Abstract
Mammalian RNA polymerase II complexes and coactivators containing homologs of yeast Srb/Med proteins have been isolated recently from tissue culture cells. The yeast Srb/Med complex is involved in global gene expression and is essential, but it is not yet known if its mammalian counterparts are broadly expressed in tissues or if they are essential. We have isolated the murine gene encoding Srb7, an Srb/Med complex protein whose sequence and function is highly conserved between yeast and humans. The mouse Srb7 gene is single copy, and Northern analysis showed that it is expressed in all tissues examined. Disruption of the gene in embryonic stem cells revealed that it is essential for cell viability and murine embryonic development. These results, together with evidence that murine Srb7 is associated exclusively with high molecular weight forms of RNA polymerase II in extracts, suggest that Srb7-containing polymerase complexes occur in most tissues and have essential roles in expression of protein coding genes.
Keywords: RNA polymerase II, yeast, Srb/Med, mouse Srb7 gene, tissue culture cells
Transcription initiation in eukaryotes involves the recruitment of RNA polymerase II and associated factors to promoters by gene-specific activators (Orphanides et al. 1996; Roeder 1996; Ptashne and Gann 1997; Hampsey 1998; Myer and Young 1998). Attempts to reconstitute transcription in vitro led to the identification of one set of initiation factors that are associated with RNA polymerase II at promoters, and these were called basal or general transcription factors (GTFs). The GTFs include TFIID, TFIIB, TFIIE, TFIIF, and TFIIH. Genetic and biochemical studies have identified additional factors that are necessary for appropriate regulated expression in vivo or for reconstitution of more physiologically relevant activities in vitro, among which are the yeast Srb/Med complex.
We and others have proposed that the form of RNA polymerase II that is generally recruited to promoters by transcriptional activators in yeast cells contains an Srb/med complex (for review, see Koleske and Young 1995; Ptashne and Gann 1997; Hampsey 1998; Myer and Young 1998). The genes encoding the yeast Srb proteins were discovered through a genetic screen designed to identify components of the transcription apparatus, which are involved in the response to transcriptional regulators (Nonet and Young 1989). Attempts to purify Srb proteins led to the isolation of a large complex containing core RNA polymerase II, a subset of the general transcription factors, and various regulatory proteins (Thompson et al. 1993; Koleske and Young 1994). This holoenzyme complex had the capacity to initiate transcription and respond to activators when supplemented with additional purified general transcription factors in vitro. A subcomplex that dissociated from the holoenzyme with anti-CTD antibodies was found to reconstitute the response to activators in vitro; this subcomplex contains Srb and additional proteins called Meds (Kim et al. 1994). The response to activators is critical because in vitro systems reconstituted with yeast GTFs and polymerase alone were not activator-responsive (Flanagan et al. 1991). Two of the yeast Srb proteins (Srb4 and Srb6) were found to be required for transcription of most protein coding genes in vivo (Thompson and Young 1995; Holstege et al. 1998). Because these Srb proteins are found tightly associated with the holoenzyme, it follows that the Srb-containing holoenzyme is the form of RNA polymerase II that is recruited to most promoters in yeast cells.
Transcription-competent RNA polymerase II complexes containing Srb homologs were identified in mammalian cells soon after the discovery of the yeast holoenzyme (Ossipow et al. 1995, 1999; Chao et al. 1996; Maldonado et al. 1996; Pan et al. 1997; Cho et al. 1998; Neish et al. 1998). The precise composition of these complexes was not established, due in part to technical difficulties associated with purifying intact megadalton size multiprotein complexes. However, mammalian Srb7 was found to be a component of all complexes assayed by Western blot and was shown to be tightly associated with high molecular weight forms of RNA polymerase II (Ossipow et al. 1999). More recently, investigators have described more highly defined protein complexes that were isolated using assays for cofactor function and that contain homologs of the yeast Srb/Med proteins (Jiang et al. 1998; Gu et al. 1999; Naar et al. 1999; Rachez et al. 1999; Ryu et al. 1999). These purified complexes lack RNA polymerase II but associate with the enzyme where they function as coactivators and corepressors (Hampsey and Reinberg 1999).
These recent developments suggest that Srb/Med-containing complexes have diverse and important roles in regulation of transcription initiation in mammalian cells. It would be interesting to determine whether Srb/Med genes are generally expressed in mammalian tissues and whether they are essential for viability. To address these issues, we have isolated the murine Srb7 gene and used it to determine the range of mammalian tissues that express this gene and to ascertain whether it is essential for cell viability and development.
Results and Discussion
Murine Srb7 genomic and cDNA clones were isolated from libraries by using a human SRB7 cDNA clone (Chao et al. 1996) as probe. Among the Srb/mediator proteins shared by yeast and mammalian holoenzymes, Srb7 was selected for investigation because its sequence and function is highly conserved between yeast and humans (Chao et al. 1996), it has been observed in all mammalian RNA polymerase II complexes examined thus far (Chao et al. 1996; Maldonado et al. 1996; Scully et al. 1997; Neish et al. 1998; Ossipow et al. 1999), and Srb7 is associated exclusively with the high molecular weight form of RNA polymerase II in murine cell extracts (Ossipow et al. 1999). Southern analysis of genomic DNA indicated that the Srb7 gene is single copy (data not shown). The cDNA clone and a portion of the genomic clone were sequenced, and the predicted mouse Srb7 protein sequence is shown in Figure 1. The 144-amino-acid mouse and human Srb7 proteins are identical except for a single amino acid difference at residue 36. Orthologous sequences were also identified in Caenorhabditis elegans and Schizosaccharomyces pombe (Fig. 1). Sequence homology among the Srb7 proteins was highest in the amino terminus, which is the portion of human Srb7 which can functionally substitute for the corresponding yeast sequence (Chao et al. 1996).
Figure 1.

Alignment of sequences predicted for mouse, human, worm, and yeast Srb7 proteins. Human and S. cerevisiae Srb7 protein sequences are from Chao et al. (1996) and Myers et al. (1998), respectively. The putative S. pombe and C. elegans homologs are derived from GenBank accession nos. 4007762 and Z81475 (gene C24H11.6, aligned sequence starting at conserved methionine), respectively. The asterisk (*) indicates the single amino acid difference between human and mouse Srb7. Boxed residues are identical; shaded residues are similar as determined by the SeqVu program (Riek et al. 1995).
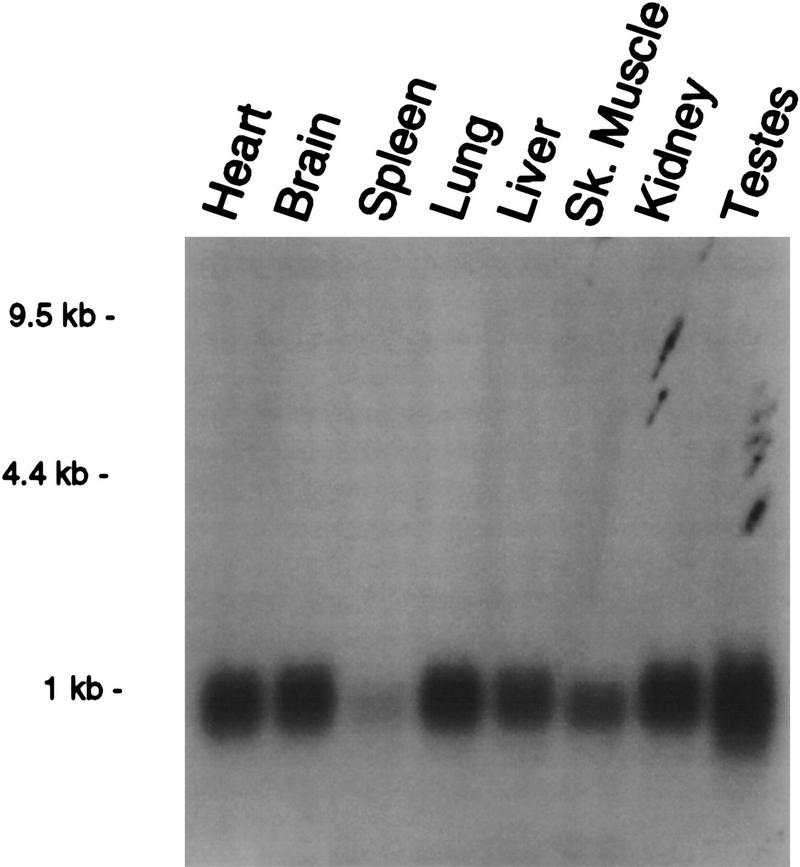
The presence of Srb7 mRNA in various tissues was investigated by probing a multi-tissue Northern blot with radiolabeled Srb7 cDNA (Fig. 2). A single band of the predicted size was observed in samples from each of the tissues examined, including heart, brain, spleen, lung, liver, muscle, kidney, and testes. We conclude that the Srb7 gene is expressed in most murine tissues.
Figure 2.
Expression of Srb7. The full-length Srb7 cDNA clone was hybridized to a multiple tissue Northern blot from Clontech.
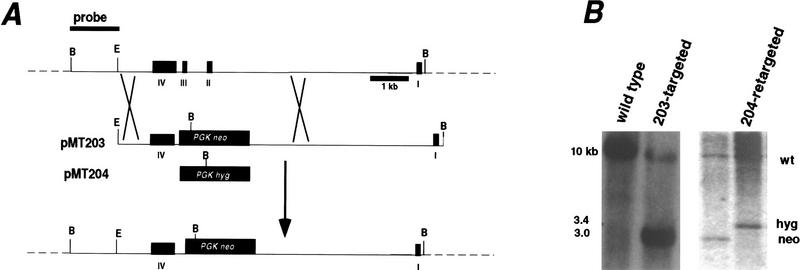
The requirement for Srb/Med components in vivo has not been established in higher eukaryotes. We disrupted the Srb7 gene in embryonic stem (ES) cells to explore the requirement for its function in the mouse. A genomic DNA clone was used to create a targeting construct that deletes the second and third exons of the Srb7 gene (Fig. 3A), eliminating sequence coding for residues 16–87 of the 144-amino-acid reading frame. ES cells were transfected with the targeting vector and drug-resistant colonies were screened for homologous integration, which occurred (Fig. 3B, left) in ∼5% of the clones. These cells showed no obvious growth phenotype. To determine if a homozygous deletion of the Srb7 gene could be achieved, heterozygous ES cell clones were retargeted with a vector containing a different drug resistance marker. The mutated allele was replaced (Fig. 3B, right) at a frequency similar to that seen in the initial targeting experiments (9/74), whereas no clones were found to be homozygously disrupted (0/74). This is consistent with inviability of homozygous ES cells and suggests that the Srb7 gene is essential for cell viability.
Figure 3.
(A) Targeted disruption of Srb7. An 8.5-kb BamHI–EcoRI (E and B) genomic fragment in pBS II KS− was the basis for the targeting construct. Exons II and III were replaced by a PGK–neo or PGK–hyg cassette. Exons are numbered I–IV; construct pMT204 is the same as pMT203, except the drug resistance marker was replaced with a PGK–hyg cassette. (B, left) BamHI-digested genomic DNA from G418-resistant ES cell clones (transformed with pMT203) were genotyped by Southern hybridization with the probe indicated in A. (Right) Clones targeted with the neo-containing vector (203-targeted) were retargeted with the hyg vector (204-targeted). Hygromycin B-resistant clones showed replacement of the neo allele but not disruption of the remaining wild-type allele.
To further explore the effects of Srb7 disruption, heterozygous ES cells were used to derive mice mutant for Srb7. Of six chimeras derived from three different ES clones, one animal, which showed 40% chimerism, gave germ-line transmission. Heterozygous animals showed no overt phenotype. In contrast, no homozygous animals were born from heterozygous intercrosses, suggesting that the absence of SRB7 is lethal.
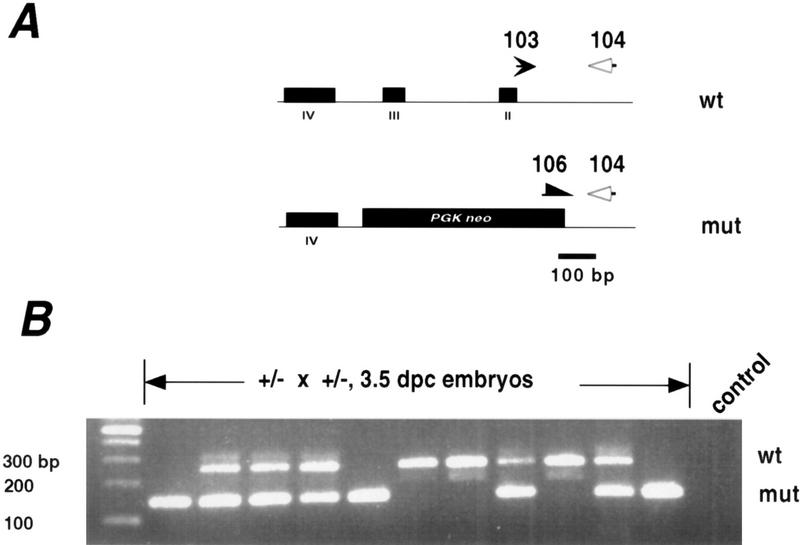
Next we examined the developmental stage at which homozygous embryos die. As embryonic transcription begins soon after cleavage commences, it would be expected that maternal stores of the Srb7 protein would allow the zygote to reach at least this stage. To test this, embryos of different developmental stages were genotyped by PCR (Fig. 4). Homozygous embryos were found in Mendelian ratios (n = 26, χ2 P < 0.05) until the blastocyst stage. Culture of blastocysts yielded no outgrowth of homozygous cells, suggesting developmental arrest at this stage (data not shown). Analysis of later embryos (7.5 and 8.5 dpc) showed numerous empty decidua and no homozygous embryos (n = 25, significant departure from Mendelian ratios, χ2 P < 0.01). We conclude that the Srb7 gene product is essential for both embryonic development and cell viability. The survival of embryos to the blastocyst stage is likely due to maternal stores of the protein. By comparison, homozygous disruptions of Rad51 die immediately after the beginning cleavage (Tsuzuki et al. 1996), whereas cyclin A2−/− embryos survive to the early postimplantation stage though the deletion is cell lethal (Murphy et al. 1997). Whereas Rad51, cyclinA2, and Srb7 disruptions are all apparently cell lethal, they have disparate developmental phenotypes, the latter being temporally intermediate. Compared to disruptions of other transcription factors, Srb7−/− embryos show earlier lethality (∼4–6 dpc) than those deficient for the abundant transcription factor Sp1 (11 dpc) (Marin et al. 1997), the transcriptional coactivator/histone acetylase p300 (9 dpc) (Yao et al. 1998), or the chromatin remodeling factor BRM (nonlethal proliferation defects) (Reyes et al. 1998).
Figure 4.
(A) Schematic depicting PCR genotyping assay. The number 104 denotes primer oMT104, common to both alleles; 103 denotes oMT103, specific to the wild-type allele; 106 denotes oMT106, specific to the mutant allele. (B) PCR genotyping of 3.5 dpc embryos (blastocysts or late morulae) derived from heterozygous crosses. Embryos were digested individually with proteinase K and subjected to PCR as detailed in Materials and Methods. Control reaction contained media alone. Wild-type, heterozygous, and homozygous mutant embryos were found in Mendelian ratios.
Recent studies have identified mammalian RNA polymerase II coactivator complexes containing Srb/Med proteins, but it has not been clear whether these complexes have general or essential roles in gene expression, as is the case in yeast cells (Holstege et al. 1998). The results described here reveal that the gene encoding Srb7 is expressed in most mouse tissues and is essential for viability, suggesting that Srb7-containing polymerase complexes exist in most tissues and play esential roles in expression of protein coding genes.
Materials and methods
Isolation of clones and expression patterns
The human cDNA clone was used to probe a mouse brain cDNA library [provided by Tony Koleske (Yale University, New Haven, CT) and David Baltimore (California Institute of Technology, Pasadena, CA)] by hybridization, and positive clones were sequenced. The cDNA was then used to probe a 129 SVJ BAC genomic library (Genome Systems) to isolate overlapping genomic clones. A 10-kb BamHI fragment was found to contain the entire coding sequence and was subcloned into pBluescript II (PM429). This segment was used for subsequent constructions. Exons were identified by restriction mapping and partial sequencing of PM429.
The cDNA clone was hybridized to a mouse multitissue Northern blot (Clontech).
Targeting
Either a PGK–neo or a PGK–hyg selection cassette was inserted between the AvrII and HpaI sites of an 8.5-kb genomic subclone, resulting in the deletion of the second and third exons of the gene (pMT203 and pMT204, respectively). Linearized vectors were electroporated into J1 ES cells (Li et al. 1992), and drug-resistant colonies were screened by PCR and Southern hybridization of BamHI-digested DNA with an external probe (pMT205). Homologously targeted clones were shown to be single copy integrants by probing with an internal probe. Retargeting experiments were analyzed by Southern hybridization. In these experiments the individual alleles could be distingushed by restriction polymorphisms. This allowed the recognition of targeting events that replaced one mutant allele with another. Heterozygous clones were injected into BALB/c blastocysts, and resulting animals chimeric by coat color (20%–90%) were mated with BALB/c and 129SvJ mice to generate mixed-background and inbred lines carrying the muation. To genotype embryos, heterozygotes were mated, embryos were collected and developmental stage noted, and were placed in 0.2-ml PCR tubes with 5 μl of proteinase K buffer (1× Promega Mg-free PCR buffer + 200 μg/ml proteinase K). After 1 hr digestion at 50°C, the proteinase K was inactivated by heating to 95°C for 10 min, a PCR master mix was added, and the samples were amplified and analyzed by gel electrophoresis. Southern hybridization with an internal oligonucleotide was used to confirm identity of PCR products. The following primers were used: oMT103, GGGAGGCATTTGAGGCTTTATTTGATTTTG (wild-type allele); oMT104, CACCACCACCACCCAGCTGCTCTA (common); oMT106, CTTAATCGCCTTGCACCACATCCC (mutant allele); and oMT108, CCTCTGGAAGATTGTGCTGT (internal hybridization oligonucleotide).
Acknowledgments
We thank Jessie Dausman and Ruth Curry for invaluable assistance. This work was supported by National Institutes of Health grants to R.J. and R.A.Y. M.T. is suported by a predoctoral fellowship from the Howard Hughes Medical Institute.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked ‘advertisement’ in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL young@wi.mit.edu; FAX (617) 258-0376.
References
- Chao DM, Gadbois EL, Murray PJ, Anderson SF, Sonu MS, Parvin JD, Young RA. A mammalian SRB protein associated with an RNA polymerase II holoenzyme. Nature. 1996;380:82–85. doi: 10.1038/380082a0. [DOI] [PubMed] [Google Scholar]
- Cho H, Orphanides G, Sun X, Yang XJ, Ogryzko V, Lees E, Nakatani Y, Reinberg D. A human RNA polymerase II complex containing factors that modify chromatin structure. Mol Cell Biol. 1998;18:5355–5363. doi: 10.1128/mcb.18.9.5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flanagan PM, Kelleher RJD, Sayre MH, Tschochner H, Kornberg RD. A mediator required for activation of RNA polymerase II transcription in vitro. Nature. 1991;350:436–438. doi: 10.1038/350436a0. [DOI] [PubMed] [Google Scholar]
- Gu W, Malik S, Ito M, Yuan CX, Fondell JD, Zhang X, Martinez E, Qin J, Roeder RG. A novel human SRB/MED-containing cofactor complex, SMCC, involved in transcription regulation. Mol Cell. 1999;3:97–108. doi: 10.1016/s1097-2765(00)80178-1. [DOI] [PubMed] [Google Scholar]
- Hampsey M. Molecular genetics of the RNA polymerase II general transcriptional machinery. Microbiol Mol Biol Rev. 1998;62:465–503. doi: 10.1128/mmbr.62.2.465-503.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampsey M, Reinberg D. RNA polymerase II as a control panel for multiple coactivator complexes. Curr Opin Genet Dev. 1999;9:132–139. doi: 10.1016/S0959-437X(99)80020-3. [DOI] [PubMed] [Google Scholar]
- Holstege FC, Jennings EG, Wyrick JJ, Lee TI, Hengartner CJ, Green MR, Golub TR, Lander ES, Young RA. Dissecting the regulatory circuitry of a eukaryotic genome. Cell. 1998;95:717–728. doi: 10.1016/s0092-8674(00)81641-4. [DOI] [PubMed] [Google Scholar]
- Jiang YW, Veschambre P, Erdjument-Bromage H, Tempst P, Conaway JW, Conaway RC, Kornberg RD. Mammalian mediator of transcriptional regulation and its possible role as an end-point of signal transduction pathways. Proc Natl Acad Sci. 1998;95:8538–8543. doi: 10.1073/pnas.95.15.8538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YJ, Bjorklund S, Li Y, Sayre MH, Kornberg RD. A multiprotein mediator of transcriptional activation and its interaction with the C-terminal repeat domain of RNA polymerase II. Cell. 1994;77:599–608. doi: 10.1016/0092-8674(94)90221-6. [DOI] [PubMed] [Google Scholar]
- Koleske AJ, Young RA. An RNA polymerase II holoenzyme responsive to activators. Nature. 1994;368:466–469. doi: 10.1038/368466a0. [DOI] [PubMed] [Google Scholar]
- ————— The RNA polymerase II holoenzyme and its implications for gene regulation. Trends Biochem Sci. 1995;20:113–116. doi: 10.1016/s0968-0004(00)88977-x. [DOI] [PubMed] [Google Scholar]
- Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- Maldonado E, Shiekhattar R, Sheldon M, Cho H, Drapkin R, Rickert P, Lees E, Anderson CW, Linn S, Reinberg D. A human RNA polymerase II complex associated with SRB and DNA-repair proteins [published erratum appears in 384: 384] Nature. 1996;381:86–89. doi: 10.1038/381086a0. [DOI] [PubMed] [Google Scholar]
- Marin M, Karis A, Visser P, Grosveld F, Philipsen S. Transcription factor Sp1 is essential for early embryonic development but dispensable for cell growth and differentiation. Cell. 1997;89:619–628. doi: 10.1016/s0092-8674(00)80243-3. [DOI] [PubMed] [Google Scholar]
- Murphy M, Stinnakre MG, Senamaud-Beaufort C, Winston NJ, Sweeney C, Kubelka M, Carrington M, Brechot C, Sobczak-Thepot J. Delayed early embryonic lethality following disruption of the murine cyclin A2 gene. Nat Genet. 1997;15:83–86. doi: 10.1038/ng0197-83. [DOI] [PubMed] [Google Scholar]
- Myer VE, Young RA. RNA polymerase II holoenzymes and subcomplexes. J Biol Chem. 1998;273:27757–27760. doi: 10.1074/jbc.273.43.27757. [DOI] [PubMed] [Google Scholar]
- Naar AM, Beaurang PA, Zhou S, Abraham S, Solomon W, Tjian R. Composite co-activator ARC mediates chromatin-directed transcriptional activation. Nature. 1999;398:828–832. doi: 10.1038/19789. [DOI] [PubMed] [Google Scholar]
- Neish AS, Anderson SF, Schlegel BP, Wei W, Parvin JD. Factors associated with the mammalian RNA polymerase II holoenzyme. Nucleic Acids Res. 1998;26:847–853. doi: 10.1093/nar/26.3.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonet ML, Young RA. Intragenic and extragenic suppressors of mutations in the heptapeptide repeat domain of Saccharomyces cerevisiae RNA polymerase II. Genetics. 1989;123:715–724. doi: 10.1093/genetics/123.4.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orphanides G, Lagrange T, Reinberg D. The general transcription factors of RNA polymerase II. Genes & Dev. 1996;10:2657–2683. doi: 10.1101/gad.10.21.2657. [DOI] [PubMed] [Google Scholar]
- Ossipow V, Tassan JP, Nigg EA, Schibler U. A mammalian RNA polymerase II holoenzyme containing all components required for promoter-specific transcription initiation. Cell. 1995;83:137–146. doi: 10.1016/0092-8674(95)90242-2. [DOI] [PubMed] [Google Scholar]
- Ossipow V, Fonjallaz P, Schibler U. An RNA polymerase II complex containing all essential initiation factors binds to the activation domain of PAR leucine zipper transcription factor thyroid embryonic factor. Mol Cell Biol. 1999;19:1242–1250. doi: 10.1128/mcb.19.2.1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan G, Aso T, Greenblatt J. Interaction of elongation factors TFIIS and elongin A with a human RNA polymerase II holoenzyme capable of promoter-specific initiation and responsive to transcriptional activators. J Biol Chem. 1997;272:24563–24571. doi: 10.1074/jbc.272.39.24563. [DOI] [PubMed] [Google Scholar]
- Ptashne M, Gann A. Transcriptional activation by recruitment. Nature. 1997;386:569–577. doi: 10.1038/386569a0. [DOI] [PubMed] [Google Scholar]
- Rachez C, Lemon BD, Suldan Z, Bromleigh V, Gamble M, Naar AM, Erdjument-Bromage H, Tempst P, Freedman LP. Ligand-dependent transcription activation by nuclear receptors requires the DRIP complex. Nature. 1999;398:824–828. doi: 10.1038/19783. [DOI] [PubMed] [Google Scholar]
- Reyes JC, Barra J, Muchardt C, Camus A, Babinet C, Yaniv M. Altered control of cellular proliferation in the absence of mammalian brahma (SNF2alpha) EMBO J. 1998;17:6979–6991. doi: 10.1093/emboj/17.23.6979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riek RP, Handschumacher MD, Sung SS, Tan M, Glynias MJ, Schluchter MD, Novotny J, Graham RM. Evolutionary conservation of both the hydrophilic and hydrophobic nature of transmembrane residues. J Theor Biol. 1995;172:245–258. doi: 10.1006/jtbi.1995.0021. [DOI] [PubMed] [Google Scholar]
- Roeder RG. The role of general initiation factors in transcription by RNA polymerase II. Trends Biochem Sci. 1996;21:327–335. [PubMed] [Google Scholar]
- Ryu S, Zhou S, Ladurner AG, Tjian R. The transcriptional cofactor complex CRSP is required for activity of the enhancer-binding protein Sp1. Nature. 1999;397:446–450. doi: 10.1038/17141. [DOI] [PubMed] [Google Scholar]
- Scully R, Anderson SF, Chao DM, Wei W, Ye L, Young RA, Livingston DM, Parvin JD. BRCA1 is a component of the RNA polymerase II holoenzyme. Proc Natl Acad Sci. 1997;94:5605–5610. doi: 10.1073/pnas.94.11.5605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson CM, Young RA. General requirement for RNA polymerase II holoenzymes in vivo. Proc Natl Acad Sci. 1995;92:4587–4590. doi: 10.1073/pnas.92.10.4587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson CM, Koleske AJ, Chao DM, Young RA. A multisubunit complex associated with the RNA polymerase II CTD and TATA-binding protein in yeast. Cell. 1993;73:1361–1375. doi: 10.1016/0092-8674(93)90362-t. [DOI] [PubMed] [Google Scholar]
- Tsuzuki T, Fujii Y, Sakumi K, Tominaga Y, Nakao K, Sekiguchi M, Matsushiro A, Yoshimura Y, Morita T. Targeted disruption of the Rad51 gene leads to lethality in embryonic mice. Proc Natl Acad Sci. 1996;93:6236–6240. doi: 10.1073/pnas.93.13.6236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao TP, Oh SP, Fuchs M, Zhou ND, Ch’ng LE, Newsome D, Bronson RT, Li E, Livingston DM, Eckner R. Gene dosage-dependent embryonic development and proliferation defects in mice lacking the transcriptional integrator p300. Cell. 1998;93:361–372. doi: 10.1016/s0092-8674(00)81165-4. [DOI] [PubMed] [Google Scholar]