Abstract
The adaptation of growth in response to nutritional changes is essential for the proper development of all organisms. Here we describe the identification of the Drosophila homolog of the target of rapamycin (TOR), a candidate effector for nutritional sensing. Genetic and biochemical analyses indicate that dTOR impinges on the insulin signaling pathway by autonomously affecting growth through modulating the activity of dS6K. However, in contrast to other components in the insulin signaling pathway, partial loss of dTOR function preferentially reduces growth of the endoreplicating tissues. These results are consistent with dTOR residing on a parallel amino acid sensing pathway.
Keywords: Growth, insulin, starvation, S6K
During the development of unicellular and multicellular organisms, growth is dependent on the integration of diverse intracellular signals, which are triggered by patterning and/or environmental cues (Oldham et al. 2000). The availability of nutrients strongly influences growth of single cells and multicellular organisms and in some cases specifies alternative developmental programs. For example, in yeast, nitrogen or carbon deprivation leads to withdrawal from vegetative growth, arrest in G0, and up-regulation of autophagy (Johnston et al. 1977; Ohsumi 1999). Likewise, in the nematode Caenorhabditis elegans, limiting nutrients or overcrowding elicits an alternative developmental program termed the dauer stage (Riddle 1997). In this stage, the animal remains sexually immature and stockpiles additional lipids for survival during unfavorable growth periods (Kimura et al. 1997). In more complex organisms such as insects and mammals, an alternative developmental program is lacking. Instead, nutrient limitation is managed by delaying development and, in severe cases, reducing the final body size of the organism. In the case of Drosophila, when nutrients become limiting, available resources are mobilized toward maintaining the growth of the mitotic tissues, which during metamorphosis form smaller, yet still fertile, flies (Robertson 1963).
In metazoans, one component of the complex physiological response of energy homeostasis and growth control is the insulin and IGF signaling system (Efstratiadis 1998; Patti and Kahn 1998). Insulin and IGF activate two main signaling pathways via the insulin receptor substrates (IRS1–4): the Ras/MAPK pathway, which is involved in proliferation, and the phosphatidyl-inositol 3-kinase (PI3K) signaling pathway, which is involved in cell growth, survival, and metabolic homeostasis (White 1998; Rameh and Cantley 1999). PI3K mediates its effects on downstream signaling components through the production of phosphatidylinositol 3,4,5-tris phosphate (PIP3), which acts to recruit pleckstrin homology (PH) domain–containing proteins like protein kinase B (PKB) (Chan et al. 1999; Meier and Hemmings 1999). The actions of activated PI3K are antagonized by the 3′-phospho-inositol specific lipid phosphatase encoded by the tumor suppressor gene PTEN (Maehama and Dixon 1999). An important protein implicated in the PI3K signaling pathway is the target of rapamycin (TOR). TOR has been reported to be regulated by PKB (Scott et al. 1998). Initially, TOR1 and TOR2 were identified in yeast as mutations that conferred resistance to the antiproliferative effects of rapamycin (Heitman et al. 1991). Rapamycin is an antibiotic that inhibits both yeast TOR and mammalian TOR (mTOR, also known as FRAP or RAFT) function by forming an inhibitory complex with the immunophilin, FK506 binding protein-12 (FKBP12), that binds to a region adjacent to the kinase domain termed the FKBP12-rapamycin binding domain (FRB; Brown et al. 1994; Sabatini et al. 1994; Thomas and Hall 1997; Cutler et al. 1999). TOR is most related to the ATM/DNA-PK family of checkpoint protein kinases and is more distantly related to the PI3K family (Thomas and Hall 1997; Cutler et al. 1999).
A key downstream target of mTOR function is protein synthesis. In part, mTOR positively mediates protein synthesis by modulating the activities of important translational components, including the translation initiation factor 4E binding proteins (4E-BP1-3) and the ribosomal protein S6 kinases (Chou and Blenis 1995; Lawrence and Abraham 1997; Dennis et al. 1999). Under conditions of reduced nutrients, such as amino acid limitation, mTOR negatively regulates protein synthesis and positively up-regulates autophagy (Dennis et al. 1999). Thus, mTOR may serve as a nutritional checkpoint for cell growth and ultimately, proliferation.
Little is known concerning the role of TOR during metazoan development. Here we (see also, Zhang et al. 2000, this issue) describe the genetic and biochemical characterization of the Drosophila homolog of TOR (dTOR). We find that the strong dTOR mutants have similar characteristics to upstream components in the dPI3K branch of the insulin pathway. Weak dTOR mutants, however, display a unique phenotype. Unlike larvae mutant for components of the insulin signaling pathway, imaginal disks of larvae with reduced dTOR function grow to their normal size at the apparent expense of the endoreplicating tissue.
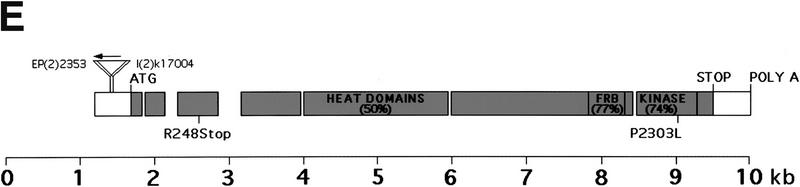
Results and Discussion
We recently established a tissue-specific genetic screen for recessive mutations affecting cell growth and proliferation in the Drosophila compound eye. In this screen, genetically mosaic flies are generated in which the eye and head capsule are homozygous for a randomly induced mutation, while the rest of the body and the germ line are heterozygous and, thus, phenotypically wild type. Remarkably, mosaic flies containing a mutation in a growth-promoting gene have eye and head structures that are strongly reduced in size relative to their wild-type sized heterozygous bodies and are termed pinhead flies (Fig. 1A). Two EMS-induced pinhead mutations (2L1 and 2L19; Fig. 1B,C) mapped to chromosomal position 34A, where the Drosophila homolog of TOR (dTOR) is located (Adams et al. 2000). These mutations fail to complement two lethal P-element insertions, EP(2)2353 and l(2)k17004 (Törok et al. 1993; Rørth et al. 1998; Spradling et al. 1999), located 262 and 211 bp, respectively, upstream of the putative translation start site of dTOR (Fig. 1E). The dTOR genomic region spans ∼10 kb and is composed of seven exons (7.4 kb cDNA) encoding a 2480-amino acid protein with a predicted Mw of 282 kD (Fig. 1E). The dTOR protein exhibits 35%, 38%, 35%, and 41% overall amino acid identity to yeast TOR1, TOR2, C. elegans TOR, and Arabidopsis thaliana TOR, respectively. The overall identity with mTOR is significantly higher (56%) and is especially conserved in the kinase and FRB domains (74% and 77%). Sequence analysis of DNA from flies heterozygous for the EMS-induced dTOR mutations revealed two nucleotide substitutions. The lesion of dTOR2L1 results in a change of a proline to a leucine at amino acid position 2303 (P2303L; Fig. 1E). The location of this mutation within a highly conserved region of the kinase domain implies that the kinase activity of dTOR is critical for its function, as has been shown for the yeast TORs (Zheng et al. 1995). In contrast, the lesion in dTOR2L19 is an arginine changed to a nonsense mutation at amino acid residue 248 (R248Stop), giving rise to a stop codon. This mutation would be predicted to result in a short, truncated protein and should thus be a complete loss-of-function mutation.
Figure 1.
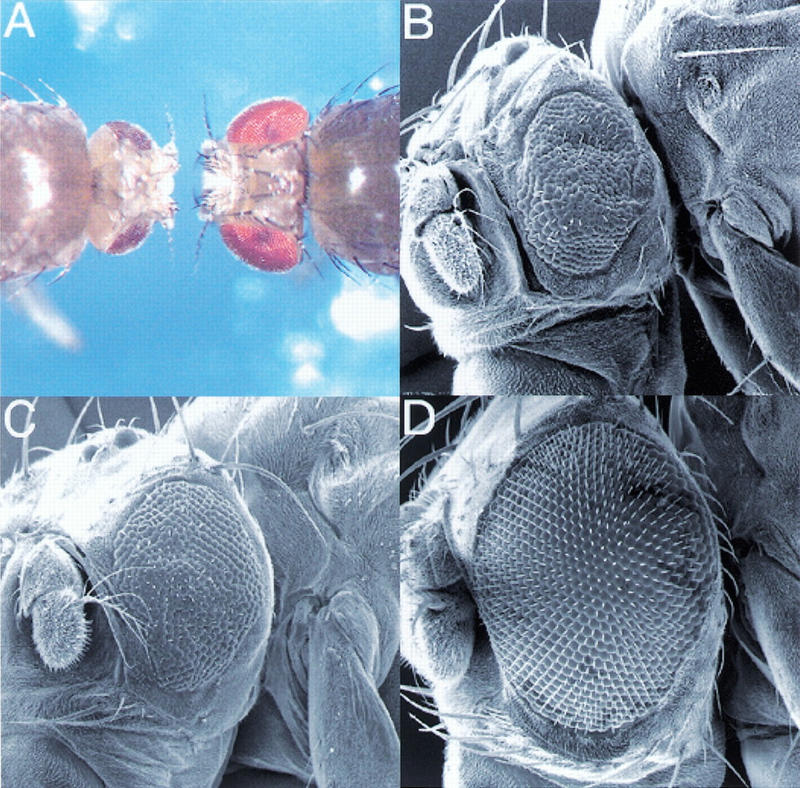
Identification of dTOR. Tissue-specific induction of dTOR2L1 and dTOR2L19 mutant clones using the ey-Flp technique (Newsome et al. 2000) produced flies with reduced head size relative to their body. (A) Dorsal view of y w ey-Flp; P(w+) l(2)2L-3.1 FRT40/CyO y+ (right) and y w ey-Flp; dTOR2L1 FRT40/P(w+) l(2)2L-3.1 FRT40 (left). Flies of the following genotype were examined by SEM: (B) y w ey-Flp; dTOR2L19 FRT40/P(w+) l(2)2L-3.1 FRT40; (C) y w ey-Flp; dTOR2L1 FRT40/P(w+) l(2)2L-3.1 FRT40; (D) wild-type eye. Bar, 100 μm. (E) Genomic structure and mutants of dTOR. The grey region consists of the coding region, and the gaps indicate introns. The percentage amino acid identity of each domain to human TOR is shown in parentheses.
The phenotypes associated with the complete loss of dTOR function are remarkably similar to phenotypes associated with mutations in the insulin receptor (Inr) pathway. First, strong dTOR mutants arrest development at a similar stage as do strong mutants in the Inr pathway or amino acid–starved larvae with little detectable imaginal tissue (see below). Second, dTOR mutant clones have a significant proliferative disadvantage similar to Inr pathway mutant clones (data not shown). Clones of dTOR null mutant cells, although severely affected, are not cell lethal. Similarly, in most mammalian cell types, rapamycin decreases but does not abolish cell growth, except for IL2-mediated T-cell proliferation (Chou and Blenis 1995). Third, the strict autonomous control of cell growth without disturbing the specification and differentiation is also seen with the dTOR mutants. Indeed, loss of dTOR function in clones of homozygous mutants cells in the adult eye showed that only the mutant cells, as exemplified by the dark, circular rhabdomeres, are severely reduced in size (Fig. 2C). Analysis of imaginal wing disk cells at the end of the third larval instar by fluorescence-activated cell sorting (FACS), confirmed that cells from the weak heteroallelic combination, dTOR2L1/dTORl(2)k17004, are smaller than those of wild type (Fig. 2B). The effect on cell size is more pronounced in G1 than G2 (data not shown), consistent with mTOR function having a predominate role on cell growth during G1 (Terada et al. 1993; Barbet et al. 1996). Despite this observation, there is no apparent difference between the distribution of dTOR mutant and wild-type cells within each phase of the cell cycle (Fig. 2A). Although the similarities of the loss-of-function mutant phenotypes of dTOR and other components of the Inr pathway are consistent with a model in which dTOR acts downstream of dPI3K, the analysis of partial loss-of-function dTOR mutants and the biochemical analysis of dS6K activity (see below) indicates a more complex relationship between dTOR and the Inr pathway.
Figure 2.
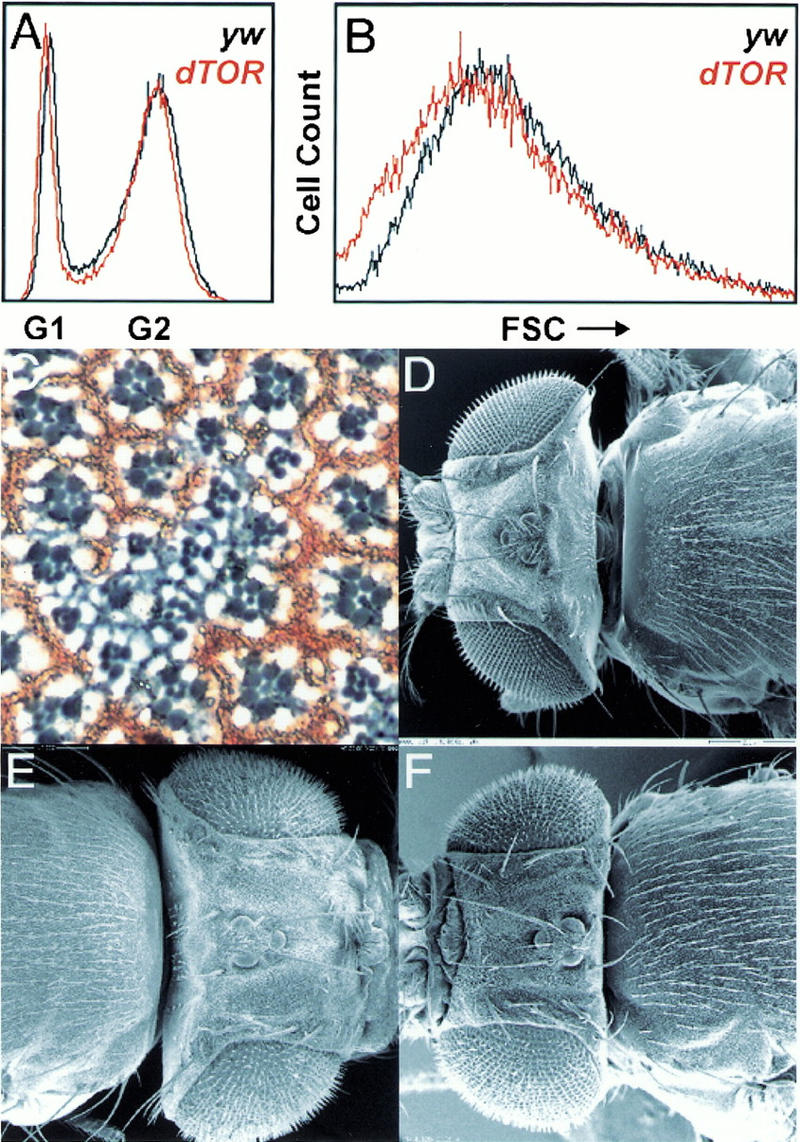
dTOR acts autonomously in the control of cell growth in the eye. (A,B) FACS analysis of dTOR2L1/dTORl(2)k17004 mutant wing disks showing forward scatter (FSC) and cell cycle distribution. (C) Section through an eye containing a mitotic clone of dTORl(2)k17004. The area of the mutant clone is demarcated by the absence of red pigment. At the boundaries of the clone, ommatidia consist of mixed wild-type heterozygous and homozygous mutant cells. (D–F) Flies were examined by SEM. (D) y w control. (E). Loss of dPTEN function in the eye and head capsule results in a fly with a big head. (F) The large head caused by loss of dPTEN function in the eye can be suppressed by homozygosity for dTOREP(2)2353. Bar, 200 μm.
We examined genetic interactions between dTOR mutants and other mutations in the insulin pathway. dPTEN encodes a negative effector of insulin signaling (Goberdhan et al. 1999; Huang et al. 1999), and eyes and heads lacking dPTEN function are significantly larger than wild-type heads (Fig. 2D,E; Gao et al. 2000). Removal of dTOR function strongly reduced the size of dPTEN mutant heads (Fig. 2F), suggesting that dTOR is required for the increased growth generated by the loss of dPTEN function.
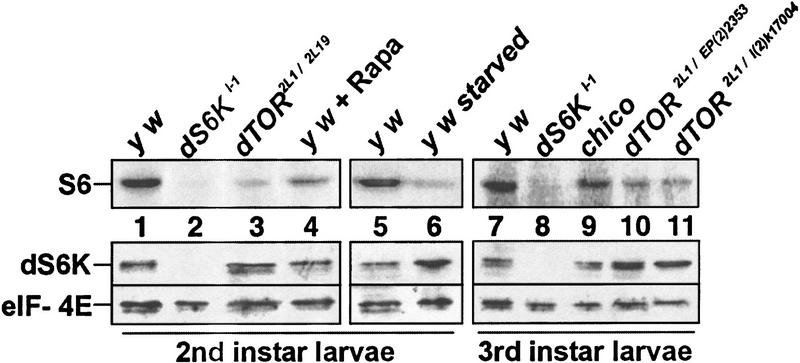
In vertebrates, S6K activity is blocked by rapamycin, an inhibitor of TOR. Therefore, we analyzed dS6K activity in immunoprecipitates of extracts from larvae mutant for dTOR, dS6K, chico, and larvae treated with rapamycin or deprived of amino acids. A severe reduction in the phosphorylation of ribosomal protein S6 was observed in extracts from strong dTOR2L1/dTOR2L19 mutant larvae (Fig. 3, cf. lanes 1 and 3). This was not caused by a reduction in dS6K protein as shown by Western blotting of these extracts (Fig. 3). In addition, we consistently noted that the dS6K protein was up-regulated in the dTOR mutant larvae and amino acid–starved larvae. In all cases, Western blot analysis showed equivalent amounts of initiation factor 4E (eIF-4E; Fig. 3). dS6K activity was not detected in dS6Kl-1 null mutants and was severely reduced when wild-type larvae were starved for amino acids or treated with rapamycin (Fig. 3, lanes 2,4,6). Higher doses of rapamycin blocked development during early larval stages, leading to lethality (data not shown). Analysis of the weak dTOR2L1/dTORl(2)k17004 or dTOR2L1/dTOREP(2)2353 heteroallelic combinations also revealed a reduction in dS6K activity in the third larval instar and an up-regulation of the protein as compared with wild-type flies (Fig. 3, cf. lane 7 with lanes 10 and 11). The surprising fact that dTOR mutants and amino acid starvation result in an up-regulation of dS6K levels suggests that dTOR and amino acids may negatively control the protein levels of dS6K. Unexpectedly, dS6K activity as well as protein levels were unaffected in chico mutants (Fig. 3, lane 9). It may be that Inr does not signal to dS6K or that dS6K resides on a parallel pathway that bifurcates upstream of Chico. In support of the latter possibility, Inr has been shown to genetically interact with dPI3K independently of Chico (Boehni et al. 1999), presumably through docking sites for the p60 adaptor of dPI3K in the Inr C-terminal tail. This result suggests that there is a dS6K independent pathway for growth control and that the reduced Inr-mediated dPI3K signaling in a chico mutant is sufficient for dS6K activation.
Figure 3.
dTOR mutations and amino acid starvation affect the level and activity of dS6K. In vitro kinase assay to assess the levels of dS6K activity (top) and Western blot of the protein levels of dS6K and Drosophila eIF-4E (bottom).
The biochemical differences between the ability of Chico and dTOR to activate dS6K argue for a more complex relationship between the Inr pathway and dTOR. Given the low number of pharate adults, we compared the weight of dTOR, dS6Kl-1, and chico mutants at an early pupal stage (Fig. 4A). The weight of the dTOR mutant pupae is more similar to dS6K than to chico mutant pupae (Fig. 4B). Thus, in the absence of dS6K function or the presence of reduced dTOR levels, cellular growth rates are diminished but larvae pupariate at a larger size as a result of a longer developmental delay. Importantly, dS6K mutant flies have cells that are smaller but of the normal number (Montagne et al. 1999). However, in chico mutants, pupariation is initiated at a much smaller size. The result is that chico mutants emerge after only a 2-d delay and are smaller than dTOR and dS6K mutants because of fewer and smaller cells (Boehni et al. 1999; Montagne et al. 1999; data not shown). Therefore, while insulin signaling controls cell size and cell number, dS6K primarily controls cell size. It will be of interest to know whether dTOR is also limited to controlling only cell size.
Figure 4.
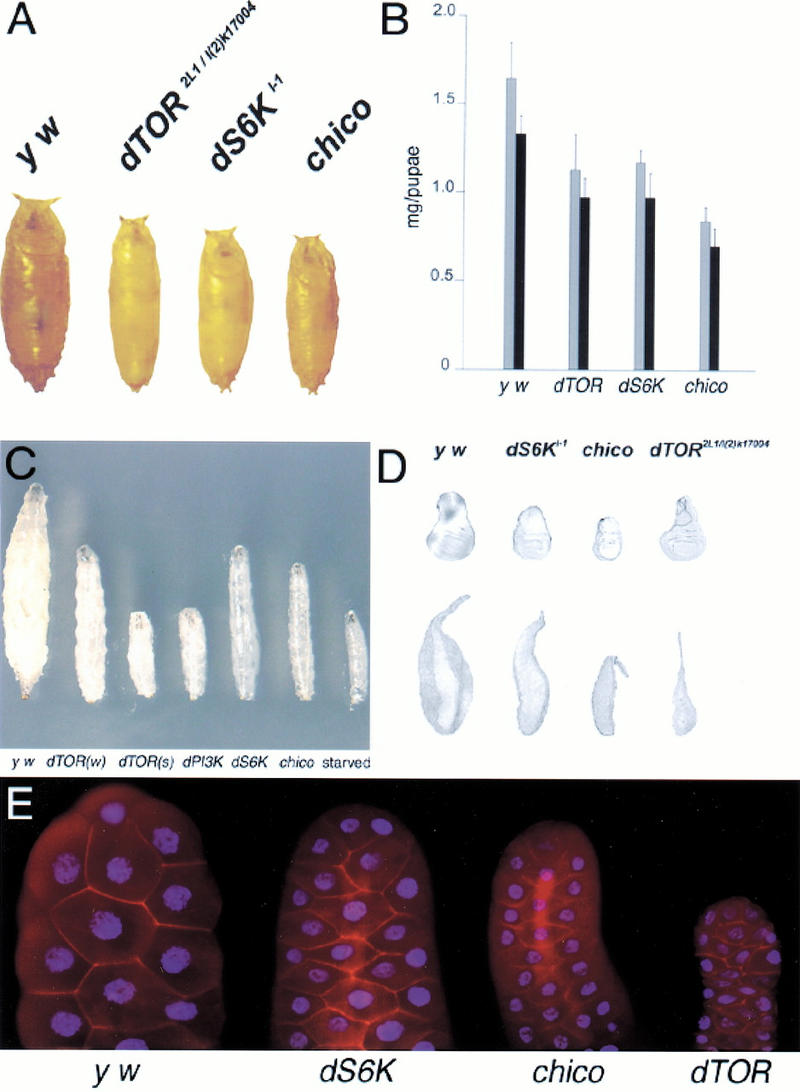
dTOR larval and pupal mutant phenotypes. (A) Pupal sizes of the various mutant combinations. Pupae were collected just after puparium formation. (B) Pupal weights of the various mutant combinations. Gray bars are female, black bars are male. (C) Heteroallelic combinations of dTOR mutant larvae compared with mutants of other components in the Inr pathway. All larvae were taken from a staged collection at day 5. dTOR(w) is dTOR2L1/dTORl(2)k17004, dTOR(s) is dTOR2L1/dTOR2L19, dPI3K is a null mutation (Weinkove et al. 1999), and dS6K is dS6Kl-1. (D) The size of mutant disks and salivary glands of hypomorphic dTOR mutants are distinct from dS6K and chico. (E) DAPI and phalloidin staining of salivary glands from y w, dS6Kl-1, chico, and dTOR2L1/dTORl(2)k17004 mutant larvae. All disks and salivary glands were taken from larvae that had everted their spiracles just before pupariation.
Larvae are composed of mitotic cells, largely represented by the imaginal disks, and of endoreplicating tissues, which form larval structures like the gut, fat body, and salivary glands. An increase in DNA ploidy of larval cells is required for the ∼200-fold increase in mass obtained by the larvae during the 5-d period between the completion of embryogenesis and the beginning of pupation (Church and Robertson 1966). During starvation, larvae sacrifice their endoreplicating tissue to maintain the growth and proliferation of the mitotic cells that are required to form the reproductive adult (Britton and Edgar 1998). Furthermore, dS6K activity is reduced in starved larvae and dTOR mutants (Fig. 3). These observations prompted us to analyze the mitotic and endoreplicating tissues of dTOR, dS6K, and chico mutant larvae just before pupariation. Strong dTOR and dPI3K mutants, as well as amino acid–starved larvae, are incapable of growth and have barely detectable imaginal and endoreplicative tissues (Fig. 4C; Weinkove et al. 1999; data not shown). Surprisingly, the wing disks of the weak dTOR heteroallelic combination were of approximately equivalent size to that of wild-type larvae, whereas those of dS6Kl-1 mutants were reduced (Fig. 4D). However, the amount of endoreplicating tissue in the dTOR mutant as compared to wild-type larvae was severely decreased. This is clearly demonstrated by comparing the salivary glands of dTOR mutant and wild-type larvae (Fig. 4D). In contrast, the size of endoreplicating tissue and imaginal disks in dS6K null mutants as well as chico null mutants was reduced in size to approximately the same extent (Fig. 4D). Staining of the salivary glands with DAPI and phalloidin revealed that the size of the nuclei and, thus, the degree of endoreplication was severely reduced in dS6K, chico, and dTOR mutants (Fig. 4E). The difference in size between dTOR and dS6K mutant salivary glands is largely caused by a very pronounced reduction in cytoplasmic volume in dTOR mutants. The nuclear to cytoplasmic ratio was higher in dTOR salivary glands than in y w, dS6K, or chico mutant salivary glands (4.5, 2.25, and 2.1 times, respectively). Thus, it appears that partial loss of dTOR function permits the growth of imaginal tissue to wild-type size, while endoreplicating tissue is disproportionally reduced, a phenotype distinct from dS6K mutants. Consistent with this finding, we were unable to rescue the lethality of the different dTOR mutants by constitutive expression of a S6K1 variant, D3E-E389, which exhibits high basal activity in the absence of mitogens (Dennis et al. 1996) under the control of the α-tubulin promoter, which rescues all aspects of the dS6Kl-1 null phenotype (Montagne et al. 1999; data not shown). Therefore, dS6K-independent processes must contribute to the weak dTOR phenotype.
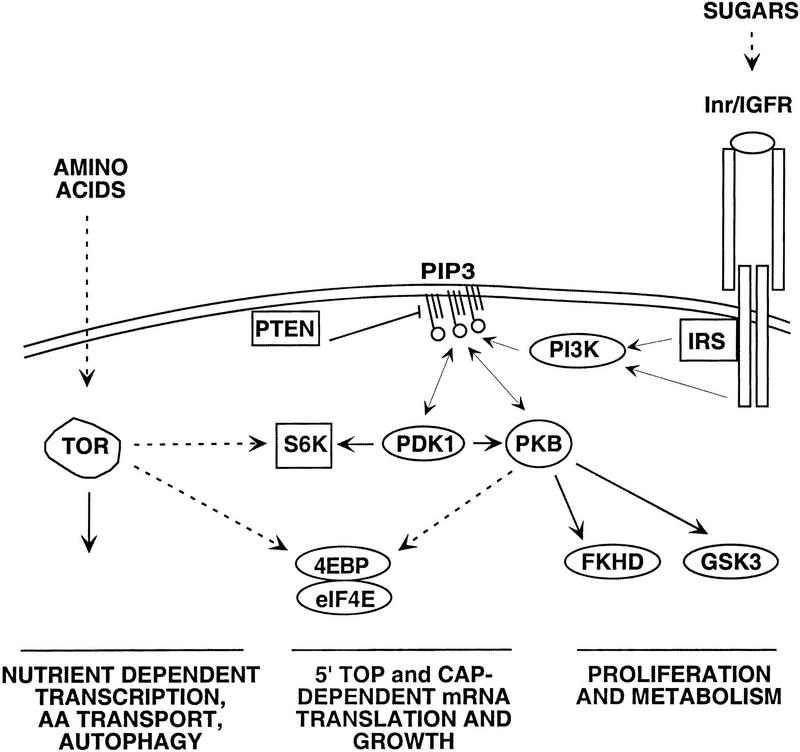
The effect of rapamycin and amino acids on translation is mediated through the S6Ks and the 4E-BPs (Dennis et al. 1999). Unlike the other elements in the PI3K signaling pathway, absence of amino acids blocks both S6K activation and 4E-BP phosphorylation (Blommaart et al. 1995; Hara et al. 1998). Indeed, a mutant of S6K1, lacking a portion of both its amino and carboxyl termini, is resistant to rapamycin but still sensitive to the fungal metabolite wortmannin, an inhibitor of PI3K. This suggests that the PI3K-dependent signal to S6K activation does not involve TOR. This same mutant is also unaffected by amino acid withdrawal, consistent with the role of mTOR as an amino acid checkpoint in S6K activation (Hara et al. 1998; Dennis et al. 1999). Although there is some controversy concerning the ability of mitogens to activate mTOR, we find, as have others (Gingras et al. 1999; cited in Hara et al. 1998), that the in vitro activity of mTOR from cultured cells toward either itself, S6K1, or 4E-BP1 is unaffected by mitogens (P.B. Dennis and G. Thomas, unpubl.). Thus, mTOR may act as a permissive signal that primes 4E-BP phosphorylation and S6K activation by the PI3K signaling pathway if amino acids, and possibly other nutrients, are at sufficient levels (Hara et al. 1998; Gingras et al. 1999). Likewise, in Drosophila larvae, amino acids are necessary, but not sufficient, for imaginal disk and endoreplicating tissue proliferation (Britton and Edgar 1998), compatible with dTOR acting in a parallel pathway involved in amino acid sensing (Fig. 5). The fact that chico mutant larvae have normal levels of dS6K activity and that the dTOR larval phenotypes with respect to the imaginal disks and endoreplicating tissues are so distinct compared with other mutants in the Inr pathway, supports the possibility that dTOR is not responsive to insulin signaling.
Figure 5.
Model showing the possible relationship of dTOR with the insulin pathway. The unexpected finding that hypomorphic mutations of dTOR, but not null mutations of chico, strongly inhibit dS6K activity may be explained by a two-threshold model. The threshold for dS6K inactivation by dTOR is low, whereas that for dS6K inactivation by the Inr pathway is high. Thus, a low level of PI3K signaling from Inr may be sufficient for dS6K activation in the absence of Chico. Dashed arrow indicates indirect or unclear interaction. Solid arrow indicates direct interaction.
It is well established in yeast that TOR is an important mediator of nutrient limitation, and it has been proposed that TOR acts as an amino acid effector to coordinate the response of yeast to different nutritional conditions (Barbet et al. 1996). Indeed, the similarities between dTOR mutant larvae and larvae deprived of amino acids are striking. Therefore, it is likely that dTOR also functions as an amino acid sensor in multicellular organisms. The fact that yeast and Arabidopsis do not have an insulin system suggests that TOR may be an ancestral and widespread nutritional sensor. To provide additional levels of control, it may have been integrated into the insulin system later to respond to different modes of nutrient deprivation with different developmental responses.
Materials and methods
Molecular characterization of dTOR
We used the ey-FLP method (Newsome et al. 2000) for a tissue-specific F1 screen for recessive mutations affecting cell growth and proliferation. Two EMS-induced alleles (dTOR2L1 and dTOR2L19) that produce flies with small heads relative to their heterozygous, phenotypically wild-type sized bodies were identified and failed to complement two P-element insertions in the 5′ region of dTOR. The genomic structure was predicted by GENSCAN and GADFLY (GENSCAN, http://genes.mit.edu/GENSCAN.html; BDGP, http://www.fruitfly.org).
Clonal and larval analysis
Clonal analysis was done as described (Boehni et al. 1999). The following genotypes were used to compare mosaic clones of dPTEN versus dPTEN–dTOR mutants: y w ey-Flp; P(w+) l(2)2L-3.1 FRT40/FRT40 dPTEN117 and y w ey-Flp; P(w+) l(2)2L-3.1 FRT40/FRT40 dPTEN117, dTOREP(2)2353. For imaginal disk/salivary gland and FACS analysis, late third instar larvae just before pupariation were examined as previously described (Montagne et al. 1999). Imaginal disks and salivary glands were dissected and fixed in paraformaldehyde as described (Montagne et al. 1999). DAPI and phalloidin staining were performed following standard procedure and analyzed by confocal imaging. The nuclear and cytoplasmic volumes were derived from nuclear radial and internuclei measurements, respectively, from 10 independent salivary glands. These measurements were converted to volumes using the formula V = 4/3πr3. The cytoplasmic volume was determined by subtracting the nuclear volume from the total cell volume.
S6K assay
Amino acid–starved larvae were obtained as described (Britton and Edgar 1998). For the rapamycin (Calbiochem) feeding, 24 h after egg laying, 100 μL of a 40 μM rapamycin solution was added once a day for up to 6 d. Five days AED, rapamycin-fed larvae were developmentally delayed at the second instar. Larvae were extensively washed, frozen in liquid nitrogen, and squashed in extraction buffer (Stewart et al. 1996). Cell debris was removed by centrifugation and for each dS6K activity assay, 40 μg of total protein extract was immunoprecipitated using 10 μL of a monoclonal antibody generated by NOVARTIS, using a fusion protein between maltose binding protein (New England Biolabs) and the carboxy-terminal amino acids 435–490 of dS6K. After incubation with 20 μL of protein-G Sepharose (Amersham Pharmacia), immuno complexes were sedimented, extensively washed, and assayed for [32P] Pi incorporation as previously described (Stewart et al. 1996). Western blotting was performed with polyclonal antibodies directed against the amino terminus of dS6K (Stewart et al. 1996) or the Drosophila eIF4E (provided by J.M. Sierra, Centro de Biologia Molecular Severo Ochoa).
Acknowledgments
We thank B. Froesch, P. Gallant, K. Nairz, L. MacDougall, H. Stocker, and S. Graham for critical reading of the manuscript; T. Gutjahr for SEM; M. Hall and T. Schmelzle for helpful suggestions; J. Mestan for generating the monoclonal dS6K antibody and T. Hayden for FACS analysis; and D. Agee, F. Evans, S. Messenger, S. Anderson, K. Schaubmayer, and F. Oldham for encouragement. S.O. and T.R. are the recipients of long-term fellowships from the Human Frontiers Science Program and the Roche Research Foundation. This work was supported in part by a grant from the Swiss Cancer League to G.T. and E.H.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL hafen@zool.unizh.ch; FAX 41-1-635-68-20.
Article and publication are at www.genesdev.org/cgi/doi/10.1101/gad.845700.
References
- Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, Amanatides PG, Scherer SE, Li PW, Hoskins RA, Galle RF, et al. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- Barbet NC, Schneider U, Helliwell SB, Stansfield I, Tuite MF, Hall MN. TOR controls translation initiation and early G1 progression in yeast. Mol Biol Cell. 1996;7:25–42. doi: 10.1091/mbc.7.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blommaart EF, Luiken JJ, Blommaart PJ, van Woerkom GM, Meijer AJ. Phosphorylation of ribosomal protein S6 is inhibitory for autophagy in isolated rat hepatocytes. J Biol Chem. 1995;270:2320–2326. doi: 10.1074/jbc.270.5.2320. [DOI] [PubMed] [Google Scholar]
- Boehni R, Riesgo-Escovar J, Oldham S, Brogiolo W, Stocker H, Andruss BF, Beckingham K, Hafen E. Autonomous control of cell and organ size by CHICO, a Drosophila homolog of vertebrate IRS1–4. Cell. 1999;97:865–875. doi: 10.1016/s0092-8674(00)80799-0. [DOI] [PubMed] [Google Scholar]
- Britton JS, Edgar BA. Environmental control of the cell cycle in Drosophila: Nutrition activates mitotic and endoreplicative cells by distinct mechanisms. Development. 1998;125:2149–2158. doi: 10.1242/dev.125.11.2149. [DOI] [PubMed] [Google Scholar]
- Brown EJ, Albers MW, Shin TB, Ichikawa K, Keith CT, Lane WS, Schreiber SL. A mammalian protein targeted by G1-arresting rapamycin-receptor complex. Nature. 1994;369:756–758. doi: 10.1038/369756a0. [DOI] [PubMed] [Google Scholar]
- Chan TO, Rittenhouse SE, Tsichlis PN. AKT/PKB and other D3 phosphoinositide-regulated kinases: Kinase activation by phosphoinositide-dependent phosphorylation. Annu Rev Biochem. 1999;68:965–1014. doi: 10.1146/annurev.biochem.68.1.965. [DOI] [PubMed] [Google Scholar]
- Chou MM, Blenis J. The 70 kDa S6 kinase: Regulation of a kinase with multiple roles in mitogenic signaling. Curr Opin Cell Biol. 1995;7:806–814. doi: 10.1016/0955-0674(95)80064-6. [DOI] [PubMed] [Google Scholar]
- Church RB, Robertson FW. Biochemical analysis of genetic differences in the growth of Drosophila. Genet Res. 1966;7:383–407. doi: 10.1017/s0016672300009836. [DOI] [PubMed] [Google Scholar]
- Cutler NS, Heitman J, Cardenas ME. TOR kinase homologs function in a signal transduction pathway that is conserved from yeast to mammals. Mol Cell Endocrinol. 1999;155:135–142. doi: 10.1016/s0303-7207(99)00121-5. [DOI] [PubMed] [Google Scholar]
- Dennis PB, Pullen N, Kozma SC, Thomas G. The principal rapamycin-sensitive p70s6k phosphorylation sites T229 and T389 are differentially regulated by rapamycin-insensitive kinase-kinases. Mol Cell Biol. 1996;16:6242–6251. doi: 10.1128/mcb.16.11.6242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis PB, Fumagalli S, Thomas G. Target of rapamycin (TOR): Balancing the opposing forces of protein synthesis and degradation. Curr Opin Genet Dev. 1999;9:49–54. doi: 10.1016/s0959-437x(99)80007-0. [DOI] [PubMed] [Google Scholar]
- Efstratiadis A. Genetics of mouse growth. Int J Dev Biol. 1998;42:955–976. [PubMed] [Google Scholar]
- Gao X, Neufeld TP, Pan D. Drosophila PTEN regulates cell growth and proliferation through PI3K-dependent and -independent pathways. Dev Biol. 2000;221:404–418. doi: 10.1006/dbio.2000.9680. [DOI] [PubMed] [Google Scholar]
- Gingras AC, Gygi SP, Raught B, Polakiewicz RD, Abraham RT, Hoekstra MF, Aebersold R, Sonenberg N. Regulation of 4E-BP1 phosphorylation: A novel two-step mechanism. Genes & Dev. 1999;13:1422–1437. doi: 10.1101/gad.13.11.1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goberdhan DC, Paricio N, Goodman EC, Mlodzik M, Wilson C. Drosophila tumor suppressor PTEN controls cell size and number by antagonizing the Chico/PI3-kinase signaling pathway. Genes & Dev. 1999;13:3244–3258. doi: 10.1101/gad.13.24.3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara K, Yonezawa K, Weng QP, Kozlowski MT, Belham C, Avruch J. Amino acid sufficiency and mTOR regulate p70 S6 kinase and eIF-4E BP1 through a common effector mechanism. J Biol Chem. 1998;273:14484–14494. doi: 10.1074/jbc.273.23.14484. [DOI] [PubMed] [Google Scholar]
- Heitman J, Movva NR, Hall MN. Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science. 1991;253:905–909. doi: 10.1126/science.1715094. [DOI] [PubMed] [Google Scholar]
- Huang H, Potter CJ, Tao W, Li DM, Brogiolo W, Hafen E, Sun H, Xu T. PTEN affects cell size, cell proliferation and apoptosis during Drosophila eye development. Development. 1999;126:5365–5372. doi: 10.1242/dev.126.23.5365. [DOI] [PubMed] [Google Scholar]
- Johnston GC, Pringle JR, Hartwell LH. Coordination of growth with cell division in the yeast Saccharomyces cerevisiae. Exp Cell Res. 1977;105:79–98. doi: 10.1016/0014-4827(77)90154-9. [DOI] [PubMed] [Google Scholar]
- Kimura KD, Tissenbaum HA, Liu Y, Ruvkun G. daf-2, an insulin receptor–like gene that regulates longevity and diapause in Caenorhabditis elegans. Science. 1997;277:942–946. doi: 10.1126/science.277.5328.942. [DOI] [PubMed] [Google Scholar]
- Lawrence JC, Jr, Abraham RT. PHAS/4E-BPs as regulators of mRNA translation and cell proliferation. Trends Biochem Sci. 1997;22:345–349. doi: 10.1016/s0968-0004(97)01101-8. [DOI] [PubMed] [Google Scholar]
- Maehama T, Dixon JE. PTEN: A tumour suppressor that functions as a phospholipid phosphatase. Trends Cell Biol. 1999;9:125–128. doi: 10.1016/s0962-8924(99)01519-6. [DOI] [PubMed] [Google Scholar]
- Meier R, Hemmings BA. Regulation of protein kinase B. J Recept Signal Transduct Res. 1999;19:121–128. doi: 10.3109/10799899909036639. [DOI] [PubMed] [Google Scholar]
- Montagne J, Stewart MJ, Stocker H, Hafen E, Kozma SC, Thomas G. Drosophila S6 kinase: A regulator of cell size. Science. 1999;285:2126–2129. doi: 10.1126/science.285.5436.2126. [DOI] [PubMed] [Google Scholar]
- Newsome TP, Asling B, Dickson BJ. Analysis of Drosophila photoreceptor axon guidance in eye-specific mosaics. Development. 2000;127:851–860. doi: 10.1242/dev.127.4.851. [DOI] [PubMed] [Google Scholar]
- Ohsumi Y. Molecular mechanism of autophagy in yeast, Saccharomyces cerevisiae. Phil Trans R Soc Lond B. 1999;354:1577–1581. doi: 10.1098/rstb.1999.0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldham S, Boehni R, Stocker H, Brogiolo W, Hafen E. Genetic control of size in Drosophila. Phil Trans R Soc Lond B. 2000;355:945–952. doi: 10.1098/rstb.2000.0630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patti ME, Kahn CR. The insulin receptor—A critical link in glucose homeostasis and insulin action. J Basic Clin Physiol Pharmacol. 1998;9:89–109. doi: 10.1515/jbcpp.1998.9.2-4.89. [DOI] [PubMed] [Google Scholar]
- Rameh LE, Cantley LC. The role of phosphoinositide 3-kinase lipid products in cell function. J Biol Chem. 1999;274:8347–8350. doi: 10.1074/jbc.274.13.8347. [DOI] [PubMed] [Google Scholar]
- Riddle DL. Genetic and environmental regulation of dauer larva development. In: Riddle DL, Blumenthal T, Meyer B, Priess J, editors. C. elegans. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1997. pp. 739–768. [PubMed] [Google Scholar]
- Robertson FW. The ecological genetics of growth in Drosophila. VI. The genetic correlation between the duration of the larval period and body size in relation to the larval diet. Genet Res. 1963;4:74–92. [Google Scholar]
- Rørth P, Szabo K, Bailey A, Laverty T, Rehm J, Rubin GM, Weigmann K, Milan M, Benes V, Ansorge W, et al. Systematic gain-of-function genetics in Drosophila. Development. 1998;125:1049–1057. doi: 10.1242/dev.125.6.1049. [DOI] [PubMed] [Google Scholar]
- Sabatini DM, Erdjument-Bromage H, Lui M, Tempst P, Snyder SH. RAFT1: A mammalian protein that binds to FKBP12 in a rapamycin-dependent fashion and is homologous to yeast TORs. Cell. 1994;78:35–43. doi: 10.1016/0092-8674(94)90570-3. [DOI] [PubMed] [Google Scholar]
- Scott PH, Brunn GJ, Kohn AD, Roth RA, Lawrence JC., Jr Evidence of insulin-stimulated phosphorylation and activation of the mammalian target of rapamycin mediated by a protein kinase B signaling pathway. Proc Natl Acad Sci. 1998;95:7772–7777. doi: 10.1073/pnas.95.13.7772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart MJ, Berry CO, Zilberman F, Thomas G, Kozma SC. The Drosophila p70s6k homolog exhibits conserved regulatory elements and rapamycin sensitivity. Proc Natl Acad Sci. 1996;93:10791–10796. doi: 10.1073/pnas.93.20.10791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terada N, Franklin RA, Lucas JJ, Blenis J, Gelfand EW. Failure of rapamycin to block proliferation once resting cells have entered the cell cycle despite inactivation of p70 S6 kinase. J Biol Chem. 1993;268:12062–12068. [PubMed] [Google Scholar]
- Thomas G, Hall MN. TOR signaling and control of cell growth. Curr Opin Cell Biol. 1997;9:782–787. doi: 10.1016/s0955-0674(97)80078-6. [DOI] [PubMed] [Google Scholar]
- Törok T, Tick G, Alvarado M, Kiss I. P-lacW insertional mutagenesis on the second chromosome of Drosophila melanogaster: Isolation of lethals with different overgrowth phenotypes. Genetics. 1993;135:71–80. doi: 10.1093/genetics/135.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinkove D, Neufeld TP, Twardzik T, Waterfield MD, Leevers SJ. Regulation of imaginal disc cell size, cell number and organ size by Drosophila class I(A) phosphoinositide 3-kinase and its adaptor. Curr Biol. 1999;9:1019–1029. doi: 10.1016/s0960-9822(99)80450-3. [DOI] [PubMed] [Google Scholar]
- White MF. The IRS-signaling system: A network of docking proteins that mediate insulin and cytokine action. Recent Prog Horm Res. 1998;53:119–138. [PubMed] [Google Scholar]
- Zheng XF, Florentino D, Chen J, Crabtree GR, Schreiber SL. TOR kinase domains are required for two distinct functions, only one of which is inhibited by rapamycin. Cell. 1995;82:121–130. doi: 10.1016/0092-8674(95)90058-6. [DOI] [PubMed] [Google Scholar]