Abstract
The activity of the stress-responsive σ factor, σE, is induced by the extracytoplasmic accumulation of misfolded or unfolded protein. The inner membrane protein RseA is the central regulatory molecule in this signal transduction cascade and acts as a σE-specific anti-σ factor. Here we show that σE activity is primarily determined by the ratio of RseA to σE. RseA is rapidly degraded in response to extracytoplasmic stress, leading to an increase in the free pool of σE and initiation of the stress response. We present evidence that the putative inner membrane serine protease, DegS, is responsible for this regulated degradation of RseA.
Keywords: σE, RseA, periplasm, stress response, proteolysis
The cellular response to stress is one of the most highly conserved regulatory responses among all organisms. The exposure of cells to stresses such as heat shock leads to the accumulation of partially and fully denatured proteins that interfere with normal cellular function. This accumulation of misfolded and unfolded protein leads to the transcriptional induction of a conserved set of proteins known as heat shock proteins (Bukau 1993; Yura et al. 1993; Becker and Craig 1994; Georgopoulos et al. 1994; Gross 1996). The heat shock proteins include chaperones, folding catalysts, and proteases that act to promote the refolding or degradation of misfolded proteins (Bukau 1993; Yura et al. 1993; Becker and Craig 1994; Georgopoulos et al. 1994; Gross 1996). Whereas the activities and transcriptional regulation of the heat shock proteins have been well characterized, comparatively little is known about the exact nature of the signal generated by stress and how this signal is conveyed to the transcriptional regulators of the stress response.
The hallmark of the Gram-negative bacterial cell is the existence of two membrane-bound subcellular compartments—the cytoplasm and the periplasm. Conditions in each of these compartments differ markedly. The cytoplasm is energy rich, reducing, and osmotically stable, whereas the periplasm lacks ATP, is oxidizing, and is in contact with the external milieu (Nikaido 1994). Because optimal cellular growth requires that the cell be able to sense and respond to changes in these disparate subcellular compartments, it is not surprising that the stress response in the Gram-negative bacterium Escherichia coli is compartmentalized into cytoplasmic and extracytoplasmic responses. The cytoplasmic response is governed by the alternate σ factor σ32 (Grossman et al. 1984; Landick et al. 1984; Yura et al. 1984), whereas the extracytoplasmic response is controlled by two partially overlapping signal transduction cascades, the σE and Cpx systems (Erickson and Gross 1989; Wang and Kaguni 1989; Mecsas et al. 1993; Danese et al. 1995; Connolly et al. 1997).
Cells are believed to sense stress by monitoring the protein folding pathways in various cellular compartments. This is often achieved by incorporating chaperones and/or proteases directly into the signal transduction cascade (McMillan et al. 1994). For example, the activity of σ32 is controlled by both chaperone-mediated inactivation and regulated proteolysis. The chaperones DnaK and DnaJ bind reversibly to σ32 to inhibit its function (Straus et al. 1989), and cellular stress is thought to be monitored by the competition between σ32 and misfolded or unfolded proteins for binding to this chaperone complex (Straus et al. 1990; Craig and Gross 1991; Bukau 1993). In addition, the regulated degradation of σ32 by the essential inner membrane protease HflB has a key role in the initial response to stress. Interaction with chaperones may be responsible for targeting σ32 for degradation by HflB (Tomoyasu et al. 1993, 1995; Herman et al. 1995). Release of σ32 from chaperone-mediated inactivation in combination with stabilization of the σ factor lead to an increase in both the activity and amount of σ32 in the cell in response to heat shock. Temperature itself also has a role in the regulation. Inhibitory secondary structures in σ32 mRNA are relieved by high temperature, thereby selectively increasing the translation of σ32 (Morita et al. 1999).
In contrast to the regulation of the σ32-mediated response, relatively little is known about the regulation of the σE-mediated stress response. Stresses that result in general protein unfolding (e.g., heat) activate both σ32 and σE (Grossman et al. 1984; Erickson et al. 1987; Straus et al. 1987; Erickson and Gross 1989; Wang and Kaguni 1989; Rouvière et al. 1995), whereas stresses that result in the specific accumulation of misfolded proteins in the periplasmic space (e.g., overexpression of outer membrane proteins), uniquely induce σE (Mecsas et al. 1993; Raina et al. 1995; Missiakas et al. 1996; Rouvière and Gross 1996). The mechanism by which the periplasmic accumulation of unfolded protein is conveyed across the inner membrane to σE is largely unknown. To date, two main regulators of σE have been identified. The inner membrane protein RseA is the central regulator of σE activity. Deletion of the gene encoding RseA leads to maximal induction of σE under nonstress conditions, and RseA is capable of signaling periplasmic stress in the absence of the other known regulator of σE (De Las Peñas et al. 1997b; Missiakas et al. 1997). The cytoplasmic face of RseA binds to σE and inhibits σE-directed transcription under nonstress conditions (De Las Peñas et al. 1997b; Missiakas et al. 1997). The periplasmic face of RseA binds to a second negative regulator of σE, RseB (De Las Peñas et al. 1997b; Missiakas et al. 1997). Deletion of the gene encoding RseB leads to only a slight (twofold) increase in σE activity, and it is thought that the interaction between RseB and RseA acts to fine-tune RseA activity (De Las Peñas et al. 1997b; Missiakas et al. 1997). The genes encoding σE, RseA, and RseB lie in a single operon and are transcribed primarily from a σE-dependent promoter (De Las Peñas et al. 1997b; Missiakas et al. 1997).
The central event in the initiation of the σE-mediated stress response is the inactivation of RseA. Here we show that σE activity is determined by the relative levels of σE and RseA in the cell and that RseA is regulated by controlled proteolysis. We present evidence that the putative inner membrane serine protease DegS is responsible for the regulated degradation of RseA.
Results
Changes in σE activity result from changes in the cellular levels of σE and RseA
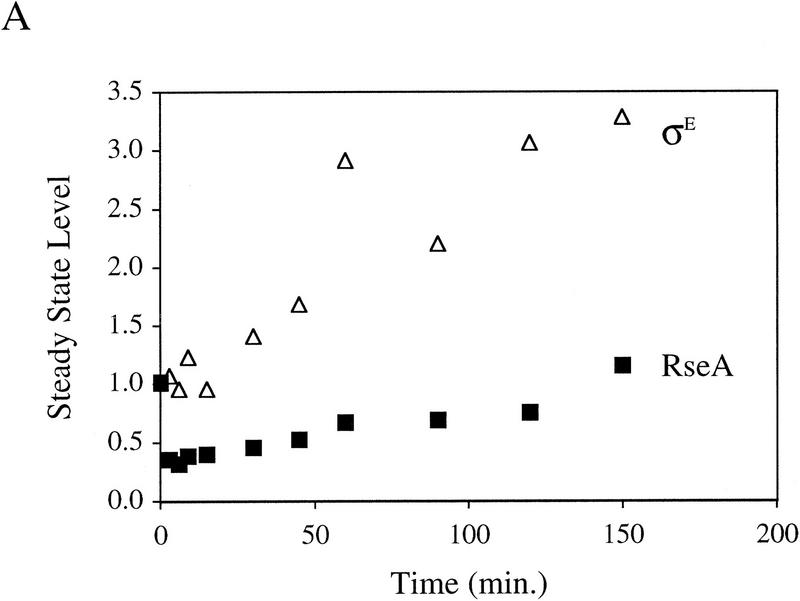
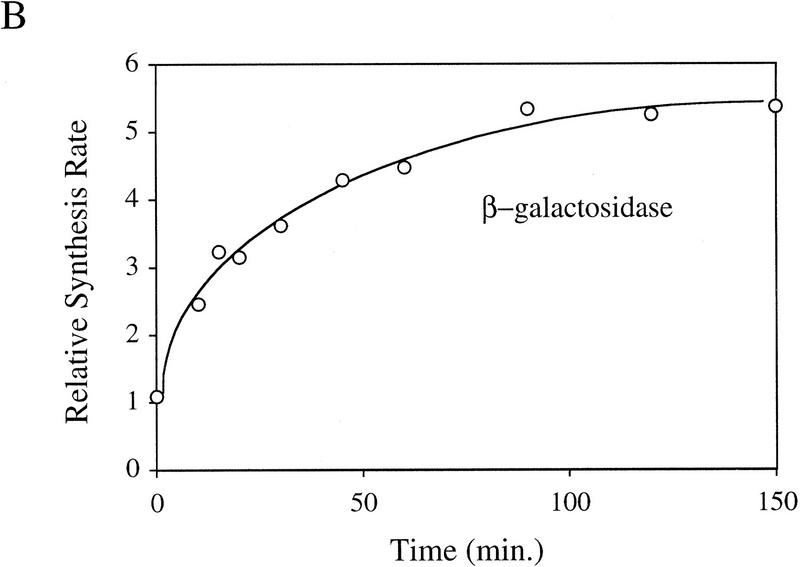
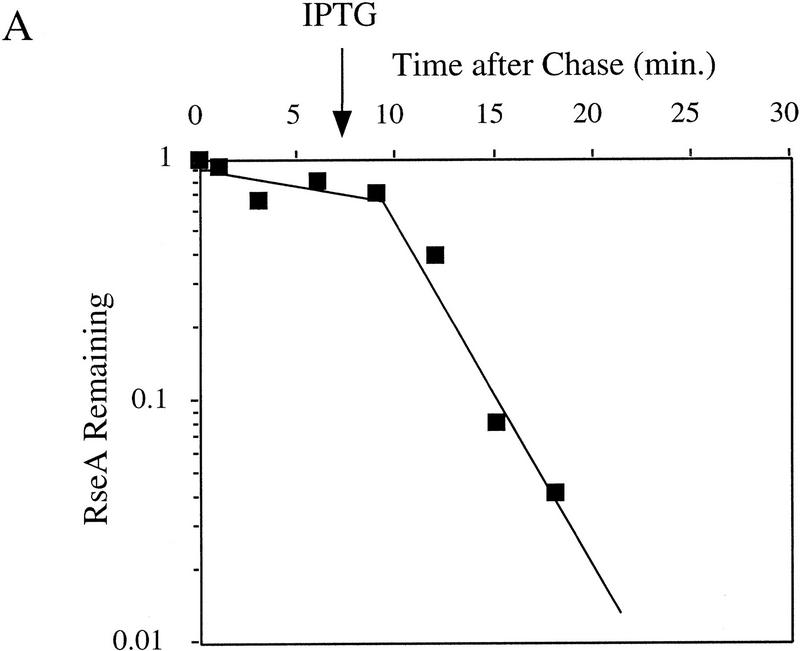
It is likely that the activity of σE is determined by the relative levels of σE and RseA in the cell because RseA binds directly to σE to inhibit the activity of the σ factor (De Las Peñas et al. 1997b; Missiakas et al. 1997). To test this idea, we used Western blot analysis to determine whether the levels of RseA or σE change upon induction of the stress response. The stress response was induced in these experiments by overexpression of the outer membrane protein OmpC (Mecsas et al. 1993). Within 3 min of initiating the stress response, the level of RseA drops ∼2.5-fold and then increases slowly over the next 150 min (Fig. 1A). In contrast, the level of σE increases steadily (Fig. 1A). In unstressed cells, the levels of RseA and σE remain constant over the time course of the experiment (data not shown). σE activity, as measured by the rate of synthesis of β-galactosidase from a single-copy σE-dependent lacZ reporter gene (Mecsas et al. 1993), correlates with these alterations in levels of RseA and σE over the course of the stress response. σE activity initially increases rapidly and then continues to increase more slowly for the first 60 min, eventually reaching rates 5- to 10-fold greater than those in unstressed cells (Fig. 1B; data not shown). The initial rapid increase in σE activity correlates with the immediate decrease in RseA levels following induction, and the slow rise that follows correlates with the increase in σE levels (Fig. 1, cf. A and B). These results show that both the decreased level of RseA and the increased level of σE contribute to the increase in σE activity observed following initiation of the stress response.
Figure 1.
Steady-state levels of σE and RseA, and EσE activity during the stress response. (A) Steady-state levels of RseA and σE during the extracytoplasmic stress response. Wild-type cells containing pompC (CAG45025) were grown to early-log phase and IPTG added at t = 0 to induce overexpression of OmpC. The levels of RseA (█) and σE (▵) were determined by Western blot analysis as described in Materials and Methods. The amount of each protein at a given time was normalized to the amount of that protein at time t = 0. A representative data set is shown. (B) EσE activity during the extracytoplasmic stress response. Wild-type cells containing pompC (CAG45025) were grown to early-log phase and IPTG was added at t = 0. EσE activity was determined at various time points by measuring the rate of synthesis of β-galactosidase (○) from a single-copy σE-dependent lacZ reporter gene in Φλ[rpoHP3–lacZ] using the pulse-labeling protocol described in Materials and Methods. The synthesis rate shown for β-galactosidase is normalized to the synthesis rate of the protein at t = 0. A representative data set is shown.
Induction of the stress response is not due to changes in the relative rates of synthesis of σE and RseA
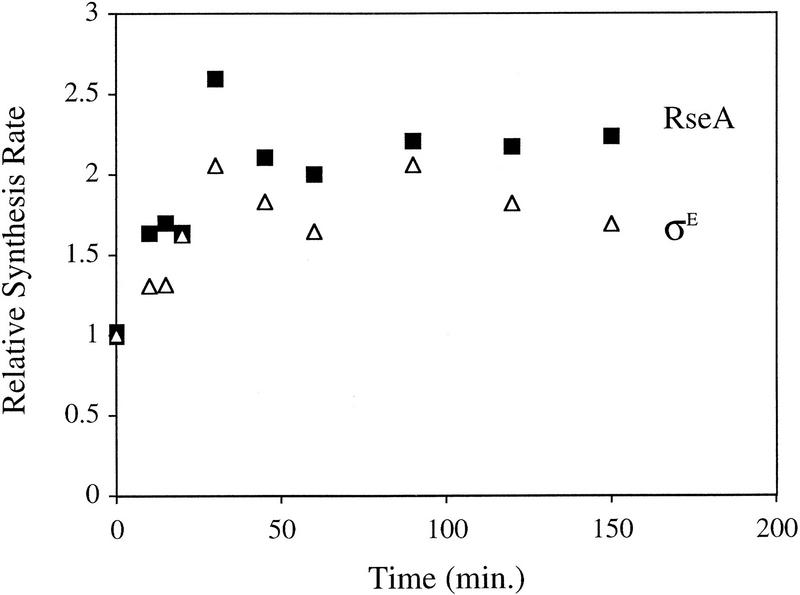
Changes in the cellular levels of RseA and σE after overexpression of OmpC could result either from altered rates of synthesis, degradation, or both. Given the very rapid decrease in RseA levels, it seems unlikely that alterations in synthesis alone account for this dramatic change. Examination of the rates of synthesis of σE and RseA by pulse-labeling immunoprecipitation revealed that the rates of synthesis of both proteins increase over the first 60 min after induction, with a 2- to 4-fold increase in RseA synthesis and a 1.5- to 4-fold increase in σE synthesis (Fig. 2; data not shown). Because the rates of synthesis of σE and RseA increase simultaneously, the change in their relative cellular levels following induction is not achieved by differential synthesis.
Figure 2.
Synthesis of σE and RseA during the extracytoplasmic stress response, induced in wild-type cells containing pompC (CAG45025) by the addition of IPTG at t = 0. The rates of synthesis of σE (▵) and RseA (█) at various time points after induction were determined by pulse labeling as described in Materials and Methods. The rate at each time point for a given protein was normalized to the initial rate for that protein at t = 0. A representative data set is shown.
Pre-existing and newly synthesized RseA are degraded rapidly under stress conditions
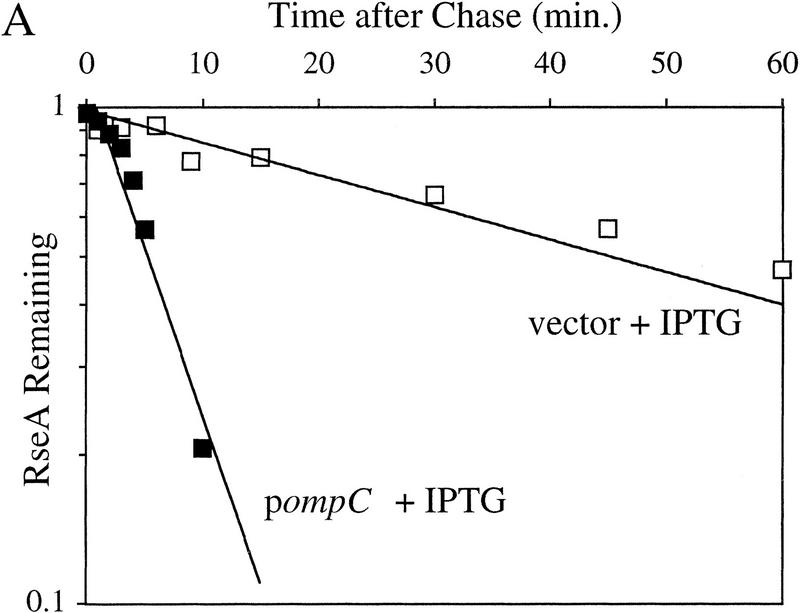
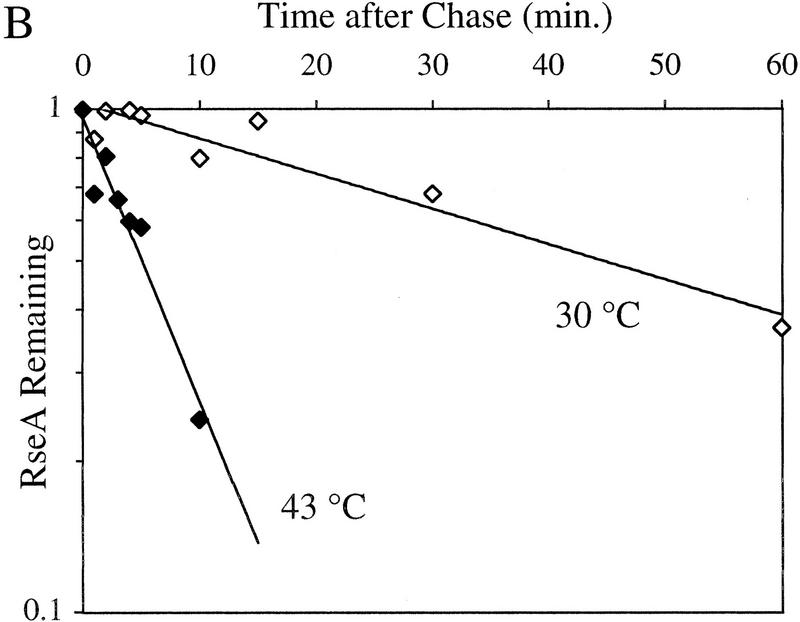
RseA is likely to be preferentially degraded in response to extracytoplasmic stress because changes in its rate of synthesis cannot explain the drop in cellular level of RseA observed following induction. To examine this possibility, we compared the half-life of RseA before and after initiation of the stress response. The stress response was initiated by either the overexpression of OmpC at 30°C or temperature upshift from 30°C to 43°C. The half-life of RseA in unstressed cells growing at 30°C is 44.3 ± 6.1 min (Fig. 3A; Table 1) and is dramatically reduced upon exposure to stress. Overexpression of OmpC leads to an 8.6-fold decrease in the half-life of RseA (t1/2 = 5.2 ± 2.7 min; Fig. 3A; Table 1). RseA continues to be degraded rapidly up to 150 min after the induction of the stress response (data not shown). Similarly, temperature upshift leads to a 14.3-fold decrease in the half-life of RseA (t1/2 = 3.1 ± 1.4 min; Fig. 3B; Table 1). These results show that RseA synthesized after initiation of the stress response is degraded rapidly.
Figure 3.
RseA stability under stress and nonstress conditions. (A) The stability of RseA synthesized after initiation of the stress response in cells overexpressing OmpC. Wild-type cells containing pompC (CAG45025, █) or vector alone (CAG45027, □) were grown to early log phase, and the stress response was initiated by the addition of IPTG to induce expression of OmpC. After 10 min of induction, the cells were pulse-labeled with [35S]methionine followed by a chase of cold methionine and cysteine. The stability of RseA was determined as described in Materials and Methods. A representative data set is shown. (B) The stability of RseA synthesized after temperature upshift. Wild-type cells (CAG16037) were grown to early log phase and either left at 30°C (⋄) or shifted to 43°C (♦). Five minutes after shift to high temperature the cells were pulse-labeled with [35S]methionine, followed by a chase of cold methionine and cysteine. The stability of RseA was determined as described in Materials and Methods. A representative data set is shown.
Table 1.
Half-lives of RseA under stress and nonstress conditions
| Relevant genotype
|
Growth condition
|
Half-life of RseAa (min)
|
|---|---|---|
| Wild type | no stress | 44.3 ± 6.1 |
| Wild type pompC | overproduction | 5.2 ± 2.7 |
| of OmpC | ||
| Wild type | 30°C → 43°C | 3.1 ± 1.4 |
| surA- | no stress | 8.9 ± 1.0 |
| ΔrseB | no stress | 18.6 ± 3.1 |
Half-lives were determined from a minimum of three independent experiments.
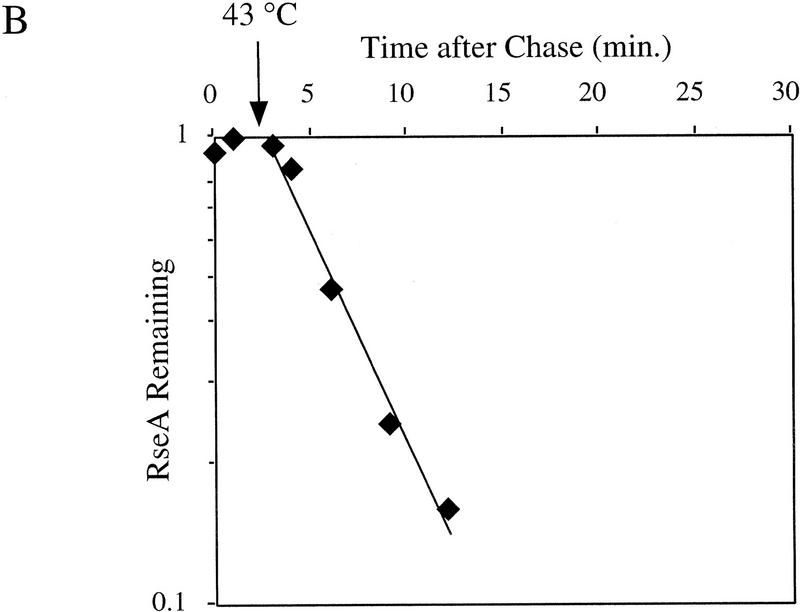
To determine whether pre-existing RseA is also degraded, we asked whether RseA that is labeled prior to initiation of the stress response is degraded rapidly following exposure to stress. Pre-existing RseA is degraded at approximately the same rate as newly synthesized RseA whether the stress response is induced by overexpression of OmpC or shift to high temperature (Fig. 4A,B). These results indicate that the immediate decrease in the cellular concentration of RseA following stress is a consequence of its rapid degradation. σE, on the other hand, is stable under nonstress conditions, and its stability remains unchanged after temperature upshift (data not shown). Therefore, increased cellular levels of σE following stress are a consequence of increased synthesis. In addition, these experiments allow us to estimate that the lag between induction of the stress response and degradation of RseA is <3 min.
Figure 4.
The stability of RseA synthesized prior to initiation of the stress response. Cells containing pompC (CAG45025, █) or wild-type cells (CAG16037, ♦) were grown to early log phase and pulse-labeled with [35S]methionine, followed by a chase of cold methionine and cysteine. At the indicated times, IPTG was added to induce overexpression of OmpC (A) or the cells were shifted from 30°C to 43°C (B). The stability of RseA was determined as described in Materials and Methods. A representative data set is shown.
RseA stability is altered under conditions of constitutive stress
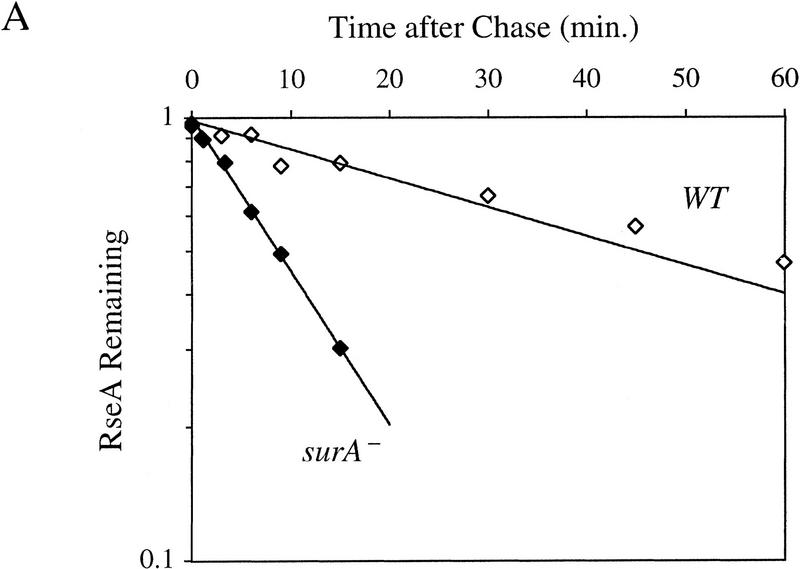
If RseA stability is the major determinant of σE activity in the cell, then the half-life of RseA might be decreased in cells in which σE activity is constitutive. Deletion of the gene encoding the periplasmic peptidyl prolyl isomerase SurA leads to a constitutive five- to sevenfold induction of σE activity (Missiakas et al. 1996; Rouvière and Gross 1996). RseA is unstable in surA− cells, with a half-life of 8.9 ± 1.0 min (Fig. 5A; Table 1). This 5.0-fold decrease in the half-life of RseA reflects the five- to sevenfold increase in σE activity observed in surA− strains (Missiakas et al. 1996; Rouvière and Gross 1996) and supports the idea that the regulated stability of RseA is the major mechanism for adjusting σE activity in the cell.
Figure 5.
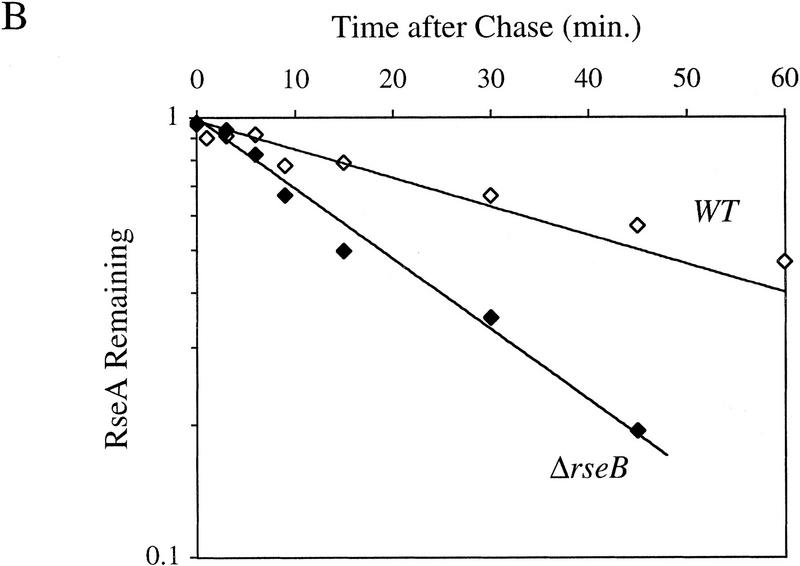
RseA stability under conditions of constitutive activation of σE. (A) RseA stability in cells lacking the periplasmic peptidyl prolyl isomerase SurA. surA− (CAG24029, ♦) or isogenic wild-type cells (CAG16037, ⋄) were grown to early log phase and then pulse-labeled with [35S]methionine, followed by a chase of cold methionine and cysteine. The stability of RseA was then determined as described in Materials and Methods. A representative data set is shown. (B) RseA stability in cells lacking RseB. ΔrseB cells (CAG22951, ♦) or isogenic wild-type cells (CAG16037, ⋄) were grown to early log phase and then pulse-labeled with [35S]methionine, followed by a chase of cold methionine and cysteine. The stability of RseA was then determined as described in Materials and Methods. A representative data set is shown.
RseB stabilizes RseA
It is likely that RseB exerts a negative regulatory effect on σE by modulating RseA activity through its interaction with the periplasmic domain of RseA. One possibility is that this interaction protects RseA from degradation. The half-life of RseA in ΔrseB cells is 18.6 ± 3.1 min (Fig. 5B; Table 1) or 2.4-fold lower than in wild-type cells. The magnitude of this effect is consistent with the twofold increase in σE activity observed in ΔrseB cells (De Las Peñas et al. 1997b; Missiakas et al. 1997). These results indicate that RseB stabilizes RseA under nonstress conditions.
DegS is required for wild-type σE activity
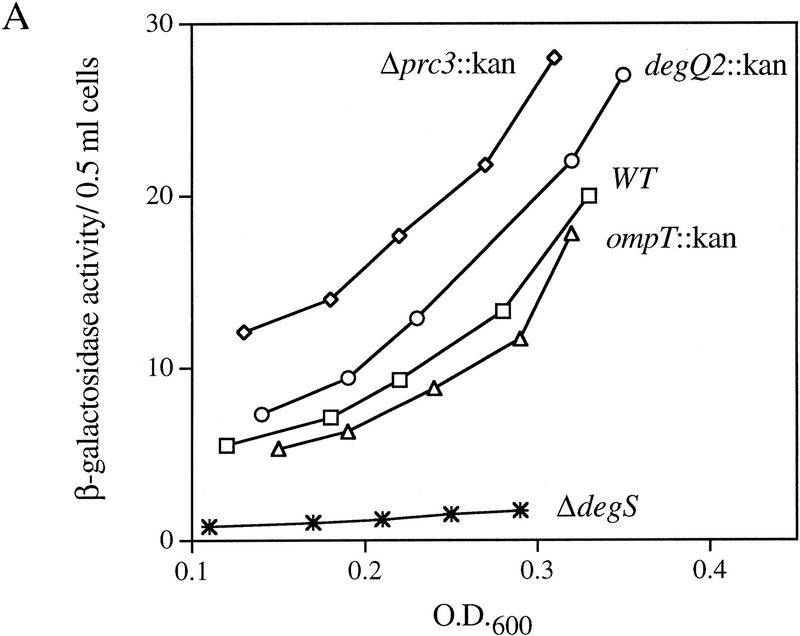
It is likely that an extracytoplasmic protease initiates the degradation of RseA because the signal initiating the σE-dependent response is generated in the periplasmic compartment. Loss of this protease should stabilize RseA, causing an increase in the intracellular concentration of RseA and a concomitant decrease in σE activity. Because RseA is degraded even under nonstress conditions, loss of the protease that degrades RseA should decrease the basal activity of σE. To identify the protease responsible for the degradation of RseA, we asked whether deletions of any of the known extracytoplasmic proteases altered σE activity under nonstress conditions. σE activity was determined by monitoring β-galactosidase expression from a single-copy σE-dependent lacZ reporter gene. Deletion of degP has been shown previously to have no effect on σE activity (Mecsas et al. 1993), and deletion of the genes encoding the Prc (Tsp), OmpT, and DegQ proteases similarly showed little or no effect on basal σE activity (Fig. 6A). In contrast, deletion of the gene encoding the putative protease DegS resulted in a fivefold decrease in basal σE activity in early log phase cells and eliminated the growth phase regulation that leads to a large increase in σE activity as cells approach stationary phase (Fig. 6A,B). These results show that DegS has a role in regulating the basal activity of σE.
Figure 6.
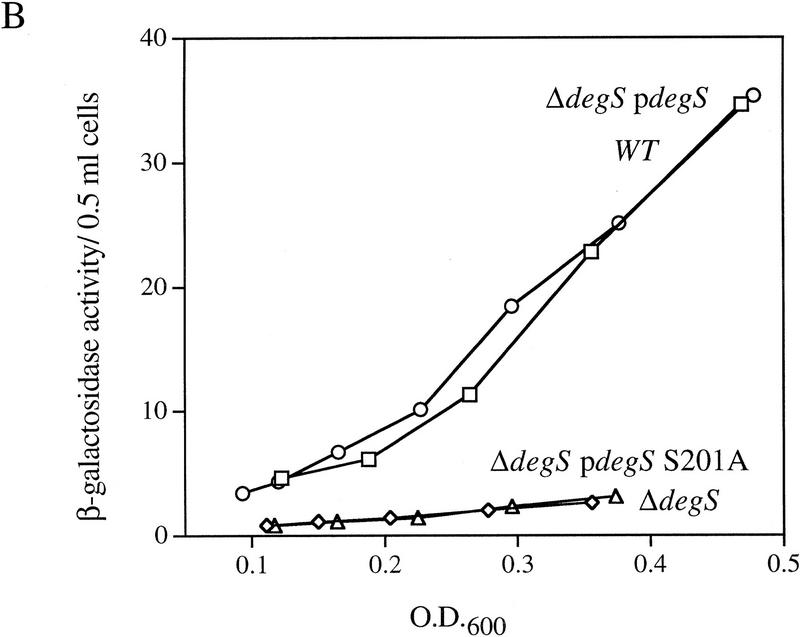
EσE activity in cells lacking various extracytoplasmic proteases. (A) EσE activity in strains lacking the extracytoplasmic proteases Prc (Tsp), DegQ, OmpT, or DegS. EσE activity was assayed by monitoring β-galactosidase activity produced from a single copy Φλ[rpoHP3–lacZ] fusion in wild-type (CAG16037, □), and the isogenic Δprc3::Km (CAG33314, ⋄), degQ2::Km (CAG33397, ○), ompT::Km (CAG33263, ▵), and ΔdegS (CAG33315, ✠ ) strains. β-Galactosidase assays were performed as described in Materials and Methods. A representative data set is shown. (B) EσE activity in cells lacking DegS. EσE activity was assayed in wild-type (CAG33385, □) and isogenic ΔdegS (CAG33391, ⋄) strains carrying vector alone as well as in ΔdegS cells carrying the plasmid pdegS (CAG33393, ○) or pdegS S201A (CAG33382, ▵). A representative data set is shown.
DegS is a putative inner membrane protein and shows a high degree of homology to the large family of HtrA serine proteases (Bass et al. 1996; Waller and Sauer 1996; Pallen and Wren 1997). Although it has not been shown formally to possess proteolytic activity, it is likely that DegS is a protease, as the sequences surrounding the predicted catalytic triad are highly conserved (Bass et al. 1996; Waller and Sauer 1996; Pallen and Wren 1997). To determine whether the putative proteolytic activity of DegS is required for σE activity, we asked whether an active-site mutant of degS could restore σE activity to cells lacking degS. Introduction of an alanine residue in place of the putative active-site serine of a plasmid-encoded degS gene resulted in a mutant protein that failed to complement the loss of σE activity observed in ΔdegS cells (Fig. 6B). In contrast, a plasmid encoding wild-type DegS restored σE activity to these cells (Fig. 6B). Western blot analysis confirmed that the mutant DegS protein accumulated to a level similar to that of the plasmid-expressed wild-type protein (data not shown), indicating that its inability to restore σE activity is not due to rapid degradation of the mutant protein. These results suggest that the putative proteolytic activity of DegS is required for σE activity under nonstress conditions but do not rule out the possibility that DegS may possess other important biochemical activities.
Cells lacking degS show an obvious slow growth phenotype, accumulate rapidly growing suppressors, and grow poorly in minimal medium (Waller and Sauer 1996; B.M. Alba, unpubl.). Because a plasmid encoding degS fully restores σE activity to ΔdegS cells, alterations in σE activity result from the ΔdegS allele rather than some other mutation present in these cells. However, the growth properties of ΔdegS strains render them exceedingly difficult to work with so we sought conditions under which the ΔdegS allele is stable. Because σE is an essential σ factor (De Las Peñas et al. 1997a) and degS is required for wild-type σE activity, we hypothesized that the instability of ΔdegS strains results from the fact that their low level of σE activity is barely sufficient for viability. A suppressor that allows cells lacking σE to grow has been identified previously (De Las Peñas et al. 1997a), and this suppressor might stabilize cells lacking degS by lowering their requirement for σE. ΔdegS cells containing this suppressor grow well in minimal media and do not readily accumulate additional rapidly growing suppressors (B.M. Alba, unpubl.). This result provides a background in which the ΔdegS allele is stable and supports the idea that DegS is involved in the regulation of σE. All subsequent experiments involving the ΔdegS allele were performed in sup strains.
DegS is required for activation of σE activity under stress conditions
The observation that DegS is required for σE activity under nonstress conditions supports a role for DegS in the basal degradation of RseA but does not indicate whether DegS is similarly involved in the regulated degradation of RseA in response to stress. To determine whether DegS is required for signal transduction to σE, we monitored σE activity in ΔdegS cells in response to overexpression of OmpC (Table 2). σE activity is induced in response to overexpression of OmpC in sup strains, indicating that the signal transduction pathway is intact in cells containing the suppressor. In contrast, isogenic ΔdegS cells show no induction of σE in response to overexpression of OmpC. A plasmid encoding wild-type DegS, but not the putative active site mutant, restores σE induction significantly to these cells. Cell fractionation experiments confirmed that OmpC is overexpressed and properly localized to the outer membrane in cells lacking DegS, arguing that the failure to induce σE is not due to aberrant sorting of the signal molecule in this genetic background (data not shown).
Table 2.
DegS is required for signal transduction to σE
| Strain
|
Relevant genotypea
|
Induction of OmpC
|
EσE activity
|
Fold induction
|
|---|---|---|---|---|
| CAG41073 | degS+ | − | 8 | |
| + | 306 | 38 | ||
| CAG43150 | ΔdegS | − | 2 | |
| + | 3 | 1.5 | ||
| CAG43133 | ΔdegS pdegS | − | 6 | |
| + | 64 | 11 | ||
| CAG43134 | ΔdegS pdegS S201A | − | 2 | |
| + | 1 | 0.5 | ||
| CAG43132 | ΔdegS pSU21 | − | 3 | |
| + | 1 | 0.3 | ||
| CAG43126 | degS+ pSU21 | − | 35 | |
| + | 202 | 6b |
EσE activity was measured by monitoring β-galactosidase activity from a single copy σE-dependent lacZ reporter gene in Φλ[rpoHP3–lacZ] as described in Materials and Methods. OmpC expression was induced by the addition of IPTG as indicated, and fold induction determined by calculating the ratio of β-galactosidase activity in the presence and absence of IPTG.
All strains carry the suppressor of ΔrpoE (sup) and the plasmid pompC.
The basal level of β-galactosidase activity is increased in this strain. This effect is dependent on the presence of pompC, the vector plasmid pSU21, and a chromosomal copy of degS. As a result the fold induction is lower in this strain than in strains lacking the vector plasmid pSU21 (see first row). Interaction between the two plasmids in the suppressor strain may also account for the lack of complete induction in the pdegS strain.
DegS is required for the transduction of periplasmic stress to σE
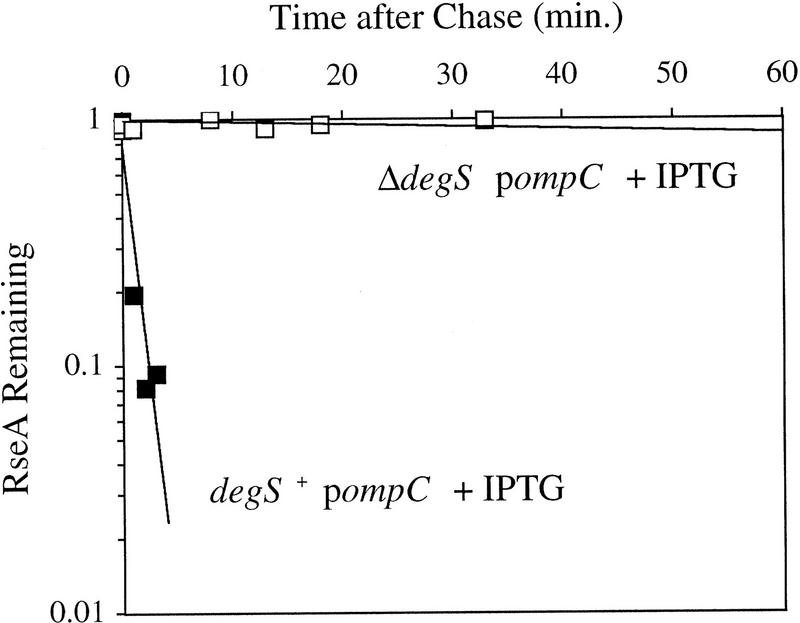
The above results show that DegS is required for signal transduction to σE but do not address the exact role of this protein in the signal transduction cascade. One possibility is that DegS has a role in regulating the stability of RseA and RseA is stabilized in cells lacking DegS. Consistent with this idea, RseA is stable under nonstress conditions and is not degraded in response to overexpression of OmpC in cells lacking DegS (Fig. 7). In contrast, isogenic cells possessing DegS show regulated proteolysis of RseA (Fig. 7). These results show that DegS is required for both the basal and regulated degradation of RseA.
Figure 7.
RseA stability in cells lacking DegS. sup degS+ (CAG41073, █) or sup ΔdegS (CAG43150, □) cells were grown to early log phase, and the stress response was initiated by the addition of IPTG to induce expression of OmpC. Five minutes after induction of the stress response the cells were pulse-labeled with [35S]methionine, followed by a chase of cold methionine and cysteine. The stability of RseA was determined as described in Materials and Methods. A representative data set is shown. The stability of RseA in sup-containing cells is greater than that in wild-type cells, consistent with the lower basal activity of σE in sup-containing cells (data not shown).
Discussion
In the Gram-negative bacterium E. coli, the extracytoplasmic accumulation of misfolded and unfolded protein generates a signal that is transmitted across the inner membrane to induce the activity of the stress-responsive σ factor σE. Induction of σE leads to the increased production of several periplasmic proteins, including the periplasmic protease DegP (Erickson and Gross 1989) and the periplasmic peptidyl-prolyl isomerase FkpA (Missiakas et al. 1996; Danese and Silhavy 1997), which help to maintain cellular integrity under stress conditions. The central regulatory molecule in this signal transduction cascade is the inner membrane anti-σ factor RseA. Here we show that RseA is degraded rapidly upon initiation of the stress response, leading to an increase in the free pool of σE and transcriptional induction of σE and its regulon. Furthermore, we present several lines of evidence that the regulated proteolysis of RseA is initiated by the recently described putative inner membrane protease DegS.
Mechanism of σE activation
Our experiments indicate that regulated proteolysis of RseA initiates the cascade of events that results in increased σE activity during the stress response. RseA is degraded rapidly following heat shock, a general inducer of the stress response, or overexpression of OmpC, a specific inducer of the extracytoplasmic stress response. This result implies either that both types of inducers generate the same signal or that the two pathways converge immediately prior to or at the level of RseA degradation.
Data presented here also indicate that the observed increase in σE activity after stress can be explained by an increase in the free pool of σE that is transcriptionally active because it is not complexed with RseA. According to this model, the majority of σE in the unstressed cell is present in an inactive RseA/σE complex. Immediately upon induction of the stress response, the pool of free σE increases rapidly due to loss of RseA. At later times, the free pool of σE is sustained by the increased synthesis of σE. Immediately following initiation of the stress response, the cellular level of RseA in stressed cells is two- to fourfold lower than in unstressed cells. This is a consequence of a ninefold increase in its rate of degradation coupled with a two- to fourfold increase in its rate of synthesis. Concomitantly, the cellular level of σE increases about threefold as a result of increased synthesis. Taken together, this leads to a 5- to 10-fold increase in the amount of free σE in the cell, consistent with the 5- to 10-fold increase in σE activity observed after induction of the stress response.
There may be additional levels of regulation. The rpoEP2 promoter, which controls the synthesis of σE and RseA shows lower induction than other σE-dependent promoters (Rouvière et al. 1995). This is true whether protein synthesis or RNA transcript level is measured (Rouvière et al. 1995, this work). Further studies will be necessary to determine whether poor induction of rpoEP2 is simply a consequence of its intrinsic strength or reflects additional levels of regulation at this promoter.
Mechanism of RseA degradation
It is likely that RseA is degraded from or initially cleaved at its periplasmic side because the signal responsible for activating the σE-mediated response is generated in the periplasm. No major degradation fragments of RseA have been detected in vivo, suggesting that degradation of the periplasmic domain also triggers proteolysis of the cytoplasmic domain of RseA. Previous work has shown that the isolated cytoplasmic domain of RseA acts as an effective anti-σ factor (De Las Peñas et al. 1997b; Missiakas et al. 1997), further supporting the notion that the entire protein needs to be degraded to induce the stress response. The observation that truncation of the periplasmic domain of the Pseudomonas aeruginosa RseA homolog MucA leads to constitutive activity of its cognate σ factor (Martin et al. 1993; Boucher et al. 1997) further supports the idea that clipping of the periplasmic domain is sufficient to inactivate this anti-σ factor, potentially by triggering proteolysis of the entire protein. It is possible that RseA is pulled from the membrane during proteolysis and degraded completely by a single protease. Alternatively, a second protease could degrade the cytoplasmic domain of RseA. Either the clipped fragment of RseA is a good target for cytoplasmic proteases or the activity of such a cytoplasmic protease must be regulated in concert with the activity of the protease responsible for the initial periplasmic clipping of RseA.
Cellular role of DegS
Data presented here indicate that DegS is required for both steady-state and regulated degradation of RseA because RseA is stable in cells lacking DegS under both stress and nonstress conditions. As a consequence, unstressed cells lacking DegS exhibit low σE activity, and σE can no longer be induced in response to extracytoplasmic stress. The simplest model explaining these observations is that DegS degrades RseA directly. Although DegS has not been shown formally to possess proteolytic activity, the finding that its putative active site serine is required for wild-type regulation of σE argues strongly that the role of DegS in this signal transduction cascade is proteolytic. Consistent with this idea, topology predictions suggest that the active site of DegS faces the periplasm, perfectly situating the catalytic triad to carry out the proposed initial cleavage of RseA that triggers the stress response. We cannot rule out the possibility that DegS affects RseA stability indirectly. For example, DegS could regulate the level of extracytoplasmic signal or the activity of a second protease. Unambiguous identification of the protease responsible for the regulated degradation of RseA awaits direct evidence of a functional interaction between such a protease and RseA.
Many of the well-studied cytoplasmic proteases carry dual regulatory and housekeeping functions (Gottesman 1996). In addition to the regulatory function identified here, DegS may also possess housekeeping functions. The identification of additional DegS substrates will help to further define the cellular role(s) of this protein.
Mechanism of action of RseB
Our results suggest a potential mechanism of action for RseB, an additional regulator of σE that binds to the periplasmic domain of RseA. RseB may act to block access of the protease to RseA, leading to stabilization of the anti-σ factor. In a manner similar to the titration of DnaK and DnaJ away from σ32 by the accumulation of misfolded proteins in the cytoplasm (Straus et al. 1990; Craig and Gross 1991; Bukau 1993), RseB may be titrated off of RseA by misfolded proteins accumulating in the periplasm, triggering the degradation of RseA and initiation of the σE-directed stress response. Because deleting RseB has only a small (twofold) effect on the stability of RseA, additional mechanisms must destabilize RseA upon induction. Identification of other molecules that interact with RseB and determination of the fate of RseB after initiation of the stress response will help to clarify the role of this protein in the signal transduction cascade.
Mechanism by which extracytoplasmic stress is sensed
The exact nature of the signal that initiates the extracytoplasmic stress response and how this signal results in the regulated degradation of RseA are unknown. Proteolysis of RseA is maximal by 3 min after overexpression of OmpC, at which time very little OmpC has accumulated in the periplasm. On the other hand, by 3 min, the flux of OmpC through the inner membrane is maximal as the export of secreted proteins is rapid, with a half-life of <30 sec (Randall and Hardy 1986). These observations suggest that at least one signal for the response is coupled to an event that happens during or immediately following transit of outer membrane proteins through the inner membrane. Overexpression of periplasmic proteins in general does not induce the σE-mediated stress response (Mecsas et al. 1993), arguing that titration of the secretion machinery is not the mechanism by which the response is initiated. Studies of the outer membrane protein folding pathway in cells lacking the periplasmic peptidyl-prolyl isomerase SurA suggest that the signal is generated after translocation and prior to formation of the folded monomer species (Rouvière and Gross 1996). Further elucidation of the outer membrane protein folding pathway will help to identify additional steps in this process that might be monitored by the extracytoplasmic stress response.
The signal leading to RseA degradation could be sensed directly by RseA or the protease responsible for RseA degradation. Interaction of the signal molecule with RseA could render the protein susceptible to proteolysis by modification, conformational change, or a change in oligomeric state. For example, overexpression of outer membrane proteins might lead to an accumulation of unfolded precursors exiting the inner membrane, and these unfolded species might interact directly with RseA to promote its degradation. Alternatively, or in addition to interacting directly with RseA, these unfolded signal molecules could also activate the protease directly as they transit the membrane.
Rather than being sensed directly by RseA or the protease, the signal leading to RseA degradation could be sensed indirectly. In a manner similar to the σ32-mediated cytoplasmic response, cells may indirectly monitor extracytoplasmic protein folding and the accumulation of misfolded molecules by sensing the levels of free periplasmic folding agents such as SurA, FkpA, and PpiD, as well as RseB. These proteins could be titrated off of RseA by the accumulation of misfolded substrates, rendering RseA susceptible to degradation. Alternatively, or in addition to interacting with RseA, these folding proteins might bind to the protease responsible for RseA degradation, inhibiting its proteolytic activity under nonstress conditions. According to this model, titration of these folding molecules by misfolded substrates would then lead to activation of the protease.
These titration models accommodate the observed rapidity of the response and allow for the response to be varied under different conditions. Periplasmic folding agents are likely to interact with outer membrane proteins as they leave the inner membrane to traffic them properly. Those protein-folding agents bound to RseA or the protease could be in spatial proximity to the exiting outer membrane proteins and interact with them preferentially, leading to the rapid destabilization of RseA. If several proteins must be titrated, a variable response could be generated. The fact that deletion of the genes encoding SurA, FkpA, PpiD, and RseB leads to an induction of σE activity is consistent with their involvement in the signal transduction cascade and the observation that the effects of double mutants in several of these proteins are additive further supports a titration model (Raina et al. 1995; Dartigalongue and Raina 1998). Identification of molecules interacting with RseA or DegS under nonstress and stress conditions will help to differentiate among the various models of signal transduction.
Implications for related signal transduction pathways
σE belongs to a family of σ70-like factors known as extracytoplasmic function (ECF) σ, whose cellular functions are related to extracytoplasmic processes (Lonetto et al. 1994), and homologs of the σE operon are found in many other Gram-negative bacteria, including several human pathogens. Salmonella typhimurium lacking σE are more susceptible to antimicrobial peptides and have reduced ability to colonize host organs (Humphreys et al. 1999), suggesting that the σE regulon plays a role in bacterial survival in the host. In P. aeruginosa, the σE homolog AlgU transcribes genes necessary for the production of the exopolysaccharide alginate and is negatively regulated by the Rse homologs MucA and MucB (Martinez-Salazar et al. 1996; Schurr et al. 1996; Xie et al. 1996). Chronic P. aeruginosa lung infection in patients with cystic fibrosis is associated with the development of a mucoid bacterial phenotype that is caused by overproduction of alginate. Interestingly, MucA is mutationally inactivated in the majority of isolates obtained from the lungs of these patients (Martin et al. 1993; Boucher et al. 1997). The resulting high activity of AlgU leads to very high levels of alginate and the mucoid phenotype, which has been proposed to protect P. aeruginosa from both exogenous antibiotics and host immune defenses (Anwar et al. 1992; Deretic et al. 1994). Given the similarities between the σE and AlgU operons and what is known about their regulation, it is likely that the signal transduction pathways are conserved. Light-induced carotenogenesis in Myxococcus xanthus is controlled by the ECF σ CarQ, whose activity, in turn, appears to be negatively regulated by the inner membrane protein CarR (Gorham et al. 1996). The finding that a CarR–LacZ fusion protein is detectable only in dark-grown and not light-grown cells suggests that, like RseA, the activity of CarR might be regulated at the level of stability (Gorham et al. 1996). This observation is particularly intriguing as RseA and CarR share no obvious sequence or structural similarities and suggests that proteolysis may be a common mechanism for regulating the activity of ECF anti-σ factors.
Materials and methods
Media, reagents, and enzymes
Luria–Bertani (LB) and M9 minimal medium were prepared as described (Sambrook et al. 1989). M9 was supplemented with 0.2% glucose, 1 mm MgSO4, 2 μg/ml thiamine, and all amino acids (40 μg/ml), except for media used in pulse-labeling experiments, in which methionine and cysteine were not added. In addition, ΔdegS cells were grown in M9 titrated to pH 7.5 with Tris base. Where needed, medium was supplemented with 30 μg/ml kanamycin (Km), 20 μg/ml chloramphenicol (Cm), and/or 100 μg/ml ampicillin (Ap).
Strains
Bacterial strains used in this study are described in Table 3. Their construction is described briefly below.
Table 3.
Strains and plasmids used in this study
| Strain/plasmid
|
Relevant genotype
|
Source/reference
|
|---|---|---|
| Strain | ||
| BL21(DE3) | hsdS gal ompTrB−mB− (DE3 PlacUV5::T7 polymerase) | Studier and Moffatt (1986) |
| MC1061 | araD Δ(ara–leu)7697 ΔlacX74 galU galK hsr hsn strA | Casadaban and Cohen (1980) |
| CAG16037 | MC1061 Φλ[rpoH P3::lacZ] | Mecsas et al. (1993) |
| CAG22951 | 16037 ΔrseB nadB-3140::Tn10, TcR | De Las Peñas et al. (1997b) |
| CAG24029 | MC1061 surA::Tn10dCm, CmR | Rouvière and Gross (1996) |
| CAG33149 | BL21(DE3) pLC234, KmR | DeLas Peñas et al. (1997b) |
| PW149 | MC1061 ΔdegS pPW142 | Waller and Sauer (1996) |
| CAG33304 | PW149 Φλ[rpoHP3::lacZ], CmR | this work |
| CAG33263 | 16037 ompT::Km, KmR | this work |
| CAG33314 | 16037 Δprc3::Km, KmR | this work |
| CAG33315 | MC1061 Φλ[rpoH P3::lacZ] ΔdegS | this work |
| CAG33382 | 33315 pPLT13 pLC261, KmR CmR | this work |
| CAG33385 | 16037 pPLT13 pSU21, KmR CmR | this work |
| CAG33391 | 33315 pPLT13 pSU21, KmR CmR | this work |
| CAG33393 | 33315 pPLT13 pLC259, KmR CmR | this work |
| CAG33397 | MC1061 Φλ[rpoH P3::lacZ] degQ::Km, KmR | this work |
| CAG41073 | 16037 sup pPLT13 pEMC1, KmR ApR | this work |
| CAG43097 | 33315 argR::trimethoprim, TriR | this work |
| CAG43126 | 41073 pSU21, ApR KmR TriR CmR | this work |
| CAG43132 | 43150 pSU21, ApR KmR TriR CmR | this work |
| CAG43133 | 43150 pLC259, ApR KmR TriR CmR | this work |
| CAG43134 | 43150 pLC261, ApR KmR TriR CmR | this work |
| CAG43150 | 41073 ΔdegS argR::trimethoprim, ApR KmR TriR | this work |
| CAG45025 | 16037 pPLT13 pEMC1, KmR ApR | this work |
| CAG45027 | 16037 pPLT13 pKK223-3, KmR ApR | this work |
| Plasmid | ||
| pKK223-3 | IPTG-inducible expression vector with tac promoter, ApR | Amersham Pharmacia Biotechnology |
| pEMC1a | ompC in pKK223-3, ApR | Catron and Schnaitman (1987) |
| pLC234 | periplasmic domain of rseA fused to an amino-terminal 6-histidine tag in pET28b, KmR | De Las Peñas et al. (1997b) |
| pSU21 | cloning vector, p15A ori, lac promoter, CmR | Bartolomé et al. (1991) |
| pLC259b | degS and degS promoter in pSU21, CmR | this work |
| pLC261c | degS S201A and degS promoter in pSU21, CmR | this work |
| pPW142 | degS in pMAK705, CmR | Waller and Sauer (1996) |
| pPLT13 | mini-F carrying lacIq, KmR | Tavormina et al. (1996) |
Referred to as pompC in text.
Referred to as pdegS in text.
Referred to as pdegS S201A in text.
ompT::Km (Akiyama and Ito 1996) and Δprc3::Km (Silber and Sauer 1994) alleles were moved by P1 transduction into strain CAG16037 as described previously (Miller 1972). The degQ2::Km strain carrying the σE-dependent reporter gene (CAG33397) was created by lysogenizing PW152 (Waller and Sauer 1996) with Φλ[rpoHP3–lacZ] (Mecsas et al. 1993). A ΔdegS strain (CAG33315) carrying the σE-dependent reporter gene Φλ[rpoHP3–lacZ] was constructed in two steps. PW149 (Waller and Sauer 1996) was first lysogenized with Φλ[rpoHP3–lacZ], creating CAG33304. CAG33304 was then cured of the degS-encoding plasmid pPW142 (Waller and Sauer 1996), which contains a temperature-sensitive origin of replication by the following procedure: CAG33304 was grown overnight at 37°C in LB and the resulting culture plated on LB at 30°C for single colonies. Single colonies were then screened for plasmid loss (Cm sensitivity) at 30°C on LB Cm plates. Cm-sensitive colonies were then screened by PCR to confirm loss of the wild-type allele of degS. The sup pompC ΔdegS strain (CAG43150) was constructed as follows: The ΔdegS allele of CAG33315 was tightly (∼90%) linked to argR::trimethoprim (Tian and Maas 1994) through P1 transduction, resulting in CAG43097. Next, sup pompC cells (CAG41073) were transduced with a CAG43097 P1 lysate, and trimethoprim-resistant transductants were selected on M9 minimal media plates lacking l-arginine and supplemented with 5 μg/ml trimethoprim. Colonies were screened by PCR to verify cotransduction of the ΔdegS allele.
Plasmids
Plasmids used in this study are listed in Table 3. pEMC1 (Catron and Schnaitman 1987) contains the ompC gene cloned into the expression vector pKK223-3 (Amersham Pharmacia Biotech). pLC259 was created by PCR amplification of the degS gene and its promoter from MC1061 chromosomal DNA using Pfu polymerase and the primers DEGS74 (5′-GCTCTAGATGTCGTAAACCGGGCATCAGG) and DEGS75 (5′-GGGGTACCGAGCGCACGACTTAATTGGTTG). The resulting fragments were then digested at restriction sites XbaI and KpnI (indicated by underlining) and cloned into the corresponding sites of the general cloning vector pSU21 (Bartolomé et al. 1991). pLC261 was constructed by creating a point mutation at codon 201 of degS, altering serine 201 to alanine. Primer pairs DEGS78 (5′-CCACGGTAACGCTGGCGGCGC)/DEGS69 (5′-CAGTTGATCTATACCACCGC) and DEGS79 (5′-GCGCCGCCAGCGTTACCGTGG)/DEGS68 (5′-TTGACAGTACCGATGAGACG) were amplified from pLC259, creating two separate PCR fragments containing the point mutation. These fragments were then joined by PCR using primers DEGS69 and DEGS68. A 370-bp BamHI–BssHII fragment containing the point mutation was then subcloned from the joined PCR fragment back into pLC259 to create pLC261. The point mutation creates a MwoI restriction site, and the presence of the mutation was confirmed by restriction digest. The sequences of degS in pLC259 and pLC261 were confirmed by DNA sequencing.
Western blot detection of σE and RseA
Cells were grown in M9 minimal media at 30°C and 1-ml samples taken in duplicate at various time points. Proteins were precipitated by the addition of TCA to a final concentration of 5% and then resuspended directly in Laemmli sample buffer. Samples were loaded onto Tris-glycine SDS gels such that extracts from equal numbers of cells were loaded in each lane. Western transfer was done according to standard methods (Harlow and Lane 1988). The blots were probed with 1:10,000 dilutions of polyclonal antibodies to the following: σE, the periplasmic domain of RseA, the cytoplasmic domain of RseA, and maltose binding protein (MBP). Anti-rabbit immunoglobulin radiolabeled with 35S (Amersham Pharmacia Biotech) was used as the secondary antibody. Bands were visualized using the Molecular Dynamics Storm 560 PhosphorImager scanning system, and the intensity of the bands quantified using the program ImageQuant 1.2. Band intensities of duplicate samples were averaged. To control for loading inconsistencies, this average was then normalized to the intensity of MBP in those lanes.
Determination of the half-life of RseA by pulse-chase-immunoprecipitation
All strains to be assayed were grown at 30°C to an OD450 of 0.15–0.3 in M9 minimal media lacking methionine and cysteine except ΔdegS strains, which were grown in M9 (pH 7.5) lacking methionine and cysteine. Cells were pulse-labeled for 1 min with EasyTag Expre35S35S protein labeling mix (NEN Life Sciences Products). An 800-μl sample was then removed and a cold chase of methionine and cysteine was added to a final concentration of 0.1%. Samples of 900 μl were removed at indicated times after the chase. To initiate the extracytoplasmic stress response at the appropriate time either before or after addition of the protein labeling mix, the temperature was shifted to 43°C or ompC expression was induced by the addition of IPTG to 2 mm. All samples were added to 100 μl of ice-cold 50% TCA and incubated on ice for >15 min. Samples were centrifuged to pellet the precipitated proteins and pellets were resuspended by vortexing and boiling for 5 min in 50 μl of 50 mm Tris-HCl (pH 7.5), 2% SDS, 1 mm PMSF, and 10 mm EDTA. The amount of 750 μl of RIPA buffer (50 mm Tris at pH 7.5, 150 mm NaCl, 1% sodium deoxycholate, 1% Triton X-100, 0.1% SDS) was then added to the samples, and a 5-μl aliquot of each was counted in a scintillation counter. To normalize the samples to the total amount of protein, equal numbers of counts per minute were immunoprecipitated in a total volume of 500 μl using 2 μl of a polyclonal antibody raised against the periplasmic domain of RseA and 25 μl of a 1:1 suspension of protein A-conjugated Sepharose beads in RIPA buffer. As an internal control, an aliquot of an extract derived from pulse-labeled cells overexpressing the periplasmic domain of RseA (strain CAG33149) was added to each sample prior to immunoprecipitation. The samples were rocked at 4°C for 1–3 hr, and the beads and associated immune complexes were harvested by centrifugation and washed three times with 900 μl of RIPA buffer. Immunoprecipitated proteins were eluted from the beads by the addition of 30 μl of Laemmli sample buffer and boiling for 5 min. The entire sample was then loaded onto 15% SDS Tris-glycine gels and immunoprecipitated proteins were visualized with the Molecular Dynamics Storm 560 PhosphorImager scanning system. The intensity of the band corresponding to full-length RseA protein was normalized to the intensity of the RseA periplasmic domain standard in each lane after background correction using the program ImageQuant 1.2. Half-lives were determined by fitting to the exponential decay equation. Experiments were conducted a minimum of two times each.
Determination of synthesis rates by pulse-labeling immunoprecipitation
Cells to be assayed were grown at 30°C in M9 minimal media lacking methionine and cysteine to OD450 of 0.15–0.3. At various times, 1 ml of the growing culture was added to EasyTag Expre35S35S protein labeling mix and incubated for 1 min. A 30-sec chase was then done with the addition of 100 μl of a 1% solution of cold methionine and cysteine to ensure the completion of protein synthesis. Ice-cold 50% TCA (100 μl) was added and samples were incubated on ice for >15 min, processed, and immunoprecipitated using polyclonal antisera directed against σE or RseA, or monoclonal antibody against β-galactosidase (5 Prime → 3 Prime, Inc.) as described above for the determination of the half-life of RseA. Equal numbers of counts per minute from each sample were immunoprecipiated to control for changes in cell density over the growth phase. Synthesis rate experiments were repeated a minimum of two times each.
β-Galactosidase assays
σE activity was assayed by monitoring β-galactosidase activity from a chromosomal σE-dependent lacZ reporter gene in Φλ[rpoHP3–lacZ] as described previously (Miller 1972; Mecsas et al. 1993). Cells to be assayed were grown at 30°C in LB medium and single point determinations were made at the indicated optical densities. Because σE activity and thus the rate of β-galactosidase synthesis from the σE-dependent reporter gene increases as the cells enter mid-log phase when grown in LB (J. Mecsas and C.A. Gross, unpubl.), fold effects were determined by comparing the slopes of the initial linear portion of the plots of β-galactosidase activity/0.5 ml cells versus OD600. Slopes were determined by linear regression analysis.
Acknowledgments
We thank Eric Roche and Bob Sauer for sharing strains and plasmids and communicating unpublished results. We also thank Christina Onufryk for communicating unpublished results, Werner Maas for sharing strains, and Ken Keiler, Christophe Herman, Jon Tupy and José de la Torre for critical reading of the manuscript. This work was supported by U.S. Public Health Service Grant GM36278 from the National Institute of Health, Training Grant T32CA09043 from the National Cancer Institute, and National Science Foundation Minority Graduate Fellowship NSF-99/404908-21320 awarded to B.M.A.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked ‘advertisement’ in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL cgross@cgl.ucsf.edu; FAX (415) 476-4204.
References
- Akiyama Y, Ito K. SecY protein, a membrane embedded secretion factor of E. coli, is cleaved by the OmpT protease in vitro. Biochem Biophys Res Commun. 1996;167:711–715. doi: 10.1016/0006-291x(90)92083-c. [DOI] [PubMed] [Google Scholar]
- Anwar H, Strap JL, Costeron JW. Establishment of aging biofilms: Possible mechanism of bacterial resistance to antimicrobial therapy. Antimicrob Agents Chemother. 1992;36:1347–1351. doi: 10.1128/aac.36.7.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartolomé B, Jubete Y, Martinez E, de la Cruz F. Construction and properties of a family of pACYC184-derived cloning vectors compatible with pBR322 and its derivatives. Gene. 1991;102:75–78. doi: 10.1016/0378-1119(91)90541-i. [DOI] [PubMed] [Google Scholar]
- Bass S, Gu Q, Christen A. Multicopy suppressors of prc mutant Escherichia coli include two HtrA (DegP) protease homologs (HhoAB), DksA, and a truncated RlpA. J Bacteriol. 1996;178:1154–1161. doi: 10.1128/jb.178.4.1154-1161.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker J, Craig EA. Heat-shock proteins as molecular chaperones. Eur J Biochem. 1994;219:11–23. doi: 10.1007/978-3-642-79502-2_2. [DOI] [PubMed] [Google Scholar]
- Boucher JC, Yu H, Mudd MH, Deretic V. Mucoid Pseudomonas aeruginosa in cystic fibrosis: Characterization of muc mutations in clinical isolates and analysis of clearance in a mouse model of respiratory infection. Infect Immun. 1997;65:3838–3846. doi: 10.1128/iai.65.9.3838-3846.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukau B. Regulation of the Escherichia coli heat-shock response. Mol Microbiol. 1993;9:671–680. doi: 10.1111/j.1365-2958.1993.tb01727.x. [DOI] [PubMed] [Google Scholar]
- Casadaban MJ, Cohen SN. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J Mol Biol. 1980;138:179–207. doi: 10.1016/0022-2836(80)90283-1. [DOI] [PubMed] [Google Scholar]
- Catron KM, Schnaitman CA. Export of protein in Escherichia coli: A novel mutation in ompC affects expression of other major membrane proteins. J Bacteriol. 1987;169:4327–4334. doi: 10.1128/jb.169.9.4327-4334.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly L, De Las Peñas A, Alba BM, Gross CA. The response to extracytoplasmic stress in Escherichia coli is controlled by partially overlapping pathways. Genes & Dev. 1997;11:2012–2021. doi: 10.1101/gad.11.15.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig AE, Gross CA. Hsp70-the cellular thermometer? Trends Biochem Sci. 1991;16:135–140. doi: 10.1016/0968-0004(91)90055-z. [DOI] [PubMed] [Google Scholar]
- Danese P, Silhavy T. The σE and Cpx signal transduction systems control the synthesis of periplasmic protein-folding enzymes in Escherichia coli. Genes & Dev. 1997;11:1183–1193. doi: 10.1101/gad.11.9.1183. [DOI] [PubMed] [Google Scholar]
- Danese PN, Snyder WB, Cosma CL, Davis LJ, Silhavy TJ. The Cpx two-component signal transduction pathway of Escherichia coli regulates transcription of the gene specifying the stress-inducible periplasmic protease, DegP. Genes & Dev. 1995;9:387–398. doi: 10.1101/gad.9.4.387. [DOI] [PubMed] [Google Scholar]
- Dartigalongue C, Raina S. A new heat shock gene, ppiD, encodes a peptidyl-prolyl isomerase required for folding of outer membrane proteins in Escherichia coli. EMBO J. 1998;17:3968–3980. doi: 10.1093/emboj/17.14.3968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Las Peñas A, Connolly L, Gross CA. σE is an essential sigma factor in Escherichia coli. J Bacteriol. 1997a;179:6862–6864. doi: 10.1128/jb.179.21.6862-6864.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— The σE-mediated response to extracytoplasmic stress in Escherichia coli is transduced by RseA and RseB, two negative regulators of σE. Mol Microbiol. 1997b;24:373–386. doi: 10.1046/j.1365-2958.1997.3611718.x. [DOI] [PubMed] [Google Scholar]
- Deretic V, Schurr MJ, Boucher JC, Martin DW. Conversion of Pseudomonas aeruginosa to mucoidy in cystic fibrosis: Environmental stress and regulation of bacterial virulence by alternative sigma factors. J Bacteriol. 1994;176:2773–2780. doi: 10.1128/jb.176.10.2773-2780.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson JW, Gross CA. Identification of the σE subunit of Escherichia coli RNA polymerase: A second alternate σ factor involved in high-temperature gene expression. Genes & Dev. 1989;3:1462–1471. doi: 10.1101/gad.3.9.1462. [DOI] [PubMed] [Google Scholar]
- Erickson JW, Vaughn V, Walter WA, Neidhardt FC, Gross CA. Regulation of the promoters and transcripts of rpoH, the Escherichia coli heat-shock regulatory gene. Genes & Dev. 1987;1:419–432. doi: 10.1101/gad.1.5.419. [DOI] [PubMed] [Google Scholar]
- Georgopoulos C, Liberek K, Zylicz M, Ang D. Properties of the heat shock proteins of Escherichia coli and the autoregulation of the heat shock response. In: Morimoto R, Tissières A, Georgopoulos C, editors. The biology of heat shock proteins and molecular chaperones. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1994. pp. 209–249. [Google Scholar]
- Gorham HC, McGowan SJ, Robson PRH, Hodgson DA. Light-induced carotenogenesis in Myxococcus xanthus: Light-dependent membrane sequestration of ECF sigma factor CarQ by anti-sigma factor CarR. Mol Microbiol. 1996;19:171–186. doi: 10.1046/j.1365-2958.1996.360888.x. [DOI] [PubMed] [Google Scholar]
- Gottesman S. Proteases and their targets in Escherichia coli. Annu Rev Genet. 1996;30:465–506. doi: 10.1146/annurev.genet.30.1.465. [DOI] [PubMed] [Google Scholar]
- Gross CA. Function and regulation of the heat shock proteins. In: Neidhardt FC, et al., editors. Escherichia coli and Salmonella: Cellular and molecular biology. Vol. 1. Washington, D.C.: ASM Press; 1996. pp. 1382–1399. [Google Scholar]
- Grossman AD, Erickson JW, Gross CA. The htpR gene product of E. coli is a sigma factor for heat-shock promoters. Cell. 1984;38:383–390. doi: 10.1016/0092-8674(84)90493-8. [DOI] [PubMed] [Google Scholar]
- Harlow E, Lane D. Antibodies: A laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1988. pp. 471–510. [Google Scholar]
- Herman C, Thevenet D, D’Ari R, Bouloc P. Degradation of σ32, the heat shock regulator in Escherichia coli during growth at different temperatures. Proc Natl Acad Sci. 1995;92:3516–3520. doi: 10.1073/pnas.92.8.3516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphreys S, Stevenson A, Bacon A, Weinhardt AB, Roberts M. The alternative sigma factor, σE, is critically important for the virulence of Salmonella typhimurium. Infect Immun. 1999;67:1560–1568. doi: 10.1128/iai.67.4.1560-1568.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landick R, Vaughn V, Lau ET, VanBogelen RA, Erickson JW, Neidhardt FC. Nucleotide sequence of the heat shock regulatory gene of E. coli suggests its protein product may be a transcription factor. Cell. 1984;38:175–182. doi: 10.1016/0092-8674(84)90538-5. [DOI] [PubMed] [Google Scholar]
- Lonetto MA, Brown KL, Rudd KE, Buttner MJ. Analysis of the Streptomyces coelicolor sigE gene reveals the existence of a subfamily of eubacterial RNA polymerase σ factors involved in the regulation of extracytoplasmic functions. Proc Natl Acad Sci. 1994;91:7573–7577. doi: 10.1073/pnas.91.16.7573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin DW, Schurr MJ, Mudd MH, Govan JR, Holloway BW, Deretic V. Mechanism of conversion to mucoidy in Pseudomonas aeruginosa infecting cystic fibrosis patients. Proc Natl Acad Sci. 1993;90:8377–8381. doi: 10.1073/pnas.90.18.8377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Salazar JM, Moreno S, Najera R, Boucher JC, Espin G, Soberon-Chavez G, Deretic V. Characterization of the genes coding for the putative sigma factor AlgU and its regulators MucA, MucB, MucC, and MucD in Azotobacter vinelandii and evaluation of their roles in alginate biosynthesis. J Bacteriol. 1996;178:1800–1808. doi: 10.1128/jb.178.7.1800-1808.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMillan DR, Gething MJ, Sambrook J. The cellular response to unfolded proteins: Intercompartmental signaling. Curr Opin Biotechnol. 1994;5:540–545. doi: 10.1016/0958-1669(94)90071-x. [DOI] [PubMed] [Google Scholar]
- Mecsas J, Rouvière PE, Erickson JW, Donohue TJ, Gross CA. The activity of σE, an Escherichia coli heat-inducible σ-factor, is modulated by expression of outer membrane proteins. Genes & Dev. 1993;7:2618–2628. doi: 10.1101/gad.7.12b.2618. [DOI] [PubMed] [Google Scholar]
- Miller JH. Experiments in molecular genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1972. pp. 274–281. [Google Scholar]
- Missiakas D, Betton J-M, Raina S. New components of protein folding in extracytoplasmic compartments of Escherichia coli SurA, FkpA and Skp/OmpH. Mol Microbiol. 1996;21:871–884. doi: 10.1046/j.1365-2958.1996.561412.x. [DOI] [PubMed] [Google Scholar]
- Missiakas D, Lemaire M, Mayer M, Georgopoulos C, Raina S. Modulation of the Escherichia coli σE (RpoE) heat shock transcription factor activity by the RseA, RseB, and RseC proteins. Mol Microbiol. 1997;24:355–372. doi: 10.1046/j.1365-2958.1997.3601713.x. [DOI] [PubMed] [Google Scholar]
- Morita M, Kanemori M, Yanagi H, Yura T. Heat-induced synthesis of σ32 in Escherichia coli: Structural and functional dissection of rpoH mRNA secondary structure. J Bacteriol. 1999;181:401–410. doi: 10.1128/jb.181.2.401-410.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikaido H. Porins and specific diffusion channels in bacterial outer membranes. J Biol Chem. 1994;269:3905–3908. [PubMed] [Google Scholar]
- Pallen MJ, Wren BW. The HtrA family of serine proteases. Mol Microbiol. 1997;26:209–221. doi: 10.1046/j.1365-2958.1997.5601928.x. [DOI] [PubMed] [Google Scholar]
- Raina S, Missiakas D, Georgopoulos C. The rpoE gene encoding the σE (σ24) heat shock sigma factor of Escherichia coli. EMBO J. 1995;14(5):1043–1055. doi: 10.1002/j.1460-2075.1995.tb07085.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randall LL, Hardy SJ. Correlation of competence for export with lack of tertiary structure of the mature species: A study in vivo of maltose-binding protein in E. coli. Cell. 1986;46:921–928. doi: 10.1016/0092-8674(86)90074-7. [DOI] [PubMed] [Google Scholar]
- Rouvière P, Gross CA. SurA, a periplasmic protein with peptidyl prolyl isomerase activity, participates in the assembly of outer membrane porins. Genes & Dev. 1996;10:3170–3182. doi: 10.1101/gad.10.24.3170. [DOI] [PubMed] [Google Scholar]
- Rouvière PE, De Las Peñas A, Mecsas J, Lu CZ, Rudd KE, Gross CA. rpoE, the gene encoding the second heat-shock sigma factor, σE, in Escherichia coli. EMBO J. 1995;14:1032–1042. doi: 10.1002/j.1460-2075.1995.tb07084.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch E, Maniatis T. Molecular cloning: A laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Schurr MJ, Yu H, Martinez-Salazar JM, Boucher JC, Deretic V. Control of AlgU, a member of the σE-like family of stress sigma factors, by the negative regulators MucA and MucB and Pseudomonas aeruginosa conversion to mucoidy in cystic fibrosis. J Bacteriol. 1996;178:4997–5004. doi: 10.1128/jb.178.16.4997-5004.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silber KR, Sauer RT. Deletion of the prc (tsp) gene provides evidence for additional tail-specific proteolytic activity in Escherichia coli K-12. Mol & Gen Genet. 1994;242:237–240. doi: 10.1007/BF00391018. [DOI] [PubMed] [Google Scholar]
- Straus DB, Walter WA, Gross CA. The heat shock response of E. coli is regulated by changes in the concentration of σ32. Nature. 1987;329:348–351. doi: 10.1038/329348a0. [DOI] [PubMed] [Google Scholar]
- ————— The activity of σ32 is reduced under conditions of excess heat shock protein production in Escherichia coli. Genes & Dev. 1989;3:2003–2010. doi: 10.1101/gad.3.12a.2003. [DOI] [PubMed] [Google Scholar]
- ————— DnaK, DnaJ, and GrpE heat shock proteins negatively regulate heat shock gene expression by controlling the synthesis and stability of σ32. Genes & Dev. 1990;4:2202–2209. doi: 10.1101/gad.4.12a.2202. [DOI] [PubMed] [Google Scholar]
- Studier FW, Moffatt BA. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986;189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- Tavormina PL, Reznikoff WS, Gross CA. Identifying interacting regions in the beta subunit of Escherichia coli RNA polymerase. J Mol Biol. 1996;258:213–223. doi: 10.1006/jmbi.1996.0244. [DOI] [PubMed] [Google Scholar]
- Tian G, Maas WK. Mutational analysis of the arginine repressor of Escherichia coli. Mol Microbiol. 1994;13:599–608. doi: 10.1111/j.1365-2958.1994.tb00454.x. [DOI] [PubMed] [Google Scholar]
- Tomoyasu T, Yamanaka K, Murata K, Suzaki T, Bouloc P, Kato A, Niki H, Hiraga S, Ogura T. Topology and subcellular localization of FtsH protein in Escherichia coli. J Bacteriol. 1993;175:1352–1357. doi: 10.1128/jb.175.5.1352-1357.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomoyasu T, Gamer J, Bukau B, Kanemori M, Mori H, Rutman AJ, Oppenheim AB, Yura T, Yamanaka K, Niki H, Higara S, Ogura T. Escherichia coli FtsH is a membrane-bound, ATP-dependent protease which degrades the heat-shock transcription factor σ32. EMBO J. 1995;14:2551–2560. doi: 10.1002/j.1460-2075.1995.tb07253.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waller PR, Sauer RT. Characterization of degQ and degS, Escherichia coli genes encoding homologs of the DegP protease. J Bacteriol. 1996;178:1146–1153. doi: 10.1128/jb.178.4.1146-1153.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang QP, Kaguni JM. A novel sigma factor is involved in expression of the rpoH gene of Escherichia coli. J Bacteriol. 1989;171:4248–4253. doi: 10.1128/jb.171.8.4248-4253.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Hershberger CD, Shankar S, Ye RW, Chakrabarty AM. Sigma factor-anti-sigma factor interaction in alginate synthesis: Inhibition of AlgT by MucA. J Bacteriol. 1996;178:4990–4996. doi: 10.1128/jb.178.16.4990-4996.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yura T, Tobe T, Ito K, Osawa T. Heat shock regulatory gene (htpR) of Escherichia coli is required for growth at high temperature but is dispensable at low temperature. Proc Natl Acad Sci. 1984;81:6803–6807. doi: 10.1073/pnas.81.21.6803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yura T, Nagai H, Mori H. Regulation of the heat-shock response in bacteria. Annu Rev Microbiol. 1993;47:321–350. doi: 10.1146/annurev.mi.47.100193.001541. [DOI] [PubMed] [Google Scholar]