Abstract
Background
Most of the non-B HIV-1 subtypes are predominant in Sub-Saharan Africa and India although they have been found worldwide. In the last decade, immigration from these areas has increased considerably in Spain. The objective of this study was to evaluate the prevalence of non-B subtypes circulating in a cohort of HIV-1-infected immigrants in Seville, Southern Spain and to identify drug resistance-associated mutations.
Methods
Complete protease and first 220 codons of the reverse transcriptase coding regions were amplified and sequenced by population sequencing. HIV-1 subtypes were determined using Stanford University Drug Resistance Database, and phylogenetic analysis was performed comparing multiple reported sequences. Drug resistance mutations were defined according to the International AIDS Society-USA.
Results
From 2000 to 2010 a total of 1,089 newly diagnosed HIV-1-infected patients were enrolled in our cohort. Of these, 121 were immigrants, of which 98 had ethical approval and informed consent to include in our study. Twenty-nine immigrants (29/98, 29.6%) were infected with non-B subtypes, of which 15/29 (51.7%) were CRF02-AG, mostly from Sub-Saharan Africa, and 2/29 (6.9%) were CRF01-AE from Eastern Europe. A, C, F, J and G subtypes from Eastern Europe, Central-South America and Sub-Saharan Africa were also present. Some others harboured recombinant forms CRF02-AG/CRF01-AE, CRF2-AG/G and F/B, B/C, and K/G, in PR and RT-coding regions. Patients infected with non-B subtypes showed a high frequency of minor protease inhibitor resistance mutations, M36I, L63P, and K20R/I. Only one patient, CRF02_AG, showed major resistance mutation L90M. Major RT inhibitor resistance mutations K70R and A98G were present in one patient with subtype G, L100I in one patient with CRF01_AE, and K103N in another patient with CRF01_AE. Three patients had other mutations such as V118I, E138A and V90I.
Conclusions
The circulation of non-B subtypes has significantly increased in Southern Spain during the last decade, with 29.6% prevalence, in association with demographic changes among immigrants. This could be an issue in the treatment and management of these patients. Resistance mutations have been detected in these patients with a prevalence of 7% among treatment-naïve patients compared with the 21% detected among patients under HAART or during treatment interruption.
Keywords: non-B HIV-1 subtypes, immigrant, Spain, resistance mutation
Background
Human immunodeficiency virus type 1 (HIV-1) is the major pathogen responsible for the AIDS pandemic. Several genetic variants can be recognized within HIV-1 group M, including nine subtypes (A through K), at least 43 major circulating recombinants forms (CRFs), and multiple unique recombinant forms (URFs) (http://www.hiv.lanl.gov). The prevalence of HIV-1 subtypes varies greatly depending on the geographic region. Subtype B is predominant in North America and Western Europe, including Spain, although is responsible for only 10% of global infections [1]. Non-B HIV-1 subtypes and its recombinants, such as subtype C, A, CRF01_AE or CRF02_AG, are prevalent in Sub-Saharan Africa, Asia and Eastern Europe [2]. These subtypes cause up to 90% of the 36 million estimated infections, playing an important role in the HIV-1 pandemic [3,4]. Human migration produced in the last decade has contributed to the current spread of non-B subtypes in developed countries [5-7].
The extensive variability of HIV-1 has a potential impact on epidemiology, diagnosis, therapy and prevention of infection. In fact, faster progression to AIDS among individuals infected with non-B subtypes or recombinant variants enhances the importance of identifying these strains [8,9]. In addition, differences in the sensitivity to antiretrovirals in patients infected with non-B subtypes due to the high prevalence of polymorphisms in protease (PR) and/or reverse transcriptase (RT) associated with resistance to antiretroviral therapy has to be taken into account [10-16].
Finally, diagnostic tests, including viral load measurements, might be affected by the diversity of HIV-1 strains [17,18]. Therefore, HIV-1 subtype characterization is becoming an important aspect to adequate clinical management of HIV-1-infected individuals [3]. Sub-Saharan population in Spain has increased in recent years. Around 10-15% of HIV-1-infected immigrants are characterized with non-B subtype during the first medical evaluation [19-21].
Thus, the objective of this study was to analyse the prevalence of non-B subtypes in a cohort of HIV-1-infected immigrants in Southern Spain from 2000 to 2010, and to characterize drug resistance mutations associated to PR and RT.
Methods
Patients
From January 2000 to December 2010 a total of 1,089 new HIV-1-infected patients were included in the dynamic open cohort of the Infectious Diseases Service at the University Hospital Virgen del Rocio located in Seville, Southern Spain. Of these, 121 (11.1%) were HIV-1 infected immigrants. For the present retrospective study, immigrant patients of any nationality with available samples were selected. The study was approved by the Ethical Committee of the Hospital. A total of 98 patients were included and all signed an informed consent.
Laboratory determinations
CD4+ T cell count was determined in fresh samples by flow cytometry. Plasma HIV-1 RNA was measured by quantitative PCR (HIV Monitor™ Test Kit, Roche Molecular System, Hoffman-La Roche, Basel, Switzerland), according to the manufacturer's instructions. This assay has a lower detection limit of 50 HIV-1 RNA copies/ml.
Subtyping and drug resistance profile
For genetic analysis, in patients with detectable viral load, HIV-1 RNA was extracted from plasma using a viral RNA purification kit (Qiagen, Diagnostics, Barcelona, Spain). cDNA was synthesized by avian myeloblastosis virus (AMV) reverse transcriptase. In patients with undetectable viral load and undergoing antiretroviral treatment at the time of the study, HIV-1 proviral DNA from cryopreserved peripheral blood mononuclear cells (PBMCs) isolated from heparinized blood by Ficoll density-gradient centrifugation, were used for DNA extraction (Qiagen DNA blood kit, Diagnostics, Barcelona, Spain), following the manufacturer's instructions.
PCR reactions were carried out in 50 μL final volumes containing 10 mM Tris HCl pH 8.8, 50 mM KCl, 1.5 mM MgCl2, 0.1% Triton X-100, 0,2 mM of each dNTP, 0.7 units Taq polymerase (Eurotaq, Euroclone, S.p.A, Syziano, Italy), 0.5 μM of each primer, and 1 μg purified DNA. Cycling parameters were 94°C for 5 min followed by 35 cycles of 94°C for 15 s, 55°C for 15 s and 72°C for 30 s, followed by a 10 min hold at 72°C. Identical conditions were used for the nested PCR. Outer primers PRF2 (5'-cagaagagagtctcaggtttggg-3') and PRF5 (5'-tggagtattgtatggattttcagg-3') for the PR-coding region, and 47RV (5'-gtattagtaggacctacacct-3') and Pol18 (5'-agactcacaatatgca-3') for the RT-coding region, were used. Inner primers Pol10 (5'-ccctcaagggcaggagc-3') and Pol14 (5'-gggccatccattcctgg-3') for the PR-coding region, and A35 (5'-ttggttgcactttaaattttcccattagtcctatt-3') and Lp2 (5'-atcaggatgagattcataacccatcca-3') for RT-coding region, were used. A fragment of 450 bp encompassing the entire PR-coding region and a fragment of 670 bp that includes the first 220 codons of the RT-coding region were generated by nested PCR.
Sequencing of PCR purified amplicons was performed using Applied Biosystems 310 Sequencer and BigDye deoxy terminator procedure as specified by the manufacturer.
Subtyping of the PR and RT sequences was determined using the Stanford University Drug Resistance Database (http://hivdb.stanford.edu). Afterwards, the PR and RT sequences were aligned together with other HIV-1 group M reported sequences available in the GenBank by computer software CLUSTAL W. Phylogenetic trees were generated using the Neighbour-Joining method and bootstrap re-sampling of multiple alignments (1,000 data sets), included in the CLC DNA Workbench software, was employed to test the tree robustness. Drug resistance profiles in PR and RT-coding regions were defined according to the International AIDS Society-USA.
Statistical Analysis
Continuous variables are shown as median [interquartile range (IQR)], and categorical variables as number of cases (percentage). Chi square test were used to analyze differences between categorical variables. The statistical analysis was performed using the Statistical Package for the Social Sciences software (SPSS 16.0, Chicago, Illinois, USA).
Sequence Accession Number
GenBank accession numbers for consensus sequences: AM000054 (A); K03455 (B); AY563172 (C); DQ189088 (F); AY017457 (G); EF614151 (J); AJ249239 (K); FM252023 (CRF01_AE); FN557324 (CRF02_AG). Protease sequences: JF338626-JF338629 (PR_SE10-PR_SE13), JF338630-JF338633 (PR_SE15-PR_SE18), JF338634-JF338641 (PR_SE20-PR_SE27), JF338642-JF338653 (PR_SE29-PR_SE40), JF338653 (PR_SE40), JF338654-JF338658 (PR_SE42-PR_SE46), JF338659-JF338663 (PR_SE48-PR_SE52), JF338664-JF338686 (PR_SE54-PR_SE76), JF338687-JF338690 (PR_SE78-PR_SE81), JF338691-JF338716 (PR_SE90-PR_SE115). RT sequences: JF338717-JF338719 (RT_SE10-RT_SE12), JF338720-JF338731 (RT_SE14-RT_SE25), JF338732-JF338744 (RT_SE27-RT_SE39), JF338745-JF338761 (RT_SE41-RT_SE57), JF338762-JF338780 (RT_SE59-RT_SE77), JF338781 (RT_SE79), JF338782-JF338807 (RT_SE90-RT_SE115).
Results and discussion
Patients' characteristics
Baseline characteristics of the 98 immigrant individuals studied are summarized in Table 1. The study population was grouped into seven categories according to their home countries: Central-South America (Argentina, Peru, Brazil, Colombia, Cuba, Bolivia, Ecuador, El Salvador, Guatemala, Panama, Paraguay, Dominican Republic and Venezuela), Western Europe (France, Italy, Portugal, Belgium, Germany and Ireland), Sub-Saharan Africa (Nigeria, Cameroon, Angola, Uganda, Sierra Leone, Guinea, Cote d'Ivoire), Northern Africa (Morocco), Eastern Europe (Romania, Ukraine and Russia), North America (United States), and Asia (India). The largest immigrant group in Southern Spain was from Central-South America (47/98, 47.9%). In most cases, these individuals were infected with HIV-1 before moving to Spain.
Table 1.
Characteristics of the 98 HIV-1-infected immigrant individuals studied
| Male gender (%) | 68 (69.4) |
| Age (years) | 35 [30-38] |
| CD4+ count (cell/mm3) | 321 [212-491] |
| HIV-1 viral load (log10IU/mL) | 4.28 [1.6-4.8] |
| Naïve ART patients (%) | 62 (63,2) |
| Time in treatment (months) | 25.1 [14.2-48.1] |
| Risk factor (%) | |
| Heterosexual | 41 (41.8) |
| Bisexual | 42 (42,8) |
| Injecting drug users | 7 (7.1) |
| Others | 8 (8.1) |
| Geographical Region (%) | |
| Central-South America | 47 (47.9) |
| Western Europe | 14 (14,3) |
| Sub-Saharan Africa | 19 (19.4) |
| Northern Africa | 5 (5.1) |
| Eastern Europe | 10 (10.2) |
| North America | 1 (1) |
| Asia | 1 (1) |
| Not determined | 1 (1) |
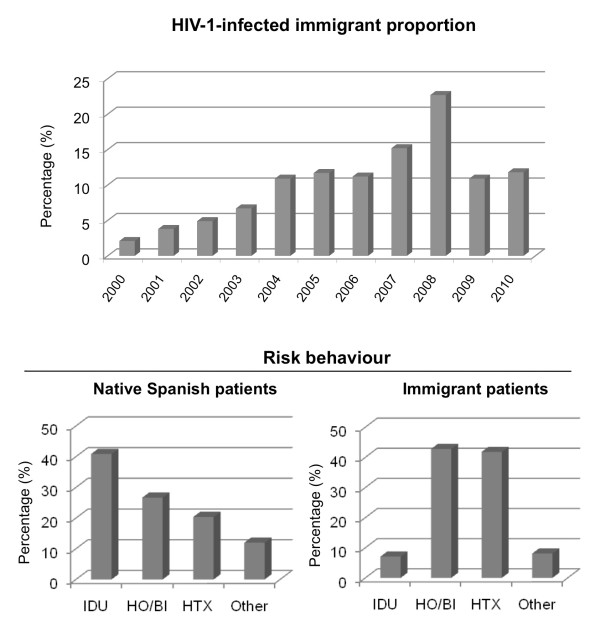
The proportion of immigrants in our cohort of HIV-1-infected patients from 2000 to 2010 is shown in Figure 1. The proportion has been rising year by year from 2.1% in 2000 to around 12% in 2010. Notably, there has been a marked progressive increase up to 2008, which exceeded 20% (Figure 1). Regarding risk behaviour, injecting drug use was the most common route of transmission among native Spanish patients (around 40% vs. 7% among immigrants, p < 0.001 Chi Square test), while sexual transmission (hetero plus homosexual) was predominant (80% vs. 42% among native Spanish patients, p < 0.001 Chi Square test) among immigrants (Figure 1).
Figure 1.
Proportion of HIV-1-infected immigrants from 2000 to 2010 in Southern Spain, and comparison of risk behavior between native Spanish and immigrant patients. IDU, injecting drug users; HO/BI, homosexual/bisexual; HTX, heterosexual.
Subtype analysis
Both PR and RT sequences were obtained for 87 patients, while only PR or RT sequences were obtained for seven and four individuals, respectively. Twenty-nine of the overall sequences analyzed belonged to non-B subtypes (29.6%, Table 2). These included A, C, F, G, J, and K subtypes, and recombinant forms, such as CRF02_AG, as well as CRF01_AE. Besides, seven patients harbored different subtypes for each gene, including recombinants B/C, F/B, K/G, CRF02_AG/CRF01_AE, and CRF02_AG/G in PR and RT respectively. According to the geographical origin, the non-B subtype was detected in a total of 79% (15/19) of Sub-Saharan Africans, followed by 60% Eastern Europeans (6/10), 20% Northern Africans (1/5), 10.6% Central-South Americans (5/47) and 7.1% Western Europeans (1/14).
Table 2.
Characteristics of the immigrant patients infected with HIV-1 non-B subtypes (N = 29)
| Patient code | Origin | Treatment | Subtype in PR gene |
Protease inhibitor resistance mutations |
Subtype in RT gene |
RT inhibitor resistance mutations |
|---|---|---|---|---|---|---|
| SE58 | EE | HAART | A | M36I, V77I | N.D. | N.D. |
| SE115 | CSA | Naive | B | M36I | C | None |
| SE71 | CSA | HAART | C | M36I | C | V118I |
| SE73 | EE | Naive | F | L10V, K20R, M36I | F | None |
| SE43 | CSA | HAART | F | M36I | B | None |
| SE99 | EE | Naive | F | K20R, M36I | F | None |
| SE108 | CSA | Naive | F | K20R, M36I | F | None |
| SE37 | SSA | Naive | G | K20I, M36I | G | K70R, A98G |
| SE103 | SSA | Naive | G | K20I, M36I | G | None |
| SE31 | SSA | Naive | J | K20I, M36I, V77I | J | None |
| SE91 | SSA | Naive | K | M36I, L63P | G | E138A |
| SE11 | SSA | HAART | CRF02_AG | K20I, M36I, L63P | G | None |
| SE16 | SSA | TI | CRF02_AG | K20I, M36I | G | None |
| SE113 | N.D. | Naive | CRF02_AG | K20I, M36I | G | None |
| SE66 | WE | HAART | CRF02_AG | K20I, M36I | CRF02_AG | None |
| SE59 | NAF | HAART | CRF02_AG | K20I, M36I, L90M | CRF02_AG | None |
| SE67 | SSA | HAART | CRF02_AG | K20I, M36I, L63P | CRF02_AG | None |
| SE49 | SSA | Naive | CRF02_AG | K20I, M36I, L63P | CRF02_AG | None |
| SE50 | SSA | TI | CRF02_AG | K20I, M36I, L63P, V77I | CRF02_AG | None |
| SE33 | SSA | TI | CRF02_AG | L10V, K20I, M36I | CRF02_AG | None |
| SE36 | CSA | HAART | CRF02_AG | K20I, M36I | CRF02_AG | None |
| SE101 | SSA | Naive | CRF02_AG | K20I, M36I | CRF02_AG | None |
| SE110 | EE | Naive | CRF02_AG | K20I, M36I | CRF02_AG | None |
| SE111 | SSA | HAART | CRF02_AG | K20I, M36I | CRF02_AG | 90I |
| SE34 | SSA | TI | CRF02_AG | K20I, M36I | CRF01_AE | L100I |
| SE96 | SSA | Naive | CRF02_AG | K20I, M36I | CRF02_AG | None |
| SE62 | EE | Naive | CRF01_AE | M36I | CRF01_AE | None |
| SE60 | EE | HAART | CRF01_AE | M36I | CRF01_AE | K103N |
| SE19 | SSA | HAART | N.D | N.D. | CRF02_AG | None |
Protease and RT inhibitor resistance mutations.
CSA, Central-South America; SSA, Sub-Sahara Africa; EE, Eastern Europe; WE, Western Europe; NAF, Northern Africa. HAART, Highly Active Antiretroviral Therapy. TI, treatment interruption. Bold, major resistance. N.D., not determined.
Phylogenetic analysis
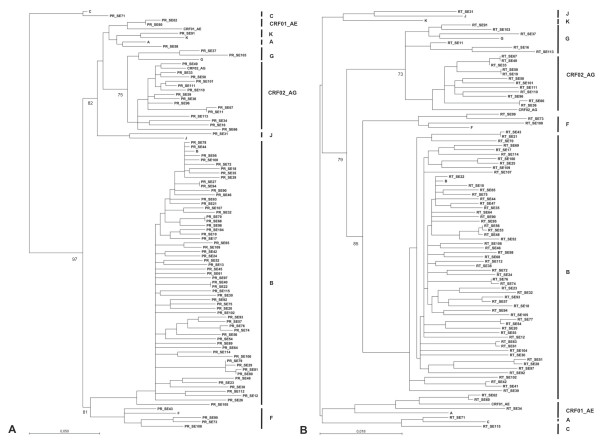
Phylogenetic trees were generated based on PR and RT sequences and included reference sequences representing different HIV-1 subtypes. All sequences clustered within the correspondent subtype, which was concordant with the data provided by Stanford University (Figure 2). Only three of the RT sequences: SE16, SE113, and SE11 corresponding to subtype G were located slightly apart from this subtype although within the same branch. Noteworthy, no sub-clusters were formed based on the geographical origin of the patients. Sequences PR_SE15, PR_SE25, PR_SE51, PR_SE55, RT_SE14, RT_SE15, RT_SE27, RT_SE29, RT_SE45, RT_SE79, were excluded from the phylogenetic analysis due to the limited length that could have influenced the analysis.
Figure 2.
Phylogenetic analyses derived from protease (A) and reverse transcriptase (B) sequences from HIV-1-infected immigrants from Southern Spain. Trees were constructed using the neighbor-joining method. Consensus sequences of subtypes A, B, C, F, G, J, K, and circulating recombinant forms CRF02_AG and CRF02_AE are included. Bootstrap values at nodes are shown as percentage of 1000 resamplings (only values greater than 70% are shown). Central-South America (CSA); Sub-Saharan Africa (SSA); Western Europe (WE); Northern Africa (NAF); Eastern Europe (EE); North America (NAM); not determined (ND).
Prevalence of resistance mutations
All patients infected with non-B HIV-1 showed at least one polymorphic mutation in PR (Table 2). Most of these mutations have been frequently found and reported amongst non-B HIV-1 subtypes [14,16]. The M36I mutation in protease was observed in all patients, whilst K20I/R, L63P, V77I and L10V/I mutations were present in 75%, 17.8%, 10.7% and 7.1% of patients, respectively. However, only patient SE59 had the major mutation L90M (1/28; 3.5%). This patient was infected with the recombinant form CRF02_AG and had been under HAART for at least two years.
On the contrary, only six patients showed RT mutations. Three of them (SE37, SE34 and SE60) showed major RT mutations (3/28, 10.7%, table 2). Patient SE37, naïve for antiretroviral treatment, had A98G and K70R major mutations associated with resistance to non-nucleoside RT inhibitors (NNRTI), and nucleoside RT inhibitors (NRTI), respectively. Patient SE34, in treatment interruption, had L100I mutation associated with NNRTI resistance, and finally patient SE60, receiving HAART, had the NNRTI resistance mutation K103N. Minor mutations or polymorphisms in RT were observed in patients SE71, SE91 and SE111 i.e.: V118I, E138A, and V90I mutations respectively.
Discussion
This study evaluates the prevalence of HIV-1 subtypes and PR and RT drug resistance mutations among HIV-1-infected immigrants living in Southern Spain. Immigration had been gradually increasing since 2000, with a peak of 22% in 2008. Since then, it has been slowly decreasing to 11.8% in 2010. The largest immigrant group was form Central-South America (47.9%) most of them infected with the HIV-1 B subtype. However, up to 25% of immigrants were from countries where HIV-1 non-B subtypes are predominant, as Sub-Saharan Africa with 79% of HIV-1 non-B subtypes (subtypes A, G, J or CRF02_AG), or Eastern Europe with 60% (subtypes A, F, CRF02_AG or CRF01_AE). This marked increase in individuals infected with non-B HIV-1 subtype in our society may have a direct impact on the spread of these subtypes among Spanish native individuals in addition to the changes in appropriate treatment regimes due to differences in genetic sequences amongst the different HIV-1 strains. These results are similar to those found in studies conducted in other regions in Spain, Madrid and Canary Islands [19,20,22].
Previous studies reported a high prevalence of minor resistance mutations in naïve patients infected with HIV-1 non-B subtype that could facilitate the emergence of major mutations [16,23,24]. However, the clinical relevance of minor mutations is unclear. Our results indicated that patients infected with HIV-1 non-B subtypes showed a high frequency of minor PR mutations (polymorphisms) as M36I, L63P, and K20R/I, in contrast to the low proportion of major resistance mutations. In fact, only one patient, subtype CRF02_AG, had L90M mutation probably as a result of receiving antiretroviral therapy during two years (1/28, 3.5%). No difference between the number of patients with major or minor RT mutations (3/28, 10.7% in both cases) was found. Nevertheless, the proportion of patients with major RT mutations was higher compared to patients with major PR mutations.
It is of note that one treatment naïve patient, with G subtype, showed major RT mutations associated with resistance to NRTI (K70R) and NNRTI (A98G). This patient might have acquired these resistance mutations at the moment of infection. One limitation of our study was the impossibility to test plasma samples of antiretroviral-experienced patients before the initiation of antiretroviral therapy to determine primary resistance mutations.
An increasing effort has been done to characterize the difference in antiretroviral therapy responses within HIV-1 non-B subtypes. Although clinical evidence remains limited, response to antiretroviral therapy does not appear to differ significantly among subtypes [3]. In our study, 29 patients showed HIV-1 non-B subtype in PR and/or RT. Eleven of them were under antiretroviral therapy and had good virological and immunological responses. Nevertheless, larger cohorts are necessary to confirm our results. On the other hand, 14 immigrants infected by HIV-1 non-B subtype were naïve for antiretroviral therapy. Most of them with viral load around 4 log HIV-1 copies/ml, and two patients had undetectable viral load for a long period of time (one of them at least for one year). As previous studies have described the failure of laboratory techniques to detect HIV-1 non-B subtypes [25-27], however it would be interesting to establish whether the failure of the diagnosis may be due to a phenotype characteristic of viral load controller in this population.
Conclusions
The circulation of HIV-1 non-B subtypes among immigrants, with a prevalence of 29.6%, has significantly increased in Southern Spain in the last decade, in association with demographic changes. Spread of CRF02-AG, G and F subtypes, and also some recombinant forms appear to predominate, which is consistent with previous findings [19,20,22,28]. This fact may affect the treatment and management of these patients, and the spread of these subtypes among native Spanish population. Furthermore, a low percentage of the immigrants with the non-B subtypes had major resistance mutations. Prevalence of these resistance mutations varies from 7% among treatment-naïve patients to 21% among patients under HAART or during treatment interruption.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
BF made most of the amplifications, sequencing, drug resistance profiles, sequence alignments and wrote a first draft of the manuscript. PPR, FJMF and FFC made some amplifications and sequencing. MAF helped in alignments of the sequences and revising the manuscript. MT and RCM helped in the management of the patients and collecting samples. LLC, ML, and PV recruited all the patients. AV designed and coordinated the study, helped in the alignment of the sequences and finalized the manuscript in its final form. All authors read and approved the final manuscript.
Contributor Information
Beatriz de Felipe, Email: beadefelipe@gmail.com.
Pilar Pérez-Romero, Email: mariap.perez.exts@juntadeandalucia.es.
María Abad-Fernández, Email: macarju@hotmail.com.
Felipe Fernandez-Cuenca, Email: felip254@gmail.com.
Francisco J Martinez-Fernandez, Email: francisco@gmail.com.
Mónica Trastoy, Email: m34tras@yahoo.es.
Rosario del Carmen Mata, Email: rosmat@yahoo.es.
Luis F López-Cortés, Email: lflopez@telefonica.net.
Manuel Leal, Email: maleal@telefonica.net.
Pompeyo Viciana, Email: pompeyo@telefonica.net.
Alejandro Vallejo, Email: alejandro.vallejo@salud.madrid.org.
Acknowledgements
We thank all the participants of this study for their cooperation. This study was supported in part by Fondo de Investigación Sanitaria (FIS) 07/0070, Redes Temáticas de Investigación Cooperativa en Salud (RETICS) Red de Investigación en SIDA (RIS) RD06/0006/0021 and RD/06/0006/0034, and Servicio Andaluz de Salud, Consejería de Salud 0407/2007.
References
- Thomson M, Najera R. Increasing HIV-1 genetic diversity in Europe. J Infect Dis. 2007;196:1120–1124. doi: 10.1086/521683. [DOI] [PubMed] [Google Scholar]
- Vidal N, Peeters M, Mulanga-Kabeya C, Nzilambi N, Robertson D, Ilunga W, Sema H, Tshimanga K, Bongo B, Delaporte E. Unprecedented degree of human immunodeficiency virus type 1 (HIV-1) group M genetic diversity in the Democratic Republic of Congo suggests that the HIV-1 pandemic originated in Central Africa. J Virol. 2000;74:10498–10507. doi: 10.1128/JVI.74.22.10498-10507.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geretti AM. HIV-1 subtypes: epidemiology and significance for HIV management. Curr Opin Infect Dis. 2006;19:1–7. doi: 10.1097/01.qco.0000200293.45532.68. [DOI] [PubMed] [Google Scholar]
- Hemelaar J, Gouws E, Ghys PD, Osmanov S. Global and regional distribution of HIV-1 genetic subtypes and recombinants in 2004. AIDS. 2006;20:W13–23. doi: 10.1097/01.aids.0000247564.73009.bc. [DOI] [PubMed] [Google Scholar]
- Balotta C, Facchi G, Violin M, Van Dooren S, Cozzi-Lepri A, Forbici F, Bertoli A, Riva C, Senese D, Caramello P, Carnevale G, Rizzardini G, Cremonini L, Monno L, Rezza G, Perno CF, Ippolito G, d'Arminio-Monforte A, Vandamme AM, Moroni M. ICONA Study Group. Increasing prevalence of non-clade B HIV-1 strains in heterosexual men and women, as monitored by analysis of reverse transcriptase and protease sequences. J Acquir Immune Defic Syndr. 2001;27:499–505. doi: 10.1097/00126334-200108150-00012. [DOI] [PubMed] [Google Scholar]
- De Paschale M, Cagnin D, Cerulli T, Mena M, Magnani C, Perini P, Re T, Villa M, Viganò P, Maltempo C, Manco MT, Agrappi C, Mirri P, Gatti A, Rescaldani C, Clerici P. Epidemiology of HIV-1 subtypes in an urban area of northern Italy. Clin Microbiol Infect. 2011;17:935–940. doi: 10.1111/j.1469-0691.2010.03382.x. [DOI] [PubMed] [Google Scholar]
- Habekova M, Takacova M, Lysy J, Mokras M, Camacho R, Truska P, Stanekova D. Genetic Subtypes of HIV Type 1 Circulating in Slovakia. AIDS Res Human Retroviruses. 2010;26:1103–1107. doi: 10.1089/aid.2009.0220. [DOI] [PubMed] [Google Scholar]
- Vasan A, Renjifo B, Hertzmark E, Chaplin B, Msamanga G, Essex M, Fawzi W, Hunter D. Different rates of disease progression of HIV type 1 infection in Tanzania based on infecting subtype. Clin Infect Dis. 2006;42:843–852. doi: 10.1086/499952. [DOI] [PubMed] [Google Scholar]
- Kiwanuka N, Laeyendecker O, Quinn TC, Wawer MJ, Shepherd J, Robb M, Kigozi G, Kagaayi J, Serwadda D, Makumbi FE, Reynolds SJ, Gray RH. HIV-1 subtypes and differences in heterosexual HIV transmission among HIV-discordant couples in Rakai, Uganda. AIDS. 2009;18:2479–2484. doi: 10.1097/QAD.0b013e328330cc08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Descamps D, Chaix ML, Montes B, Pakianather S, Charpentier C, Storto A, Barin F, Dos Santos G, Krivine A, Delaugerre C, Izopet J, Marcelin AG, Maillard A, Morand-Joubert L, Pallier C, Plantier JC, Tamalet C, Cottalorda J, Desbois D, Calvez V, Brun-Vezinet F, Masquelier B, Costagliola D. the ANRS AC11 Resistance Study Group. Increasing prevalence of transmitted drug resistance mutations and non-B subtype circulation in antiretroviral-naive chronically HIV-infected patients from 2001 to 2006/2007 in France. J Antimicrob Chemother. 2010;65:2620–2627. doi: 10.1093/jac/dkq380. [DOI] [PubMed] [Google Scholar]
- Eshleman SH, Laeyendecker O, Parkin N, Huang W, Chappey C, Paquet AC, Serwadda D, Reynolds SJ, Kiwanuka N, Quinn TC, Gray R, Wawer M. Antiretroviral drug susceptibility among drug-naive adults with recent HIV infection in Rakai, Uganda. AIDS. 2009;27:845–852. doi: 10.1097/QAD.0b013e328327957a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu LY, Subramaniam R, Bacheler L, Paton NI. Characterization of mutations in CRF01_AE virus isolates from antiretroviral treatment-naive and -experienced patients in Singapore. J Acquir Immune Defic Syndr. 2005;38:5–13. doi: 10.1097/00126334-200501010-00002. [DOI] [PubMed] [Google Scholar]
- Kantor R, Katzenstein D. Drug resistance in non-subtype B HIV-1. J Clin Virol. 2004;29:152–159. doi: 10.1016/S1386-6532(03)00115-X. [DOI] [PubMed] [Google Scholar]
- Peeters M, Aghokeng AF, Delaporte E. Genetic diversity among human immunodeficiency virus-1 non-B subtypes in viral load and drug resistance assays. Clin Microbiol Infection. 2010;16:1525–1531. doi: 10.1111/j.1469-0691.2010.03300.x. [DOI] [PubMed] [Google Scholar]
- Wheeler WH, Ziebell RA, Zabina H, Pieniazek D, Prejean J, Bodnar UR, Mahle KC, Heneine W, Johnson JA, Hall HI. the Variant and Resistant HIV Surveillance Group. Prevalence of transmitted drug resistance associated mutations and HIV-1 subtypes in new HIV-1 diagnoses, U.S.-2006. AIDS. 2010;24:1203–1212. doi: 10.1097/QAD.0b013e3283388742. [DOI] [PubMed] [Google Scholar]
- Vergne L, Stuyver L, Van Houtte M, Butel C, Delaporte E, Peeters M. Natural polymorphism in protease and reverse transcriptase genes and in vitro antiretroviral drug susceptibilities of non-B HIV-1 strains from treatment-naive patients. J Clin Virol. 2006;36:43–49. doi: 10.1016/j.jcv.2006.01.012. [DOI] [PubMed] [Google Scholar]
- Gueudin M, Plantier JC, Lemée V, Schmitt MP, Chartier L, Bourlet T, Ruffault A, Damond F, Vray M, Simon F. Evaluation of the Roche Cobas TaqMan and Abbott RealTime extraction-quantification systems for HIV-1 subtypes. J Acquir Immune Defic Syndr. 2007;44:500–505. doi: 10.1097/QAI.0b013e31803260df. [DOI] [PubMed] [Google Scholar]
- Rouet F, Chaix ML, Nerrienet E, Ngo-Giang-Huong N, Plantier JC, Burgard M, Peeters M, Damond F, Ekouevi DK, Msellati P, Ferradini L, Rukobo S, Maréchal V, Schvachsa N, Wakrim L, Rafalimanana C, Rakotoambinina B, Viard JP, Seigneurin JM, Rouzioux C. Impact of HIV-1 genetic diversity on plasma HIV-1 RNA Quantification: usefulness of the Agence Nationale de Recherches sur le SIDA second-generation long terminal repeat-based real-time reverse transcriptase polymerase chain reaction test. J Acquir Immune Defic Syndr. 2007;45:380–388. doi: 10.1097/QAI.0b013e3180640cf5. [DOI] [PubMed] [Google Scholar]
- Holguín A, Alvarez A, Pena MJ, Artiles F, Molina L, Soriano V. HIV-positive immigrants in the Canary Islands, Spain: implications for public health in Europe. HIV Clin Trials. 2003;4:184–192. doi: 10.1310/9X1G-MJ6L-PBJN-1PVE. [DOI] [PubMed] [Google Scholar]
- Holguín A, de Mulder M, Yebra G, López M, Soriano V. Increase of non-B subtypes and recombinants among newly diagnosed HIV-1 native Spaniards and immigrants in Spain. Curr HIV Res. 2008;6:327–334. doi: 10.2174/157016208785132455. [DOI] [PubMed] [Google Scholar]
- Lospitao E, Alvarez A, Soriano V, Holguín A. HIV-1 subtypes in Spain: a retrospective analysis from 1995 to 2003. HIV Med. 2005;6:313–320. doi: 10.1111/j.1468-1293.2005.00313.x. [DOI] [PubMed] [Google Scholar]
- Treviño A, Soriano V, Rodriguez C, Arredondo M, Rivas P, Herrero-Mendoza D, Parra P, del Romero J, Anta L, Puente S, de Mendoza C. Changing rate of non-B subtypes and coinfection with hepatitis B/C viruses in newly diagnosed HIV type 1 individuals in Spain. AIDS Res Hum Retroviruses. 2011;27:633–638. doi: 10.1089/aid.2010.0247. [DOI] [PubMed] [Google Scholar]
- Bennett DE, Camacho RJ, Otelea D, Kuritzkes DR, Fleury H, Kiuchi M, Heneine W, Kantor R, Jordan MR, Schapiro JM, Vandamme AM, Sandstrom P, Boucher CA, van de Vijver D, Rhee SY, Liu TF, Pillay D, Shafer RW. Drug resistance mutations for surveillance of transmitted HIV-1 drug-resistance: 2009 update. PLoS One. 2009;4:e4724. doi: 10.1371/journal.pone.0004724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montes B, Vergne L, Peeters M, Reynes J, Delaporte E, Segondy M. Comparison of drug resistance mutations and their interpretation in patients infected with non-B HIV-1 variants and matched patients infected with HIV-1 subtype B. J Acquir Immune Defic Syndr. 2004;35:329–336. doi: 10.1097/00126334-200404010-00001. [DOI] [PubMed] [Google Scholar]
- Drexler JF, de Souza Luna LK, Pedroso C, Pedral-Sampaio DB, Queiroz AT, Brites C, Netto EM, Drosten C. Rates of and reasons for failure of commercial human immunodeficiency virus type 1 viral load assays in Brazil. J Clin Microbiol. 2007;45:2061–2063. doi: 10.1128/JCM.00136-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geelen S, Lange J, Borleffs J, Wolfs T, Weersink A, Schuurman R. Failure to detect a non-B HIV-1 subtype by the HIV-1 Amplicor Monitor test, version 1.5: a case of unexpected vertical transmission. AIDS. 2003;17:781–778. doi: 10.1097/00002030-200303280-00027. [DOI] [PubMed] [Google Scholar]
- McConnell MJ, Docobo-Pérez F, Mata RC, Fernandez-Cuenca F, Viciana P, López-Cortés LF, Trastoy M, Pachón J, Pérez-Romero P. Molecular epidemiology of HIV type 1 in newly diagnosed patients in southern Spain. AIDS Res Hum Retroviruses. 2008;24:881–887. doi: 10.1089/aid.2008.0051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García F, Pérez-Cachafeiro S, Guillot V, Alvarez M, Pérez-Romero P, Pérez-Elías MJ, Viciana I, Blanco JR, López-Dieguez M, de Mendoza C. Cohort of the Spanish AIDS Research Network (CoRIS) Transmission of HIV drug resistance and non-B subtype distribution in the Spanish cohort of antiretroviral treatment naïve HIV-infected individuals (CoRIS) Antiviral Res. 2011;91:150–153. doi: 10.1016/j.antiviral.2011.05.010. [DOI] [PubMed] [Google Scholar]