Abstract
In positive-stranded viruses, the genomic RNA serves as a template for both translation and RNA replication. Using poliovirus as a model, we examined the interaction between these two processes. We show that the RNA polymerase is unable to replicate RNA templates undergoing translation. We discovered that an RNA structure at the 5′ end of the viral genome, next to the internal ribosomal entry site, carries signals that control both viral translation and RNA synthesis. The interaction of this RNA structure with the cellular factor PCBP up-regulates viral translation, while the binding of the viral protein 3CD represses translation and promotes negative-strand RNA synthesis. We propose that the interaction of 3CD with this RNA structure controls whether the genomic RNA is used for translation or RNA replication.
Keywords: PCBP, positive-stranded virus, RNA replication, translational control, viral polymerase
The genomes of positive-stranded RNA viruses are important in at least three major processes: They act as mRNAs to direct the synthesis of viral proteins; they serve as templates for genome replication; and they are packaged along with structural proteins during viral assembly. The balance between these processes must be properly maintained to allow efficient viral proliferation. Thus, once the viral RNA-dependent RNA polymerase and other essential proteins are synthesized, the genomic RNA must be used as a template for negative-strand RNA replication (Pogue et al. 1994). In principle, this creates a conflict between the translation and replication machinery: While the ribosomes are moving along the viral RNA in the 5′ to 3′ direction, the polymerase initiates replication at the 3′ end of the same RNA and moves in the opposite direction as it synthesizes the complementary negative strand. It is not known whether the arrangement of ribosomes and polymerases on the RNA template allows both processes to occur simultaneously or whether translation and RNA replication interfere with each other.
To examine the interplay between translation and RNA replication, we have used poliovirus as a model, because both in vitro and in vivo systems are available to dissect the viral cycle (Molla et al. 1991; Barton and Flanegan 1993; Gamarnik and Andino 1996). Several experiments have suggested that the highly conserved 5′-untranslated region (5′UTR) of the poliovirus genome plays an important role in the regulation of both translation and RNA replication (Rohll et al. 1994). Two functional domains have been defined within this region: a short 5′-terminal element involved in RNA replication (Andino et al. 1990a, 1993; Harris et al. 1994; Roehl et al. 1997) and a longer element, termed the internal ribosomal entry site (IRES), involved in viral translation (Jang et al. 1988; Pelletier et al. 1988; Trono et al. 1988). It was originally thought that these two elements were independent; however, recent evidence suggests a functional overlap between them (Simoes and Sarnow 1991; Borman et al. 1994; Shiroki et al. 1995).
The regulatory function of the 5′ UTR is probably mediated through its interactions with cellular and viral proteins (Ehrenfeld and Semler 1995; Jackson and Kaminski 1995; Belsham and Sonenberg 1996). The short 5′-terminal element of the 5′ UTR folds into a cloverleaf-like structure and forms a ribonucleoprotein complex with the cellular poly(C)-binding proteins 1 and 2 (referred to as PCBP in this report, but also known as hnRNP E or αCP), and the viral protein 3CD, which is the uncleaved precursor of the viral protease (3Cpro) and polymerase (3Dpol) (Gamarnik and Andino 1997; Parsley et al. 1997). Mutations that disrupt the formation of this ribonucleoprotein complex impair viral RNA replication (Andino et al. 1990a; Rohll et al. 1994). On the other hand, the IRES element enables ribosomes to internally enter the RNA without scanning from the 5′ end (Jang et al. 1988; Pelletier et al. 1988; Trono et al. 1988; Chen and Sarnow 1995). The mechanism by which the translation apparatus recognizes IRES sequences is unknown, but it has been proposed that many canonical initiation factors as well as other specific cellular proteins participate in the process (Meyer et al. 1995; Pestova et al. 1996). Currently, three noncanonical factors that bind to the IRES have been identified: polypyrimidine tract binding protein (PTB; Hellen et al. 1993); La autoantigen (Meerovitch et al. 1993); and PCBP (Blyn et al. 1996). Interestingly, PCBP appears to participate in both viral translation and RNA replication (Blyn et al. 1996, 1997; Gamarnik 1997; Parsley et al. 1997).
We have analyzed the interplay between poliovirus translation and RNA replication and the contribution of specific ribonucleoprotein complexes to the regulation of both processes. We show that the viral polymerase is unable to use the genomic RNA as a template for RNA synthesis while it is being used by translating ribosomes. We found that the cloverleaf RNA at the 5′ end of the viral genome is a bifunctional element involved in the regulation of both viral translation and RNA replication. The binding of the cellular protein PCBP to the cloverleaf enhances viral translation, while the binding of the viral protein 3CD represses translation and facilitates negative-strand synthesis. Thus, we propose that overlapping translation and replication signals within the cloverleaf function as a strategy to coordinate the use of the genomic RNA for translation or RNA replication.
Results
Actively translating ribosomes inhibit the elongation activity of the poliovirus RNA polymerase 3Dpol
Poliovirus translation and negative-strand RNA synthesis use the same RNA template, but each process starts at different ends and proceeds in opposite directions. To investigate whether a poliovirus RNA template can support translation and RNA replication simultaneously, we measured the elongation activity of a partially purified viral polymerase (3Dpol) added to a translation extract programmed with poliovirus RNA. A subgenomic poliovirus replicon (Polio–Luc) carrying a luciferase gene as a reporter was used as the template (Andino et al. 1993). Translation was measured by the amount of luciferase activity produced, and RNA synthesis was measured by the incorporation of [32P]UMP into the poliovirus RNA by elongation of a specific primer complementary to the 3′ UTR.
When the ribosomes were actively translating, [32P]UMP incorporation was almost undetectable (Fig. 1A), suggesting that the viral polymerase was unable to synthesize RNA. In contrast, when translation was inhibited by cycloheximide, which stalled the ribosomes at the beginning of the coding region, 3Dpol was able to incorporate [32P]UMP into RNA (Fig. 1B). These results indicate that poliovirus RNA synthesis occurs only when the RNA is free of translating ribosomes. Therefore, the virus should have a mechanism to clear the genome of ribosomes to produce a viral RNA competent for RNA replication.
Figure 1.
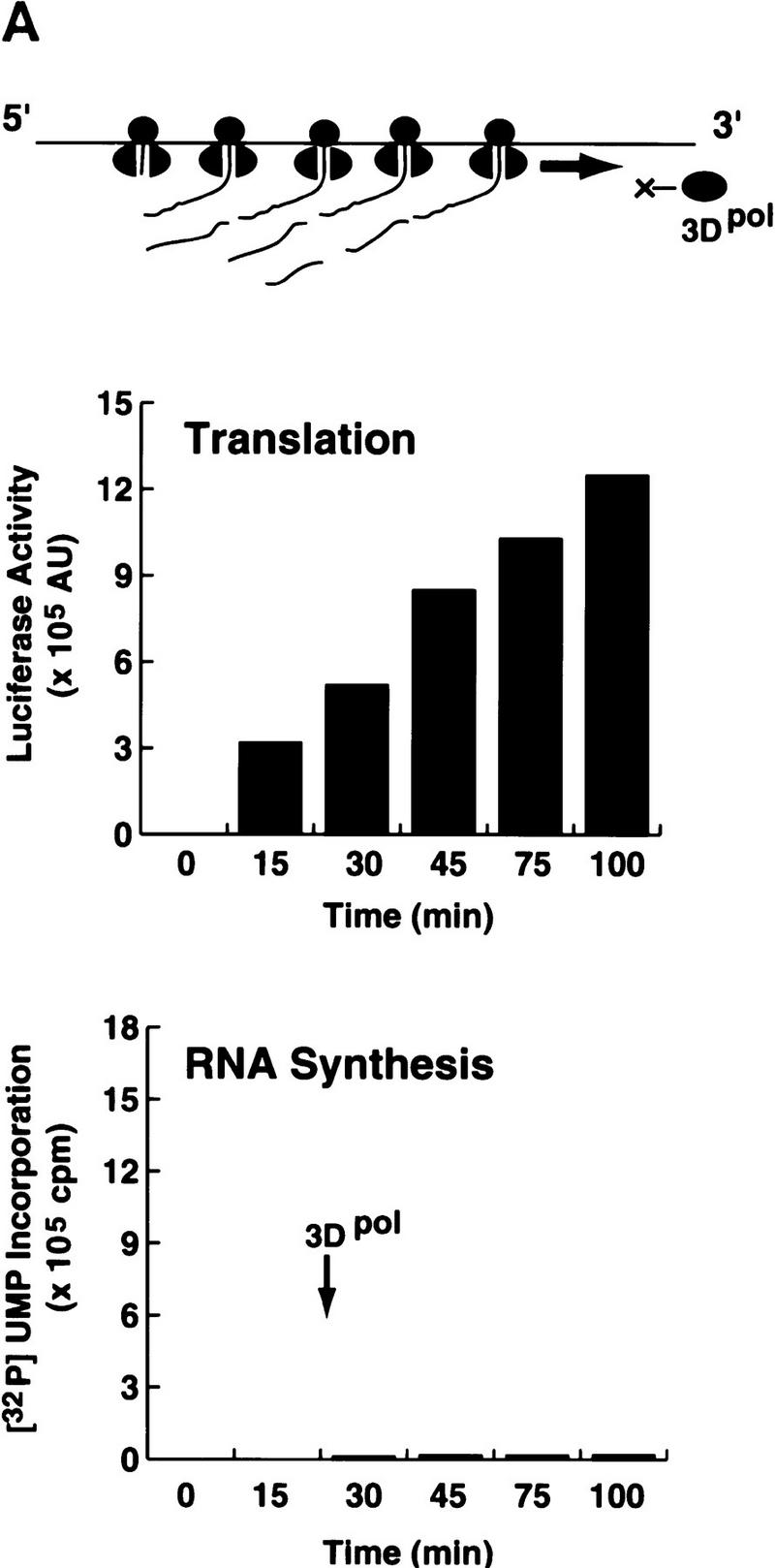
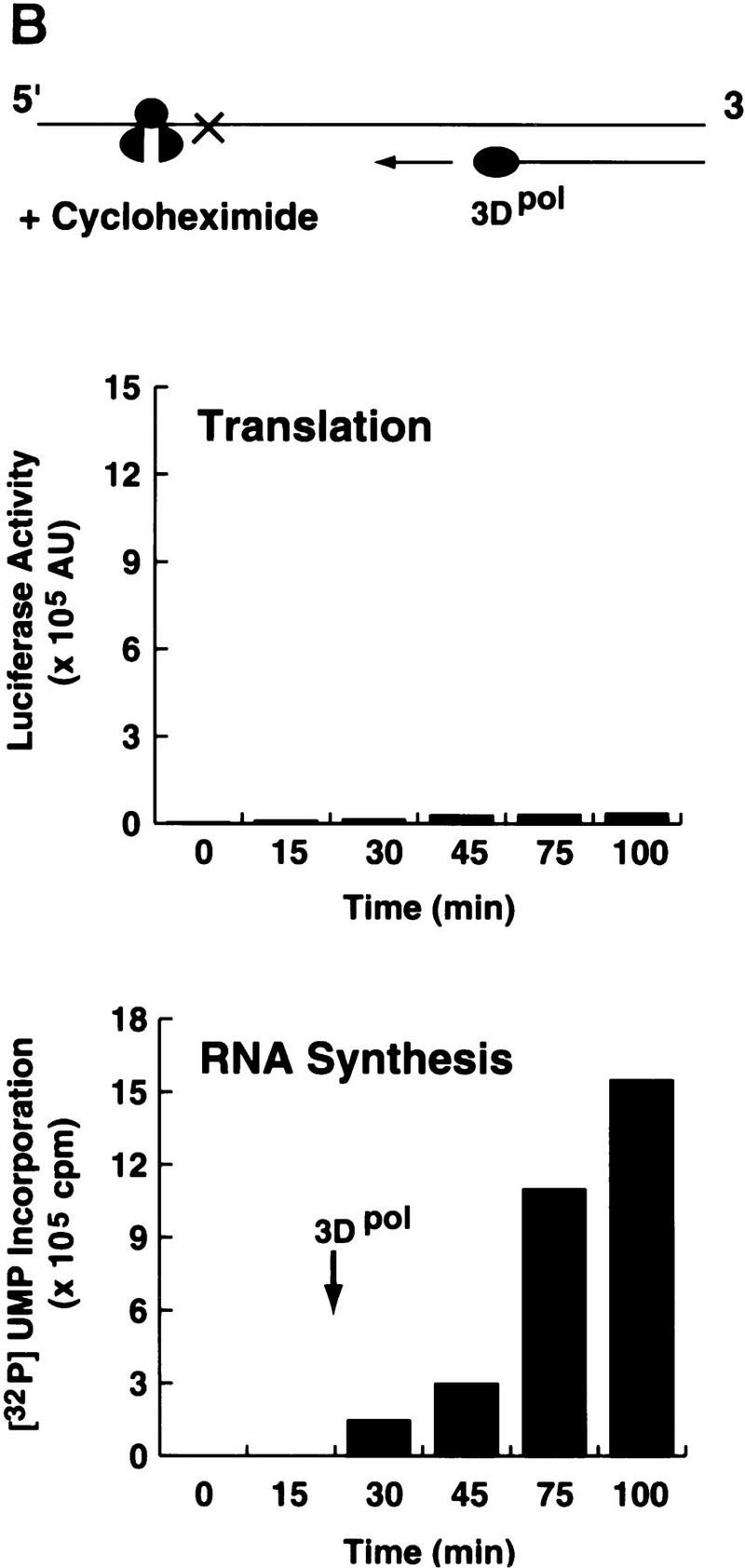
Poliovirus translation inhibits 3Dpol RNA elongation activity. (A) A poliovirus replicon (Polio–Luc, Fig. 2A) was in vitro-translated in a translation system supplemented with purified poliovirus 3D polymerase (3Dpol). Translation (top) was determined by the amount of luciferase activity produced and was expressed as arbitrary units (AU). RNA synthesis (bottom) was measured by [32P]UMP incorporation into an acid-insoluble fraction. (B) The same experiment described in A was performed in the presence of cycloheximide (100 μg/μl).
Poliovirus-infected extracts contain an activity that inhibits viral translation
It is possible that during the course of infection the virus induces activities that control initiation of translation when RNA replication must begin. To examine this possibility, we analyzed the effect of extracts from poliovirus-infected cells on translation of a Polio–Luc RNA in Xenopus oocytes. We have showed previously that Xenopus oocytes can support poliovirus replication (Gamarnik and Andino 1996). Microinjection of poliovirus RNA together with a cytoplasmic HeLa cell extract initiates a replication cycle, in which viral translation, genome replication, and assembly closely resemble the processes observed in mammalian cells.
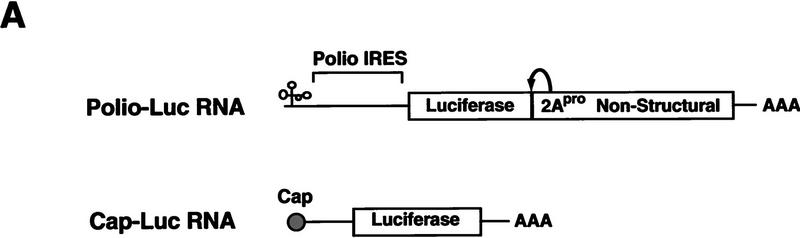
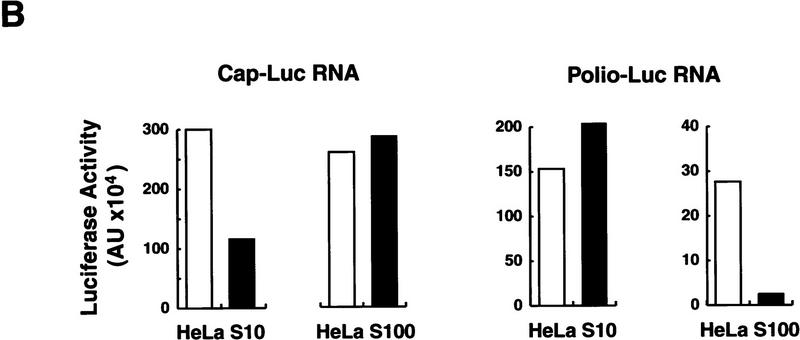
Two types of RNA were microinjected into Xenopus oocytes: a poliovirus replicon (Polio–Luc) in which the sequences encoding capsid proteins were replaced by a luciferase reporter gene; and, as a control, a capped RNA encoding luciferase (Cap–Luc) that included the 5′ and 3′ UTRs of the β-globin mRNA (Fig. 2A). Each construct was coinjected with cytoplasmic fractions obtained from either uninfected or poliovirus-infected HeLa cells. It is known that poliovirus induces the selective inhibition of host cell translation, which has been associated with the cleavage of the initiation factor eIF-4G by the viral protease 2Apro (for review, see Mathews 1996). As expected, crude S10 extracts from infected HeLa cells strongly inhibited cap-dependent translation, but stimulated Polio–Luc RNA translation by 30% (Fig. 2B). Interestingly, a further purified fraction obtained from poliovirus-infected cells (HeLa S100) contained an activity that strongly inhibited viral translation (Fig. 2B, right). This negative effect was specific for poliovirus cap-independent translation, because Cap–Luc RNA was efficiently translated after coinjection with either infected or uninfected S100 HeLa cell extracts (Fig. 2B, left).
Figure 2.
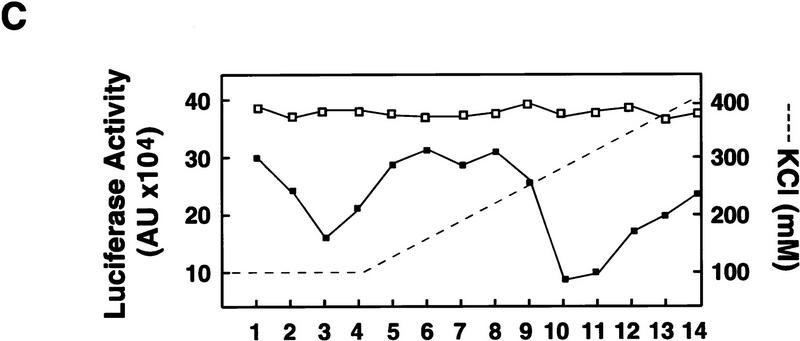
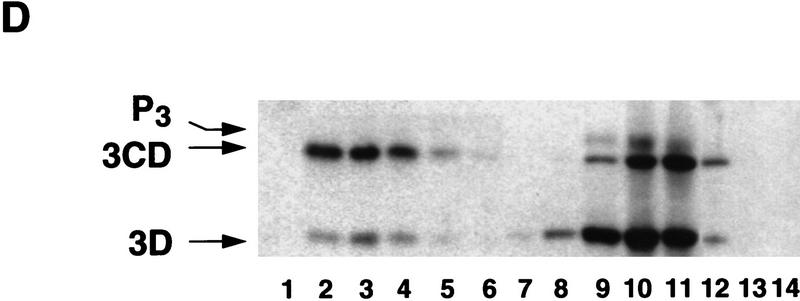
Poliovirus-infected cell extracts contain an activity that specifically inhibits poliovirus translation. (A) Schematic representation of the chimeric poliovirus luciferase RNA (Polio–Luc) and capped luciferase RNA (Cap–Luc). In Polio–Luc, the coding region of the poliovirus capsid proteins was replaced by the luciferase reporter gene, and a cleavage site for 2Apro has been introduced between luciferase and 2Apro (represented by the arrow). The Cap–Luc RNA consists of the luciferase gene flanked by the 5‘ and 3‘ uncoding regions of the β-globin mRNA. (B) Microinjection of infected S100 HeLa cell extract into Xenopus oocytes specifically inhibits poliovirus cap-independent translation. Polio–Luc or Cap–Luc RNA was injected into oocytes together with uninfected (open bars) or poliovirus-infected S10 or S100 HeLa cell fractions (solid bars) as indicated in each case. Luciferase activity was determined in oocytes after 3 hr of incubation at 22°C and expressed in arbitrary units (AU). (C) Elution profile of the translation inhibitory activity after ion-exchange chromatography. Infected S100 HeLa cell extract was loaded onto a HiTrap SP column (Pharmacia) and eluted with a KCl gradient, as indicated at right. The translation inhibitory activity was determined by coinjection of 20 nl of each fraction (1–14) together with 5 nl of HeLa S10 (to provide the cellular factor essential for poliovirus translation in oocytes, PTF) and 20 ng of Polio–Luc RNA (▪) or Cap–Luc RNA ( ) into oocytes. Luciferase activity was determined in oocyte extracts after 3 hr of incubation at 22°C and expressed in AU. (D) Viral proteins 3Dpol, 3CD, and P3 copurified with the viral translation inhibitory activity. Western blot analysis of fractions 1–14 eluted from the HiTrap SP column is shown. Two microliters of each fraction was resolved in a 10% SDS–polyacrylamide gel, transferred to nitrocelluose membrane, and probed with specific anti-3CD antibodies. The electrophoretic mobility of P3, 3CD, and 3D is indicated at left.
Because the inhibitory effect on translation was observed only with injection of infected extracts, the inhibitory activity must involve either a viral factor or a virally modified cellular factor. To characterize this activity further, we fractionated the infected S100 extract by chromatography on a HiTrap SP column. Part of the inhibitory activity was recovered in the flowthrough, while a larger part was retained in the column and eluted at 250 mm KCl (Fig. 2C). As observed with the total extract, microinjection of the fractions had no effect on translation of Cap–Luc RNA (Fig. 2C). Then, we analyzed whether the fractions showing inhibitory activity contained any viral protein. Western blot analysis revealed that the inhibitory activity copurified with the viral proteins 3Dpol, 3CD, and partially with their precursor P3 (Fig. 2D), but not with other viral proteins (data not shown). Interestingly, 3Dpol is the RNA-dependent RNA polymerase that is synthesized as a fusion protein with 3Cpro, a protease that also binds to specific sequences of the viral 5′ UTR (Andino et al. 1990b, 1993).
The viral protein 3CD represses poliovirus translation
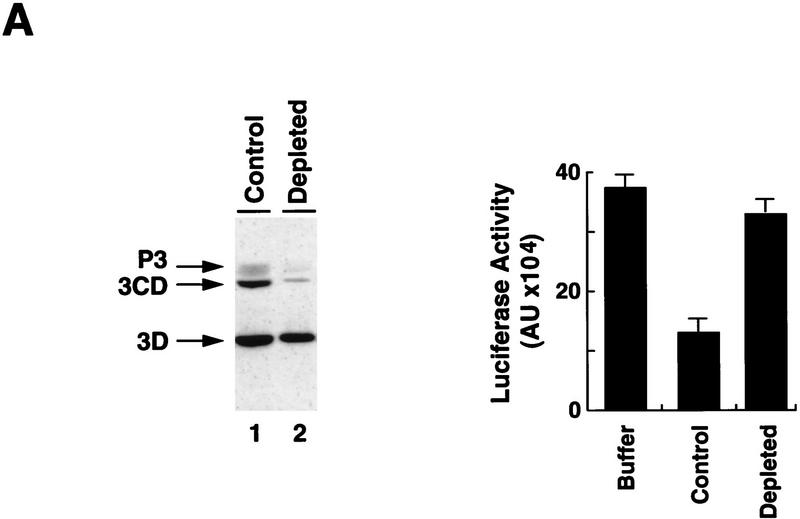
To determine whether any of the 3D-containing proteins were responsible for the inhibitory effect on translation, we took advantage of the RNA-binding properties of 3Cpro. Affinity chromatography with an immobilized RNA was used to deplete 3CD from the most active column fractions. The treated fraction retained most of the 3Dpol protein but <10% of the original amounts of 3CD and P3 (Fig. 3A, left). Significantly, the depleted fraction lacked the ability to repress viral translation when microinjected into Xenopus oocytes (Fig. 3A, right), suggesting that the 3Cpro domain is required for this inhibitory activity.
Figure 3.
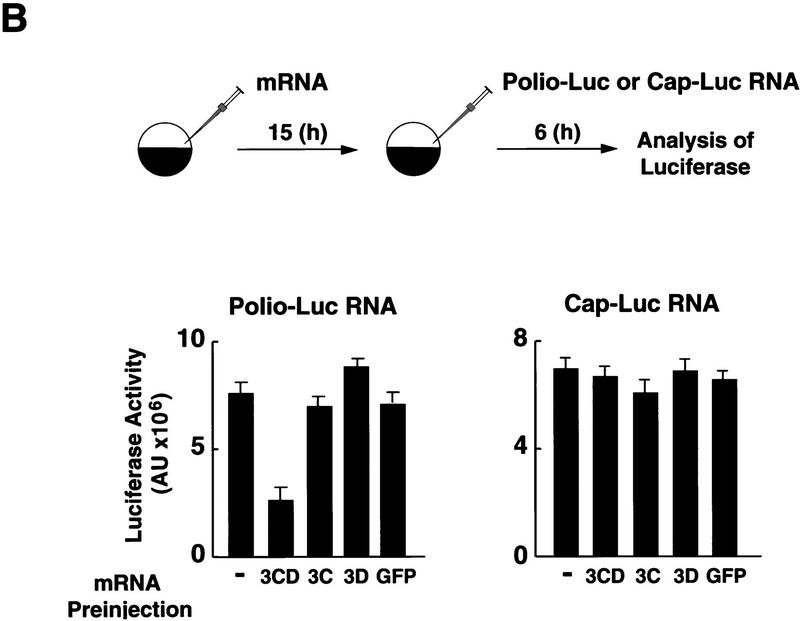
The polymerase–protease precursor, 3CD, represses viral translation. (A) Depletion of 3CD from infected cell extracts correlates with loss of translation inhibition. The viral protein 3CD was depleted from a partially purified infected HeLa fraction by affinity chromatography by use of an immobilized cloverleaf RNA (see Materials and Methods). (Left) Two microliters of 3CD-depleted extract (lane 2) and 2 μl of a nondepleted control (lane 1) were subjected to Western blot analysis as described for Fig. 2C. (Right) Luciferase activity was determined in oocyte extracts 3 hr after coinjection of Polio–Luc RNA with buffer, nondepleted control, or 3CD-depleted fractions as indicated in the bottom. (B) Overexpression of mutated 3CD (Gln-182 → Asn) in Xenopus oocytes inhibits poliovirus translation. (Top) Schematic diagram of the microinjection protocol. Oocytes were injected with 4 ng of a capped RNA encoding for 3CD, 3C, 3D, or an unrelated RNA encoding GFP, and incubated at 17°C for 15 hr. Then, oocytes were microinjected a second time with 40 ng of Polio–Luc or Cap–Luc RNA. Luciferase expression in oocytes was measured by enzymatic activity 6 hr after the second microinjection. Translation of Polio–Luc and Cap–Luc RNA was determined in oocytes that were preinjected with the mRNAs or with buffer control (−) as indicated at the bottom. Luciferase activity was expressed in arbitrary units (AU).
We next determined whether 3CD alone is sufficient to inhibit poliovirus translation. Four different proteins were expressed in Xenopus oocytes by preinjection of synthetic mRNAs encoding the corresponding polypeptide: 3Cpro, 3Dpol, a mutated 3CD with an alteration at the cleavage site between 3Cpro and 3Dpol (Gln 182 to Asn; Andino et al. 1993) that completely eliminates the autoproteolytic processing of the precursor 3CD; and an unrelated mRNA control encoding green fluorescent protein (GFP). Because expression of each protein reached the highest level between 10 and 15 hr after injection, as monitored by Western blot analysis (data not shown), we injected Polio–Luc and Cap–Luc RNA 15 hr after injection of the mRNAs. While translation of Cap–Luc RNA proceeded normally in oocytes expressing 3CD (Fig. 3B, right), the translation of Polio–Luc RNA was inhibited by 60% (Fig. 3B, left). In contrast, none of the other preinjections (3C, 3D, or GFP) had a significant effect on luciferase expressed by either Cap–Luc or Polio–Luc. Taken together, these results strongly suggest that the protease–polymerase fusion 3CD specifically inhibits viral cap-independent translation.
The cloverleaf RNA controls poliovirus translation
Because 3CD is a known RNA-binding protein that binds to the cloverleaf domain of the poliovirus 5′ UTR, we reasoned that 3CD might exert its inhibitory effect by interacting with this or other regulatory RNA elements. We have demonstrated previously that 3CD interacts specifically with the isolated cloverleaf to form a ternary ribonucleoprotein complex with a ribosome-associated cellular factor, PCBP (Andino et al. 1993; Gamarnik and Andino 1997; Parsley et al. 1997); but it was unknown whether 3CD interacts with other regions of the viral RNA. To examine other possible sites of 3CD interactions with the viral UTRs, we performed mobility-shift experiments using several defined domains of the poliovirus RNA as probes. The results obtained indicated that 3CD binds only to the cloverleaf RNA (A. Gamarnik and R. Andino, in prep.).
To charaterize further the regulatory role of 3CD and the cloverleaf, we examined whether the cloverleaf RNA directly participates in poliovirus translation. We have shown previously that disrupting the interaction of 3CD with the cloverleaf RNA affects positive-strand RNA synthesis without impairing viral translation (Andino et al. 1993). In that previous study, we observed a small enhancement of translation for mutants in which 3CD was unable to interact with the cloverleaf RNA. Those differences were originally interpreted as insignificant. However, because the results presented here strongly implicate 3CD in translational control, and because it has been shown previously that PCBP is a positive regulator of poliovirus translation (Gamarnik and Andino 1997; Parsley et al. 1997), we re-examined the importance of these RNA–protein interactions in the translation process. To this end, we designed Polio–Luc constructs containing cloverleaf mutations that specifically disrupted the binding of either 3CD or PCBP.
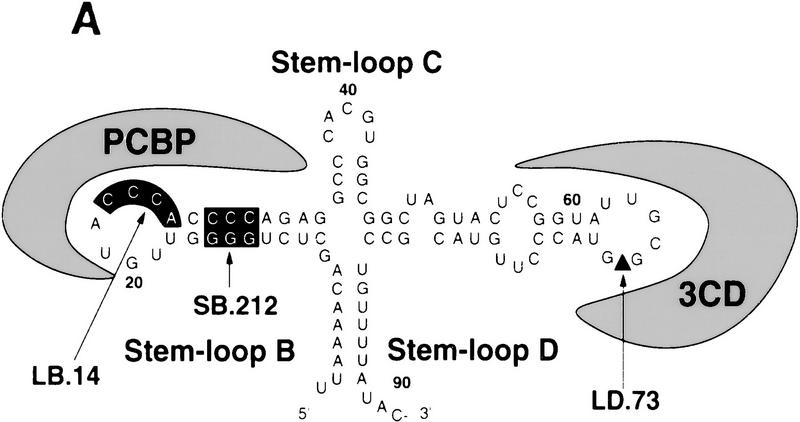
The polymerase precursor 3CD binds to stem–loop D of the cloverleaf RNA, whereas PCBP specifically interacts with stem–loop B (Fig. 4A) (Gamarnik and Andino 1997; Parsley et al. 1997). Three types of mutant RNAs were constructed: one with the entire cloverleaf deleted (ΔCL); a second type in which the interaction of the RNA with PCBP was either abolished by a 4-nucleotide deletion at the top of stem–loop B (LB.14) or reduced by a substitution in stem B (SB.212); and a third type in which the interaction of 3CD with the RNA was partially disrupted by a 4-nucleotide insertion at the top of stem–loop D (LD.73).
Figure 4.
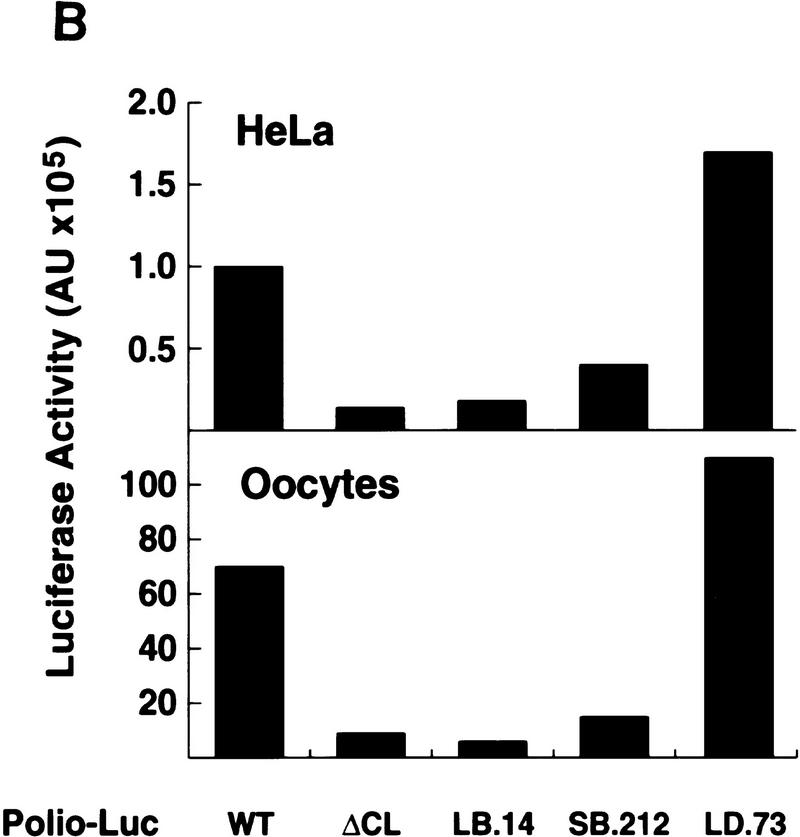
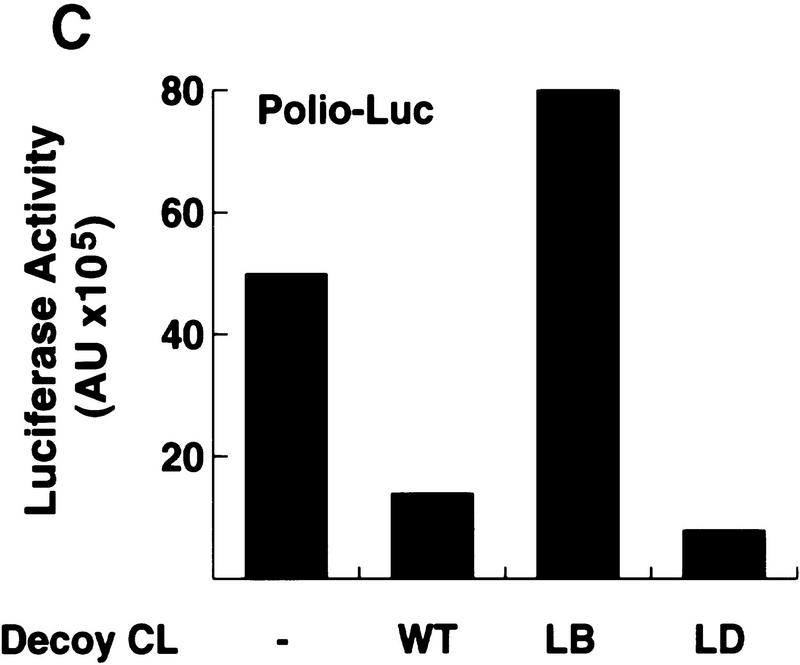
The cloverleaf structure formed at the 5′ end of the viral genome controls viral translation. (A) Schematic representation of the ribonucleoprotein complex formed around the cloverleaf RNA. The predicted cloverleaf structure is composed of stem–loop B (nucleotides 10–34), stem–loop C (nucleotides 35–45), and stem–loop D (nucleotides 51–78). Viral factor 3CD and cellular protein PCBP are shown interacting with their specific target sequences. The locations of the mutations introduced into the cloverleaf structure of the Polio–Luc RNAs are indicated by arrows: LB.14 (nucleotides 23–26, CCCA, were deleted in loop B); SB.212 (nucleotides 14–16, GGG, and nucleotides 28–30, CCC, were replaced with AAA and UUU, respectively, which maintain the stem B structure); and LD.73 (nucleotides GUAC were inserted in position 70 of loop D). (B) Luciferase activity produced by Polio–Luc constructs carrying wild-type or mutated cloverleaf structures. In vitro-transcribed Polio–Luc RNAs were either transfected into HeLa cells (top) or microinjected into Xenopus oocytes (bottom). The RNAs are indicated as WT (wild-type), ΔCL (cloverleaf-deleted), LB.14 (loop B muted), SB.212 (stem B mutated), and LD.73 (loop D mutant). Luciferase activity was measured in HeLa cell extracts 2 hr after electroporation and in oocyte extracts 10 hr after injection, and expressed in arbitrary units (AU). (C) Microinjection of decoy cloverleaf RNAs into Xenopus oocytes interferes with poliovirus translation. Wild-type Polio-Luc RNA was coinjected with buffer (−), 30 ng of wild-type cloverleaf (WT), or 30 ng of mutant cloverleaf decoys (LB, nucleotides C23 to A26 deleted, or LD, nucleotides GUAC inserted in position 70). Luciferase activity was determined in oocyte extracts 10 hr after injection.
Translation efficiencies of wild-type and mutant Polio–Luc RNAs were evaluated by measurement of luciferase activity produced as a function of time after transfection into HeLa cells or microinjection into Xenopus oocytes. These experiments were carried out under conditions in which luciferase activity was produced only by the input RNA. For the transfections into HeLa cells, luciferase activity was measured prior to RNA replication (Andino et al. 1993), while in Xenopus oocytes, the amount of newly synthesized RNA was negligible in comparison to the injected RNA. The Polio–Luc RNA construct with the deleted cloverleaf (ΔCL) or with the mutation that abolished PCBP binding (LB.14) translated at 10% of the efficiency of wild type (Fig. 4B). The Polio–Luc mutant with reduced binding to PCBP (SB. 212) translated at 40% of the efficiency of wild type. In contrast, the mutant with a deficiency in 3CD-cloverleaf interaction (LD.73) showed a substantial increase in viral translation (Fig. 4B). These results suggest that the binding site for PCBP within the cloverleaf structure is necessary for efficient viral translation. The involvement of the cloverleaf in translation was first postulated by Simoes and Sarnow (1991). In agreement with our results, these authors reported a poliovirus mutant with a 6-nucleotide insertion at the top of stem–loop B, which resulted in a significant decrease in viral translation (Simoes and Sarnow 1991). Furthermore, the increase of translation that we observed with LD.73 suggests that the inhibitory effect of 3CD may involve its binding to the cloverleaf structure.
The role of 3CD and PCBP in viral translation was evaluated further by competition experiments. We microinjected an excess of wild-type or mutated cloverleaf competitor together with the Polio–Luc reporter construct into Xenopus oocytes. We hypothesized that the free cloverleaf RNAs would interact with 3CD and/or PCBP, sequestering the proteins from their normal function in translation. Indeed, when wild-type or stem–loop D mutant RNA decoys were coinjected with Polio–Luc, we observed an 80% inhibition of luciferase production (Fig. 4C). Because both decoys have intact PCBP-binding sites, this result suggests further that PCBP is required for efficient translation. In contrast, a cloverleaf competitor carrying the stem–loop B mutation (unable to bind PCBP but fully capable of binding 3CD) did not decrease but rather stimulated viral translation, presumably by titrating out 3CD expressed by the Polio–Luc RNA. Taken together, these results indicate that the cloverleaf is a bifunctional element: In addition to its previously described function in RNA replication, it plays a central role in the regulation of poliovirus translation.
Synthesis of poliovirus negative-strand RNA in Xenopus oocytes
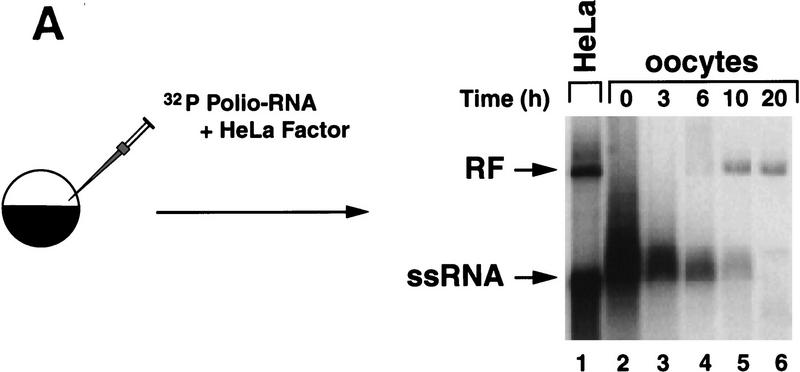
In previous studies, we analyzed the role of the cloverleaf structure in RNA synthesis using mutants that had defects in 3CD binding to the cloverleaf. These mutations yielded viable viruses that displayed a general reduction of RNA accumulation, in which the levels of positive-strand RNA seemed to be more compromised than those of negative strand (Andino et al. 1990a). Furthermore, we found that viruses carrying mutations in the cloverleaf that completely abrogated 3CD binding were unable to synthesize detectable quantities of either negative- or positive-strand RNA in HeLa cells (R. Andino, unpubl.). For these mutants, it was difficult to determine whether the synthesis of negative strand was affected directly by the disruption of the ternary complex or indirectly as a consequence of the lack of positive-strand synthesis. To overcome this problem, we developed a novel method using Xenopus oocytes that permits the analysis of negative-strand synthesis in the absence of positive-strand amplification. Briefly, 32P-labeled synthetic poliovirus RNA is microinjected into Xenopus oocytes, and the fate of the labeled positive-strand RNA is analyzed by native agarose gel electrophoresis. During viral replication three classes of RNAs accumulate in infected cells: single-stranded RNA (ssRNA), replicative intermediate (RI), and the fully double-stranded replicative form (RF) (for review, see Johnson et al. 1995). Because RI and RF are composed of positive- as well as negative-strand RNAs, the conversion of the input 32P-labeled RNA into these forms can be taken as an indicator of negative-strand synthesis.
When 32P-labeled positive-strand wild-type RNA was microinjected into oocytes, the input molecule was converted to a DNase- and RNase A-resistant RNA form with identical mobility to the RF obtained from HeLa cell crude replication complexes (Fig. 5A, lanes 5,6). Although the 32P-labeled input RNA was degraded over time, the amount of mononucleosides released is not sufficient to yield detectable newly synthesized ssRNA (Gamarnik and Andino 1996, and data not shown). Thus, the labeled RNA observed at later time points is presumably composed of the input 32P-labeled positive-strand and newly synthesized unlabeled negative-strand RNA.
Figure 5.
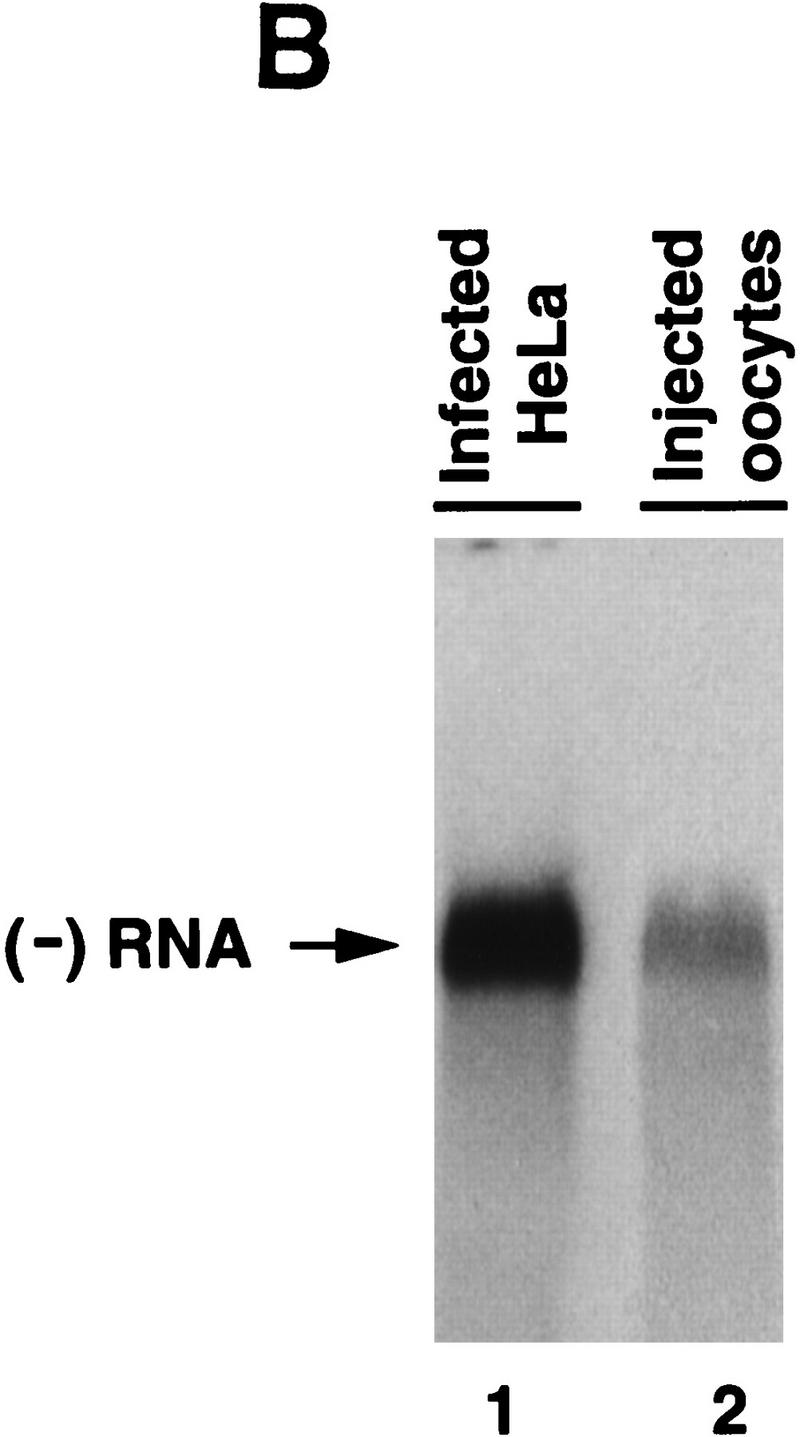
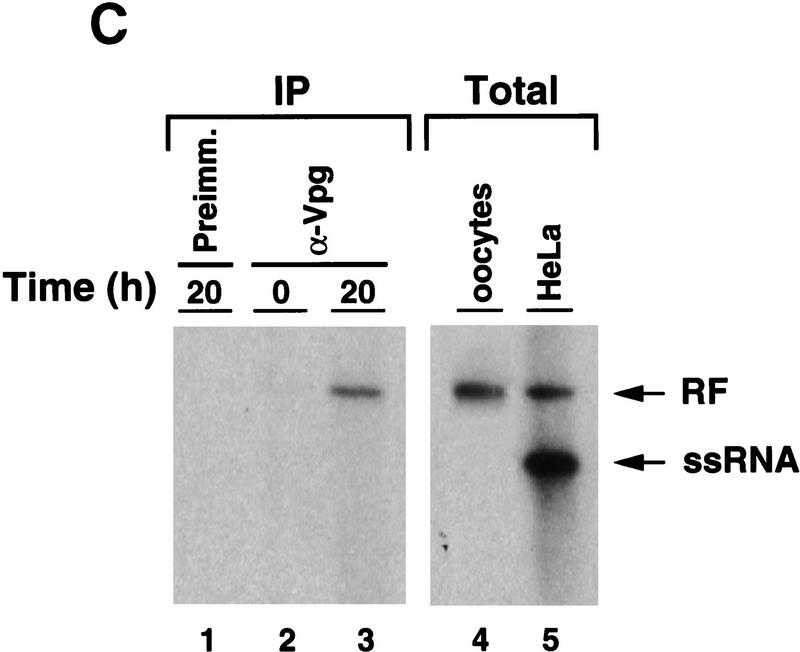
Synthesis of viral negative-strand RNA in Xenopus oocytes. (A) Oocytes were microinjected with in vitro-transcribed 32P-labeled poliovirus RNA together with HeLa S10 extracts (Gamarnik and Andino 1996), and the conversion of the input RNA into a double-stranded form was analyzed as a function of the time in 1% native agarose gels electrophoresis, as indicated on the top. For comparison, 32P-labeled poliovirus RNA was synthesized in crude replication complexes (CRC) obtained from poliovirus-infected HeLa cells (lane 1). Single stranded viral RNA (ssRNA) and the double stranded replicative form (RF), are indicated at left. (B) Northern blot analysis confirms that the oocytes produce negative-strand RNA. Oocyte extracts obtained from 200 oocytes at 20 hr after microinjection of unlabeled viral RNA were used to detect newly synthesized negative-strand RNA (lane 2). DNase- and RNase-treated samples were extracted with phenol–chloroform, ethanol precipitated, and resolved under denaturing conditions through 1% agarose gel electrophoresis. Then, RNA was transferred to a nylon filter, and hybridized with a specific probe complementary to the viral negative strand. As a control, infected HeLa extracts treated in similar conditions was analyzed (lane 1). (C) The replicative form synthesized in oocytes contains a covalently linked Vpg molecule. Oocytes injected with 32P-labeled poliovirus RNA were lysed at 0 and 20 hr post-injection (as described in Materials and Methods), immunoprecipitated with anti-Vpg antibodies (α-Vpg) or preimmune sera (Preimm.), and analyzed in 1% native agarose gel (lanes 1–3). Total RNA from oocytes injected with 32P-labeled poliovirus RNA obtained 20 hr after incubation (lane 4) and 32P-labeled RNA synthesized in crude replication complexes obtained from infected HeLa cells (lanes 5) are shown.
Next, we studied whether the newly synthesized double-stranded RNA in fact contains authentic poliovirus negative strand. RNA obtained from a large number of oocytes microinjected with unlabeled positive-strand RNA or from virus-infected HeLa cells was RNase and DNase treated and analyzed by Northern blotting after denaturing agarose gel electrophoresis. Both samples displayed a single RNA band of identical electrophoretic mobility that hybridized specifically with a poliovirus positive-strand RNA probe (Fig. 5B).
To characterize further the RF RNA produced in oocytes, we examined whether this molecule contains Vpg, a genome-linked viral peptide. During RNA replication, Vpg is added to the 5′ end of the growing RNA chains at a very early stage, possibly as a primer of RNA synthesis. This protein is found in both positive- and negative-strand RNAs, suggesting that a similar mechanism is responsible for initiation of synthesis of both strands (Flanegan et al. 1977; Pettersson et al. 1978). Therefore, in infected cells, the RF RNA contains Vpg linked to the 5′ end of both strands. However, the RF synthesized after one round of replication in oocytes should only carry Vpg attached to the 5′ end of the negative strand, because the positive strand is the product of T7 RNA polymerase transcription. As expected, phenol- and SDS-treated double-stranded RNA produced in oocytes was immunoprecipitated by antibodies directed against Vpg but not by pre-immune sera (Fig. 5C, lanes 1,3). In addition, anti-Vpg antibodies did not precipitate 32P-labeled RNA at early time points before double-stranded RNA was observed (Fig. 5C, lane 2), showing that the input ssRNA cannot be precipitated by anti-Vpg antibodies. These results indicate that the double-stranded RNA formed in oocytes contains negative-strand RNA covalently linked to Vpg, which closely resembles that produced in HeLa infected cells.
Binding of 3CD to the cloverleaf RNA is required for negative-strand RNA synthesis
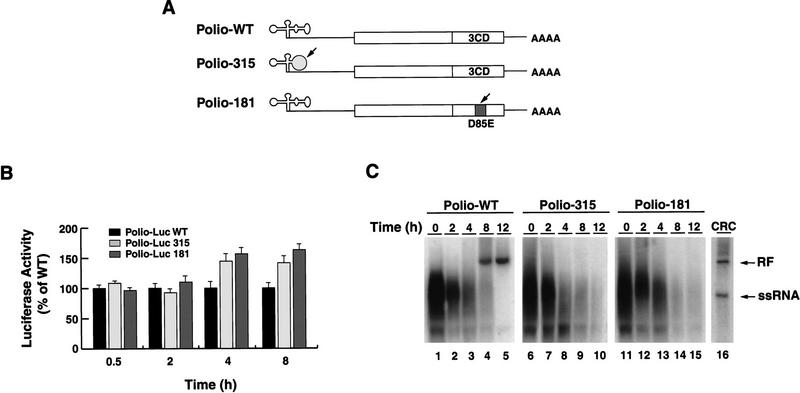
Using the method described in the previous section, we determined whether negative-strand RNA synthesis is affected by mutations that completely disrupt 3CD interaction with the cloverleaf. Two mutants were used (schematically represented in Fig. 6A), one in which the cloverleaf structure was modified by altering 2 bp at the top of stem–loop D (Polio-315), and the other with an alteration in the putative RNA-binding domain of 3CD, which was unable to bind to the cloverleaf RNA but displayed normal proteolysis (Polio-181). These mutants yielded no virus after transfection into HeLa cells, indicating that the mutation severely compromised viral replication.
Figure 6.
The interaction between 3CD and the cloverleaf RNA is required for negative strand RNA synthesis. (A) Schematic representation of wild-type poliovirus genome (Polio-WT), a mutant in the cloverleaf structure in which two Us in positions 60 and 68 were replaced by Cs to disrupt stem–loop D (Polio-315), and a mutant in 3CD-coding sequence in which the Asp-85 was replaced by Glu, which abrogates RNA binding (Polio-181). (B) Disrupting 3CD–cloverleaf interaction increases viral translation. Oocytes were microinjected with Polio–Luc WT (black bars), Polio–Luc 315 (white bars), or Polio-Luc 181 (gray bars) RNAs and incubated at 22°C. Cytoplasmic extracts were obtained, and luciferase activity was measured at the indicated times (0.5, 2, 4, and 8 hr). (C) Disruption of 3CD–cloverleaf interaction abolishes negative-strand RNA synthesis. Oocytes were microinjected with 32P-labeled Polio–WT (lanes 1–5), mutant Polio-315 (lanes 6–10), or mutant Polio-181 (lanes 11–15) and incubated at 30°C. Total RNA was extracted at 0 , 2, 4, 8, and 12 hr, and analyzed on 1% agarose gels. For comparison, 32P-labeled poliovirus RNA was synthesized in crude replication complexes (CRC) obtained from poliovirus-infected HeLa cells (lane 16). Double-stranded (ds) replicative form and single-stranded (ss) poliovirus RNAs are indicated.
We first studied the ability of these mutants to direct translation in the oocyte system. The amount of luciferase activity produced by the mutants during the first 2 hr postinjection was similar to that of wild type (Fig. 6B). In contrast, at 4 and 8 hr post-injection, both mutated RNAs were translated at higher levels than the wild-type RNA, once again indicating that the inability of 3CD to interact with the cloverleaf structure results in an increase in protein synthesis.
We next examined negative-strand RNA synthesis in both wild type and mutants. When 32P-labeled wild-type RNA was microinjected into oocytes, the input molecules were readily converted to an RF form (Fig. 6C, lanes 4,5). However, the RNAs corresponding to the mutants that were defective in 3CD binding to the cloverleaf were unable to synthesize RF (lanes 9,10,14,15), indicating that the interaction of 3CD with the cloverleaf RNA not only down-regulates viral translation but is essential for negative-strand RNA synthesis.
Discussion
The replication of positive-stranded RNA viruses presents an unresolved conundrum: How is the negative-strand RNA synthesized in the face of a wave of translating ribosomes moving in the opposite direction? We have studied this problem in the context of poliovirus replication and have found that actively translating ribosomes prevent RNA synthesis. Because each molecule of genomic RNA must be used for translation prior to RNA replication (Kuge et al. 1986; Collis et al. 1992; Novak and Kirkegaard 1994), the virus should have a mechanism to down-regulate translation to begin RNA synthesis. We found that an RNA element at the 5′ end of the viral genome, the cloverleaf RNA, contains overlapping signals for translation and RNA replication. The binding of the cellular protein PCBP to this RNA greatly enhances viral translation, while the binding of the viral polymerase precursor, 3CD, represses viral translation and promotes the synthesis of negative-strand RNA. We propose that these RNA–protein interactions determine the switch from translation to RNA replication.
Role of the cloverleaf RNA in translation and RNA replication
The results described here suggest that the cloverleaf coordinates the use of the viral RNA as a template for translation or RNA replication. We propose that after viral entry, the genomic RNA interacts with translation initiation factors to begin protein synthesis. Once a critical concentration of viral proteins is reached, 3CD binds to the cloverleaf RNA, shuts down viral translation and promotes negative-strand synthesis.
We examined the effects of mutations in the 3CD binding site in the cloverleaf or in the RNA-binding domain in 3CD on translation and negative-strand RNA synthesis. The results showed that the mutated viral RNAs, while translating more efficiently than wild type, do not accumulate negative-strand RNA (Fig. 6). This phenotype could result from the inability of these mutants to shut down translation, as predicted by our model. However, on the basis of the dramatic effect on negative-strand synthesis, it seems more likely that the cloverleaf participates in both repression of translation and initiation of RNA synthesis.
The cloverleaf structure was originally described as an element required for positive-strand RNA synthesis. This conclusion was drawn from the study of mutants with defects either in 3CD or in the cloverleaf that debilitated complex formation, which reduced the ratio of positive- to negative-strand RNA accumulated in infected cells (Andino et al. 1990a). In that previous study, we did not analyze nonviable mutants (unable to form 3CD–cloverleaf complexes) because of the lack of a sensitive method to study RNA synthesis. Here, using a novel assay that allows us to measure negative-strand synthesis independently of positive-strand amplification, we found that mutations that completely abrogate binding of 3CD to the cloverleaf impaired negative-strand synthesis.
It is possible that the defect on positive-strand synthesis detected previously was a consequence of a primary effect on negative-strand synthesis. On the other hand, it could be that, although, the interaction of the cloverleaf with 3CD is required for both initiation of positive- and negative-strand synthesis, each process has a different degree of dependence on complex formation. Thus, a partial defect in ribonucleoprotein complex formation could have a more profound effect on positive-strand than on negative-strand synthesis, resulting in the decreased ratio of positive to negative strands observed.
Repression of viral translation by the binding of 3CD to the cloverleaf RNA
Two sets of experiments presented in this report demonstrate that the binding of 3CD to stem–loop D of the cloverleaf RNA represses viral translation. First, we show that providing an excess of 3CD in trans specifically inhibits poliovirus translation without interfering with cap-dependent translation (Figs. 2 and 3). Second, we examined the effect of 3CD in cis produced by Polio–Luc RNA replicons. Mutations either in 3CD or the 3CD-binding site of the cloverleaf, resulted in an increase of the level of viral translation compare to those of wild type (Figs. 4 and 6).
In addition, we demonstrate that the cloverleaf RNA is involved in the positive regulation of viral translation. We show that the interaction of PCBP with stem–loop B of the cloverleaf enhances translation 10-fold (Fig. 4). This result is in agreement with recent studies that showed that PCBP is required for efficient poliovirus translation in HeLa cells and oocytes (Blyn et al. 1996, 1997; Gamarnik and Andino 1997; Parsley et al. 1997). Furthermore, there is increasing evidence that PCBP participates in translational control of cellular mRNAs (Holcik and Liebhaber 1997). PCBP is a component of an RNP complex that forms at the 3′ UTR of the human α-globin mRNA and determines its stability (Kiledjian et al. 1995). Also, PCBP appears to be responsible for translational silencing of the 15-lipoxigenase mRNA (Ostareck et al. 1997).
The molecular mechanism by which PCBP participates in cap-independent translation remains unknown. Perhaps the binding of PCBP to the viral 5′ UTR mediates interactions between the viral RNA and the translation machinery during the internal entry of ribosomes. How could 3CD repress viral translation? It is possible that the binding of 3CD to the cloverleaf alters the interaction of PCBP with this RNA element, which could interfere with the ability of PCBP to promote translation. Thus, the elucidation of the role of PCBP and 3CD in the regulation of viral translation may also clarify cellular pathways of translational control.
In addition, other viral and cellular factors may participate in the regulation of viral translation. For instance, it has been showed that the viral protein 3AB interacts in solution with 3CD and that the complex 3AB/3CD tightly binds to the cloverleaf (Harris et al. 1994). The relevance of this interaction in the regulation of viral translation and RNA replication warrants study.
Cell compartmentalization and translational control
Given that viral translation must stop to allow RNA synthesis to proceed, it is intriguing that viral protein synthesis in infected cells continues for several hours after RNA replication has already started (Levintow 1974). Poliovirus RNA is synthesized on membrane-associated structures. It has been speculated that membrane compartmentalization may sequester the replication machinery from the rest of the cytoplasm, thereby providing an adequate environment for RNA synthesis (Caliguiri and Tamm 1969; Bienz et al. 1987; Irurzun et al. 1992). According to our model, the local 3CD concentration in a given compartment could determine whether translation or replication will be favored. Can this compartmentalization maintain two separate pools of genomic RNAs, one used only for translation and the other for RNA replication? Previous experiments suggested that this is not the case. Viral RNA replication depends on translation of the genome in cis, that is, a particular viral genome must be used first as a template for translation to become competent for RNA synthesis (Novak and Kirkegaard 1994). Therefore, each molecule of viral RNA must be used as a template for both processes and regulation of the use of the RNA template would be need throughout the entire replicative cycle.
Control of translation and RNA replication in positive-stranded RNA viruses
In the proposed model, the repression of viral translation must ensure that all viral proteins required for RNA replication have been produced in sufficient quantities. Poliovirus proteins are expressed as part of a large polyprotein. Thus, each individual polypeptide accumulates in equimolar concentrations and, in principle, could act as a translation shut-off factor. Our results show that 3CD, the precursor of the poliovirus RNA polymerase, inhibits viral translation. Interestingly, the RNA phage Qβ uses the interaction of its RNA polymerase with Shine–Dalgarno sequences to control translation of the core protein (Kolakofsky and Weissmann 1971; Weber et al. 1972; Meyer et al. 1981), suggesting that animal RNA viruses and bacterial phages might use similar mechanisms to down-regulate translation.
Could this strategy be used by other eukaryotic RNA viruses? The entire poliovirus IRES can be replaced with corresponding sequences of different members of the Picornaviridae family such as coxsackievirus B3, rhinovirus 14, mengovirus, encephalomyocarditis virus, without major consequences for viral replication (Alexander et al. 1994; Rohll et al. 1994). Moreover, the same sequences can be replaced by the IRESs of other positive-stranded RNA viruses such as hepatitis C virus, a member of the Flaviviridae family (Lu and Wimmer 1996). These observations suggest that the mechanisms and factors that control the switch from translation to RNA replication in these viruses have been conserved. Furthermore, it is reasonable to speculate that even for viruses with capped genomic positive-stranded RNAs, translation and negative-strand synthesis are antagonistic. For these viruses however, a different mechanism for translational control is probably used. In conclusion, the results presented here provide insight into a general strategy by which positive-stranded RNA viruses might use common RNA structures for translation and initiation of RNA replication to coordinate these two processes.
Materials and methods
HeLa cell transfections, infections, and cytoplasmic extract preparation
To test the translation efficiencies of wild-type and mutant Polio–Luc RNAs, 100-mm dishes containing ∼3 × 106 HeLa cells were trypsinized and transfected with 10 μg of in vitro transcribed RNA per plate by standard electroporation procedures. After 2 hr of incubation at 37°C, cells were washed with PBS, scraped from the plates, and lysed in 200 μl of lysis buffer (Promega). Luciferase activity was measured in 10 μl of extract with a luciferase system as recommended by the manufacturer (Promega) and quantified by use of an Optocomp I luminometer.
For poliovirus infection, ∼4 × 108 HeLa cells grown in suspension were mock infected or infected with wild-type poliovirus at a multiplicity of infection of 30 pfu per cell. After absorption at room temperature for 30 min, 1 liter of fresh medium was added, and the cultures were incubated at 37°C for 6 hr. Then, the cells were collected by centrifugation and washed three times with cold PBS. The pellet was resuspended in 2 vols of hypotonic buffer (20 mm HEPES at pH 7.4, 10 mm KCl, 1.5 mm Mg(CH3CO2)2, 2 mm dithiothreitol), incubated on ice for 20 min, and homogenized by 20 strokes with a glass Dounce homogenizer. A postnuclear supernatant was obtained by centrifugation at 5000g for 10 min at 4°C. This supernatant was submitted to a second centrifugation (15,000g for 20 min) to obtain S10 cytoplasmic extract. Further centrifugation yielded a postribosomal supernatant (S100) and a ribosomal pellet (P100) as described previously (Brown and Ehrenfeld 1979). The fractions were supplemented with 5% glycerol and stored at −70°C.
Partially purified S100 fractions were depleted of 3CD by use of a biotinylated cloverleaf RNA. The cloverleaf RNA was transcribed in vitro in the presence of limiting concentrations of biotin–16-UTP to incorporate 2–3 biotinylated nucleotides per molecule of RNA. Thirty micrograms of this RNA was incubated with 40 μl of streptavidin beads and washed five times with PBS. Finally, 200 μl of the partially purified fraction containing 3CD was incubated with the beads for 1 hr on ice, and, after centrifugation, the supernatant was injected directly into Xenopus oocytes or analyzed by Western blotting with anti-3CD antibodies. The control sample was treated under the same conditions, except that biotin was not added during transcription of the cloverleaf RNA.
Microinjections in Xenopus oocytes
Oocytes were surgically isolated and enzymatically defolliculated as described previously (Gamarnik and Andino 1996). Manually sorted stage VI oocytes were injected with 20 nl of in vitro-transcribed Polio–Luc or Cap–Luc RNA (1 μg/μl) and 20 nl of the HeLa cell fraction, to provide the cellular factor essential for poliovirus translation in oocytes (PTF; Gamarnik and Andino 1996). Expression of 3C, 3D, 3CD, or GFP proteins in Xenopus oocytes was carried out by injection of a capped RNA encoding for the respective protein. The capped RNAs were obtained by in vitro-transcription with T7 polymerase. Injected oocytes were incubated for 15 hr at 17°C and injected a second time with 40 ng of Polio–Luc RNA or Cap–Luc RNA together with 20 nl of uninfected S10 HeLa cell extract to provide PTF. The effect of decoy cloverleaf RNAs (wild-type, loop B mutant, and loop D mutant) on viral translation was determined by coinjection of 10 nl of decoy RNA (3 μg/μl) or buffer control with 20 nl of Polio–Luc RNA together with 20 nl of S10 HeLa cell proteins. For measurement of luciferase expression, 10 oocytes were lysed in lysis buffer (20 μl per oocyte; Promega) and centrifuged for 5 min at 10,000 g. The supernatant (5 μl) was assayed by use of a luciferase system as described above.
To analyze negative-strand RNA synthesis, in vitro-transcribed 32P-labeled poliovirus RNA (30 ng) was microinjected into Xenopus oocytes together with 100 ng of S10 HeLa cell proteins. Oocytes were incubated at 30°C in a media containing 50 μg/ml of actinomycin D (Buller and White 1990). Thirty oocytes were lysed at various times in 400 μl of TENSK buffer (50 mm Tris-HCl at pH 7.5, 5 mm EDTA, 100 mm NaCl, 1% SDS, 200 μg/ml proteinase K), incubated at 37°C for 1 hr, extracted with phenol–chloroform, and precipitated with ethanol. Samples were resuspended in 50 μl of TE, treated with DNases and analyzed by electrophoresis through 1% native agarose gels and autoradiographed. rRNA, visualized by ethidium bromide, was used as an internal control for RNA extraction. Crude replication complexes were prepared as described previously (Takeda et al. 1986).
To analyze the presence of Vpg-linked RNA, 60 oocytes were injected with in vitro-transcribed 32P-labeled poliovirus RNA and processed as described above with the exception that proteinase K was omitted in the TENSK buffer and the incubation at 37°C was not performed. After phenol-chloroform extraction and ethanol precipitation (to remove noncovalently bound Vpg), the samples were diluted to 0.5 ml with NE buffer (50 mm Tris-HCl at pH 7.4, 100 mm NaCl, 0.02% NP-40), plus 10 μl of preimmune or anti-Vpg antibodies, and the mixture was incubated for 1 hr on ice. Then, 50 μl of protein A–agarose (Boehringer) equilibrated in NE buffer was added, and the mixture was incubated for 1 hr rocking at 4°C. After incubation, the samples were centrifuged at maximum speed for 10 sec, and the beads were washed four times with 1 ml of NE buffer. After the final wash, the beads were resuspended in NE buffer containing 1% SDS and removed by centrifugation. Ten micrograms of glycogen was added, and the samples were phenol extracted, ethanol precipitated, analyzed through 1% native agarose gels and autoradiographed.
For Northern blot analysis, 200 oocytes were injected with unlabeled poliovirus RNA (30 ng) together with 100 ng of S10 HeLa cell proteins. Oocytes were incubated at 30°C for 20 hr, lysed in TENSK buffer and incubated for 1 hr at 37°C. Then, the samples were extracted with phenol-chloroform, precipitated with ethanol, treated with DNases, separated on a denaturing agarose gel, transferred to a nylon filter, and hybridized with a specific probe complementary to the poliovirus negative-strand RNA. As a control an infected HeLa cell extract was treated under the same conditions as the oocyte extracts.
Translation/replication
Reticulocyte translation lysates were obtained from Promega. Thirty-five microliters of lysate was supplemented with 4 μg of S10 HeLa cell extract, a mixture of the 20 amino acids at 50 μm final concentration, and 4 μl of buffer 3D (50 mm HEPES at pH 8.0, 4 mm DTT, 3 mm Mg(CH3CO2)2, 5 μm ZnCl2, 0.1% NP-40). One microliter of Polio–Luc RNA was used as a template. A primer complementary to the 3′ UTR (CAATCCAATTCGACT) was annealed to the template by 5 min of incubation at 60°C. The translation reaction was initiated by incubating the mixture at 30°C with or without cycloheximide. After 15 min of incubation to allow for translation to begin, one-half of the translation reaction was combined with ATP, GTP, and CTP (0.25 mm), [32P]UTP (0.3 μCi, 25 μm final concentration), and 3 μl of a partially purified poliovirus polymerase. Both reactions (translation and RNA replication) were allowed to proceed at 30°C for 90 min; samples were removed every 15 min, nucleotide incorporation into RNA was determined by TCA precipitation, and translation was monitored by luciferase activity produced over time. Poliovirus polymerase was obtained from poliovirus-infected HeLa cells as described (Hey et al. 1986) and partially purified by means of a HiTrapQ chromatography (Pharmacia).
Acknowledgments
We are grateful to Judith Frydman, Alan Frankel, Elizabeth Blackburn, and members of Andino’s laboratory particularly to Nina Boddeker, Shane Crotty, and Debbie Silvera for their useful comments on the manuscript; and Amy Corder for graphics. This work was supported by funds provided by the Department of Microbiology and Immunology, University of California, San Francisco and U.S. Public Health Service grant AI40085 to R.A.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL andino@cgl.ucst.edu; FAX (415) 476-0939.
References
- Alexander L, Lu HH, Wimmer E. Polioviruses containing picornavirus type 1 and/or type 2 internal ribosomal entry site elements: Genetic hybrids and the expression of a foreign gene. Proc Natl Acad Sci. 1994;91:1406–1410. doi: 10.1073/pnas.91.4.1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andino R, Rieckhof GE, Baltimore D. A functional ribonucleoprotein complex forms around the 5′ end of poliovirus RNA. Cell. 1990a;63:369–380. doi: 10.1016/0092-8674(90)90170-j. [DOI] [PubMed] [Google Scholar]
- Andino R, Rieckhof GE, Trono D, Baltimore D. Substitutions in the protease (3Cpro) gene of poliovirus can suppress a mutation in the 5′ noncoding region. J Virol. 1990b;64:607–612. doi: 10.1128/jvi.64.2.607-612.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andino R, Rieckhof GE, Achacoso PL, Baltimore D. Poliovirus RNA synthesis utilizes an RNP complex formed around the 5′-end of viral RNA. EMBO J. 1993;12:3587–3598. doi: 10.1002/j.1460-2075.1993.tb06032.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton DJ, Flanegan JB. Coupled translation and replication of poliovirus RNA in vitro: Synthesis of functional 3D polymerase and infectious virus. J Virol. 1993;67:822–831. doi: 10.1128/jvi.67.2.822-831.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belsham GJ, Sonenberg N. RNA-protein interactions in regulation of picornavirus RNA translation. Microbiol Rev. 1996;60:499–511. doi: 10.1128/mr.60.3.499-511.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bienz K, Egger D, Pasamontes L. Association of polioviral proteins of the P2 genomic region with the viral replication complex and virus-induced membrane synthesis as visualized by electron microscopic immunocytochemistry and autoradiography. Virology. 1987;160:220–226. doi: 10.1016/0042-6822(87)90063-8. [DOI] [PubMed] [Google Scholar]
- Blyn LB, Swiderek KM, Richards O, Stahl DC, Semler BL, Ehrenfeld E. Poly(rC) binding protein 2 binds to stem-loop IV of the poliovirus RNA 5′ noncoding region: Identification by automated liquid chromatography-tandem mass spectrometry. Proc Natl Acad Sci. 1996;93:11115–11120. doi: 10.1073/pnas.93.20.11115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blyn LB, Towner JS, Semler BL, Ehrenfeld E. Requirement of poly(rC) binding protein 2 for translation of poliovirus RNA. J Virol. 1997;71:6243–6246. doi: 10.1128/jvi.71.8.6243-6246.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borman AM, Deliat FG, Kean KM. Sequences within the poliovirus internal ribosome entry segment control viral RNA synthesis. EMBO J. 1994;13:3149–3157. doi: 10.1002/j.1460-2075.1994.tb06613.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown BA, Ehrenfeld E. Translation of poliovirus RNA in vitro: Changes in cleavage pattern and initiation sites by ribosomal salt wash. Virology. 1979;97:396–405. doi: 10.1016/0042-6822(79)90350-7. [DOI] [PubMed] [Google Scholar]
- Buller AL, White MM. Functional acetylcholine receptors expressed in Xenopus oocytes after injection of Torpedo beta, gamma, and delta subunit RNAs are a consequence of endogenous oocyte gene expression. Mol Pharmacol. 1990;37:423–428. [PubMed] [Google Scholar]
- Caliguiri LA, Tamm I. Membranous structures associated with translation and transcription of poliovirus RNA. Science. 1969;166:885–886. doi: 10.1126/science.166.3907.885. [DOI] [PubMed] [Google Scholar]
- Chen CY, Sarnow P. Initiation of protein synthesis by the eukaryotic translational apparatus on circular RNAs. Science. 1995;268:415–417. doi: 10.1126/science.7536344. [DOI] [PubMed] [Google Scholar]
- Collis PS, O’Donnell BJ, Barton DJ, Rogers JA, Flanegan JB. Replication of poliovirus RNA and subgenomic RNA transcripts in transfected cells. J Virol. 1992;66:6480–6488. doi: 10.1128/jvi.66.11.6480-6488.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrenfeld E, Semler BL. Anatomy of the poliovirus internal ribosome entry site. Curr Top Microbiol Immunol. 1995;203:65–83. doi: 10.1007/978-3-642-79663-0_3. [DOI] [PubMed] [Google Scholar]
- Flanegan JB, Petterson RF, Ambros V, Hewlett NJ, Baltimore D. Covalent linkage of a protein to a defined nucleotide sequence at the 5′-terminus of virion and replicative intermediate RNAs of poliovirus. Proc Natl Acad Sci. 1977;74:961–965. doi: 10.1073/pnas.74.3.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamarnik AV, Andino R. Replication of poliovirus in Xenopus oocytes requires two human factors. EMBO J. 1996;15:5988–5998. [PMC free article] [PubMed] [Google Scholar]
- ————— Two functional complexes formed by KH domain containing proteins with the 5′ noncoding region of poliovirus RNA. RNA. 1997;3:882–892. [PMC free article] [PubMed] [Google Scholar]
- Harris KS, Xiang W, Alexander L, Lane WS, Paul AV, Wimmer E. Interaction of poliovirus polypeptide 3CDpro with the 5′ and 3′ termini of the poliovirus genome. Identification of viral and cellular cofactors needed for efficient binding. J Biol Chem. 1994;269:27004–27014. [PubMed] [Google Scholar]
- Hellen CU, Witherell GW, Schmid M, Shin SH, Pestova TV, Gil A, Wimmer E. A cytoplasmic 57-kDa protein that is required for translation of picornavirus RNA by internal ribosomal entry is identical to the nuclear pyrimidine tract-binding protein. Proc Natl Acad Sci. 1993;90:7642–7646. doi: 10.1073/pnas.90.16.7642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hey TD, Richards OC, Ehrenfeld E. Synthesis of plus- and minus-strand RNA from poliovirion RNA template in vitro. J Virol. 1986;58:790–796. doi: 10.1128/jvi.58.3.790-796.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holcik M, Liebhaber SA. Four highly stable eukaryotic mRNAs assemble 3′ untranslated region RNA- protein complexes sharing cis and trans components. Proc Natl Acad Sci. 1997;94:2410–2414. doi: 10.1073/pnas.94.6.2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irurzun A, Perez L, Carrasco L. Involvement of membrane traffic in the replication of poliovirus genomes: Effects of brefeldin A. Virology. 1992;191:166–175. doi: 10.1016/0042-6822(92)90178-r. [DOI] [PubMed] [Google Scholar]
- Jackson RJ, Kaminski A. Internal initiation of translation in eukaryotes: The picornavirus paradigm and beyond. RNA. 1995;1:985–1000. [PMC free article] [PubMed] [Google Scholar]
- Jang SK, Krausslich HG, Nicklin MJ, Duke GM, Palmenberg AC, Wimmer E. A segment of the 5′ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J Virol. 1988;62:2636–2643. doi: 10.1128/jvi.62.8.2636-2643.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson K, Sarnow P. Viral RNA synthesis. In: Rotbart H, editor. Human enterovirus infections. Washington, DC.: ASM; 1995. pp. 95–112. [Google Scholar]
- Kiledjian M, Wang X, Liebhaber SA. Identification of two KH domain proteins in the alpha-globin mRNP stability complex. EMBO J. 1995;14:4357–4364. doi: 10.1002/j.1460-2075.1995.tb00110.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolakofsky D, Weissmann C. Q replicase as repressor of Q RNA-directed protein synthesis. Biochim Biophys Acta. 1971;246:596–599. doi: 10.1016/0005-2787(71)90799-4. [DOI] [PubMed] [Google Scholar]
- Kuge S, Saito I, Nomoto A. Primary structure of poliovirus defective-interfering particle genomes and possible generation mechanisms of the particles. J Mol Biol. 1986;192:473–487. doi: 10.1016/0022-2836(86)90270-6. [DOI] [PubMed] [Google Scholar]
- Levintow L. The reproduction of picornaviruses. In: Fraenkel-Conrat H, Wagner R, editors. Comprehensive virology. New York, NY: Plenum Press; 1974. pp. 109–164. [Google Scholar]
- Lu HH, Wimmer E. Poliovirus chimeras replicating under the translational control of genetic elements of hepatitis C virus reveal unusual properties of the internal ribosomal entry site of hepatitis C virus. Proc Natl Acad Sci. 1996;93:1412–1417. doi: 10.1073/pnas.93.4.1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews MB. Interaction between viruses and the cellular machinery for protein synthesis. In: Hershey JW, Mathews MB, Sonenberg N, editors. Translational control. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1996. pp. 505–548. [Google Scholar]
- Meerovitch K, Svitkin YV, Lee HS, Lejbkowicz F, Kenan DJ, Chan EK, Agol VI, Keene JD, Sonenberg N. La autoantigen enhances and corrects aberrant translation of poliovirus RNA in reticulocyte lysate. J Virol. 1993;67:3798–3807. doi: 10.1128/jvi.67.7.3798-3807.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer F, Weber H, Weissmann C. Interactions of Q beta replicase with Q beta RNA. J Mol Biol. 1981;153:631–660. doi: 10.1016/0022-2836(81)90411-3. [DOI] [PubMed] [Google Scholar]
- Meyer K, Petersen A, Niepmann M, Beck E. Interaction of eukaryotic initiation factor eIF-4B with a picornavirus internal translation initiation site. J Virol. 1995;69:2819–2824. doi: 10.1128/jvi.69.5.2819-2824.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molla A, Paul AV, Wimmer E. Cell-free, de novo synthesis of poliovirus. Science. 1991;254:1647–1651. doi: 10.1126/science.1661029. [DOI] [PubMed] [Google Scholar]
- Novak JE, Kirkegaard K. Coupling between genome translation and replication in an RNA virus. Genes & Dev. 1994;8:1726–1737. doi: 10.1101/gad.8.14.1726. [DOI] [PubMed] [Google Scholar]
- Ostareck DH, Ostareck-Lederer A, Wilm M, Thiele BJ, Mann M, Hentze MW. mRNA silencing in erythroid differentiation: hnRNP K and hnRNP E1 regulate 15-lipoxygenase translation from the 3′ end. Cell. 1997;89:597–606. doi: 10.1016/s0092-8674(00)80241-x. [DOI] [PubMed] [Google Scholar]
- Parsley TB, Towner JS, Blyn LB, Ehrenfeld E, Semler BL. Poly (rC) binding protein 2 forms a ternary complex with the 5′- terminal sequences of poliovirus RNA and the viral 3CD proteinase. RNA. 1997;3:1124–1134. [PMC free article] [PubMed] [Google Scholar]
- Pelletier J, Kaplan G, Racaniello VR, Sonenberg N. Cap-independent translation of poliovirus mRNA is conferred by sequence elements within the 5′ noncoding region. Mol Cell Biol. 1988;8:1103–1112. doi: 10.1128/mcb.8.3.1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova TV, Hellen CU, Shatsky IN. Canonical eukaryotic initiation factors determine initiation of translation by internal ribosomal entry. Mol Cell Biol. 1996;16:6859–6869. doi: 10.1128/mcb.16.12.6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersson RF, Ambros V, Baltimore D. Identification of a protein linked to nascent poliovirus RNA and to the polyuridylic acid of negative-strand RNA. J Virol. 1978;27:357–365. doi: 10.1128/jvi.27.2.357-365.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogue GP, Huntley CC, Hall TC. Common replication strategies emerging from the study of diverse groups of positive-strand RNA viruses. Arch Virol Suppl. 1994;9:181–194. doi: 10.1007/978-3-7091-9326-6_18. [DOI] [PubMed] [Google Scholar]
- Roehl HH, Parsley TB, Ho TV, Semler BL. Processing of a cellular polypeptide by 3CD proteinase is required for poliovirus ribonucleoprotein complex formation. J Virol. 1997;71:578–585. doi: 10.1128/jvi.71.1.578-585.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohll JB, Percy N, Ley R, Evans DJ, Almond JW, Barclay WS. The 5′-untranslated regions of picornavirus RNAs contain independent functional domains essential for RNA replication and translation. J Virol. 1994;68:4384–4391. doi: 10.1128/jvi.68.7.4384-4391.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiroki K, Ishii T, Aoki T, Kobashi M, Ohka S, Nomoto A. A new cis-acting element for RNA replication within the 5′ noncoding region of poliovirus type 1 RNA. J Virol. 1995;69:6825–6832. doi: 10.1128/jvi.69.11.6825-6832.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simoes EA, Sarnow P. An RNA hairpin at the extreme 5′ end of the poliovirus RNA genome modulates viral translation in human cells. J Virol. 1991;65:913–921. doi: 10.1128/jvi.65.2.913-921.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda N, Kuhn RJ, Yang CF, Takegami T, Wimmer E. Initiation of poliovirus plus-strand RNA synthesis in a membrane complex of infected HeLa cells. J Virol. 1986;60:43–53. doi: 10.1128/jvi.60.1.43-53.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trono D, Pelletier J, Sonenberg N, Baltimore D. Translation in mammalian cells of a gene linked to the poliovirus 5′ noncoding region. Science. 1988;241:445–448. doi: 10.1126/science.2839901. [DOI] [PubMed] [Google Scholar]
- Weber H, Billeter MA, Kahane S, Weissmann C, Hindley J, Porter A. Molecular basis for repressor activity of Q replicase. Nat New Biol. 1972;237:166–170. doi: 10.1038/newbio237166a0. [DOI] [PubMed] [Google Scholar]