Abstract
Receptor activator of nuclear factor-kappa B ligand (RANKL) is a member of tumor necrosis factor (TNF) superfamily that plays a key role in the regulation of differentiation, activation and survival of osteoclasts and also in tumor cell migration and bone metastasis. Osteoclast activation induced by RANKL regulates hematopoietic stem cell mobilization as part of homeostasis and host defense mechanisms thereby linking regulation of hematopoiesis with bone remodeling. Binding of RANKL to its receptor, Receptor activator of nuclear factor-kappa B (RANK) activates molecules such as NF-kappa B, mitogen activated protein kinase (MAPK), nuclear factor of activated T cells (NFAT) and phosphatidyl 3-kinase (PI3K). Although the molecular and cellular roles of these molecules have been reported previously, a systematic cataloging of the molecular events induced by RANKL/RANK interaction has not been attempted. Here, we present a comprehensive reaction map of the RANKL/RANK-signaling pathway based on an extensive manual curation of the published literature. We hope that the curated RANKL/RANK-signaling pathway model would enable new biomedical discoveries, which can provide novel insights into disease processes and development of novel therapeutic interventions.
Database URL: http://www.netpath.org/pathways?path_id=NetPath_21
Introduction
The paracrine system involved in many tissues includes numerous cytokines and growth factors necessary for the differentiation and maturation of cells of which amongst tumor necrosis factor (TNF) family assumes a vital role. The TNF superfamily consists of numerous ligand/receptor proteins involved in various cellular processes such as development, homeostasis and apoptosis. RANKL, also known as TNFSF11, osteoprotegerin ligand, TNF-related activation-induced cytokine (TRANCE) or osteoclast differentiation factor, is a member of the TNF superfamily (1). The membrane bound form of RANKL is a type II transmembrane glycoprotein with an extracellular region, a transmembrane domain and an intracellular cytoplasmic region (2). RANKL also exist in a soluble form derived either through an alternative splicing event or by the proteolytic cleavage of the membrane anchoring domain of RANKL by a membrane-associated metalloprotease, such as TNF converting enzyme, matrix metalloproteinase 14, matrix metalloproteinase 7 or Disintegrin and metalloproteinase domain-containing protein 19 (3–6). As a member of TNF family, RANK is expressed mainly on cells of monocyte/macrophage lineage such as pro-osteoclasts and osteoclasts, whereas the RANKL is synthesized by and expressed on cell membrane of bone marrow stromal cell and osteoblasts. Expression of RANKL has also been detected in a wide variety of tissues and specific cells including spleen, placenta, heart, stomach, thyroid gland, lung, brain, thymus, lymph nodes, osteoclasts and peripheral blood leukocytes (7–10). In bone metabolism, osteoclasts play primary role by involving in bone resorption. In brain, RANKL is expressed in the lateral septal nucleus and RANK is expressed specifically in the neurons and astrocytes of the preoptic area and medial septal nucleus in the hypothalamus. Hanada et al. (11) have shown a novel function of RANKL/RANK signaling in brain that plays a significant role in the regulation of body temperature and fever response in inflammation. Prolactin, progesterone and parathyroid hormone-like related protein stimulate RANKL expression on mammary gland epithelial cells whereas RANK is constitutively expressed in these cells. Knockout studies in mice have shown that RANKL and RANK are required for the development, proliferation and survival of lactating mammary gland during pregnancy. RANKL/RANK-signaling system is critical for the induction of lactation by mediating lobulo-alveloar morphogenesis. Kim et al., have also shown that RANK activation by RANKL is crucial to control proliferation of mammary epithelial cells (12,13). Furthermore, RANKL is reported to be upregulated in primary malignant bone tumors including osteosarcoma, chondrosarcoma, giant cell tumor (14) and multiple myeloma (15). RANKL has also been shown to be overexpressed in >90% of metastatic tumor cells in adenocarcinoma lesions of prostate, breast, lung and thyroid origin (16) and also in breast cancer stromal cells (17).
RANKL binds to type I transmembrane protein known as RANK (TNFRSF11A) to form a functional hexamer complex containing trimeric RANKL and RANK (18–20). The RANKL/RANK interaction is essential to osteoclastogenesis as it stimulates the differentiation, maturation and survival of osteoclasts (21–25). In the immune system, RANKL expressed by activated T cells interacts with RANK expressed on dendritic cells in order to regulate survival and function of dendritic cells (10,26–28). RANKL/RANK system plays an important role in controlling the development of AIRE+ thymic medullary epithelial cells in the thymus (29) and in lymph node organogenesis (30). Osteoprotegerin (OPG), also known as (TNFRSF11B) or osteoclastogenesis inhibitory factor, is a decoy receptor that modulates signaling by RANKL during osteoclastogenesis. OPG binds to RANKL and neutralizes its function thereby negatively regulating osteoclast differentiation, maturation and survival (31,32). Undifferentiated pre-osteoblastic bone marrow stromal cells were known to express high levels of RANKL along with relatively low levels of OPG. The ratio of RANKL/OPG increases during their activation and differentiation into osteoclasts. This ratio declines during differentiation of undifferentiated pre-osteoblastic cells into mature osteoblasts during which their osteoclastogenic activity is suppressed (33). Thus the RANKL/OPG ratio plays a significant role in determining bone mass and skeletal integrity. RANK, being a member of TNF receptor superfamily, lacks any catalytic activity and therefore recruits adaptor molecules such as TNFR-associated factors (TRAFs) to transduce signals.
Several pathways have been shown to be activated by RANKL/RANK-signaling system. These pathways include, NF-kappa B, mitogen activated protein kinases (MAPK's), protein kinase C (PKC), Ca2+/Calcineurin/nuclear factor of activated T cells (NFAT) and phosphoinositide-3-kinase (PI3K). The functional importance of NF-kappa B (34–40), JNK (41–44), SRC-PI3K-AKT (45,46) and p38MAPK (47) pathways in osteoclast differentiation and osteoclastogenesis have been extensively studied. Although the RANKL/RANK pathway has been well studied, a comprehensive map of this signaling pathway is not available in any of the publicly available signal transduction pathway resources. Our group has previously developed a signaling pathway resource called NetPath (http://www.netpath.org), which currently comprises of 20 human signaling pathways involved in immune- and cancer-signaling pathways (48). With the goal of creating manually curated individual pathways relevant to molecular biologists and clinicians alike, we describe here the development of a comprehensive resource of RANKL/RANK-signaling pathway. Similar approaches were previously undertaken by various groups to provide comprehensive maps of epidermal growth factor receptor (49), interleukin-1 receptor (50) and toll-like receptor (51) signaling pathways. RANKL/RANK-signaling pathway resource is a NetPath module freely available at http://www.netpath.org/pathways?path_id=NetPath_21.
Curation of RANKL pathway reactions
An extensive curation of scientific literature was carried out to catalog RANKL/RANK stimulated reactions. The reactions in NetPath collectively represent individual biochemical events such as protein–protein interactions (PPIs), enzyme–substrate reactions or protein translocation events. We have cataloged these reactions which are proved to be induced or enhanced by the RANKL–RANK interaction. The curation of RANKL pathway reactions was based on the following criteria: (i) reactions should be stimulated by RANKL/RANK system, (ii) reactions must be either induced or enhanced in vivo, (iii) proteins involved in reactions should be from human system, however other mammalian proteins were annotated if they were not reported from human system (The mammalian proteins should have orthologous proteins identified in human), (iv) reactions if stimulated by multiple ligands were not considered and (v) only experiments carried out in cell lines of mammalian origin were considered. We used PathBuilder (52), a curation tool previously developed by our group, to annotate pathway reactions captured in this curation process.
PPIs
PPIs are important mediators and regulators of signaling. We have curated PPIs reported in literature for RANKL/RANK-signaling system as direct or complex interactions. Direct interactions represent interaction between two molecules whereas complex interactions represent the existence of more than two molecules as a complex. We have also curated various features pertaining to these PPIs including domains and motifs involved in the interaction, dependence of the reactions on various post-translational modifications (PTMs), subcellular location of the interaction and also the cell type in which the reactions were reported.
Enzyme–substrate reactions
PTMs are known to regulate signaling cascades by altering functional properties and recruiting proteins to subcellular compartments. We have captured several events of phosphorylation, dephosphorylation, ubiquitination and proteolytic cleavage in RANKL/RANK-signaling pathway. We could not capture any study specific to other PTMs such as sumoylation, desumoylation, deubiquitination, acetylation, deacetylation, methylation, demethylation, palmitoylation and glycosylation reactions in RANKL/RANK-signaling pathway. We cataloged these reactions as either direct or indirect catalytic events. Whenever the upstream enzymes responsible for modification were reported in the literature, PTMs are curated as direct. However, induced/indirect represents PTMs on proteins for which no report is available on the upstream enzyme responsible for the modification. We documented the enzyme–substrate reactions responsible for direct catalysis events also as PPIs. Whenever available in the literature, we have also cataloged the site and residue information of the PTMs and mapped these on to the respective protein sequences provided in RefSeq database. The molecules which are reported to be activated or inhibited by various assays such as GTPase assay or kinase activity assay but their PTM status has not been reported in RANKL signaling are cataloged into activation/inhibition reactions.
Protein translocation events
During signal transduction, proteins translocate across subcellular compartments or are secreted into the extracellular space, a fate determined by interaction partners, various PTMs, or a specific regulatory event. We have curated translocation events in RANKL pathway along with their PTM dependence and description of the experimental system of observation. We used Gene Ontology (GO) to define subcellular localization (53).
Gene regulation
We have cataloged transcriptionally regulated genes by RANKL/RANK signaling from normal human cells. The cataloging involves the identification and curation of genes differentially regulated by RANKL signaling reported in literature as demonstrated using diverse experimental strategies including DNA microarrays, northern blot, serial analysis of gene expression and quantitative RT–PCR. The cells/cell types in which the genes are identified to be differentially regulated are also curated. In addition, we have also documented transcriptional regulators of the differentially regulated genes.
Summary of RANKL/RANK pathway curation
Our manual literature survey on RANKL/RANK-signaling pathway has cataloged 88 molecules involved in 73 PPIs, 59 enzyme–substrate reactions and 10 protein translocation events. PPIs include 58 binary interactions and 15 protein complexes. The 59 enzyme–substrate reactions comprise of 14 direct reactions which were also considered as direct PPIs and 45 RANKL induced catalytic reactions. We have also cataloged 74 genes which are differentially regulated at the mRNA level by RANKL signaling in various cell types in human system. The interaction domains or motifs for 14 proteins in this pathway have also been documented. The site and residue information for PTMs were captured for 17 proteins. We annotated transcriptional regulators for eight differentially regulated genes from our literature survey pertaining to studies on RANKL pathway. Each and every pathway reactions annotated in NetPath were linked to scientific articles. Six PPIs which were not available in HPRD were submitted to HPRD (54). All the RANK receptor activated reactions were reviewed by a pathway authority (http://www.netpath.org/pathway_authority?path_id=NetPath_21). We have captured several molecules associated with RANKL signaling but not included in the review articles on this pathway to the best of our knowledge. These molecules include FHL2, LYN, FYN, PTK2, ATP6V1E1, ETV5, CYLD, SQSTM1 and LIMD1. We hope that future review articles will consider the addition of these proteins while describing RANKL/RANK signaling.
Protein complexes in RANKL/RANK-signaling pathway
Multiprotein complexes, not individual proteins, are increasingly being recognized as the molecular basis of cellular fluxes of signals. Thus, the identification of dynamic protein complexes is of major interest to the experimental as well as computational biologists. The current effort on RANKL/RANK-signaling system has cataloged several such protein complexes. These complexes are enlisted in Table 1. There were seven protein complexes which were associated with RANK and seven complexes with TRAF6 as a constituent.
Table 1.
Protein complexes curated in RANKL/RANK signaling
| Protein complex | References |
|---|---|
| TNFRSF11A–TRAF6–TAB2– MAP3K7–TAB1 | Mizukami et al., 2002 (67) |
| TNFRSF11A–TRAF1–TRAF3–TRAF6 | Wong et al., 1999 (68) |
| RUNX1–TRAF6–FHL2 | Bai et al., 2008 (69) |
| TNFRSF11A–SRC–TRAF6 | Wong et al., 1999 (68) |
| TNFRSF11A–CBL–PIK3R1–PIK3R2–TRAF6 | Arron et al., 2001 (70) |
| TNFRSF11A–TAB1–MAP3K7 | Mizukami et al., 2002 (67) |
| TNFRSF11A–TAB2–TRAF6 | Mizukami et al., 2002 (67) |
| SQSTM1–TRAF6–CYLD | Jin et al., 2008 (71) |
| LYN–TNFRSF11A–PTPN6–GAB2 | Kim et al., 2009 (72) |
| IKBKG–IKBKG–IKBKG | Darwech et al., 2009 (73) |
The table contains a list of proteins which are reported to form complexes when RANK (TNFRSF11A) is stimulated by RANKL (TNFSF11). Five of the protein complexes which contained RANK also had TRAF6 as a constituent molecule, indicating a central role of TRAF6 in RANKL/RANK signaling.
Data formats and availability
The RANKL pathway data described in this work is integrated into NetPath which is publicly available at (http://www.netpath.org). NetPath RANKL pathway page provides a summary of the pathway, statistics of the number of molecules and total reactions curated under different data categories such as physical interactions, enzyme catalysis, transport and gene regulation. An overview of the RANKL/RANK pathway page in NetPath with the curated information on PPIs, enzyme catalysis and PTMs, protein translocation reactions and gene regulation is provided in Figure 1. The annotated data can be downloaded in various community standard data exchange formats. These include Proteomics Standards Initiative for Molecular Interaction (PSI-MI version 2.5), Biological PAthway eXchange (BioPAX level 3) and Systems Biology Markup Language (SBML level 2.1). PSI-MI is a community standard for molecular interaction data which facilitates data comparison, exchange and verification (55,56). BioPAX is an emerging standard community language that enables integration, exchange, visualization and analysis of biological pathway data which also incorporates information that can be stored in the PSI-MI format (57). SBML is a machine-readable format for representing models which facilitates mathematical simulation (58). An excel file is also provided to download the list of upregulated and downregulated genes.
Figure 1.
An overview of the RANKL pathway page in NetPath. RANKL pathway page in NetPath hosts the information on the number of molecules curated for RANKL pathway, statistics of the total number of molecules, link to RANKL pathway reactions and the list of genes which are differentially upregulated by RANKL/RANK pathway. Every molecule in the pathway page is linked to the corresponding NetPath molecule page which is further linked to Entrez gene, HPRD, OMIM and Swiss-Prot identifiers. The reaction page of the RANKL pathway contains the list of each type of reactions such as physical interactions, enzyme catalysis and transport with a brief description about the reactions with their PTM dependence or interacting regions/domains/motifs whenever it was available in literature. The list of curators and reviewers are provided in the RANKL pathway page with the details of the pathway authority. A comments tab is provided in the pathway page to invite the queries and suggestions from the community so as to update and improve RANKL pathway alike other pathways in NetPath.
Visualization of the RANKL/RANK-signaling pathway
RANKL pathway data which are made available using different community standards allows interoperability with other data visualization and analysis software such as Cytoscape (59), Ingenuity pathway analysis, Visualization and layout services for BioPAX pathway models (VISIBIOweb) (60) or Chisio BioPAX Editor (ChiBE) (61). Moreover, various software tools are currently under development which converts user-defined formats into these standard formats. For example, conversion between GenMAPP Pathway Markup Language (GPML) and standard pathway exchange formats such as SBML and BioPAX are under development using Paxtools (62,63). A signaling network of RANKL pathway can be created using cytoscape by importing the BioPAX file. The number of steps which leads to any reaction from the stimulated receptor currently depends on the interconnectivity of molecules and their reactions reported in a pathway. Thus, we hope that the RANKL pathway data available in various data formats will help the computational as well as basic biologist to visualize their own custom maps using different pathway visualization tools available in the community.
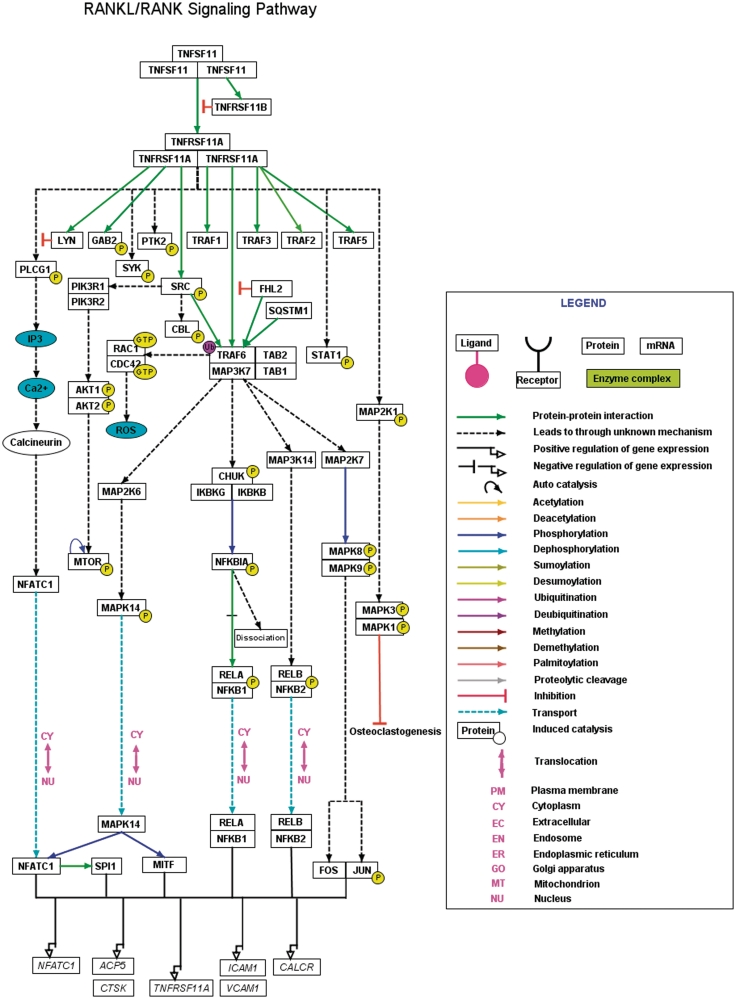
However, taking into account the heterogeneity and the necessity for experimental validation of independent pathway reactions, we have applied a set of stringent criteria to select pathway reactions from the curated data to be depicted in a pathway map. The selection of such reactions stimulated by RANK was based on any of the following criteria: (i) availability of two or more research articles which established a reaction, (ii) availability of two or more different cell types in which a particular reaction was demonstrated, (iii) proof for a PPIs as component of known protein complexes, (iv) Whether reactions reported using high-throughput experiments have a low-throughput experimental support and (v) whether reactions were represented in expert review articles with their biological significance. A basic network model of the RANKL pathway applying the above criteria was manually generated using PathVisio software (Figure 2). A .gpml file with citation containing description of each of the reactions, PTM status, site and residue of modification mapped to specific RefSeq accession and PubMed identifiers are also available to download at http://www.netpath.org/netslim/rankl_pathway.html. A full view of RANKL pathway image in html format is also available at the above link with each node and edge hyperlinked to their corresponding NetPath molecule page and literature evidences respectively.
Figure 2.
A high-confidence RANKL/RANK pathway reaction map using PathVisio. The RANKL/RANK pathway reaction map represents the high-confidence molecular reactions induced by the binding of RANKL homotrimer to its homotrimer receptor complex. The presented map of RANKL/RANK contains 43 proteins involved in 17 molecular associations, 24 enzyme catalysis reactions and four translocation events. The nodes and edges represent the molecules and their reactions respectively. A detailed legend representative of the information of different types of edges distinguished with various colors is provided. A downloadable version of this map is available with the description for each of the reactions and the inclusion criteria for selection of the reaction in the map at http://www.netpath.org/netslim/rankl_pathway.html.
Update process
With the involvement of the pathway authority, the RANKL/RANK pathway will be updated periodically as more reactions become available. The proteins and the microRNAs which are differentially regulated by activated RANK will be curated in the future. We hope to get the active participation from the scientific community to update and improve the RANKL pathway reactions.
Conclusions
Curated and organized data pertaining to signaling pathways are crucial to understand biological processes and gene regulation. This is especially true in the wake of increasing use of high-throughput technologies to study gene and protein expression as well as PTMs (64–66). Combining such a pathway map with high-throughput data can be used to study perturbations using systems biology approaches. We anticipate that the RANKL/RANK signaling pathway resource described here will help accelerate research in both normal physiology as well as disease mechanisms. The RANKL/RANK pathway reactions are available to download at (http://www.netpath.org/pathways?path_id=NetPath_21). We encourage interested members of the scientific community to help us improve the quality of the information in NetPath by submitting their suggestions and critical comments through http://www.netpath.org/comments.
Funding
Department of Biotechnology (DBT), Government of India to the Institute of Bioinformatics; Senior Research Fellowship from Council of Scientific and Industrial Research (CSIR), New Delhi, India (to R.R., J.S. and B.M.); Department of Biotechnology, Young Investigator award (research grants to T.S.K.P.); Wellcome Trust-DBT India Alliance Early Career Fellowship (to H.G.). Funding for open access charge: Wellcome Trust/DBT India Alliance.
Conflict of interest. None declared.
References
- 1.Collin-Osdoby P, Rothe L, Anderson F, et al. Receptor activator of NF-kappa B and osteoprotegerin expression by human microvascular endothelial cells, regulation by inflammatory cytokines, and role in human osteoclastogenesis. J. Biol. Chem. 2001;276:20659–20672. doi: 10.1074/jbc.M010153200. [DOI] [PubMed] [Google Scholar]
- 2.Wright HL, McCarthy HS, Middleton J, et al. RANK, RANKL and osteoprotegerin in bone biology and disease. Curr. Rev. Musculoskelet. Med. 2009;2:56–64. doi: 10.1007/s12178-009-9046-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lum L, Wong BR, Josien R, et al. Evidence for a role of a tumor necrosis factor-alpha (TNF-alpha)-converting enzyme-like protease in shedding of TRANCE, a TNF family member involved in osteoclastogenesis and dendritic cell survival. J. Biol. Chem. 1999;274:13613–13618. doi: 10.1074/jbc.274.19.13613. [DOI] [PubMed] [Google Scholar]
- 4.Schlöndorff J, Lum L, Blobel CP, et al. Biochemical and pharmacological criteria define two shedding activities for TRANCE/OPGL that are distinct from the tumor necrosis factor alpha convertase. J. Biol. Chem. 2001;276:14665–14674. doi: 10.1074/jbc.M010741200. [DOI] [PubMed] [Google Scholar]
- 5.Chesneau V, Becherer JD, Zheng Y, et al. Catalytic properties of ADAM19. J. Biol. Chem. 2003;278:22331–22340. doi: 10.1074/jbc.M302781200. [DOI] [PubMed] [Google Scholar]
- 6.Hikita A, Kadono Y, Chikuda H, et al. Identification of an alternatively spliced variant of Ca2+-promoted Ras inactivator as a possible regulator of RANKL shedding. J. Biol. Chem. 2005;280:41700–41706. doi: 10.1074/jbc.M507000200. [DOI] [PubMed] [Google Scholar]
- 7.Kartsogiannis V, Zhou H, Horwood NJ, et al. Localization of RANKL (receptor activator of NF kappa B ligand) mRNA and protein in skeletal and extraskeletal tissues. Bone. 1999;25:525–534. doi: 10.1016/s8756-3282(99)00214-8. [DOI] [PubMed] [Google Scholar]
- 8.Wong BR, Josien R, Lee SY, et al. TRANCE (tumor necrosis factor [TNF]-related activation-induced cytokine), a new TNF family member predominantly expressed in T cells, is a dendritic cell-specific survival factor. J. Exp. Med. 1997;186:2075–2080. doi: 10.1084/jem.186.12.2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wong BR, Rho J, Arron J, et al. TRANCE is a novel ligand of the tumor necrosis factor receptor family that activates c-Jun N-terminal kinase in T cells. J. Biol. Chem. 1997;272:25190–25194. doi: 10.1074/jbc.272.40.25190. [DOI] [PubMed] [Google Scholar]
- 10.Anderson DM, Maraskovsky E, Billingsley WL, et al. A homologue of the TNF receptor and its ligand enhance T-cell growth and dendritic-cell function. Nature. 1997;390:175–179. doi: 10.1038/36593. [DOI] [PubMed] [Google Scholar]
- 11.Hanada R, Leibbrandt A, Hanada T, et al. Central control of fever and female body temperature by RANKL/RANK. Nature. 2009;462:505–509. doi: 10.1038/nature08596. [DOI] [PubMed] [Google Scholar]
- 12.Fata JE, Kong YY, Li J, et al. The osteoclast differentiation factor osteoprotegerin ligand is essential for mammary gland development. Cell. 2000;103:41–50. doi: 10.1016/s0092-8674(00)00103-3. [DOI] [PubMed] [Google Scholar]
- 13.Kim NS, Kim HJ, Koo BK, et al. Receptor activator of NF-kappaB ligand regulates the proliferation of mammary epithelial cells via Id2. Mol. Cell. Biol. 2006;26:1002–1013. doi: 10.1128/MCB.26.3.1002-1013.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grimaud E, Soubigou L, Couillaud S, et al. Receptor activator of nuclear factor kappaB ligand (RANKL)/osteoprotegerin (OPG) ratio is increased in severe osteolysis. Am. J. Pathol. 2003;163:2021–2031. doi: 10.1016/s0002-9440(10)63560-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Giuliani N, Bataille R, Mancini C, et al. Myeloma cells induce imbalance in the osteoprotegerin/osteoprotegerin ligand system in the human bone marrow environment. Blood. 2001;98:3527–3533. doi: 10.1182/blood.v98.13.3527. [DOI] [PubMed] [Google Scholar]
- 16.Huang L, Cheng YY, Chow LT, et al. Tumour cells produce receptor activator of NF-kappaB ligand (RANKL) in skeletal metastases. J. Clin. Pathol. 2002;55:877–878. doi: 10.1136/jcp.55.11.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kitazawa S, Kitazawa R. RANK ligand is a prerequisite for cancer-associated osteolytic lesions. J. Pathol. 2002;198:228–236. doi: 10.1002/path.1199. [DOI] [PubMed] [Google Scholar]
- 18.Lam J, Nelson CA, Ross FP, et al. Crystal structure of the TRANCE/RANKL cytokine reveals determinants of receptor-ligand specificity. J. Clin. Invest. 2001;108:971–979. doi: 10.1172/JCI13890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ito S, Wakabayashi K, Ubukata O, et al. Crystal structure of the extracellular domain of mouse RANK ligand at 2.2-A resolution. J. Biol. Chem. 2002;277:6631–6636. doi: 10.1074/jbc.M106525200. [DOI] [PubMed] [Google Scholar]
- 20.Liu C, Walter TS, Huang P, et al. Structural and functional insights of RANKL-RANK interaction and signaling. J. Immunol. 2010;184:6910–6919. doi: 10.4049/jimmunol.0904033. [DOI] [PubMed] [Google Scholar]
- 21.Lacey DL, Timms E, Tan HL, et al. Osteoprotegerin ligand is a cytokine that regulates osteoclast differentiation and activation. Cell. 1996;93:165–176. doi: 10.1016/s0092-8674(00)81569-x. [DOI] [PubMed] [Google Scholar]
- 22.Yasuda H, Shima N, Nakagawa N, et al. Osteoclast differentiation factor is a ligand for osteoprotegerin/osteoclastogenesis-inhibitory factor and is identical to TRANCE/RANKL. Proc. Natl Acad. Sci. USA. 1998;95:3597–3602. doi: 10.1073/pnas.95.7.3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tat SK, Pelletier JP, Velasco CR, et al. New perspective in osteoarthritis: the OPG and RANKL system as a potential therapeutic target? Keio. J. Med. 2009;58:29–40. doi: 10.2302/kjm.58.29. [DOI] [PubMed] [Google Scholar]
- 24.Lean JM, Matsuo K, Fox SW, et al. Osteoclast lineage commitment of bone marrow precursors through expression of membrane-bound TRANCE. Bone. 2000;27:29–40. doi: 10.1016/s8756-3282(00)00306-9. [DOI] [PubMed] [Google Scholar]
- 25.Fuller K, Wong B, Fox S, et al. TRANCE is necessary and sufficient for osteoblast-mediated activation of bone resorption in osteoclasts. J. Exp. Med. 1998;188:997–1001. doi: 10.1084/jem.188.5.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cremer I, Dieu-Nosjean MC, Maréchal S, et al. Long-lived immature dendritic cells mediated by TRANCE-RANK interaction. Blood. 2002;100:3646–3655. doi: 10.1182/blood-2002-01-0312. [DOI] [PubMed] [Google Scholar]
- 27.Wiethe C, Dittmar K, Doan T, et al. Enhanced effector and memory CTL responses generated by incorporation of receptor activator of NF-kappa B (RANK)/RANK ligand costimulatory molecules into dendritic cell immunogens expressing a human tumor-specific antigen. J. Immunol. 2003;171:4121–4130. doi: 10.4049/jimmunol.171.8.4121. [DOI] [PubMed] [Google Scholar]
- 28.Yu Q, Gu JX, Kovacs C, et al. Cooperation of TNF family members CD40 ligand, receptor activator of NF-kappa B ligand, and TNF-alpha in the activation of dendritic cells and the expansion of viral specific CD8+ T cell memory responses in HIV-1-infected and HIV-1-uninfected individuals. J. Immunol. 2003;170:1797–1805. doi: 10.4049/jimmunol.170.4.1797. [DOI] [PubMed] [Google Scholar]
- 29.Rossi SW, Kim MY, Leibbrandt A, et al. RANK signals from CD4q3-inducer cells regulate development of Aire-expressing epithelial cells in the thymic medulla. J. Exp. Med. 2007;204:1267–1272. doi: 10.1084/jem.20062497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sobacchi C, Frattini A, Guerrini MM, et al. Osteoclast-poor human osteopetrosis due to mutations in the gene encoding RANKL. Nat. Genet. 2007;39:960–962. doi: 10.1038/ng2076. [DOI] [PubMed] [Google Scholar]
- 31.Simonet WS, Lacey DL, Dunstan CR, et al. Osteoprotegerin: a novel secreted protein involved in the regulation of bone density. Cell. 1997;89:309–319. doi: 10.1016/s0092-8674(00)80209-3. [DOI] [PubMed] [Google Scholar]
- 32.Yasuda H, Shima N, Nakagawa N, et al. Identity of osteoclastogenesis inhibitory factor (OCIF) and osteoprotegerin (OPG): a mechanism by which OPG/OCIF inhibits osteoclastogenesis in vitro. Endocrinology. 1998;139:1329–1337. doi: 10.1210/endo.139.3.5837. [DOI] [PubMed] [Google Scholar]
- 33.Hofbauer LC, Heufelder AE. Role of receptor activator of nuclear factor-kappaB ligand and osteoprotegerin in bone cell biology. J. Mol. Med. 2001;79:243–253. doi: 10.1007/s001090100226. [DOI] [PubMed] [Google Scholar]
- 34.Ruocco MG, Maeda S, Park JM, et al. IkappaB kinase (IKK) beta, but not IKK alpha, is a critical mediator of osteoclast survival and is required for inflammation-induced bone loss. J. Exp.Med. 2005;201:1677–1687. doi: 10.1084/jem.20042081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chaisson ML, Branstetter DG, Derry JM, et al. Osteoclast differentiation is impaired in the absence of inhibitor of kappa B kinase alpha. J. Biol. Chem. 2004;279:54841–54848. doi: 10.1074/jbc.M406392200. [DOI] [PubMed] [Google Scholar]
- 36.Aya K, Alhawagri M, Hagen-Stapleton A, et al. NF-(kappa) B-inducing kinase controls lymphocyte and osteoclast activities in inflammatory arthritis. J. Clin. Invest. 2005;115:1848–1854. doi: 10.1172/JCI23763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yamashita T, Yao Z, Li F, et al. NF-kappaB p50 and p52 regulate receptor activator of NF-kappaB ligand (RANKL) and tumor necrosis factor-induced osteoclast precursor differentiation by activating c-Fos and NFATc1. J. Biol. Chem. 2007;282:18245–18253. doi: 10.1074/jbc.M610701200. [DOI] [PubMed] [Google Scholar]
- 38.Tucker E, O'Donnell K, Fuchsberger M, et al. A novel mutation in the Nfkb2 gene generates an NF-kappa B2 “super repressor”. J. Immunol. 2007;179:7514–7522. doi: 10.4049/jimmunol.179.11.7514. [DOI] [PubMed] [Google Scholar]
- 39.Vaira S, Alhawagri M, Anwisye I, et al. RelA/p65 promotes osteoclast differentiation by blocking a RANKL-induced apoptotic JNK pathway in mice. J. Clin. Invest. 2008;118:2088–2097. doi: 10.1172/JCI33392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vaira S, Johnson T, Hirbe AC, et al. RelB is the NF-kappaB subunit downstream of NIK responsible for osteoclast differentiation. Proc. Natl Acad. Sci. USA. 2008;105:3897–3902. doi: 10.1073/pnas.0708576105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yamamoto A, Miyazaki T, Kadono Y, et al. Possible involvement of IkappaB kinase 2 and MKK7 in osteoclastogenesis induced by receptor activator of nuclear factor kappaB ligand. J. Bone Miner. Res. 2002;17:612–621. doi: 10.1359/jbmr.2002.17.4.612. [DOI] [PubMed] [Google Scholar]
- 42.David JP, Sabapathy K, Hoffmann O, et al. JNK1 modulates osteoclastogenesis through both c-Jun phosphorylation-dependent and -independent mechanisms. J. Cell Sci. 2002;115:4317–4325. doi: 10.1242/jcs.00082. [DOI] [PubMed] [Google Scholar]
- 43.Ikeda F, Nishimura R, Matsubara T, et al. Critical roles of c-Jun signaling in regulation of NFAT family and RANKL-regulated osteoclast differentiation. J. Clin. Invest. 2004;114:475–484. doi: 10.1172/JCI19657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ikeda F, Matsubara T, Tsurukai T, et al. JNK/c-Jun signaling mediates an anti-apoptotic effect of RANKL in osteoclasts. J. Bone Miner. Res. 2008;23:907–914. doi: 10.1359/jbmr.080211. [DOI] [PubMed] [Google Scholar]
- 45.Soriano P, Montgomery C, Geske R, Bradley A. Targeted disruption of the c-src proto-oncogene leads to osteopetrosis in mice. Cell. 1991;64:693–702. doi: 10.1016/0092-8674(91)90499-o. [DOI] [PubMed] [Google Scholar]
- 46.Wong BR, Besser D, Kim N, et al. TRANCE, a TNF family member, activates Akt/PKB through a signaling complex involving TRAF6 and c-Src. Mol. Cell. 1999;4:1041–1049. doi: 10.1016/s1097-2765(00)80232-4. [DOI] [PubMed] [Google Scholar]
- 47.Matsumoto M, Sudo T, Saito T, et al. Involvement of p38 mitogen-activated protein kinase signaling pathway in osteoclastogenesis mediated by receptor activator of NF-kappa B ligand (RANKL) J. Biol. Chem. 2000;275:31155–31161. doi: 10.1074/jbc.M001229200. [DOI] [PubMed] [Google Scholar]
- 48.Kandasamy K, Mohan SS, Raju R, et al. NetPath: a public resource of curated signal transduction pathways. Genome Biol. 2010;11:R3. doi: 10.1186/gb-2010-11-1-r3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Oda K, Matsuoka Y, Funahashi A, et al. A comprehensive pathway map of epidermal growth factor receptor signaling. Mol. Syst. Biol. 2005;1:2005.0010. doi: 10.1038/msb4100014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wasiliew P, Weber A, Dittrich-Breiholz O, et al. A comprehensive map of the IL-1R signalling network. Cell Commun. Signaling. 2009;7:A25. [Google Scholar]
- 51.Oda K, Kitano H. A comprehensive map of the toll-like receptor signaling network. Mol. Syst. Biol. 2006;2 doi: 10.1038/msb4100057. 2006.0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kandasamy K, Keerthikumar S, Raju R, et al. PathBuilder–open source software for annotating and developing pathway resources. Bioinformatics. 2009;25:2860–2862. doi: 10.1093/bioinformatics/btp453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Carbon S, Ireland A, Mungall CJ, et al. AmiGO: online access to ontology and annotation data. Bioinformatics. 2009;25:288–289. doi: 10.1093/bioinformatics/btn615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Keshava Prasad TS, Goel R, Kandasamy K, Keerthikumar S, et al. Human protein reference database–2009 update. Nucleic Acids Res. 2009;37:D767–D772. doi: 10.1093/nar/gkn892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hermjakob H, Montecchi-Palazzi L, Bader G, et al. The HUPO PSI's molecular interaction format–a community standard for the representation of protein interaction data. Nat. Biotechnol. 2004;22:177–183. doi: 10.1038/nbt926. [DOI] [PubMed] [Google Scholar]
- 56.Orchard S, Kerrien S. Molecular interactions and data standardisation. Methods Mol. Biol. 2010;604:309–318. doi: 10.1007/978-1-60761-444-9_21. [DOI] [PubMed] [Google Scholar]
- 57.Demir E, Cary MP, Paley. S, et al. The BioPAX community standard for pathway data sharing. Nat. Biotechnol. 2010;28:935–942. doi: 10.1038/nbt.1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hucka M, Finney A, Sauro HM, et al. The systems biology markup language (SBML): a medium for representation and exchange of biochemical network models. Bioinformatics. 2003;19:524–531. doi: 10.1093/bioinformatics/btg015. [DOI] [PubMed] [Google Scholar]
- 59.Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dilek A, Belviranli ME, Dogrusoz U. VISIBIOweb: visualization and layout services for BioPAX pathway models. Nucleic Acids Res. 2010;38:W150–W154. doi: 10.1093/nar/gkq352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Babur O, Dogrusoz U, Demir E, Sander C. ChiBE: interactive visualization and manipulation of BioPAX pathway models. Bioinformatics. 2010;26:429–431. doi: 10.1093/bioinformatics/btp665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Salomonis N, Hanspers K, Zambon AC, et al. GenMAPP 2: new features and resources for pathway analysis. BMC Bioinformatics. 2007;8:217. doi: 10.1186/1471-2105-8-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Evelo,C. (2009) Community curation on WikiPathways: How we assist knowledge collection. Available from Nature Precedings < http://dx.doi.org/10.1038/npre.2009.3115.1> (2009)
- 64.Kashyap M, Marimuthu A, Kishore CJH, et al. Genomewide mRNA profiling of esophageal squamous cell carcinoma for identification of cancer biomarkers. Cancer Biol. Therapy. 2009;8:16–26. doi: 10.4161/cbt.8.1.7090. [DOI] [PubMed] [Google Scholar]
- 65.Amanchy R, Zhong J, Hong R, et al. Identification of c-Src tyrosine kinase substrates in platelet-derived growth factor receptor signaling. Mol. Oncol. 2009;3:439–450. doi: 10.1016/j.molonc.2009.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bose R, Molina H, Patterson AS, et al. Phosphoproteomic analysis of Her2/neu signaling and inhibition. Proc. Natl Acad. Sci. USA. 2006;103:9773–9778. doi: 10.1073/pnas.0603948103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mizukami J, Takaesu G, Akatsuka H, et al. Receptor activator of NF-kappaB ligand (RANKL) activates TAK1 mitogen-activated protein kinase kinase kinase through a signaling complex containing RANK, TAB2, and TRAF6. Mol Cell Biol. 2002;22:992–1000. doi: 10.1128/MCB.22.4.992-1000.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wong BR, Besser D, Kim N, et al. TRANCE, a TNF family member, activates Akt/PKB through a signaling complex involving TRAF6 and c-Src. Mol Cell. 1999;4:1041–1049. doi: 10.1016/s1097-2765(00)80232-4. [DOI] [PubMed] [Google Scholar]
- 69.Bai S, Zha J, Zhao H, et al. Tumor necrosis factor receptor-associated factor 6 is an intranuclear transcriptional coactivator in osteoclasts. J Biol Chem. 2008;283:30861–30867. doi: 10.1074/jbc.M802525200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Arron JR, Vologodskaia M, Wong BR, et al. A positive regulatory role for Cbl family proteins in tumor necrosis factor-related activation-induced cytokine (trance) and CD40L-mediated Akt activation. J Biol Chem. 2001;276:30011–30017. doi: 10.1074/jbc.M100414200. [DOI] [PubMed] [Google Scholar]
- 71.Jin W, Chang M, Paul EM, et al. Deubiquitinating enzyme CYLD negatively regulates RANK signaling and osteoclastogenesis in mice. J Clin Invest. 2008;118:1858–1866. doi: 10.1172/JCI34257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kim HJ, Zhang K, Zhang L, et al. The Src family kinase, Lyn, suppresses osteoclastogenesis in vitro and in vivo. Proc Natl Acad Sci U S A. 2009;106:2325–2330. doi: 10.1073/pnas.0806963106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Darwech I, Otero J, Alhawagri M, et al. Impediment of NEMO oligomerization inhibits osteoclastogenesis and osteolysis. J Cell Biochem. 2009;108:1337–1345. doi: 10.1002/jcb.22364. [DOI] [PMC free article] [PubMed] [Google Scholar]