Abstract
Cohesin complex acts in the formation and maintenance of sister chromatid cohesion during and after S phase. Budding yeast Scc1p/Mcd1p, an essential subunit, is cleaved and dissociates from chromosomes in anaphase, leading to sister chromatid separation. Most cohesin in higher eukaryotes, in contrast, is dissociated from chromosomes well before anaphase. The universal role of cohesin during anaphase thus remains to be determined. We report here initial characterization of four putative cohesin subunits, Psm1, Psm3, Rad21, and Psc3, in fission yeast. They are essential for sister chromatid cohesion. Immunoprecipitation demonstrates stable complex formation of Rad21 with Psm1 and Psm3 but not with Psc3. Chromatin immunoprecipitation shows that cohesin subunits are enriched in broad centromere regions and that the level of centromere-associated Rad21 did not change from metaphase to anaphase, very different from budding yeast. In contrast, Rad21 containing similar cleavage sites to those of Scc1p/Mcd1p is cleaved specifically in anaphase. This cleavage is essential, although the amount of cleaved product is very small (<5%). Mis4, another sister chromatid cohesion protein, plays an essential role for loading Rad21 on chromatin. A simple model is presented to explain the specific behavior of fission yeast cohesin and why only a tiny fraction of Rad21 is sufficient to be cleaved for normal anaphase.
Keywords: Sister chromatid cohesion, cohesin, SMC, Rad21, Psc3, S. pombe
Sister chromatid cohesion is an important cellular process essential for accurate chromosome segregation (Miyazaki and Orr-Weaver 1994; Yanagida 1995; Nasmyth 1999). If the sisters were not linked, they would diffuse apart, which makes it difficult for cells to handle chromosomes for accurate segregation. In fact, aberration in this linkage may lead to chromosomal instability and aneuploidy, which are seen in the majority of cancers (Lengauer et al. 1998). This linkage between sister chromatids also allows them to align properly, which may help in repairing chromosomal DNA damage by homologous recombination.
Sister chromatid cohesion is established during DNA replication and is assumed to be maintained until anaphase, a mitotic stage when sister chromatid separation takes place. Recent studies have demonstrated the presence of a multiprotein complex required for sister chromatid cohesion, called cohesin (Hirano 1999; Nasmyth et al. 2000). In Saccharomyces cerevisiae, the cohesin subunits are Smc1p, Smc3p, Scc1p/Mcd1p, and Scc3p, whose defects induce premature sister chromatid separation (Guacci et al. 1997; Michaelis et al. 1997; Toth et al. 1999). In frog, homologs of Smc1p, Smc3p, Scc1p/Mcd1p, and Scc3p form the complex with an additional subunit. Immunodepletion of the frog subunits causes defects in sister chromatid cohesion (Losada et al. 1998, 2000).
Two subunits of the cohesin complex belong to members of the structural maintenance of chromosomes (SMC) superfamily (e.g., Hirano 1999). SMC has the globular domains at the N and C termini and a central hinge region consisting of long coiled coils. These globular domains interact with ATP and possibly also with DNA. The function of the hinge region is unknown. SMC proteins are also essential for mitotic chromosome condensation (Sutani and Yanagida 1997; Kimura et al. 1997; Sutani et al. 1999) and sex chromosome dosage compensation (Lieb et al. 1998), and DNA repair (Lehmann et al. 1995). These proteins are mostly bound to other non-SMC subunits. The budding yeast cohesin contains two non-SMC subunits, Scc1p/Mcd1p and Scc3p. Scc1p/Mcd1p is similar to Schizosaccharomyces pombe Rad21, originally identified as a protein involved in DNA repair (Birkenbihl and Subramani 1992, 1995; Tatebayashi et al. 1998).
Besides the cohesin complex, several other proteins are known to be required for sister chromatid cohesion. S. pombe Mis4, homologous to Scc2p in S. cerevisiae and to Nipped-B in Drosophila (Rollins et al. 1999), collectively called adherin, is needed for cohesion and becomes essential during the S phase (Furuya et al. 1998). S. cerevisiae Eco1p/Ctf7p is necessary for the establishment of cohesion only in the S phase (Skibbens et al. 1999; Toth et al. 1999). A fission yeast homolog Eso1 is also needed for sister chromatid cohesion (Tanaka et al. 2000). Other proteins that may interact with cohesin subunits and play a possible role in cohesion are Trf4p in S. cerevisiae (Castano et al. 1996) and BimD in Aspergillus nidulans (Holt and May 1996). These sister chromatid cohesion proteins are implicated in the normal progression of the S phase.
Existence of sister chromatid cohesion proteins was first presumed by the observation that sister chromatid separation may depend on the destruction of proteins other than mitotic cyclins (Holloway et al. 1993; Surana et al. 1993; Irniger et al. 1995). Proteins such as a glue or cohesion protein were thus considered (Miyazaki and Orr-Weaver 1994). Very recent findings in budding yeast suggest that dissociation of the cohesin complex from the chromosomes requires the cleavage of Scc1p by a protease under the control of Esp1p (Ciosk et al. 1998; Uhlmann et al. 1999). Esp1p, which is similar to fission yeast Cut1, is bound to Pds1p, a presumed inhibitor of anaphase similar to fission yeast Cut2. On anaphase proteolysis of Pds1p, which is ubiquitinated by anaphase promoting complex (APC)/cyclosome, Esp1p is liberated to degrade Scc1p, thus resulting in sister chromatid separation (Uhlmann et al. 1999). We are interested in whether the same mechanisms can be applied to fission yeast, as the same set of separin/Cut1 and securin/Cut2 complexes exists (Funabiki et al. 1996; Kumada et al. 1998). On the contrary, most of the cohesin complex was found to be dissociated from chromosomes at prophase, well before anaphase, in Xenopus (Losada et al. 1998). Also, in human and mouse, most of the cohesin exists outside the chromosomes before metaphase (Schmiesing et al. 1998; Darwiche et al. 1999). In higher eukaryotes, the protein complex other than cohesin may act for sister chromatid cohesion until anaphase. Alternatively, a residual fraction of cohesin remaining on the chromosomes may be sufficient for the association of sister chromatids until anaphase.
We therefore addressed the question of whether fission yeast cohesin is dissociated in anaphase, as in budding yeast, or is largely dissociated from chromosomes before mitosis, as in vertebrate cells. To answer this, the cohesin complex had to be identified in fission yeast first. However, only Rad21, a homolog of Scc1p, had been previously investigated (Birkenbihl and Subramani 1995; Tatebayashi et al. 1998) when we began this investigation. By looking for homologs of Smc1p, Smc3p, and Scc3p, all of their counterparts in S. pombe designated Psm1, Psm3, and Psc3, respectively, were identified, and the presence of the cohesin complex was established by analysis of the immunoprecipitates. Chromatin immunoprecipitation (CHIP) and immunofluorescent and green fluorescent protein (GFP)-tagging microscopy showed that the S. pombe cohesin complex differed in one important aspect from vertebrates and budding yeast; namely, the majority of cohesin was located in the nucleus and seemed to be bound to chromatin throughout the cell cycle. To our surprise, the level of associated Rad21 detected by CHIP did not change during the highly synchronous progression from metaphase to anaphase. As in budding yeast, however, a small population of Rad21 was cleaved at the onset of anaphase, and this cleavage was essential for proper progression of anaphase. These results are interpreted in the Discussion section, in which we present a simple model to explain the specific behavior of S. pombe cohesin.
Results
Identification of fission yeast cohesin genes
To characterize the cohesin complex in S. pombe, three genes homologous to S. cerevisiae (SMC1, SMC3, and SCC3) were identified. Two homologous sequences were found in the database (SPAC10F6.09C and SPAC17H9.20P). The psm3+ gene encodes a 1194–amino acid polypeptide (molecular mass = 137 kD) interrupted with one intron (indicated in Fig. 1A by the arrows). The N-terminal, central, and C-terminal regions were 40.7%–62.8% identical to budding yeast Smc3p. The Psm3 sequence was more similar to human hSMC3 in the N and C termini than to S. cerevisiae Smc3p. The sequence in the database for psc3+ similar to Scc3p was partial, so we obtained the whole genomic clone from a cosmid and then sequenced it. Psc3 consists of 978 amino acids (calculated molecular mass = 113 kD) with four putative introns. The central region is 31%–35% identical to homologous proteins. Degenerate polymerase chain reaction (PCR) was employed to isolate the other cohesin gene psm1+ (similar to budding yeast SMC1) using the C terminus as the primer. The PCR fragment obtained was used as the probe to isolate a cosmid that contained the 11.5-kb genomic DNA fragment coding for the whole protein (1228 aa and molecular mass = 140 kD) with one intron. The remaining cohesin gene rad21+ has been described previously (Birkenbihl and Subramani 1992); it is similar to Scc1p. All the postulated cohesin subunit genes could thus be studied.
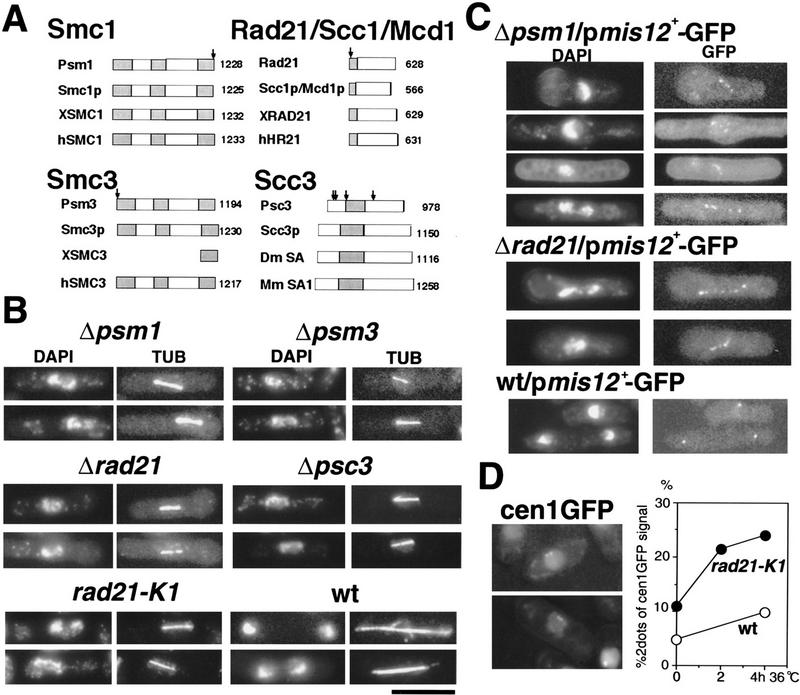
Figure 1.
S. pombe cohesin genes and their disruption phenotypes. (A) Schematic representations of cohesin subunits. S. pombe subunits designated Psm1, Psm3, Rad21, and Psc3 are homologs of budding yeast Smc1p, Smc3p, Scc1p, and Scc3p, respectively. The conserved regions are hatched. The arrows indicate the positions of introns in the S. pombe genes. Except for Rad21, cohesin subunits were first identified in this study. (X) Xenopus; (h) human, (Dm) Drosophila; (Mm) mouse. (B) Phenotypes of gene-disrupted Δpsm1, Δpsm3, Δrad21, and Δpsc3. Gene-disrupted cells were observed after germination (33°C for 10 h) using DAPI for DNA and anti-tubulin (TUB) antibody. Aberrant segregation phenotypes of disrupted cells were very similar. Temperature-sensitive rad21-K1 (36°C for 4 h) and wild-type mitotic cells (26°C) are shown as control. The bar, 10 μm. (C) Localization of Mis12–GFP, a GFP-tagged kinetochore protein, in cohesin gene disruption mutant cells. Gene-disrupted cells (Δpsm1, Δrad21) expressing Mis12–GFP were observed after DAPI staining. The GFP dots represent kinetochores, the number of which exceeded more than three, indicating that sister kinetochores were prematurely separated. (D) The LacO repeat was integrated in rad21-K1 mutant about 30 kb apart from the inner centromere region of chromosome I and the repeat was visualized by the LacI–NLS protein tagged with GFP (Straight et al. 1996; Nabeshima et al. 1997). Cells grown at 26°C were shifted to 36°C for 4 h, and interphase cells were measured for the frequencies revealing two GFP signals (two examples shown in the left panel).
Fission yeast cohesin subunits are essential for sister chromatid cohesion, and their defects lead to blocking mitosis
To examine defective phenotypes of the cohesin subunit genes, gene disruption was performed by one-step replacement using the S. pombe ura4+ gene as the marker. Disruption of psm1+, psm3+, rad21+, and psc3+ was verified by genomic Southern hybridization of heterozygous diploid cells (data not shown). Tetrad analysis of the sporulated diploids indicated that only two spores were viable, and both were Ura−. All these genes were, hence, essential for cell viability.
Gene-disrupted cells were germinated in the absence of uracil. They were observed by fluorescence microscopy using DAPI for DNA and antitubulin antibody for microtubules (Fig. 1B). The wild-type cells are shown as control at the bottom. Interestingly, all disrupted strains produced phenotypes similar to those of temperature-sensitive (ts) rad21-K21 mutant at the restrictive temperature (36°C). The phenotypes were defective in mitosis, revealing the short spindle along which condensed chromosomes were scattered. Similar phenotypes were also found in adherin mutant mis4-242 cells (Furuya et al. 1998; Goshima et al. 1999). These arrest phenotypes appeared to be caused by the Mad2-dependent spindle checkpoint, as the double mutant lacking mad2 exited mitosis without the delay (K. Furuya, Y. Toyoda, and M. Yanagida, in prep.).
To assess whether precocious separation took place, behavior of the kinetochores was examined in Δpsm1 and Δrad21 gene-disrupted cells. For this purpose, plasmid-carrying Mis12–GFP (an essential kinetochore protein tagged with GFP; Goshima et al. 1999) was introduced into the heterozygous diploids, and the GFP signals were observed in haploid segregants germinated in the absence of uracil (Fig. 1C). Mis12–GFP signals were dispersed into five or six signals in Δrad21 and Δpsm1 haploid cells (10% frequency), whereas they were not observed in the wild-type control. Sister cohesion was thus at least partly impaired in the gene-disrupted cells.
The defect of the sister chromatid cohesion was also verified in ts rad21-K21 mutant cells (Tatebayashi et al. 1998) by examining the centromere DNA. For this, the Lac repressor tagged with GFP and nuclear localization signal (NLS) was expressed and associated with the LacO DNA repeats, which were integrated into the lys1 locus near the cen1 (Straight et al. 1996; Nabeshima et al. 1997). Interphase rad21 mutant cells frequently revealed the two GFP signals, which presumably represented the separated sister pericentromere DNAs. The frequency of cells with the two GFP cen1 dots increased to 25% after 4 h at 36°C, higher than the wild-type control (Fig. 1D). These results showed that sister cohesion was impaired in ts-mutant as well as in gene-disrupted cells.
A stable complex formation by Psm1, Psm3, and Rad21 but not by Psc3
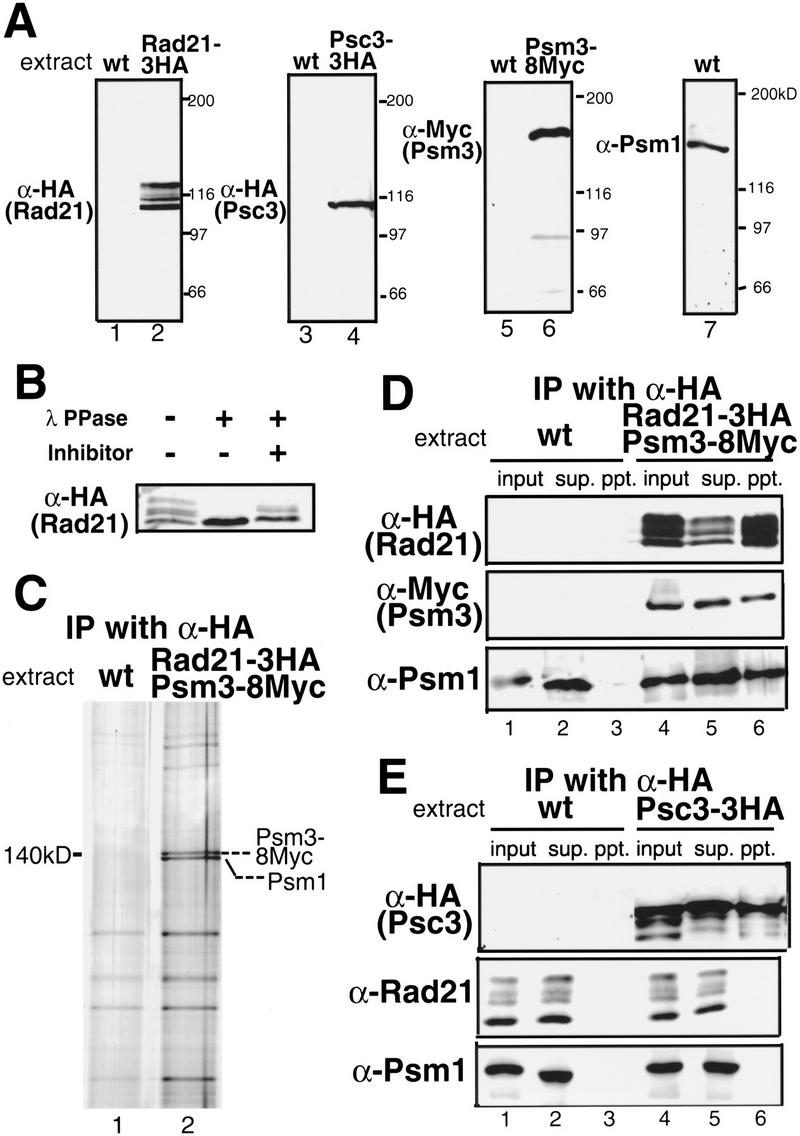
To examine whether Psm1, Psm3, Rad21, and Psc3 formed the complex in vivo, strains were constructed in which the genes for Rad21, Psc3, and Psm3 were tagged with 3xHA or 8xMyc, integrated into the chromosome under the native promoter. These tagged proteins were identified by immunoblotting (Fig. 2A). Polyclonal antibodies raised against the recombinant protein (aa 1–631) were used to detect Psm1. Rad21–3HA produced multiple phosphorylated bands over 100 kD (Birkenbihl and Subramani 1995), whereas Psc3–3HA, Psm1, and Psm3–8Myc showed the expected 116, 140, and 148 kD bands, respectively. No band was detected in the control wild-type cells (wt) when antibodies against HA or Myc were used. Upper bands of Rad21 were produced by phosphorylation because they were lost after phosphatase treatment (Fig. 2B), as reported previously (Birkenbihl and Subramani 1995).
Figure 2.
Stable complex formation of cohesin subunits. (A) Detection of Rad21, Psm1, Psm3, and Psc3 by immunoblot using anti-HA(12CA5), anti-Myc(9E10) monoclonal antibodies or polyclonal antibody against the recombinant Psm1 protein. Extracts of the strains chromosomally integrated with Rad21–3HA, Psc3–3HA, or Psm3–8Myc under the native promoter were used. Wild-type control produced no band by anti-HA or anti-Myc antibodies. (B) Extracts of cells expressing tagged Rad21–3HA were treated with (+) or without (−) λ protein phosphatase and a phosphatase inhibitor (10 mM sodium orthovanadate) at 30°C for 30 min. Upper modified bands were diminished and the lowest bands were intensified after treatment. (C) To identify stable complex formation of cohesin subunits, a strain chromosomally integrated with Rad21–3HA and Psm3–8Myc under the native promoter was constructed and employed for immunoprecipitation using antibodies against HA. Silver staining of the precipitated proteins is shown. Proteins present in the precipitates from the integrant strain but not from the wild-type corresponded to Psm3–8Myc and Psm1. (D) Extracts of cells prepared in (C) were examined by immunoblot using antibodies against anti-HA, anti-Myc, and anti-Psm1 antibodies. Psm1 and Psm3–8Myc were coprecipitated with Rad21–3HA. (E) Extracts of cells expressing tagged Psc3–3HA were prepared, and immunoprecipitation was performed using anti-HA antibodies. Precipitates obtained were immunoblotted using anti-HA, anti-Rad21, and anti-Psm1 antibodies. Neither Psm1 nor Rad21 was coprecipitated with Psc3–3HA.
Rad21 was immunoprecipitated by anti-HA antibody using extracts of the doubly integrated strain with Rad21–3HA and Psm3–8Myc under their native promoters. Coimmunoprecipitated proteins were silver stained (Fig. 2C). Two intense silver bands around 140 kD, which were not present in the wild-type extracts, were identified as Psm1 and Psm3–8Myc by immunoblotting (Fig. 2D). Although precipitated Rad21–3HA was clearly detected by immunoblotting, it was not seen as a single silver-stained band in immunoprecipitates, probably because it was multiply hyperphosphorylated (Fig. 2C). It was also highly susceptible to proteolysis during extract preparation.
No silver-stain band corresponding to Psc3 was detected in the immunoprecipitates. We examined by immunoprecipitation whether Psc3–3HA was bound to Rad21 and/or Psm1. Anti-HA antibody was used to immunoprecipitate Psc3–3HA produced by the integrated gene under its native promoter. Neither Psm1 nor Rad21 was coprecipitated with Psc3–3HA (Fig. 2E). This result was unexpected because Scc3p, the homolog in budding yeast, is a subunit of cohesin. In fission yeast, Psc3 might loosely bind to the cohesin complex. Another possiblity is that only the insoluble chromatin-bound fraction of Psc3 would be able to form the complex with other subunits. Alternatively, Psc3 might interact with cohesin only during a particular cell cycle stage such as the S phase. Immunoprecipitation was done for cells arrested in the presence of hydroxyurea, but Psc3 was not coprecipitated with Rad21 or Psm1 (data not shown). Psc3, thus, might form the complex with an undetectable fraction of cohesin or not be a stable member of the cohesin subunits in fission yeast.
Viability loss of rad21-K1 and phosphorylation of Rad21 protein occurs during the S phase
To examine whether Rad21 becomes essential when cells traverse the G1/S phase, rad21-K1 mutant cells were first arrested in the G1 phase by nutrient starvation at 26°C and then released to the rich medium at the restrictive temperature (36°C). Under the culture conditions, the traverse of cells from the G1 to the M phase could be monitored. Viability of rad21-K1 decreased 2 h after the release, at the timing of the S phase (Fig. 3A), as verified by the FACS pattern. The amounts of cellular DNAs in wild-type and rad21-K1 cells shown at right indicated that replication occurred between 2 and 4 h in rad21-K1. The defective phenotype observed in the subsequent mitosis was similar to that found in gene-disrupted Δrad21 cells (data not shown). These results suggested that Rad21 became essential during replication. Judging from the FACS pattern, the completion of DNA replication was somewhat delayed in rad21-K1.
Figure 3.
Essential role of Rad21 and its phosphorylation during the S phase. (A) The nitrogen-starved culture of rad21-K1 mutant at 26°C was transferred to the complete medium at 36°C for 8 h. Cell viability and the DNA content were measured by plating and FACScan analysis, respectively. The wild-type control is also shown. Cell viability in the mutant culture initially arrested at the G1 phase decreased during the S phase, which occurred 2–4 h after the shift. (B) Upper phosphorylated bands of Rad21 increased during the S phase. Wild-type cells integrated with the Rad21–8Myc gene were nitrogen starved at 26°C and shifted to the complete medium at 36°C for 8 h as in (A). Cell extracts made were immunoblotted using anti-Myc antibody. Hyperphosphorylated forms of Rad21 were abundant in 2–4 h and subsequently hypophosphorylated form increased (upper panel). Note that the single upper band faintly seen in the arrested cells was not hyperphosphorylated (see text). (C) Upper phosphorylation bands of Rad21 were examined in rad21-K1 mutant. Wild-type cells (lane 1), rad21-K1 at 26°C (lane 2) and at 36°C for 4 h (lane 3). Upper phosphorylated forms were greatly diminished in rad21 mutant cells.
Rad21 was phosphorylated, producing multiple bands, and this phosphorylation was proposed to be functionally relevant as the phosphorylated form was diminished in a radiosensitive mutant rad21-45 (Birkenbihl and Subramani 1995). It was also shown that phosphorylation of Rad21 was extensive in the S/G2 phase (Birkenbihl and Subramani 1995), which indicates that the hyperphosphorylated form of Rad21 is abundant during S/G2. We examined the degree of phosphorylation of Rad21 under a culture condition of wild-type cells, first nitrogen starved in the G1 phase and then released to the rich medium. As seen in the immunoblot pattern (Fig. 3B), hyperphosphorylated forms of Rad21 were already abundant 2 h after the release, corresponding to the onset of the S phase, and reached the maximum after 4 h (the FACS analysis shown below indicated the S phase from 2–4 h). A faint upper band seen in nitrogen-starved cells was due to an unknown cause and probably not caused by phosphorylation, as the same band was obtained after protein phosphatase treatment (data not shown). Rad21 was reported to be hypophosphorylated in the G1 arrested cdc10-129 mutant (Birkenbihl and Subramani 1995) and we obtained the same result (data not shown). It was then found that the level of hyperphosphorylated Rad21 decreased in rad21-K1 cultured at both 26° and at 36°C for 4 h (Fig. 3C). The higher degree of Rad21 phosphorylation was, thus, in parallel with more established sister chromatid cohesion.
CHIP reveals abundance of cohesin subunits in the centromere regions
To examine whether the cohesin complex interacts with specific chromosome regions, CHIP was performed using various DNA probes. Rad21–3HA, Psm1–3HA, or Psc3–3HA integrated cells under their native promoters were grown exponentially and fixed with 1% formaldehyde and then extracts were immunoprecipitated with anti-HA antibody after genomic DNAs were sheared by sonication to an average length of 800 bp. DNAs coprecipitated with Rad21–3HA, Psm1–3HA, or Psc3–3HA were amplified by the PCR method using primers whose chromosomal locations are illustrated in Figure 4A. They abundantly reside either at the centromeric and pericentromeric regions in chromosome I, but the level was much less at the arm region near the cdc2+ gene of chromosome II ( Fig. 4B). The cnt1 and dg are the probes for the inner and outer centromere regions, respectively, whereas lys1 is in the pericentric arm probe of chromosome I (Takahashi et al. 1992). Another 10 probes (a1–a10) were derived from a single cosmid c1750 (Machida et al. 2000).
Figure 4.
Interaction of Rad21 with centromere and pericentromere. (A) Left panel, schematic representation of cen1, the centromere of chromosome I. Positions of the centromere (cnt1, dg) or pericentromere (lys1) primers are indicated by the vertical lines. Right panel, ten primers derived from c1750, a cosmid covering the cdc2+ gene. (B) CHIP (chromatin immunoprecipitation) was performed using the probes described in A. Extracts of wild-type (wt, no tag), or integrants with Rad21–3HA, Psm1–3HA, Psc3–3HA, or Mis6–8Myc under the native promoter were made. Immunoprecipitation was done using anti-HA antibody (anti-Myc antibody used for the control Mis6), and PCR was done for the precipitates. Control Mis6, a kinetochore protein, was associated only with cnt1 (Saitoh et al. 1997). (C) Quantification of the PCR DNA products is shown. Serial dilutions of the template DNA were made for PCR reactions to optimize the PCR products within linear range. Coprecipitated DNAs with proteins were amplified and the percentage of precipitate DNA compared with total DNA was calculated by measuring the intensity of the PCR products.
The cohesin subunits were clearly distinguished from Mis6, an authentic centromere protein, which was restricted only in the inner centromere region essential for chromosome segregation (Saitoh et al. 1997; Takahashi et al. 2000). In these experiments, the amount of template DNA was optimized to generate the PCR products within a linear range by serial dilutions of total or coprecipitated DNAs (Tanaka et al. 1999). The amplified coprecipitated DNAs with Rad21–3HA, Psm1–3HA, or Psc3–3HA were then examined by calculating the percentage of coprecipitated DNA compared with total DNA (Fig. 4C). The maximum distribution of the cohesin subunits was found in the repetitive outer centromere region, containing dg and dh repeats, which is not vital to the centromere function in chromosome segregation (Takahashi et al. 1992). The low levels of coprecipitated DNA derived from the arm region were variable depending on the primers used. More extensive analysis is needed to determine how these proteins are distributed along the entire arms.
Intracellular location of cohesion proteins
Intracellular location of Rad21 was examined in the wild-type strain integrated with Rad21–Myc expressed from the native promoter. Fixed cells were observed by immunofluorescence microscopy using anti-Myc and SPB antibodies (Fig. 5A). Rad21–Myc was located mainly in the nuclear chromatin region from interphase through mitosis. Centromere-specific staining was not observed. GFP-tagged Rad21 integrated on a chromosome with the native promoter also did not show specific centromere localization in living and fixed cells (see below). No significant change was recognized in cells at different cell cycle stages, consistent with the previous study (Birkenbihl and Subramani 1995). Basically identical nuclear localization was obtained for Psm1 and Psm3 (data not shown).
Figure 5.
Rad21 localizes in the nucleus throughout the cell cycle (A) Fixed wild-type cells expressing integrated Rad21–8Myc were stained by DAPI for DNA, by anti-Sad1 antibody against Sad1, an SPB protein, and by anti-Myc antibodies against Rad21-8Myc. Rad21 was localized in the nuclear chromatin region showing the punctate signals. (B) Localization of Psc3–HA and Rad21 was observed by immunofluorescence microscopy. The merged image is produced by red color Psc3–HA and green color Rad21. They were similar but did not appear to be identical. (C) The detergent-washing procedures (in situ chromatin binding assay; Kearsey et al. 2000) recently developed for visualising the S phase-specific nuclear retention of MCM protein essential for replication (Cdc21 is one of them) were applied to Rad21–GFP. Cdc21–GFP showed the nuclear signal only in the S phase cells, while Rad21–GFP signals were seen in all the cell cycle stages.
Immunoprecipitation experiments suggested that Psc3 might not be a subunit of the stable complex. We therefore determined whether Rad21 and Psc3 were distinctly distributed in the nucleus. Psc3–HA integrant cells were doubly stained as shown in Figure 5B. The individual and merged images taken in the same cells indicated that the location of Psc3–HA was very similar but did not appear to be identical to that of Rad21. Basically identical results were obtained under different staining conditions.
Although Rad21 was found in the nucleus throughout the cell cycle, its interaction with chromatin might be altered during the cell cycle. The procedure developed for visualizing cell cycle–dependent location of MCM proteins in S. pombe (Kearsey et al. 2000) was applied to Rad21 tagged with GFP. Cdc21, an MCM protein essential for replication, remained specifically in the nucleus only during the G1/S phase when the detergent washing step was introduced (Fig. 5C, top). If the detergent step was not used, Cdc21 remained in the nucleus throughout the cell cycle (data not shown). After detergent washing, however, no cell cycle–dependent nuclear localization was observed for Rad21–GFP. Rad21–GFP was found in the nucleus throughout the cell cycle regardless of the absence or presence of the detergent washing procedures.
Cleavage and phosphorylation change of Rad21 during metaphase–anaphase transition
To examine whether the chromatin interaction of Rad21–HA altered during the progression from mitotic metaphase to anaphase, the CHIP experiment was performed using extracts of cold-sensitive β-tubulin mutant, nda3-KM311 (Fig. 6A). Mutant cells were first arrested at mitotic prophase by culturing them at 20°C for 8 h and then released to the rapid progression into metaphase followed by anaphase using the temperature shift to 36°C, the permissive temperature. A highly synchronous cell cycle advance took place from the mitotic prophase to anaphase within 10 min (Hiraoka et al. 1984). The transition from metaphase to anaphase occurs very rapidly and highly synchronously at ∼6 min as seen by the destruction of mitotic cyclin Cdc13 (Fig. 6A, α-Cdc13) and the peak of cells showing a dividing nucleus in the early anaphase (filled circles in the bottom time course of Fig. 6A). Under these culture conditions, the levels of three probes, cnt1, dg, and lys1, coprecipitated with antibodies against Rad21–HA, did not significantly alter during the transition from mitotic prophase to anaphase (0–9 min). The results were strikingly different from the behavior of Scc1p in budding yeast (Tanaka et al. 1999), suggesting that the bulk of Rad21 bound to these probe DNAs did not dissociate from chromatin during the mitotic transition in nda3-KM311 cells. Basically the same result was obtained for other synchronous cultures by the release of G2-arrested cdc25 mutant cells (data not shown).
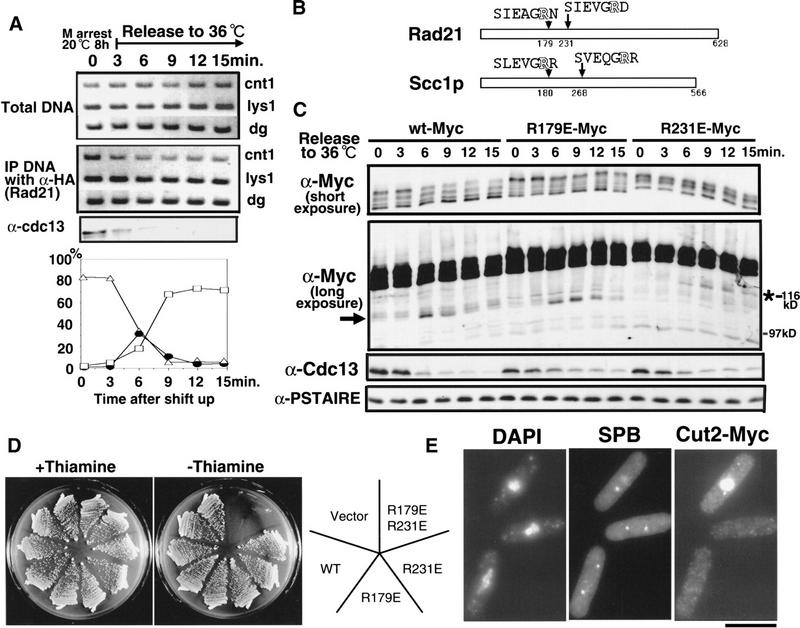
Figure 6.
Behavior of wild-type and substitution mutant Rad21 proteins during the metaphase–anaphase transition in nda3-KM311. (A) nda3-KM311 strain integrated with the tagged Rad21–3HA gene under the native promoter was arrested at 20°C for 8 h, then shifted to 36°C, the permissive temperature, and aliquots of the culture were taken every 3 min. Extracts prepared were immunoprecipitated using anti-HA antibodies, and coprecipitated DNAs were amplified by PCR using the three different primers indicated. The level of Cdc13 is shown as the control of timing for cyclin destruction. The frequency of cells showing dividing nucleus in early anaphase (filled circles) peaked at 6 min when Cdc13 was rapidly degraded. Open triangles, cells containing a single nucleus; open rectangles, cells containing two nuclei. (B) Schematic drawing of the two putative arginine R cleavage sites in Rad21. (C) Prophase-arrested nda3-KM311 cells integrated with the wild type Rad21–8Myc (wt-Myc), single substitution mutants Rad21 R179E–8Myc or Rad21 R231E–8Myc under the native promoter were released to the permissive temperature as in A. Aliquots of the culture were taken every 3 min. Extracts were then prepared and immunoblotted using anti-Myc, anti-Cdc13 (mitotic cyclin) and anti-PSTAIRE antibodies. Top panel, immunoblot patterns taken after a brief exposure (α-Myc short exposure). Second panel, immunoblot pattern taken after the long exposure (α-Myc long exposure). The arrow and asterisk show the positions of cleaved fragments in wt-Myc, R179E-Myc and R231E-Myc. These cleaved products lack the N-terminal 231 and 179 amino acids, respectively. (D) Wild-type cells carrying plasmid alone (vector), the wild-type rad21+ gene, the single-cleavage mutants (R179E and R231E) or the double mutant R179ER231E under the REP41 promoter were streaked on the synthetic EMM2 plates with (promoter off) or without (promoter on) thiamine. Plates were incubated at 33°C. (E) Plasmid containing the double cleavage mutant gene R179ER231E under the REP81 promoter was introduced in the wild type strain integrated with the tagged Cut2–8Myc gene. Cells were grown in the liquid EMM2 medium in the presence of 2 μM thiamine at 33°C, followed by the removal of thiamine for 14 h in order to induce the expression of the toxic double-mutant protein R179ER231E. Cells were fixed and stained by DAPI for DNA, by anti-Sad1 antibodies for the SPB, and by anti-Myc antibodies for Cut2-Myc. The bar indicates 10 μm.
We then addressed the question of whether Rad21 was cleaved during the mitotic transition as found for budding yeast Scc1p. Rad21 contains two potential cleavage R residues at the 179 and 231 amino acid positions (Fig. 6B; Uhlmann et al. 1999). It was examined whether the cleavage could be detected in the synchronous culture of nda3-KM311, and the cleavage pattern would be altered if the noncleavable mutation of Rad21 was introduced. Wild-type and two-substitution Rad21 mutants (the change to E from R) were tagged with Myc (wt-Myc, R179E-Myc, R231E-Myc) integrated into a chromosome of the nda3-KM311 strain with the native promoter. Immunoblot patterns of those integrated nda3-KM311 cells during the synchronous mitotic progression are shown in Figure 6C.
The cleaved product was seen only after rather prolonged exposure of the immunoblot patterns (indicated by the arrow in the middle panel). In the short exposure (top panel), however, only multiply phosphorylated Rad21 bands were seen: No cleaved band was detected in the molecular mass range between 20 and 110 kD (data not shown). The amount was <5% of the total Rad21, judging from the exposure time required for revealing the cleaved band. The intensity of the hyperphosphorylated uppermost band decreased ∼6 min after the shift in nda3-KM311 integrated with the wild-type Rad21 tagged with Myc (wt-Myc), whereas it remained significantly high in nda3-KM311 integrated with mutant R179E-Myc or R231E-Myc. Note that the hypophosphorylated lowest band increased the intensity.
The cleaved product appeared only transiently in anaphase (6 min) in wt-Myc. It was ∼25 kD smaller than the full-length hypophosphorylated Rad21–Myc in either wt-Myc or R179E-Myc (the position indicated by the arrow) but 20 kD smaller in R231E-Myc (the position indicated by the asterisk), suggesting that Rad21–Myc was cleaved at the presumed sites. This was confirmed by our observation that the cleaved bands of R179E-Myc and R231E-Myc migrated at the same positions as the N-terminally truncated 231 and 179 aa fragments, respectively (data not shown). Taken together, uppermost hyperphosphorylated Rad21 appeared to be cleaved during the metaphase–anaphase progression at the presumed sites, because the decrease of the band intensities was retarded in the noncleavable mutants of Rad21. Dephosphorylation or very fast de novo synthesis of Rad21 occurred in anaphase. Note that single cleavage mutants grew normally at any temperature but introduction of the double-mutant gene with the native promoter has not been successful, probably because of the toxic effect of the product.
Requirement of the cleavage for normal progression of anaphase
To determine if the cleavage of Rad21 is essential for sister chromatid separation, the single-cleavage mutants (R179E and R231E) or the double mutant (R179E R231E) were moderately overexpressed under the REP41 promoter in the wild-type cells (Fig. 6D). Neither the wild-type protein nor the single mutant proteins (R179E, R231E) affected cell proliferation when Rad21 under the inducible promoter was synthesized by depleting thiamine from the media. In contrast, overexpression of the double mutant Rad21 (R179ER231E) using the same inducible promoter became lethal (Fig. 6D). To investigate its cellular phenotype, liquid cell cultures were made in the absence of thiamine. Sister chromatid separation was indeed blocked in cells expressing the double mutant Rad21, even though the degradation of Cut2/securin normally took place, which was the indication of cell cycle progression into anaphase (two lower cells in Fig. 6E). Thus, expression of the double mutant Rad21 was highly inhibitory against sister chromatid separation in anaphase but not against Cut2 destruction. These results indicate that the cleavage of Rad21 occurring in anaphase may be essential for proper anaphase.
Recruitment of Rad21 requires fully functional Mis4
To examine whether location of Rad21 protein might be altered in mis4-242 mutant, rad21+ gene was tagged with GFP at the C terminus and integrated into the chromosome under the native promoter, and its fluorescence was observed in living cells (Fig. 7A). The GFP signal was in the nuclear chromatin regions with punctate spots in the wild-type cells. However, in mis4-242 mutant cells, the punctate signals were present but significantly reduced even at the permissive temperature and virtually disappeared after 4 h at 36°C.
Figure 7.
Recruitment and phosphorylation of Rad21 requires fully functional Mis4. (A) Punctate appearance of Rad21–GFP seen in the wild-type nucleus was diminished in mis4-242 at 26°C and completely vanished at 36°C for 4 h. Rad21–GFP was chromosomally integrated and expressed under the native promoter. Punctate staining did not correspond to localization of the centromeres. (B) Extracts of wild type cells (lanes 1,5), mis4-242 mutant cells integrated with Rad21–3HA grown at 26°C or 36°C (lanes 2,3,6,7) or the wild-type cells integrated with Rad21–3HA (lanes 4,8) were prepared and used for CHIP with anti-HA antibodies. Total DNA before immunoprecipitation (lanes 1–4) and coprecipitated DNA (lanes 5–8) were amplified by PCR using the probes, cnt1, dg, and lys1. Association of Rad21 with chromatin was greatly reduced at cnt1, dg locus and almost diminished at lys1 locus in mis4-242 mutant cells even at 26°C, the permissive temperature when compared with the untagged wild type. (C) Upper phosphorylation bands of Rad21 were examined in mis4-242 mutant. Wild-type cells at 26°C (lane 1), mis4-242 mutant cells at 26°C (lane 2), and at 36°C for 4 h (lane 3). Upper phosphorylated forms were greatly diminished in the mutant cells.
The CHIP experiment was done to determine whether the DNA probes cnt1, dg, and lys1 were coprecipitated in the wild-type and mis4-242 mutant equally well with anti-HA antibodies against Rad21–3HA. Both wild-type and mis4 strains were integrated with Rad21–3HA with the native promoter into the chromosome. Neither the centromere DNA fragments, cnt1 and dg, nor the pericentric arm DNA fragment, lys1, were amplified at all in mis4-242 after 4 h at 36°C (Fig. 7B, lane 7). Surprisingly, the amplified DNA fragments at cnt1 and dg were significantly reduced and the lys1 DNA fragment was hardly amplified even at the permissive temperature (Fig. 7B, lane 6). These results suggest not only that fully functional Mis4 is required for normal loading of Rad21 onto chromatin but also that a relatively low level of Rad21 association with chromatin could be sufficient for cells to survive. It should be stressed that mis4-242 could grow nearly normally at 26°C, whereas cohesin was only weakly bound to the chromatin region. Finally, hyperphosphorylated Rad21 in mis4-242 was decreased at 26°C, the permissive temperature, and greatly diminished at 36°C, the restrictive temperature; this result is similar to that in the rad21-K1 mutant (Figs. 3C, 7C).
Discussion
This article reports that all four genes of fission yeast cohesin subunits, Rad21, Psm1, Psm3, and Psc3, are essential for cell viability. Their gene disruption phenotypes are highly similar, which is consistent with the notion that they form the same essential complex reported in other organisms (Michaelis et al. 1997; Losada et al. 1998; Toth et al. 1999). We were able to demonstrate that Rad21, Psm1, and Psm3 formed the complex. Precocious sister chromatid separation was observed in cohesin subunit mutants, strongly suggesting that fission yeast cohesin was also required to establish sister chromatid cohesion. As ts rad21-K1 mutant cells lost viability during the S phase, sister cohesion was likely to be established by the complex during the S phase. However, the cleavage of Rad21, although its level is rather fractional, must take place in anaphase, and overproduction of the noncleavable double Rad21 mutant protein blocks sister chromatid separation, strongly suggesting that Rad21 has a role in the anaphase as well as in the S phase.
Immunoprecipitation failed to show that Psc3, a homolog of Scc3p, was a part of the complex. Immunolocalization and CHIP showed that Psc3 behaved similarly (but not identically) to other cohesin subunits. Certain diversifications may thus exist in the subunit compositions of cohesin. In frog, a total of five cohesin subunits were reported (Losada et al. 1998, 2000). Gene disruption results presented in this paper showed that the loss of any four subunits led to the same phenotypes, suggesting that the complex acts as a functional entity and that each of the subunits is essential for its function. Psc3 is probably functionally related to cohesin, and it might perform its essential function without complex formation. Alternatively, a small population of cohesin that carries essential function may consist of the four subunits. These are equally possible scenarios.
Rad21 was characterized by its hyperphosphorylation: Multiple bands produced became a single band after protein phosphatase treatment. Human and frog Scc1p/Rad21 homologs, in contrast, were detected as a single band by polyclonal antibodies (Losada et al. 1998; T. Matsusaka and M. Yanagida, unpubl.). Whether vertebrate Rad21/Scc1p homologs are actually phosphorylated remains to be determined. Hyperphosphorylation of fission yeast Rad21 was a cell cycle–dependent phenomenon. It appeared to occur at the onset of S phase when the nitrogen-starved G1 cells were released in the rich medium.
Is this phosphorylation of Rad21 functionally relevant? It appears to be the target of proteolysis, but we have no definitive evidence that phosphorylation is functionally essential. The previous (Birkenbihl and Subramani 1995) and the present studies showed that hyperphosphorylated Rad21 was diminished both in radiosensitive rad21-45 and ts rad21-K1. In addition, phosphorylation was greatly reduced in mis4 mutant, in which recruitment of Rad21 to punctate nuclear signals and chromatin precipitates were greatly diminished. This hyperphosphorylation might be directly related to the activation of Rad21 for cohesion. Alternatively, phosphorylated Rad21 was possibly recruited to proper chromatin sites. Later in anaphase, phosphorylated Rad21 might be degraded. The previous work indicated that the phosphorylated region resided within the N-terminal 215 amino acids (Birkenbihl and Subramani 1995); this region contains many S and T residues and has one Cdc2 site. A protein kinase (or kinases) responsible for hyperphosphorylation of Rad21 is likely to be active at the onset of the S phase. Its identification is of considerable interest.
CHIP experiment showed that Rad21, Psm1, and Psm3 were bound to centromere and pericentromere DNA probes much more abundantly than the probes present in the arm. It is very unlikely, however, that the cohesin complex functions as an essential kinetochore component, as do Mis6, Mis12, or Cnp1 in fission yeast (Saitoh et al. 1997; Goshima et al. 1999; Takahashi et al. 2000). These kinetochore chromatin proteins are present exclusively in the central innermost centromere regions essential for maintaining chromosomes, whereas the cohesin proteins exist in a much broader region including the outer centromeric repeats dg and pericentric arm probes such as lys1 not required for correct chromosomal segregation. Indeed, centromere-specific chromatin (Takahashi et al. 2000) was maintained in rad21-K1 mutant cells (K. Furuya, K. Takahashi, and M. Yanagida, in prep.).
Important behavior of Rad21 was revealed during the metaphase–anaphase transition in nda3-KM311 mutant. First, chromatin-bound Rad21 did not seem to alter abruptly from metaphase (3 min) to anaphase (6 min) within the resolution of detection. Second, the same nuclear localization of Rad21 was observed during the cell cycle by regular and detergent-treated fluorescence microscopy that was successful in demonstrating the S phase–specific nuclear localization of MCM proteins in S. pombe. In contrast to the above two unchanged properties, the state of hyperphosphorylated Rad21 was grossly altered. Its level was dramatically decreased from metaphase to anaphase in nda3-KM311 mutant. This rapid decline was shown to be partly caused by proteolysis and also probably by protein dephosphorylation, as the intensity of the hypophosphorylated form increased from metaphase to anaphase. Among many phosphatases studied, only Dis2 PP1 phosphatase has been known to be activated from metaphase to anaphase because of the down-regulation of Cdc2 kinase (Ishii et al. 1996).
It is unknown whether only single protease is responsible for the decrease of Rad21. In budding yeast, Esp1 seems to be the sole protease for anaphase destruction of Scc1p (Uhlmann et al. 1999). Because the cleavage sites of Rad21 were similar to those of Scc1p, the protease responsible for this cleavage is probably Cut1, a homolog to Esp1p. We, however, have not been able to examine whether proteolysis of Rad21 was delayed in cut1 mutant cells, as inactivation of mutant Cut1 protein in the temperature-shift experiment requires extensively long time, whereas the cleavage took place nearly immediately after the temperature shift.
Is the cleavage of Rad21 in anaphase functionally essential? As only a very small fraction was cleaved, such cleavage might not affect overall function of Rad21. This cleavage, however, was most likely essential for a proper anaphase, as in budding yeast, because overexpression of noncleavable double mutant Rad21 protein (R179ER231E) in wild-type cells was highly toxic and prevented sister chromatid separation. In budding yeast, the cleavage of Scc1p appeared to be sufficient not only for sister chromatid separation but also for triggering the metaphase-to-anaphase transition (F. Uhlmann and K. Nasmyth, unpubl.). The cleavage of Rad21/Scc1p might be a general phenomenon because a small amount of human SCC1 is cleaved at the onset of anaphase (J.M. Peters, pers. comm.).
We propose a simple model to explain the specific behavior of Rad21 in fission yeast. First, Rad21 may be highly abundant in this organism, and the presence of its tiny fraction becomes functionally essential in anaphase. It is unknown whether this essential Rad21 fraction is entirely destroyed during the metaphase–anaphase progression. Second, the bulk of chromatin-associated Rad21 remains along chromosomes in anaphase without causing any harm to cells. It might be necessary to remain bound to chromatin for the establishment of cohesion at the next S phase because the duration of G1/S is very short in fission yeast. Third, the difference between the bulk and the fractional Rad21 that undergoes proteolysis may be caused by localization or protein modification or both. These hypotheses can explain many of the results that we obtained. The same localization and CHIP results during the mitotic transition are likely to represent the behavior of the bulk fractions of Rad21. Behavior of the functionally essential Rad21 may be obscured by its paucity. Note that the C-terminal fragment can associate with the other two cohesin subunits (A. Murakami and M. Yanagida, unpubl.), so that Rad21 could remain bound to chromatin even after it is cleaved on anaphase.
Another attractive hypothesis is that the essential population of Rad21 is located in restricted chromosome regions such as the centromeres. This would explain the paucity of functionally essential Rad21. Indeed, in human cells, a small amount of SCC1 remains bound to centromeric regions until the onset of anaphase (J.M. Peters, pers. comm.). Likewise, in fission yeast, cohesin subunits are abundantly present in the centromere regions, as they are in budding yeast. Furthermore, Rad21 hardly binds to the pericentric arm region, but little remains associated with centromere in mis4 mutant cells at the permissive temperature. Therefore, it may be possible that proteolysis of a very small fraction of Rad21 located at the centromere is essential for sister chromatid separation. No further results, however, support this hypothesis because no variation in the centromere regions was yet obtained from the CHIP experiments of the synchronous nda3-KM311 culture and localization of GFP-tagged Rad21.
There has been no definitive experimental evidence reported for the direct structural role of the cohesin complex for bridging the sister chromatids (Nasmyth 1999; Yanagida 2000). If cohesin is a catalyst rather than a structural link or glue between sister chromatids, the form free from chromatin may still be able to execute its function. In fact, factors interacting with SMC proteins have been reported to show various enzymatic activities. Bovine recombination complex-1 (RC-1) contains DNA polymerase ɛ and ligase III as well as SMC1 and SMC3 (Jessberger et al. 1996). SbcC, an SMC family protein in Escherichia coli, interacts with SbcD, a nuclease (Connelly et al. 1998). In addition, Trf4p in budding yeast, which is required for sister chromatid cohesion, was recently found as DNA polymerase κ (Wang et al. 2000). The presumed synthetic lethality of the rad21-K1 mutation with mis5-268 or cdc17-K42 (data not shown), which are defective, respectively, in an MCM protein (Takahashi et al. 1994) having DNA helicase (Ishimi 1997) and ligase activity, indicates the S phase role of Rad21 and also its implication in DNA metabolism.
Material and methods
Strains and media
The S. pombe haploid strains used were wild-type 972h− and its derivative strains: nda3-KM311 (Hiraoka et al. 1984), mis4-242 (Furuya et al. 1998), and rad21-K1 (Tatebayashi et al. 1998). For gene disruption, a diploid strain was made by crossing haploid ST485 (h+ leu1 ura4 ade6-210) and ST486 (h− leu1 ura4 ade6-216). The culture media used were complete YPD (1% yeast extract, 2% Bactopeptone, and 2% glucose) and the minimal EMM2.
Epitope tagging, plasmid, and construction of Rad21 mutant strains
NotI restriction sites were introduced at the C termini of the coding regions of psm1+, psm3+, rad21+, psc3+, and cut2+ genes by PCR. The NotI restriction sites were ligated with the 3xHA or 8xMyc or GFP tag sequence, and resulting genes were cloned into an integration vector, pYC6 carrying the S. pombe ura4+ gene or pYC11 carrying the S. cerevisiae LEU2 gene. Resulting plasmids were introduced to either h− ura4 or h− leu strain, respectively. The arginine at aa 179 and 231 of Rad21 were replaced to glutamic acid by site-directed mutagenesis. The resulting mutated rad21 genes were cloned into plasmids pREP41 or pREP81 carrying the nmt1 promoter to induce overexpression of the mutant Rad21. The mutated rad21 genes were also introduced together with 654-nt upstream promoter, the S. pombe leu1+ gene and 8xMyc tag into the leu1-32 locus of heterozygous rad21+ gene-disrupted diploid cells. Correct integration was confirmed by Southern blotting.
Antibodies
To obtain anti-Psm1 antibody, the N-terminal 1894-bp sequence of psm1+ gene was inserted in frame into pGEX-6p (Pharmacia). Glutathione S-transferase (GST)–Psm1 fusion protein was produced in E. coli by inducing expression with 1 mM of isopropyl-β-D-thiogalactoside (IPTG) for 4 h at 36°C. The fusion protein recovered as insoluble inclusion bodies was sonicated, separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and electroeluted. The purified protein was used for immunization, and polyclonal antiserum was obtained. Anti-Rad21 antibody was a gift of Dr. S. Subramani (Birkenbihl and Subramani 1995).
Isolation of psm1+, psm3+, and psc3+
To isolate the psm1+ gene, mixed oligonucleotides based on the conserved C-terminal amino acid sequences of S. cerevisiae, Xenopus, and human SMC1 homologs were made. They were 5′-GA(AG)AA(AG)ACIGTIGCIGCI(CT)TIGC-3′ (encoding EKTVAAL) and 5′-TTIA(GA)I(GC)(TA)(TGA)ATIAC(TG A)AT(GA)AA(TC)TG-3′ (encoding QFIVISL on the complementary strand), and used for degenerated PCR. The resulting 176-bp PCR fragment, which coded for the amino acid sequence 78% identical to the C terminus of S. cerevisiae Smc1p, was employed as the hybridization probe to screen an S. pombe cosmid library, and cosmid 196 located on the long arm of chromosome II was obtained. An 11.5-kb XhoI–SacI fragment of cosmid 196 contained the full-length coding sequence of the psm1+ gene, and its nucleotide sequence was determined. A partial sequence of the psm3+ gene was determined in the database of the Sanger Center as an S. pombe hypothetical ORF and SPAC10F6.09C as a homolog of S. cerevisiae Smc3p. The partial sequence of the psc3+ gene was located on a cosmid SPAC17H9.20P. Using the N terminus 521-bp fragment in the cosmid as a probe, the rest of the genomic coding region of psc3+ was obtained from cosmid 456 located on the long arm of chromosome I and, its nucleotide sequence was determined.
Gene disruption
One-step gene replacement (Rothstein 1983) was employed. The 1.8-kb S. pombe ura4+ gene was replaced with the following coding fragments of the cohesin subunit genes: 0.75 kb EcoRV of psm1+, 2.25kb HpaI–BglII of psm3+, 0.7 kb KpnI–HincII of rad21+ , and 1.5 kb NcoI–EcoT22I of psc3+. The resulting disrupted genes were introduced into the chromosomes of diploid cells by homologous recombination. Gene disruptions were verified by Southern hybridization. Gene-disrupted cells were sporulated and dissected by tetrad analysis. The phenotypes of gene-disrupted cells were examined after germination. Heterozygous diploid cells were sporulated at 26°C, and Ura+ spores were germinated in EMM2 liquid medium lacking uracil at 33°C. Germinated cells were fixed and stained with DAPI and anti-tubulin TAT1 antibody.
Immunoprecipitation
Exponentially growing cells (3 × 108) of wild-type, Rad21–3HA/Psm3–8Myc, or Psc3–3HA integrated strains were disrupted with glass beads in modified TEG buffer (40 mM Tris-HCl at pH 7.5, 1 mM EDTA, 10% Glycerol +0.1% NP-40, 150 mM NaCl, and 1 mM PMSF) containing protease inhibitors (1 μg/mL leupeptin, 1μg/mL aprotinin, 1μg/mL pepstatin A). Extracts were then centrifuged (17,000g, 20 min) and treated with protein A-sepharose beads (50 μL) for 1 h, then incubated for 3 h with anti-HA monoclonal antibody 12CA5 (Boehringer) cross-linked to the protein A-sepharose beads (10 μL). Beads were washed five times with the modified TEG buffer. Proteins precipitated with the beads were separated by SDS-PAGE and detected by silver staining or immunoblotting.
Microscopy
The centromere DNA of chromosome I (cen1) in a living cell was visualized as described previously (Straight et al. 1996; Nabeshima et al. 1997). Immunofluorescence microscopy was done using anti-Sad1, anti-tubulin TAT1, anti-HA 12CA5 (BAbCO), anti-Myc 9E10 (Calbiochem), and anti-Rad21 antibodies (Hagan and Hyams 1988; Birkenbihl and Subramani 1995; Hagan and Yanagida 1995). Strains expressing GFP-fused proteins were observed either in living or fixed cells with methanol as described previously by Nabeshima et al. (1997). In situ chromatin binding assay was performed as described (Kearsey et al. 2000).
CHIP
CHIP was performed as described previously (Saitoh et al. 1997). The primers used are available upon request.
Acknowledgments
We thank Dr. S. Subramani for the anti-Rad21 antibody and Dr. H. Masukata for sharing reagents. This work was supported by the CREST Research Project of Japan Science Technology (JST) Corporation.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL yanagida@kozo.biophys.kyoto-u.ac.jp; FAX 81-75-753-4208.
Article and publication are at www.genesdev.org/cgi/doi/10.1101/gad.832000.
References
- Birkenbihl RP, Subramani S. Cloning and characterization of rad21 an essential gene of Schizosaccharomyces pombe involved in DNA double-strand-break repair. Nucleic Acids Res. 1992;20:6605–6611. doi: 10.1093/nar/20.24.6605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkenbihl RP, Subramani S. The rad21 gene product of Schizosaccharomyces pombe is a nuclear, cell cycle-regulated phosphoprotein. J Biol Chem. 1995;270:7703–7711. doi: 10.1074/jbc.270.13.7703. [DOI] [PubMed] [Google Scholar]
- Castano IB, Brzoska PM, Sadoff BU, Chen H, Christman MF. Mitotic chromosome condensation in the rDNA requires TRF4 and DNA topoisomerase I in Saccharomyces cerevisiae. Genes & Dev. 1996;10:2564–2576. doi: 10.1101/gad.10.20.2564. [DOI] [PubMed] [Google Scholar]
- Ciosk R, Zachariae W, Michaelis C, Shevchenko A, Mann M, Nasmyth K. An ESP1/PDS1 complex regulates loss of sister chromatid cohesion at the metaphase to anaphase transition in yeast. Cell. 1998;93:1067–1076. doi: 10.1016/s0092-8674(00)81211-8. [DOI] [PubMed] [Google Scholar]
- Ciosk R, Shirayama M, Shevchenko A, Tanaka T, Toth A, Shevchenko A, Nasmyth K. Cohesin's binding to chromosomes depends on a separate complex consisting of Scc2 and Scc4 proteins. Mol Cell. 2000;5:243–254. doi: 10.1016/s1097-2765(00)80420-7. [DOI] [PubMed] [Google Scholar]
- Connelly JC, Kirkham LA, Leach DR. The SbcCD nuclease of Escherichia coli is a structural maintenance of chromosomes (SMC) family protein that cleaves hairpin DNA. Proc Natl Acad Sci. 1998;95:7969–7974. doi: 10.1073/pnas.95.14.7969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darwiche N, Freeman LA, Strunnikov A. Characterization of the components of the putative mammalian sister chromatid cohesion complex. Gene. 1999;233:39–47. doi: 10.1016/s0378-1119(99)00160-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funabiki H, Kumada K, Yanagida M. Fission yeast Cut1 and Cut2 are essential for sister chromatid separation, concentrate along the metaphase spindle and form large complexes. EMBO J. 1996;15:6617–6628. [PMC free article] [PubMed] [Google Scholar]
- Furuya K, Takahashi K, Yanagida M. Faithful anaphase is ensured by Mis4, a sister chromatid cohesion molecule required in S phase and not destroyed in G1 phase. Genes & Dev. 1998;12:3408–3418. doi: 10.1101/gad.12.21.3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G, Saitoh S, Yanagida M. Proper metaphase spindle length is determined by centromere proteins Mis12 and Mis6 required for faithful chromosome segregation. Genes & Dev. 1999;13:1664–1677. doi: 10.1101/gad.13.13.1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guacci V, Koshland D, Strunnikov A. A direct link between sister chromatid cohesion and chromosome condensation revealed through the analysis of MCD1 in S. cerevisiae. Cell. 1997;91:47–57. doi: 10.1016/s0092-8674(01)80008-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagan IM, Hyams JS. The use of cell division cycle mutants to investigate the control of microtubule distribution in the fission yeast Schizosaccharomyces pombe. J Cell Sci. 1988;89:343–357. doi: 10.1242/jcs.89.3.343. [DOI] [PubMed] [Google Scholar]
- Hagan I, Yanagida M. The product of the spindle formation gene sad1+ associates with the fission yeast spindle pole body and is essential for viability. J Cell Biol. 1995;129:1033–1047. doi: 10.1083/jcb.129.4.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T. SMC-mediated chromosome mechanics: A conserved scheme from bacteria to vertebrates? Genes & Dev. 1999;13:11–19. doi: 10.1101/gad.13.1.11. [DOI] [PubMed] [Google Scholar]
- Hiraoka Y, Toda T, Yanagida M. The NDA3 gene of fission yeast encodes β-tubulin: A cold-sensitive nda3 mutation reversibly blocks spindle formation and chromosome movement in mitosis. Cell. 1984;39:349–358. doi: 10.1016/0092-8674(84)90013-8. [DOI] [PubMed] [Google Scholar]
- Holloway SL, Glotzer M, King RW, Murray AW. Anaphase is initiated by proteolysis rather than by the inactivation of maturation-promoting factor. Cell. 1993;73:1393–1402. doi: 10.1016/0092-8674(93)90364-v. [DOI] [PubMed] [Google Scholar]
- Holt CL, May GS. An extragenic suppressor of the mitosis-defective bimD6 mutation of Aspergillus nidulans codes for a chromosome scaffold protein. Genetics. 1996;142:777–787. doi: 10.1093/genetics/142.3.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irniger S, Piatti S, Michaelis C, Nasmyth K. Genes involved in sister chromatid separation are needed for B-type cyclin proteolysis in budding yeast. Cell. 1995;81:269–278. doi: 10.1016/0092-8674(95)90337-2. [DOI] [PubMed] [Google Scholar]
- Ishii K, Kumada K, Toda T, Yanagida M. Requirement for PP1 phosphatase and 20S cyclosome/APC for the onset of anaphase is lessened by the dosage increase of a novel gene sds23+. EMBO J. 1996;15:6629–6640. [PMC free article] [PubMed] [Google Scholar]
- Ishimi Y. A DNA helicase activity is associated with an MCM4, -6, and -7 protein complex. J Biol Chem. 1997;272:24508–24513. doi: 10.1074/jbc.272.39.24508. [DOI] [PubMed] [Google Scholar]
- Jessberger R, Riwar B, Baechtold H, Akhmedov AT. SMC proteins constitute two subunits of the mammalian recombination complex RC-1. EMBO J. 1996;15:4061–4068. [PMC free article] [PubMed] [Google Scholar]
- Kearsey SE, Montgomery S, Labib K, Lindner K. Chromatin binding of the fission yeast replication factor mcm4 occurs during anaphase and requires ORC and cdc18. EMBO J. 2000;19:1681–1690. doi: 10.1093/emboj/19.7.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura K, Hirano T. ATP-dependent positive supercoiling of DNA by 13S condensin: A biochemical implication for chromosome condensation. Cell. 1997;90:625–634. doi: 10.1016/s0092-8674(00)80524-3. [DOI] [PubMed] [Google Scholar]
- Kumada K, Nakamura T, Nagao K, Funabiki H, Nakagawa T, Yanagida M. Cut1 is loaded onto the spindle by binding to Cut2 and promotes anaphase spindle movement upon Cut2 proteolysis. Curr Biol. 1998;8:633–641. doi: 10.1016/s0960-9822(98)70250-7. [DOI] [PubMed] [Google Scholar]
- Lehmann AR, Walicka M, Griffiths DJ, Murray JM, Watts FZ, McCready S, Carr AM. The rad18 gene of Schizosaccharomyces pombe defines a new subgroup of the SMC superfamily involved in DNA repair. Mol Cell Biol. 1995;15:7067–7080. doi: 10.1128/mcb.15.12.7067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengauer C, Kinzler KW, Vogelstein B. Genetic instabilities in human cancers. Nature. 1998;396:643–649. doi: 10.1038/25292. [DOI] [PubMed] [Google Scholar]
- Lieb JD, Albrecht MR, Chuang PT, Meyer BJ. MIX-1: An essential component of the C. elegans mitotic machinery executes X chromosome dosage compensation. Cell. 1998;92:265–277. doi: 10.1016/s0092-8674(00)80920-4. [DOI] [PubMed] [Google Scholar]
- Losada A, Hirano M, Hirano T. Identification of Xenopus SMC protein complexes required for sister chromatid cohesion. Genes & Dev. 1998;12:1986–1997. doi: 10.1101/gad.12.13.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losada A, Yokochi T, Kobayashi R, Hirano T. Identification and characterization of SA/Scc3p subunits in the Xenopus and human cohesin complexes. J Cell Biol. 2000;150:405–416. doi: 10.1083/jcb.150.3.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machida M, Yamazaki S, Kunihiro S, Tanaka T, Kushida N, Jinnno K, Haikawa Y, Yamazaki J, Yamamoto S, Sekine M, et al. A 38 kb segment containing the cdc2 gene from the left arm of fission yeast chromosome II: Sequence analysis and characterization of the genomic DNA and cDNAs encoded on the segment. Yeast. 2000;16:71–80. doi: 10.1002/(SICI)1097-0061(20000115)16:1<71::AID-YEA505>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- Michaelis C, Ciosk R, Nasmyth K. Cohesins: Chromosomal proteins that prevent premature separation of sister chromatids. Cell. 1997;91:35–45. doi: 10.1016/s0092-8674(01)80007-6. [DOI] [PubMed] [Google Scholar]
- Miyazaki WY, Orr-Weaver TL. Sister-chromatid cohesion in mitosis and meiosis. Annu Rev Genet. 1994;28:167–187. doi: 10.1146/annurev.ge.28.120194.001123. [DOI] [PubMed] [Google Scholar]
- Nabeshima K, Saitoh S, Yanagida M. Use of green fluorescent protein for intracellular protein localization in living fission yeast cells. Methods Enzymol. 1997;283:459–471. doi: 10.1016/s0076-6879(97)83037-6. [DOI] [PubMed] [Google Scholar]
- Nasmyth K. Separating sister chromatids. Trends Biochem Sci. 1999;24:98–104. doi: 10.1016/s0968-0004(99)01358-4. [DOI] [PubMed] [Google Scholar]
- Nasmyth K, Peters JM, Uhlmann F. Splitting the chromosome: Cutting the ties that bind sister chromatids. Science. 2000;288:1379–1385. doi: 10.1126/science.288.5470.1379. [DOI] [PubMed] [Google Scholar]
- Rollins RA, Morcillo P, Dorsett D. Nipped-B, a Drosophila homologue of chromosomal adherins, participates in activation by remote enhancers in the cut and Ultrabithorax genes. Genetics. 1999;152:577–593. doi: 10.1093/genetics/152.2.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothstein RJ. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- Saitoh S, Takahashi K, Yanagida M. Mis6, a fission yeast inner centromere protein, acts during G1/S and forms specialized chromatin required for equal segregation. Cell. 1997;90:131–143. doi: 10.1016/s0092-8674(00)80320-7. [DOI] [PubMed] [Google Scholar]
- Schmiesing JA, Ball AR, Jr, Gregson HC, Alderton JM, Zhou S, Yokomori K. Identification of two distinct human SMC protein complexes involved in mitotic chromosome dynamics. Proc Natl Acad Sci. 1998;95:12906–12911. doi: 10.1073/pnas.95.22.12906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skibbens RV, Corson LB, Koshland D, Hieter P. Ctf7p is essential for sister chromatid cohesion and links mitotic chromosome structure to the DNA replication machinery. Genes & Dev. 1999;13:307–319. doi: 10.1101/gad.13.3.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straight AF, Belmont AS, Robinett CC, Murray AW. GFP tagging of budding yeast chromosomes reveals that protein–protein interactions can mediate sister chromatid cohesion. Curr Biol. 1996;6:1599–1608. doi: 10.1016/s0960-9822(02)70783-5. [DOI] [PubMed] [Google Scholar]
- Surana U, Amon A, Dowzer C, McGrew J, Byers B, Nasmyth K. Destruction of the CDC28/CLB mitotic kinase is not required for the metaphase to anaphase transition in budding yeast. EMBO J. 1993;12:1969–1978. doi: 10.1002/j.1460-2075.1993.tb05846.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutani T, Yanagida M. DNA renaturation activity of the SMC complex implicated in chromosome condensation. Nature. 1997;388:798–801. doi: 10.1038/42062. [DOI] [PubMed] [Google Scholar]
- Sutani T, Yuasa T, Tomonaga T, Dohmae N, Takio K, Yanagida M. Fission yeast condensin complex: Essential roles of non-SMC subunits for condensation and Cdc2 phosphorylation of Cut3/SMC4. Genes & Dev. 1999;13:2271–2283. doi: 10.1101/gad.13.17.2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Murakami S, Chikashige Y, Funabiki H, Niwa O, Yanagida M. A low copy number central sequence with strict symmetry and unusual chromatin structure in fission yeast centromere. Mol Biol Cell. 1992;3:819–835. doi: 10.1091/mbc.3.7.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Yamada H, Yanagida M. Fission yeast minichromosome loss mutants mis cause lethal aneuploidy and replication abnormality. Mol Biol Cell. 1994;5:1145–1158. doi: 10.1091/mbc.5.10.1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Chen ES, Yanagida M. Requirement of Mis6 centromere connector for localizing a CENP-A-like protein in fission yeast. Science. 2000;288:2215–2219. doi: 10.1126/science.288.5474.2215. [DOI] [PubMed] [Google Scholar]
- Tanaka T, Cosma MP, Wirth K, Nasmyth K. Identification of cohesin association sites at centromeres and along chromosome arms. Cell. 1999;98:847–858. doi: 10.1016/s0092-8674(00)81518-4. [DOI] [PubMed] [Google Scholar]
- Tanaka K, Yonekawa T, Kawasaki Y, Kai M, Furuya K, Iwasaki M, Murakami H, Yanagida M, Okayama H. Fission yeast Eso1p is required for establishing sister chromatid cohesion during S phase. Mol Cell Biol. 2000;20:3459–3469. doi: 10.1128/mcb.20.10.3459-3469.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatebayashi K, Kato J, Ikeda H. Isolation of a Schizosaccharomyces pombe rad21ts mutant that is aberrant in chromosome segregation, microtubule function, DNA repair and sensitive to hydroxyurea: Possible involvement of Rad21 in ubiquitin-mediated proteolysis. Genetics. 1998;148:49–57. doi: 10.1093/genetics/148.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth A, Ciosk R, Uhlmann F, Galova M, Schleiffer A, Nasmyth K. Yeast cohesin complex requires a conserved protein, Eco1p(Ctf7), to establish cohesion between sister chromatids during DNA replication. Genes & Dev. 1999;13:320–333. doi: 10.1101/gad.13.3.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlmann F, Lottspeich F, Nasmyth K. Sister-chromatid separation at anaphase onset is promoted by cleavage of the cohesin subunit Scc1. Nature. 1999;400:37–42. doi: 10.1038/21831. [DOI] [PubMed] [Google Scholar]
- Wang Z, Castano IB, De Las Penas A, Adams C, Christman MF. Pol κ: A DNA polymerase required for sister chromatid cohesion. Science. 2000;289:774–779. doi: 10.1126/science.289.5480.774. [DOI] [PubMed] [Google Scholar]
- Yanagida M. Frontier questions about sister chromatid separation in anaphase. Bioessays. 1995;17:519–526. doi: 10.1002/bies.950170608. [DOI] [PubMed] [Google Scholar]
- ————— Cell cycle mechanisms of sister chromatid separation; roles of Cut1/separin and Cut2/securin. Genes Cells. 2000;5:1–8. doi: 10.1046/j.1365-2443.2000.00306.x. [DOI] [PubMed] [Google Scholar]