Abstract
Ssn6–Tup1 regulates multiple genes in yeast, providing a paradigm for corepressor functions. Tup1 interacts directly with histones H3 and H4, and mutation of these histones synergistically compromises Ssn6–Tup1-mediated repression. In vitro, Tup1 interacts preferentially with underacetylated isoforms of H3 and H4, suggesting that histone acetylation may modulate Tup1 functions in vivo. Here we report that histone hyperacetylation caused by combined mutations in genes encoding the histone deacetylases (HDACs) Rpd3, Hos1, and Hos2 abolishes Ssn6–Tup1 repression. Unlike HDAC mutations that do not affect repression, this combination of mutations causes concomitant hyperacetylation of both H3 and H4. Strikingly, two of these class I HDACs interact physically with Ssn6–Tup1. These findings suggest that Ssn6–Tup1 actively recruits deacetylase activities to deacetylate adjacent nucleosomes and promote Tup1–histone interactions.
Keywords: Chromatin, transcription, nucleosome, yeast, acetylation
The importance of chromatin organization to transcriptional regulation has become increasingly clear with the discovery of multiple ATP-dependent nucleosome remodeling activities (such as Swi/Snf), histone acetyltransferases (HATs), and histone deacetylases (HDACs; for review, see Kingston and Narlikar 1999). The HATs and HDACs in particular are now known to serve as important cofactors for a specific transcriptional activator (Brownell and Allis 1996) and repressor proteins (Laherty et al. 1997; Nagy et al. 1997; Kadosh and Struhl 1998; Luo et al. 1998). Despite the definition of these chromatin remodeling activities, little is yet known about how the opposing activities of HATs and HDACs are balanced to create open or closed chromatin structures at individual promoters. Although part of the answer likely lies in the direct recruitment of these activities, the average half-life of acetyl moieties in bulk chromatin is quite short, suggesting that additional factors may be required to stabilize individual histone acetylation patterns.
The yeast Tup1 repressor interacts directly with the amino-terminal tail domains of histones H3 and H4 that are subject to acetylation (Edmondson et al. 1996). Tup1 is part of a corepressor complex comprising three or four molecules of Tup1 and one molecule of Ssn6 (Varanasi et al. 1996; Redd et al. 1997). Ssn6–Tup1 does not bind to DNA directly but is apparently recruited to specific promoters by DNA binding proteins such as α2, Mig1, and Crt1 (Treitel and Carlson 1995; Wahi and Johnson 1995; Huang et al 1998). The Ssn6–Tup1 complex has been termed a global repressor because it is required for the repression of multiple families of genes (DeRisi et al 1997; Wahi et al. 1998). These include cell type specific genes as well as genes responsive to different physiological conditions.
The H3 and H4 tail domains are both necessary and sufficient for interaction with Tup1 in vitro (Edmondson et al. 1996, 1998). Moreover, histone mutations that compromise Tup1 binding also reduce repression of multiple Tup1-regulated reporter genes and the histone binding domain within Tup1 (Edmondson et al. 1996) overlaps the repression domain (Tzamarias and Struhl 1994). These findings indicate that Tup1-histone interactions are important to Ssn6–Tup1 repression in vivo and also indicate a functional redundancy for the H3 and H4 tails in the repression mechanism.
Genetic studies have identified a number of other factors required for Ssn6–Tup1 functions, including Sin4 (Jiang and Stillman 1992), Srb10, Srb11 (Wahi and Johnson 1995; Carlson 1997), Srb8 (Wahi et al. 1998), and Med3 (Papamichos-Chronakis et al. 2000). These proteins are all found in subcomplexes associated with the RNA polymerase II holoenzyme (Carlson 1997), suggesting that Ssn6–Tup1 may interact with specific transcription proteins to effect repression. A modest amount of repression (two- to fourfold) can be achieved in vitro in the presence of the basal transcription components alone (Herschbach et al. 1994; Redd et al. 1997). Thus, Ssn6–Tup1 may be multifunctional, interacting with basal transcription proteins to halt transcription and interacting with histones to maintain the repressed state.
Tup1 binds poorly to hyperacetylated H3 and H4 in vitro (Edmondson et al. 1996), predicting that increased histone acetylation should negatively influence repression in vivo. If so, then particular HDAC activities might be required for repression. Two major HDAC complexes have been isolated from yeast (HDA and HDB) that contain Hda1 or Rpd3, respectively, as catalytic subunits (Rundlett et al. 1996). Three other putative yeast HDAC genes have been identified, HOS1, HOS2, and HOS3 (Carmen et al. 1996; Rundlett et al. 1996). Of these, Hos3 has been confirmed to have HDAC activity in vitro (Carmen et al. 1999). Several mammalian HDACs are homologous to Rpd3 or Hda1, and the overall HDAC family can be categorized by size and sequence similarities into two subclasses (Grozinger et al. 1999). Class I enzymes include Rpd3, Hos1, Hos2, and the mammalian HDACs 1, 2, and 3. Class II enzymes are more similar to Hda1 and include HDACs 4, 5, and 6.
To test the hypothesis that particular HDAC activities are required for Ssn6–Tup1 repression, we examined repression in strains carrying disruptions of various HDAC genes. Mutation of class I HDACs results in a dramatic hyperacetylation of both H3 and H4. This hyperacetylation accompanies a substantial loss of Ssn6–Tup1-mediated repression. Strikingly, Ssn6–Tup1 interacts directly with at least two of these three HDACs, Rpd3 and Hos2, suggesting that targeting of these activities is an important component of the repression mechanism.
Results
Ssn6–Tup1-mediated repression is compromised in rpd3 hos1 hos2 mutant cells
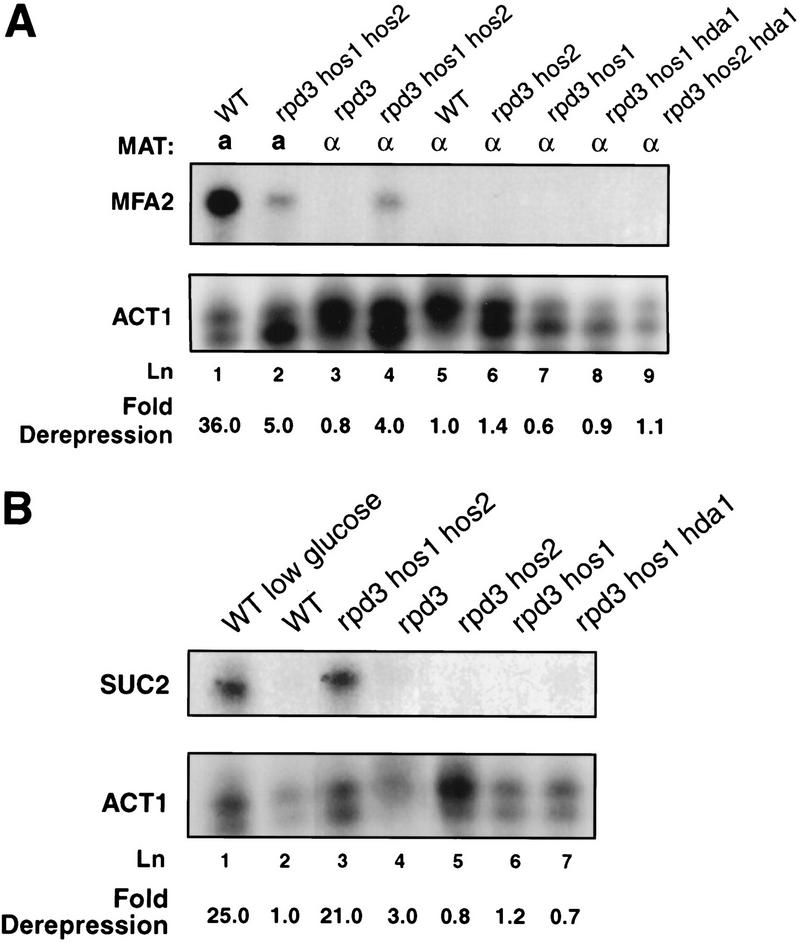
We examined the effects of multiple HDAC mutations on the expression levels of two Ssn6–Tup1 regulated genes, MFA2 and SUC2. MFA2 is an a cell type-specific gene. Repression of MFA2 in α cells is dependent on recruitment of Ssn6–Tup1 by α2/MCM1 (Wahi and Johnson 1995). As expected, MFA2 RNA was undetectable by nuclease protection assays of samples prepared from wild-type α cells (Fig. 1A, lane 5). Repression was maintained in rpd3 α cells as well as in rpd3 hos1 or rpd3 hos2 α cells (Fig. 1A, lanes 3,6,7). However, MFA2 repression was compromised fourfold in α cells carrying combined mutations in RPD3, HOS1, and HOS2 (Fig. 1A, lane 4), consistent with our previous observation of loss of repression of an a cell-specific reporter gene in these cells (Edmondson et al. 1998). In contrast to the loss of repression observed in the rpd3 hos1 hos2 cells, MFA2 repression was maintained in α cells bearing two other combinations of three mutant HDAC alleles, rpd3 hos1 hda1 or rpd3 hos2 hda1 (Fig. 1A, lanes 8,9).
Figure 1.
Ssn6–Tup1-mediated repression is abolished in rpd3 hos1 hos2 cells. (A) Endogenous MFA2 RNA levels in the indicated wild-type (WT) or mutant a and α strains were assayed by S1 nuclease protection. A representative gel and averages of MFA2 RNA levels normalized to ACT1 RNA levels from three independent experiments are shown. Fold derepression values reflect the normalized MFA2 signals relative to those observed in wild-type α cells. (B) Endogenous SUC2 mRNA levels were assayed by S1 nuclease protection and normalized to ACT1 RNA levels as in (A). Fold derepression values reflect the amount of SUC2 signal relative to that observed in wild-type cells under fully repressing conditions (lane 2). Values shown are averaged from three independent experiments.
Strikingly, the level of MFA2 expression detected in the rpd3 hos1 hos2 α cells is almost equivalent to that observed in a cells bearing these mutations (Fig. 1A, cf. lanes 2 and 4). These data indicate that RPD3, HOS1, and HOS2 are also important for activation of MFA2, as noted previously in rpd3 cells (Vidal and Gaber 1991). The similar levels of expression observed in the rpd3 hos1 hos2 a and α cells indicate that Ssn6–Tup1 repression is largely reversed in the α cells. Tup1 and Ssn6 protein levels are not affected by loss of these HDAC activities (Edmondson et al. 1998), and nuclease protection experiments indicate that expression levels of MCM1, SRB10, and SRB11 are unchanged in rpd3 hos1 hos2 cells (data not shown). Thus, loss of repression in these cells is not caused by decreased expression of these repressive factors.
To determine if loss of RPD3, HOS1, and HOS2 affects other genes regulated by Ssn6–Tup1, we examined repression of SUC2 in the mutant HDAC strains. SUC2 is expressed when cells are grown in media containing low levels of glucose and is repressed in high levels of glucose (Trumbly 1992; Carlson 1997). As was the case for MFA2, we found that SUC2 RNA levels are elevated in rpd3 hos1 hos2 cells under repressing (high-glucose) conditions (Fig. 1B, lane 3), reaching levels comparable to those observed in wild-type cells grown under derepressing conditions (Fig. 1B, lanes 1,3). Loss of repression is again specific for combined mutations in RPD3, HOS1, and HOS2 (Fig. 1B, lanes 4–7). Thus, at least two separate classes of genes regulated by Ssn6–Tup1 exhibit compromised repression in rpd3 hos1 hos2 cells, indicating that loss of these deacetylase activities affects a central aspect of Ssn6–Tup1 functions.
Increased histone acetylation at target promoters is associated with loss of Ssn6–Tup1 repression
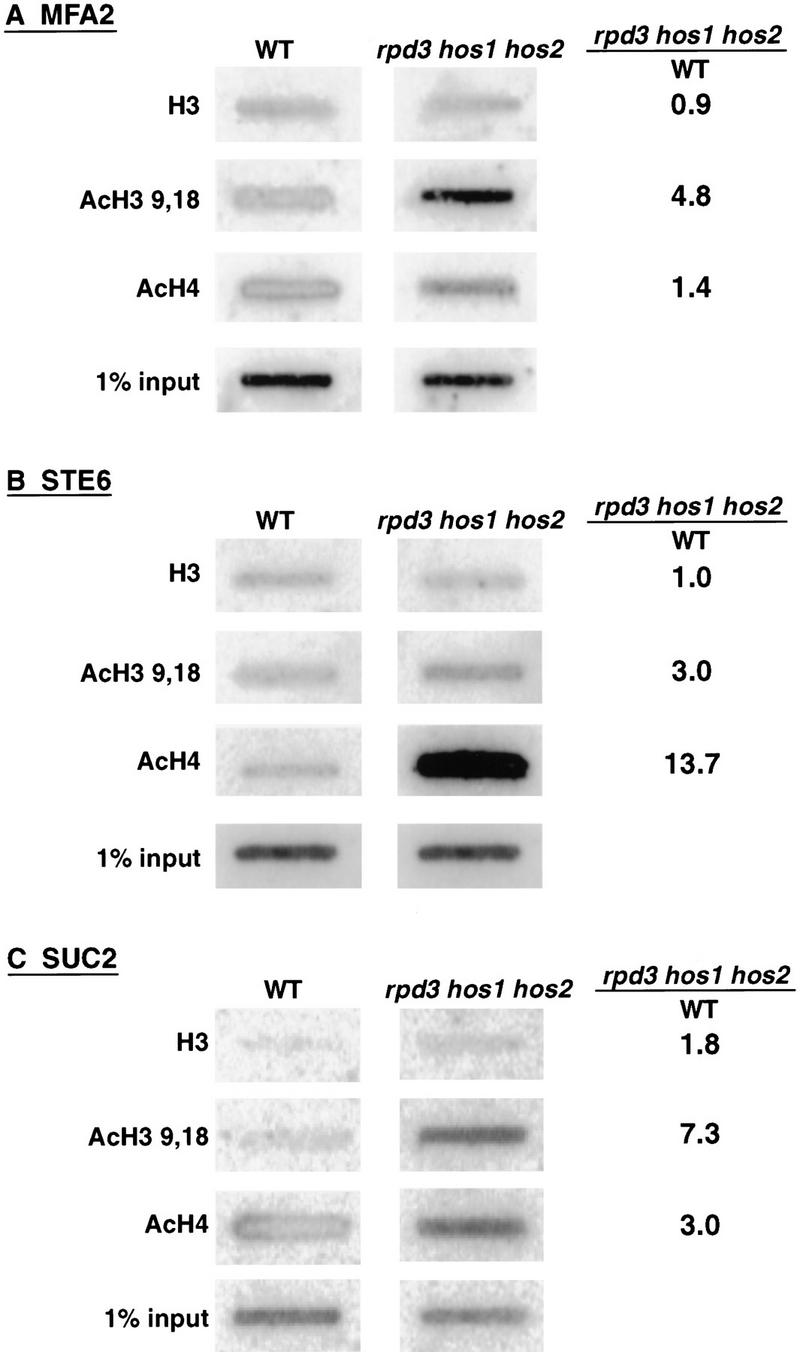
The above results suggest that Ssn6–Tup1 are not able to effect repression in the face of increased acetylation of histones associated with target promoters. To confirm that levels of histone acetylation are altered at the promoter regions of Ssn6–Tup1 regulated genes in the rpd3 hos1 hos2 cells, we immunoprecipitated chromatin fragments isolated from wild-type or HDAC mutant cells with antibodies specific for H3, for isoforms of H3 acetylated at lysines 9 and/or 18 (AcH3 9,18), or acetylated isoforms of H4 (at one or more of lysines 5, 8, 12, or 16; AcH4). DNA was isolated from the immunoprecipitates and probed for the presence of promoter sequences of the a cell-specific genes MFA2 and STE6 or for SUC2 promoter sequences. DNA precipitated by each of the antibodies was quantitated and normalized to the amount of chromatin subjected to immunoprecipitation (see “input” in Fig. 2).
Figure 2.
Acetylation of histones H3 and H4 is increased at Ssn6–Tup1 regulated promoters in rpd3 hos1 hos2 cells. Chromatin immunoprecipitations using antibodies specific for H3, AcH3 9,18, or AcH4 (as indicated) were carried out with chromatin extracted from wild-type or rpd3 hos1 hos2 cells. Immunoprecipitated DNA was applied to slot blots and probed for (A) MFA2- (B) STE6-, or (C) SUC2-specific promoter sequences. Signals were quantitated by PhosphorImage analysis and normalized to the amounts of promoter sequences detected in the chromatin input. The ratio of these normalized values is presented in the right-hand column.
We observed increased acetylation of H3 (4.8-fold) at the MFA2 promoter in the rpd3 hos1 hos2 α cells relative to wild-type α cells (Fig. 2A). We also saw a slight increase in H4 acetylation at this promoter (Fig. 2A). H3 acetylation was increased at the STE6 promoter in the mutant cells (threefold) as well, and an even greater increase in H4 acetylation (∼14-fold) occurred at this promoter (Fig. 2B). Changes in acetylation of both histones were also observed at SUC2, with a 7.3-fold increase in H3 acetylation and a threefold increase in H4 acetylation. Although the degree of change varied between the individual promoters examined, in each case, acetylation of H3 and H4 was increased on loss of RPD3, HOS1, and HOS2, corresponding to the loss of repression observed above.
Loss of histone deacetylase functions leads to hyperacetylation of histones in vivo
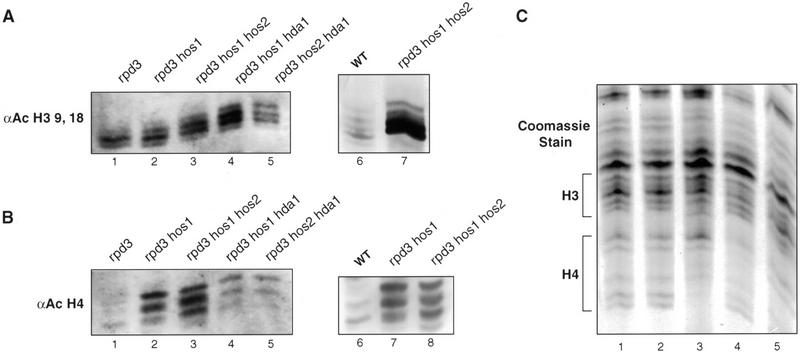
Our observation that Ssn6–Tup1 repression is disrupted only on combined loss of Rpd3, Hos1, and Hos2 suggests that these HDACs may share redundant substrate specificities that affect Ssn6–Tup1 functions. The fact that other combinations of three HDAC mutations do not affect Ssn6–Tup1 repression suggests that other mutations create changes in acetylation patterns different than those created by loss of Rpd3, Hos1, and Hos2, and that these changes are not disruptive to Ssn6–Tup1 functions. To further investigate the nature of changes caused by these mutations, we resolved histones isolated from strains carrying single, double, or triple HDAC mutations by acid urea electrophoresis, which separates differently modified histone isoforms (Allis et al. 1980; Krajewski and Luchnik 1991). The acetylation state of the isolated histones was examined further on immunoblots probed with antibodies specific for acetylated H3 or acetylated H4.
Progressive increases in H3 acetylation occur on disruption of increasing numbers of HDAC genes (Fig. 3A). For example, an enrichment of the more slowly migrating, highly acetylated H3 isoforms were observed in rpd3 hos1 cells than in rpd3 cells (Fig. 3A, cf. lanes 1 and 2), and an even greater enrichment of di-, tri-, and tetra-acetylated H3 was observed in the rpd3 hos1 hos2 cells (Fig. 3A, lane 3). Other triple combinations of deacetylase mutants (rpd3 hda1 hos1 and rpd3 hda1 hos2) caused a similar increase in the more highly acetylated isoforms of H3 (Fig. 3A, lanes 4,5).
Figure 3.
Bulk histone acetylation in wild-type or HDAC mutant cells. Yeast histones isolated from wild-type or various HDAC mutant strains (as indicated) were resolved by acid-urea gel electrophoresis. Immunoblots were performed using antibodies specific for acetylated isoforms of H3 (A) or H4 (B). Two separate blots are shown in each panel. Lanes 1–5 in A and B are from a single immunoblot of a sister to the gel shown in C, probed with the different antibodies. Equal amounts of protein were loaded into each lane, as confirmed by Coomassie blue staining (C).
In contrast to this progressive increase in H3 acetylation, only two mutant strains exhibited a marked increase in H4 acetylation relative to the others. Histones isolated from rpd3 hos1 and rpd3 hos1 hos2 cells reacted more strongly with the anti-AcH4 antibodies, and tri- and tetra-acetylated isoforms were more prevalent in these samples (Fig. 3B, lanes 2,3). Tri- and tetra-acetylated H4 isoforms are also evident in the other triple mutants, but to a lesser degree.
Of all the HDAC mutants we examined, the greatest combined change in the acetylation states of both H3 and H4 occurred in the rpd3 hos1 hos2 cells. The loss of Ssn6–Tup1 repression in these cells, but not that in other mutants, is consistent with our previous observations that these histones provide redundant functions in the repression mechanism and that high levels of acetylation are required to prevent Tup1 binding (Edmondson et al. 1996, 1998).
Ssn6–Tup1 interacts with Hos2 and Rpd3
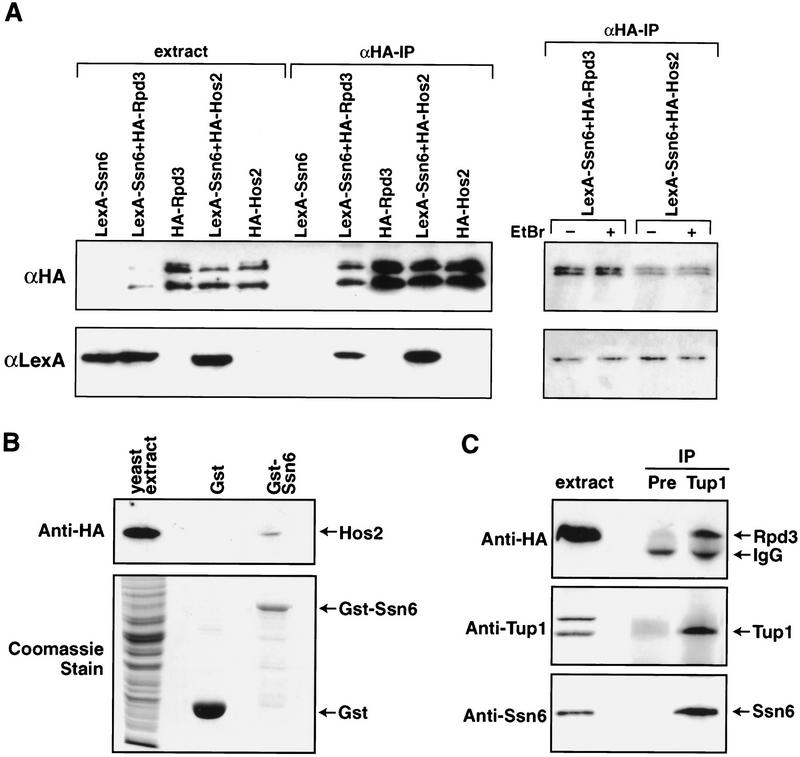
Rpd3 is recruited together with Sin3 to some target genes in yeast through association with DNA-bound repressors such as Ume6 (Rundlett et al. 1996; Kadosh and Struhl 1997). Similarly, mammalian homologs of Rpd3 serve as corepressors for unliganded nuclear hormone receptors, MAD/MAX heterodimers, and the Rb tumor suppressors (Laherty et al. 1997; Nagy et al. 1997; Luo et al. 1998). We, therefore, asked whether HDACs required for Ssn6–Tup1 repression interact directly with this corepressor complex. Using a two-hybrid assay, we saw a reproducible but weak signal indicating interaction between the TPR domain of Ssn6 (amino acids 1–398) and the HDACs Rpd3 and Hos2 (data not shown). We hypothesized that the weakness of this transcription-based assay might reflect the repressive properties of Ssn6, Rpd3, and Hos2. Thus, we examined interactions between the two hybrid fusion proteins directly by coimmunoprecipitation. HA-tagged fusion proteins were immunoprecipitated from whole-cell extracts using HA-specific antibodies. The immunoprecipitated proteins were then examined by immunoblot using lexA-specific antibodies (Fig. 4A). As expected, the HA-Rpd3 and HA-Hos2 proteins were immunoprecipitated with the HA-specific antibody (Fig. 4A, upper panel). LexA–Ssn6 coimmunoprecipitated with HA-Rpd3 and HA-Hos2 (Fig. 4A, lower panel) but not with the HA-Gal4 activation domain alone (data not shown). Reciprocal immunoprecipitations using LexA-specific antibodies also corroborated interaction between LexA–Ssn6 and the Rpd3 and Hos2 fusion proteins (data not shown). The interactions observed between LexA–Ssn6 and HA-Hos2 or HA-Rpd3 are not mediated by DNA, as these interactions are not affected by the addition of ethidium bromide to the immunoprecipitation (Fig. 4A, far right panel).
Figure 4.
Ssn6–Tup1 interacts with HDACs in vivo and in vitro. (A) Anti-HA immunoprecipitations were performed on cell extracts from yeast strains carrying the indicated expression plasmids and the presence of tagged fusion proteins determined by Western blot. Shown are cell extracts and immunoprecipitated fractions. The upper panel was probed with the anti-HA antibody and the lower panel with the anti-LexA antibody. LexA–Ssn6 coimmunoprecipitates with HA-Gal4–Rpd3 and HA-Gal4–Hos2. In the far right panel, the experiment was repeated in the presence of ethidium bromide (50 μg/mL) to demonstrate that the interactions observed are not mediated by DNA. (B) Purified recombinant GST–Ssn6 (amino acids 1–398) or GST alone was incubated with extracts from yeast expressing HA-tagged Hos2 from the native HOS2 locus. The top panel shows an anti-HA antibody immunoblot. HA-Hos2 interacts with GST–Ssn6, but not with GST alone. The lower panel shows a Coomassie blue stain of extract (30% of input) and the recombinant proteins. (C) Extracts from yeast expressing HA-tagged Rpd3 from the native RPD3 locus were immunoprecipitated with anti-Tup1 antibodies. The upper panel is an anti-HA immunoblot showing that HARpd3 coimmunoprecipitates with anti-Tup1 antisera (Tup1) but not with preimmune sera (PRE). The middle and lower panels show parallel blots probed with anti-Tup1 (middle) or anti-Ssn6 (lower) antisera.
To confirm interactions between the corepressor and Hos2, we purified a GST fusion protein containing a fragment of Ssn6 containing the TPR domain from E. coli and mixed this protein with whole-cell extracts prepared from yeast expressing an integrated, HA-tagged version of Hos2. HA-Hos2 bound to the GST–Ssn6 fusion protein (Fig. 4B) but did not bind to GST alone (Fig. 4B). These data confirm the above in vivo interaction.
Finally, we immunoprecipitated native Ssn6–Tup1 complexes from extracts of yeast expressing an integrated, HA-tagged Rpd3 using anti-Tup1 specific antisera. HA-Rpd3 and Ssn6 were easily detected in the anti-Tup1 immunoprecipitate (Fig. 4C) but not in the control immunoprecipitate.
Together, these experiments demonstrate that the Ssn6–Tup1 complex can interact with at least two different HDAC proteins, Rpd3 and Hos2. At present, we cannot distinguish whether Ssn6 or Tup1 (or both proteins) interact with these HDACs, as both the Gst–Ssn6 and lexA-Ssn6 fusions contain the Tup1 interaction domain and both Ssn6 and Tup1 are present in our immunoprecipitates. Nevertheless, these interactions provide a molecular explanation for our genetic experiments, which indicate that these HDAC activities are required for Ssn6–Tup1 functions.
Discussion
Our previous experiments demonstrated that the repression domain of Tup1 binds to the amino terminal tails of histones H3 and H4 in vitro. These histone domains are subject to multiple posttranslational modifications, and Tup1 binds preferentially to underacetylated isoforms of H3 and H4 (Edmondson et al. 1996). Other findings (J.R. Bone and S.Y. Roth, unpubl.) indicate that Ssn6–Tup1 regulated genes are normally associated with lesser-acetylated histones under conditions of repression. These data predict that increased histone acetylation should relieve Ssn6–Tup1 repression and that HDAC activities will be important for Ssn6–Tup1 functions in vivo. Consistent with this prediction, our current experiments indicate both that Ssn6–Tup1 functions are significantly compromised in cells deficient in particular HDAC activities and that at least two HDAC proteins interact with the corepressor.
Interestingly, we observe significant changes in Ssn6–Tup1-mediated repression only when we simultaneously disrupt RPD3, HOS1, and HOS2. Although we have not yet tested every possible combination of HDAC mutations, our results suggest some specificity in these effects, as two other triple mutant combinations did not compromise Ssn6–Tup1 repression. Our data indicate that the high levels of H3 and H4 acetylation that occur in the rpd3 hos1 hos2 cells antagonize Ssn6–Tup1 repression, which is consistent with our previous findings that high levels of acetylation are needed to prevent Tup1 binding and that H3 and H4 serve redundant functions in repression (Edmondson et al. 1996). Moreover, our data indicate that Rpd3, Hos1, and Hos2 have at least partially overlapping substrate specificities, possibly reflecting evolutionary conservation of class I HDAC functions. Rpd3 has been implicated in the regulation of several genes, and interactions between Rpd3 and several other proteins have been described (Kadosh and Struhl 1997, 1998). However, our studies are the first to identify gene targets for Hos1 and Hos2, and they establish a novel association of Hos2 with Ssn6–Tup1.
In vivo, histone acetylation states are in constant flux, reflecting the combined action of HATs and HDACs (for examples, see Waterborg 2000). Our data indicate that Ssn6–Tup1 directly alter this flux at specific promoters by recruiting one or more HDAC activities. In this way, Ssn6–Tup1 might be similar to the mammalian corepressors SMRT and NcoR, which contain multiple repressor domains, each associated with a different HDAC (Kao et al. 2000). However, we do not yet know whether Ssn6–Tup1 interacts directly or indirectly with Rpd3 and Hos2, as endogenous factors in our cell extracts might mediate the interactions we detect. In any case, decreased acetylation of H3 and H4 would facilitate chromatin folding and stabilize interactions between these histones and Tup1. Indeed, Tup1 has been shown to spread along the length of a repressed a cell-specific gene, indicating that the corepressor may serve an architectural function (Ducker and Simpson 2000). The interactions of Tup1 with the histone tails might also sterically limit reacetylation of H3 and H4, stabilizing the underacetylated, repressed state.
Interestingly, multiple proteins have been identified that share some structural and functional similarity with Tup1. A closely related Schizosaccharomyces pombe Tup1 homolog functions as a repressor and interacts with H3 and H4 (Mukai et al. 1999). Groucho, a transcriptional corepressor important to Drosophila development, has some sequence similarity to Tup1 and directly recruits Rpd3 for repression (Guoqing et al. 1999). The mammalian TLE proteins show sequence similarity to both Tup1 and Groucho (Palaparti et al. 1997). These proteins also act as repressors and interact with histones, as well as with a mammalian homolog of Ssn6 (Grbavec et al. 1999). Thus, the ability of Tup1-like corepressors to interact with and modulate chromatin structure is conserved across evolution, underscoring the importance of these functions to the regulation of gene expression.
Materials and methods
Yeast strains
All yeast strains except DY5329 and DY5330 are isogenic in the W303 strain background (Table 1). HDA1, HOS1, and HOS2 were disrupted in diploid strains using plasmids pB93TRP (hda1::TRP1), M3349 (hos1::HIS3), and M3354 (hos2::TRP1). Plasmid pB93TRP (Rundlett et al. 1996) was provided by M. Grunstein (UCLA), and details on construction of M3349 and M3354 are available on request. To construct strains DY5329 and DY5330, an in-frame 3 X HA epitope tag and a LEU2 marker were integrated into strain BJ5459 after the RPD3 and HOS2 genes, respectively, using plasmids pDM180 and pDM181 (from D. Moazed, Harvard Medical School, MA).
Table 1.
Yeast strains
| Strain
|
MAT
|
Genotype
|
|---|---|---|
| DY150 | a | ade2 can1 his3 leu2 trp1 ura3 |
| DY151 | α | ade2 can1 his3 leu2 trp1 ura3 |
| DY2395 | a | ade2 can1 his3 leu2 lys2 trp1 ura3 |
| DY2390 | α | ade2 can1 his3 leu2 lys2 trp1 ura3 |
| DY4548 | α | rpd3::LEU2 ade2 can1 his3 leu2 lys2 trp1 ura3 |
| DY4555 | α | rpd3::LEU2 hos2::TRP1 ade2 can1 his3 leu2 lys2 trp1 ura3 |
| DY4558 | α | rpd3::LEU2 hos1::HIS3 ade2 can1 his3 leu2 lys2 trp1 ura3 |
| DY4562 | α | hos1::HIS3 hos2::TRP1 ade2 can1 his3 leu2 lys2 trp1 ura3 |
| DY4565 | α | rpd3::LEU2 hos1::HIS3 hos2::TRP1 ade2 can1 his3 leu2 lys2 trp1 ura3 |
| DY5138 | α | rpd3::LEU2 hos1::HIS3 hda1::URA3 ade2 can1 his3 leu2 lys2 trp1 ura3 |
| DY5140 | α | rpd3::LEU2 hos2::TRP1 hda1::URA3 ade2 can1 his3 leu2 lys2 trp1 ura3 |
| DY5329 | a | RPD3::HA3 tag::LEU2 pep4::HIS3 prb1 can1 his3 leu2 lys2 trp1 ura3 |
| DY5330 | a | HOS2::HA3 tag::LEU2 pep4::HIS3 prb1 can1 his3 leu2 lys2 trp1 ura3 |
Plasmids
DNA fragments corresponding to RPD3, HOS2, SSN6 (amino acids 1–398), or TUP1 (amino acids 7–253) were generated by PCR. Oligonucleotide primer sequences are available on request. PCR fragments were cloned into pACTII (Clontech), pGEX-2T (Pharmacia), or pBTM116a. pBTM116a was made from pBTM116 (Bartel and Fields 1995) by inserting a BamHI linker into the SmaI site in the polylinker.
Yeast RNA isolation
Cells were grown to a density of 2 × 107 cells/mL. Cycloheximide was added to 50 mg/mL, and cells were cultured an additional 15 min, transferred to prechilled centrifuge bottles, placed on ice, and pelleted by centrifugation at 3000g for 5 min (4°C). RNA extraction was performed according to Rose et al. (1990).
Sl Nuclease Analysis
End-labeled oligonucleotides (0.2 pM) complementary to MFA2, SUC2, and ACT1 (sequences available on request) were hybridized with 75 μg of total RNA. Hybridizations and S1 nuclease digestions were performed as in Iyer and Struhl (1996) using 50 U of nuclease (GIBCO BRL).
Chromatin immunoprecipitations
Chromatin immunoprecipitations were done as described (Kuo and Allis 1999) with slight modifications. Lysates from 250 mL of cells at a density of 5 × 107 cells/mL were sonicated on ice 5 × 10 sec at 30% output, 90% duty cycle using a Heat System UltraSonicator fitted with a microtip. After clarification, 1 mL of extract was placed in an microfuge tube. CaCl2 was added to 10 mM. The extract was prewarmed to 37° C for 5 min and then digested with micrococcal nuclease (300 U/mL) for 5 min, followed by addition of EDTA to 25 mM. Extract corresponding to 3.5 × 108 cells was transferred to a new microfuge tube. Antibodies specific to H3 Ac9,18 (15 μL; Edmondson et al. 1996), unacetylated H3 (20 μL; Edmondson et al. 1996), or acetylated H4 (10 μL; ‘penta’ antibody form, C.D. Allis, University of Virginia) were added and volumes adjusted to 200 μL. Immunoprecipitations were conducted (rotating) at 4° C overnight. Then 20 μg of sonicated salmon sperm DNA and 60 μL of a 1:1 suspension of protein A sepharose beads (Pharmacia) were added. After rotation for 1 h at 4° C, beads were collected in a microcentrifuge. Antigens were eluted and cross-links were reversed as in Kuo and Allis (1999), except incubation at 65° C was extended to overnight. Slot blots were prehybridized in RapidHyb (Amersham) for 3 h and then hybridized with an α-dCTP32–dATP32 double-labeled probe at 55° C overnight. Probes correspond to 180–200-bp promoter fragments (sequences provided on request).
Histone purification and gels
Histones were isolated and analyzed as described in Edmondson et al. (1996).
Western blots
Proteins were transferred to PVDF membranes, which were blocked for 2 h in 1%–5% nonfat dry milk in TBST (10 mM Tris at pH 7.5, 150 mM NaCl, 0.1% Tween 20) and then incubated with primary antibody for 2 h at room temperature or overnight at 4° C. Primary antibodies used were antiacetylated H3 (Edmondson et al. 1996) 1:2000, antiacetylated H4 (Upstate Biotechnology) 1:1000, anti-HA (BAbCO) 1:1000, anti-LexA (Upstate) 1:10,000, anti-Ssn6 (this lab) 1:1000, and anti-Tup1 (this lab) 1:1000. Blots were incubated with HRP-conjugated secondary antibody (Pierce; 1:25,000 dilution) and developed with Super Signal (Pierce).
Coimmunoprecipitation and GST-pulldown assays
Log phase cultures (25 mL) were pelleted and resuspended in 1 mL of extraction buffer (125 mM NaCl, 25 mM Tris at pH 7.5, 15 mM EGTA, 15 mM MgCl2, 0.1% Tween 20, 5% glycerol, 1 mM PMSF, 1 μM leupeptin, 1 μM pepstatin). Cell extracts for anti-HA immunoprecipitation or for GST-pulldown assays were made by glass-bead breaking (5 min at 4° C). For anti-Tup1 immunoprecipitations, yeast were frozen in liquid nitrogen and broken with a mortar and pestle. All extracts were clarified by in a microcentrifuge. For immunoprecipitations, 5–10 μl of antibody was added and samples were rotated overnight at 4°C. Protein A-sepharose beads (25 μL of 1:1 slurry) were then added for an additional 2 h of rotation. Beads were recovered by low-speed centrifugation, washed four times in extraction buffer, and resuspended in SDS-PAGE loading buffer.
GST fusion proteins were purified as in Mukai et al. (1999). Yeast extracts were incubated with purified proteins bound to glutathione beads 2 h to overnight at 4°C. The beads were then washed four times in extraction buffer and resuspended in SDS-PAGE loading buffer.
Acknowledgments
We thank C.D. Allis for antibodies to acetylated H4 and D. Moazed and M. Grunstein for plasmids as indicated in Materials and Methods. This work was supported by grants from the NIH (GM51189) and the Robert A. Welch Foundation to S.Y.R., a grant from the NIH (GM39067) to D.J.S., and an ACS fellowship (PF4398) to J.R.B. DNA sequencing was carried out by the UTMDACC core.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL syr@mdacc.tmc.edu; FAX (713) 790-0329.
Article and publication are at www.genesdev.org/cgi/doi/10.1101/gad.829100.
References
- Allis CD, Glover CV, Bowen JK, Gorovsky MA. Histone variants specific to the transcriptionally active, amitotically dividing macronucleus of the unicellular eucaryote, Tetrahymena thermophila. Cell. 1980;20:609–617. doi: 10.1016/0092-8674(80)90307-4. [DOI] [PubMed] [Google Scholar]
- Bartel PL, Fields S. Analyzing protein–protein interactions using two-hybrid system. Methods Enzymol. 1995;254:241–263. doi: 10.1016/0076-6879(95)54018-0. [DOI] [PubMed] [Google Scholar]
- Brownell JE, Allis CD. Special HATs for special occasions: Linking histone acetylation to chromatin assembly and gene activation. Curr Opin Genet Dev. 1996;6:176–184. doi: 10.1016/s0959-437x(96)80048-7. [DOI] [PubMed] [Google Scholar]
- Carlson M. Genetics of transcriptional regulation in yeast: Connections to the RNA polymerase II CTD. Annu Rev Cell Biol. 1997;13:1–23. doi: 10.1146/annurev.cellbio.13.1.1. [DOI] [PubMed] [Google Scholar]
- Carmen AA, Rundlett SE, Grunstein M. HDA1 and HDA3 are components of a yeast histone deacetylase (HDA) complex. J Biol Chem. 1996;271:15837–15844. doi: 10.1074/jbc.271.26.15837. [DOI] [PubMed] [Google Scholar]
- Carmen AA, Griffin PR, Calaycay JR, Rundlett SE, Suka Y, Grunstein M. Yeast HOS3 forms a novel trichostatin A-insensitive homodimer with intrinsic histone deacetylase activity. Proc Natl Acad Sci. 1999;96:12356–12361. doi: 10.1073/pnas.96.22.12356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeRisi JL, Iyer VR, Brown PO. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science. 1997;278:680–686. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- Ducker CE, Simpson RT. The organized chromatin domain of the repressed yeast a cell-specific gene STE6 contains two molecules of the corepressor Tup1p per nucleosome. EMBO J. 2000;19:400–409. doi: 10.1093/emboj/19.3.400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmondson DG, Smith MM, Roth SY. Repression domain of the yeast global repressor Tup1 interacts directly with histones H3 and H4. Genes & Dev. 1996;10:1247–1259. doi: 10.1101/gad.10.10.1247. [DOI] [PubMed] [Google Scholar]
- Edmondson DG, Zhang W, Watson A, Xu W, Bone JR, Yu Y, Stillman D, Roth SY. In vivo functions of histone acetylation/deacetylation in Tup1p repression and Gcn5p activation. Cold Spring Harbor Symp Quant Biol. 1998;63:459–468. doi: 10.1101/sqb.1998.63.459. [DOI] [PubMed] [Google Scholar]
- Grbavec D, Lo R, Liu Y, Greenfield A, Stifani S. Groucho/transducin-like enhancer of split (TLE) family members interact with the yeast transcriptional co-repressor SSN6 and mammalian SSN6-related proteins: Implications for evolutionary conservation of transcription repression mechanisms. Biochem J. 1999;337:13–17. [PMC free article] [PubMed] [Google Scholar]
- Grozinger CM, Hassig CA, Schreiber SL. Three proteins define a class of human histone deacetylases related to yeast Hda1p. Proc Natl Acad Sci. 1999;96:4868–4873. doi: 10.1073/pnas.96.9.4868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guoqing C, Joseph F, Sheenah M, Albert C. A functional interaction between the histone deacetylase Rpd3 and the corepressor Groucho in Drosophila development. Genes & Dev. 1999;13:2218–2230. doi: 10.1101/gad.13.17.2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herschbach BM, Arnaud MB, Johnson AD. Transcriptional repression directed by the yeast α 2 protein in vitro. Nature. 1994;370:309–311. doi: 10.1038/370309a0. [DOI] [PubMed] [Google Scholar]
- Huang M, Zhou Z, Elledge SJ. The DNA replication and damage checkpoint pathways induce transcription by inhibition of the Crt1 repressor. Cell. 1998;94:595–605. doi: 10.1016/s0092-8674(00)81601-3. [DOI] [PubMed] [Google Scholar]
- Iyer V, Struhl K. Absolute mRNA levels and transcriptional initiation rates in Saccharomyces cerevisiae. Proc Natl Acad Sci. 1996;93:5208–5212. doi: 10.1073/pnas.93.11.5208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang YW, Stillman DJ. Involvement of the SIN4 global transcriptional regulator in the chromatin structure of Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:4503–4514. doi: 10.1128/mcb.12.10.4503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadosh D, Struhl K. Repression by Ume6 involves recruitment of a complex containing Sin3 corepressor and Rpd3 histone deacetylase to target promoters. Cell. 1997;89:365–371. doi: 10.1016/s0092-8674(00)80217-2. [DOI] [PubMed] [Google Scholar]
- ————— Targeted recruitment of the Sin3–Rpd3 histone deacetylase complex generates a highly localized domain of repressed chromatin in vivo. Mol Cell Biol. 1998;18:5121–5127. doi: 10.1128/mcb.18.9.5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao HY, Downes M, Ordentlich P, Evans RM. Isolation of a novel histone deacetylase reveals that class I and class II deacetylases promote SMRT-mediated repression. Genes & Dev. 2000;14:55–66. [PMC free article] [PubMed] [Google Scholar]
- Kingston RE, Narlikar GJ. ATP-dependent remodeling and acetylation as regulators of chromatin fluidity. Genes & Dev. 1999;13:2339–2352. doi: 10.1101/gad.13.18.2339. [DOI] [PubMed] [Google Scholar]
- Krajewski WA, Luchnik AN. Relationship of histone acetylation to DNA topology and transcription. Mol Gen Genet. 1991;230:442–448. doi: 10.1007/BF00280301. [DOI] [PubMed] [Google Scholar]
- Kuo MH, Allis CD. In vivo cross-linking and immunoprecipitation for studying dynamic protein: DNA associations in a chromatin environment. Methods. 1999;19:425–433. doi: 10.1006/meth.1999.0879. [DOI] [PubMed] [Google Scholar]
- Laherty CD, Yang WM, Sun JM, Davie JR, Seto E, Eisenman RN. Histone deacetylases associated with the mSin3 corepressor mediate mad transcriptional repression. Cell. 1997;89:349–356. doi: 10.1016/s0092-8674(00)80215-9. [DOI] [PubMed] [Google Scholar]
- Luo RX, Postigo AA, Dean DC. Rb interacts with histone deacetylase to repress transcription. Cell. 1998;92:463–473. doi: 10.1016/s0092-8674(00)80940-x. [DOI] [PubMed] [Google Scholar]
- Mukai Y, Matsuo E, Roth SY, Harashima S. Conservation of histone binding and transcriptional repressor functions in a Schizosaccharomyces pombe Tup1p homolog. Mol Cell Biol. 1999;19:8461–8468. doi: 10.1128/mcb.19.12.8461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy L, Kao HY, Chakravarti D, Lin RJ, Hassig CA, Ayer DE, Schreiber SL, Evans RM. Nuclear receptor repression mediated by a complex containing SMRT, mSin3A, and histone deacetylase. Cell. 1997;89:373–380. doi: 10.1016/s0092-8674(00)80218-4. [DOI] [PubMed] [Google Scholar]
- Palaparti A, Baratz A, Stifani S. The Groucho/transducin-like enhancer of split transcriptional repressors interact with the genetically defined amino-terminal silencing domain of histone H3. J Biol Chem. 1997;272:26604–26610. doi: 10.1074/jbc.272.42.26604. [DOI] [PubMed] [Google Scholar]
- Papamichos-Chronakis M, Conlan RS, Gounalaki N, Copf T, Tzamarias D. Hrs1/Med3 is a Cyc8–Tup1 corepressor target in the RNA polymerase II holoenzyme. J Biol Chem. 2000;275:8397–8403. doi: 10.1074/jbc.275.12.8397. [DOI] [PubMed] [Google Scholar]
- Redd MJ, Arnaud MB, Johnson AD. A complex composed of Tup1 and Ssn6 represses transcription in vitro. J Biol Chem. 1997;272:11193–11197. doi: 10.1074/jbc.272.17.11193. [DOI] [PubMed] [Google Scholar]
- Rose M, Winston F, Hieter P. Methods in yeast genetics: A laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1990. [Google Scholar]
- Rundlett SE, Carmen AA, Kobayashi R, Bavykin S, Turner BM, Grunstein M. HDA1 and RPD3 are members of distinct yeast histone deacetylase complexes that regulate silencing and transcription. Proc Natl Acad Sci. 1996;93:14503–14508. doi: 10.1073/pnas.93.25.14503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treitel MA, Carlson M. Repression by SSN6–TUP1 is directed by MIG1, a repressor/activator protein. Proc Natl Acad Sci. 1995;92:3132–3136. doi: 10.1073/pnas.92.8.3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trumbly RJ. Glucose repression in the yeast Saccharomyces cerevisiae. Mol Microbiol. 1992;6:15–21. doi: 10.1111/j.1365-2958.1992.tb00832.x. [DOI] [PubMed] [Google Scholar]
- Tzamarias D, Struhl K. Functional dissection of the yeast Cyc8–Tup1 transcriptional co-repressor complex. Nature. 1994;369:758–761. doi: 10.1038/369758a0. [DOI] [PubMed] [Google Scholar]
- Varanasi US, Klis M, Mikesell PB, Trumbly RJ. The Cyc8 (Ssn6)–Tup1 corepressor complex is composed of one Cyc8 and four Tup1 subunits. Mol Cell Biol. 1996;16:6707–6714. doi: 10.1128/mcb.16.12.6707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal M, Gaber RF. RPD3 encodes a second factor required to achieve maximum positive and negative transcriptional states in Saccharomyces cerevisiae. Mol Cell Biol. 1991;11:6317–6327. doi: 10.1128/mcb.11.12.6317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahi M, Johnson AD. Identification of genes required for alpha 2 repression in Saccharomyces cerevisiae. Genetics. 1995;140:79–90. doi: 10.1093/genetics/140.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahi M, Komachi K, Johnson AD. Gene regulation by the yeast Ssn6–Tup1 corepressor. Cold Spring Harbor Symp Quant Biol. 1998;63:447–457. doi: 10.1101/sqb.1998.63.447. [DOI] [PubMed] [Google Scholar]
- Waterborg JH. Steady-state levels of histone acetylation in Saccharomyces cerevisiae. J Biol Chem. 2000;275:13007–13011. doi: 10.1074/jbc.275.17.13007. [DOI] [PubMed] [Google Scholar]