Abstract
The EIR1 gene of Arabidopsis is a member of a family of plant genes with similarities to bacterial membrane transporters. This gene is expressed only in the root, which is consistent with the phenotypes of the eir1 mutants—the roots are agravitropic and have a reduced sensitivity to ethylene. The roots of eir1 mutants are also insensitive to the excess auxin produced by alf1-1 and fail to induce an auxin-inducible gene in the expansion zone. Although they fail to respond to internally generated auxin, they respond normally to externally applied auxin. Expression of the EIR1 gene in Saccharomyces cerevisiae confers resistance to fluorinated indolic compounds. Taken together, these data suggest that the EIR1 protein has a root-specific role in the transport of auxin.
Keywords: Root gravitropism, auxin, membrane protein, auxin transport inhibitors, ethylene
Plant tropisms, growth toward or away from a stimulus such as light or gravity, have been ascribed to differential growth—one side of the organ elongates to a greater extent than the other, resulting in a curvature toward or away from the stimulus (Darwin 1880; Poff et al. 1994). Gravitropism in roots can be demonstrated by manipulating plants so that they lie horizontal to the surface of the earth (Okada and Shimura 1992). Within a short time, the roots curve downward, exhibiting a positive gravitropic growth response.
Although it is not known how roots sense gravity, there is considerable evidence that the auxin, indole-3-acetic acid (IAA), is involved in the response to gravity. Transport studies suggest that IAA is redistributed in response to gravity so that it accumulates along the lower side of the root tip (Young et al. 1990). This redistribution of IAA in the root tip may be critical for gravitropism because removal of the root tip abolishes the response (Blancaflor et al. 1998). These studies are consistent with earlier views (the Cholodny-Went hypothesis, see Estelle 1996), which suggested that when roots are oriented horizontally, IAA accumulates along the lower side of the elongating zone, resulting in inhibition of cell elongation in those cells while those on top elongate, a process that eventuates in the downward bending of the root.
Mutants resistant to IAA have phenotypes that also support the involvement of IAA in gravitropism. Several auxin-resistant mutants are agravitropic (Estelle and Klee 1994). Many of these mutants are pleiotropic and have defects in both the root and the shoot. agr1 and eirl1 (ethylene insensitive root 1), however, exhibit a root-specific phenotype in gravitropism (Bell and Maher 1990; Roman et al. 1995). Interestingly, eir1 root growth also appears to be less sensitive to ethylene than that of the wild type, suggesting an involvement of ethylene in the regulation of root tropic responses.
If gravitropism involves the redistribution of IAA in response to gravity, then there must be transporters that alter the distribution of IAA within the root tissue (Goldsmith 1977; Lomax et al. 1995). IAA is thought to be transported down to the root tip via the vascular system of the root, and then transported up from the root tip to the elongation zone where it is probably localized in the epidermis. This latter polarized cell-to-cell transport could be explained by a chemiosmotic model (Goldsmith 1977; Lomax et al. 1995). This model posits that uncharged IAA in the acidic extracellular space enters a cell either by passive diffusion or facilitated transport. In the more basic cytosol, IAAH dissociates to form IAA−. Transport of IAA− out of the cell and on to the next is thought to depend on an efflux carrier (Jacobs and Gilbert 1983; Lomax et al. 1995). Therefore, gravitropism could result from the differential activity of an IAA efflux carrier in response to gravity.
Support for the chemiosmotic model comes from the effects of auxin transport inhibitors such as 2,3,5-triiodobenzoic acid (TIBA) and N-1-naphtylphtalamic acid (NPA) (Thomson et al. 1973; Katekar and Geissler 1980), which interfere with auxin efflux (Sussman and Goldsmith 1981). Plants grown in the presence of TIBA or NPA are agravitropic (e.g., Mulkey and Evans 1982; Lee et al. 1984). Moreover, mutants with altered response to these auxin transport inhibitors have phenotypes consistent with the hypothesis that transport of auxin is critical for the gravitropic response. AUX1 is resistant to auxin and defective in gravitropism (Maher and Martindale 1980). Recently, aux1 has also been shown to be resistant to auxin transport inhibitors (Fujita and Syõno 1996). Despite the connection between auxin transport inhibitors and gravitropism, the targets of these inhibitors and the molecules involved in directed auxin transport have not yet been identified.
In this report we explore the isolation and characterization of EIR1, a gene whose function is required for gravitropism. Genetic and physiological analyses of the gene and eir1 mutants suggest that EIR1 is involved in root-specific auxin transport. Furthermore, the EIR1 protein may be a target for regulation of auxin transport both by ethylene and synthetic transport inhibitors. These findings provide molecular evidence for the critical role of auxin transport in gravitropism.
Results
Isolation and phenotypic characterization of eir1-3
We isolated a plant from the Ac transposon pool B222-24 (Keller et al. 1992) whose root did not respond positively to gravity. This agravitropism segregated as if it resulted from a mutation in a single gene. A comparison of DNA from the mutant line B222-24 with the untransposed parental line B222 on Southern blots revealed that the mutant contained an additional copy of the transposon. This extra Ac element cosegregated with the mutant phenotype, suggesting that the mutation, designated eir1-3 was caused by the transposon.
This agravitropic mutation, eir1-3, is allelic to two previously described mutations, wav6-52 (allelic with agr1), which was isolated as an agravitrophic mutant (Bell and Maher 1990; Okada and Shimura 1990) and eir1-1, which was isolated as an ethylene insensitive mutant (Roman et al. 1995). The new mutation, eir1-3, fails to complement wav6-52 and eir1-1 showing that all three are alleles of EIR1. All three mutants have similar phenotypes with the severity of the mutant phenotype in the order eir1-3 = eir1-1 > wav6-52.
eir1 mutant roots do not respond to gravity when germinated and grown on agar plates oriented vertically. Instead, eir1 roots grow in random directions, whereas EIR1 roots grow downward. If the seedlings are reoriented so that the roots are now parallel to the surface of the earth, after 24 hr, the roots of the wild type reorient downward (roughly 90°), whereas roots of eir1 fail to reorient their growth (Fig. 1A). These severe defects in gravitropism appear to be restricted to the root, as the hypocotyl in all three eir1 mutant strains tested, still reorients when germinated in the dark. In another assay, seedlings were kept on 2% agar plates that were tilted vertically at an angle of less than 90°. Under these conditions, EIR1 roots do not penetrate the agar but grow on the surface in a wavy pattern, which is caused by reversible turns of the root tip (Okada and Shimura 1990). In contrast, eir1 roots exhibit a roughly linear growth pattern interrupted by random turns (Fig. 1B). When wild-type seeds are germinated on plates whose surface is parallel to the surface of the earth, they enter the agar and form a characteristic array of almost concentric curls (Garbers et al. 1996). eir1 mutant roots, however, failed to curl on the bottom of the plate and grew out in irregular patterns (Fig. 1C,D).
Figure 1.
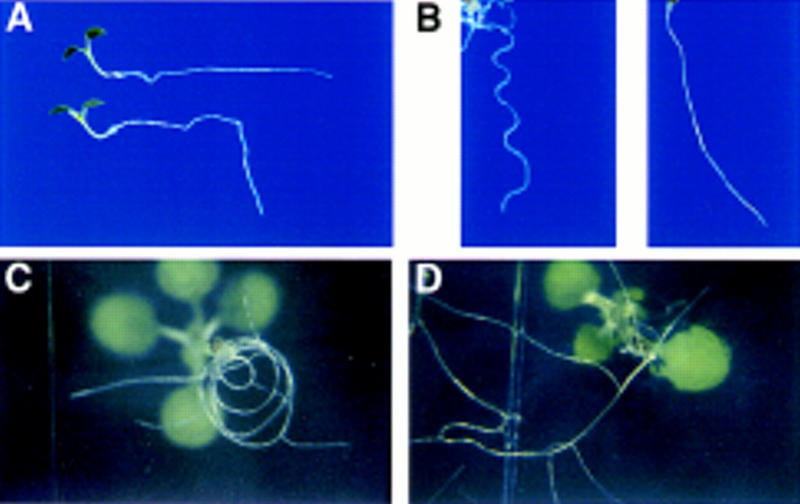
Tropic responses are blocked in roots of eir1-3. (A) Seedlings were grown on plates placed at 90° for 5 days, and then manually reoriented so that the roots were parallel to the surface of the earth. The direction of new root growth was monitored after 48 hr. Wild type (bottom) reorients root growth in response to gravity. eir1-3 (top) no longer responds to such changes. (B) Growth pattern of wild type (left) and eir1-3 (right) primary roots on 2% agar plates. Wavy growth on the surface is no longer visible in eir1-3. On reaching the bottom of the plate, wild-type roots (C) grow in a spiral pattern; eir1-3 (D) grows irregularly with random turns in direction.
eir1 mutants are less sensitive to both auxin transport inhibitors and ethylene. eir1 roots have a phenotype that is similar to EIR1 roots grown in the presence of NPA and TIBA, inhibitors of auxin transport that block cell elongation (Sussman and Goldsmith 1981). Moreover, eir1 root elongation was much more resistant than EIR1 to NPA and TIBA (Fig. 2; data not shown). In contrast, these auxin transport inhibitors inhibit lateral root formation to the same extent in both wild type and eir1 mutants. Also, eir1 root growth is more resistant than wild type to 1-aminocyclopropane-1-carboxylic acid (ACC), the immediate biosynthetic precursor of the growth regulator ethylene (Fig. 2). The root growth inhibition of eir1 mutants, however, is no different from EIR1 with respect to other growth regulators (abscissic acid, gibberellic acid, kinetin), the auxin-analog NAA (α-napthaleneacetic acid), and 2,4-d (2,4-dichloro-phenoxyacetic acid).
Figure 2.
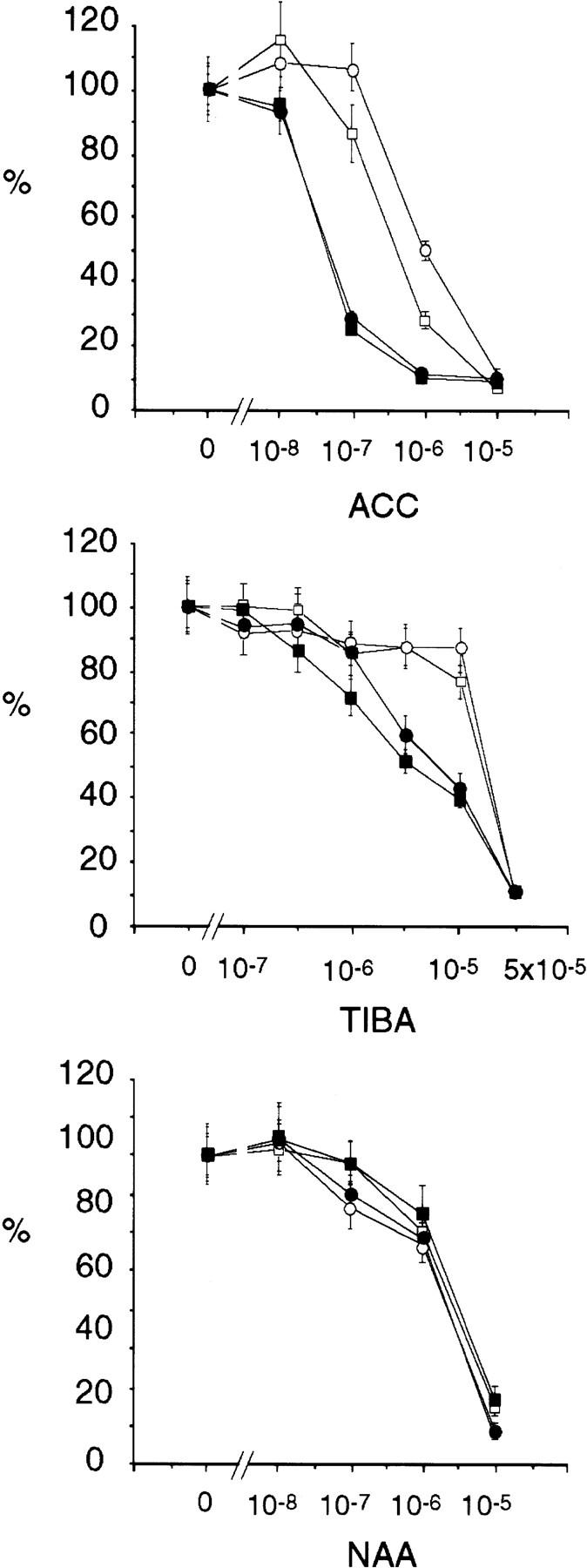
Dose response curves of root growth from wild-type plants and eir1 mutants [(•) Col-O; (▪) Ws; (○) eir1-1; ( ) eir1-3]. Root elongation determined at 12 DAG was normalized to root growth on unsupplemented medium (100%). Standard deviations are shown as bars; molarities used are indicated.
The eir1 mutants have longer roots than wild-type plants (Table 1), which could be due to an increased rate of cell division and/or to greater elongation of individual root cells. Direct measurement showed that eir1-3 root cells were longer than wild-type cells (Table 1). We have not, however, ruled out the possibility that increased cell division contributes to the increased length as well.
Table 1.
Root growth and cell elongation
Length of primary roots was determined at ∼12 DAG.
Elongation of 35–40 young trichoblasts was determined on images. Root lengths are indicated in mm; cell length in μm.
Cloning of EIR1
We cloned the eir1-3 allele using an inverse PCR approach. A 600-bp fragment amplified from eir1-3 DNA hybridized to the additional band caused by the Ac insertion in eir1-3. This subcloned fragment was used to screen an Arabidopsis thaliana genomic phage library. Three genomic clones of the putative EIR1 gene (λ5-3, λ6-1, and λ6-3) had the same restriction pattern. We used the subcloned insert of λ5-3 for screening cDNA libraries. Eight hybridizing phage clones were isolated from ∼5 × 105 plaques screened. These clones all show similar restriction patterns. Two inserts of ∼2.2 kb were completely sequenced. The largest cDNA clone contained a continous open reading frame (ORF) starting 29 bp downstream of its 5′end. Comparison of the cDNA with the genomic clone revealed that the ORF is split into nine exons coding for a predicted protein of 69.3 kD.
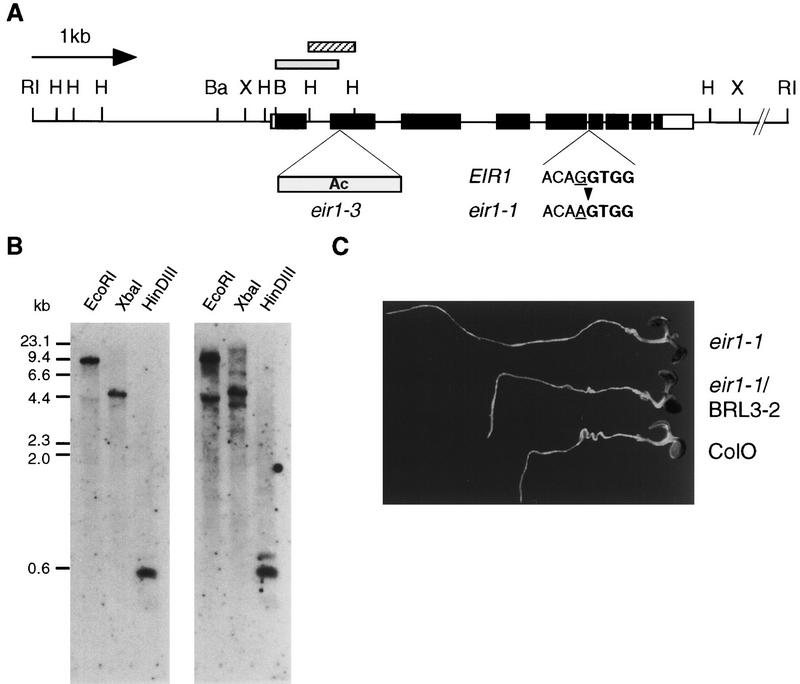
The Ac insertion in eir1-3 is located after codon 113 in exon 2 (Fig. 3). The insertion is flanked by a perfect 8-bp direct repeat and probably results in a null allele of the affected gene. We found that eir1-1 (as compared with the progenitor Columbia wild type) contains a transition mutation at the intron 5/exon 6 border that replaces the absolutely conserved G at splice position −1 (Brown 1996). The eir1-1 mutation presumably results in a truncated EIR1 protein that would lack a conserved portion of the molecule (Fig. 3).
Figure 3.
The EIR1. (A) Schematic representation of an EcoRI fragment isolated from phage λ5-3 (see text for details). The bars indicate the nine exons of EIR1. Those segments presumed to be translated are black. Two mutations are indicated beneath the line: Insertion of Ac in eir1-3 after amino acid 133 and base substitution of the intron/exon junction in eir1-1. The shaded bar above the line indicates the genomic fragment amplified by inverse PCR (see text). (RI) EcoRI; (H) HinDIII; (Ba) BamHI; (X) XbaI; (B) BclI. (B) Southern blot made with genomic DNA from ecotype Ws hybridized with a 600-bp HinDIII fragment (hatched bar above the line in A). (Left) High stringency wash; (right) low stringency wash. (C) The gravitropism defect of eir1-1 is suppressed by transformation with the EcoRI fragment shown in A. The plant at the top is the recipient, eir1-1/BRL3-2 in the middle is the transformant, and Col-O at the bottom is wild type.
To determine whether our cloned segment was the EIR1 gene, we transformed eir1-1 with the putative EIR1 ORF and >2 kb of upstream sequences. All five independent hygromycin-resistant transformants of eir1-1 tested had a root growth phenotype typical of wild type (Fig. 3C). Therefore, the defects of the eir1-1 mutant were complemented by the genomic fragment. As there were no other large ORFs present on the genomic fragment used in the transformation, we have designated the ORF as the coding region of EIR1.
EIR1, a highly conserved plant gene family with similarities to bacterial transporters
Several lines of evidence suggest that EIR1 belongs to a highly conserved gene family. Arabidopsis has several genes with considerable homology to EIR. In addition to several Arabidopsis expressed sequence tags (ESTs) (GenBank accession nos. T04468, T43636, R84151, and Z38079), similar ORFs were found in database entries of the Arabidopsis Genome Initiative. Two close relatives dubbed AEH1 and AEH2 (for Arabidopsis EIR1 Homolog) were located on clones T26J12 and MKQ4 on chromosome 1 and 5 respectively. These relatives probably account for the extra restriction fragments that hybridize to the EIR1 probe under conditions of high stringency (Fig. 3B). We have also identified and sequenced a related rice EST (GenBank accession no. D25054), which is derived from root-specific cDNA (Fig.4). No other closely related sequences could be found outside the plant kingdom, suggesting that EIR1 and its homologs represent a family of genes unique to higher plants.
Alignment of the deduced amino acid sequences of EIR1, AEH1, AEH2, and REH1 (Rice EIR1 Homolog) revealed that the regions of identity are restricted to the amino and carboxyl termini (Fig. 4A). Hydropathy plots (Fig. 4B) and topology predictions identified 10 potential transmembrane domains shared by the four members of the gene family. The transmembrane domains are located in the highly conserved portions of the proteins—five at the amino terminus and five at the carboxyl terminus (Fig. 4A).
Figure 4.
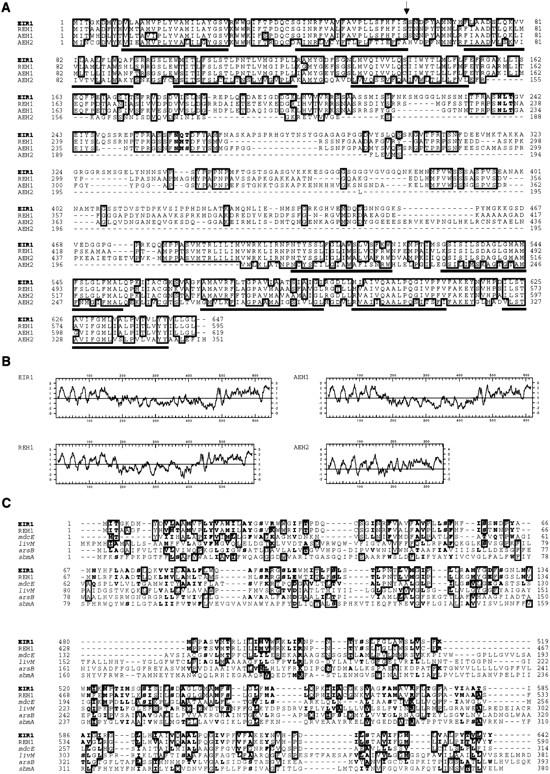
(A) Alignment of deduced amino acid sequences of EIR1, the rice homolog REH1, and the two putative Arabidopsis homologs AEH1 and AEH2. For EIR1 and REH1, ORFs of the cDNAs were deduced. The protein sequences of AEH1 and AEH2 were deduced from the genomic sequences by identifying canonical splice donor and acceptor sites. Identical residues are boxed and dashes indicate gaps in the sequence. Black lines correspond to the 10 potential transmembrane domains shared by all four proteins. Potential, conserved N-glycosylation sites are in bold letters. An arrow indicates the cleavage site of a potential amino-terminal signal peptide found for EIR1, REH1, and AEH1. (B) Hydrophobicity plots of EIR1, REH1, AEH1, and AEH2 according to Kyte and Doolittle (1982). Positive values indicate hydrophobic areas. (C) Alignment of the conserved amino-terminal (top) and carboxy-terminal (bottom) transmembrane domains of EIR1 and REH1 with a number of selected bacterial transporters (for more details, see text). Identical residues are boxed. The bold letters represent positions where exchanges are conservative (L,I,V,M; A,S,T; F,W,Y; N,Q; D,E; and K,R) and shared by EIR1 and at least two other sequences. Dashes indicate gaps in the alignment.
The internal segment of the protein, though less conserved in sequence than the putative membrane spanning domains, exhibits a number of similarities. This segment is enriched for hydrophilic amino acids with a remarkably high content of serines and prolines. EIR1 possesses a number of potential N-glycosylation sites, two of which are also found in REH1 and AEH1 (Fig. 4A). As EIR1 has no endoplasmic reticulum retention signal but does have a potential amino-terminal signal peptide (von Heijne 1986), the protein could transit the secretory pathway to the plasma membrane.
The two hydrophobic portions of EIR1 show restricted similarity to a number of bacterial membrane proteins (Fig. 4C). The mdcE (U95087) protein is a potential malonate transporter from Klebsiella pneumoniae (Hoenke et al. 1997), whereas livM (P22729) is involved in high affinity uptake of leucine into Escherichia coli (Adams et al. 1990).
Particularly noteworthy is the similarity of EIR1 to the class of efflux carriers that remove toxic compounds from the interior of the cell. For example, E. coli arsB (P52146) represents a part of the arsenic efflux system (Diorio et al. 1995). sbmA (X54153), another integral membrane protein of E. coli, has been shown to be necessary for uptake of the antibiotic Microcin 25 (Salomon and Farias 1995). Portions of EIR1 show 35%–40% similarity to these proteins. Our finding that the amino and the carboxyl terminus of EIR1 exhibit similarities to the corresponding parts of bacterial transporters, suggests that EIR1 may be a membrane protein with a related function.
EIR1 affects the root-specific response to endogenous ethylene
The reduced sensitivity of eir1 roots to inhibition by ethylene suggested that EIR1 might be a gene involved in regulation of ethylene responses specific to the root. To test this hypothesis, we examined the response of the entire eir1 mutant plant to endogenous ethylene by constructing double mutants of eir1 with eto3 and ctr1. eto3 causes overproduction of ethylene, giving rise to the typical triple response (the hypocotyl of plants germinated in the dark remains short and undergoes radial swelling, and apical hook formation is exaggerated). Mutations in the Raf-like protein kinase CTR1 phenocopy the ethylene-grown phenotype without elevating endogenous ethylene concentrations, suggesting that CTR1 acts as a negative regulator of ethylene signal transduction (Kieber et al. 1993).
The double mutants eir1-3 eto3-1 and eir1-3 ctr1-1 were germinated both in the dark and under constant illumination. Dark-germinated plants still undergo the triple response, indicating that the eir1 mutation has no influence on germination and early development of the aerial parts of the seedling (Fig. 6A,B, below). The inhibition of root elongation caused by eto3 and ctr1 mutations, however, is considerably reduced in the double mutants (Fig. 5A,B).
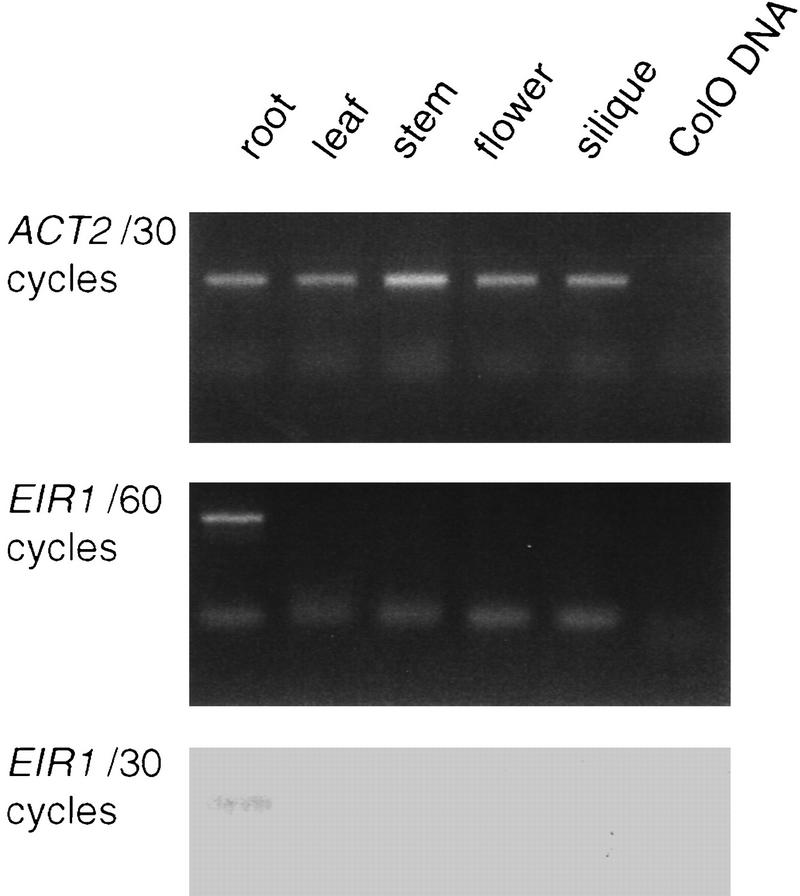
Figure 6.
RS–PCR performed with tissue-specific cDNA of Col-O plants. (Top) Expression of ACT2 is detectable in all tissues tested after 30 cycles. (Middle) An EIR1-specific amplification product derived from root cDNA after 60 cycles. No signal was detectable in any other tissues tested. (Bottom) Southern blot of EIR1-specific amplification products after 30 cycles using a transcript-specific oligonucleotide as a probe.
Figure 5.
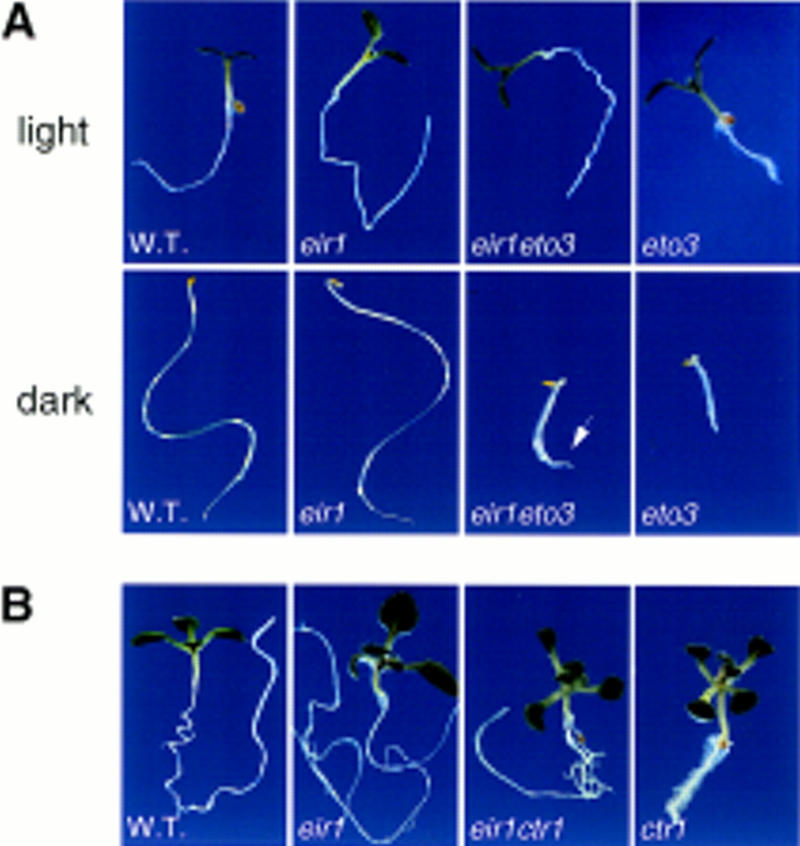
The phenotype of eir1 is restricted to the root. (A) From left to right: Wild type (Col-O), eir1-3, eto3-1, and eir1-3 eto3-1 were germinated under constant illumination or in the dark. Pictures were taken at 5 DAG (light) and at 2–3 DAG (dark), respectively. The image of the eir1 eto3 mutant grown in the dark reveals the similarity of its hypocotyl and that of eto3. With time the root of eir1 eto3 (arrow) becomes much longer than that of eto3. Average primary root lengths of light-grown plants are as follows: Col-O, 37 ± 3 mm; eir1-3, 44 ± 4 mm; eto3-1, 8 ± 1 mm; eir1-3 eto3-1, 22 ± 4 mm. (B) From left to right: Wild type, ctr1-1, eir1-3, and eir1-3 ctr1-1 at ∼10 DAG. Note the intermediate root elongation phenotype of the double mutant. The aerial part of eir1-3 ctr1-1 still resembles ctr1-1. Average primary root lengths were as follows: ctr1-1, 6 ± 1 mm; eir1-3 ctr1-1, 16 ± 2 mm.
These results suggest that reduced ethylene sensitivity of the eir1 mutant is completely restricted to the root. Moreover, the phenotype is not caused by a block in biosynthesis or transport of ethylene because eir1-3 bypasses the root phenotypes of ctr1-1, a mutation thought to be constitutive for the transduction of the ethylene signal.
EIR1 expression is localized to the root
We used RNA-specific-PCR (RS–PCR) to analyze EIR1 expression in the plant. Primers located on the 5′ end of the EIR1–cDNA were used to amplify transcripts from reverse-transcribed poly(A)+ RNA derived from roots, leaves, stems, flowers, and siliques. Primers for first strand cDNA synthesis were chimeric, having a 5′ extension with no complementary sequences in the Arabidopsis genome. This sequence extension was used for subsequent PCR to avoid contamination. Genomic DNA from ecotype Col-O served as a negative control. The experiment (Fig. 6) revealed a specific RS–PCR product in the root, but not in any other tissues. The root-specificity of EIR1 expression correlates well with the root-specific alterations detected in eir1 mutants, suggesting that these defects are likely to be a consequence of the absence of EIR1 function in the roots.
EIR1 function is required for auxin homeostasis in root cells
The involvement of EIR1 in root-specific auxin distribution was tested by analysis of the expression pattern of an auxin-inducible gene, AtIAA2. The expression of AtIAA2 has been shown to be strongly induced within a few minutes after exposure to auxin (Abel et al. 1995). The AtIAA2 expression pattern was visualized using a reporter construct, PIG4::GUS, a transgene expressing β-glucuronidase under control of the AtIAA2-promoter. AttIAA2 expression is strongest in the root meristem in wild type and eir1-3. When wild type is gravistimulated, expression of AtIAA2 extends into the elongation and differentiation zone. Moreover, the expression is asymmetric with the lower portion of the elongation zone showing more intense staining than the upper (Fig. 7). This asymmetric staining suggests that the lower portion of the elongation zone has elevated auxin levels as compared with the upper level. In contrast, reporter expression in eir1-3 does not respond to the gravistimulus and remains restricted to the root tip (Fig. 7).
Figure 7.
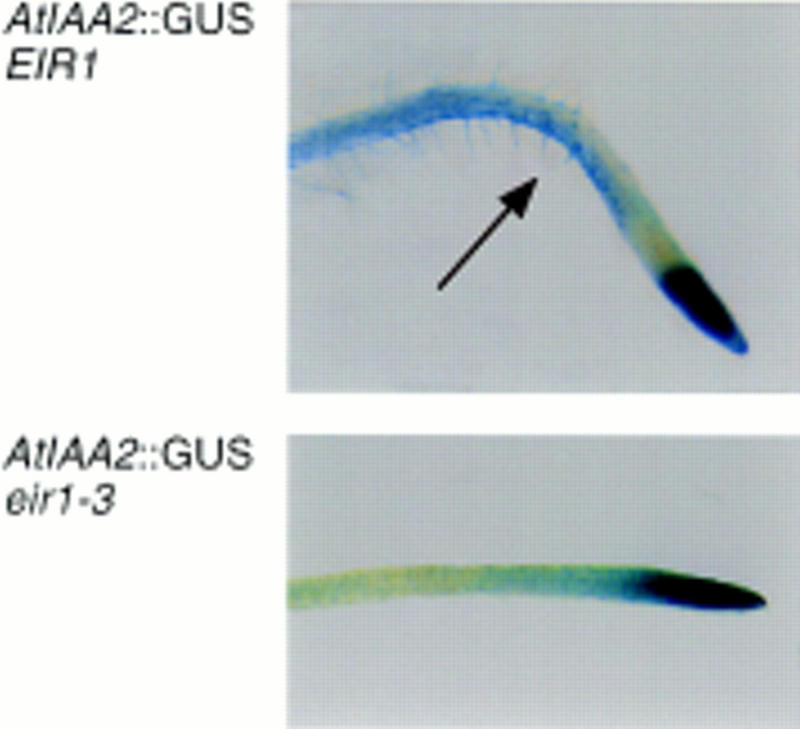
AtIAA2::GUS expression in primary roots: Wild-type roots exhibit asymmetric GUS-staining in the elongation zone on root bending. In eir1-3, expression of the auxin reporter no longer responds to gravistimuli.
The eir1 root is less sensitive to ethylene and shows increased resistance to synthetic auxin transport inhibitors. These phenotypes could be explained if ethylene, like auxin transport inhibitors, interferes with tissue distribution of auxin. We therefore determined the effect of exogenous auxin on PIG4::GUS. Expression of AtIAA2 has been shown to be induced strongly within a few minutes after exposure to auxin (Abel et al. 1995). In plants grown on regular medium, GUS staining is found in the root meristem and in the stele proximal to the root meristem (Fig. 8A). Addition of NAA to the medium induces reporter gene expression in both the root meristem and elongation zone of the root tip in wild type and the eir1 mutant (Fig.8A). Therefore, eir1 mutants retain their ability to respond to exogenous auxin.
Figure 8.
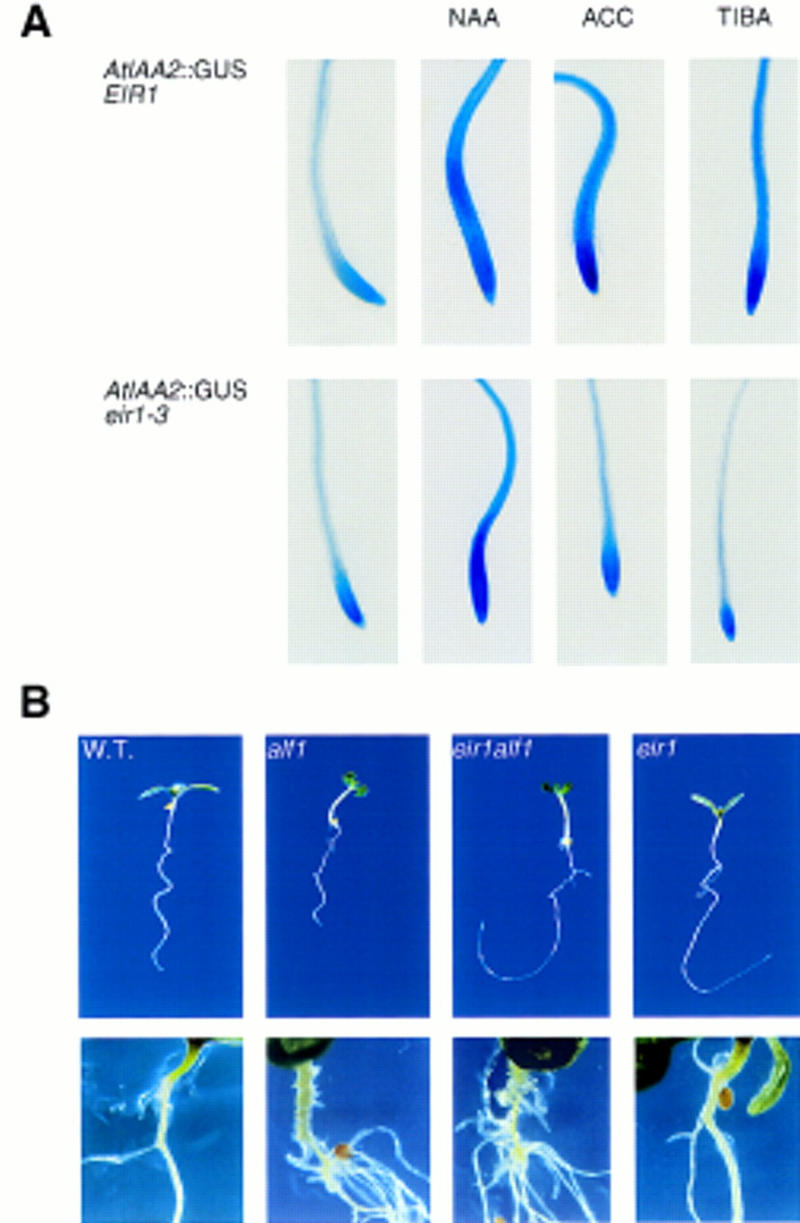
(A) AtIAA2::GUS expression upon hormone and TIBA treatment. From left to right: wild type (top) and eir1-3 (bottom) plants were grown on standard medium (PNA), or for 24 hr on PNA supplemented with 1 μm NAA, 1 μm ACC, or 5 μm TIBA. In untreated plants expression of the auxin reporter is restricted to the tip and the stele of the roots. eir1-3 no longer responds to ACC and TIBA treatment. (B) eir1-3 suppresses the inhibition of root elongation in alf1-1. (Top) The plants are ∼6 DAG. From left to right: wild type, alf1-1, eir1-1 alf1-1, and eir1-1. (Bottom) Comparison of the primary root–hypocotyl junction of older plants. Excessive lateral and adventitious root formation is not suppressed in the eir1-1 alf1-1 double mutant.
Plants with the reporter responded quite differently to growth in ACC (1 μm ACC for 24 hr). In wild type, the entire elongation and differentiation zone shows considerable GUS staining on ACC treatment. Furthermore, expression of GUS in the the cell division zone appeared to be enhanced (Fig. 8A). In striking contrast, eir1-3 roots grown in ACC shows virtually no response in these tissues. Expression is restricted to the root tip at an intensity similar to that of plants grown in the absence of ACC (Fig. 8A).
The results with the auxin transport inhibitor TIBA are similar to those obtained with exogenous ACC. The reporter construct is induced in wild type, but the mutant has a very reduced response (Fig. 8A). As auxin is the only known endogenous inducer of AtIAA2 (Abel et al. 1996), ectopic expression of AtIAA2 in wild-type roots treated with auxin transport inhibitors should be a consequence of elevated auxin concentrations in those cells that express the reporter. Unaltered AtIAA2 expression in TIBA- and ACC-treated eir1-3 roots suggests that auxin concentrations in cells of the root elongation zone remain unaffected when treated with these compounds.
eir1 blocks the inhibition of root growth caused by high endogenous levels of auxin
If EIR1 is responsible for the redistribution of endogenous auxin, then the eir1 mutation should block the defects in strains producing high levels of auxin. The effect of endogenous auxin was examined in eir1-1 alf1-1 double mutants. The alf1 mutation results in an enormously increased concentration of internal auxin, which leads to severe morphological alterations, which include the development of numerous short adventitous and lateral roots (Boerjan et al. 1995; Celenza et al. 1995). The short root phenotype is caused by inhibition of cell elongation. The eir1-1 mutation completely suppresses the short root phenotype caused by alf1-1, and retains the agravitropic phenotype, whereas the aerial portion of the eir1 alf1 double mutant resembles alf1. Furthermore, the increased adventitious and lateral root formation, typical of alf1, is not blocked by eir1-3 suggesting that eir1 represents a root-tip-specific suppressor of the elevated auxin concentrations present in alf1 (Fig. 8B).
EIR1 in Saccharomyces cerevisiae confers increased resistance to indolic compounds
We analyzed the growth of yeast strains that express a plasmid-borne Arabidopsis EIR1 gene under the control of the ADH1 promoter. Wild-type yeast strains are only slightly sensitive to fluorinated indolic compounds such as 5-dl-fluoro-tryptophan or 5-fluoro-indole, toxic analogs of potential precursors of IAA (Bartel 1997). Strains that carry the Δgef1 deletion [a mutant that alters ion homeostasis in yeast (Gaxiola et al. 1998)], however, are much more sensitive to 5-fluoro-indole, 5-fluoro-dl-tryptophan, and 5-fluoro-indoleacetic acid. Remarkably, gef1 strains that contain the EIR1 gene were much more resistant to these indolic compounds than isogenic gef1 strains with only a vector (Fig. 9A). The increased resistance conferred by EIR1 can also be observed in wild type, but the effect is more subtle because of the greater intrinsic resistance of strains with a functional GEF1 gene.
Figure 9.
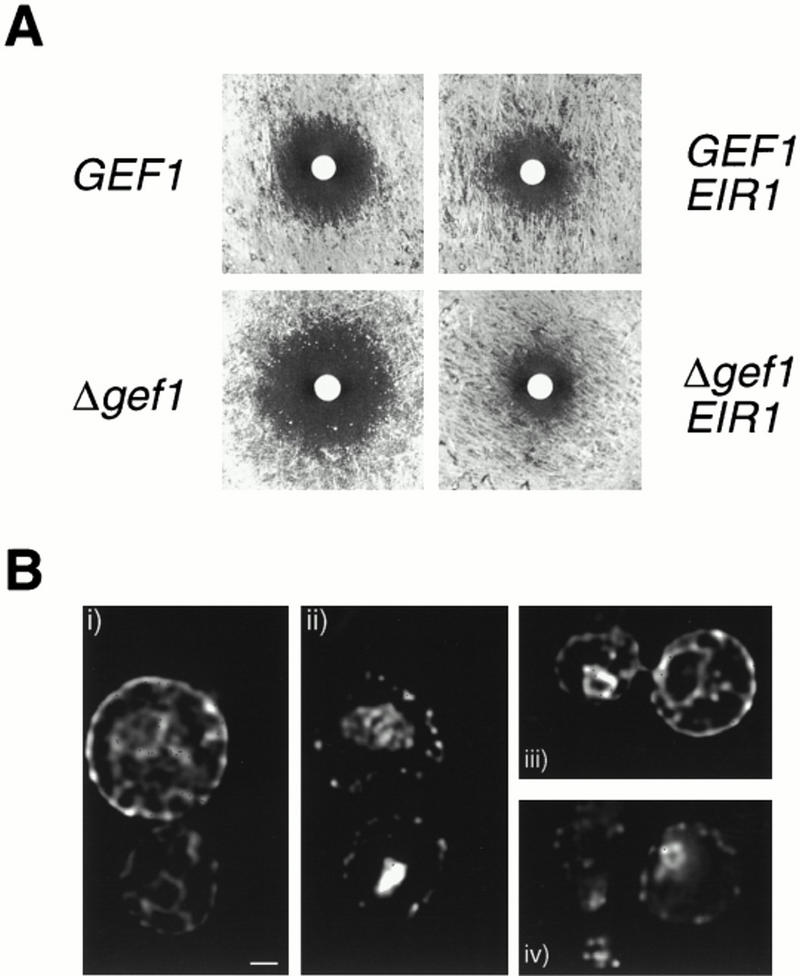
EIR1 expressed in S. cerevisiae. (A) Growth of both wild-type and gef1 strains on 5-fluoro-indole (30 μl of a 100 mm stock solution) is rescued by constitutive expression of EIR1. Similar experiments expressing either ClC-0 or a mutated version of EIR1 failed to restore yeast growth in the presence of fluorinated indolic compounds. (B) Immunolocalization of HA-tagged EIR1 protein (i, iii). Most of the signal is localized to the plasma membrane. Weaker signals proximal to the nucleus (indicated by the DAPI staining in ii and iv) could be ER localization (bar, 1 μm). Controls were performed with strains expressing EIR1 without HA tag and by omitting the primary antibody during immunostaining. No specific signals could be detected in these experiments.
Expression of the EIR1 gene is required for this resistance because yeast strains containing a mutant form of the EIR1 gene (a frameshift in the EIR1 ORF, plasmid pADE1-H) fail to show the increased resistance to fluoro-indoles. Moreover, this resistance is specific to these indolic compounds because strains carrying the EIR1 gene are no more resistant than controls to fluconazole, another inhibitor of yeast growth. In addition, the increased resistance is not simply the consequence of expression of a foreign transporter in yeast. Expression of the Torpedo marmorata chloride channnel (ClC-0), which suppresses many of the gef1 defects, failed to confer increased resistance to indolic compounds.
To localize the EIR1 protein in yeast, we introduced a functional, hemagglutinin (HA) epitope-tagged version of EIR1 into S. cerevisiae. Examination of immunodecorated yeast cells using charge-coupled microscopy localized the most intense staining of EIR1 to the plasma membrane (Fig. 9B). This membrane localization is consistent with a role for EIR1 in excluding compounds from the cell and, thereby, preventing the toxicity of the indolic compounds.
Discussion
Our isolation of eir1-3, a new transposon-tagged allele of EIR1, permitted the cloning and characterization of both the mutant and wild-type genes. Sequence analysis shows that eir1-3 is an Ac insertion in the second of nine exons and eir1-1 is a base substitution at a conserved splice site junction. Both of these mutations are likely to be null alleles because they should result in completely defective proteins. Expression of EIR1 appears to be restricted to the root, which is consistent with the finding that all of the eir1 mutant phenotypes, the most striking of which is agravitropism, affect the root and not other parts of the plant.
The phenotypes of the eir1 mutant suggest a block in root-specific auxin transport
The expression pattern of the auxin-inducible fusion, AtIAA2::GUS, in eir1-3 is consistent with a block in auxin transport in the roots of this mutant. In wild-type and eir1-3 plants, this reporter is expressed in root tips and at a low level in the younger parts of the vascular tissue. Wild-type plants in the presence of ethylene show increased expression of the reporter in the elongation zone, suggesting that these cells have an increased level of IAA.
The expression of the auxin-inducible reporter on gravistimulation supports and extends these results. In wild type, the auxin reporter is expressed asymmetrically, with more intense GUS staining localized to the lower side of the elongation zone. This distribution is consistent with a model that proposes an inhibitory role for auxin in the regulation of root cell elongation and differential inhibition as the basis for gravitropism. Consistent with this interpretation, the agravitropic eir1-3 mutant grown under the same conditions fails to show differential staining or induction of the reporter in the elongation zone.
The failure of cells in the elongation zone of eir1 roots to respond to IAA could be a consequence either of a failure to synthesize or to redistribute this growth regulator in response to ethylene and gravistimulation. The effect of the eir1 mutation on the root phenotype of the alf1 mutant supports the redistribution hypothesis. The alf1 mutation results in an ∼10-fold increase in the endogenous concentration of IAA (Boerjan et al. 1995). The high auxin level enhances the formation of lateral and adventitious roots but, also inhibits root elongation. Primary root growth in the eir1 alf1 double mutant is not inhibited, showing that eir1 suppresses the inhibitory effect of IAA on root elongation caused by the alf1 mutation. eir1 does not block the hyper-induction of lateral roots caused by alf1, however, showing that there are high levels of auxin in the root of the eir1 alf1 double mutant.
These data are consistent with a model in which EIR1 functions in auxin distribution in the root elongation zone. Two directions of auxin transport have been suggested for roots (Estelle 1996 and references therein)—acropetal transport in the central cylinder from the base to the tip of the root and basipetal transport from the root tip to the elongation zone. If the inhibition of root growth in the alf1-1 mutant results from the inhibition of cell expansion by excess auxin in the cells of the elongation zone, then the suppression of alf1 by eir1 is a consequence of eir1’s defect in basipetal auxin transport into the elongation zone.
The root phenotype of eto3 and ctr1, like that of the alf1 mutant, is also suppressed by eir1. In both mutants, the entire plant exhibits a strong ethylene response. eto3 causes ethylene overproduction, whereas ctr1 is probably a negative regulator of the ethylene response because ctr1 strains act as if they were in the presence of high ethylene although they do not have elevated ethylene concentrations (Kieber et al. 1993). The eir1 mutant partially suppresses the ctr1 phenotypes, suggesting that EIR1 acts either downstream of ETO3 and CTR1 or in a pathway parallel to that in which ETO3 and CTR1 function (Roman et al. 1995).
eir1 is more resistant to both auxin transport inhibitors and ethylene
The decreased sensitivity of the eir1 root to the inhibitory effects of ethylene as well as to the synthetic auxin transport inhibitors TIBA and NPA suggests a connection between auxin and ethylene. This behavior is similar to that of the HOOKLESS1 (HLS1) mutants of Arabidopsis (Lehmann et al. 1996). HLS1 is thought to control bending in the apical tip of the hypocotyl because hls1 mutants fail to form the apical hook during germination. Expression of the HLS1 gene and enhanced hook formation are induced by treatment of plants with ethylene, which causes differential cell elongation. Remarkably, wild-type seedlings grown in the presence of NPA have the same effect on apical hook formation and tissue distribution of auxin-induced genes as does the hls1 mutant. Therefore, auxin transport inhibitors phenocopy the hls1 mutant, which is defective in the response of the apical hook to ethylene. These observations led to the speculation (Lehmann et al. 1996) that an ethylene response gene may control differential cell growth by regulating auxin activity or distribution.
The growth characteristics of the eir1 mutants also suggest a connection between auxin and ethylene. The eir1 mutant root, like the apical hook of the hls1 mutant is less sensitive to both exogenous and endogenous ethylene. Growth of wild type in the presence of auxin transport inhibitors blocks apical hook formation and the negative gravitropic response of the root. Moreover, like hls1, the eir1 roots are resistant to auxin transport inhibitors. In fact, this cross-resistance to both ethylene and auxin transport inhibitors is characteristic of mutants defective for auxin and ethylene responses (Fujita and Syõno 1996; C. Luschnig and G.R. Fink, unpubl.). This phenomenon probably represents an underlying mechanistic connection between the ethylene response and the auxin response, which is not yet understood.
The properties of EIR1 are consistent with a role in transport of auxin
The amino acid sequence of EIR1 is consistent with a role for this protein in transport of IAA. EIR1 is predicted to be an integral membrane protein. The presence of potential N-glycosylation sites and a potential amino-terminal signal peptide suggests a possible localization in the plasma membrane. EIR1 also has similarities to several membrane proteins involved in translocation of a variety of different substances across the plasma membrane. The transporters related to EIR1 are diverse in their substrate specificity and translocate amino acids, heavy metals, antibiotics, and dicarboxylic acids.
Perhaps the most compelling evidence that EIR1 has a role in transport is that expression of EIR1 in S. cerevisiae confers increased resistance to fluorinated analogs of indolic compounds. The resistance phenotypes are strongest in the gef1 mutant, which has increased sensitivity to various compounds, probably as a result of altered ion homeostasis (Gaxiola et al. 1998). Resistance to these indoles is completely dependent on a functional EIR1 gene product, as neither ClC-O, nor a mutated version of EIR1 were capable of restoring yeast growth in the presence of fluorinated indolic compounds.
The EIR1 protein could prevent the inhibition of yeast by these compounds either by preventing their uptake or facilitating their efflux from the cytosol. The preferential localization of EIR1 in the plasma membrane of yeast is consistent with either of these mechanisms.
Why is eir1 agravitropic?
Gravitropism, the curvature of the root in response to gravity, results from greater elongation of the upper side of the root than the lower side. Differential root elongation has been postulated to arise as the consequence of a gravity-induced auxin gradient with more auxin on the lower than the upper side (Kaufman et al. 1995). The factors responsible for creating the auxin gradient are not known.
The simplest model to explain the phenotypes of the eir1 mutant is that EIR1 is required for efflux of auxin from the cells of the root tip into the elongation zone. If the root is oriented so that there is an increase in the auxin concentration on one side of the root tip, then EIR1 would pump auxin into the adjacent elongation zone with the concomitant inhibition of cell elongation. In eir1 mutants, the increased auxin in the lower portion of the root tip would fail to be transported into the elongation zone, and there would be no differential elongation. The predicted phenotypes of such a defect agree with those we observe for an eir1 mutation. The root should be agravitropic and longer overall than an EIR1 root. Furthermore, as we observed, increased levels of internal auxin should fail to inhibit the root or to induce root-specific auxin-inducible transcripts. The insensitivity of the eir1 root to ethylene can be reconciled with the model if ethylene inhibits root growth by increasing the internal auxin concentrations (Suttle 1991).
Our model is also consistent with the response of eir1 mutants to externally added auxins. If the eir1 block were not in efflux, but rather in uptake of auxin, as has been proposed for aux1 mutants (Bennett et al. 1996), then like the aux1 mutants, the eir1 mutants should be resistant to external auxin. The eir1 mutants, however, respond normally to external auxin. Root elongation is inhibited as in wild type, and induction of the AtIAA2-reporter construct appears to be unaffected. This model in its simplest form does not explicitly account for the increased resistance of the eir1 mutants to the auxin transport inhibitors. An understanding of this resistance as well as the other phenotypes of the eir1 mutants will require an investigation of EIR1 expression, localization, and regulation in Arabidopsis.
Materials and methods
Plant strains and growth conditions
Plants were grown aseptically on unsupplemented plant nutrient agar (PNA) without sucrose (Haughn and Somerville 1986). Growth responses were tested by adding various supplements to the medium as indicated. Plates were wrapped in gas-permeable surgical tape and kept under continuous illumination. For gravitropic response experiments, plates were kept in a vertical position. For the “root waving assay” plates were kept at an angle of about 30°. Root elongation was assayed at 10–12 days after germination (DAG). Formation of lateral roots was compared by counting lateral roots on both wild-type and mutant plants grown under conditions as described in the text.
Seed stocks for eir1-1 and eto3-1 were obtained from the Arabidopsis Biological Resource Center (Ohio State University, Columbus). ctr1-1 was a kind gift from J. Hua (Caltech, Pasadena, CA). wav6-52 was obtained from K. Okada (National Institute for Basic Biology, Okazaki, Japan). PIG4::GUS was a kind gift from J. Normanly (University of Massachusetts, Amherst). Transposon line B222 was obtained from DNA Plant Technology Corporation (Oakland, CA).
Inverse PCR, cloning, and structural analysis of EIR1
Genomic DNA was prepared according to a protocol from Quiagen. After grinding the frozen tissue, the resulting powder was incubated at 74°C for 20 min in lysis buffer (100 mm Tris/HCl at pH 9.5, 1.4 m NaCl, 0.02 m EDTA, 2% cetyltrimethyl-ammonium bromide (CTAB), 1% PEG 8000). After extraction with an equal amount of chloroform, DNA was precipitated with isopropanol. After resuspension in 1 m NaCl and treatment with RNase A, the DNA was loaded onto equilibrated Quiagen columns and purified according to the manufacturer’s instructions. DNA extracted from the Ac line B222 and eir1-3 was digested with EcoRI and BclI. The ends of the DNA were made blunt with Klenow fragment. This DNA was religated and used for inverse PCR performed with oligonucleotides CCTCGGGTTCGAAATCG and GGGGAAGAACTAATGAAGTGTG. After 40 cycles of amplification at 60°C annealing temperature, the products were separated on 1% agarose gels. A fragment specific for eir1-3 DNA was cloned into pGEMT (Promega) to give pGsac1 and used for Southern hybridization on eir1-3 and wild-type DNA.
Phage genomic and cDNA libraries of A. thaliana (Kieber et al. 1993; obtained from the Arabidopsis Biological Resource Center) were probed with pGsac1 using standard techniques (Ausubel et al. 1987). Genomic clone λ5-3, which hybridized to pGsac1, was subcloned into pBluescriptII (Stratagene) to give pB5-3. The sequence of an EcoRI fragment ∼9 kb in length was determined on an ABI automated DNA sequencer. For sequence analysis of eir1-1, the coding region of this allele and its corresponding wild type (Col-O) were amplified with PCR. The point mutation in eir1-1 was confirmed by subsequent PCR amplification of sequences covering the mutation.
Two full-length cDNA clones subcloned into pBSII (pBc5-2 and pBc6-1) were completely sequenced. The rice EST (D25054) homologous to EIR1 was obtained from MAFF DNA Bank [National Institute of Agrobiological Resources (NIAR), Japan].
Sequence comparisons with database entries were performed using Gapped BLAST and PSI-BLAST algorithms (Altschul et al. 1997). Multiple alignments and structural predicitions were performed using the algorithms at BCM Search Launcher.
Complementation of eir1-1 in transgenic plants
An EcoRI fragment of the genomic clone pB5-3, which carries the entire coding region and >2 kb of upstream sequences of the EIR1 gene, was subcloned into pBIBhyg (Becker 1990). The resulting T-DNA vector pBRL was transformed into Agrobacterium tumefaciens strain GV3101 via electroporation, and used for subsequent vacuum infiltration of eir1-1 plants (Bechtold et al. 1993). Correct integration of the full-length transgene was confirmed on Southern blots.
RS–PCR
For expression analysis, total RNA from tissue of sterile grown plants was isolated (Niyogi et al. 1992). Vegetative tissue isolated from plants 15 DAG was used. Flower-specific RNA was isolated at ∼20 DAG and silique-specific RNA at ∼25 DAG. Poly(A)+ RNA was isolated with the polyATract kit from Promega. About 50 ng of poly(A)+ RNA of each tissue was used for RS–PCR.
RS–PCR with slight modifications was performed as described by
Shuldiner et al. (1993). Oligonucleotides
GAACATCGATGACCAAGCTTAGGTATCGATAGCCCCACGGAACTCAAA
(underscored bases correspond to nucleotides 454–470 of the
EIR1 coding region) and
CTTATACGGATATCCTGGCAATTCGGACTTGTTAGCTTTAGGGTTAA
(underscored bases are complementary to nucleotides 335–351 of
ACT2 coding region) were added to poly(A)+ RNA to a final
concentration of 2 μm in a volume of 10 μl. The tubes
were placed at 65°C for 10 min and allowed to cool down to 37°C.
First-strand cDNA synthesis was performed using GIBCO BRL AMV reverse
transcriptase. Primer pairs GAACATCGATGACCAAGCTTAGGTATCGATA and
GGCAAAGACATGTACGATGTTTTAGCGG (bases 10–37 of EIR1 coding
region) or CTTATACGGATATCCTGGCAATTCGGACTT and
GTCTGTGACAATGGAACTGGAATG (bases 31–54 of ACT2 coding region)
were used in a standard PCR for 30 cycles with 40 sec at 94°C, 40
sec at 60°C, and 1 min at 72°C. For EIR1,
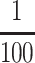 of this reaction was used for reamplification under the same
conditions. 32P-End-labeled oligonucleotide
GTGAAAAGAGCGTTATCATCCATTCTAG (complementary to bases 292–319 of
EIR1 coding region) allowed us to verify the identity of the
EIR1-specific band on a Southern blot.
of this reaction was used for reamplification under the same
conditions. 32P-End-labeled oligonucleotide
GTGAAAAGAGCGTTATCATCCATTCTAG (complementary to bases 292–319 of
EIR1 coding region) allowed us to verify the identity of the
EIR1-specific band on a Southern blot.
Preparation and microscopic analysis of roots
Whole plants were incubated twice in methanol/glacial acetic acid (3:1) and rinsed several times in PBT (130 mm NaCl, 10 mm sodium phosphate at pH 7.0, 0.1% Tween 20). Then, roots were mounted onto microscope slides into clearing solution (stock solution: 8 grams of chloralhydrate in 2.5 ml of 20% glycerol). After 10 min, roots were viewed under a Zeiss microscope using Nomarski Optics. Dark-field photographs of live plants were made using a Wild M5-A microscope. These images were used for determination of root cell length. GUS stainings were performed as described (Lehmann et al. 1996). Images were recorded on Kodak Ektachrome 160T film and processed using Adobe Photoshop.
Complementation analysis and construction of double mutants
For complementation analysis of the three putative eir1 alleles, we crossed eir1-3/eir1-3 plants (eir1-3 still contains the Ac donor T-DNA construct conferring hygromycin resistance) into plants homozygous for either eir1-1 or wav6-52.
Heterozygous F1 plants (eir1-3/wav6-52 and eir1-3/eir1-1) identified as resistant to hygromycin were defective in root gravitropism, giving evidence for the allelism of the three mutants analyzed. F2 plants derived from each of the F1 heterozygotes were all Eir1−, whereas the hygromcin resistance marker segregated as a single, dominant locus.
Double mutants (e.g., ctr1-1/ctr1-1 eir1-3/eir1-3) were derived from crosses of homozygous single mutant lines and scored for segregation in the F2 generation of the initial crosses. Double mutant candidates were backcrossed into their two parental single mutant lines and their genotype verified by complementation with parental testers. For eir1-1 alf1-1 double mutants, we used eir1-1/eir1-1 plants for pollination of alf1-1/ALF1 heterozygotes. F2 seeds were scored for segregation of Eir1− and Alf1− phenotypes. The double mutant was verified by segregation of the aerial Alf1− phenotype in Eir1− F3 plants derived from the initial cross.
Yeast manipulations and constructs
All experiments were carried out in W303 (a/α ura3-1 can1-100 leu2-3, 112 trp1-1 his3-11, 15). For disruption of GEF1, we used plasmid pRG52. For analysis of CLC-0 we used the vector PRS1024 (for more details, see Gaxiola et al. 1998).
Yeast strains were grown overnight at 30°C in synthetic complete (SC) medium, and ∼2 × 106 cells were plated onto SC plates. Solutions of inhibitors used in the filter growth assays were spotted onto Schleicher & Schuell filter paper 740. After they dried, the filters were transferred onto the yeast plates, which then were incubated at 25°C in the dark for 2–5 days. After that, yeast growth was monitored and documented.
For expression of EIR1 in S. cerevisiae the insert of pBc5-2 was cloned into pAD4M (described in Ballester et al. 1989) to give pAD-E1. A frameshift mutation in EIR1 was obtained by filling in the internal HindIII site resulting in a nonsense mutation after codon 178 (plasmid pADE1-H). For construction of the HA-tagged version of EIR1, we used primers GGGTCTAGAGTACTCTACTACGTTCTTTTGGGGCTTTACCCATACGATGGTCCTGAC and GGGTCTAGAGTCGACGCACTGAGCAGCGTAAT for PCR amplification of a fragment encoding three copies of the HA epitope. The PCR product was ligated into pAD-E1 resulting in pAD-E1HA coding for a protein with the 3×HA tag fused to the authentic carboxyl terminus of EIR1. Immunostaining of the tagged protein in haploid and diploid cells was performed as described by Gaxiola et al. (1998). Cells were viewed by using charge-coupled device microscopy and sectioned by using Scanalytics (Billerica, MA)
Acknowledgments
We thank Jian Hua and Andrew Diener for critical reading of the manuscript, and members of the Fink laboratory for helpful suggestions. C.L. thanks Marie-Theres Hauser for suggestions on root preparation. C.L. was supported by Schroedinger Fellowship J01414-Gen from the FWF, Austria, and by the National Science Foundation (NSF). R.A.G. was supported by the PEW Latin American Fellows Program and by the European Community. Work was supported by NSF grant MCB9317175 to G.R.F. G.R.F. is an American Cancer Society Professor of Genetics.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Note
Sequence data described in this paper have been submitted to GenBank under accession nos. AF056026 and AF056027.
Footnotes
E-MAIL fink@wi.mit.edu; FAX (617) 258-9872.
References
- Abel S, Ballas N, Wong L-M, Theologis A. DNA elements responsive to auxin. BioEssays. 1996;18:647–654. doi: 10.1002/bies.950180808. [DOI] [PubMed] [Google Scholar]
- Abel S, Nguyen MD, Theologis A. The PS-IAA4/5-like family of early auxin-inducible mRNAs in Arabidopsis thaliana. J Mol Biol. 1995;251:533–549. doi: 10.1006/jmbi.1995.0454. [DOI] [PubMed] [Google Scholar]
- Adams MD, Wagner LM, Graddis TJ, Landick R, Antonucci TK, Gibson AL, Oxender DL. Nucleotide sequence and genetic characterization reveal six essential genes for the LIV-I and LS transport systems of Escherichia coli. J Biol Chem. 1990;265:11436–11443. [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current protocols in molecular biology. New York, NY: John Wiley/Greene; 1987. [Google Scholar]
- Ballester R, Michaeli T, Ferguson K, Xu HP, McCormick F, Wigler M. Genetic analysis of mammalian GAP expressed in yeast. Cell. 1989;59:681–686. doi: 10.1016/0092-8674(89)90014-7. [DOI] [PubMed] [Google Scholar]
- Bartel B. Auxin biosynthesis. Annu Rev Plant Physiol Plant Mol Biol. 1997;48:51–66. doi: 10.1146/annurev.arplant.48.1.51. [DOI] [PubMed] [Google Scholar]
- Bechtold N, Ellis J, Pelletier G. In planta Agrobacterium mediated gene transfer by infiltration of adult Arabidopsisplants. CR Jeances Acad Sci Ser III Sci Vie. 1993;316:1194–1199. [Google Scholar]
- Becker D. Binary vectors which allow the exchange of plant selectable markers and reporter genes. Nucleic Acids Res. 1990;18:203. doi: 10.1093/nar/18.1.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell CJ, Maher PE. Mutants of Arabidopsis thalianawith abnormal gravitropic responses. Mol & Gen Genet. 1990;220:289–293. [Google Scholar]
- Bennett MJ, Marchant A, Green HG, May ST, Ward SP, Millner PA, Walker AR, Schulz B, Feldmann KA. Arabidopsis AUX1gene: A permease-like regulator of root gravitropism. Science. 1996;273:948–950. doi: 10.1126/science.273.5277.948. [DOI] [PubMed] [Google Scholar]
- Blancaflor EB, Fasano JM, Gilroy S. Mapping the functional roles of cap cells in the response of Arabidopsisprimary roots to gravity. Plant Physiol. 1998;116:213–222. doi: 10.1104/pp.116.1.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boerjan W, Cervera M-T, Delarue M, Beeckman T, Dewitte W, Bellini C, Caboche M, Van Onckelen H, Van Montagu M, Inze D, et al. superroot, a recessive mutation in Arabidopsis,confers auxin overproduction. Plant Cell. 1995;7:1405–1419. doi: 10.1105/tpc.7.9.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JWS. Arabidopsisintron mutations and pre-mRNA splicing. Plant J. 1996;10:771–780. doi: 10.1046/j.1365-313x.1996.10050771.x. [DOI] [PubMed] [Google Scholar]
- Celenza JL, Grisafi PL, Fink GR. A pathway for lateral root formation in Arabidopsis thaliana. Genes & Dev. 1995;9:2131–2142. doi: 10.1101/gad.9.17.2131. [DOI] [PubMed] [Google Scholar]
- Darwin C. Power of movements in plants. London, UK: John Murray; 1880. [Google Scholar]
- Diorio C, Cai J, Marmor J, Shinder R, DuBow MS. An Escherichia colichromosomal are operon homolog is functional in arsenic detoxification and is conserved in gram-negative bacteria. J Bacteriol. 1995;177:2050–2056. doi: 10.1128/jb.177.8.2050-2056.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estelle M. Plant tropisms: The ins and outs of auxin. Curr Biol. 1996;6:1589–1591. doi: 10.1016/s0960-9822(02)70780-x. [DOI] [PubMed] [Google Scholar]
- Estelle M, Klee HJ. Auxin and cytokinin in Arabidopsis. In: Meyerowitz EM, Somerville CR, editors. Arabidopsis. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1994. pp. 555–578. [Google Scholar]
- Fujita H, Syõno K. Genetic analysis of the effects of polar auxin transport inhibitors on root growth in Arabidopsis thaliana. Plant Cell Physiol. 1996;37:1094–1101. doi: 10.1093/oxfordjournals.pcp.a029059. [DOI] [PubMed] [Google Scholar]
- Garbers C, DeLong A, Deruére J, Bernasconi P, Söll D. A mutation in protein phospatase 2A regulatory subunit A affects auxin transport in Arabidopsis. EMBO J. 1996;15:2215–2124. [PMC free article] [PubMed] [Google Scholar]
- Gaxiola RA, Yuan DS, Klausner RD, Fink GR. The yeast CLCchloride channel functions in cation homeostasis. Proc Natl Acad Sci. 1998;95:4046–4050. doi: 10.1073/pnas.95.7.4046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldsmith MHM. The polar transport of auxin. Annu Rev Plant Physiol. 1977;28:439–478. [Google Scholar]
- Haughn GW, Somerville C. Sulfonylurea-resistant mutants of Arabidopsis thaliana. Mol & Gen Genet. 1986;204:430–434. [Google Scholar]
- Hoenke S, Schmid M, Dimroth P. Sequence of a gene cluster from Klebsiella pneumoniae encoding malonate decarboxylase and expression of the enzyme in Escherichia coli. Eur J Biochem. 1997;246:530–538. doi: 10.1111/j.1432-1033.1997.00530.x. [DOI] [PubMed] [Google Scholar]
- Jacobs M, Gilbert SF. Basal localization of the presumptive auxin transport carrier in pea stems. Science. 1983;220:1297–1300. doi: 10.1126/science.220.4603.1297. [DOI] [PubMed] [Google Scholar]
- Katekar GF, Geissler AE. Auxin transport inhibitors. Plant Physiol. 1980;66:1190–1195. doi: 10.1104/pp.66.6.1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman PB, Wu L-L, Brock TG, Kim D. Plant hormones. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1995. Hormones and the orientation of growth. [Google Scholar]
- Keller J, Lim E, James Jr DW, Dooner HK. Germinal and somatic activity of the maize element Activator (Ac) in Arabidopsis. Genetics. 1992;131:449–459. doi: 10.1093/genetics/131.2.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieber JJ, Rothenberg M, Roman G, Feldmann KA, Ecker JA. CTR1, a negative regulator of the ethylene response pathway in Arabidopsis,encodes a member of the Raf family of protein kinases. Cell. 1993;72:427–441. doi: 10.1016/0092-8674(93)90119-b. [DOI] [PubMed] [Google Scholar]
- Kiss JR, Hertel R, Sack FD. Amyloplasts are necessary for full gravitropic sensitivity in roots. Planta. 1989;177:198–206. [PubMed] [Google Scholar]
- Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Lee JS, Mulkey TJ, Evans ML. Inhibition of polar calcium movement and gravitropism in roots treated with auxin-transport inhibitors. Planta. 1984;160:536–543. [PubMed] [Google Scholar]
- Lehman A, Black R, Ecker JR. HOOKLESS1, an ethylene response gene, is required for differential cell elongation in the Arabidopsishypocotyl. Cell. 1996;85:183–194. doi: 10.1016/s0092-8674(00)81095-8. [DOI] [PubMed] [Google Scholar]
- Lomax TL, Muday GK, Rubery PH. Auxin Transport. In: Davies PK, editor. Plant hormones. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1995. pp. 509–530. [Google Scholar]
- Maher EP, Martindale SJB. Mutants of Arabidopsis thalianawith altered responses to auxin and gravity. Biochem Genet. 1980;18:1041–1054. doi: 10.1007/BF00484337. [DOI] [PubMed] [Google Scholar]
- Mulkey TJ, Evans ML. Suppression of asymmetric acid efflux and gravitropism in maize roots treated with auxin transport inhibitors or sodium orthovanadate. J Plant Growth Regul. 1982;1:259–265. [Google Scholar]
- Niyogi KK, Fink GR. Two anthranilate synthase genes in Arabidopsis:Defense-related regulation of the tryptophan pathway. Plant Cell. 1992;4:721–733. doi: 10.1105/tpc.4.6.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada K, Shimura Y. Reversible root tip rotation in Arabidopsisseedlings induced by obstacle-touching stimulus. Science. 1990;250:274–276. doi: 10.1126/science.250.4978.274. [DOI] [PubMed] [Google Scholar]
- ————— Mutational analysis of root gravitropism and phototropism of Arabidopsisthaliana seedlings. Aust J Plant Physiol. 1992;19:439–448. [Google Scholar]
- Poff KL, Janoudi A-K, Rosen ES, Orbovic V, Konjevic R, Fortin MC, Scott TK. The physiology of tropisms. In: Meyerowitz EM, Somerville CR, editors. Arabidopsis. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1994. pp. 639–664. [Google Scholar]
- Roman G, Lubarsky B, Kieber JJ, Rotheneberg M, Ecker JR. Genetic analysis of ethylene signal transduction in Arabidopsis thaliana:Five novel mutant loci integrated into a stress response pathway. Genetics. 1995;139:1393–1409. doi: 10.1093/genetics/139.3.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano CP, Cooper ML, Klee HJ. Uncoupling auxin and ethylene effects in transgenic tobacco and Arabidopsisplants. Plant Cell. 1993;5:181–189. doi: 10.1105/tpc.5.2.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomon RA, Farias RN. The peptide antibiotic Microcin 25 is imported through the TonB pathway and the sbmAprotein. J Bacteriol. 1995;177:3323–3325. doi: 10.1128/jb.177.11.3323-3325.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuldiner AR, Perfetti R, Roth J. RNA template-specific polymerase chain reaction (RS-PCR) In: White BA, editor. Methods in molecular biology: PCR protocols: Current methods and applications. Totowa, NJ: Humana Press; 1993. pp. 169–176. [DOI] [PubMed] [Google Scholar]
- Sussman MR, Goldsmith MHM. The action of specific inhibitors of auxin transport on uptake of auxin and binding of N-1-naphtylphtalamic acid to a membrane site in maize coleoptyles. Planta. 1981;152:13–18. doi: 10.1007/BF00384978. [DOI] [PubMed] [Google Scholar]
- Suttle JC. Biochemical bases for the loss of basipetal IAA transport with advancing physiological age in etiolated Helianthushypocotyls. Plant Physiol. 1991;96:875–880. doi: 10.1104/pp.96.3.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson K-S, Hertel R, Müller S, Tavares JE. 1-n-Naphtylphtalamic acid and 2,3,5-Triiodobenzoic acid invitro binding to particulate cell fractions and action on auxin transport in corn coleoptyles. Planta. 1973;109:337–352. doi: 10.1007/BF00387102. [DOI] [PubMed] [Google Scholar]
- von Heijne G. A new method for predicting signal sequence cleavage sites. Nucleic Acids Res. 1986;14:4683–4690. doi: 10.1093/nar/14.11.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young LM, Evans ML, Hertel R. Correlations between gravitropic curvature and auxin movement across gravistimulated roots of Zea mays. Plant Physiol. 1990;92:792–796. doi: 10.1104/pp.92.3.792. [DOI] [PMC free article] [PubMed] [Google Scholar]