Abstract
It is now possible to slow and trap a single molecule of double-stranded DNA (dsDNA), by stretching it using a solid-state nanopore smaller in diameter than the double helix. By applying an electric force larger than the threshold for stretching, dsDNA can be impelled through the pore. Once a current blockade associated with a translocating molecule is detected, the electric field in the pore is switched in an interval less than the translocation time to a value below the threshold for stretching. According to molecular dynamics (MD) simulations, this leaves the dsDNA stretched in the pore constriction, while the B-form canonical structure is preserved outside the pore. In this configuration, the translocation velocity is substantially reduced from 1bp/10ns to ~1bp/2ms in the extreme, potentially facilitating high fidelity reads for sequencing, precise sorting, and high resolution (force) spectroscopy.
Keywords: nanopore, nanotube, DNA, single-molecule technique
1. Introduction
Atoms can be slowed and subsequently trapped for precise characterization and manipulation using optical intensity gradients and the corresponding light pressure forces.1 Molecules, on the other hand, have a more complex energy-level structure. Lacking a closed, two-level energy system, the molecular population dynamics are more difficult to control, which generally precludes manipulation by optical trapping directly. Instead, to manipulate them researchers have resorted to electrostatic fields defined by microfabrication on a chip2,3. Thus, just like for an optical trap, strong field gradients in this case controlled by the lithography are used to slow and trap molecules.
We are motivated to slow and trap a single DNA molecule for sequencing applications4 and for measurements of the electromechanics of DNA on the scale of a protein binding site (3–10nm)5,6—providing information that is vital for understanding how gene transcription, replication and silencing work. Applications like these, along with the structure of DNA, impose especially stringent specifications on the trap that can only be addressed with nanofabrication. B-form double-stranded DNA (dsDNA) is a stiff, highly charged polymer with a twisting, propeller-like, helical structure ~2nm in diameter and having an axial rise of 0.34nm per base-pair.7 By scrupulous design, the electric potential gradient in a nanometer-diameter pore can have comparable dimensions to dsDNA. Moreover, the force associated with the electric potential can be used to impel a dsDNA molecule to translocate through a pore, even if the diameter is smaller than the double helix.
It is especially timely to consider the prospects for trapping a single molecule of DNA in a nanopore. With the advent of next-generation-sequencing technologies, it has become exponentially harder to assemble a genome from short reads as the number of repeats grows. Nanopore sequencing is advantageous above all because it has the potential for very long reads, reducing the computational burden posed by alignment and genome assembly, while at the same time eliminating logistically challenging and error-prone amplification and library formation due to its exquisite single molecule sensitivity. Nanopore sequencing relies on the electrolytic current that develops when a DNA molecule, immersed in electrolyte, is forced by an electric field to translocate through a pore. Each nucleotide in the pore presents an energy barrier to the passage of ions, which blocks the current through the pore in a characteristic way. Prior work has established that there is a voltage threshold for permeation of dsDNA through d<3nm pores, corresponding to the force required to stretch the leading nucleotides in the pore.8, 9 For a bi-conical pore geometry, the leading edge of the dsDNA penetrates into the membrane to a constriction about d~2.5nm. If the differential force acting on the leading nucleotides is insufficient to stretch the helix, the translocation stalls there. It stalls because, while dsDNA has a helical structure ~2 nm in diameter, the solvated structure is about 2.6–2.9 nm in diameter (according to neutron scattering).7 As the bias increases and the differential force on the leading nucleotides exceeds that required to stretch dsDNA, the molecule is pulled towards the center of and eventually through the membrane. The confinement in pores having diameters 1.6<d<2.5nm causes the base-pairs to tilt.10
Following preliminary reports described elsewhere,11,12 we have developed a method that uses a time-varying electric field and the stretching transition to slow and trap a single molecule of dsDNA in a nanopore in a solid-state membrane. It is now possible to trap a single dsDNA molecule in a nanopore <3nm in diameter up to 2 minutes by first applying a voltage larger than this threshold and forcing the molecule to translocate through the pore. If the electric field is then rapidly switched to a value below threshold during the translocation, a single λ-DNA molecule becomes trapped for seconds to minutes in the pore, whereas the λ-DNA translocates in <1ms if the field is maintained above threshold. Molecular dynamics (MD) simulations of these experiments reveal that, when trapped, the dsDNA is stretched in the pore constriction in a specific tilted orientation, depending on the orientation of the leading nucleotides, while the B-form canonical structure is preserved outside the pore. Moreover, commensurate with the bandwidth of the nanopore, if the duration of the trap is extremely long, we observe in the ion current what may be signatures of the base-pairs translocating through the pore.
2. Experimental and Simulation Details
The fabrication of a nanopore in silicon nitride membranes 50μm×50μm is described elsewhere.13 Nominally, the thickness of the membrane is 30nm. To reduce the thickness to about 15nm, the membrane is uniformly etched in 20:1 H2O:49% HF for 30–40 min at room temperature. After a 10s O2 plasma clean, the thickness of the exposed nitride membrane is measured in-situ using Electron Energy Loss Spectroscopy, and then a nanometer-size pore is sputtered in it using a tightly focused (1.3nm spot-size) 9°α (cone angle), high energy (200kV) electron beam emanating from a JEOL 2010-F transmission electron microscope (TEM) operating in convergent beam diffraction mode, biased at 170μA emission current, using a 150mm condenser aperture. The sputtering time is typically about ~30s. Using TEM images taken at different tilt angles, we model the pore geometry as two intersecting cones (bi-conical) each with >20° cone angle.13
After sputtering, a membrane with a pore in it is subjected to a standard clean in (7:3) H2SO4:H2O2 for 3 minutes. Subsequently, the membrane chip is mounted in a two-chamber acrylic holder. Silicone Orings are used to seal the chip into the holder between the two chambers, leaving the nanopore as the only connection between the two chambers. The cis chamber has a volume of 100μL; the trans chamber has a volume of approximately 13mL. Voltage is applied and the DC, AC, current response and noise characteristics of the nanopore are measured in 100mM KCl solution at 23±1°C using Ag/AgCl electrodes (Warner) in each chamber. The electrolytic current is measured using an Axopatch 200B.
Switching the electric field at high-speed demands a nanopore with a high frequency response.14 So, we measured the current response of the pore and used it to create a comprehensive, small signal model derived from the nanostructure. The frequency response of nanopores was measured using a Signal Recovery 7280 lock-in amplifier. A small AC voltage signal (50 mV-rms amplitude) at various frequencies is applied to the cis chamber, and the in-phase and out-of-phase components of the membrane current or voltage are measured by a phase-sensitive lock-in technique. In the supplement, Fig. S1(a) shows the current frequency response of a 15nm thick membrane with a 2.5nm diameter pore in it. A detailed model that accurately accounts for the frequency response of the nanopore, which is rooted in the physical structure and reflects in a limited way the distributed nature of the electrical parameters is shown in Fig. S1(b).
From this analysis we deduce that the frequency response of these membranes cannot be accurately represented by the single capacitor model as suggested by Smeets et al15, but rather consists of a parallel combination of three high pass filters along with the pore resistance. Relying on the model, it is also possible to describe the relationship between the AC pore current and the AC voltage responses across the membrane. In the supplement, we show measurements and simulations of the voltage transient associated with an input voltage step from 1V to 0 applied across the Ag/AgCl electrodes spanning the membrane. The voltage decays with a time constant of about τ=210ns. According to the model, the membrane voltage tracks the input transient, but with a slower time constant: i.e. τ=460ns associated with the electrolyte resistance Rel=4.6kΩ and an effective capacitance Ceff=118pF, which is derived from the membrane window capacitance, Cwin=11.9pF in parallel with the series combination of the Si depletion capacitances Cdt =127pF and Cdb = 666pF.
MD simulations were performed following the protocols described elsewhere.8–10,16,17 All MD simulations were performed using the program NAMD2, a 1.0fs time step, periodic boundary conditions, and particle mesh Ewald electrostatics (grid spacing <0.15nm). In the simulations employing the CHARMM27 and BKS force fields, the van der Waals energies were computed using smooth 1.0–1.2 nm and 0.54–0.55 nm cutoffs, respectively.
Two nanopore systems with different cross sections were created. The first nanopore system used a modified version of the silica model described in Cruz-Chu et al.18 To produce the first nanopore system, a block of crystalline silica containing 13,000 silicon and 26,000 oxygen atoms was melted in an MD simulation using the modified BKS force field;19,20 an external potential expelled atoms from the region that would become the pore.21 The silica was annealed for 50ps at 7000K, 50ps at 500K, 50ps at 2000K, and 50ps at 300K; the temperature was controlled using Langevin thermostat with a damping coefficient of 5ps−1. The resulting pore had a shape of two intersecting cones each making a 20° angle with the pore axis. The minimum cross-section of the pore was a 2.6nm × 2.1nm ellipse located at the center of the 10-nm-thick membrane. In all further simulations, the atoms of the membrane were restrained to their positions at the end of the annealing process. Harmonic restraints of 13.9nN/nm were used to implement the restraints; additional harmonic bonds of 695pN/nm were applied between silicon and oxygen atoms within 0.22nm of each other. These bond and restraint force constants gave the membrane a dielectric constant of ~5. The charges of the silicon and oxygen atoms and their Lennard-Jones parameters were taken from Cruz-Chu et al.18 A 60-base-pair fragment of B-form dsDNA was inserted halfway through the pore. The 3′ and 5′ ends of the DNA fragment were covalently linked across the periodic boundaries, making the molecule effectively infinite.
The second nanopore system used a silicon nitride model described previously9,10. A β-Si3N4 crystal was replicated to produce a hexagonal prism of crystalline material with hexagonal side lengths of 4.56nm and a thickness of 10.5nm. A double cone pore having a circular cross-section with a minimum diameter of 2.0nm was cut into the hexagonal prism by removal of atoms.10 The cones made an angle of 10° with the pore axis. The charges of the silicon and nitrogen atoms and their Lennard-Jones parameters were taken from Wendel and Goddard.22 Bonds were added between adjacent atoms with the bond constants of 3470pN/nm.10 The atoms were constrained to their initial positions by restraints of 695pN/nm to obtain a dielectric constant of 7.5. To obtain a realistic conformation of dsDNA confined in the 2.0-nm pore, a 58-base-pair fragment of B-form dsDNA was simulated in solution under the influence of a phantom pore9 with the minimum radius being reduced from 2.8nm to 2.0nm over 6 ns. The ends of the molecule were not covalently linked as above.
Both DNA–nanopore structures were solvated with 100mM KCl electrolyte, forming a systems of about 120,000 atoms. The systems underwent 2000 steps of energy minimization and a 300-ps equilibrations with the pressure maintained at 1atm and the temperature maintained at 295°K. To ensure that the DNA conformation in the pore was fully relaxed, the 2.0nm silicon nitride pore system was further equilibrated at fixed volume for 7ns.
Both nanopore systems were then simulated with various external electrostatic biases applied. During these simulations, the volume of the system was fixed and the temperature was maintained at 295K by applying the Langevin thermostat to only atoms of the membrane with a damping coefficient of 1ps−1.
3. Results and Discussion
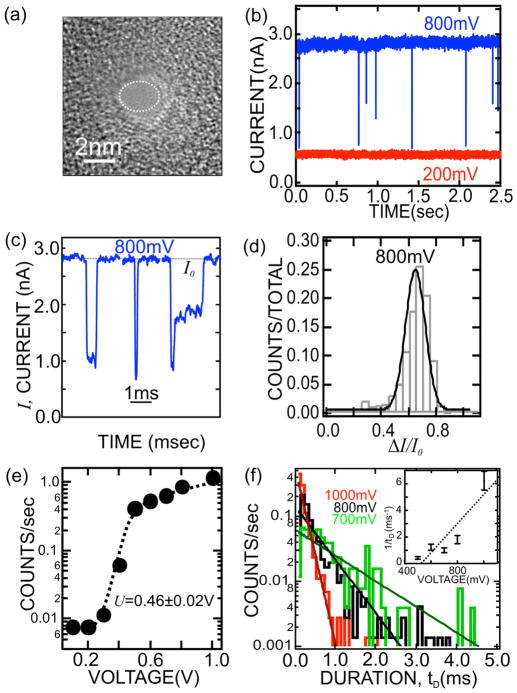
A TEM image of a nanopore with a 2.5×2.0±0.2nm cross-section—smaller than the DNA double helix—in a membrane 15.0±2.2 nm thick is shown in Figure 1(a). When λ–DNA is injected into the electrolyte at the negative (cis) electrode and 800mV applied across the membrane, current transients like those shown in Figure 1(b) are observed. The transients occur randomly as a function of time, but the inter-arrival time decreases with increasing concentration of DNA on the cis side of the bi-cell. There are a variety of current transients, some of which are illustrated in Figure 1(c). We speculate that the disparity in shape and duration of the current transients reflects the molecular configuration in the neighborhood of the pore and the time required to disentangle the DNA in solution into the single file of base-pairs required to permeate the pore.10 The distribution of transients can be represented by a blockade with a peak value at ΔI/I0=0.62±0.11 of the open pore current as shown in Figure 1(d). A blockade is caused by the reduction of the electrolytic current through the pore due to the translocation of DNA. In support of this interpretation, MD estimates also indicate a ΔI/I0=0.71±0.07 blockade through a 2.0nm diameter pore in 100mM KCl.
FIGURE 1.
(a) A TEM image of a 2.5×2.0nm cross-section pore in a silicon nitride membrane 15.0nm thick. (b) Electrolytic current measured in 100mM KCl at 800mV (blue) and 200mV (red) through the pore shown in (a) as a function of time. The frequency of blockades decreases dramatically with voltage; at 0.2V no transients are observed. (c) Three examples of current blockades observed in the pore shown in (a) at V=800mV under the same conditions as (b). These blockades are all associated with λ-DNA translocating through the pore. (d) The frequency of blockades observed at 800mV with a particular change in current normalized to the open pore current in the same pore. (e) The frequency of blockades observed with the 2.5nm pore as a function of membrane voltage, illustrating the frequency drop as voltage decreases below 0.5V. The dotted line represents a fit to the data. (f) Distributions illustrating the frequency as a function of the duration of a current blockade, tD, above threshold at 1.0V (red) 800mV(black) and 700mV(green). The distribution depends sensitively on the voltage. Inset: The reciprocal of the duration, tD−1, as a function of the applied voltage.
Figure 1(b) illustrates threshold behavior, showing a dearth of transients found in a current trace measured at 200mV compared to an 800mV trace. Figure 1(e) summarizes the voltage dependence of the frequency of blockade events over the range from 100mV to 1V. Generally, we observe an abrupt rise in the number of blockades over a range of ~200mV near a threshold that is sensitive to the pore diameter. If we assume each blockade corresponds to dsDNA permeating the pore, then the permeation rate can be described by the transition-state relation of the Kramers type: R=R0V/(1+exp[q*(U−V)/kT]), where R0 is a frequency factor, q*U is the effective barrier height, q*V is the reduction in the energy barrier due to the applied potential, and kT is the thermal energy.23 Using this relation, the data was fit and the results were overlaid on the scatter plot in Figure 1(e). We find a threshold of U=0.46±0.02V with q*=0.8±0.2e, which presumably corresponds to the force required to stretch the leading nucleotides in the pore—below this voltage DNA is not supposed to permeate the membrane.
The dependence of the blockade duration on the membrane voltage offers more support to the interpretation of the blockade current as a translocation across the membrane. Figure 1(f) shows the frequency of current transients associated with λ-DNA as a function of duration with the voltage as a parameter. If the blockade duration corresponds with the interval that DNA blocks the pore, then the average transient width tD signifies the time required for 48.502 kbp λ-DNA to translocate through the pore. We find that tD=0.16±0.01ms, 0.53±0.06ms, 1.1±0.1ms, 0.82±0.07ms and 2.49±0.25ms for voltages of 1.0V, 800mV, 700mV, 600mV and 500mV, respectively. The corresponding translocation velocity is 1bp/3.3ns at 1V and 1bp/11ns at 800mV to 1bp/50ns at 500mV, which is consistent with MD (see Figure 5). The inset to Figure 1(f) shows a plot of the voltage dependence of the reciprocal of the average transient width, i.e. 1/tD, measured above threshold. 1/tD vanishes near the threshold value. The line in the inset is a least-square fit to the data. It has a slope of 11V−1s−1 with a voltage-intercept of 0.53V that is comparable to the threshold voltage inferred from Figure 1(e).
FIGURE 5. MD simulation of the nanopore trap.
(a,b) Snapshots of the simulated systems that include dsDNA and water and ions (not shown) as well as a 2.6nm×2.1nm-cross section (a) or a 2.0nm-diameter (b) pore. The dsDNA within the constriction is stretched beyond 0.34nm/base-pair by 0–9% (larger pore) or 20–30% (smaller pore) depending on the external bias voltage. (c,d) The number of base-pairs permeating through the larger (c) or the smaller (d) pore in four MD simulations carried out at different biases. The simulations predict a voltage threshold between 500mV and 1.0V in both cases. (Insets) Normalized histogram of the DNA displacement in the pore constriction under a 0V bias. The solid line shows the distribution expected for a particle restrained by a harmonic spring of a 3.0nN/nm (larger pore) or a 7.2 nN/nm (smaller pore) spring constant.
These observations are in sharp contrast with prior work on larger diameter pores, and measurements that we performed on a pore with cross-section 3.6×3.2±0.2nm in a 31.5±2.0nm thick nitride membrane. In the supplement, we show that, if there is a threshold for permeating a 3.6×3.2nm pore, it is very low (U<17±11mV) and the transient widths tD=0.031±0.007ms, 0.032±0.007ms, 0.068±0.003ms, and 0.403±0.260ms for measured for transmembrane voltages of 600mV, 400mV, 200mV and 100mV respectively, correspond to translocation velocities ranging from 1bp/0.6ns to 1bp/8.3ns, which exceed the velocities found using smaller pores, and yet are consistent with prior estimates.24,25
The voltage-intercept for 1/tD near the threshold suggests that dsDNA can be trapped in a nanopore that is smaller in diameter than the double helix. To test this hypothesis first we forced dsDNA into a 2.5nm pore and then, when a blockade in the current is detected, reduced the transmembrane bias while the DNA was still in the pore. Once the dsDNA is in the pore, if the bias is reduced below the stretching threshold, the pore is supposed to act as a trap resisting the motion of the molecule.
Figures 2(a,b) show data demonstrating that it is possible to slow or weakly trap a single λ-DNA molecule in the pore by switching the electric field. First, a transmembrane bias of 800mV, which is above threshold according to Figure 1(d), is applied. Once the onset of a blockade like that shown in the inset to Figure 2(a) is detected by the differentiator, a programmed delay of 200μs is introduced before a latch switches the voltage from 800mV to 200mV—a value well below the threshold. All the while, the pore current is monitored. In the supplement we show that the transmembrane voltage tracks the voltage applied to the Ag/AgCl electrodes, but with a longer time constant (<500ns). Eventually, the blockade ends and the current returns to the open pore value, I0, but not before we observe a sharp transient like that shown near t=7.709 s in Figure 2(a).
FIGURE 2. Trapping a λ-DNA molecule in a nanopore.

(a) Upon detection of a current blockade indicating that dsDNA is translocating through the 2.5nm pore (shown in Figure 1(a)), the voltage across the pore is switched from 800mV (above the stretching threshold) to 200mV (below threshold). As a result, the duration of the current transient (blue) increases from ~200μs to about 6.79s. The transient observed at 7.709s is supposed to indicate a single λ-DNA molecule exiting the pore. The grey trace represents data taken with a bandwidth of 10kHz; the blue trace is the same data with a 1kHz (8-pole Bessel) filter. Inset: Detail showing the onset of a current transient and the 200μs built-in delay executed before triggering the voltage pulse. (b) Histograms showing the distribution of dwell times observed at 800mV (grey) and the distribution of elapsed time spanning the instant when a blockade event triggers the voltage switch from 800mV to 200mV to the return of the current to the open pore value seconds later (blue). The translocation time increases 1500x. (c) A magnified view of the pore current observed during (left) and after (right) the blockade showing the change in the magnitude of the current fluctuations. Histograms quantifying the fluctuations in the current distribution of the normalized current during the blockade (d) and for the open pore (e) at 200mV. The blockade current and open pore current fluctuations can be fit with Gaussians that have widths of σ=33.6pA and σ=18.3pA respectively.
We supposed that transients like these are indicative of the λ-DNA molecule exiting the pore after 6.8s. So, we reasoned that the current blockade observed during the time interval from t=0.918 to 7.709s must be evidence of a weakly trapped λ–DNA molecule in the pore. We do not observe a current blockade at 200mV if there is no onset of a blockade at 800mV. Moreover, as shown in the supplement, a larger (3.6×3.2nm) pore at a constant bias of 200mV shows a distribution of blockade durations that peaks near 200μs but does not exceed 1ms, making the long (6.8s) duration shown in Figure 2(a) extraordinary. If the molecule is weakly trapped in the pore, then the average translocation velocity must have slowed substantially to a value of about (48.5kbp – 200μs×1bp/11ns)/6.8s=1bp/224μs which is about 20,000x slower than the velocity estimate for tD obtained at 800mV.
After repeating this type of measurement hundreds of times on the same pore, a comparison between the distribution of the duration of blockades observed with a transmembrane bias of 800mV, and the time that expires between the triggered voltage switch from 800mV to 200mV and the return of the current to the open pore value reveals the dichotomy illustrated in Figure 2(b). While the peak in the distribution obtained at a constant bias of 800mV occurs near tD=200μs, the peak of the distribution found when the voltage is switched from 800mV to 200mV occurs near tD~500–600ms. This suggests that the translocation velocity at the peak must have slowed substantially to a value of about (48.5kbp – 200μs×1bp/11ns)/500ms=1bp/17μs, which is about 1500x slower than the velocity obtained at 800mV. In comparison, we do not observe blockades >4ms at constant bias of 800mV. Comparing the distribution of the normalized blockade current ΔI/I0 obtained at a constant bias of 800mV with the blockades measured after switching the voltage from 800mV to 200mV, we find that they overlap within the error. The mean blockade signal fit to a Gaussian is ΔI/I0=0.62±0.11 at 800mV constant bias and 0.46±0.07 when the bias is switched from 800mV to 200mV.
The fluctuations illustrated in Figures 2(c,d) supports the hypothesis that the pore current is blockaded by λ-DNA. We observed that for t<7.709s the amplitude of the current fluctuations increases relative to the open pore value found for t>7.709s. Figure 2(c) differentiates the current fluctuations during the blockade from the open pore current. Focusing on the data filtered with a low-pass (8 pole Bessel) 1kHz filter, the width of the histogram shown in Figure 2(d) taken from the blockaded current is σ=33pA (0.28I0) with χ2=0.52, while the open pore histogram measured over the same 0.5s interval is only σ=17pA (0.16I0) wide with χ2=0.43, indicating a signal beyond the noise for t<7.709 that is due to base-pairs translocating through the pore. To corroborate this assertion, we and others14,26 have measured the low frequency current noise spectral density associated with a pore in a membrane in the absence of DNA. We have determined that the noise spectrum scales with the pore resistance according to: , where I is the current through the device, f is the frequency, β ≈ 1 is an exponent and Rp is the pore resistance with n=1.06±0.15. Accounting for the blockade of ΔI/I0=0.5 and the change in pore resistance, the current noise should decrease overall from the open pore value, yet we observe an increase by 2x instead. Thus, the large fluctuating current is due to the trapped dsDNA.
We have made similar findings on six pores with similar geometries. Figure 3 summarizes the results obtained on a 2.6×2.0±0.2nm pore. The threshold voltage for this pore derived from the fit to the data shown in Fig. 3(b) is estimated to be about U=0.30±0.04V (with q*=1.1±0.2). Figure 3(c) shows an extraordinary event obtained from this pore, which represents a λ-DNA molecule trapped by first applying a bias of 600mV (above threshold) across the membrane and then, once the onset of a blockade is detected, switching the voltage to 100mV (below threshold). Eventually, the current returns to the open pore value, near t=58.8s in Figure 3(c). We assume that the transient at t=58.8s is indicative of the molecule exiting the pore after 56s, corresponding to a translocation velocity of >1bp/1.8ms—about 97,000x slower than the peak value obtained at a constant bias of 600mV.
FIGURE 3. Trapping a λ-DNA molecule in a nanopore.
(a) A TEM micrograph of a 2.6×2.0nm cross-section pore in a silicon nitride membrane 15nm thick. (b) The frequency of blockades observed with this pore as a function of membrane voltage illustrating the frequency drop as voltage decreases below 0.3V. The dotted line represents a fit to the data. (c) Triggered by the onset of a current blockade indicating that dsDNA is translocating through the pore of (a), the voltage is switched from 600mV (above the stretching threshold) to 100mV (below threshold). As a result the molecule is trapped in the pore till t=58.8s. (d) Histograms showing the distribution of dwell times observed at 600mV(grey) and the distribution of elapsed time spanning the instant when a blockade event triggers the voltage switch from 600mV to 150mV (blue) or 100mV(red) to the return of the current to the open pore value seconds later. The peak in the distribution of current blockade durations (blue) increases from about 900μs to about 200ms, increasing 200x. (e),(f) Histograms showing the distribution of the current during the blockade in the interval 14–14.5s, when λ-DNA is trapped (e), and the open pore for t>58.8s (f). The distribution for the trapped molecule must be fit to at least two Gaussians: one (solid blue) offset from the median (ΔI=0) by Δ I=+2.86pA with a width of σ=7.8pA; and another (solid red line) offset by −6.51pA with a width of σ=5.9pA. The black line represents the sum. In contrast, the data in (f) representing the open pore can be fit by a single Gaussian with a width σ=4.26pA.
For this pore, we also measured the blockade duration distribution at a constant bias of 600mV and when the voltage is switched from 600mV to 150mV and from 600mV to 100mV, 200μs after detecting the onset of a blockade. Similar to the data presented in Figure 2, we observed that the duration of the blockade is extended from an average of 900μs to 200ms—the velocity is ~200x slower as illustrated by Figure 3(d). In this case, the distribution of the normalized blockade current ΔI/I0 with a constant bias of 600mV and the blockades measure after switching the voltage from 600mV to 150mV or 100mV closely resemble each other—the former peaks at ΔI/I0=0.62±0.11, while the latter peak near ΔI/I0=0.62±0.15 and ΔI/I0=0.59±0.15 respectively. The apparent translocation velocity depends on the voltage applied after switching: i.e. the distribution found when we switch from 600mV to 150mV(100mV) has a peak duration near 150ms(200ms) respectively.
Trapping a DNA molecule should facilitate the high fidelity reads required for sequencing it. With the molecule trapped (i.e. for t<58.8s in Fig. 3(c)), we filtered the current data using a 10–600Hz bandpass filter and formed histograms of the current fluctuations using 0.5s windows. Each window in the trapped data (t<58.8s) shows a histogram similar to that shown in Figure 3(e), which can be represented by the superposition of two Gaussian distributions: one (solid blue) offset from the median (ΔI=0) by ΔI=+2.86pA with a width of σ=7.8pA; and another (solid red) offset by ΔI= −6.51pA with a width of σ=5.9pA. The goodness of fit, measured by the reduced chi-squared statistic, improves from χ2=5.04 for a single Gaussian fit to χ2=0.71 for two Gaussians, indicating that two Gaussians represent a superior model. This is in contrast to the data shown in Fig. 3(f), which represents the open pore current fit by a single Gaussian with a width σ=4.26pA with χ2=0.67 measured at the same voltage.
We tentatively attribute the separate peaks in Fig. 3(e) at ΔI=+2.86pA and ΔI= −6.51pA to partially resolved signals associated with C-G/G-C and A-T/T-A base-pairs, respectively. This identification is supported by the observation that λ-DNA has a nearly uniform distribution of base-pairs T-A, A-T, C-G, G-C. Thus, the currents corresponding to each type of base-pair should translate to approximately equal, distinct distributions in the blockade current convolved with the electrical noise. The difference between C-G and A-T base-pairs can be resolved in this case because of the correspondingly longer (~2ms) time each base-pair spends in the constriction compared with the (220μs) peak dwell time for the molecule in pore of Figure 2(a). However, apparently the signal-to-noise ratio is inadequate for discriminating C-G from G-C or A-T from T-A, despite the slowing. This assertion is also corroborated by even longer duration measurements of blockade currents associated with streptavidin bound, 100bp long, C-G and AT biotinylated duplexes trapped by the electric field in a pore in a configuration described elsewhere.27
While a smaller diameter increases the signal (i.e. blockade current), there is a concomitant increase in noise as well. Figure 4 summarizes the results obtained on the smaller pore, 2.2×1.8±0.2nm in cross-section, shown in (a). The threshold estimated from the voltage dependence of the frequency of blockade events given in Figure 4(b) is about U=0.48±0.04V, which is not substantially larger than the pore of Figure 1. The large blockade current estimated to be ΔI/I0=0.65±0.11 supports our contention that the pore diameter is smaller. In Figure 4(c) we show an analysis of an extraordinary event obtained from this pore, which represents λ-DNA trapped by first applying a bias of 500mV (near threshold) across the membrane, and then, once the onset of a blockade is detected, the voltage is switched to 100mV (below threshold). We observe that the current returns to the open pore value near t=70.6s and the molecule exits the pore after about 56.7s. With the molecule trapped, we filtered the current data using a 10–600Hz bandpass filter and formed histograms of the current fluctuations using 0.5s windows. The trapped data show in Figure 4(c) can be represented by the two Gaussian distributions: one (solid blue) offset from the median (ΔI=0) by ΔI=+7.01pA with a width of σ=17.8pA; and another (solid red) offset by ΔI=−6.92pA with a width of σ=18.0pA. While the offset are comparable to those observed in the 2.6nm pore, the width of the distributions are ~2.4x larger. The data shown in Figure 4(d), which represents the open pore current data fit by a single Gaussian with a width σ=10.1pA measured at the same voltage shows the same noisy trend.
FIGURE 4. Trapping a λ-DNA molecule in a nanopore.
(a) A TEM micrograph of a 2.2×1.8nm cross-section pore in a silicon nitride membrane 15nm thick. (b) The frequency of blockades observed with this pore as a function of membrane voltage illustrating the frequency drop as voltage decreases below 0.5V. The dotted line represents a fit to the data. (c) Histograms showing the distribution of dwell times observed at 500mV(grey) and the distribution of elapsed time spanning the instant when a blockade event triggers the voltage switch from 500mV to 100mV (blue) to the return of the current to the open pore value seconds later. The peak in the distribution of current blockade durations (blue) increases from about 900μs to about 1.5s, increasing 1500x. (d),(e) Histograms showing the distribution of the current during the blockade in the interval when λ-DNA is trapped (e), and the open pore (f). The distribution for the trapped molecule must be fit to at least two Gaussians: one (solid blue) offset from the median (ΔI=0) by ΔI=+7.01pA with a width of σ=17.7pA; and another (solid red line) offset by −6.92pA with a width of σ=18.0pA. The black line represents the sum. In contrast, the data in (f) representing the open pore can be fit by a single Gaussian with a width σ=10.1pA.
After repeating trapping experiment hundreds of times on the same pore, a comparison between the distribution of the duration of blockades observed at 500mV, and the time that expires between the triggered voltage switch from 500mV to 100mV and the return of the current to the open pore value reveals the dichotomy illustrated in Figure 4(e). This distribution is generally shifted toward an even longer time in this smaller pore in comparison with the larger pores of Figures 2(b) and 3(d), with blockades most frequently taking tD=1.5s, as opposed to 500ms or 200ms. This suggests that the translocation velocity at the peak must have slowed substantially to a value of about (48.5kbp – 200μs×1bp/19ns)/2s=1bp/53μs, which is about 1500x slower than the velocity obtained at 500mV.
MD simulations corroborate our interpretation of the experiments by showing that the motion of the dsDNA can be slowed or effectively stopped when the driving voltage is turned off. We created two models of nanopore systems to explore the differences between silica and silicon nitride pore surfaces that may be produced different experimental fabrication techniques, each having different pore dimensions. Figure 5(a) and (b) illustrate the simulated systems, which include fragments of dsDNA, 100mM KCl, and a nanopore having an ellipical 2.6nm×2.1nm cross-section (Figure 5(a)) or a circular 2.0nm diameter cross-section (Figure 5(b)). The molecular conformation within the 2.6nm×2.1nm pore was nearly unstretched at a bias of 0V, having a length per basepair that deviated little from its equilibrium value of 0.34nm. In contrast, the dsDNA within the 2.0nm pore was substantially stretched to 0.41nm per base-pair even at 0V. The distortion from the B-form dsDNA structure can be clearly seen in Figure 5(b). Under application of a 500mV transmembrane bias, the dsDNA was stretched to 0.37nm/bp in the larger pore and 0.44nm/bp in the smaller pore. As illustrated in Figure 5(c), at 250 and 500mV, the dsDNA’s motion in the 2.6nm×2.1nm pore was arrested following a small initial displacement allowed by stretching. Only when a bias of 1000mV was applied dsDNA transport was observed. Likewise, transport of dsDNA in the smaller pore was only observed for biases of 1000mV and above as shown in Figure 5(d). At the same bias, the rate of dsDNA transport was much higher for the larger pore.
The insets of Figures 5(c) and (d) characterize displacements of the base pair nearest to the pore constriction in the simulations performed at a 0V bias. The figures show normalized histograms of the displacement from the average position in the pore. The histograms have a Gaussian form, which implies that the pores act as harmonic traps for the dsDNA near the center of the pore with effective spring constants of 3.0±0.8 and 7.2±0.8nN/nm for the larger and smaller pores, respectively. The probability of an escape from a trap depends sharply on the force applied to the molecule, explaining the thresholds in Figures 5(c) and (d). The force required to restart the motion is essentially determined by the product of the spring constant, k, and the distance over which escape of a base-pair from the trap occurs x0: i.e. q*E ~kx0. The trap profile should be invariant for displacing dsDNA by one base-pair, so x0=L/2, where L is the length per base-pair. Therefore, we estimate kx0=510pN for the 2.6nm×2.1nm pore and kx0=1500pN for the 2.0nm pore.
4. Conclusions
In summary, it is now possible to slow a single molecule dsDNA by stretching it using a nanopore smaller in diameter than the double helix. According to MD simulations, the dsDNA is stretched in the pore constriction with the base-pairs tilted, while the B-form canonical structure is preserved outside the pore. In this configuration, the translocation velocity can be substantially reduced from 1bp/10ns to ~1bp/2ms in the extreme. We find that temporal current fluctuations associated with the molecule trapped in the pore are larger than the 1/f noise associated with electrolyte and unlike the open current; it cannot be fit to a single Gaussian. These fluctuations are tentatively attributed to the difference in current between C-G(G-C) and A-T(T-A) base-pairings in λ-DNA. One obvious problem with sequencing this way is determining which nucleotide is on which strand, e.g. distinguishing not just C-G from T-A, but C-G from G-C. MD simulations show that the base-pair tilt caused by the confinement is maintained during a translocation with the nucleotides of one strand always lagging their partners on the other.10 At low bias, the electric potential of the nucleotides in the tilted configuration presents a distinct energy barrier to the ions with a passage rate that is exponentially related to the height. So, the height difference for different sequences should have substantial effects on the current. However, work presented elsewhere27 indicates that it is difficult to discriminate A-T from T-A, and C-G from G-C for the conditions tested here.
Supplementary Material
Acknowledgments
We gratefully acknowledge frequent technical discussions with Winston Timp and Deqiang Wang and their critical reading of this manuscript. This work was funded by grants from National Institutes of Health [R01 HG003713A, PHS 5 P41-RR05969], Large Resource Allocation Committee [MCA05S028], the Petroleum Research Fund (48352-G6), and the National Science Foundation [TH 2008-01040 ANTC, PHY-0822613 and DMR-0955959].
References
- 1.Ashkin A. Optical trapping and manipulation of neutral particles using lasers. National Acad Sciences. 1997;94:4853–4860. doi: 10.1073/pnas.94.10.4853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Krüger P, Luo X, Klein MW, Brugger K, Haase A, Wildermuth S, Groth S, Bar-Joseph I, Folman R, Schmiedmayer J. Trapping and Manipulating Neutral Atoms with Electrostatic Fields. Physical Review Letters. 2003;91:233201. doi: 10.1103/PhysRevLett.91.233201. [DOI] [PubMed] [Google Scholar]
- 3.Meek SA, Bethlem HL, Conrad H, Meijer G. Trapping Molecules on a Chip in Traveling Potential Wells. Physical Review Letters. 2008;100:153003. doi: 10.1103/PhysRevLett.100.153003. [DOI] [PubMed] [Google Scholar]
- 4.Branton D, Deamer DW, Marziali A, Bayley H, Benner SA, Butler T, Di Ventra M, Garaj S, Hibbs A, Huang XH, Jovanovich SB, Krstic PS, Lindsay S, Ling XSS, Mastrangelo CH, Meller A, Oliver JS, Pershin YV, Ramsey JM, Riehn R, Soni GV, Tabard-Cossa V, Wanunu M, Wiggin M, Schloss JA. The potential and challenges of nanopore sequencing. Nat Biotechnol. 2008;26:1146–53. doi: 10.1038/nbt.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mirsaidov U, Timp W, Zou X, Dimitrov V, Schulten K, Feinberg AP, Timp G. Nanoelectromechanics of Methylated DNA in a Synthetic Nanopore. Biophys J. 2009;96:L32–L43. doi: 10.1016/j.bpj.2008.12.3760. [DOI] [PMC free article] [PubMed]
- 6.Zhao Q, Sigalov G, Dimitrov V, Dorvel B, Mirsaidov U, Sligar S, Aksimentiev A, Timp G. Stretching and Unzipping Nucleic Acid Hairpins Using a Synthetic Nanopore. Nano Lett. 2007;7(6):1680–1685. doi: 10.1021/nl070668c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lederer H, May RP, Kjems JK, Baer G, Heumann H. Solution Structure of a Short DNA Fragrment Studied by Neutron-Scattering. Eur J Biochem. 1986;161:191–196. doi: 10.1111/j.1432-1033.1986.tb10141.x. [DOI] [PubMed] [Google Scholar]
- 8.Heng JB, Aksimentiev A, Ho C, Marks P, Grinkova YV, Sligar S, Schulten K, Timp G. Stretching DNA using the electric field in a synthetic nanopore. Nano Lett. 2005;5:1883–1888. doi: 10.1021/nl0510816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heng JB, Aksimentiev A, Ho C, Marks P, Grinkova YV, Sligar S, Schulten K, Timp G. The electromechanics of DNA in a synthetic nanopore. Biophys J. 2006;90:1098–1106. doi: 10.1529/biophysj.105.070672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Comer J, Dimitrov V, Zhao Q, Timp G, Aksimentiev A. Microscopic Mechanics of Hairpin DNA Translocation through Synthetic Nanopores. Biophys J. 2009;96:593–608. doi: 10.1016/j.bpj.2008.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Timp W, Mirsaidov UM, Wang D, Comer J, Aksimentiev O, Timp G. Nanopore Sequencing: Electrical Measurements of the Code of Life. IEEE Trans Nanotechnology. 2010;9:281–294. doi: 10.1109/TNANO.2010.2044418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mirsaidov UM, Wang D, Timp W, Timp G. Molecular Diagnostics for Personal Medicine Using a Nanopore. WIRES Review Nanomedicine Nanobiotechnology. 2010;2:367–381. doi: 10.1002/wnan.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ho C, Qiao R, Heng JB, Chatterjee A, Timp RJ, Aluru NR, Timp G. Electrolytic transport through a synthetic nanometer-diameter pore. Proc Natl Acad Sci U S A. 2005;102:10445–50. doi: 10.1073/pnas.0500796102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dimitrov V, Mirsaidov U, Wang D, Sorsch T, Mansfield W, Miner J, Klemens F, Cirelli R, Yemenicioglu S, Timp G. Nanopores in solid-state membranes engineered for single molecule detection. Nanotechnology. 2010;21:065502. doi: 10.1088/0957-4484/21/6/065502. [DOI] [PubMed] [Google Scholar]
- 15.Smeets RMM, Keyser UF, Dekker NH, Dekker C. Noise in solid-state nanopores. Proc Natl Acad Sci U S A. 2008;105:417–421. doi: 10.1073/pnas.0705349105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aksimentiev A, Heng JB, Timp G, Schulten K. Microscopic kinetics of DNA translocation through synthetic nanopores. Biophys J. 2004;87:2086–97. doi: 10.1529/biophysj.104.042960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aksimentiev A. Deciphering ionic current signatures of DNA transport through a nanopore. Nanoscale. 2010;2:468–483. doi: 10.1039/b9nr00275h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cruz-Chu ER, Aksimentiev A, Schulten K. Water-silica force field for simulating nanodevices. Journal of Physical Chemistry B. 2006;110:21497–508. doi: 10.1021/jp063896o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vanbeest BWH, Kramer GJ, Vansanten RA. Force-Fields for Silicas and Aluminophosphates Based on Abinitio Calculations. Physical Review Letters. 1990;64:1955–8. doi: 10.1103/PhysRevLett.64.1955. [DOI] [PubMed] [Google Scholar]
- 20.Vollmayr K, Kob W, Binder K. Cooling-rate effects in amorphous silica: A computer-simulation study. Physical Review B. 1996;54:15808–27. doi: 10.1103/physrevb.54.15808. [DOI] [PubMed] [Google Scholar]
- 21.Wells DB, Abramkina V, Aksimentiev A. Exploring transmembrane transport through alpha-hemolysin with grid-steered molecular dynamics. Journal of Chemical Physics. 2007;127:125101. doi: 10.1063/1.2770738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wendel J, Goddard W. The Hessian biased force field for silicon nitride ceramics: predictions of thermodynamic and mechanical properties for a- and β-Si3N4. The Journal of chemical physics. 1992;97:5048–62. [Google Scholar]
- 23.Goychuk I, Hanggi P. Ion channel gating: A first-passage time analysis of the Kramers type. Proc Natl Acad Sci U S A. 2002;99:3552–3556. doi: 10.1073/pnas.052015699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fologea D, Uplinger J, Thomas B, McNabb DS, Li JL. Slowing DNA translocation in a solid-state nanopore. Nano Lett. 2005;5:1734–7. doi: 10.1021/nl051063o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Storm AJ, Storm C, Chen JH, Zandbergen H, Joanny JF, Dekker C. Fast DNA translocation through a solid-state nanopore. Nano Lett. 2005;5:1193–7. doi: 10.1021/nl048030d. [DOI] [PubMed] [Google Scholar]
- 26.Smeets R, Dekker N, Dekker C. Low-frequency noise in solid-state nanopores. Nanotechnology. 2009;20:095501. doi: 10.1088/0957-4484/20/9/095501. [DOI] [PubMed] [Google Scholar]
- 27.Wang D, Shim J, Timp W, Mirsaidov U, Comer J, Aksimentiev A, Timp G. Nature Nanotechnology submitted to. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.