]
- Chen F, Weinberg RA. Biochemical evidence for the autophosphorylation and transphosphorylation of transforming growth factor β receptor kinases. Proc Natl Acad Sci. 1995;92:1565–1569. doi: 10.1073/pnas.92.5.1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Weisberg E, Fridmacher V, Watanabe M, Naco G, Whitman M. Smad4 and FAST-1 in the assembly of activin-responsive factor. Nature. 1997;389:85–89. doi: 10.1038/38008. [DOI] [PubMed] [Google Scholar]
- Datto M, Li Y, Panus J, Howe D, Xiong Y, Wang X-F. Transforming growth factor β induces the cyclin-dependent kinase inhibitor p21 through a p53-independent mechanism. Proc Natl Acad Sci. 1995;92:5545–5549. doi: 10.1073/pnas.92.12.5545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derynck R, Zhang Y. Intracellular signaling: The Mad way to do it. Curr Biol. 1996;6:1226–1229. doi: 10.1016/s0960-9822(96)00702-6. [DOI] [PubMed] [Google Scholar]
- Derynck R, Feng X-H. TGF-β receptor signaling. Biochim Biophy Acta (Rev Cancer) 1997;1333:F105–F150. doi: 10.1016/s0304-419x(97)00017-6. [DOI] [PubMed] [Google Scholar]
- Feng X-H, Derynck R. Ligand-independent activation of TGF-β signaling pathways by heteromeric cytoplasmic domains of TGF-β receptors. J Biol Chem. 1996;271:13123–13129. doi: 10.1074/jbc.271.22.13123. [DOI] [PubMed] [Google Scholar]
- Feng X-H, Derynck R. A kinase subdomain of transforming growth factor-β (TGF-β) type I receptor determines the TGF-β intracellular signaling specificity. EMBO J. 1997;16:3912–3923. doi: 10.1093/emboj/16.13.3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng X-H, Filvaroff EH, Derynck R. TGF-β-induced downregulation of cyclin A expression requires a functional TGF-β receptor complex. Characterization of chimeric and truncated type I and type II receptors. J Biol Chem. 1995;270:24237–24245. doi: 10.1074/jbc.270.41.24237. [DOI] [PubMed] [Google Scholar]
- Goldman P, Tran V, Goodman RH. The multifunctional role of the co-activator CBP in transcriptional regulation. Recent Prog Hormone Res. 1997;52:103–119. [PubMed] [Google Scholar]
- Goyette MC, Cho K, Fasching CL, Levy DB, Kinzler KW, Paraskeva C, Vogelstein B, Stanbridge E. Progression of colorectal cancer is associated with multiple tumor suppressor gene defects but inhibition of tumorigenicity is accomplished by correction of any single defect via chromosome transfer. Mol Cell Biol. 1992;12:1387–1395. doi: 10.1128/mcb.12.3.1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graycar JL, Miller DA, Arrick BA, Lyons RM, Moses HL, Derynck R. Human transforming growth factor-β 3: Recombinant expression, purification, and biological activities in comparison with transforming growth factors-β1 and -β2. Mol Endocrinol. 1989;3:1977–1986. doi: 10.1210/mend-3-12-1977. [DOI] [PubMed] [Google Scholar]
- Gyuris J, Golemis E, Chertkov H, Brent R. Cdi1, a human G1 and S phase protein phosphatase that associates with Cdk2. Cell. 1993;75:791–803. doi: 10.1016/0092-8674(93)90498-f. [DOI] [PubMed] [Google Scholar]
- Hata A, Lo R, Wotton D, Lagna G, Massagué J. Mutations increasing autoinhibition inactivate tumour suppressors Smad2 and Smad4. Nature. 1997;88:82–87. doi: 10.1038/40424. [DOI] [PubMed] [Google Scholar]
- Heldin C-H, Miyazono K, ten Dijke P. TGF-β signalling from cell membrane to nucleus through SMAD proteins. Nature. 1997;390:465–471. doi: 10.1038/37284. [DOI] [PubMed] [Google Scholar]
- Hoodless P, Haerry T, Abdollah S, Stapleton M, O’Connor MB, Attisano L, Wrana JL. MADR1, a MAD-related protein that functions in BMP2 signaling pathways. Cell. 1996;85:489–500. doi: 10.1016/s0092-8674(00)81250-7. [DOI] [PubMed] [Google Scholar]
- Horvai A, Xu L, Korzus E, Brard G, Kalafus D, Mullen T-M, Rose D, Rosenfeld MG, Glass CK. Nuclear integration of JAK/STAT and Ras/AP-1 signaling by CBP and p300. Proc Natl Acad Sci. 1997;94:1074–1079. doi: 10.1073/pnas.94.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamei Y, Xu L, Heinzel T, Torchia J, Kurokawa R, Gloss B, Lin S-C, Heyman R, Rose D, Glass C, Rosenfeld MG. A CBP integrator complex mediates transcriptional activation and AP-1 inhibition by nuclear receptors. Cell. 1996;85:403–414. doi: 10.1016/s0092-8674(00)81118-6. [DOI] [PubMed] [Google Scholar]
- Keeton M, Curriden S, van Zonneveld A-J, Loskutoff DJ. Identification of regulatory sequences in the type 1 plasminogen activator inhibitor gene responsive to transforming growth factor β. J Biol Chem. 1991;266:23048–23052. [PubMed] [Google Scholar]
- Kim J, Johnson K, Chen H, Carroll S, Laughon A. Drosophila Mad binds to DNA and directly mediates activation of vestigial by Decapentaplegic. Nature. 1997;388:304–308. doi: 10.1038/40906. [DOI] [PubMed] [Google Scholar]
- Kraus V, Moran E, Nevins JR. Promoter-specific trans-activation by the adenovirus E1A12S product involves separate E1A domains. Mol Cell Biol. 1992;12:4391–4399. doi: 10.1128/mcb.12.10.4391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretzschmar M, Liu F, Hata A, Doody J, Massagué J. The TGF-β family mediator Smad1 is phosphorylated directly and activated functionally by the BMP receptor kinase. Genes & Dev. 1997;11:984–995. doi: 10.1101/gad.11.8.984. [DOI] [PubMed] [Google Scholar]
- Kwok R, Lundblad J, Chrivia J, Richards J, Bächinger H, Brennan R, Roberts S, Green M, Goodman RH. Nuclear protein CBP is a coactivator for the transcription factor CREB. Nature. 1994;370:223–226. doi: 10.1038/370223a0. [DOI] [PubMed] [Google Scholar]
- Lagna G, Hata A, Hemmati-Brivanlou A, Massagué J. Partnership between DPC4 and SMAD proteins in TGF-β signalling pathways. Nature. 1996;383:832–836. doi: 10.1038/383832a0. [DOI] [PubMed] [Google Scholar]
- Lill N, Grossman S, Ginsberg D, DeCaprio J, Livingston DM. Binding and modulation of p53 by p300/CBP coactivators. Nature. 1997;387:823–827. doi: 10.1038/42981. [DOI] [PubMed] [Google Scholar]
- Liu F, Hata A, Baker JC, Doody J, Cárcamo J, Harland RM, Massagué J. A human Mad protein acting as a BMP-regulated transcriptional activator. Nature. 1996;381:620–623. doi: 10.1038/381620a0. [DOI] [PubMed] [Google Scholar]
- Macías-Silva M, Abdollah S, Hoodless P, Pirone R, Attisano L, Wrana JL. MADR-2 is a substrate of the TGF-β receptor and its phosphorylation is required for nuclear accumulation and signaling. Cell. 1996;87:1215–1224. doi: 10.1016/s0092-8674(00)81817-6. [DOI] [PubMed] [Google Scholar]
- Massagué J, Hata A, Liu F. TGF-β signalling through the Smad pathway. Trends Cell Biol. 1997;7:187–192. doi: 10.1016/S0962-8924(97)01036-2. [DOI] [PubMed] [Google Scholar]
- Nakao A, Imamura T, Kawabata M, Ishisaki A, Oeda E, Tamaki K, Hanai J, Heldin C-H, Miyazono K, ten Dijke P. TGF-β receptor-mediated signalling through Smad2, Smad3 and Smad4. EMBO J. 1997;16:5353–5362. doi: 10.1093/emboj/16.17.5353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Hata A, Lo R, Massagué J, Pavletich N. A structural basis for mutational inactivation of the tumour suppressor Smad4. Nature. 1997;388:87–93. doi: 10.1038/40431. [DOI] [PubMed] [Google Scholar]
- Shikama N, Lyon J, La Thangue N. The p300/CBP family: Integrating signals with transcription factors and chromatin. Trends Cell Biol. 1997;7:230–236. doi: 10.1016/S0962-8924(97)01048-9. [DOI] [PubMed] [Google Scholar]
- Silver P, Keegan L, Ptashne M. Amino terminus of the yeast GAL4 gene product is sufficient for nuclear localization. Proc Natl Acad Sci. 1984;81:5951–5955. doi: 10.1073/pnas.81.19.5951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souchelnytskyi S, Tamaki K, Engström U, Wernstedt C, ten Dijke P, Heldin C-H. Phosphorylation of Ser465 and Ser467 in the C terminus of Smad2 mediates interaction with Smad4 and is required for transforming growth factor-β signaling. J Biol Chem. 1997;272:28107–28115. doi: 10.1074/jbc.272.44.28107. [DOI] [PubMed] [Google Scholar]
- Wrana JL, Attisano L. MAD-related proteins in TGF-β signalling. Trends Genet. 1996;12:493–496. doi: 10.1016/s0168-9525(96)30109-1. [DOI] [PubMed] [Google Scholar]
- Wrana JL, Attisano L, Wieser R, Ventura F, Massagué J. Mechanism of activation of the TGF-β receptor. Nature. 1994;370:341–347. doi: 10.1038/370341a0. [DOI] [PubMed] [Google Scholar]
- Wu R-Y, Zhang Y, Feng X-H, Derynck R. Homomeric and heteromeric interactions are required for signaling activity and functional cooperativity of Smad-3 and -4. Mol Cell Biol. 1997;17:2521–2528. doi: 10.1128/mcb.17.5.2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yingling J, Datto M, Wong C, Frederick J, Liberati N, Wang X-F. Tumor suppressor Smad4 is a transforming growth factor β-inducible DNA binding protein. Mol Cell Biol. 1997;17:7019–7028. doi: 10.1128/mcb.17.12.7019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Feng X-H, Wu R-Y, Derynck R. Receptor-associated Mad homologues synergize as effectors of the TGF-β response. Nature. 1996;383:168–172. doi: 10.1038/383168a0. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Musci T, Derynck R. The tumor suppressor Smad4/DPC4 as a central mediator of Smad function: Cooperativity with other Smads and interference of a Smad4/DPC4 mutant with nuclear translocation of Smads. Curr Biol. 1997;7:270–276. doi: 10.1016/s0960-9822(06)00123-0. [DOI] [PubMed] [Google Scholar]