Abstract
NEDD8 is a ubiquitin (Ub)-like protein. Here we report a novel ubiquitinylation-related pathway for modification by NEDD8. NEDD8 was activated by an E1 (Ub-activating enzyme)-like complex, consisting of APP–BP1 and hUba3 with high respective homologies to the amino- and carboxy-terminal regions of E1 and then linked to hUbc12 (a human homolog of yeast Ub-conjugating enzyme Ubc12p). The major target protein modified by NEDD8 was found to be Hs–cullin-4A (Cul-4A), a member of the family of human cullin/Cdc53 proteins functioning as an essential component of a multifunctional Ub–protein ligase E3 complex that has a critical role in Ub-mediated proteolysis.
Keywords: Ubiquitin, ubiquitin-like protein, NEDD8, SUMO-1, cullin-4A, Cdc53
Ubiquitin (Ub), an 8.6-kD highly conserved protein, is covalently attached to target proteins by a multienzymatic system consisting of E1 (Ub-activating), E2 (Ub-conjugating), and E3 (Ub-ligating) enzymes to form the degradation signal for proteolytic attack by proteasomes (for review, see Hershko and Ciechanover 1992; Coux et al. 1996; Hochstrasser 1997; Varshavsky 1997). Moreover, various Ub-like proteins are present universally in eukaryotes (for review, see Johnson and Hochstrasser 1997; Saitoh et al. 1997). Of them, SUMO-1 is capable of modifying RanGAP1 (Mahajan et al. 1997) or the PML protein (Müller et al. 1998). Intriguingly, its yeast homolog Smt3p was found to link covalently to target proteins by a new pathway related to the Ub system, activated by an Aos1p/Uba2p heterodimeric complex (Johnson et al. 1997) and conjugated by Ubc9p (Johnson and Blobel 1997; Schwarz et al. 1998) as the E1- and E2-like enzymes, respectively. The gene encoding NEDD8, another mammalian Ub-like protein, was identified as one of multiple neural precursor cell-expressed developmentally down-regulated genes in mice (Kumar et al. 1993; Kamitani et al. 1997). NEDD8 has the highest identity to Ub among many Ub-like proteins. The mechanism for ligation of NEDD8 to appropriate proteins, however, remains largely unknown. In this study we report a novel modification system of NEDD8, consisting of the APP–BP1/hUba3 heterodimer and hUbc12 as the E1- and E2-like enzymes, respectively, which resembles that of the recently described Smt3p/SUMO-1, as mentioned above (Johnson and Blobel 1997; Johnson et al. 1997; Schwarz et al. 1998), indicating that three distinct pathways for modification of Ub and Ub-like proteins exist in cells. Moreover, we report that NEDD8 is likely to be conjugated to Cul-4A via the carboxy-terminal Gly residue in a manner analogous to ubiquitinylation. Cul-4A is one member of a family of human cullin proteins (Kipreos et al. 1996). Yeast Cdc53p (a homolog of Hs–Cul-1) functions as a common subunit of the large Ub–protein ligase E3 complex responsible for a ubiquitinylation-dependent proteolytic pathway that regulates various biologically important processes, such as the cell cycle, metabolism (Mathias et al. 1996), and gene expression (for review, see Jackson 1996; Hershko 1997; Hoyt 1997). Therefore, the results obtained in this study suggest that modification of Cul-4A by NEDD8 has an important role for regulation of the cell cycle.
Results and Discussion
Identification of a NEDD8-activating enzyme
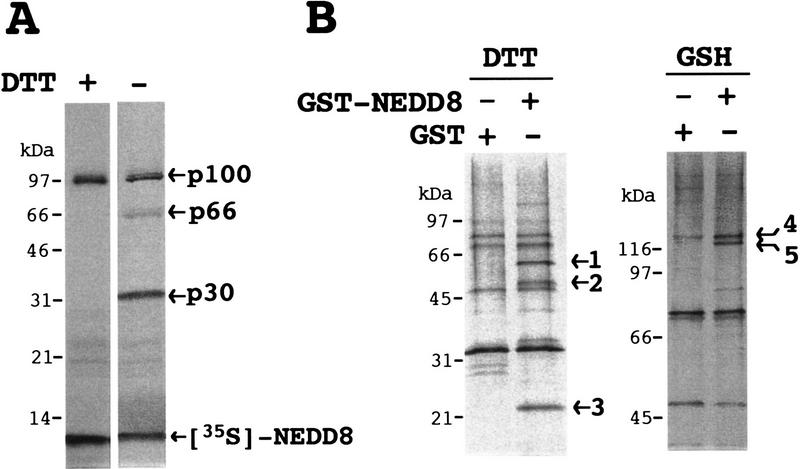
To explore the conjugating mechanism of NEDD8, we first attempted to identify the protein(s) that can interact with human 35S-labeled NEDD8 in rabbit reticulocyte lysates. Three major bands of 100, 66, and 30 kD were reproducibly detected in addition to unmodified 35S-labeled NEDD8, and the 66- and 30-kD bands disappeared by treatment with the reducing reagent dithiothreitol (DTT) (Fig. 1A). To characterize these proteins, we isolated them from reticulocyte lysates by affinity chromatography with GST–NEDD8 fused protein as a ligand. Five proteins, 1–5 (see Fig. 1B), were eluted from the GST–NEDD8 column but not from the control GST resin. Proteins 1–3 were eluted by DTT (Fig. 1B, left); and proteins 4 and 5 by subsequent treatment with reduced glutathione, GSH (Fig. 1B, right).
Figure 1.
Analysis of proteins interacting with NEDD8 in rabbit reticulocyte lysates. (A) Analysis of proteins linked to 35S-labeled NEDD8. After 35S-labeled NEDD8 had been synthesized for 60 min at 30°C in 5 μl of a reticulocyte lysate transcription/ translation system, samples (2.5 μl) of the resultant translational products were subjected directly to SDS-PAGE in the presence (+) or absence (−) of DTT and autoradiographed. (B) SDS-PAGE analysis of proteins purified by GST–NEDD8–GSH–Sepharose 4B chromatography. After reticulocyte lysates had been applied to GSH–Sepharose 4B resin bearing GST or GST–NEDD8, the bound materials were eluted with 40 mm DTT (left) and subsequently with 30 mm GSH (right). Samples were subjected to SDS-PAGE and stained with silver. Proteins bound specifically to GST–NEDD8 are numbered at right (1–5).
Sequence analysis of the 62-kD protein 1 eluted from the GST–NEDD8 affinity column showed that it was almost identical to the known protein, APP–BP1 (Fig. 2A), which had been found to interact with the APP, or β-amyloid precursor protein (Chow et al. 1996). APP–BP1 showed strong similarity to the amino-terminal region of the Ub-activating enzyme E1; but this presumptive E1-like protein lacked the carboxy-terminal region containing the conserved Cys residue required for the formation of a thioester bond with Ub (Hatfield and Vierstra 1992; Dohmen et al. 1995), indicating that this protein perhaps differs from the DTT-sensitive 66-kD band shown in Figure 1A. In considering that the E1-like enzyme for Smt3p is a heterodimer of Aos1p and Uba2p, which correspond to the amino- and carboxy-terminal regions, respectively, homologous to E1 (Johnson et al. 1997), we predicted that a hypothetical protein with a similarity to the carboxy-terminal region of E1 that is capable of interacting with APP–BP1 must be present. We believed that Uba3p deposited in the yeast genome database (PIR accession no. S54087), with unknown function (Hochstrasser 1997), might be a possible candidate. By computer analysis in public databases we found a cDNA (GenBank accession no. AA336365) encoding a human protein with a high homology to Uba3p and deduced its complete primary structure by cDNA sequencing (Fig. 2B). The hUba3 includes the consensus sequence for a nucleotide binding site, GXGXXG (position 55–60), which is present in Uba2p and E1 enzymes (Dohmen et al. 1995; Hass and Siepmann 1997) and also the consensus sequence PZCTXXXXP (Z is a nonpolar residue; position 214–222) around the essential Cys residue in E1 enzymes that becomes linked to Ub in an E1–Ub thioester linkage (Hatfield and Vierstra 1992; Dohmen et al. 1995). The protein, having 43% overall identity with yeast Uba3p, was thought to be a presumptive human homolog of the yeast Uba3p; therefore, we named it, tentatively, hUba3. We assume that protein 2 of ∼50 kD in Figure 1B may be hUba3, judging from its size and sensitivity to DTT; however, no sequence information is available.
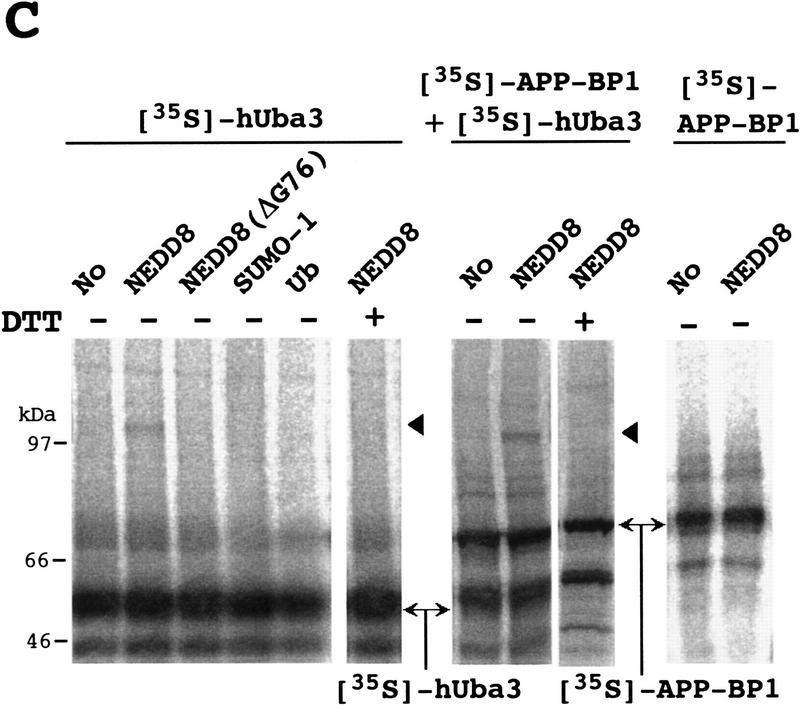
Figure 2.

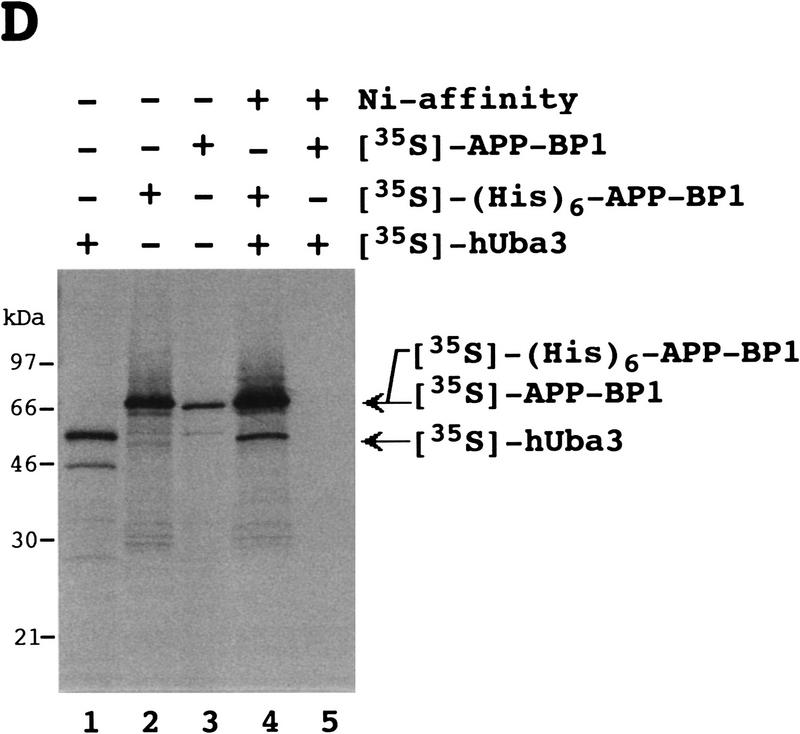
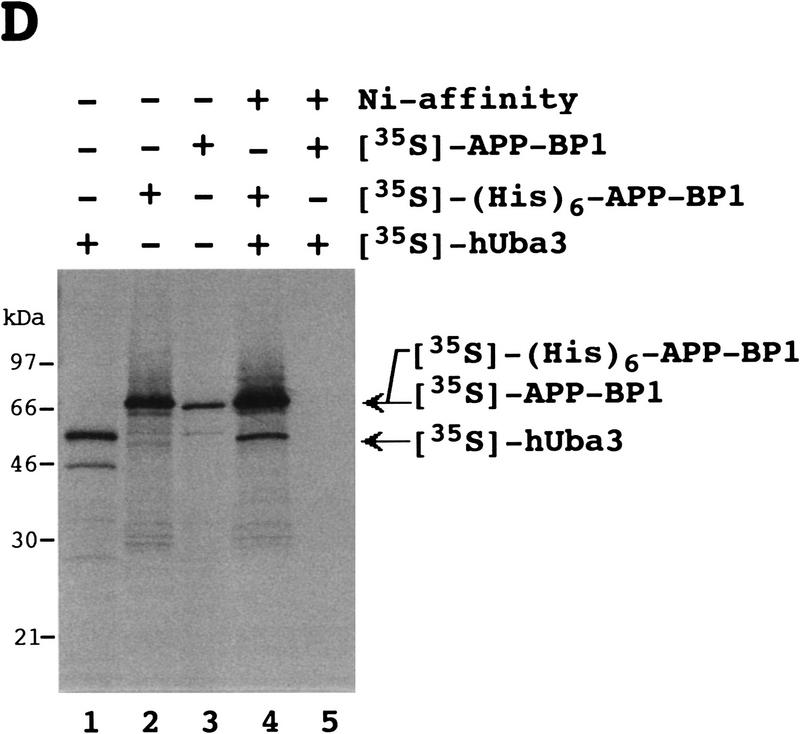
Identification of an E1-like APP–BP1/hUba3 heterodimer for activation of NEDD8. (A) Sequence alignment of two peptides of the rabbit 62-kD protein associated with GST–NEDD8 (band 1 in Fig. 1B) with human APP–BP1 (Chow et al. 1996). Partial amino acid sequences of the fragments of the 62-kD band digested with lysylendopeptidase were determined with a protein sequencer and are shown above the APP–BP1 sequences in italics. (X) An unidentified residue. The identical amino acids are boxed in black. (B) Primary structure of human Uba3 (Hs) deduced from the nucleotide sequence of a human cDNA and its sequence alignment with yeast Uba3p (Sc) (PIR accession no. S54087). Amino acid residues are numbered from the amino terminus. Identical amino acids are boxed in black. The motif shown (•) is the consensus sequence for a nucleotide binding site. The asterisk indicates the essential Cys residue conserved in E1 enzymes that becomes linked to Ub in an E1–Ub thioester linkage. (C) Thioester linkage between 35S-labeled hUba3 and GST–NEDD8. 35S-Labeled hUba3 and/or 35S-labeled APP–BP1 were cosynthesized for 60 min at 30°C in vitro in a 5 μl reticulocyte lysate transcription/translation system in the presence of 0.35 μg of unlabeled GST–NEDD8, GST–NEDD8(Δ76G), GST–SUMO-1, or GST-Ub, and a part of each (2.5 μl) was analyzed as in Fig. 1A. Arrowheads indicate the 35S-labeled hUba3–GST–NEDD8 complex. (D) Complex formation between 35S-labeled (His)6–APP–BP1 and 35S-labeled hUba3. 35S-labeled APP–BP1, 35S-labeled (His)6–APP–BP1, and 35S-labeled hUba3 were synthesized individually for 60 min at 30°C in vitro, and 1 μl of each was analyzed (lanes 1–3), as described in Fig. 1A. After samples (5 μl) of 35S-labeled (His)6–APP–BP1 or 35S-labeled APP–BP1 had been incubated with 35S-labeled hUba3 (5 μl) for 30 min at 30°C, half of each sample was applied onto a Ni-chelate column. After the column had been washed with 50 mm Na-phosphate buffer (pH 8.0) containing 0.5 m NaCl, the materials eluted with 100 mm EDTA (lanes 4,5) were analyzed as in Fig. 1A.
To examine whether NEDD8 is linked to hUba3, 35S-labeled hUba3 was synthesized in an in vitro transcription–translation sytem in the presence of an excess amount of GST–NEDD8. A new band of ∼110 kD was identified in addition to the unmodified 35S-labeled hUba3 (Fig. 2C), the intensity of which was increased by adding 35S-labeled APP–BP1. We concluded that this 110-kD band represented a complex formed by a thioester linkage between GST–NEDD8 and 35S-labeled hUba3 because it was lost by treatment with DTT and no significant band was observed when the mutated GST–NEDD8, in which the carboxy-terminal Gly residue was deleted [termed NEDD8(Δ76G)], was used instead of unmodified GST–NEDD8 (Fig. 2C). We also found that GST–SUMO-1 and GST–Ub did not link to 35S-labeled hUba3 and that GST–NEDD8 did not form a linkage with 35S-labeled APP–BP1 directly.
It was of interest to test whether APP–BP1 and hUba3 form heterodimeric complex. To test this possibility, we carried out purification by Ni-chelate column chromatography after 35S-labeled (His)6–APP–BP1 had been incubated with 35S-labeled hUba3 in reticulocyte lysates, and analyzed the eluates by SDS–PAGE and subsequent autoradiography. Besides 35S-labeled (His)6–APP–BP1, the band of 35S-labeled hUba3 was recovered, which was not evident when 35S-labeled APP–BP1 was used instead of 35S-labeled (His)6–APP–BP1 (Fig. 2D), suggesting strongly that both 35S-labeled (His)6–APP–BP1 and 35S-labeled hUba3 were coeluted from the Ni-chelate column by their complex formation. Therefore the APP–BP1 and hUba3 form a complex, presumably a heterodimer, which is assumed to function as an E1-like enzyme for the activation of NEDD8.
Identification of a NEDD8-conjugating enzyme
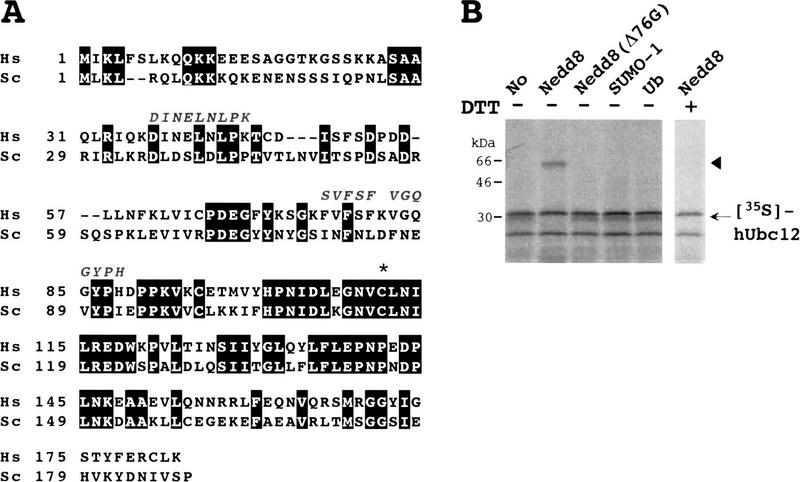
We next carried out sequence analysis of the peptide fragments derived from the 22-kD protein 3 that interacted with GST–NEDD8 (see Fig. 1B) and found that three of the fragments obtained had high similarity to a cDNA clone deposited in GenBank (accession no. T48884). We sequenced the cDNA clone entirely and found that it encodes a protein that has 42% identity to Ubc12p, a member of the Ub-conjugating enzyme E2 family in yeast (SWISS-PROT accession no. P52491) (Fig. 3A). Their sequence alignment showed that this newly identified protein is a human counterpart of yeast Ubc12p. Therefore, we named it hUbc12 and predicted that hUbc12 is a presumptive conjugating enzyme for NEDD8. Actually, the presumptive active Cys residue, required for the formation of a thioester bond between Ub and a family of E2 enzymes (Jentsch 1992; Hass and Siepmann 1997), is conserved in hUbc12 (see asterisk in Fig. 3A).
Figure 3.
Identification of hUbc12 as a conjugating enzyme for NEDD8. (A) Primary structure of human Ubc12 (Hs) deduced from the nucleotide sequence of a human cDNA and sequence alignment with yeast Ubc12p (Sc) (SWISS-PROT accession no. P52491). The identical amino acids are boxed in black. The amino acid sequences of three peptides of the rabbit 22-kD protein associated with GST–NEDD8 (band 3 in Fig. 1B) are shown above the sequence of hUbc12. The asterisk indicates the essential Cys residue conserved in a family of Ub-conjugating E2 enzymes that becomes linked to Ub in an E2–Ub thioester linkage. (B) Thioester linkage between 35S-labeled hUbc12 and GST–NEDD8. 35S-Labeled hUbc12 synthesized in vitro in the presence of unlabeled GST–NEDD8, GST–NEDD8(Δ76G), GST–SUMO-1, or GST–Ub was analyzed as in Fig. 2. The arrowhead indicates the 35S-labeled hUbc12–GST–NEDD8 complex.
To validate this assumption, we examined whether 35S-labeled hUbc12 forms a thioester linkage with GST–NEDD8 in reticulocyte lysates. When 35S-labeled hUbc12 was incubated with GST–NEDD8, a new larger band was evident besides 35S-labeled hUbc12 but was not observed by treatment with DTT or when the mutated GST–NEDD8(Δ76G) mentioned above was used (Fig. 3B). Moreover, neither GST–Ub nor GST–SUMO-1 linked to hUbc12. These results indicate that hUbc12 presumably acts as an E2-like enzyme specific for the conjugation of NEDD8.
Covalent modification of Hs–cullin-4A by NEDD8
Finally, we attempted to clarify the nature of the 100-kD component linked to 35S-labeled NEDD8 that was resistant to treatment with DTT (see Fig. 1A, left lane). We detected two bands in the GSH eluate from the GST–NEDD8 column (Fig. 1B, right, bands 4 and 5). We found that protein 5, but not 4, a protein of ∼120 kD, contained NEDD8 (perhaps GST–NEDD8) by immunoblotting with anti-NEDD8 antibody (data not shown) and therefore used it for chemical sequence analysis. Surprisingly it had a striking homology with Hs–cullin-4A (called simply Cul-4A), reported recently as a member of the cullin/Cdc53 family of proteins (Kipreos et al. 1996; Fig. 4A). We found the cDNA fragment of Cul-4A in our human cDNA bank (Kato et al. 1994) and called it Cul-4A(524C), because it covered the carboxy-terminal 524 amino acid residues, but lacked the short amino-terminal region. We next examined whether Cul-4A(524C) is modified by NEDD8. 35S-Labeled Cul-4A(524C) was modified by GST–NEDD8 in reticulocyte lysates, which was insensitive to treatment with DTT (Fig. 4B). We also found that the 20-kD fragment of Cul-4A covering the carboxy-terminal 171 amino acid residues, designated Cul-4A(171C), was sufficient for the formation of the linkage with GST–NEDD8 (Fig. 4C), indicating that NEDD8 is covalently linked to the carboxy-terminal region of Cul-4A. Moreover, no complex with 35S-labeled Cul-4A was formed when the mutated GST–NEDD8(Δ76G), GST–SUMO-1, or GST–Ub was used instead of GST–NEDD8, implying that NEDD8 is conjugated to Cul-4A via the carboxy-terminal Gly residue in a manner analogous to ubiquitinylation (Hershko and Ciechanover 1992) and that the presently described novel ligation pathway for NEDD8 did not catalyze formation of a linkage between Cul-4A and Ub or SUMO-1. These findings indicate strongly that Cul-4A is a major target protein for modification by NEDD8. Recently, Kamitani et al. (1997) also found that an ∼90-kD NEDD8-modified protein, differing from RanGAP1, was detected in all mammalian cell lines tested. We presume that this protein is Cul-4A or is in its family of proteins, although the nature of this 90-kD protein has not yet been characterized.
Figure 4.
Linkage of NEDD8 to Cul-4A in a DTT-insensitive fashion. (A) Alignment of amino acid sequences of two peptides of the rabbit 120-kD protein associated with GST–NEDD8 (protein 5 band in Fig. 1B) and human Cul-4A that we had cloned. Identical amino acids are boxed in black. The Cul-4A cDNA was named Cul-4A(524C), as described in the text. (B) Linkage between 35S-labeled Cul-4A(524C) and GST–NEDD8. 35S-labeled Cul-4A(524C) synthesized in vitro in the presence of unlabeled GST–NEDD8, GST–NEDD8(Δ76G), GST–SUMO-1, or GST–Ub was analyzed as in Fig. 2C. The arrowhead indicates the 35S-labeled Cul-4A(524C)–GST–NEDD8 complex. (C) Linkage between 35S-Labeled Cul-4A(171C) and GST–NEDD8. 35S-labeled Cul-4A(171C) that corresponds to the carboxy-terminal 171 amino acid residues (see Materials and methods) was treated or not with unlabeled GST–NEDD8 and analyzed as in B. The arrowhead indicates the 35S-labeled Cul-4A(171C)–GST–NEDD8 complex.
In the present study, we reported a novel modification system of NEDD8, consisting of the APP–BP1/hUba3 complex and hUbc12, which are related to E1 and E2 enzyme, respectively, in the ubiquitinylation pathway. This NEDD8-ligation pathway resembles that of Smt3p/SUMO-1, as mentioned in the introductory section (Johnson and Blobel 1997; Johnson et al. 1997; Schwarz et al. 1998). Intriguingly, quite recently Rub1p, a presumptive yeast homolog of mammalian NEDD8 displaying 59% amino acid identity to human NEDD8, was found to be ligated to target protein through Ula1p/Uba3p and Ubc12p as the E1- and E2-like enzymes, respectively (Liakopoulos et al. 1998). In addition to the similarities in Uba3 and Ubc12 proteins between humans and yeast (Figs. 2B and 3A), APP–BP1 and Ula1p show high sequence similarity, that is, 26% amino acid identity, indicating evolutional conservation of the post-translational protein-modifying system for Rub1p/NEDD8 and a common role of this system in eukaryotes.
So far, it can be concluded that three different systems operate for activation of Ub and Ub-like proteins: A Ub-activating enzyme, E1, consisting of a single polypeptide, and two heterodimeric E1-like complexes, Aos1p/Uba2p and APP–BP1/hUba3 for activation of Smt3p/SUMO-1 and Rub1p/NEDD8, respectively (this study; Johnson et al. 1997; Liakopoulos et al. 1998; for review, see Hochstrasser et al. 1998), although there is no direct evidence that APP–BP1 and hUba3 form a heterodimer. It is notable that Ub is conjugated by multiple species of Ubc, whereas Smt3p/SUMO-1 and NEDD8 each use a specific conjugating enzyme, Ubc9 and hUbc12, respectively (this study; Johnson and Blobel 1997; Schwarz et al. 1998). Taken together, it is conceivable that three distinct pathways for modification of Ub and Ub-like proteins exist in both yeast and mammalian cells.
Here, we reported that NEDD8 was likely to be conjugated to Cul-4A via the carboxy-terminal Gly residue in a manner analogous to ubiquitinylation (Fig. 4B). Moreover, we observed that the carboxy terminal domain of Cul-4A, which has been conserved in various species (Kipreos et al. 1996), was sufficient for the conjugation of NEDD8 (Fig. 4C), indicating that the sites accepting NEDD8 exist in this region. One interesting aspect is that Cul-4A is one member of a family of human cullin proteins that have high sequence homologies (Kipreos et al. 1996). So far, 6 species of human cullin family proteins including Cul-1, Cul-2, Cul-3, Cul-4B, and Cul-5 in addition to Cul-4A have been reported (Kipreos et al. 1996). Whether NEDD8 is also ligated to other cullin family proteins awaits further study.
The ligation of NEDD8 to Cul-4A was essentially the same as the recently observed conjugation of Rub1p to yeast Cdc53p (homolog of Hs–Cul-1) (Lammer et al. 1998; Liakopoulos et al. 1998). Yeast Cdc53p functions as a common component of a large Ub–protein ligase E3 complex (called SCF Ub ligase) that regulates multiple cellular functions, such as G1/S progression of the cell cycle (Mathias et al. 1996; for review, see Jackson 1996; Hershko 1997: Hoyt 1997), gene expression (Li and Johnston 1997), and methionine biosynthesis (Patton et al. 1998). Of them, it is of interest to consider a relationship between the NEDD8 ligation system and the function of the SCF Ub ligase responsible for the ubiquitinylation of cell-cycle factors involved in the G1/S transition of the cell cycle (for review, see Jackson 1996; Hershko 1997; Hoyt 1997). Recently, Liakopoulos et al. (1998) reported that modification of yeast Cdc53p by Rub1p may affect optimal assembly or function of the SCF complex, although RUB1, ULA1, UBA3, and UBC12, all components of the NEDD8 ligating system, are not essential for viability (Lammer et al. 1998; Liakopoulos et al. 1998). Moreover, a deletion of ENR2 (equivalent to ULA1) is synthetic lethal with temperature-sensitive alleles of cdc34 (i.e., UBC3) and enhances the phenotypes of cdc4, cdc53, and skp1, all of which are components of the SCF Ub–ligase complex, implying that the Rub1p ligation pathway is linked closely to cell-cycle regulation (Lammer et al. 1998). Consistent with this notion, the mutation of hamster SMC, encoding a protein nearly identical to APP–BP1, is responsible for cell-cycle defects in the ts41 cell line (Handel and Weintraub 1992; Hochstrasser 1998). In considering these observations, the novel pathway for the ligation of NEDD8 to Cul-4A described here may provide new insight into understanding of the regulatory mechanism for ubiquitinylation mediated by the SCF Ub ligase.
Materials and methods
Biochemical analysis
Chemical analysis of proteins that interacted with the GST–NEDD8 fused protein was performed as follows: Fifty milliliters of rabbit reticulocyte lysate including 1 mm ATP, 1 mm MgCl2, 0.5 mm DTT, 2 mm PMSF, 20 μg/ml aprotinin, and 20 μm leupeptin was incubated for 15 min at 30°C with 5 mg of GST–NEDD8 and then mixed at 4°C for 1 hr with 2 ml of GSH–Sepharose 4B (Pharmacia). The materials were then loaded onto a column and washed with 20 ml of 50 mm Tris-HCl (pH 8.0) containing 0.5 m NaCl. After washing, the absorbed materials were eluted with 20 mm Tris-HCl (pH 8.0), 40 mm DTT, and subsequently with 20 mm Tris-HCl (pH 8.0), 30 mm GSH. The eluate was concentrated with a Microcon (Amicon). After SDS-PAGE, the protein fragments obtained by digestion with lysylendopeptidase were resolved by reverse-phase HPLC and sequenced by automated Edman degradation, as reported previously (Kawasaki et al. 1990).
Molecular–biological analysis
The cDNAs encoding human NEDD8 (accession no. D23662), SUMO-1, and Cul-4A were found in our human full-length cDNA library prepared with a multifunctional shuttle vector, pKA1 (Kato et al. 1994). Note that the Cul-4A cDNA lacked the short amino-terminal region and so was designated Cul-4A(524C), because it covered the carboxy-terminal 524 amino acid residues (see Fig. 4A). To make deletion mutants of the Cul-4A cDNA, we digested the cDNA with EcoRI to remove the amino-terminal region, which left the carboxy-terminal 171 amino acid residues; thus, we termed the deleted cDNA Cul-4A(171C). The cDNA clones for hUba3 and hUbc12 were obtained from ATCC and sequenced by a double-strand strategy in an automatic DNA sequencer. Various GST-fused proteins were synthesized in Escherichia coli using the pGEX–2TK expression vector (Pharmacia). The [35S]-methionine-labeled APP–BP1, 35S-hUba3, 35S-labeled NEDD8, 35S-labeled (His)6–APP–BP1, 35S-labeled Cul-4A(524C), and 35S-labeled Cul-4A(171C) proteins were synthesized by an in vitro transcription/translation system according to the manufacturer’s recommendations (Promega). cDNAs of APP–BP1 and GST–NEDD8(Δ76G) were prepared by PCR. The nucleotide sequence data reported in this paper will appear in the GSDB, DDBJ, EMBL, and NCBI nucleotide sequence databases under the following accession numbers: AB012190 for hUba3, AB012191 for hUbc12, and AB012193 for Hs–Cullin-4A.
Acknowledgments
We thank Kazuo Kamemura, Akihiko Komuro, Toshiaki Suzuki, Nobuyuki Tanahashi, and our colleagues for advice throughout this study.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL seishi@sagami.or.jp; FAX 81-427-49-7631.
References
- Chow N, Korenberg JR, Chen X-N, Neve RL. APP-BP1, a novel protein that binds to the carboxyl-terminal region of the amyloid precursor protein. J Biol Chem. 1996;271:11339–11346. doi: 10.1074/jbc.271.19.11339. [DOI] [PubMed] [Google Scholar]
- Coux O, Tanaka K, Goldberg AL. Structure and functions of the 20S and 26S proteasomes. Annu Rev Biochem. 1996;65:801–847. doi: 10.1146/annurev.bi.65.070196.004101. [DOI] [PubMed] [Google Scholar]
- Dohmen RJ, Sappen R, McGrath JP, Forrova H, Kolarov J, Goffeau A, Varshavsky A. An essential yeast gene encoding a homolog of ubiquitin-activating enzyme. J Biol Chem. 1995;270:18099–18109. doi: 10.1074/jbc.270.30.18099. [DOI] [PubMed] [Google Scholar]
- Handeli S, Weintraub H. The ts41 mutation in Chinese hamster cells leads to successive S phases in the absence of intervening G2, M, and G1. Cell. 1992;71:599–611. doi: 10.1016/0092-8674(92)90594-3. [DOI] [PubMed] [Google Scholar]
- Hass AL, Siepmann TJ. Pathways of ubiquitin conjugation. FASEB J. 1997;11:1257–1268. doi: 10.1096/fasebj.11.14.9409544. [DOI] [PubMed] [Google Scholar]
- Hatfield PM, Vierstra RD. Multiple forms of ubiquitin-activating enzyme E1 from wheat. J Biol Chem. 1992;267:14799–14803. [PubMed] [Google Scholar]
- Hershko A. Roles of ubiquitin-dependent proteolysis in cell cycle control. Curr Opin Cell Biol. 1997;9:788–799. doi: 10.1016/s0955-0674(97)80079-8. [DOI] [PubMed] [Google Scholar]
- Hershko A, Ciechanover A. The ubiquitin system for protein degradation. Annu Rev Biochem. 1992;61:761–807. doi: 10.1146/annurev.bi.61.070192.003553. [DOI] [PubMed] [Google Scholar]
- Hochstrasser M. Ubiquitin-dependent protein degradation. Annu Rev Genet. 1997;30:405–439. doi: 10.1146/annurev.genet.30.1.405. [DOI] [PubMed] [Google Scholar]
- Hochstrasser M. There’s the Rub: A novel ubiquitin-like modification linked to cell cycle regulation. Genes & Dev. 1998;12:901–907. doi: 10.1101/gad.12.7.901. [DOI] [PubMed] [Google Scholar]
- Hoyt MA. Eliminating obstacles: Regulated proteolysis in the eukaryotic cell cycle. Cell. 1997;91:149–151. doi: 10.1016/s0092-8674(00)80396-7. [DOI] [PubMed] [Google Scholar]
- Jackson PK. Cell cycle: Cull and destroy. Curr Biol. 1996;6:1209–1212. doi: 10.1016/s0960-9822(96)00697-5. [DOI] [PubMed] [Google Scholar]
- Jentsch S. The ubiquitin-conjugation system. Annu Rev Genet. 1992;26:179–207. doi: 10.1146/annurev.ge.26.120192.001143. [DOI] [PubMed] [Google Scholar]
- Johnson ES, Blobel G. Ubc9p is the conjugating enzyme for the ubiquitin-like protein Smt3p. J Biol Chem. 1997;272:26799–26802. doi: 10.1074/jbc.272.43.26799. [DOI] [PubMed] [Google Scholar]
- Johnson ES, Schwienhorst I, Dohmen RJ, Blobel G. The ubiquitin-like protein Smt3p is activated for conjugation to other proteins by an Aos1p/Uba2p heterodimer. EMBO J. 1997;16:5509–5519. doi: 10.1093/emboj/16.18.5509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson PR, Hochstrasser M. SUMO-1: Ubiquitin gains weight. Trends Cell Biol. 1997;7:408–413. doi: 10.1016/S0962-8924(97)01132-X. [DOI] [PubMed] [Google Scholar]
- Kamitani T, Kito K, Nguyen HP, Yeh ET. Characterization of NEDD8, a developmentally down-regulated ubiquitin-like protein. J Biol Chem. 1997;272:28557–28562. doi: 10.1074/jbc.272.45.28557. [DOI] [PubMed] [Google Scholar]
- Kato S, Sekine S, Oh S-W, Kim N-S, Umezawa Y, Abe N, Yokoyama-Kobayashi M, Aoki T. Construction of a human full-length cDNA bank. Gene. 1994;150:243–250. doi: 10.1016/0378-1119(94)90433-2. [DOI] [PubMed] [Google Scholar]
- Kawasaki H, Emori Y, Suzuki K. Production and separation of peptides from proteins stained with Coomassie Brilliant Blue R-250 after separation by sodium dodecyl sulphate-polyacrylamide gel electrophoresis. Anal Biochem. 1990;191:332–336. doi: 10.1016/0003-2697(90)90227-z. [DOI] [PubMed] [Google Scholar]
- Kipreos ET, Lander LE, Wing JP, He WW, Hedgecock EM. cul-1 is required for cell cycle exit in C. elegansand identifies a novel gene family. Cell. 1996;85:829–839. doi: 10.1016/s0092-8674(00)81267-2. [DOI] [PubMed] [Google Scholar]
- Kumar S, Yoshida Y, Noda M. Cloning of a cDNA which encodes a novel ubiquitin-like protein. Biochem Biophys Res Commun. 1993;195:393–399. doi: 10.1006/bbrc.1993.2056. [DOI] [PubMed] [Google Scholar]
- Lammer D, Mathias N, Laplaza JM, Jiang W, Liu Y, Callis J, Goebl M, Estelle M. Modification of yeast Cdc53p by the ubiquitin-related protein Rub1p affects function of the SCFCdc4complex. Genes & Dev. 1998;12:914–926. doi: 10.1101/gad.12.7.914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li FN, Johnston M. Grr1 of Saccharomyces cerevisiae is connected to the ubiquitin proteolysis machinery through Skp1: Coupling glucose sensing to gene expression and the cell cycle. EMBO J. 1997;16:5629–5638. doi: 10.1093/emboj/16.18.5629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liakopoulos D, Doenges G, Matuschewski K, Jentsch S. A novel protein modification pathway related to the ubiquitin system. EMBO J. 1998;17:2208–2214. doi: 10.1093/emboj/17.8.2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan R, Delphin C, Guan T, Gerace L, Melchior F. A small ubiquitin-related polypeptide involved in targeting RanGAP1 to nuclear pore complex protein RanBP2. Cell. 1997;88:97–107. doi: 10.1016/s0092-8674(00)81862-0. [DOI] [PubMed] [Google Scholar]
- Mathias N, Johnson SL, Winey M, Adams AM, Goetsch L, Pringle JR, Byers B, Goebl MG. Ced53p acts in concert with Cdc4p and Cdc34p to control the G1-to-S-phase transition and identifies a conserved family of proteins. Mol Cell Biol. 1996;16:6634–6643. doi: 10.1128/mcb.16.12.6634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller S, Matunis MJ, Dejean A. Conjugation with the ubiquitin-related modifier SUMO-1 regulates the partitioning of PML within the nucleus. EMBO J. 1998;17:61–70. doi: 10.1093/emboj/17.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patton EE, Willems AR, Sa D, Kuras L, Thomas D, Craig KL, Tyers M. Cdc53 is a scaffold protein for multiple Cdc34/Skp1/F-box protein complexes that regulate cell division and methionine biosynthesis in yeast. Genes & Dev. 1998;12:692–705. doi: 10.1101/gad.12.5.692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh H, Pu RT, Dasso M. SUMO-1: Wrestling with a new ubiquitin-related modifier. Trends Biochem Sci. 1997;22:374–376. doi: 10.1016/s0968-0004(97)01102-x. [DOI] [PubMed] [Google Scholar]
- Schwarz SE, Matuschewski K, Liakopoulos D, Scheffner M, Jentsch S. The ubiquitin-like proteins SMT3 and SUMO-1 are conjugated by the UBC9 E2 enzyme. Proc Natl Acad Sci. 1998;95:560–564. doi: 10.1073/pnas.95.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshavsky A. The ubiquitin system. Trends Biochem Sci. 1997;22:383–387. doi: 10.1016/s0968-0004(97)01122-5. [DOI] [PubMed] [Google Scholar]