Abstract
The family of Toll-like receptors (TLRs) is critical in linking innate and acquired immunity. Polymorphisms in the genes encoding TLRs have been associated with autoimmune diseases and cancer. We investigated the genetic variation of TLR genes and its potential impact on melanoma susceptibility and patient survival. The study included 763 cutaneous melanoma cases recruited in Germany and 736 matched controls that were genotyped for 47 single nucleotide polymorphisms (SNPs) in 8 TLR genes. The relationship between genotype, disease status and survival was investigated taking into account patient and tumor characteristics, and melanoma treatment. Analysis of 7 SNPs in TLR2, 7 SNPs in TLR3 and 8 SNPs in TLR4 showed statistically significant differences in distribution of inferred haplotypes between cases and controls. No individual polymorphism was associated with disease susceptibility except for the observed tendency for TLR2-rs3804099 (odds ratio OR = 1.15, 95% CI 0.99–1.34, p = 0.07) and TLR4-rs2149356 (OR = 0.85, 95% CI 0.73–1.00, p = 0.06). Both polymorphisms were part of the haplotypes associated with risk modulation. An improved overall survival (Hazard ratio HR 0.53, 95% CI 0.32–0.88) and survival following metastasis (HR 0.55, 95% CI 0.34–0.91) were observed in carriers of the variant allele (D299G) of TLR4-rs4986790. In addition various TLR2, TLR4 and TLR5 haplotypes were associated with increased overall survival. Our results point to a novel association between TLR gene variants and haplotypes with melanoma survival. Our data suggest a role for the D299G polymorphism in the TLR4 gene in overall survival and a potential link with systemic treatment at stage IV of the disease. The polymorphic amino acid residue, located in the ectodomain of TLR4, can have functional consequences.
Introduction
Malignant melanoma, with its propensity to metastasize and chemoresistance, remains a fatal neoplasm. Spontaneous regression, a phenomenon likely mediated by the immune system, is more common in melanoma than in most other cancers. The role of host immune system in melanoma is evidenced by the association of autoimmune conditions such as thyroiditis, vitiligo or the appearance of autoantibodies with an improved prognosis in patients treated with interleukin 2 and/or interferon α therapy [1], [2], [3], [4]. Variations in genes involved in critical pathways of the immune system might therefore influence the susceptibility to autoimmune diseases and subsequently melanoma susceptibility, progression and prognosis.
Toll-like receptors (TLR), the mammalian homologues of the Drosophila Toll protein, are perceived to be sensors for recognition of a number of invading pathogens.
TLR are critical in linking innate and acquired immunity and in serving as detectors of infectious pathogens and cancer debris [5], [6], [7]. TLRs belong to the family of pattern-recognition receptors (PRRs) which are expressed, among others, on antigen-presenting cells like dendritic cells or T-cells. After induction through pathogen-associated molecules, TLRs transduce signals via distinct intracellular pathways leading to the activation of transcription factors such as NFkB, interferon regulatory factors (IRFs) or AP-1. Finally these transcription receptors trigger inflammatory responses such as the release of inflammatory cytokines and type I interferons [7], [8], [9].
Polymorphisms in genes encoding TLR associated with infectious and non-infectious diseases, including autoimmunity and cancer, have been studied extensively [10], [11], [12], [13]. However, the variants in the genes have not been investigated for association with either melanoma susceptibility or disease outcome. In order to investigate the effect of polymorphisms in the TLR genes on melanoma susceptibility and survival, we genotyped 47 single nucleotide polymorphisms (SNP) in eight TLR genes in 763 cases from Germany and 736 ethnically matched controls.
Results
The two multiplex reactions contained 373 replicates from different SNPs that showed a concordance rate over >99%. The rate of undetermined samples by the Sequenom procedure was between 6 and 33% of the individual SNPs. Genotype distributions of all polymorphisms in controls did not show statistically significant deviation from Hardy-Weinberg equilibrium.
Out of 47TLR SNPs analyzed by Sequenom mass spectrometry TLR1-rs3923647 and TLR1-rs5743613, TLR2-rs5743704 and -rs5743708, TLR4-rs11536869 and -rs11536897 and TLR6-rs5743815 had minor allele frequencies <0.05; no carrier of variant allele for TLR9-rs5743846 was detected. In total, the genotyping results of 47 SNPs represent 99 polymorphisms in the selected TLR genes.
Case-control study
We did not observe any differences in the distribution of allele and genotype frequencies between melanoma cases and controls (Table S1, online only). The variant allele for TLR2-rs3804099 was associated with an increased risk (OR = 1.15, 95% CI 0.99–1.34, p = 0.07) and for TLR4-rs2149356 with a decreased risk (OR = 0.85, 95% CI 0.73–1.00, p = 0.06) of melanoma. However, for both, the association was not statistically significant.
Haplotype analysis
Analysis of 7 SNPs in TLR2, 7 SNPs in TLR3 and 8 SNPs in TLR4 showed statistically significant differences in distribution of inferred haplotypes between cases and controls (Table 1). The haplotype A-C-C-C-C-G-T of the TLR2 gene locus was associated with an increased risk compared to the consensus haplotype A-C-C-C-T-G-T (OR 1.31, 95%CI 1.03–1.65). This haplotype included the SNP that showed individually an association with increased risk. A haplotype for the TLR3 gene (C-G-G-T-A-G-G) was associated with a decreased susceptibility to melanoma (OR 0.57, 95%CI 0.35–0.95) compared to the most frequent haplotype (C-G-A-C-C-C-A). The haplotype A-T-A-G-A-G-G-C of the TLR4 gene locus associated with a decreased risk for melanoma compared to the most frequent haplotype A-T-C-A-A-G-G-T (OR 0.72, 95%CI 0.57–0.91). The haplotype includes the minor allele of TLR2-rs2149356 that associated individually with a protective effect. Haplotype analysis of the other TLRs did not identify associations with melanoma risk (data not shown).
Table 1. Distribution of the most common haplotypes within TLR2, TLR3 and TLR4 in German melanoma patients and German controls.
| Gene | Haplotype | Cases (%) | Controls (%) | OR (95% CI) | P |
| TLR2 | A-C-C-C-T-G-T | 197 (12.9) | 220 (15.0) | Reference | 1.0 |
| T-C-A-C-T-G-T | 491 (32.3) | 496 (33.8) | 1.11 (0.88–1.39) | ||
| A-C-C-C-C-G-T | 457 (30.0) | 391 (26.7) | 1.31 (1.03–1.65) | ||
| T-T-C-C-C-G-T | 140 (9.2) | 141 (9.6) | 1.11 (0.82–1.50) | ||
| A-C-C-C-C-G-C | 94 (6.2) | 85 (5.8) | 1.23 (0.87–1.75) | ||
| A-C-C-A-T-G-T | 48 (3.2) | 51 (3.5) | 1.05 (0.68–1.63) | ||
| T-C-A-C-T-A-T | 38 (2.5) | 33 (2.2) | 1.29 (0.78–2.13) | ||
| T-C-C-C-T-G-T | 29 (1.9) | 24 (1.6) | 1.35 (0.76–2.40) | ||
| T-T-C-C-T-G-T | 12 (0.8) | 11 (0.7) | 1.22 (0.53–2.82) | ||
| other | 16 (1.0) | 15 (1.0) | − | ||
| TLR3 | C-G-A-C-C-C-A3 | 268 (17.6) | 256 (17.5) | Reference | 0.94 |
| C-A-G-C-C-G-G | 240 (15.8) | 211 (14.4) | 1.09 (0.84–1.40) | ||
| C-G-G-T-A-C-G | 205 (13.5) | 187 (12.8) | 1.05 (0.81–1.36) | ||
| C-G-G-C-A-C-G | 153 (10.1) | 176 (12) | 0.83 (0.63–1.09) | ||
| T-G-G-C-C-C-G | 215 (14.2) | 175 (12) | 1.17 (0.90–1.53) | ||
| C-G-G-C-C-C-A | 123 (8.1) | 128 (8.7) | 0.92 (0.68–1.24) | ||
| C-A-G-C-C-C-G | 91 (6.0) | 77 (5.3) | 1.13 (0.80–1.60) | ||
| C-G-A-C-C-C-G | 71 (4.7) | 75 (5.1) | 0.90 (0.63–1.31) | ||
| C-G-G-T-A-G-G | 27 (1.8) | 45 (3.1) | 0.57 (0.35–0.95) | ||
| C-G-G-C-C-C-G | 44 (2.9) | 33 (2.3) | 1.27 (0.79–2.06) | ||
| C-G-G-C-C-G-G | 16 (1.1) | 20 (1.4) | 0.76 (0.39–1.51) | ||
| T-G-G-C-C-C-A | 13 (0.9) | 14 (1.0) | 0.89 (0.41–1.92) | ||
| C-G-G-C-C-G-A | 16 (1.1) | 13 (0.9) | 1.18 (0.55–2.49) | ||
| other | 37 (2.4) | 53 (3.6) | − | ||
| TLR4 | A-T-C-A-A-G-G-T3 | 457 (30) | 405 (27.5) | Reference | 0.95 |
| A-C-C-G-A-G-G-T | 371 (24.4) | 357 (24.3) | 0.92 (0.76–1.12) | ||
| A-T-A-G-A-G-G-C | 192 (12.6) | 237 (16.1) | 0.72 (0.57–0.91) | ||
| A-C-C-G-A-C-G-T | 209 (13.7) | 215 (14.6) | 0.86 (0.68–1.09) | ||
| A-T-A-G-G-G-G-C | 85 (5.6) | 77 (5.2) | 0.98 (0.7–1.37) | ||
| G-T-A-G-A-G-G-T | 43 (2.8) | 52 (3.5) | 0.73 (0.48–1.12) | ||
| A-T-A-G-A-G-G-T | 60 (3.9) | 40 (2.7) | 1.33 (0.87–2.03) | ||
| A-T-A-G-A-G-A-C | 50 (3.3) | 39 (2.6) | 1.14 (0.73–1.76) | ||
| A-C-A-G-A-G-G-T | 8 (0.5) | 10 (0.7) | 0.71 (0.28–1.81) | ||
| other | 46 (3.0) | 40 (2.7) | − |
Polymorphisms in TLR genes and survival
Differences in survival according to genotype were investigated based on 622 patients with melanoma of the skin with known primary at first diagnosis without detectable metastasis (stage I/II) (339 males and 283 females out of 763 patients in total).
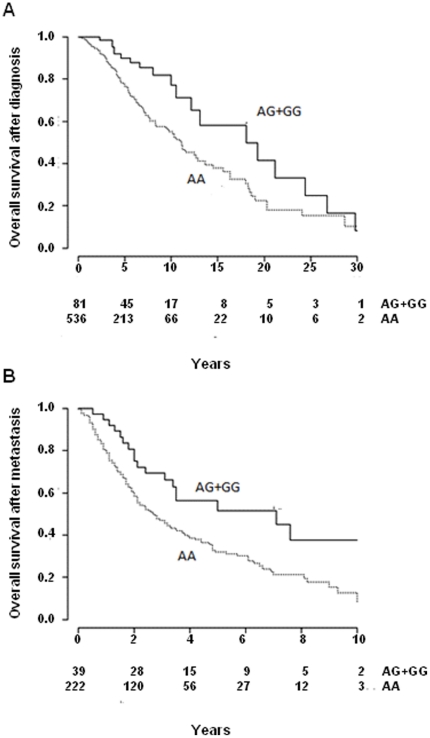
The OS from primary diagnosis was better in carriers of the minor allele (n = 81) of the TLR4-rs4986790 polymorphism than among non-carriers (n = 536). The median OS in carriers was 18.1 years (95% CI 12.1–24.4) compared to 11.0 years (95% CI 9.5–12.9) in non-carriers (log-rank p = 0.005, Figure 1A and Table 2). Cox regression adjusted for age, gender and Breslow thickness showed a decreased survival with a HR of 0.53, 95% CI 0.32–0.88, p = 0.01, Table 2) for carriers of the minor allele. Adjustment for any kind of therapy in stage IV showed no effect on this estimate (HR 0.53, 95% CI 0.32–0.87, p = 0.01, Table 2). Stratification according to therapy showed that the HR was 0.41 (95% CI 0.24–0.69, p = 0.001) among patients receiving any therapy, compared to HR 0.35 (95% CI 0.05–2.59, p = 0.30) for patients that did not receive any therapy. The statistical non-significance in the latter could be attributed to small patient numbers.
Figure 1. Overall survival (OS) and survival after metastasis (SFM) in German cutaneous melanoma patients according to theTLR4-rs4986790 (D299G) genotype.
The Kaplan-Meier curves differentiate between carriers of the minor allele (AG+GG) and homozygote genotypes (AA) for (A) overall survival, log-rank P = 0.005 and (B) survival following the first metastasis, log-rank P = 0.01. Number of patients, both carriers and non-carriers, is given below the x-axis.
Table 2. TLR4-rs4986790 genotype and overall survival (OS), survival following the first metastasis (SFM) and metastasis free survival (MFS).
| Genotype | Individuals | Median survival in years (95% CI) | Uncensored events (%) | Hazard Ratio (95% CI) | P | |
| OS | AA | 536 | 11.0 (9.5–12.9) | 161 (30.0) | Reference | |
| AG | 81 | 18.1 (12.1–24.4) | 19 (23.5) | 0.53 (0.32–0.88) | 0.01 | |
| 0.53 (0.32–0.87) | 0.01 | |||||
| MFS | AA | 536 | 7.2 (5.5–8.3) | 239 (44.6) | Reference | |
| AG | 81 | 10.6 (3.2–20.3) | 39 (48.1) | 0.81 (0.55–1.18) | 0.27 | |
| SFM | AA | 222 | 2.8 (2.1–3.5) | 144 (64.9) | Reference | |
| AG | 39 | 7.1 (3.3–11.1) | 19 (46.2) | 0.55 (0.33–0.91) | 0.02 | |
| 0.44 (0.27–0.73) | 0.001 |
The TLR4-rs4986790 polymorphism was also associated with survival time following first metastasis (SFM). Carriers of the minor allele displayed a median SFM of 7.1 years (95% CI 3.3–11.1) compared to 2.8 years (95% CI 2.1–3.5) for non-carrier patients (log-rank p = 0.01, Figure 1B and Table 2). The effect was reflected in the Cox regression model with a HR of 0.55 (95% CI 0.33–0.91, p = 0.02) and adjustment for any therapy slightly decreased the estimate (HR 0.44, 95% CI 0.27–0.73, p = 0.001, Table 2). The stratification for therapy showed a statistically significant effect (HR 0.39, 95% CI 0.23–0.66, p = 0.001) only in patients that received therapy and not in patient that did not receive any therapy (HR 0.62, 95% CI 0.08–4.86, p = 0.65). The TLR4-rs4986790 polymorphism did not associate with time from primary diagnosis to first metastasis (Table 2).
Carriers of the variant allele of TLR2-rs3804100 (TC/CC) also showed a reduced OS (HR 0.55, 95% CI 0.29–1.05, p = 0.07). Adjustment for any therapy at any clinical stage did not show an effect (HR 0.52, 95% CI 0.27–1.01, p = 0.05). We also evaluated the possible interaction between polymorphisms TLR4-rs4986790 and TLR2-rs3804100 regarding OS. Patients with the commonest genotype (rs4986790 = AA, rs3804100 = TT) showed the poorest prognosis. Among 13 patients with rs4986790 = AG and rs3804100 = TC/CC, no death was observed ten years after diagnosis. The survival heterogeneity among the four categories of individuals resulted in a probability value from the log-rank test of 0.01, which increased to 0.03 after inclusion of age, gender and Breslow thickness in a subsequent Cox regression. Other investigated polymorphisms did not associate with OS, SFM or MFS.
Association of TLR haplotypes with overall survival
Several haplotypes inferred based on polymorphisms in TLR2, TLR4 and TLR5 showed association with OS (Table 3). In the TLR2 gene, a minor haplotype A-C-C-C-C-G-C (5.5%) was significantly associated with an increased survival compared to the consensus haplotype A-C-C-C-T-G-T (HR 0.46, 95% CI 0.22 - 0.95). Another haplotype in TLR4, A-T-A-G-A-G-G-T, associated with OS (HR 1.77, 95% CI 1.02–3.08) in comparison to the most frequent haplotype. A frequent haplotype of TLR4 (A-T-A-G-A-G-G-C, 13.9%) also associated with increased OS (HR 1.4, 95% CI 0.95–2.06). Out of the 5 major TLR5 haplotypes, one associated with enhanced OS (T-T-G-C, HR 1.6, 95% CI 1.05–2.43). The remaining TLR haplotypes did not show significant association with OS (data not shown).
Table 3. Overall survival according to the commonest TLR2, TLR4 and TLR5 haplotypes.
| Gene | Haplotype | Frequency (%) | Uncensored events (%) | HR (95% CI) | P |
| TLR2 | A-C-C-C-T-G-T | 121 (11.2) | 37 (3.4) | Reference | 0.11 |
| T-C-A-C-T-G-T | 367 (33.9) | 105 (9.7) | 0.86 (0.60–1.24) | ||
| A-C-C-C-C-G-T | 345 (31.9) | 100 (9.2) | 0.84 (0.58–1.24) | ||
| T-T-C-C-C-G-T | 104 (9.6) | 32 (3.0) | 1.07 (0.67–1.73) | ||
| A-C-C-C-C-G-C | 59 (5.5) | 9 (0.8) | 0.46 (0.22–0.95) | ||
| A-C-C-A-T-G-T | 33 (3.0) | 8 (0.7) | 0.62 (0.28–1.36) | ||
| T-C-A-C-T-A-T | 20 (1.8) | 1 (0.1) | 0.24 (0.03–1.78) | ||
| T-C-C-C-T-G-T | 20 (1.8) | 4 (0.4) | 0.58 (0.21–1.65) | ||
| other | 13 (1.2) | 6 (0.6) | − | ||
| TLR4 | A-T-C-A-A-G-G-T3 | 323 (29.9) | 82 (7.6) | Reference | 0.12 |
| A-C-C-G-A-G-G-T | 268 (24.8) | 85 (7.9) | 1.09 (0.77–1.53) | ||
| A-T-A-G-A-G-G-C | 150 (13.9) | 47 (4.3) | 1.40 (0.95–2.06) | ||
| A-C-C-G-A-C-G-T | 139 (12.9) | 35 (3.2) | 1.01 (0.67–1.50) | ||
| A-T-A-G-G-G-G-C | 68 (6.3) | 17 (1.6) | 0.60 (0.35–1.04) | ||
| A-T-A-G-A-G-G-T | 40 (3.7) | 16 (1.5) | 1.77 (1.02–3.08) | ||
| A-T-A-G-A-G-A-C | 32 (3.0) | 5 (0.5) | 0.44 (0.18–1.10) | ||
| G-T-A-G-A-G-G-T | 29 (2.7) | 9 (0.8) | 1.73 (0.83–3.60) | ||
| other | 33 (3.1) | 6 (0.6) | − | ||
| TLR5 | C-T-G-C3 | 241 (22.3) | 61 (5.7) | Reference | 0.32 |
| C-T-C-C | 395 (36.6) | 108 (10.0) | 1.18 (0.85–1.63) | ||
| C-G-G-C | 287 (26.6) | 86 (8.0) | 1.33 (0.91–1.93) | ||
| T-T-G-C | 96 (8.9) | 35 (3.2) | 1.60 (1.05–2.43) | ||
| C-T-C-T | 53 (4.9) | 12 (1.1) | 0.96 (0.51–1.83) | ||
| other | 8 (0.7) | 1 (0.1) | − |
TLR Haplotype associations with survival time following first metastasis
The TLR2 haplotype T-T-C-C-C-G-T (9.6%) was associated with an increased SFM compared to the common haplotype (HR 1.54 95% CI 1.04–2.28). Other significant associations with decreased SFM were seen for TLR4- A-T-A-G-A-G-A-C and TLR4-G-T-A-G-A-G-G-T (HR 0.36, 95% CI 0.14–0.90 and HR 2.41, 95% CI 1.10–5.24), which were both rare (3.1% and 2.7%, respectively). Two frequent haplotypes A-C-C-G-A-G-G-T and A-T-A-G-A-G-G-C (26.2% and 14.1%, respectively) in this gene were associated with decreased SFM (HR 1.36, 95% CI 0.97–1.92 and HR 1.47, 95% CI 0.99–2.19, respectively). No other of the TLR haplotypes showed association with SFM.
TLR Haplotype associations with metastasis free survival (MFS) after diagnosis of primary tumor
Patients that harbored the TLR4 haplotype A-T-A-G-A-G-G-T were at an increased risk for MFS compared to those carrying the most frequent haplotype A-T-C-A-A-G-G-T (HR 2.02, 95% CI 1.28–3.17). Significant association with MFS was also seen for one of the most frequent haplotypes (C-G-G-C, frequency 26.5%) of the TLR5 gene when compared to the consensus haplotype C-T-G-C (HR 1.39, 95% CI 1.03–1.88). Other associations of TLR haplotypes with MFS were not statistically significant.
Discussion
TLRs, the mammalian homologues of Drosophila toll protein, play a role as agents of innate and adaptive immunity in various diseases. We assumed that genetic polymorphisms in various TLRs were functional and influenced melanoma susceptibility and disease outcome. Our data showed that melanoma patients carrying the variant allele for the rs4986790 polymorphism in the TLR4 gene were associated with a prolonged OS. The increase in survival was confined to the period after the detection of a first metastasis, which implied that the effect of the polymorphism might possibly modulated dependent on therapy or alternatively by biological processes associated with melanoma cell migration and/or invasion. The patients in stage IV melanoma in most instances received a chemotherapy containing DTIC/temozolomide and interferons, which is known for long time not having any clinical effect on overall survival of the entire metastatic melanoma population [14], [15].
One can speculate that the prolonged survival after first metastasis, which clinically is in most cases regionally spread, is mediated by more efficacious tumor control at the lymph node level and by interaction with the immune system.
TLR4-rs4986790 polymorphism results in a change of amino acid D299G and it co-segregates with a T399I exchange, which are both located in the ectodomain of TLR4 [16]. The ectodomain of TLR4 is responsible for detecting of lipopolysaccharides from Gram negative bacteria in cooperation with an accessory molecule, myeloid differentiation factor 2 (MD-2). MD-2 forms a 1∶1 complex with lipopolysaccharides, which is detected by TLR4 [16]. Interestingly, the amino acid residues 299 and 399 of TLR4 are not located either in the recognition surface of TLR4 for the MD-2-lipopolysaccharide complex or on the surface required for receptor dimerization. Nevertheless, both polymorphisms reduce responsiveness to lipopolysaccharides as shown in tissue culture systems with re-constituted TLR4 mutants [17]. Apart from bacterial lipopolysaccharides, TLR4 was shown to be activated by host high mobility group box 1 protein (HMGB-1) released as a result of chemo- or radiotherapy, and potential tissue damage [18]. In that study, on French breast cancer patients, the 299G form of TLR4 showed reduced binding to HMGB1 and a correlation with faster relapse after radiotherapy [18]. It is interesting to speculate whether HMGB1, released as a consequence of tissue necrosis or therapy, results in genotype dependent differential activation of TLR4, which contributes to the observed differences in survival. Further patient-based and functional studies, will be required to confirm the correlation between polymorphisms and survival. It is also worth noting that colon cancer patients carrying the minor allele of TLR4-rs4986790 exhibited worse progression free survival and OS after treatment with chemotherapy [19]. On the other hand, a comprehensive Swedish study on 20 TLR signaling pathway genes found significant association between two polymorphisms with prostate cancer mortality that did not include TLR4-rs4986790 [20]. Thus, even if modulated HMGB-1 signaling might be due to the effects of TLR4-rs4986790, the outcome in terms of OS is difficult to predict as the TLR ligation can have pleiotropic effects on cellular differentiation and growth.
With the exception of two polymorphisms, TLR2-rs3804099 and TLR4-rs2149356, none other of the 47 variants in eight TLR genes included in this study showed association with risk of melanoma in comparison to the healthy controls. The variant alleles of the two polymorphisms showed a tendency towards association with susceptibility but not strong enough to rule out a chance finding. Nevertheless, previous reports have shown associations of the rs3804099 polymorphism in the TLR2 gene with sepsis in preterm infants [21]. It was also associated with reversal reaction in leprosy and tuberculosis in Ethiopian and Vietnamese populations [22], [23]. On the other hand no association of this SNP could be found with normal tension glaucoma in a Japanese population or asthma in a German and a mixed German/Austrian population or type I diabetes mellitus in a Basque population [24], [25], [26], [27]. Since the polymorphism is synonymous (N199N) and not linked with any other variant in the gene, the association with various diseases could be due to linkage with other SNPs outside the TLR2 gene region. The most commonly discussed polymorphisms in TLR2, the R677W and R753Q, have been shown to be associated, amongst other diseases, with leprosy, tuberculosis, staphylococcal infections, coronary restenosis and sepsis [11], [12]. In our study the R677W could not be included due to immeasurable genotype, probably caused by a pseudogene located upstream of the true gene [28]. The R753Q variant was rather rare and did not show any association with melanoma susceptibility.
The variant allele of TLR4-rs2149356, exhibited association with decreased risk of melanoma that was not statistically significant. However, the haplotype containing the same variant allele was associated with statistically significant decreased risk. TLR4-rs2149356 is located in intron 2 of the gene and is linked with four other polymorphisms at the locus. One of which is located in intron 1 (rs1927911) and three (rs10116253, rs2737190, rs1927914) in the 5′end of the gene. Therefore it is possible that SNPs in the 5′ end could possibly effect transcriptional regulation. In previous studies variants in the TLR4 gene have been shown to be associated with infectious and non-infectious diseases but rather inconsistently [10], [29], [30], [31].
In conclusion, on the role of variants in eight TLR genes in malignant melanoma show an association of one variant in the TLR4 gene with the disease survival. Though we did not find any clear association for any polymorphism with the disease susceptibility, for two SNPs we did find a tendency for such an association that also extended into a frequent haplotype in TLR4 gene.
Materials and Methods
Patients and controls
763 patients of German origin (418 male, 345 female) with cutaneous melanoma were recruited at the Skin Cancer Unit at the Mannheim University Hospital, Germany, according to eligibility criteria that included histologically confirmed melanoma of the skin; AJCC stage 0 (in situ melanoma, 10), I (358), II (253), III (112) and IV (11) disease and 19 tumors had an unknown stage (Table 4). Median and mean age of the melanoma cases at diagnosis was 55 and 54.1 years, respectively. Mean Breslow thickness was 2.14 mm (95% confidence interval 1.97 to 2.31 mm).
Table 4. Characteristics of the German patients included in the study.
| All patients | Patients with AJCC stage 0, I or II at first diagnosis (FD) | |
| No. | 763 | 622 |
| Gender | ||
| Male | 418 (54.8%) | 341 (38.8%) |
| Female | 345 (45.2%) | 281 (45.2%) |
| Age at first diagnosis(FD) | ||
| Median | 55 | 54 |
| Mean | 54.1 | 54.9 |
| Breslow thickness (mm) | ||
| Mean (95% CI) | 2.14 (1.97–2.31) | 1.81 (1.66–1.96) |
| Ulceration of primary tumor | ||
| Yes | 130 (17.0)%) | 92 (14.8%) |
| No | 185 (24.3%) | 159 (25.6%) |
| unknown | 448 (58.7) | 371 (59.7%) |
| AJCC stage at FD | ||
| 0 | 10 (1.3%) | 10 (1.6%) |
| I | 396 (51.9%) | 396 (63.7%) |
| II | 216 (28.3%) | 216 (34.7%) |
| III | 111 (14.6%) | − |
| IV | 11 (1.4%) | − |
| unknown | 19 (2.5%) | − |
| Metastasized during follow-up | 237 (31.1%) | 237 (38.1%)%) |
| Deceased during follow-up | 170 (22.3%) | 127 (20.4%) |
| Therapy | ||
| No therapy | 408 (53.5%) | 364 (58.5%) |
| Any therapy | 353 (46.3%) | 258 (41.5%) |
| Chemotherapy | 28 (3.7%) | 17 (2.7%) |
| Immunotherapy | 129 (16.9) | 98 (15.8%) |
| All other therapy combinations | 199 (26.1%) | 143 (23.0%) |
Blood samples from case subjects were taken during the first appointment at the Skin Cancer Unit. Controls included 736 healthy individuals (368 male and 368 female) recruited at the Institute of Transfusion Medicine and Immunology in Mannheim, Germany. Median and mean age of the controls were 61 and 60.3 years, respectively. The ethical approval for the study was granted by Ethics Commission of the Faculty for Clinical Medicine of Ruprecht-Karls-Universität, Heidelberg and written informed consent was obtained from all study participants. DNA was extracted from blood samples using Qiagen mini-preparation kits.
Selection of SNPs
Selection of SNPs in eight human TLR genes was done by inclusion of known non-synonymous SNPs, those in regulatory regions or reported by other investigators. Additionally tagging SNPs for each gene region were selected from HapMap data using the criteria, a) exclusion of individuals with >50% missing genotypes, b) pair-wise r2>0.8 for each SNP pair and c) minor allele frequencies >5%. Of the eight TLR genes, three were located in a gene cluster on chromosome 4p14 (TLR6-TLR1-TLR10). For tagging analysis, the region from rs10024216 to rs6531668 (69.5 kb) was selected (Figure S1, online only). TLR2 and TLR3 are located on chromosome arm 4q; these two genes were about 32 Mb apart from each other and were not linked. TLR4 is on chromosome 9q23; TLR5 on 1q41 and TLR9 on 3p21. In total we selected 47 SNPs that were informative for 99 polymorphisms in 8 genes. The selected SNPs were grouped into 2 multiplex assays (W1 and W2) for the Sequenom mass array platform (Sequenom, San Diego, CA, USA, Table S2 and S3, online only). In addition, the TLR4-rs4986790 (D299G) SNP was selected for genotyping using an allelic discrimination method.
Genotyping
Genotyping was carried out with the Sequenom method. In two multiplex PCR (with the 25 or 23 primer pairs, respectively) DNA fragments with SNPs were amplified using 10 ng DNA as template in 5 µl volume. Genotype analysis was performed by the Sequenom TYPER 4.0 software. Validation of genotype data for SNPs that showed statistically significant association (TLR2-rs3804099 and TLR4-rs2149356) was done by allelic discrimination method and DNA sequencing.
Statistical analysis
All statistical calculations were carried out using SAS version 9.2 (SAS Institute, Cary, NC, USA). Odds ratios (OR) with the corresponding 95% confidence intervals (95% CI) for assessment of the association between risk and genotype were based on logistic regression adjusted for age, gender and Breslow thickness. The haplotype procedure of SAS genetics was used to estimate haplotype frequencies in cases and controls separately, and to infer possible haplotype combinations for each individual. The evidence of association between genotype/haplotype and melanoma risk were summarized by probability values.
Overall survival (OS) was defined as the time (in years) from date of first diagnosis until death or last patient contact. Metastasis free survival (MFS) was the time from diagnosis of primary melanoma until the first metastasis (either regional or distant). Survival following first metastasis (SFM) was defined as the time from first metastasis to death or last patient contact. Alive patients at the latest visit/contact were considered censored. Univariate survival curves were based on the Kaplan-Meier method and statistical significance was quantified by log-rank tests. Genotype-specific survival differences were adjusted for age, gender and Breslow thickness, based on a proportional hazard regression (Cox) model for each of the polymorphisms. Ulceration status was not included in the multivariate analysis due to unavailability of complete data. Subsequently, the analysis of the potential association between genotype and survival was adjusted for therapy administered in stage IV. We considered the start time and duration of therapy, treating therapy as a time-dependent covariate. The possible interaction between therapy and genotype was explored by including the interaction in the Cox regression and, additionally, by stratification of patients into therapy groups.
Association of haplotypes with survival time was carried out by Cox regression. Thereby, we assumed that individuals carry the most likely combination of possible haplotypes and that the effects of haplotypes interact multiplicatively. Hazard ratios (HR) were calculated taking the consensus haplotype as reference. If the frequency of the consensus haplotype was low, the most common haplotype was taken as reference.
Supporting Information
Linkage disequilibrium (LD) plot of the 69-kb TLR6-TLR1-TLR10 gene cluster on chromosome 4p14. The plot was drawn using Haploview software 4.2 and based on HapMap homepage. It shows r2−values, the higher r2, the darker the box. LD blocks are depicted as triangles. Positions of the TLR genes and SNPs are shown in the upper part of the figure.
(DOC)
Genotype distributions of TLR SNPs in malignant melanoma of the skin in patients and controls.
(DOC)
Toll-like receptor genes and polymorphisms.
(DOC)
List of iplex primer and masses.
(XLS)
Acknowledgments
We thank Sigrid Claus and Oliver Mücke for technical assistance.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by the Initiative and Networking Fund of the Helmholtz Association within the Helmholtz Alliance on Immunotherapy of Cancer. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Kalialis LV, Drzewiecki KT, Klyver H. Spontaneous regression of metastases from melanoma: review of the literature. Melanoma Res. 2009;19:275–282. doi: 10.1097/CMR.0b013e32832eabd5. [DOI] [PubMed] [Google Scholar]
- 2.Atkins MB, Mier JW, Parkinson DR, Gould JA, Berkman EM, et al. Hypothyroidism after treatment with interleukin-2 and lymphokine-activated killer cells. N Engl J Med. 1988;318:1557–1563. doi: 10.1056/NEJM198806163182401. [DOI] [PubMed] [Google Scholar]
- 3.Gogas H, Ioannovich J, Dafni U, Stavropoulou-Giokas C, Frangia K, et al. Prognostic significance of autoimmunity during treatment of melanoma with interferon. N Engl J Med. 2006;354:709–718. doi: 10.1056/NEJMoa053007. [DOI] [PubMed] [Google Scholar]
- 4.Phan GQ, Attia P, Steinberg SM, White DE, Rosenberg SA. Factors associated with response to high-dose interleukin-2 in patients with metastatic melanoma. J Clin Oncol. 2001;19:3477–3482. doi: 10.1200/JCO.2001.19.15.3477. [DOI] [PubMed] [Google Scholar]
- 5.Akira S, Takeda K, Kaisho T. Toll-like receptors: critical proteins linking innate and acquired immunity. Nat Immunol. 2001;2:675–680. doi: 10.1038/90609. [DOI] [PubMed] [Google Scholar]
- 6.Krieg AM, Vollmer J. Toll-like receptors 7, 8, and 9: linking innate immunity to autoimmunity. Immunol Rev. 2007;220:251–269. doi: 10.1111/j.1600-065X.2007.00572.x. [DOI] [PubMed] [Google Scholar]
- 7.Wang RF, Miyahara Y, Wang HY. Toll-like receptors and immune regulation: implications for cancer therapy. Oncogene. 2008;27:181–189. doi: 10.1038/sj.onc.1210906. [DOI] [PubMed] [Google Scholar]
- 8.Kumar H, Kawai T, Akira S. Toll-like receptors and innate immunity. Biochem Biophys Res Commun. 2009;388:621–625. doi: 10.1016/j.bbrc.2009.08.062. [DOI] [PubMed] [Google Scholar]
- 9.Pasare C, Medzhitov R. Toll-like receptors: linking innate and adaptive immunity. Adv Exp Med Biol. 2005;560:11–18. doi: 10.1007/0-387-24180-9_2. [DOI] [PubMed] [Google Scholar]
- 10.Misch EA, Hawn TR. Toll-like receptor polymorphisms and susceptibility to human disease. Clin Sci (Lond) 2008;114:347–360. doi: 10.1042/CS20070214. [DOI] [PubMed] [Google Scholar]
- 11.Schwartz DA, Cook DN. Polymorphisms of the Toll-like receptors and human disease. Clin Infect Dis. 2005;41(Suppl 7):S403–407. doi: 10.1086/431985. [DOI] [PubMed] [Google Scholar]
- 12.El-Omar EM, Ng MT, Hold GL. Polymorphisms in Toll-like receptor genes and risk of cancer. Oncogene. 2008;27:244–252. doi: 10.1038/sj.onc.1210912. [DOI] [PubMed] [Google Scholar]
- 13.Marshak-Rothstein A. Toll-like receptors in systemic autoimmune disease. Nat Rev Immunol. 2006;6:823–835. doi: 10.1038/nri1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eigentler TK, Caroli UM, Radny P, Garbe C. Palliative therapy of disseminated malignant melanoma: a systematic review of 41 randomised clinical trials. Lancet Oncol. 2003;4:748–759. doi: 10.1016/s1470-2045(03)01280-4. [DOI] [PubMed] [Google Scholar]
- 15.Agarwala SS. Current systemic therapy for metastatic melanoma. Expert Rev Anticancer Ther. 2009;9:587–595. doi: 10.1586/era.09.25. [DOI] [PubMed] [Google Scholar]
- 16.Park BS, Song DH, Kim HM, Choi BS, Lee H, et al. The structural basis of lipopolysaccharide recognition by the TLR4-MD-2 complex. Nature. 2009;458:1191–1195. doi: 10.1038/nature07830. [DOI] [PubMed] [Google Scholar]
- 17.Rallabhandi P, Bell J, Boukhvalova MS, Medvedev A, Lorenz E, et al. Analysis of TLR4 polymorphic variants: new insights into TLR4/MD-2/CD14 stoichiometry, structure, and signaling. J Immunol. 2006;177:322–332. doi: 10.4049/jimmunol.177.1.322. [DOI] [PubMed] [Google Scholar]
- 18.Apetoh L, Ghiringhelli F, Tesniere A, Criollo A, Ortiz C, et al. The interaction between HMGB1 and TLR4 dictates the outcome of anticancer chemotherapy and radiotherapy. Immunol Rev. 2007;220:47–59. doi: 10.1111/j.1600-065X.2007.00573.x. [DOI] [PubMed] [Google Scholar]
- 19.Tesniere A, Schlemmer F, Boige V, Kepp O, Martins I, et al. Immunogenic death of colon cancer cells treated with oxaliplatin. Oncogene. 2010;29:482–491. doi: 10.1038/onc.2009.356. [DOI] [PubMed] [Google Scholar]
- 20.Stark JR, Wiklund F, Gronberg H, Schumacher F, Sinnott JA, et al. Toll-like receptor signaling pathway variants and prostate cancer mortality. Cancer Epidemiol Biomarkers Prev. 2009;18:1859–1863. doi: 10.1158/1055-9965.EPI-08-0981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Abu-Maziad A, Schaa K, Bell EF, Dagle JM, Cooper M, et al. Role of Polymorphic Variants as Genetic Modulators of Infection in Neonatal Sepsis. Pediatr Res. 2010 doi: 10.1203/PDR.0b013e3181e6a068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bochud PY, Hawn TR, Siddiqui MR, Saunderson P, Britton S, et al. Toll-like receptor 2 (TLR2) polymorphisms are associated with reversal reaction in leprosy. J Infect Dis. 2008;197:253–261. doi: 10.1086/524688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thuong NT, Hawn TR, Thwaites GE, Chau TT, Lan NT, et al. A polymorphism in human TLR2 is associated with increased susceptibility to tuberculous meningitis. Genes Immun. 2007;8:422–428. doi: 10.1038/sj.gene.6364405. [DOI] [PubMed] [Google Scholar]
- 24.Nakamura J, Meguro A, Ota M, Nomura E, Nishide T, et al. Association of toll-like receptor 2 gene polymorphisms with normal tension glaucoma. Mol Vis. 2009;15:2905–2910. [PMC free article] [PubMed] [Google Scholar]
- 25.Kormann MS, Depner M, Hartl D, Klopp N, Illig T, et al. Toll-like receptor heterodimer variants protect from childhood asthma. J Allergy Clin Immunol. 2008;122:86–92, 92 e81–88. doi: 10.1016/j.jaci.2008.04.039. [DOI] [PubMed] [Google Scholar]
- 26.Eder W, Klimecki W, Yu L, von Mutius E, Riedler J, et al. Toll-like receptor 2 as a major gene for asthma in children of European farmers. J Allergy Clin Immunol. 2004;113:482–488. doi: 10.1016/j.jaci.2003.12.374. [DOI] [PubMed] [Google Scholar]
- 27.Santin I, Bilbao JR, de Nanclares GP, Calvo B, Castano L. No association of TLR2 and TLR4 polymorphisms with type I diabetes mellitus in the Basque population. Ann N Y Acad Sci. 2006;1079:268–272. doi: 10.1196/annals.1375.040. [DOI] [PubMed] [Google Scholar]
- 28.Malhotra D, Relhan V, Reddy BS, Bamezai R. TLR2 Arg677Trp polymorphism in leprosy: revisited. Hum Genet. 2005;116:413–415. doi: 10.1007/s00439-004-1249-9. [DOI] [PubMed] [Google Scholar]
- 29.Chen YC, Giovannucci E, Lazarus R, Kraft P, Ketkar S, et al. Sequence variants of Toll-like receptor 4 and susceptibility to prostate cancer. Cancer Res. 2005;65:11771–11778. doi: 10.1158/0008-5472.CAN-05-2078. [DOI] [PubMed] [Google Scholar]
- 30.Cheng I, Plummer SJ, Casey G, Witte JS. Toll-like receptor 4 genetic variation and advanced prostate cancer risk. Cancer Epidemiol Biomarkers Prev. 2007;16:352–355. doi: 10.1158/1055-9965.EPI-06-0429. [DOI] [PubMed] [Google Scholar]
- 31.Wang MH, Helzlsouer KJ, Smith MW, Hoffman-Bolton JA, Clipp SL, et al. Association of IL10 and other immune response- and obesity-related genes with prostate cancer in CLUE II. Prostate. 2009;69:874–885. doi: 10.1002/pros.20933. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Linkage disequilibrium (LD) plot of the 69-kb TLR6-TLR1-TLR10 gene cluster on chromosome 4p14. The plot was drawn using Haploview software 4.2 and based on HapMap homepage. It shows r2−values, the higher r2, the darker the box. LD blocks are depicted as triangles. Positions of the TLR genes and SNPs are shown in the upper part of the figure.
(DOC)
Genotype distributions of TLR SNPs in malignant melanoma of the skin in patients and controls.
(DOC)
Toll-like receptor genes and polymorphisms.
(DOC)
List of iplex primer and masses.
(XLS)