Abstract
Background
Single-nucleotide polymorphisms within TP53 gene (codon 72 exon 4, rs1042522, encoding either arginine or proline) and MDM2 promoter (SNP309; rs2279744), have been independently associated with increased risk of several cancer types. Few studies have analysed the role of these polymorphisms in the development of hepatocellular carcinoma.
Methods
Genotype distribution of TP53 codon 72 and MDM2 SNP309 in 61 viral hepatitis-related hepatocellular carcinoma cases and 122 blood samples (healthy controls) from Italian subjects were determined by PCR and restriction fragment length polymorphism (RFLP).
Results
Frequencies of TP53 codon 72 alleles were not significantly different between cases and controls. A significant increase of MDM2 SNP309 G/G and T/G genotypes were observed among hepatocellular carcinoma cases (Odds Ratio, OR = 3.56, 95% Confidence Limits, 95% CI = 1.3-9.7; and OR = 2.82, 95% CI = 1.3-6.4, respectively).
Conclusions
These results highlight a significant role of MDM2 SNP309 G allele as a susceptibility gene for the development of viral hepatitis-related hepatocellular carcinoma among Italian subjects.
Keywords: Genetic Polymorphisms, TP53 codon 72, MDM2 SNP309, hepatocellular carcinoma
Introduction
Hepatocellular carcinoma (HCC) is the most common liver malignancy as well as the third and the fifth cause of cancer death in the world in men and women, respectively [1]. HCC is an infrequent cancer in developed countries, with the exception of Southern Europe where the incidence in men (ASR 9.8 per 100,000) is significantly higher than in other developed regions [2]. The HCC incidence, however, has substantially increased in Japan during the past three decades [3], and slight increases have been reported in France and in the United Kingdom [4,5]. The incidence rates of HCC in the United States have historically been lower than in many countries. However, in recent decades, HCC age-adjusted incidence rates have doubled [6].
Etiological factors associated with the development of the disease are well known and include infection with the hepatitis C virus or hepatitis B virus, heavy alcohol intake, prolonged dietary exposure to aflatoxin B1 and primary hemochromatosis [7-9]. As for other types of cancer, the etiology and pathogenesis of HCC is multifactorial and multistep [10]. The prevalence of HCC in Italy, and particularly in Southern Italy, is significantly higher compared with other Western countries. Hepatitis virus infection, long-term alcohol and tobacco consumption account for 87% of HCC cases in the Italian population and, among these, 61% are attributable to HCV infection. A recent seroprevalence survey conducted in the general population of Campania Region (Southern Italy) reported a 7.5% positivity for HCV infection which peaked at 23.2% in the 65 years or older age groups [11]. Thus, although the virus is highly prevalent only a small percentage of infected individuals will develop liver cancer suggesting that other genetic or environmental cofactors are needed for tumor progression.
Several studies have analyzed distribution of single nucleotide polymorphism in gene encoding for cell cycle regulatory proteins and susceptibility to malignant diseases. A common polymorphism located in exon 4 of TP53 gene, resulting in a non-conservative arginine to a proline change at codon 72, is known to be important for growth suppression and apoptotic functions [12,13]. Additionally, a naturally occurring G to T sequence variation (single nucleotide polymorphism - SNP309) in the second promoter-enhancer region of the MDM2 gene has been shown to increase the binding affinity of the transcriptional activator Sp1 resulting in high levels of MDM2 protein, formation of transcriptionally inactive p53-MDM2 complexes and alteration of the p53 pathway [14-16]. These observations are consistent with an oncogenic function for the variant SNP309 [17].
Polymorphisms of TP53 codon 72 and MDM2 SNP309 have been previously investigated in Korean population in which they are associated with the early development of HCC in patients with chronic HBV infection [18].
No studies have been conducted in HCC cases of the Italian population. In the present study, we evaluated the distribution of the TP53 codon 72 genotypes and MDM2 SNP309 in a series of 61 hepatocellular carcinoma cases comprising HCV and HBV infected patients, and compared them with healthy controls to possibly identify host genetic factors associated with the increased risk of developing hepatocellular carcinoma.
Materials and methods
Patient and Tissue Samples
Liver biopsies from 53 HCV-positive, 7 HBV-positive, one HCV/HBV-positive HCC patients and 122 negative non-liver cancer control patients (blood samples) were obtained with informed consent at the liver unit of the INT "Pascale" in Naples. Each liver biopsy was divided in two sections: the first section was stored in RNA Later at -80°C (Ambion, Austin, TX), the second was subjected to histopathologic examination. Only liver biopsies determined to be hepatocellular carcinoma were included in the study.
Genomic DNA was extracted according to published procedures [19]. Tissue samples were digested with by proteinase K treatment (150 μg ml-1 at 56°C for 2 hours) in 100 - 500 μl of lysis buffer (10 mM Tris-HCl, pH 7.6, 5 mM EDTA, 150 mM NaCl, 1% SDS), followed by DNA purification by phenol-chloroform-isoamyl alcohol (25:24:1) extraction and ethanol precipitation in 0.3 M sodium acetate (pH 4.6). Genomic DNA from 122 blood samples from healthy controls were previously extracted and subjected to allele specific PCR of exon 4 of TP53 and PCR-RFLP of intron 1 of MDM2 [20,21].
TP53 codon 72 and MDM2 SNP309 polymorphisms analysis
The analysis of TP53 genotype at codon 72 was carried out by performing an allele specific PCR in which two independent reactions were performed for each sample: one specific for codon CCC (encoding proline) using the p53ProPlus (5'-GCCAGAGGCTGCTCCCCC-3') with p53Minus oligoprimers; the other specific for CGC (encoding arginine), using the p53Plus with p53ArgMinus (5'-CTGGTGCAGGGGCCACGC-3') oligoprimers. PCR amplification reactions were carried out on 100 to 200 ng genomic DNA in 50-μl reaction mixture following amplification procedures described previously [20].
The MDM2 SNP309 was determined by PCR and restriction fragment length polymorphism (RFLP) analysis of amplified products. A 174 base pairs (bp) amplimer of the MDM2 intron 1 region, containing the MspA1 polymorphic site at nucleotide 309, was amplified with the B-MDM2-309F (5'- GGGAGTTCAGGGTAAAGG-3') and the B-MDM2-309R (5'- GACCAGCTCAAGAGGAAA-3') oligoprimers, designed using the Beacon Designer 2.0 software (Premier Biosoft International, Palo Alto, CA, USA). PCR reactions were performed in a 50-μl reaction mixture containing 100-200 ng of target DNA, 20 pmol of each primer, 2.5 mM MgCl2, 50 μM of each dNTP and 1.25 U of HotMaster Taq DNA polymerase (5 Prime GmbH, Hamburg, Germany). DNA was amplified with the following steps: an initial 1-min denaturation at 94°C, followed by 32 cycles of 55°C for 45 s, 68°C for 1 min, 94°C for 30 s and a final annealing at 55°C for 30 s with 5 min elongation at 72°C. Ten-microliter aliquots of MDM2 PCR products were digested with MspA1 restriction enzyme, fractionated by electrophoresis on a 7% polyacrylamide gel in Tris-borate-EDTA running buffer and stained with ethidium bromide for DNA band visualization by UV transillumination.
TP53 PCR amplified products from the 61 cases were further subjected to direct nucleotide sequencing by Primm Srl Laboratories (Milan, Italy) using the fluorescent dye terminator technology and ABI 3730 DNA sequencers (Applied BioSystems). Nucleotide sequences were edited with Chromas Lite 2.01 (http://www.technelysium.com.au/chromas.html).
Statistical analyses
The observed and expected genotype frequencies among the study groups were analysed using the Hardy-Weinberg equilibrium theory. A Fisher's exact test or χ2 test was used, as appropriate, to compare the proportions of TP53 and MDM2 genotypes between cases of HCC and healthy control population. An unpaired Student's t-test was used to evaluate differences between the mean age of cases and control group. All analyses were performed with Epi Info 6 Statistical Analysis System Software (6.04d, 2001, Centers for Disease Control and Prevention, USA).
Differences were considered to be statistically significant when p-values were less than 0.05.
Results
In this study were included 61 cases of hepatocellular carcinoma (48 in males and 13 in females) and 122 cancer-free control subjects (all males). Table 1 summarizes the selected characteristics of the subjects. The frequencies of TP53 codon 72 genotypes has been estimated using an allele specific PCR that specifically detects either the TP53 proline or arginine 72- encoding codons. A slight higher frequency of arginine allele was observed in HCC cases compared to the controls, however, the difference did not reach statistical significance using Yates corrected χ2 test (P = 0.696). The distribution of polymorphic alleles in cases and controls are summarized in Table 2. The frequencies of the three genotypes of the TP53 gene polymorphism at codon 72 among the 61 HCC cases were 4.9% (n = 3), 32.8% (n = 20) and 62.3% (n = 38) proline homozygous, heterozygous and arginine homozygous, respectively, and the corresponding figures among controls were 7.4% (n = 9), 34.4% (n = 42), and 58.2% (n = 71), respectively. The genotype frequencies of TP53 codon 72 of were found in Hardy-Weinberg equilibrium both in HCC cases and healthy controls (χ2 = 0.81; df = 1; p = 0.37; and χ2= 0.63; df = 1; p = 0.43, respectively).
Table 1.
Characteristics and pathological features of HCC patients and healthy controls
| Variable | Cases (n = 61) n(%) |
Controls (n = 122) n(%) |
|---|---|---|
| Mean age, years [± SD] | 68.7[± 8.87] | 59.4[± 14.3] |
| Age, years | ||
| ≤ 50 | 2(3.3) | 12(13.1) |
| 50-65 | 16(26.2) | 43(45.9) |
| > 65 | 43(70.5) | 38(41) |
| Males | 48(78.7) | 122(100) |
| Females | 13(21.3) | |
| Virus | ||
| Cases HCV-pos | 53(86.9) | |
| Cases HBV-pos | 7(11.5) | |
| Cases HCV/HBV-pos | 1(1.6) |
Table 2.
Distribution of TP53 codon 72 and MDM2 SNP309 genotypes in HCC cases and controls
| HCC Cases (n = 61) n (%) |
Controls (n = 122) n (%) |
ORa (95% CI) | P Valueb | |
|---|---|---|---|---|
| TP53 codon 72 | ||||
| Pro Allele | 27(22.1) | 60(24.6) | 1 | |
| Arg Allele | 95(77.9) | 184(75.4) | 1.15(0.7-2.0) | 0.695 |
| Pro/Pro | 3(4.9) | 9(7.4) | 1 | |
| Pro/Arg | 20(32.8) | 42(34.4) | 0.96(0.2-4.3) | 0.875 |
| Arg/Arg | 38(62.3) | 71(58.2) | 1.20(0.3-5.0) | 0.716 |
| Arg/Arg + Pro/Arg | 58(95.1) | 113(92.6) | 1.12(0.3-4.5) | 0.751 |
| MDM2 SNP309 | ||||
| T Allele | 58(47.5) | 154(63.1) | 1 | |
| G Allele | 64(52.5) | 90(36.9) | 1.89(1.2-3.0) | 0.006 |
| T/T | 13(21.3) | 55(45.1) | 1 | |
| T/G | 32(52.5) | 48(39.3) | 2.82(1.3-6.4) | 0.010 |
| G/G | 16(26.2) | 19(15.6) | 3.56(1.3-9.7) | 0.009 |
| G/G + T/G | 48(78.7) | 67(54.9) | 3.03(1.4-6.6) | 0.003 |
aT allele or T/T MDM2 SNP309 and TP53 Pro allele or Pro/Pro codon 72 genotypes were considered as the baseline when calculating the relative crude ORs.
bYates corrected P values
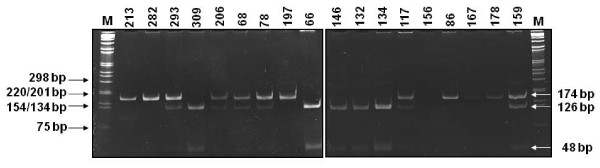
The MDM2 SNP309 genotypes were analyzed using a PCR-RFLP based assay (Figure 1). The frequencies of MDM2 SNP309 T/T, T/G and G/G genotypes among the cases were 21.3% (n = 13), 52.5% (n = 32) and 26.2% (n = 16), and among controls were 45.1% (n = 55), 39.3% (n = 48) and 15.6% (n = 19), respectively. The MDM2 SNP309 genotype frequencies were found in Hardy-Weinberg equilibrium among cases and controls group (χ2 = 0.16; df = 1; p = 0.69; and χ2= 2.33; df = 1; p = 0.13, respectively). The frequencies of MDM2 SNP309 G/G and G/T genotypes were significantly more prevalent in HCC cases (52.5%) compared with controls (36.9%), p = 0.006. When the homozygous MDM2 SNP309 T/T genotype was used as the reference group, the MDM2 SNP309 G/G and T/G genotypes were associated with a significantly increased risk for HCC (OR 3.56, 95% CI 1.3-9.7; OR 2.82, 95% CI 1.3-6.4, respectively).
Figure 1.
PCR amplification followed by restriction fragment length polymorphism (PCR-RFLP) to determine MDM2 SNP309 polymorphism. MDM2 SNP309 T allele was not cleaved by MspAI endonuclease and had a single band of 174 bp. The MDM2 SNP309 G allele was cleaved by MspAI and had two small fragments of 126 and 48 bp. The MDM2 SNP309 heterozygote had three bands of 174, 126 and 48 bp.
The combined effect of the TP53 codon72 and the MDM2 SNP309 polymorphisms was also analysed. However, no additional cancer risk effect associated with TP53 codon 72 polymorphism was identified when the risk of combined genotypes was analyzed.
Discussion
Several epidemiological studies have evaluated the association of TP53 codon 72 and MDM2 SNP309 polymorphisms and risk of or survival from several types of cancer [22-24], but few evaluated the association of both these polymorphisms in viral-related liver cancer and none in hepatocellular carcinoma from Italian patients.
In the current study the TP53 codon 72 and MDM2 nucleotide 309 polymorphisms were investigated in a series of 61 cases of hepatocellular carcinoma, mainly associated with HCV infection, and in 122 population-matched controls in order to verify the impact of these gene variants on the risk of tumour development. The TP53 codon 72 and MDM2 SNP309 genotypes were distributed in accordance with the Hardy-Weinberg equilibrium in controls and their frequencies were in broad agreement with those observed in previously published studies of Caucasian subjects [25-27]. The results showed no significant differences in the genotype distribution of the TP53 codon 72 polymorphisms between cases and controls and no increase in the frequency of the arginine or proline alleles among HCC cases. This observation is in agreement with previous studies reporting no association between codon 72 genotypes and disease severity or liver cancer in a group of 340 HCV-infected Italian subjects at different stages of disease, including 84 HCC patients, and in a group of 97 Spanish HCC patients [28,29]. Conversely, a higher frequency of Pro/Pro genotype in HCC cases (13.5%) versus control subjects (6.3%) and a significant higher risk to develop liver cancer (OR, 2.304; 95% CI, 1.014-5.234) was observed among Moroccan subjects [30]. The analysis of pooled data from six case-control studies including 836 and 279 liver cancer and 1'191 and 587 controls of Asian and Caucasian ethnicity, respectively, showed a significant higher risk (OR = 1.35, 95% CI 1.06 - 1.71) for liver cancer only among Asian patients carrying the Pro/Pro genotype [31]. A case-control study including 80 incident cases of HCC and 328 controls, nested in a cohort of 4,841 male chronic hepatitis B, showed that the lack of association between TP53 codon 72 Pro allele and HCC was apparent. Indeed, in patients with chronic liver disease there were synergistic effects between the Pro allele and HCC family history in first-degree relatives which conferred a risk of 7.60 (95% CI = 2.28-25.31) for HCC development compared with subjects without the Pro allele and without chronic liver disease [32]. These observations suggest that large case-control studies are needed in order to clarify the role of TP53 codon 72 polymorphism in the HCC pathogenesis in different populations.
In this study the frequencies of MDM2 SNP309 T/G heterozygous (52.5%) and G/G homozygous (26.2%) genotypes were significantly higher among HCC cases compared to healthy controls (P 0.010 and P = 0.009, respectively). The homozygous MDM2 SNP309 T/T genotype in HCC, on the other hand, was lower (21.3%) than that observed in controls (45.1%). The possible role of MDM2 SNP309 polymorphism in HCC development has been analyzed in few other geographical regions. In Eastern Asia MDM2 SNP309 G allele has been associated with higher risk of HCC in Japanese patients with chronic hepatitis C [33], in Korean patients with chronic HBV [18], and in Taiwanese patients infected with HBV or HCV infection [34]. Similarly, the MDM2 309 G allele was found to enhance the risk of viral-associated HCC in Moroccan (Northern Africa) [35] and in Turkish (Western Asia) patients [36]. Jin et al. (2011) performed a metanalysis on these studies, including a total of 738 cases and 1062 controls, to evaluate the reliability of the associations of MDM2 SNP309 G allele with HCC by using false-positive report probability analysis and the Venice guidelines on genetic epidemiology [37]. The pooled OR reached 1.57 (95% CI: 1.36-1.80) for G allele compared to T allele, using the fixed-effect model, indicating that this SNP may be used as a predictive molecular marker for the screening of populations with high risk of HCC [37].
Overexpression of the MDM2 protein has an oncogenic effect through the reduction of p53 levels with the consequent attenuation of the p53 DNA damage response that allows increased cell proliferation and inhibition of apoptosis, providing advantageous signals for tumor cell survival [38-40]. The study by Bond et al. (2004) showed that MDM2 protein levels in homozygous and heterozygous SNP309 G cell lines are on average 4-fold and 1.9 fold higher, respectively, than TT cell lines [14]. Furthermore, Hirata et al. reported that renal cell carcinoma tissue with G/G and G/T genotypes were more frequently positively stained for MDM2 than that with the TT genotype (50% and 26%, respectively, versus 13%) [41]. The proper regulation of MDM2 levels has been shown to be critical for p53 tumour suppression, and even a modest change in levels could affect the p53 pathway and, subsequently, increase the risk of cancer development in mouse models [42]. MDM2 binds directly to and inhibits p53 by regulating its location, stability and ability to activate transcription [14]. Several reports have shown that HCV proteins (NS3, NS5A and NS5B) can modulate activity and/or expression of p53 and may interfere with normal regulation of cell growth [43-45]. This implies that MDM2 polymorphism together with viral hepatitis protein may result in a cumulative effect on the inactivation of p53 function.
This study has limitations, including the modest sample size and the fact that cases were not well matched for age and sex with the control subjects. Although Italian blood donors may be good representatives of the general population, they are younger than HCC patients due to age limit and health requirements for blood donation. However, being fulfilled the Hardy Weinberg equilibrium for the allelic frequencies of both TP53 and MDM2 polymorphisms in the control group, it may be excluded that age and sex limitations have introduced significant bias in the study.
In conclusion, these results provide the first evidence that the MDM2 SNP309 polymorphism represents a risk factor for viral hepatitis-related HCC in the Italian population. Conversely, the TP53 polymorphism at codon 72 seems not to be a potential risk factor for development of HCC in Italian patients.
Abbreviations
HCC: Hepatocellular carcinoma; MDM2: mouse double minute-2; HBV: hepatitis B virus; HCV: hepatitis C virus.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
MLT and FMB were responsible for the overall planning and coordination of the study. FI was involved in patients enrollment and specimen collection. LB contributed to the data analysis. SL and GB carried out the histopathology evaluation of the cases. VDV was responsible for specimen processing, DNA analysis and with MLT compiled and finalized the manuscript. All authors read and approved the final manuscript.
Contributor Information
Valeria Di Vuolo, Email: vdivuolo@tin.it.
Luigi Buonaguro, Email: lbuonaguro@tin.it.
Francesco Izzo, Email: izzo@connect.it.
Simona Losito, Email: nlosito@istitutotumori.na.it.
Gerardo Botti, Email: gbotti@istitutotumori.na.it.
Franco M Buonaguro, Email: irccsvir@unina.it.
Maria Lina Tornesello, Email: mltornesello@virgilio.it.
Acknowledgements
This work was supported by grants from the Ministero della Salute (Programma Integrato Oncologia RO4/2007) and from the ICSC-World Laboratory (project MCD-2/7).
References
- Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008 2. Int J Cancer. 2010. [DOI] [PubMed]
- Cho LY, Yang JJ, Ko KP, Park B, Shin A, Lim MK. et al. Coinfection of hepatitis B and C viruses and risk of hepatocellular carcinoma: systematic review and meta-analysis 2. Int J Cancer. 2011;128:176–184. doi: 10.1002/ijc.25321. [DOI] [PubMed] [Google Scholar]
- Okuda K, Fujimoto I, Hanai A, Urano Y. Changing incidence of hepatocellular carcinoma in Japan. Cancer Res. 1987;47:4967–4972. [PubMed] [Google Scholar]
- Deuffic S, Poynard T, Buffat L, Valleron AJ. Trends in primary liver cancer. Lancet. 1998;351:214–215. doi: 10.1016/S0140-6736(05)78179-4. [DOI] [PubMed] [Google Scholar]
- Taylor-Robinson SD, Foster GR, Arora S, Hargreaves S, Thomas HC. Increase in primary liver cancer in the UK, 1979-94. Lancet. 1997;350:1142–1143. doi: 10.1016/S0140-6736(05)63789-0. [DOI] [PubMed] [Google Scholar]
- El-Serag HB, Davila JA, Petersen NJ, McGlynn KA. The continuing increase in the incidence of hepatocellular carcinoma in the United States: an update. Ann Intern Med. 2003;139:817–823. doi: 10.7326/0003-4819-139-10-200311180-00009. [DOI] [PubMed] [Google Scholar]
- Chuang SC, La VC, Boffetta P. Liver cancer: descriptive epidemiology and risk factors other than HBV and HCV infection 1. Cancer Lett. 2009;286:9–14. doi: 10.1016/j.canlet.2008.10.040. [DOI] [PubMed] [Google Scholar]
- Donato F, Gelatti U, Limina RM, Fattovich G. Southern Europe as an example of interaction between various environmental factors: a systematic review of the epidemiologic evidence 1. Oncogene. 2006;25:3756–3770. doi: 10.1038/sj.onc.1209557. [DOI] [PubMed] [Google Scholar]
- Tsukuma H, Hiyama T, Oshima A, Sobue T, Fujimoto I, Kasugai H. et al. A case-control study of hepatocellular carcinoma in Osaka, Japan. Int J Cancer. 1990;45:231–236. doi: 10.1002/ijc.2910450205. [DOI] [PubMed] [Google Scholar]
- Romeo R, Colombo M. The natural history of hepatocellular carcinoma. Toxicology. 2002;181-182:39–42. doi: 10.1016/s0300-483x(02)00252-4. [DOI] [PubMed] [Google Scholar]
- Fusco M, Girardi E, Piselli P, Palombino R, Polesel J, Maione C. et al. Epidemiology of viral hepatitis infections in an area of southern Italy with high incidence rates of liver cancer. Eur J Cancer. 2008;44:847–853. doi: 10.1016/j.ejca.2008.01.025. [DOI] [PubMed] [Google Scholar]
- Matlashewski GJ, Tuck S, Pim D, Lamb P, Schneider J, Crawford LV. Primary structure polymorphism at amino acid residue 72 of human p53. Mol Cell Biol. 1987;7:961–963. doi: 10.1128/mcb.7.2.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietsch EC, Humbey O, Murphy ME. Polymorphisms in the p53 pathway. Oncogene. 2006;25:1602–1611. doi: 10.1038/sj.onc.1209367. [DOI] [PubMed] [Google Scholar]
- Bond GL, Hu W, Bond EE, Robins H, Lutzker SG, Arva NC. et al. A single nucleotide polymorphism in the MDM2 promoter attenuates the p53 tumor suppressor pathway and accelerates tumor formation in humans. Cell. 2004;119:591–602. doi: 10.1016/j.cell.2004.11.022. [DOI] [PubMed] [Google Scholar]
- Bougeard G, Baert-Desurmont S, Tournier I, Vasseur S, Martin C, Brugieres L. et al. Impact of the MDM2 SNP309 and p53 Arg72Pro polymorphism on age of tumour onset in Li-Fraumeni syndrome. J Med Genet. 2006;43:531–533. doi: 10.1136/jmg.2005.037952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruijs MW, Schmidt MK, Nevanlinna H, Tommiska J, Aittomaki K, Pruntel R. et al. The single-nucleotide polymorphism 309 in the MDM2 gene contributes to the Li-Fraumeni syndrome and related phenotypes. Eur J Hum Genet. 2007;15:110–114. doi: 10.1038/sj.ejhg.5201715. [DOI] [PubMed] [Google Scholar]
- Bond GL, Hirshfield KM, Kirchhoff T, Alexe G, Bond EE, Robins H. et al. MDM2 SNP309 accelerates tumor formation in a gender-specific and hormone-dependent manner. Cancer Res. 2006;66:5104–5110. doi: 10.1158/0008-5472.CAN-06-0180. [DOI] [PubMed] [Google Scholar]
- Yoon YJ, Chang HY, Ahn SH, Kim JK, Park YK, Kang DR. et al. MDM2 and p53 polymorphisms are associated with the development of hepatocellular carcinoma in patients with chronic hepatitis B virus infection 1. Carcinogenesis. 2008;29:1192–1196. doi: 10.1093/carcin/bgn090. [DOI] [PubMed] [Google Scholar]
- Tornesello ML, Duraturo ML, Waddell KM, Biryahwaho B, Downing R, Balinandi S. et al. Evaluating the role of human papillomaviruses in conjunctival neoplasia. Br J Cancer. 2006;94:446–449. doi: 10.1038/sj.bjc.6602921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tornesello ML, Biryahwaho B, Downing R, Hatzakis A, Alessi E, Cusini M. et al. TP53 codon 72 polymorphism in classic, endemic and epidemic Kaposi's sarcoma in African and Caucasian patients. Oncology. 2009;77:328–334. doi: 10.1159/000260905. [DOI] [PubMed] [Google Scholar]
- Tornesello ML, Buonaguro L, Cristillo M, Biryahwaho B, Downing R, Hatzakis A. et al. MDM2 and CDKN1A gene polymorphisms and risk of Kaposi's sarcoma in African and Caucasian patients. Biomarkers. 2011;16:42–50. doi: 10.3109/1354750X.2010.525664. [DOI] [PubMed] [Google Scholar]
- Hrstka R, Coates PJ, Vojtesek B. Polymorphisms in p53 and the p53 pathway: roles in cancer susceptibility and response to treatment. J Cell Mol Med. 2009;13:440–453. doi: 10.1111/j.1582-4934.2008.00634.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whibley C, Pharoah PD, Hollstein M. p53 polymorphisms: cancer implications. Nat Rev Cancer. 2009;9:95–107. doi: 10.1038/nrc2584. [DOI] [PubMed] [Google Scholar]
- Bonds L, Baker P, Gup C, Shroyer KR. Immunohistochemical localization of cdc6 in squamous and glandular neoplasia of the uterine cervix. Arch Pathol Lab Med. 2002;126:1164–1168. doi: 10.5858/2002-126-1164-ILOCIS. [DOI] [PubMed] [Google Scholar]
- Menin C, Scaini MC, De Salvo GL, Biscuola M, Quaggio M, Esposito G. et al. Association between MDM2-SNP309 and age at colorectal cancer diagnosis according to p53 mutation status. J Natl Cancer Inst. 2006;98:285–288. doi: 10.1093/jnci/djj054. [DOI] [PubMed] [Google Scholar]
- Capasso M, Ayala F, Avvisati RA, Russo R, Gambale A, Mozzillo N, MDM2 SNP309 and p53 Arg72Pro in cutaneous melanoma: association between SNP309 GG genotype and tumor Breslow thickness. J Hum Genet. 2010. [DOI] [PubMed]
- Bonafe M, Olivieri F, Mari D, Baggio G, Mattace R, Berardelli M. et al. P53 codon 72 polymorphism and longevity: additional data on centenarians from continental Italy and Sardinia. Am J Hum Genet. 1999;65:1782–1785. doi: 10.1086/302664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leveri M, Gritti C, Rossi L, Zavaglia C, Civardi E, Mondelli MU. et al. Codon 72 polymorphism of P53 gene does not affect the risk of cirrhosis and hepatocarcinoma in HCV-infected patients. Cancer Lett. 2004;208:75–79. doi: 10.1016/j.canlet.2004.02.016. [DOI] [PubMed] [Google Scholar]
- Anzola M, Cuevas N, Lopez-Martinez M, Saiz A, Burgos JJ, de Pancorbo MM. Frequent loss of p53 codon 72 Pro variant in hepatitis C virus-positive carriers with hepatocellular carcinoma. Cancer Lett. 2003;193:199–205. doi: 10.1016/S0304-3835(03)00046-6. [DOI] [PubMed] [Google Scholar]
- Ezzikouri S, El Feydi AE, Chafik A, Benazzouz M, El KL, Afifi R. et al. The Pro variant of the p53 codon 72 polymorphism is associated with hepatocellular carcinoma in Moroccan population. Hepatol Res. 2007;37:748–754. doi: 10.1111/j.1872-034X.2007.00126.x. [DOI] [PubMed] [Google Scholar]
- Chen X, Liu F, Li B, Wei YG, Yan LN, Wen TF. p53 codon 72 polymorphism and liver cancer susceptibility: A meta-analysis of epidemiologic studies. World J Gastroenterol. 2011;17:1211–1218. doi: 10.3748/wjg.v17.i9.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu MW, Yang SY, Chiu YH, Chiang YC, Liaw YF, Chen CJ. A p53 genetic polymorphism as a modulator of hepatocellular carcinoma risk in relation to chronic liver disease, familial tendency, and cigarette smoking in hepatitis B carriers. Hepatology. 1999;29:697–702. doi: 10.1002/hep.510290330. [DOI] [PubMed] [Google Scholar]
- Dharel N, Kato N, Muroyama R, Moriyama M, Shao RX, Kawabe T. et al. MDM2 promoter SNP309 is associated with the risk of hepatocellular carcinoma in patients with chronic hepatitis C. Clin Cancer Res. 2006;12:4867–4871. doi: 10.1158/1078-0432.CCR-06-0111. [DOI] [PubMed] [Google Scholar]
- Leu JD, Lin IF, Sun YF, Chen SM, Liu CC, Lee YJ. Association between MDM2-SNP309 and hepatocellular carcinoma in Taiwanese population. World J Gastroenterol. 2009;15:5592–5597. doi: 10.3748/wjg.15.5592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezzikouri S, El Feydi AE, Afifi R, El KL, Benazzouz M, Hassar M. et al. MDM2 SNP309T > G polymorphism and risk of hepatocellular carcinoma: a case-control analysis in a Moroccan population. Cancer Detect Prev. 2009;32:380–385. doi: 10.1016/j.cdp.2009.01.003. [DOI] [PubMed] [Google Scholar]
- Akkiz H, Sumbul AT, Bayram S, Bekar A, Akgollu E. MDM2 promoter polymorphism is associated with increased susceptibility to hepatocellular carcinoma in Turkish population. Cancer Epidemiol. 2010;34:448–452. doi: 10.1016/j.canep.2010.04.008. [DOI] [PubMed] [Google Scholar]
- Jin F, Xiong WJ, Jing JC, Feng Z, Qu LS, Shen XZ. Evaluation of the association studies of single nucleotide polymorphisms and hepatocellular carcinoma: a systematic review. J Cancer Res Clin Oncol. 2011. [DOI] [PubMed]
- Arva NC, Gopen TR, Talbott KE, Campbell LE, Chicas A, White DE. et al. A chromatin-associated and transcriptionally inactive p53-Mdm2 complex occurs in mdm2 SNP309 homozygous cells 1. J Biol Chem. 2005;280:26776–26787. doi: 10.1074/jbc.M505203200. [DOI] [PubMed] [Google Scholar]
- Bond GL, Hu W, Levine A. A single nucleotide polymorphism in the MDM2 gene: from a molecular and cellular explanation to clinical effect. Cancer Res. 2005;65:5481–5484. doi: 10.1158/0008-5472.CAN-05-0825. [DOI] [PubMed] [Google Scholar]
- Michael D, Oren M. The p53-Mdm2 module and the ubiquitin system. Semin Cancer Biol. 2003;13:49–58. doi: 10.1016/S1044-579X(02)00099-8. [DOI] [PubMed] [Google Scholar]
- Hirata H, Hinoda Y, Kikuno N, Kawamoto K, Suehiro Y, Tanaka Y. et al. MDM2 SNP309 polymorphism as risk factor for susceptibility and poor prognosis in renal cell carcinoma. Clin Cancer Res. 2007;13:4123–4129. doi: 10.1158/1078-0432.CCR-07-0609. [DOI] [PubMed] [Google Scholar]
- Mendrysa SM, McElwee MK, Michalowski J, O'Leary KA, Young KM, Perry ME. mdm2 Is critical for inhibition of p53 during lymphopoiesis and the response to ionizing irradiation. Mol Cell Biol. 2003;23:462–472. doi: 10.1128/MCB.23.2.462-473.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong GZ, Jiang YF, He Y, Lai LY, Zhu YH, Su XS. HCV NS5A abrogates p53 protein function by interfering with p53-DNA binding. World J Gastroenterol. 2004;10:2223–2227. doi: 10.3748/wjg.v10.i15.2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siavoshian S, Abraham JD, Kieny MP, Schuster C. HCV core, NS3, NS5A and NS5B proteins modulate cell proliferation independently from p53 expression in hepatocarcinoma cell lines. Arch Virol. 2004;149:323–336. doi: 10.1007/s00705-003-0205-7. [DOI] [PubMed] [Google Scholar]
- Lan KH, Sheu ML, Hwang SJ, Yen SH, Chen SY, Wu JC. et al. HCV NS5A interacts with p53 and inhibits p53-mediated apoptosis. Oncogene. 2002;21:4801–4811. doi: 10.1038/sj.onc.1205589. [DOI] [PubMed] [Google Scholar]