Abstract
The SR proteins constitute a family of splicing factors, highly conserved in metazoans, that contain one or two amino-terminal RNA-binding domains (RBDs) and a region enriched in arginine/serine repeats (RS domain) at the carboxyl terminus. Previous studies have shown that SR proteins possess distinct RNA-binding specificities that likely contribute to their unique functions, but it is unclear whether RS domains have specific roles in vivo. Here, we used a genetic system developed in the chicken B cell line DT40 to address this question. Expression of chimeric proteins generated by fusion of the RS domains of heterologous SR proteins, or a human TRA-2 protein, with the RBDs of ASF/SF2 allowed cell growth following genetic inactivation of endogenous ASF/SF2, indicating that RS domains are interchangeable for all functions required to maintain cell viability. However, a chimera containing the RS domain from a related splicing factor, U2AF65, could not rescue viability and was inactive in in vitro splicing assays, suggesting that this domain performs a distinct function. We also used the DT40 system to show that depletion of ASF/SF2 affects splicing of specific transcripts in vivo. Although splicing of several simple constitutive introns was not significantly affected, the alternative splicing patterns of two model pre-mRNAs switched in a manner consistent with predictions from previous studies. Unexpectedly, ASF/SF2 depletion resulted in a substantial increase in splicing of an HIV-1 tat pre-mRNA substrate, indicating that ASF/SF2 can repress tat splicing in vivo. These results provide the first demonstration that an SR protein can influence splicing of specific pre-mRNAs in vivo.
Keywords: splicing, SR proteins, RS domains, pre-mRNA processing
A large number of protein factors play important roles in the splicing of mRNA precursors (for review, see Moore et al. 1993; Krämer 1996). Among them, a family of closely related splicing factors, collectively named SR proteins (Zahler et al. 1992), have been investigated intensively over the past several years (for review, see Fu 1995; Manley and Tacke 1996; Valcárcel and Green 1996). At least nine members of the SR family have been identified and characterized, all of which contain one or two RNP-type RNA-binding domains (RBDs) at their amino terminus and a carboxy-terminal arginine/serine-rich region (RS domain). Comparison of SR proteins from different species reveals that they are highly conserved evolutionarily throughout metazoans. SR proteins are essential splicing factors in vitro, as they are required for an early step(s) in the process of spliceosome assembly (e.g., Krainer et al. 1990a; Fu and Maniatis 1990; Fu 1993). By interacting with the U1 70K protein, a component of the U1 small nuclear ribonucleoprotein particle (snRNP), SR proteins are thought to help recruit the U1 snRNP to the 5′ splice site of the pre-mRNA to form the commitment complex (Kohtz et al. 1994; Jamison et al. 1995). In addition, SR proteins may help bring the 5′ and 3′ splice sites together by interacting simultaneously with U1 70K and U2AF35 (the small subunit of splicing factor U2AF) at the 5′ and 3′ splice sites, respectively (Wu and Maniatis 1993). RS domains have been shown to be necessary but not sufficient for these protein–protein interactions (Kohtz et al. 1994; Xiao and Manley 1997).
SR proteins also participate in splicing control. For example, they can modulate alternative splicing, when added to nuclear extracts (Ge and Manley 1990; Krainer et al. 1990b; Fu et al. 1992; Zahler et al. 1993) or transiently overexpressed in cultured cells (Cáceres et al. 1994; Screaton et al. 1995; Wang and Manley 1995; Zhang and Wu 1996). In the case of alternative 5′ splice sites, they likely function by recruiting U1 snRNP to the alternative splice sites (Eperon et al. 1993; Zahler and Roth 1995). When bound to RNA elements known as exonic splicing enhancers, SR proteins can stimulate splicing of an upstream intron (e.g., Sun et al. 1993; Tian and Maniatis 1993; Staknis and Reed 1994; Ramachatesingh et al. 1995). Evidence has been presented that this event involves stabilization of U2AF binding to the polypyrimidine tract of the intron (Wang et al. 1995; Zuo and Maniatis 1996).
The functions of SR proteins in constitutive splicing appear to be largely redundant, at least in vitro, as essentially any individual SR protein can restore splicing activity to cytoplasmic S100 extracts, which lack all SR proteins (Krainer et al. 1990a, 1991; Ge et al. 1991; Fu et al. 1992; Zahler et al. 1992). However, distinct functions of SR proteins have been described in several assays. First, certain SR proteins can commit specific pre-mRNAs to the splicing pathway, whereas others cannot (Fu 1993). Second, individual SR proteins influence selection of alternative splice sites in different ways (e.g., Kim et al. 1992; Zahler et al. 1993). Third, SR proteins recognize distinct RNA sequences. For example, SR proteins display specificity in the recognition of various splicing enhancer elements (Sun et al. 1993; Tian and Maniatis 1993; Staknis and Reed 1994; Lynch and Maniatis 1995; Ramachatesingh et al. 1995; Tacke and Manley 1995). Additionally, the RNA sequences selected by several SR proteins through in vitro enrichment experiments are distinct (Heinrichs and Baker 1995; Tacke and Manley 1995; Shi et al. 1997; Tacke et al. 1997). Finally, genetic analyses of SR proteins have demonstrated that at least two of them perform nonredundant functions in vivo that are essential for Drosophila development (Ring and Lis 1994; Peng and Mount 1995) or for viability of cultured cells (Wang et al. 1996).
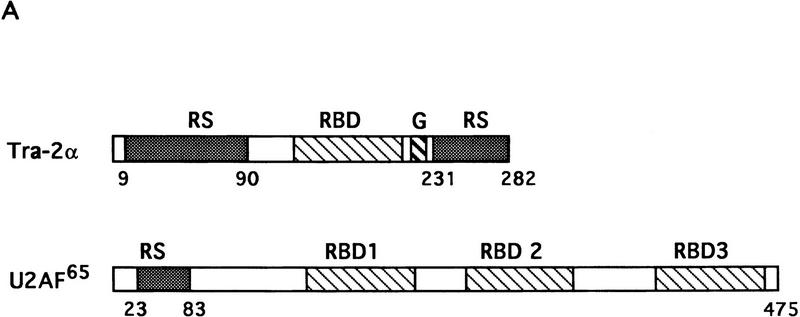
RS domains are essential for SR protein function. The ASF/SF2 RS domain is required for activation of constitutive splicing in vitro (Zuo and Manley 1993; Cáceres and Krainer 1993), and mutant ASF/SF2 proteins with partially or completely deleted RS domains could not rescue cell death following ASF/SF2 depletion, indicating that the RS domain is essential for ASF/SF2 function in vivo (Wang et al. 1996). Many splicing factors outside the SR family also contain RS-like domains (for review, see Fu 1995). For instance, the protein encoded by Drosophila transformer-2 (tra-2; Amrein et al. 1988; Goralski et al. 1989), a gene involved in the sex determination pathway (for review, see Baker 1989), contains two RS domains, one of which is essential for its function in vivo (Amrein et al. 1994). Several lines of evidence have suggested that RS domains mediate protein–protein interactions (Wu and Maniatis 1993; Kohtz et al. 1994; Amrein et al. 1994; Xiao and Manley 1997) and also direct proteins to specific subnuclear locations (Li and Bingham 1991; Hedley et al. 1995; Cáceres et al. 1997). However, U2AF65, the large subunit of U2AF, has an amino-terminal RS-like domain (Zamore et al. 1992) that appears to function by RNA–protein, as opposed to protein–protein, interactions (Valcárcel et al. 1996).
Whether the RS domains of different SR proteins perform distinct functions that contribute to the specific roles of SR proteins in vivo is not known. We tested this idea by using a genetic complementation assay in a derivative of the chicken B cell line DT40 (Wang et al. 1996). Our results show that most RS domains, despite their high degree of sequence conservation throughout evolution, are functionally interchangeable. In addition, we analyzed alternative and constitutive splicing of specific endogenous transcripts and several well-studied model pre-mRNAs in cells depleted of ASF/SF2. We provide evidence that ASF/SF2 is not essential for all splicing events, but that the levels of ASF/SF2 can modulate alternative splicing of specific pre-mRNAs in vivo, and that repression of splicing can also be a natural function of ASF/SF2.
Results
We previously constructed a DT40 cell line, DT40-ASF (formerly named #3; Wang et al. 1996), in which the ASF/SF2 gene is disrupted and expression of ASF/SF2 is driven by a tetracycline (tet)-repressible promoter. Addition of tet to the cell culture medium results in depletion of ASF/SF2 protein and rapid cell death. On the basis of the tet-induced lethality, we designed a complementation assay to test whether a given protein could provide the functions of ASF/SF2 required for cell viability (Wang et al. 1996). In brief, DT40-ASF cells are transfected with an expression vector encoding the test protein and then selected in medium containing tet. Appearance of surviving colonies indicates that the test protein can substitute for ASF/SF2. We used this strategy to examine the specificity of RS domains of different splicing factors.
The RS domains of SR proteins are interchangeable
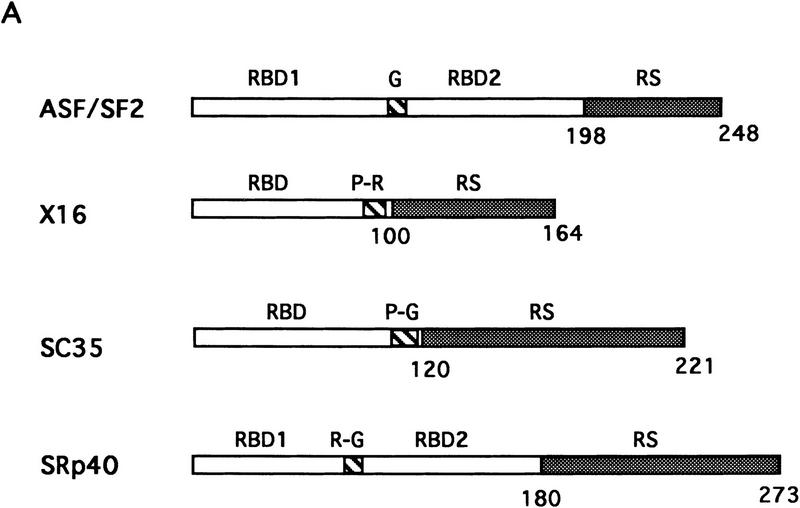
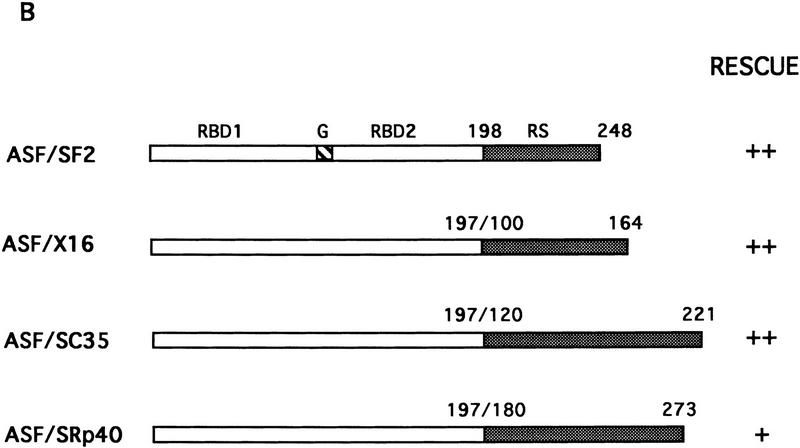
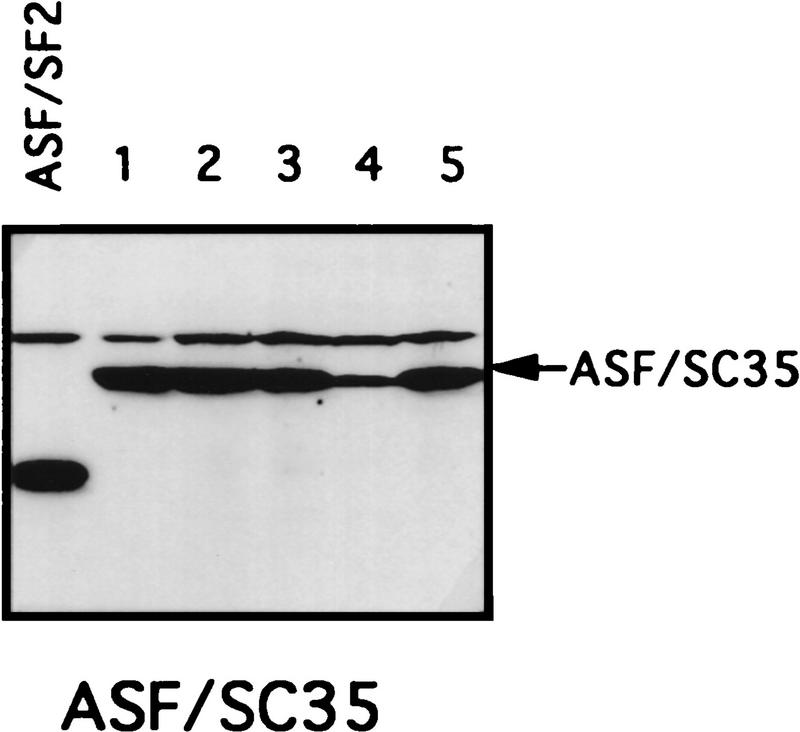
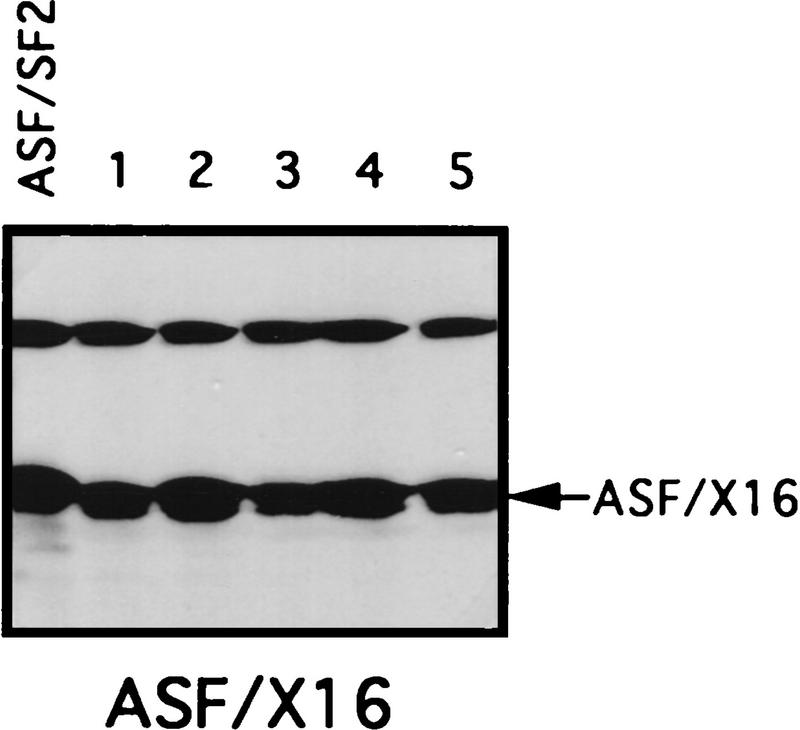
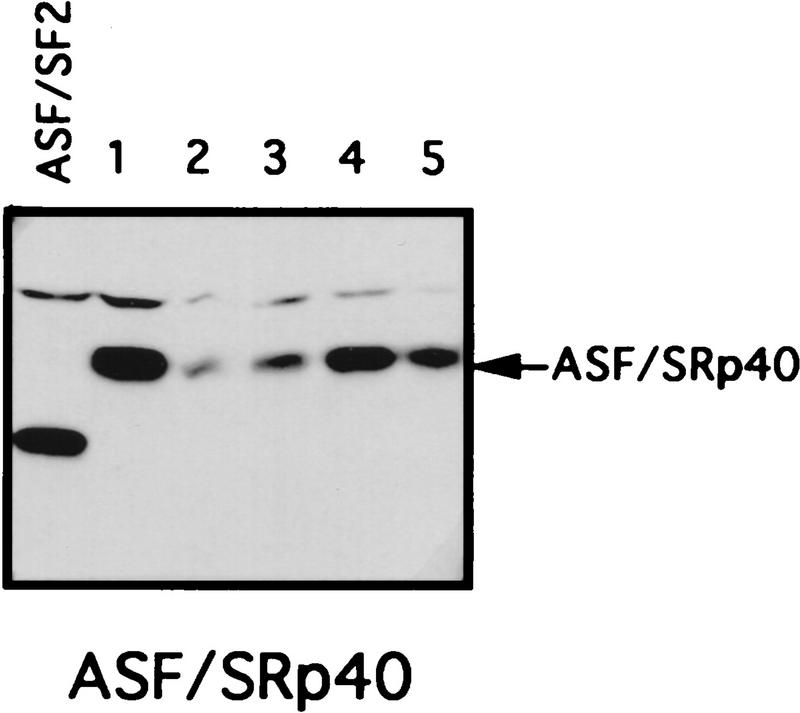
The RS domain of ASF/SF2 was first replaced by the RS domains of the SR proteins X16/SRp20 (Ayane et al. 1991), SC35 (Fu and Maniatis 1992), or SRp40 (Screaton et al. 1995) to make three chimeric proteins (see Fig. 1 for diagrams of these SR proteins and the chimeric proteins). The ability of the chimeric proteins to substitute for ASF/SF2 was then tested in the complementation assay described above. Strikingly, expression of either ASF/X16 or ASF/SC35 rescued tet-induced lethality just as well as did full-length ASF/SF2, as determined by the numbers of surviving colonies (>50 in each case; Fig. 1B). Transfection of DT40-ASF cells with the ASF/SRp40 expression vector yielded only six tet-resistant colonies (Fig. 1B), suggesting that the SRp40 RS domain may substitute less well for the ASF/SF2 RS domain. However, the recovered clones all displayed wild-type growth rates (results not shown), indicating that the SRp40 RS domain, like those of X16 and SC35, can fully substitute for the ASF/SF2 RS domain. Expression of the chimeric proteins and depletion of ASF/SF2 in the surviving clones were confirmed by Western blotting. As shown in Figure 3 (below), expression of each chimera in the surviving clones analyzed was comparable to the level of ASF/SF2 in DT40-ASF cells. Taken together, these results show that the RS domains of three different SR proteins can substitute functionally for the ASF/SF2 RS domain, indiacating that the RS domains of SR proteins are interchangeable, at least with respect to the requirements for viability of chicken DT40 cells.
Figure 1.
Fusion proteins consisting of the ASF/SF2 RBDs and an RS domain from heterologous SR proteins can rescue cell viability after ASF/SF2 depletion. (A) Schematic diagrams of the structures of ASF/SF2, X16, SC35, and SRp40. Numbers indicate amino acid residues. (RBD) RNP-type RNA-binding domain; (RS) arginine/serine-rich region; (G) glycine-rich region; (P-R) proline/arginine-rich region; (P-G) proline/glycine-rich region; (R-G) arginine/glycine-rich region. (B) Schematic diagrams of chimeric proteins. ASF/SF2 RBDs (amino acids 1–197) were fused with the RS domains of X16, SC35, and SRp40, giving ASF/X16, ASF/SC35, and ASF/SRp40, respectively. Shaded boxes indicate RS domains. The numbers above each RS domain indicate the amino acid residues in the corresponding full-length protein. DT40–ASF cells were transfected with expression vectors encoding ASF/SF2 and the chimeric proteins, and then selected in medium containing 1 μg/ml tet. (++) Appearance of >50 surviving colonies; (+) appearance of 5–20 surviving colonies.
Figure 3.
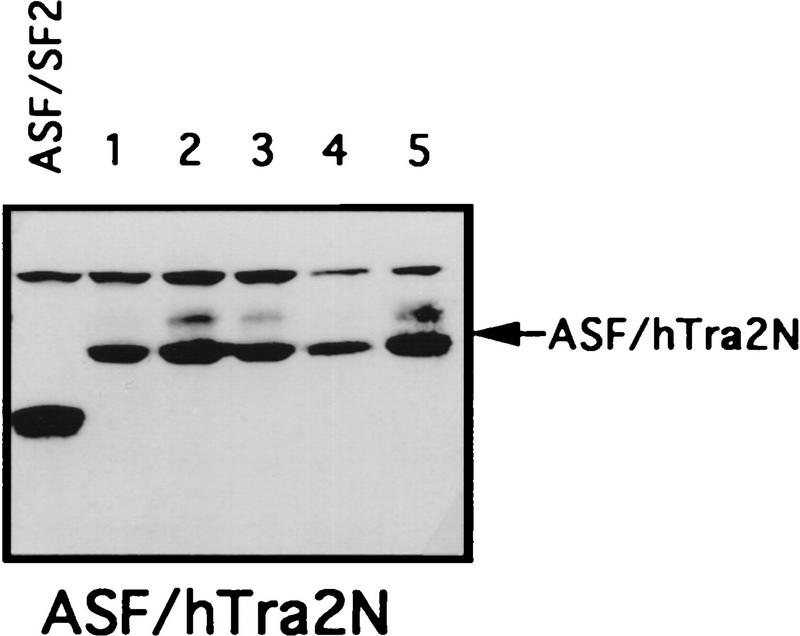
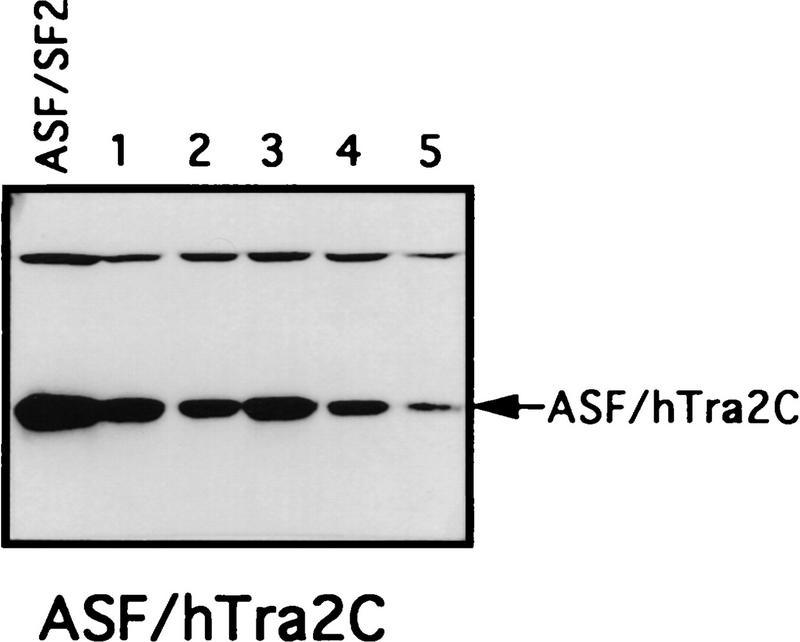
Chimeric proteins are efficiently expressed in surviving clones. Selected surviving clones from the complementation experiments were isolated and maintained in medium containing 1 μg/ml tet. Expression of the chimeric proteins in these cells was measured by Western blotting with mAb 12CA5. A DT40-ASF cell lysate was used as a control in each Western blot. Positions of the chimeric proteins are indicated by arrows. Numbers indicate independent clones analyzed.
Analysis of the RS domains of splicing factors outside the SR family
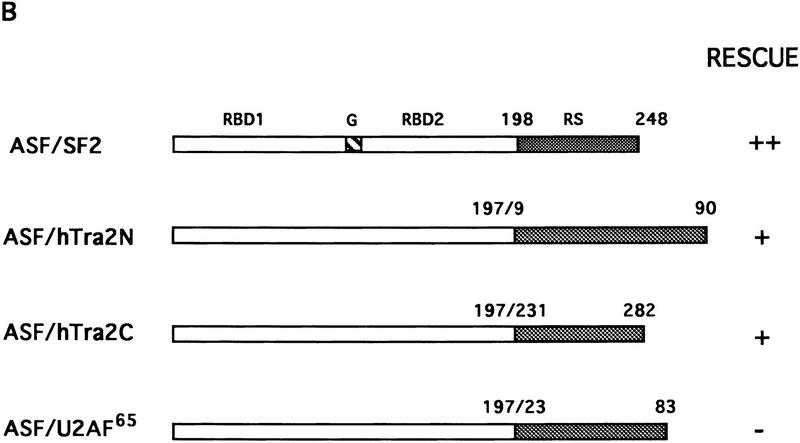
We also used the complementation assay to determine whether RS-like domains from other splicing factors can substitute for the ASF/SF2 RS domain. Recently a human homolog of Drosophila TRA-2, hTRA-2α, was identified and characterized (Dauwalder et al. 1996). Like TRA-2, hTRA-2α contains two RS domains, one at the amino- and one at the carboxyl terminus of the protein. As mentioned above, an RS-like domain is also present at the amino terminus of U2AF65, but it has been suggested to function differently from SR protein RS domains (Valcárcel et al. 1996). Therefore, we wished to test whether the two RS domains of hTRA-2α and the RS region from U2AF65 could functionally replace the ASF/SF2 RS domain. Chimeric proteins were generated by fusion of ASF/SF2 RBDs with these heterologous RS domains, and the ability of these proteins to substitute for ASF/SF2 in DT40 cells was determined as above (see Fig. 2 for diagrams). Tet-resistant clones appeared following transfection of DT40-ASF cells with either the ASF/hTRA2N or the ASF/hTRA2C expression vector, although the number of surviving clones in each case (5 or 12, respectively) was less than those obtained with ASF/SF2 (>50; Fig. 2B). Again, however, recovered clones all displayed wild-type growth rates (results not shown). Expression of the chimeric proteins and depletion of ASF/SF2 were confirmed by Western blot analysis (Fig. 3). Thus both TRA2 RS domains can functionally substitute for the ASF/SF2 RS domain. In sharp contrast to these results, transfection of DT40-ASF cells with the ASF/U2AF65 expression vector did not yield any surviving clones in multiple independent experiments (Fig. 2B). These results suggest that the U2AF65 RS-like domain performs a distinct function(s) from that of SR proteins, in keeping with previous data (e.g., Valcárcel et al. 1996).
Figure 2.
Complementation of cell death following ASF/SF2 depletion by fusion proteins containing ASF/SF2 RBDs and the RS domains of splicing factors hTRA-2α and U2AF65. (A) Schematic diagrams of hTRA-2α and U2AF65. Numbers indicate amino acid residues. Abbreviations are described in Fig. 1A. (B) Schematic diagrams of chimeric proteins. ASF/SF2 RBDs (amino acids 1–197) were fused with the amino- and carboxy-terminal RS domains of hTRA-2α and U2AF65 RS domain, giving ASF/hTRA2N, ASF/hTRA2C, and ASF/U2AF65, respectively. The numbers above each RS domain indicate the amino acid residues in the corresponding full-length protein. ASF/SF2 and the chimeric proteins were tested in the complementation assay as in Fig. 1. (++) Appearance of >50 surviving colonies; (+) appearance of 5–20 surviving colonies; (−) no surviving clones.
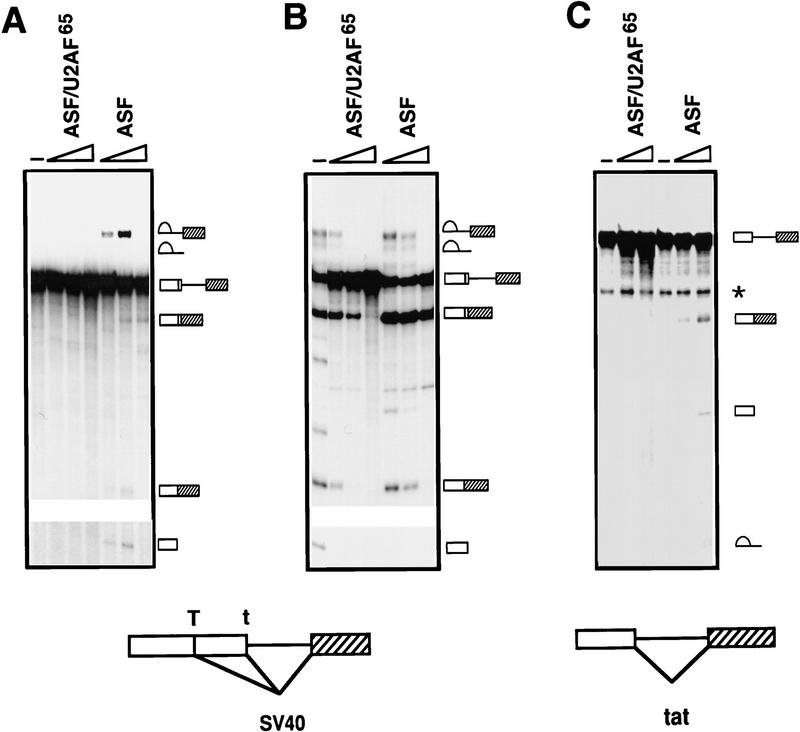
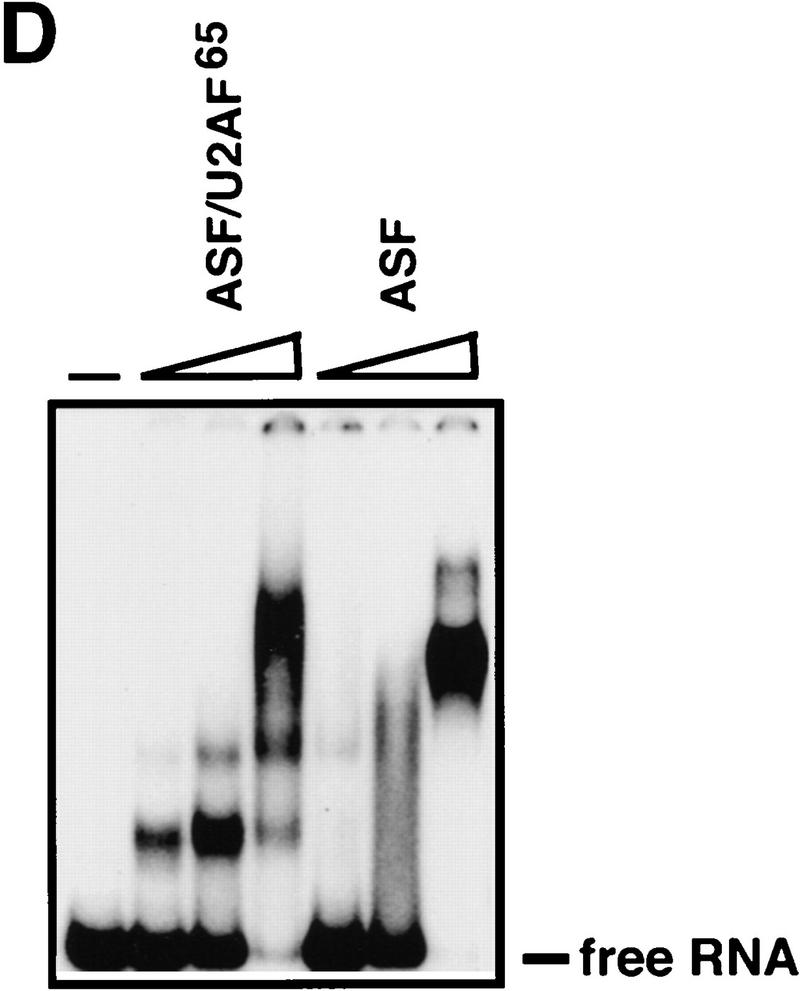
The inability of the U2AF65 RS region to substitute for the ASF/SF2 RS domain was on the one hand consistent with the different functions proposed for the two domains on the basis of in vitro studies, but on the other hand surprising given our finding that all other RS domains tested were functionally interchangeable. We therefore wished to provide evidence that the failure of ASF/U2AF65 to rescue viability was indeed attributable to a splicing defect. To this end, the chimeric protein was expressed as a His-tagged fusion in Escherichia coli and purified as described previously for ASF/SF2 (Ge et al. 1991). Its activity was then compared with that of ASF/SF2 in two in vitro assays, S100 complementation (to test general splicing activity) and nuclear extract supplementation (to measure alternative splice site switching), by use of SV40 early and HIV tat pre-mRNAs (Fig. 4). Strikingly, ASF/U2AF65 was unable to induce detectable splicing of either pre-mRNA in S100 extract (Fig. 4A,C). In contrast, ASF/SF2 not only activated splicing of both pre-mRNAs, but, with the SV40 RNA (Fig. 4A), also switched splice site selection (from large T to small t mRNA) at higher concentrations, consistent with previous results (Zuo and Manley 1993). ASF/U2AF65 was also inactive in switching splice site utilization in nuclear extract (Fig. 4B). In fact, increasing concentrations of the chimeric protein led to complete inhibition of splicing. (This also occurs with ASF/SF2, but only at higher concentrations; data not shown.) These results are particularly notable in light of previous studies showing that the ASF/SF2 RS domain can actually be dispensable for splice site switching activity (Cáceres and Krainer 1993; Zuo and Manley 1993; Wang and Manley 1995). The inability of ASF/U2AF65 to function in splicing was not attributable simply to misfolding of the chimeric protein, as ASF/U2AF65 and ASF/SF2 bound RNA with comparable efficiency and specificity in gel mobility shift assays (Fig. 4D and data not shown). Together these findings indicate not only that the U2AF65 RS-like domain is unable to provide SR protein RS domain function, but also that the ASF/U2AF65 chimeric protein can have a dominant-negative effect on splicing in vitro.
Figure 4.
ASF/U2AF65 is inactive in in vitro splicing. (A) ASF/U2AF65 is unable to activate SV40 early pre-mRNA splicing. SV40 pre-mRNA was incubated in S100 extract supplemented with 0.3, 0.6 or 1.2 μg of either His-tagged ASF/U2AF65 or ASF/SF2 for 2 hr. Splicing products were analyzed on a 5% polyacrylamide gel containing 8 m urea. (B) ASF/U2AF65 is inactive in alternative 5′ splice site selection. SV40 pre-mRNA was incubated in HeLa nuclear extract in the presence of 0.3, 0.6 or 1.2 μg of either ASF/U2AF65 or ASF/SF2 for 2 hr. RNA products were analyzed as in A. (C) ASF/U2AF65 is also unable to activate HIV tat pre-mRNA splicing. Tat pre-mRNA was incubated for 2 hr in S100 extract with 0.35 or 0.70 μg of either ASF/U2AF65 or ASF/SF2. RNA products were resolved on a 6% polyacrylamide–8 m urea gel. (Right) Symbols depicting pre-mRNA, mRNA, lariat intron–second exon intermediate, 5′ exon, and lariat intron. The asterisk indicates a cleavage product unrelated to splicing. Structures of the two pre-mRNAs are diagrammed at the bottom. (D) Gel mobility shift assay of ASF/SF2 and ASF/U2AF65 fusion proteins. 32P-Labeled A7 RNA probe (see Materials and Methods) was incubated with 1.25, 5 and 20 pmole of ASF/U2AF65 or ASF/SF2 and then resolved on a 5% native polyacrylamide gel. The unbound free RNA is indicated.
Depletion of ASF/SF2 influences alternative splicing
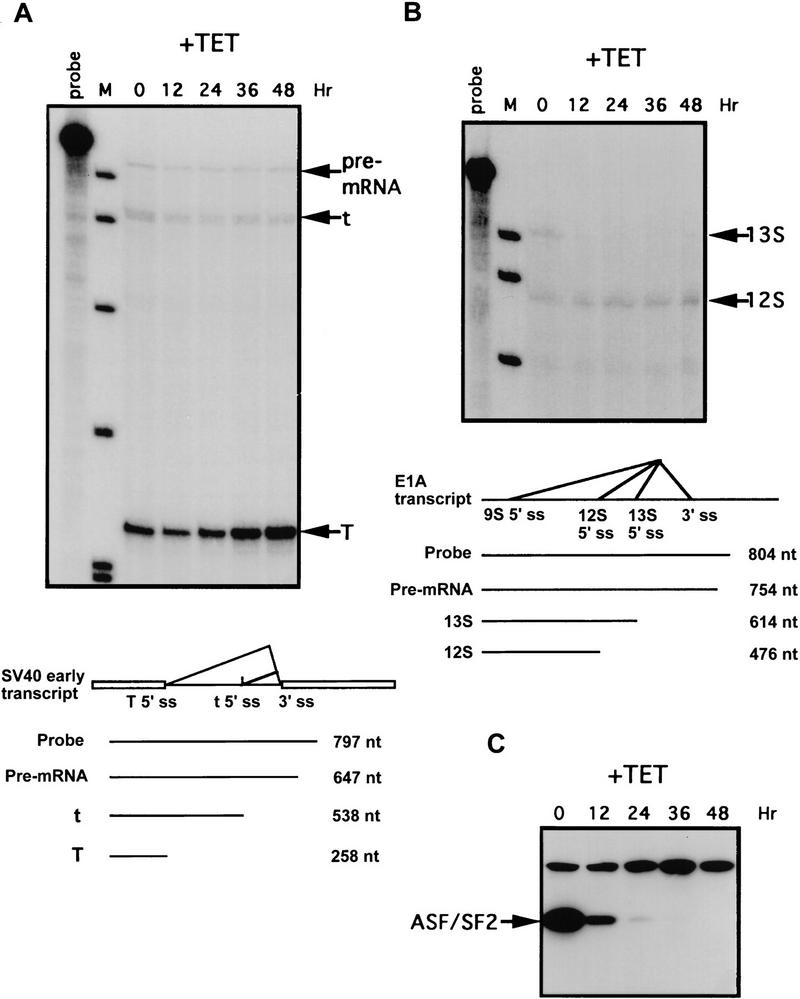
With a pre-mRNA containing an intron with multiple 5′ splice sites, increasing the ASF/SF2 concentration, either in nuclear extracts by addition of ASF/SF2 protein or in cells by transient overexpression, switches usage of the distal 5′ splice site to the proximal site (e.g., Ge and Manley 1990; Krainer et al. 1990b; Cáceres et al. 1994; Wang and Manley 1995). If ASF/SF2 functions naturally in a similar way, then depletion of ASF/SF2 in cells would be predicted to favor utilization of distal 5′ splice sites. To test this, we analyzed the effects of ASF/SF2 depletion on alternative splicing of SV40 early and adenovirus E1A pre-mRNAs, both of which contain one alternatively spliced intron with multiple 5′ splice sites (e.g., Ge et al. 1990; Harper and Manley 1992). We chose to examine the effects of ASF/SF2 depletion on these model substrates, as opposed to transcripts of endogenous genes, both because of their relative simplicity and because their response to ASF/SF2 in other assays has been well studied. DT40-ASF cells were stably transfected with vectors containing either the SV40 early or E1A gene and expressing clones were isolated. These cells were incubated with tet for different periods of time and accumulation of SV40 and E1A mRNAs was monitored by RNase protection. After 24–36 hr of tet treatment, when ASF/SF2 was barely detectable (Fig. 5C), a significant increase of SV40 large T mRNA produced by splicing from the distal 5′ splice site was seen (Fig. 5A). After ASF/SF2 was completely depleted (48 hr; Fig. 5C), large T mRNA increased by threefold. The level of small t mRNA spliced from the proximal 5′ splice site decreased slightly at all time points (Fig. 5A), so that the ratio of large T to small t mRNA increased four- to fivefold. E1A 13S mRNA, spliced from the proximal 5′ splice site, rapidly decreased after 12 hr of tet treatment (Fig. 5B), even though a considerable amount of ASF/SF2 remained (Fig. 5C), suggesting that splicing from the 13S 5′ splice site is quite sensitive to the ASF/SF2 concentration. 12S mRNA increased very slightly following ASF/SF2 depletion. 9S mRNA, produced by splicing from the most distal 5′ splice site, could not be detected because of the low expression of the E1A gene in DT40-ASF cells.
Figure 5.
ASF/SF2 depletion influences alternative splicing of SV40 early and adenovirus E1A pre-mRNAs. DT40–ASF cells stably transfected with an expression vector containing either the SV40 early gene (A) or the adenovirus E1A gene (B) were incubated in medium containing 1 μg/ml tet for 0, 12, 24, 36 and 48 hr. Total cellular RNA was isolated from these cells and analyzed by RNase protection. Schemes of alternative splicing of SV40 early and E1A pre-mRNA are illustrated at the bottom of each panel. Numbers indicate the length of probe and protected fragments. (ss) Splice site; (M) pBR322/HpaII marker. (C) The levels of ASF/SF2 proteins in DT40–ASF cells treated with tet for 0, 12, 24, 36 and 48 hr were measured by Western blotting using mAb 12CA5. The position of ASF/SF2 is indicated.
Although we cannot rule out the possibility that the above-described effects on mRNA accumulation were indirect, the fact that they are consistent with expectations from in vitro results strongly suggests they reflect changes in alternative splicing directly resulting from ASF/SF2 depletion. We also note that the cells were fully viable at all time points tested, as determined by both trypan blue exclusion and their ability to recover following withdrawal of tet (results not shown). Taken together, these results provide the first evidence that ASF/SF2, or any SR protein, can modulate alternative splicing in vivo under physiological conditions.
Depletion of ASF/SF2 does not detectably affect splicing of several mRNAs
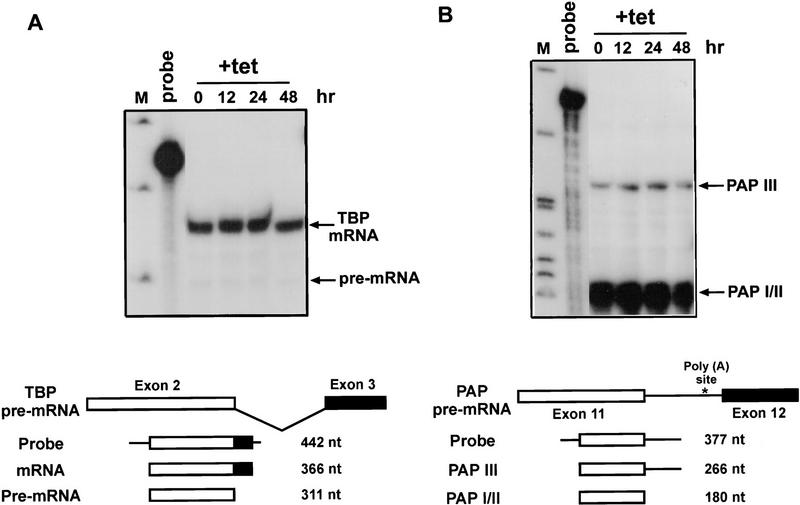
In vitro studies have shown that ASF/SF2 can on the one hand function as an essential splicing factor for all substrates tested, but on the other hand that for most introns this function is redundant and can be fulfilled by other SR proteins. Therefore, we wished to determine whether effects on splicing of transcripts with simple introns could be detected following ASF/SF2 depletion. Previous pulse-labeling experiments provided evidence that the overall rate of pre-mRNA processing was reduced following ASF/SF2 depletion, supporting the view that ASF/SF2 is required for efficient splicing of at least some, and perhaps many, endogenous transcripts (Wang et al. 1996). Here, we examined accumulation of endogenous TATA-binding protein (TBP) and poly(A) polymerase (PAP) mRNAs in response to ASF/SF2 depletion. Total RNAs were isolated from DT40-ASF cells incubated with tet for 0, 12, 24, and 48 hr, and the levels of TBP and PAP mRNAs were monitored by RNase protection. At all time points, TBP pre-mRNA and mRNA levels were both unchanged (Fig. 6A). Multiple mRNAs encoding different PAP isoforms are produced by alternative splicing (Zhao and Manley 1996). The probe used in these experiments was designed to detect PAP III, a truncated, inactive form, or the full-length, active forms (e.g., PAP I and PAP II). PAP III mRNA retains an unspliced intron so that a poly(A) site in the intron is used, while splicing is required to generate PAP I and PAP II isoforms (Fig. 6B; see Zhao and Manley 1996). Depletion of ASF/SF2 did not influence either accumulation of or the ratio between these mRNAs (Fig. 6B). In addition, we found that the level of the inner centromere protein (INCENP; Mackay et al. 1993) mRNA did not vary after ASF/SF2 depletion nor did that of the polyadenylation factor CstF-64 (data not shown). Although these experiments measured accumulated, steady-state RNA, and thus may not have detected small decreases in the rate of splicing, taken together, they suggest that ASF/SF2 is not essential for splicing of these transcripts. These findings are consistent with the idea that the ASF/SF2 function in constitutive splicing is at least partially redundant in vivo, as it is in vitro.
Figure 6.
Depletion of ASF/SF2 does not affect accumulation of TBP and PAP mRNAs. Total cellular RNA was isolated from DT40–ASF cells incubated in medium containing 1 μg/ml tet for 0, 12, 24 and 48 hr. Levels of TBP (A) or PAP (B) RNAs were measured by RNase protection. Schemes of splicing of TBP and PAP pre-mRNAs are illustrated at the bottom of each panel. Numbers indicate the length of probe and protected fragments. (M) pBR322/HpaII marker.
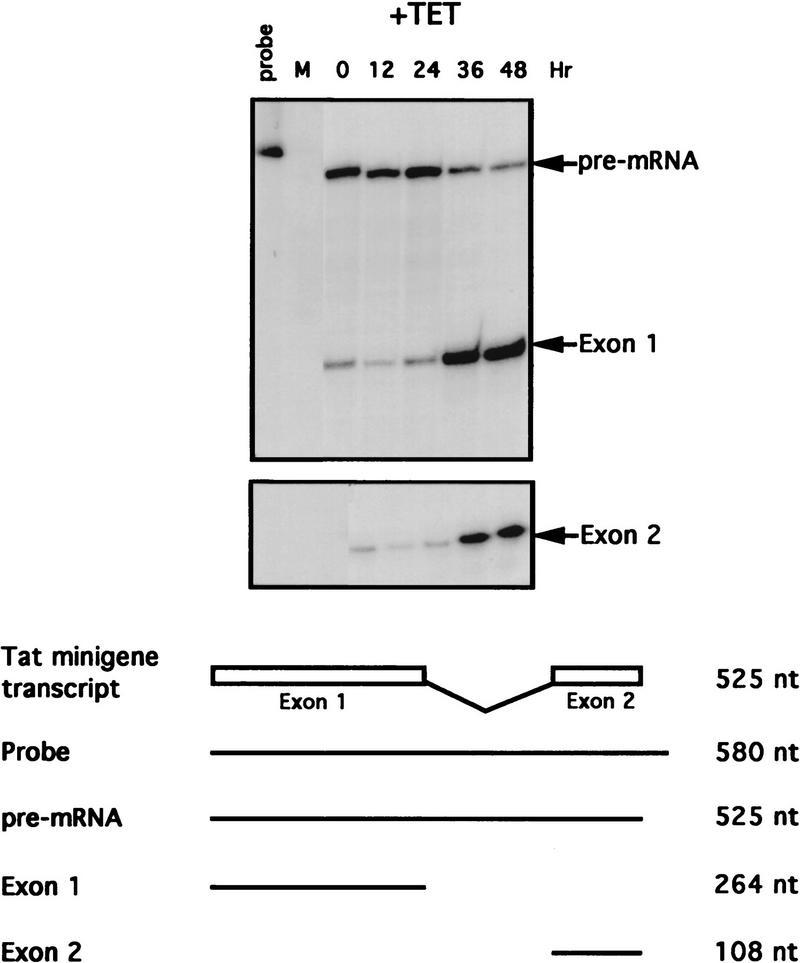
ASF/SF2 represses HIV-1 tat pre-mRNA splicing in vivo
In vitro studies have shown that efficient splicing of HIV-1 tat pre-mRNA requires addition of an excess amount of ASF/SF2 to nuclear extracts (Krainer et al. 1990a), and that ASF/SF2 can specifically commit tat pre-mRNA to splicing (Fu 1993). This likely reflects the presence of an ASF/SF2-dependent splicing enhancer in this pre-mRNA (Tacke and Manley 1995), although this has not been investigated. Therefore, we used tat pre-mRNA splicing as a model of a simple (nonalternative) splicing event that should be responsive to ASF/SF2 levels. Specifically, the in vitro experiments suggest that depletion of ASF/SF2 should result in a reduction in tat splicing. DT40-ASF cells were stably transfected with a vector containing a tat minigene essentially identical to that used in in vitro experiments (Krainer et al. 1990a), and expressing clones were isolated. Cells from one clone were incubated with tet, and accumulation of tat mRNA and unspliced pre-mRNA at different time points was monitored by RNase protection (Fig. 7). Unexpectedly, following ASF/SF2 depletion (36 and 48 hr), tat mRNA levels significantly increased, by over sixfold, with a concomitant decrease of tat pre-mRNA, indicating that ASF/SF2 depletion enhanced, rather than inhibited, tat pre-mRNA splicing. That the increased accumulation of tat mRNA was indeed caused by increased splicing is strongly supported by the corresponding decrease in pre-mRNA. On the basis of this finding, we conclude that ASF/SF2 can naturally repress splicing of certain introns, such as HIV-1 tat, in vivo. In addition, the fact that splicing can actually be enhanced following ASF/SF2 depletion argues strongly that the effects on SV40 and E1A alternative splicing were not attributable to a more general inhibition of splicing, and significantly strengthens the conclusion that ASF/SF2 function in constitutive splicing is redundant, at least for the pre-mRNAs tested.
Figure 7.
ASF/SF2 depletion activates splicing of HIV-1 tat pre-mRNA. DT40–ASF cells stably transfected with an expression vector encoding HIV-1 tat pre-mRNA were incubated with tet as in Fig. 6. Total cellular RNA was isolated from these cells and analyzed by RNase protection. Scheme of tat pre-mRNA splicing is illustrated at the bottom. Numbers indicate the length of probe and protected fragments. (M) pBR322/HpaII marker.
Discussion
We have employed a previously described genetic system (Wang et al. 1996) both to analyze systematically the specificity of RS domains of several splicing factors and to study the role of ASF/SF2 in pre-mRNA splicing under physiological conditions. Our findings have provided the first evidence that the RS domains of SR proteins are functionally interchangeable in vivo. While this property extends to the RS domains of the SR-like protein TRA-2, the U2AF65 RS-like domain was completely ineffective at providing RS domain function, in vitro as well as in vivo. Our data also provided the first evidence that an SR protein can modulate splicing of specific pre-mRNAs under physiological conditions.
One of the earliest functions suggested for RS domains was to target proteins to specific subnuclear localizations. Li and Bingham (1991) showed that the RS domains of the Drosophila proteins transformer (TRA) and SWAP (both of which participate in splicing but are not SR proteins) were both sufficient to direct a protein to subnuclear regions known as speckles, and that the TRA RS domain could substitute for the SWAP RS domain in an in vivo functional assay. However, more recent studies on the RS domains of SR proteins, while also providing evidence that they function in subnuclear targeting, suggested that several RS domains can have distinct, nonredundant functions. Cáceres et al. (1997) showed using transient transfection assays, that different RS domains could differentially target proteins to nuclear substructures. While both ASF/SF2 and SRp20 RS domains targeted a reporter protein to the nucleus, only the latter could direct it to the speckles. In a separate study, Cáceres et al. (1998) provided evidence that a subset of SR proteins are capable of shuttling between nucleus and cytoplasm. Again using transient transfections with chimeric proteins, they showed that RS domains play a dominant role in determining the shuttling properties of the protein. For example, the RS domain of ASF/SF2, a shuttling protein, could convert SRp40, a nonshuttling protein, into a shuttler, while an ASF/SF2 derivative containing the SRp40 RS domain was unable to shuttle. On the basis of these results, the authors proposed that individual RS domains have unique properties in directing distinct patterns of subcellular localization.
Although our results do not directly address the above issues, nonetheless, they have important implications regarding their general significance. For example, on the basis of the results of Cáceres et al. (1997, 1998), the ASF/SRp20 chimera we analyzed would be predicted to have a subnuclear distribution distinct from ASF/SF2, and the ASF/SRp40 protein should not be able to shuttle, unlike ASF/SF2. Assuming the same requirements for determining subcellular localization exist in transfected HeLa cells and chicken DT40 cells (the difference between chicken and human is unlikely to be important given the sequence identity of the proteins involved; see below), our findings lead to the conclusion that the proposed ability of ASF/SF2 to localize distinctively in the nucleus and to shuttle between nucleus and cytoplasm cannot be necessary for cell viability.
Given that SR protein RS domains are interchangeable, what determines the specificity of SR proteins? The answer must be the RBDs. This likely even extends to specific nuclear localization. The emerging view is that speckles may be nonspecific storage sites for SR proteins and other splicing factors and that splicing occurs at sites of transcription (e.g., Zhang et al. 1994; Misteli et al. 1997). A recent report showed that SR proteins are localized to transcriptionally active sites not necessarily coincident with speckles, and a specific SR protein, SRp20 is present in a subset of SR protein locations (Neugebauer and Roth 1997). This likely reflects the distinct RNA-binding specificities of SR proteins. These specificities appear to be determined by the RBDs with little or no contribution from RS domains, and have been shown to be functionally important in in vitro assays employing pre-mRNAs containing splicing enhancers (Tacke and Manley 1995; Tacke et al. 1997). Consistent with this observation and with the results presented here, the specificities of ASF/SF2 and SC35 in committing tat and β-globin pre-mRNAs, respectively, to splicing in vitro are determined by their RBDs, while their RS domains are interchangeable (Chandler et al. 1997). RBDs also appear to contribute to the direct interactions between SR proteins and other splicing factors (Xiao and Manley 1997; S.H. Xiao and J.L. Manley, unpubl.). It is not yet known whether RBDs contribute to the specificity of these interactions, nor is it known whether individual SR proteins discriminate in their interactions with other proteins.
Do RS domains of individual SR proteins have any specific functions? In this regard, it is noteworthy that individual RS domains are extraordinarily highly conserved throughout metazoans. For example, murine X16 is completely identical to human SRp20 (Zahler et al. 1992). SC35 and its chicken homolog PR264 are 98% identical (Fu and Maniatis 1992; Vellard et al. 1992), as are human and chicken ASF/SF2 (Wang et al. 1996). Human and chicken TRA-2 proteins are also nearly identical (T. Kashima and J.L. Manley, unpubl.). This high degree of sequence conservation in RS domains throughout evolution suggests that they possess distinct properties. Because our results have shown that RS domains are interchangeable for cell viability, perhaps different RS domains perform specific functions in higher levels of splicing regulation, such as tissue- or developmental stage-specific splicing.
Our observation that the U2AF65 RS domain could not replace the RS domain of ASF/SF2 is consistent with several distinct properties of the U2AF65 RS domain described previously. First, U2AF65 is essential for viability of flies and fission yeast (Kanaar et al. 1993; Potashkin et al. 1993). However, in contrast with the ASF/SF2 RS domain, which is required for the essential ASF/SF2 function necessary for cell viability (Wang et al. 1996), the RS domain of Drosophila U2AF65 is dispensable for the protein’s function(s) in development (Rudner et al. 1998). Second, the U2AF65 RS domain appears not to mediate protein interactions. ASF/SF2 and SC35, which bind to several splicing factors containing RS domains, including themselves, do not interact with U2AF65 (Wu and Maniatis 1993), and the regions required for the interaction between U2AF65 and U2AF35 were mapped outside the RS domains of both proteins (Zhang et al. 1992). Finally, instead of functioning as a protein-interaction domain, the U2AF65 RS domain was suggested to interact directly with the pre-mRNA branch site and promote U2 snRNP binding (Valcárcel et al. 1996). Although the molecular basis for the distinct properties of the U2AF65 RS domain is not known, our results indicate that it performs a different function(s) from many other RS domains, and cannot substitute, in vitro or in vivo, for the ASF/SF2 RS domain.
Our experiments also revealed that ASF/SF2 can function as a negative splicing regulator in vivo. Depletion of ASF/SF2 resulted in a significant increase of HIV-1 tat pre-mRNA splicing, indicating that ASF/SF2 can repress tat splicing under normal physiological conditions. Several previous studies have suggested that ASF/SF2 can inhibit splicing. ASF/SF2 has been shown to bind to a splicing repressor sequence in Rous sarcoma virus RNA (McNally and McNally 1996). ASF/SF2 can interact with a purine-rich sequence in the IIIa intron of the adenovirus L1 transcript and repress IIIa splicing in vitro, probably by blocking U2 snRNP binding to the branch site (Kanopka et al. 1996). We showed previously that transient overexpression of ASF/SF2 in cells significantly reduced the steady-state level of SV40 early mRNA, which could result from splicing inhibition (Wang and Manley 1995) and that ASF/SF2 can negatively autoregulate its expression, likely by inhibiting its own pre-mRNA splicing (Wang et al. 1996). The negative regulatory function of ASF/SF2 may be employed to maintain the balance between spliced and unspliced RNAs, which is particularly important for the life cycles of retroviruses. Given our previous finding that depletion of ASF/SF2 led to accumulation of incompletely processed pre-mRNA (Wang et al. 1996), we conclude that ASF/SF2 can play both positive and negative roles in pre-mRNA splicing in vivo. Similarly, Drosophila TRA-2, an activator of sex-specific splicing, can repress splicing of its own pre-mRNA (Mattox and Baker 1991).
Both positive and negative cis-acting regulatory sequences have been identified in the second exon of tat pre-mRNA (Amendt et al. 1995; Staffa and Cochrane 1995). ASF/SF2 likely activates tat splicing in vitro (Krainer et al. 1990a; Fu 1993; Xiao and Manley 1997) by binding to the positive regulatory sequence, or splicing enhancer, which contains a good match to the consensus ASF/SF2-binding site (Tacke and Manley 1995). Why ASF/SF2 affects tat splicing in vivo in a totally opposite way is not known. One possibility is that different combinations and/or concentrations of splicing factors exist in cells as opposed to the cell extracts used for in vitro studies, perhaps in the former situation favoring interactions with the negative regulatory element. In any case, our results indicate that in vitro studies may not necessarily reflect the exact function of SR proteins in cells.
We have provided genetic evidence that ASF/SF2 can modulate alternative splicing from multiple 5′ splice sites in vivo. Depletion of ASF/SF2 in cells switched alternative splicing of SV40 early and E1A pre-mRNAs by favoring usage of the distal 5′ splice sites, in agreement with predictions from previous studies with in vitro splicing reactions or transient transfection assays. In addition, we have recently found that ASF/SF2 can modulate alternative splicing of transcripts encoded by a cell death gene, Bcl-X (Boise et al. 1992), in a similar way, and that depletion of ASF/SF2 leads to programmed cell death (J. Wang and J.L. Manley, in prep.). Therefore, modulating levels of alternative splicing is likely an important function of ASF/SF2 in cells. However, the unchanged mRNA levels of several other transcripts tested, such as TBP and PAP, following ASF/SF2 depletion suggest that changes in the concentration or activity of ASF/SF2 will not affect all splicing events. One possibility consistent with our data and previous in vitro studies is that the functions of ASF/SF2, and likely other SR proteins, are largely redundant in splicing of many pre-mRNAs containing constitutive introns, and individual SR proteins participate specifically in splicing of a limited set of pre-mRNAs. This likely involves activation of certain splicing events, repression of others, and qualitative changes in alternative splicing patterns.
Materials and methods
Plasmid constructs
An EcoRI–ApaI DNA fragment encoding ASF/SF2 RBDs (amino acids 1–197) was ligated with a cDNA fragment encoding a heterologous RS domain to generate the chimeric protein cDNA. cDNA fragments encoding RS domains of X16, SC35, and SRp40 were obtained by restriction digestion: XhoI–BamHI fragment of X16 cDNA (amino acids 100–164; Ayane et al. 1991) encoding amino acids 100–164, AvrII–HindIII of SC35 cDNA (amino acids 120–221; Fu and Maniatis 1992) and AseI–EcoRI of SRp40 cDNA (amino acids 180–273; Screaton et al. 1995). cDNA fragments encoding RS domains of hTRA-2α and U2AF65 were obtained by PCR. Primers for PCR were as follows: for the hTRA-2α amino-terminal RS domain (amino acids 9–90), upstream 5′-CCGGAATTCGAGGGCAGAGAGTCTCGC-3′ and downstream 5′-CGCAAGCTTATTCTGGTGTATATGATCTACTTCG-3′; for the hTRA-2α carboxy-terminal RS domain (amino acids 231–282), upstream 5′-CCGGAATTCGGCAGACGTCGAGATTCTTAC-3′ and downstream 5′-CGCAAGCTTCAATAGCGTCTTGGGCTGTAG-3′; for the U2AF65 RS domain (amino acids 23–83), upstream 5′-CCGGAATTCCGGCATCGGAAGCGCAGCCAC-3′ and downstream 5′-CGCAAGCTTACTCGTGGAGGGGGGAACGAATC-3′. All three PCR products were completely sequenced, and all chimeric protein cDNAs were sequenced at the junction sites to confirm in-frame ligations. ASF/SF2 cDNA in the puro–ASF vector (Wang et al. 1996) was removed by EcoRI digestion and replaced by the chimeric protein cDNAs, giving the expression vectors encoding the chimeric proteins fused to a hemagglutin epitope at their amino-termini. To construct an E. coli expression vector to produce His-tagged ASF/U2AF65, puro–ASF/U2 was digested with KpnI and HindIII, and the isolated fragment was inserted into pDS–ASF.
The Ecogpt gene under the control of chicken β-actin promoter (Miyazaki et al. 1989) was inserted into pBSV+2 and pBSV-E1A (Wang and Manley 1995) to generate expression vectors for SV40 early and adenovirus E1A pre-mRNAs, gpt–SV40 and gpt–E1A, respectively. The HindIII (nucleotide 5171)–HpaI (nucleotide 2666) region of SV40 early gene was replaced by a BamHI–HindIII fragment containing the HIV-1 tat minigene (Krainer et al. 1990a), giving pSV–tat. Ecogpt gene was inserted into pSV–tat, producing the expression vector of tat minigene gpt–tat. HindIII–BsmI fragment of SV40 early gene, HindIII– BanII of E1A and BamHI–HindIII of tat were inserted into HindIII–SmaI site of pBluescript (Stratagene) to make constructs for labeling RNA probes by in vitro transcription.
Cell culture and transfections
DT40-ASF cells were maintained as described (Wang et al. 1996). To deplete ASF/SF2, DT40-ASF cells or derivative clones were incubated in medium containing 1 μg/ml tetracycline (Sigma). DT40-ASF cells were transfected by electroporation with expression vectors encoding ASF/SF2, chimeric proteins and reporter pre-mRNAs as appropriate, under conditions described previously (Wang et al. 1996). Cells transfected with gpt–SV40, gpt–E1A, and gpt–tat were selected in medium containing 30 μg/ml mycophenolic acid (Calbiochem). Drug-resistant clones were screened for expression of the genes by RNase protection.
Western blotting
Cells (1 × 106) were lysed in 1× SDS loading buffer and subjected to SDS-PAGE. Western blots with anti-hemagglutin monoclonal antibody (mAb) 12CA5 were performed as described (Wang and Manley 1995). In the complementation assay, the surviving clones were expanded in medium containing 1 μg/ml tetracycline and analyzed by Western blotting with mAb 12CA5 for expressions of the chimeric proteins.
In vitro splicing and RNA binding
Recombinant His-tagged ASF/U2AF65 and ASF/SF2 were expressed in E. coli and purified as described (Ge et al. 1991). Protein concentration and purity were determined by Bradford assays and SDS-PAGE. Splicing reactions with SV40 early or HIV tat pre-mRNA and HeLa cell nuclear or S100 extracts were performed as described previously (Ge et al. 1991; Zuo and Manley 1993; Xiao and Manley 1997). For RNA binding assays, purified recombinant ASF/SF2 and ASF/U2AF65 were phosphorylated and repurified as described (Xiao and Manley 1997). Gel mobility shift assays with in vitro-transcribed, 32P-labeled A7 (which contains an ASF/SF2 consensus binding site) and C2 (a non-specific control) RNAs were performed as described (Tacke and Manley 1995).
RNase protection
Radioactive RNA probes for monitoring TBP, PAP, SV40, E1A, and tat RNAs were transcribed in vitro in the presence of [32P]GTP from plasmids containing appropriate DNA fragments. Total cellular RNA was isolated from DT40-ASF or stably transfected DT40-ASF cells as described (Wang et al. 1996). In each hybridization mixture, 20 μg of each RNA sample and 200 cps of the appropriate probe were used. Hybridization and RNase protection were performed as described (Wang et al. 1996). RNase protection experiments were all repeated at least twice.
Acknowledgments
We thank Dr. Adrian Krainer for providing the tat minigene; Dr. Michael Green for providing U2AF65 cDNA; Dr. Roland Tacke for providing TRA-2α cDNA, A7 and C2 plasmids and advice; Moonkyoung Um for providing chicken TBP cDNA; Wenqing Zhao for providing chicken PAP cDNA; and Xiaohong Shi for technical assistance. We also thank members of our laboratory for helpful discussions and comments. This work was supported by National Institutes of Health grant GM-48529.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL jlm2@columbia.edu; FAX (212) 865-8246.
References
- Amendt BA, Si ZH, Stoltzfus CM. Presence of exon splicing silencers within human immunodeficiency virus type 1 tat exon 2 and tat-rev exon 3: Evidence for inhibition mediated by cellular factors. Mol Cell Biol. 1995;15:4606–4615. doi: 10.1128/mcb.15.8.4606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amrein H, Gorman M, Nöthiger R. The sex-determining gene tra-2 of Drosophila encodes a putative RNA binding protein. Cell. 1988;55:1025–1035. doi: 10.1016/0092-8674(88)90247-4. [DOI] [PubMed] [Google Scholar]
- Amrein H, Hedley ML, Maniatis T. The role of specific protein-RNA and protein-protein interactions in positive and negative control of pre-mRNA splicing by transformer 2. Cell. 1994;76:735–746. doi: 10.1016/0092-8674(94)90512-6. [DOI] [PubMed] [Google Scholar]
- Ayane M, Preuss U, Kohler G, Nielsen PJ. A differentially expressed murine RNA encoding a protein with similarities to two types of nucleic acid binding motifs. Nucleic Acids Res. 1991;19:273–278. doi: 10.1093/nar/19.6.1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker BS. Sex in flies: The splice of life. Nature. 1989;340:521–524. doi: 10.1038/340521a0. [DOI] [PubMed] [Google Scholar]
- Boise LH, González-Garcia M, Postema CE, Ding L, Lindsten T, Turka LA, Mao X, Nuñez G, Thompson CB. bcl-x, a bcl-2-related gene that functions as a dominant regulator of apoptotic cell death. Cell. 1993;74:597–608. doi: 10.1016/0092-8674(93)90508-n. [DOI] [PubMed] [Google Scholar]
- Cáceres JF, Krainer AR. Functional analysis of pre-mRNA splicing factor SF2/ASF structural domains. EMBO J. 1993;12:4715–4726. doi: 10.1002/j.1460-2075.1993.tb06160.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cáceres JF, Stamm S, Helfman DM, Krainer AR. Regulation of alternative splicing in vivo by overexpression of antagonistic splicing factors. Science. 1994;265:1706–1709. doi: 10.1126/science.8085156. [DOI] [PubMed] [Google Scholar]
- Cáceres JF, Misteli T, Screaton GR, Spector DL, Krainer AR. Role of the modular domains of SR proteins in subnuclear localization and alternative splicing specificity. J Cell Biol. 1997;138:225–238. doi: 10.1083/jcb.138.2.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cáceres JF, Screaton GR, Krainer AR. A specific subset of SR proteins shuttles continuously between the nucleus and the cytoplasm. Genes & Dev. 1998;2:55–66. doi: 10.1101/gad.12.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler SD, Mayeda A, Yeakley JM, Krainer AR, Fu XD. RNA splicing specificity determined by the coordinated action of RNA recognition motifs in SR proteins. Proc Natl Acad Sci. 1997;94:3596–3601. doi: 10.1073/pnas.94.8.3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dauwalder B, Amaya-Manzanares F, Mattox W. A human homologue of the Drosophila sex determination factor transformer-2 has conserved splicing regulatory functions. Proc Natl Acad Sci. 1996;93:9004–9009. doi: 10.1073/pnas.93.17.9004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eperon IC, Ireland DC, Smith RA, Mayeda A, Krainer AR. Pathways for selection of 5′ splice sites by U1 snRNPs and SF2/ASF. EMBO J. 1993;12:3607–3617. doi: 10.1002/j.1460-2075.1993.tb06034.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu XD. Specific commitment of different pre-mRNAs to splicing by single SR proteins. Nature. 1993;365:82–85. doi: 10.1038/365082a0. [DOI] [PubMed] [Google Scholar]
- ————— The superfamily of arginine/serine-rich splicing factors. RNA. 1995;1:663–680. [PMC free article] [PubMed] [Google Scholar]
- Fu XD, Maniatis T. Factor required for mammalian spliceosome assembly is localized to discrete regions in the nucleus. Nature. 1990;343:437–441. doi: 10.1038/343437a0. [DOI] [PubMed] [Google Scholar]
- ————— Isolation of a complementary DNA that encodes the mammalian splicing factor SC35. Science. 1992;256:535–538. doi: 10.1126/science.1373910. [DOI] [PubMed] [Google Scholar]
- Fu XD, Mayeda A, Maniatis T, Krainer A. General splicing factors SF2 and SC35 have equivalent activities in vitro, and both affect alternative 5′ and 3′ splice site selection. Proc Natl Acad Sci. 1992;89:11224–11228. doi: 10.1073/pnas.89.23.11224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge H, Manley JL. A protein factor, ASF, controls cell-specific alternative splicing of SV40 early pre-mRNA in vitro. Cell. 1990;62:25–34. doi: 10.1016/0092-8674(90)90236-8. [DOI] [PubMed] [Google Scholar]
- Ge H, Zuo P, Manley JL. Primary structure of the human splicing factor ASF reveals similarities with Drosophila regulators. Cell. 1991;66:373–382. doi: 10.1016/0092-8674(91)90626-a. [DOI] [PubMed] [Google Scholar]
- Goralski TJ, Edstrom JE, Baker BS. The sex determination locus transformer-2 of Drosophila encodes a polypeptide with similarity to RNA binding proteins. Cell. 1989;56:1011–1018. doi: 10.1016/0092-8674(89)90634-x. [DOI] [PubMed] [Google Scholar]
- Harper JE, Manley JL. Multiple activities of the human splicing factor ASF. Gene Expr. 1992;2:19–29. [PMC free article] [PubMed] [Google Scholar]
- Hedley ML, Amrein H, Maniatis T. An amino acid sequence motif sufficient for subnuclear localization of an arginine/serine-rich splicing factor. Proc Natl Acad Sci. 1995;92:11524–11528. doi: 10.1073/pnas.92.25.11524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinrichs V, Baker BS. The Drosophila SR protein RBP1 contributes to the regulation of doublesex alternative splicing by recognizing RBP1 RNA target sequences. EMBO J. 1995;14:3987–4000. doi: 10.1002/j.1460-2075.1995.tb00070.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamison SF, Pasman Z, Wang J, Will C, Lührmann R, Manley JL, Garcia-Blanco MA. U1 snRNP-ASF/SF2 interaction and 5′ splice site recognition: Characterization of required elements. Nucleic Acids Res. 1995;23:3260–3267. doi: 10.1093/nar/23.16.3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanaar R, Roche SE, Beall EL, Green MR, Rio DC. The conserved pre-mRNA splicing factor U2AF from Drosophila: Requirement for viability. Science. 1993;262:569–573. doi: 10.1126/science.7692602. [DOI] [PubMed] [Google Scholar]
- Kanopka A, Mühlemann O, Akusjärvi G. SR proteins that are essential for generic pre-mRNA splicing inhibit splicing of a regulated adenovirus pre-mRNA. Nature. 1996;381:535–538. doi: 10.1038/381535a0. [DOI] [PubMed] [Google Scholar]
- Kim YJ, Zuo P, Manley JL, Baker BS. The Drosophila RNA-binding protein RBP1 is localized to transcriptionally active sites of chromosomes and shows a functional similarity to human splicing factor ASF/SF2. Genes & Dev. 1992;6:2569–2579. doi: 10.1101/gad.6.12b.2569. [DOI] [PubMed] [Google Scholar]
- Kohtz JD, Jamison SF, Will CL, Zuo P, Lührmann R, Garcia-Blanco MA, Manley JL. Protein-protein interactions and 5′-splice site recognition in mammalian mRNA precursors. Nature. 1994;368:119–124. doi: 10.1038/368119a0. [DOI] [PubMed] [Google Scholar]
- Krainer AR, Conway GC, Kozak D. Purification and characterization of pre-mRNA splicing factor SF2 from HeLa cells. Genes & Dev. 1990a;4:1158–1171. doi: 10.1101/gad.4.7.1158. [DOI] [PubMed] [Google Scholar]
- ————— The essential pre-mRNA splicing factor SF2 influences 5′ splice site selection by activating proximal sites. Cell. 1990b;62:35–42. doi: 10.1016/0092-8674(90)90237-9. [DOI] [PubMed] [Google Scholar]
- Krainer AR, Mayeda A, Kozak D, Binns G. Functional expression of cloned human splicing factor SF2: Homology to RNA-binding proteins, U170K and Drosophila splicing regulators. Cell. 1991;66:383–394. doi: 10.1016/0092-8674(91)90627-b. [DOI] [PubMed] [Google Scholar]
- Krämer A. The structure and function of proteins involved in mammalian pre-mRNA splicing. Annu Rev Biochem. 1996;65:367–409. doi: 10.1146/annurev.bi.65.070196.002055. [DOI] [PubMed] [Google Scholar]
- Li H, Bingham PM. Arginine/serine-rich domains of the su(wa) and tra RNA processing regulators target proteins to a subnuclear compartment implicated in splicing. Cell. 1991;67:335–342. doi: 10.1016/0092-8674(91)90185-2. [DOI] [PubMed] [Google Scholar]
- Lynch KW, Maniatis T. Synergistic interactions between two distinct elements of a regulated splicing enhancer. Genes & Dev. 1995;9:284–293. doi: 10.1101/gad.9.3.284. [DOI] [PubMed] [Google Scholar]
- Mackay AM, Eckley DM, Chue C, Earnshaw WC. Molecular analysis of the INCENPs (inner centromere proteins): Separate domains are required for association with microtubules during interphase and with the central spindle during anaphase. J Cell Biol. 1993;123:373–385. doi: 10.1083/jcb.123.2.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manley JL, Tacke R. SR proteins and splicing control. Genes & Dev. 1996;10:1569–1579. doi: 10.1101/gad.10.13.1569. [DOI] [PubMed] [Google Scholar]
- Mattox W, Baker BS. Autoregulation of the splicing of transcripts from the transformer-2 gene of Drosophila. Genes & Dev. 1991;5:786–796. doi: 10.1101/gad.5.5.786. [DOI] [PubMed] [Google Scholar]
- McNally LM, McNally MT. SR protein splicing factors interact with the Rous sarcoma virus negative regulator of splicing element. J Virol. 1996;70:1163–1172. doi: 10.1128/jvi.70.2.1163-1172.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli T, Cáceres JF, Spector DL. The dynamics of a pre-mRNA splicing factor in living cells. Nature. 1997;387:523–527. doi: 10.1038/387523a0. [DOI] [PubMed] [Google Scholar]
- Miyazaki J, Takaki S, Araki K, Tashiro F, Tominaga A, Takatsu K, Yamamura K. Expression vector system based on the chicken beta-actin promoter directs efficient production of interleukin-5. Gene. 1989;79:269–277. doi: 10.1016/0378-1119(89)90209-6. [DOI] [PubMed] [Google Scholar]
- Moore MJ, Query CC, Sharp PA. Splicing of precursors to messenger RNAs by the spliceosome. In: Gesteland RF, Atkins JF, editors. The RNA world. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1993. pp. 303–358. [Google Scholar]
- Neugebauer KM, Roth MB. Distribution of pre-mRNA splicing factors at sites of RNA polymerase II transcription. Genes & Dev. 1997;11:1148–1159. doi: 10.1101/gad.11.9.1148. [DOI] [PubMed] [Google Scholar]
- Peng X, Mount SM. Genetic enhancement of RNA processing defects by a dominant mutation in B52, the Drosophila gene for an SR protein splicing factor. Mol Cell Biol. 1995;15:6273–6282. doi: 10.1128/mcb.15.11.6273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potashkin J, Naik K, Wentz-Hunter K. U2AF homolog required for splicing in vivo. Science. 1993;262:573–575. doi: 10.1126/science.8211184. [DOI] [PubMed] [Google Scholar]
- Ramchatesingh J, Zahler AM, Neugebauer KM, Roth MB, Cooper TA. A subset of SR proteins activates splicing of the cardiac troponin T alternative exon by direct interactions with an exonic enhancer. Mol Cell Biol. 1995;15:4898–4907. doi: 10.1128/mcb.15.9.4898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ring HZ, Lis JT. The SR protein B52/SRp55 is essential for Drosophila development. Mol Cell Biol. 1994;14:7499–7506. doi: 10.1128/mcb.14.11.7499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudner DZ, Breger KS, Rio DC. Molecular genetic analysis of the heterodimeric splicing factor U2AF: The RS domain on either the large or small Drosophila subunit is dispensable in vivo. Genes & Dev. 1998;12:1010–1021. doi: 10.1101/gad.12.7.1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Screaton GR, Cáceres JF, Mayeda A, Bell MV, Plebanski M, Jackson DG, Bell JI, Krainer AR. Identification and characterization of three members of the human SR family of pre-mRNA splicing factors. EMBO J. 1995;14:4336–4349. doi: 10.1002/j.1460-2075.1995.tb00108.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Hoffman BE, Lis JT. A specific RNA hairpin loop structure binds the RNA recognition motifs of the Drosophila SR protein B52. Mol Cell Biol. 1997;17:2649–2657. doi: 10.1128/mcb.17.5.2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staffa A, Cochrane A. Identification of positive and negative splicing regulatory elements within the terminal tat-rev exon of human immunodeficiency virus type 1. Mol Cell Biol. 1995;15:4597–4605. doi: 10.1128/mcb.15.8.4597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staknis D, Reed R. SR proteins promote the first specific recognition of pre-mRNA and are present together with the U1 small nuclear ribonucleoprotein particle in a general splicing enhancer complex. Mol Cell Biol. 1994;14:7670–7682. doi: 10.1128/mcb.14.11.7670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q, Mayeda A, Hampson RK, Krainer AR, Rottman FM. General splicing factor SF2/ASF promotes alternative splicing by binding to an exonic splicing enhancer. Genes & Dev. 1993;7:2598–2608. doi: 10.1101/gad.7.12b.2598. [DOI] [PubMed] [Google Scholar]
- Tacke R, Manley JL. The human splicing factors ASF/SF2 and SC35 possess different, functionally significant RNA binding specificities. EMBO J. 1995;14:3540–3551. doi: 10.1002/j.1460-2075.1995.tb07360.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tacke R, Chen Y, Manley JL. Sequence-specific RNA binding by an SR protein requires RS domain phosphorylation: Creation of an SRp40-specific splicing enhancer. Proc Natl Acad Sci. 1997;94:1148–1153. doi: 10.1073/pnas.94.4.1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian M, Maniatis T. A splicing enhancer complex controls alternative splicing of doublesex pre-mRNA. Cell. 1993;74:105–114. doi: 10.1016/0092-8674(93)90298-5. [DOI] [PubMed] [Google Scholar]
- Valcárcel J, Green MR. The SR protein family: Pleiotropic functions in pre-mRNA splicing. Trends Biochem Sci. 1996;21:296–301. [PubMed] [Google Scholar]
- ValcÁrcel J, Gaur RK, Singh R, Green MR. Interaction of U2AF65 RS region with pre-mRNA branch point and promotion of base pairing with U2 snRNA. Science. 1996;273:1706–1709. doi: 10.1126/science.273.5282.1706. [DOI] [PubMed] [Google Scholar]
- Vellard M, Sureau A, Soret J, Martinerie C, Perbal B. A potential splicing factor is encoded by the opposite strand of the trans-spliced c-myb exon. Proc Natl Acad Sci. 1992;89:2511–2515. doi: 10.1073/pnas.89.7.2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Manley JL. Overexpression of the SR proteins ASF/SF2 and SC35 influences alternative splicing in vivo in diverse ways. RNA. 1995;1:335–346. [PMC free article] [PubMed] [Google Scholar]
- Wang J, Takagaki Y, Manley JL. Targeted disruption of an essential vertebrate gene: ASF/SF2 is essential for cell viability. Genes & Dev. 1996;10:2588–2599. doi: 10.1101/gad.10.20.2588. [DOI] [PubMed] [Google Scholar]
- Wang Z, Hoffmann HM, Grabowski PJ. Intrinsic U2AF binding is modulated by exon enhancer signals in parallel with changes in splicing activity. RNA. 1995;1:21–35. [PMC free article] [PubMed] [Google Scholar]
- Wu JY, Maniatis T. Specific interactions between proteins implicated in splice site selection and regulated alternative splicing. Cell. 1993;75:1061–1070. doi: 10.1016/0092-8674(93)90316-i. [DOI] [PubMed] [Google Scholar]
- Xiao SH, Manley JL. Phosphorylation of the ASF/SF2 RS domain affects both protein-protein and protein-RNA interactions and is necessary for splicing. Genes & Dev. 1997;11:334–344. doi: 10.1101/gad.11.3.334. [DOI] [PubMed] [Google Scholar]
- Zahler AM, Roth MB. Distinct functions of SR proteins in recruitment of U1 small nuclear ribonucleoprotein to alternative 5′ splice sites. Proc Natl Acad Sci. 1995;92:2642–2646. doi: 10.1073/pnas.92.7.2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahler AM, Lane WS, Stolk JA, Roth MB. SR proteins: A conserved family of pre-mRNA splicing factors. Genes & Dev. 1992;6:837–847. doi: 10.1101/gad.6.5.837. [DOI] [PubMed] [Google Scholar]
- Zahler AM, Neugenbauer KM, Lane WS, Roth MB. Distinct functions of SR proteins in alternative pre-mRNA splicing. Science. 1993;260:219–222. doi: 10.1126/science.8385799. [DOI] [PubMed] [Google Scholar]
- Zamore PD, Patton JG, Green MR. Cloning and domain structure of the mammalian splicing factor U2AF. Nature. 1992;355:609–614. doi: 10.1038/355609a0. [DOI] [PubMed] [Google Scholar]
- Zhang G, Taneja KL, Singer RH, Green MR. Localization of pre-mRNA splicing in mammalian nuclei. Nature. 1994;372:809–812. doi: 10.1038/372809a0. [DOI] [PubMed] [Google Scholar]
- Zhang M, Zamore PD, Carmo-Fonesca M, Lamond AI, Green MR. Cloning and intracellular localization of the U2 small nuclear ribonucleoprotein auxiliary factor small subunit. Proc Natl Acad Sci. 1992;89:8769–8773. doi: 10.1073/pnas.89.18.8769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang WJ, Wu JY. Functional properties of p54, a novel SR protein active in constitutive and alternative splicing. Mol Cell Biol. 1996;16:5400–5408. doi: 10.1128/mcb.16.10.5400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W, Manley JL. Complex alternative RNA processing generates an unexpected diversity of poly(A) polymerase isoforms. Mol Cell Biol. 1996;16:2378–2386. doi: 10.1128/mcb.16.5.2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo P, Manley JL. Functional domains of the human splicing factor ASF/SF2. EMBO J. 1993;12:4727–4737. doi: 10.1002/j.1460-2075.1993.tb06161.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo P, Maniatis T. The splicing factor U2AF35 mediates critical protein-protein interactions in constitutive and enhancer-dependent splicing. Genes & Dev. 1996;10:1356–1368. doi: 10.1101/gad.10.11.1356. [DOI] [PubMed] [Google Scholar]