Abstract
Background
Chronic seronegative hepatitis C virus (HCV) infection is defined as being HCV antibody (anti-HCV) negative, but HCV RNA positivity occurs in individuals infected with human immunodeficiency virus (HIV). However, associated factors are not well established because of the small number of reported cases.
Methods
Multivariate logistic regression analysis of HIV-infected subjects from 4 cohorts (Tien et al., 2006; Bonacini et al., 2001; George et al., 2002; and Hall et al., 2004) determined factors associated with HCV RNA positivity in anti-HCV–negative subjects. HCV enzyme immunoassay 2.0 was used to determine anti-HCV status.
Results
Among 1174 anti-HCV–negative, HIV-infected subjects, the prevalence of seronegative HCV infection was 3.2% (95% confidence interval [CI], 2.2%–4.3%). History of injection drug use (IDU; OR, 5.8; 95% CI, 2.7–12.8), higher alanine aminotransferase (ALT) level (OR, 2.0 per doubling; 95% CI, 1.3–3.2), and CD4 cell count <200 cells/μL (OR, 2.3; 95% CI, 1.1–4.8) were associated with HCV RNA positivity in anti-HCV–negative subjects. Among those with a history of IDU who had either a CD4 cell count <200 cells/μL or an ALT level greater than the upper limit of normal, the prevalence of seronegative HCV infection was 24% (95% CI, 13%–39%).
Conclusions
Detectable HCV RNA in the context of a negative HCV enzyme immunoassay 2.0 result in HIV-infected patients is low, but higher than the reported prevalence in HIV-uninfected patients. Our findings suggest that HCV RNA testing should be performed in anti-HCV–negative, HIV-infected patients, especially those with a history of IDU and either a CD4 cell count <200 cells/μL or an abnormal ALT level.
Liver disease is a leading cause of mortality for HIV-infected individuals in the antiretroviral therapy era, and hepatitis C virus (HCV) coinfection is a major factor [1, 2]. The prevalence of HCV coinfection is estimated to be 15%–40% among HIV-infected individuals [3–5]. However, higher rates of HCV coinfection have been reported in specific groups, including injection drug users [6]. Given the clinical significance of HCV coinfection, national guidelines recommend HCV antibody screening for all HIV-infected patients [7, 8].
However, false-negative results of serologic tests for HCV can occur in persons with acute HCV infection(during the period from exposure to HCV antibody seroconversion) and in those with chronic HCV infection. Chronic HCV viremia, or having detectable HCV RNA in serum (HCV RNA positivity) in the absence of detectable HCV antibodies (anti-HCV negativity), has been reported in HIV-uninfected [9–12] and HIV-infected individuals [13–17]. Experimental HCV infection of chimpanzees has also resulted in chronic HCV viremia without the development of HCV antibodies [18].
HIV-infected individuals may be at particular risk for seronegative HCV infection, possibly as a result of immunosuppression, with resultant failure to mount or maintain HCV antibody titers for detection by standard serodiagnostic tests [14, 17, 19]. The reported prevalence of chronic seronegative HCV infection among anti-HCV–negative, HIV-infected patients has varied between 0% and 13.2% [13, 15, 16, 20], depending upon the population studied and the HCV EIA used. Because the total reported number of HIV-infected patients with seronegative HCV infection is small, it has been difficult to establish the risk factors for seronegative HCV infection.
We determined the prevalence of chronic seronegative HCV infection in a large, multicenter, nationally representative cohort of HIV-infected men and women. Using data combined from that cohort and data from 3 previously described cohorts, we analyzed the predictors of HCV RNA positivity among anti-HCV–negative, HIV-infected subjects to determine which groups of anti-HCV–negative subjects are at risk for having HCV RNA positivity.
METHODS
One thousand one hundred seventy-five HIV-infected individuals from 16 HIV clinics across the United States enrolled in the Fat Redistribution and Metabolic Change in HIV infection (FRAM) study were studied. The FRAM study was initiated in 2000 to investigate the prevalence and correlates of fat distribution changes, insulin resistance, and hyperlipidemia in HIV-infected men and women and a comparison group of control men and women. Informed consent was obtained from all participants in accordance with guidelines for human experimentation of the US Department of Health and Human Services and the institutional review board of each participating institution. The recruitment and data collection procedures have been previously described [21]. FRAM study researchers examined and interviewed participants using a standardized protocol. Questionnaires asked about the participants’ risk factors for HIV and HCV acquisition, socioeconomic and demographic data, and medical history.
Blood samples were collected and measurements, including transaminase levels, HIV RNA level, and CD4 cell count were collected at a central laboratory (Covance Lab; Indianapolis, IN). HCV serologic tests, using the HCV EIA 2.0 (Abbott), and determination of HCV RNA levels, using the Bayer Versant 3.0 (bDNA) assay (lower limit of detection, 615 IU/mL), were later performed for all participants with available frozen serum samples. The HCV EIA 2.0 is the standard clinical HCV screening assay used at our institution. A case of seronegative HCV infection was defined as simultaneous anti-HCV negativity and HCV RNA positivity.
To increase our sample size of seronegative HCV-infected individuals, we identified 3 other cohorts [13, 15, 16] for inclusion in our analyses through an electronic literature search in the Embase and Medline databases for published human studies in peer-reviewed journals from 1985 to September 2005 that examined the prevalence and predictors of seronegative chronic HCV infection in HIV. These 3 cohorts were included because (1) anti-HCV testing (using the HCV EIA 2.0) and simultaneous HCV RNA testing were conducted on all subjects, (2) chronic HCV infection was confirmed by longitudinal testing using the HCV EIA 2.0 and HCV RNA [15, 16] or by estimating the time of HCV infection through self-reported history of injection drug use (IDU) [13], and (3) demographic data on the patients studied were available. One study [15] performed HCV RNA testing using both a commercial and noncommercial assay; only data obtained by commercial HCV RNA testing were included in the analysis.
The original data sets were obtained from the authors of each study. Data from 2 of the studies were collected from patients seen in HIV clinics in Iowa City, Iowa [15], and Los Angeles, California [13]. The authors of the Los Angeles study provided updated data collected from subsequent patients examined at the clinic. Therefore, the data presented here were not included in their original publication [13]. Data obtained from the third study were collected from participants enrolled during 1996–2000 in the Research and Access to Care for the Homeless (REACH) cohort, which longitudinally studied the HIV-infected, marginally housed population in San Francisco, California [16]. The data obtained from these 3 cohorts, combined with the FRAM study, provided a total of 37 cases of seronegative HCV infection for analysis.
Variables common to the pooled studies were screened, including age, ethnicity, sex, alcohol use (defined by current use [yes vs. no] in 3 studies [15, 16, 21] and lifetime use [yes vs. no] in the other study [13]), history of IDU (yes vs. no), alanine aminotransferase (ALT) level, HIV RNA level, and current CD4 cell count. ORs and 95% CIs for the rate of HCV RNA positivity among anti-HCV–negative subjects were calculated using unadjusted, univariate models of the raw data from each study to identify important variables. Upon examination, ALT level was found to be right-skewed and, thus, was log-transformed for all analyses. The linearity assumption was also tested for ALT level and other numeric variables. Sex was included in analysis due to a priori interest and importance and after testing indicated that model fit was improved when sex was included. Univariate testing indicated that IDU, ALT level, and CD4 cell count were significant at the .05 level. Current CD4 cell count was modeled as <200 versus ≥200 cells/μL. A pooled data set was constructed for analysis. A multivariate logistic regression model was then run using the pooled data, with variables identified from the univariate results plus a term for study. We controlled for study because of the possibility of confounding due to duration and treatment of HIV infection, which we were not able to control for directly.
Multiple tests were performed to investigate the issues of study homogeneity and the general goodness of model fit. We tested for significant interactions between the study and the other variables in the model. No interactions reached statistical significance at the .05 level. We also tested separately for interaction between the other variables in the model (sex, CD4 cell count, IDU, and ALT level). We then ran separate models with and without study interactions in the model to calculate likelihood ratio tests. A Breslow-Day test for homogeneity of the OR was also run as a further check for study homogeneity. For each model, tests were conducted for goodness of fit using the Hosmer-Lemeshow test [22]. Because all of these checks suggested no violation of study homogeneity, for both analyses, we used a fixed effects model to obtain summary estimates from the included studies. Finally, unselected factors were added back into the final model to see whether they were significant or affected model fit. No unselected factors had P < .20 when added to the final model. SAS software, version 9.1 [23], was used for all analyses.
RESULTS
Of the 1151 HIV-infected men and women in the FRAM cohort with samples available for anti-HCV and HCV RNA testing (table 1), 869 were anti-HCV negative, and of these, 15 (1.3%) of 1151 were HCV RNA positive. HCV RNA levels were similar between those who were anti-HCV positive and anti-HCV negative. Four of the 15 patients had documentation of prior HCV infection either by serologic testing or provider chart note. In the remaining 11 patients, 4 reported a history of IDU and were therefore considered to have acquired HCV infection at, or soon after, the time of initiation of IDU. We could not rule out acute infection as a possibility in the remaining 7 patients. These patients, however, were included in the analyses, to avoid bias in assessments of associations of IDU with presence of HCV antibody.
Table 1.
Summary of cohorts included in the pooled analysis.
| Characteristic | FRAM study | Bonacini et al. [13]b | George et al. [15] | Hall et al. [16] |
|---|---|---|---|---|
| Data collection, years | 2000–2002 | 1993–1999 | 1988–2000 | 1996–2000 |
|
| ||||
| Study population | HIV-infected participants of a national multicenter cohort of HIV-infected and HIV-uninfected patients | University of Southern California HIV and Hepatitis clinic patients | University of Iowa & Iowa City VA HIV clinic patients | Participants in the San Francisco Urban Poor cohort |
|
| ||||
| HIV-infected patients included in analysisa | ||||
| All | 1151 | 345 | 130 | 229 |
| Men | 810 (70) | 294 (85) | 106 (82) | 186 (81) |
| Women | 341 (30) | 51 (15) | 24 (18) | 43 (19) |
|
| ||||
| Anti-HCV positive | ||||
| All | 282 (25) | 221 (64) | 31 (24) | 147 (64) |
| HCV RNA positive | 236 (21) | 195 (57) | 30 (23) | 119 (52) |
| HCV RNA negative | 46 (4) | 26 (8) | 1 (1) | 28 (12) |
|
| ||||
| Anti-HCV negative | ||||
| All | 869 (75) | 124 (36) | 99 (76) | 82 (36) |
| HCV RNA positive | 15 (1.3) | 7 (2) | 6 (4.6) | 9 (3.9) |
| HCV RNA negative | 854 (74) | 117 (34) | 93 (72) | 73 (32) |
NOTE. Data are no. (%) of patients, unless otherwise indicated. Anti-HCV, hepatitis C virus antibody; FRAM, Fat Redistribution and Metabolic Change in HIV infection; HCV, hepatitis C virus.
Transgendered subjects were excluded from analysis.
Table includes a larger sample size than in original publication because of additional patient data collected after submission of original manuscript.
In the data sets from the 3 other cohorts, the number of HIV-infected patients ranged from 130 to 345 (table 1). Table 2 shows the demographic and clinical characteristics of the anti-HCV–negative, HIV-infected patients in each of the cohorts. Quantitative HCV RNA data were only available for the cohorts from FRAM and Hall and colleagues, and HCV RNA levels were similar between those who were anti-HCV positive and anti-HCV negative. Among the 1174 anti-HCV–negative subjects combined from all 4 cohorts, the prevalence of seronegative HCV infection was 3.2% (95% CI, 2.2%–4.3%).
Table 2.
Demographic and clinical characteristics of anti-HCV–negative, HIV-infected subjects.
| Characteristic | FRAM study (n = 869) | Bonacini et al. [13] (n = 124) | George et al. [15] (n = 99) | Hall et al. [16] (n = 82) |
|---|---|---|---|---|
| Sex | ||||
| Men | 640 (74) | 111 (90) | 82 (83) | 72 (88) |
| Women | 229 (26) | 13 (10) | 17 (17) | 10 (12) |
|
| ||||
| Ethnicity | ||||
| African American | 303 (35) | 21 (17) | 5 (5) | 36 (44) |
| Hispanic | 80 (9) | 65 (52) | 2 (2) | 6 (7) |
| Other | 18 (2) | 6 (5) | 9 (9) | 3 (4) |
| White | 467 (54) | 32 (26) | 83 (84) | 37 (45) |
|
| ||||
| Age, median years (IQR) | 41.0 (36.0–48.0) | 37.0 (30.5–45.0 | 38.0 (32.0–44.0) | 42.2 (36.1–49.4) |
|
| ||||
| Any history of IDU | 48 (6) | 9 (7) | 10 (10) | 26 (32) |
|
| ||||
| CD4 cell count, median cells/μL (IQR) | 380 (226–561) | 86 (14–257) | 212 (68–427) | 335 (230–522) |
|
| ||||
| CD4 cell count <200 cells/μL | 187 (22) | 85 (71) | 45 (45) | 19 (23) |
|
| ||||
| HIV RNA level per 1000 copies/mL, median copies/mL (IQR) | 0.4 (0.4–10.4) | 81.1 (34.4–225.9) | 35.9 (9.0–164.9) | 14.0 (0.9–75.0) |
|
| ||||
| ALT, median U/L (IQR) | 25.0 (17.0–38.0) | 82.5 (43.0–182.0) | 23.0 (16.0–32.0) | 22.0 (17.0–32.0) |
|
| ||||
| ALT level greater than the upper limit of normala | 178 (21) | 89 (73) | 16 (16) | 10 (12) |
|
| ||||
| HCV RNA level per 1000 IU/mL, median IU/mL (IQR) | 2313.4 (1.4–7119.1) | NA | NA | 10426.3 (4075.1–16215.9) |
NOTE. Data are no. (%) of patients, unless otherwise indicated. ALT, alanine aminotransferase; anti-HCV, hepatitis C virus antibody; FRAM, Fat Redistribution and Metabolic Change in HIV infection; HCV, hepatitis C virus; IDU, injection drug use; IQR, interquartile range; NA, not available.
Defined by the central laboratory of the FRAM study.
Predictors of testing HCV RNA positivity among the anti-HCV–negative participants
Univariate analyses of demographic and potential clinical predictors of HCV RNA positivity among anti-HCV–negative subjects for the 4 cohorts are shown in table 3. A history of IDU was associated with HCV RNA positivity in the FRAM and the Hall and colleagues cohorts. An increased ALT level was associated with HCV RNA positivity in the FRAM cohort, and a CD4 cell count <200 cells/μL was associated with HCV RNA positivity in the Hall and colleagues cohort. No significant predictors were identified in the Bonacini and colleagues and George and colleagues cohorts, but the OR for the factors IDU, ALT level, and CD4 cell count were consistent across studies, providing reassurance against study heterogeneity. Although African American race was associated with HCV RNA positivity in the FRAM cohort, it was not consistently associated in the other cohorts and was also not significant on multivariate modeling.
Table 3.
Univariate logistic regression analysis of factors associated with testing HCV RNA-positive in anti–HCV–negative subjects by cohort.
| Characteristic | FRAM study (n = 869)
|
Bonacini et al. [13] (n = 124)
|
George et al. [15] (n= 99)
|
Hall et al. [16] (n= 82)
|
||||
|---|---|---|---|---|---|---|---|---|
| Proportion of HCV RNA-positive/anti-HCV–negative patients | OR (95% CI) | Proportion of HCV RNA-positive/anti-HCV–negative patients | OR (95% CI) | Proportion of HCV RNA-positive/anti-HCV–negative patients | OR (95% CI) | Proportion of HCV RNA-positive/anti-HCV–negative patients | OR (95% CI) | |
| Sex | ||||||||
| Female | 3/229 | 0.7 (0.1–2.6) | 1/13 | 1.5 (0.0–13.7) | 1/16 | 1.04 (0.02–10.3) | 2/10 | 2.3 (0.2–15.3) |
| Male | 12/640 | Reference | 6/111 | Reference | 5/83 | Reference | 7/72 | Reference |
|
| ||||||||
| Ethnicity | ||||||||
| African American | 10/303 | 4.0 (1.1–17.4) | 0/21 | 0 | 1/12 | 1.42 (0.03–14.5) | 3/36 | 0.5 (0.1–2.5) |
| Hispanic | 1/80 | 1.5 (0.0–15.0) | 6/65 | 3.2 (0.4–150) | 0/2 | 0 | 0/6 | 0 |
| White | 4/467 | Reference | 1/32 | Reference | 5/83 | Reference | 6/37 | Reference |
|
| ||||||||
| Age (per decade) | NA | 1.0 (0.9–1.1) | NA | 0.9 (0.8–1.0) | NA | 1.0 (0.9–1.1) | NA | 1.0 (0.9–1.0) |
|
| ||||||||
| Any history of IDU | ||||||||
| Yes | 5/48 | 9.4 (2.4–31.6) | 1/9 | 2.2 (0.0–22.2) | 2/10 | 5.3 (0.41–43.2) | 6/26 | 5.3 (1.0–35.1) |
| No | 10/819 | Reference | 6/113 | Reference | 4/89 | Reference | 3/56 | Reference |
|
| ||||||||
| Current CD4 cell count | ||||||||
| <200 cells/μL | 4/187 | 1.3 (0.3–4.5) | 6/85 | 2.5 (0.3–119) | 4/45 | 2.54 (0.34–29.1) | 5/19 | 5.3 (1.0–29) |
| ≥200 cells/μL | 11/676 | Reference | 1/34 | Reference | 2/54 | Reference | 4/63 | Reference |
|
| ||||||||
| HIV RNA level | ||||||||
| >400 copies/mL | 10/407 | 2.3 (0.7–8.6) | 3/101 | 8a | 6/90 | 8a | 8/67 | 1.9 (0.2–90.2) |
| ≤400 copies/mL | 5/458 | Reference | 0/4 | Reference | 0/0 | Reference | 1/15 | Reference |
|
| ||||||||
| ALT level, U/L: log (per doubling) | NA | 3.8 (1.8–8.2) | NA | 1.54 (0.8–3.0) | NA | 2.37 (0.69–8.2) | NA | 1.1 (0.4–2.9) |
NOTE. ALT, alanine aminotransferase; FRAM, Fat Redistribution and Metabolic Change in HIV infection; HCV, hepatitis C virus; IDU, injection drug use; NA, Not applicable.
OR not estimable because of zero value in the numerator of the reference group for the proportion of HCV RNA-positive individuals among anti–HCV–negative individuals.
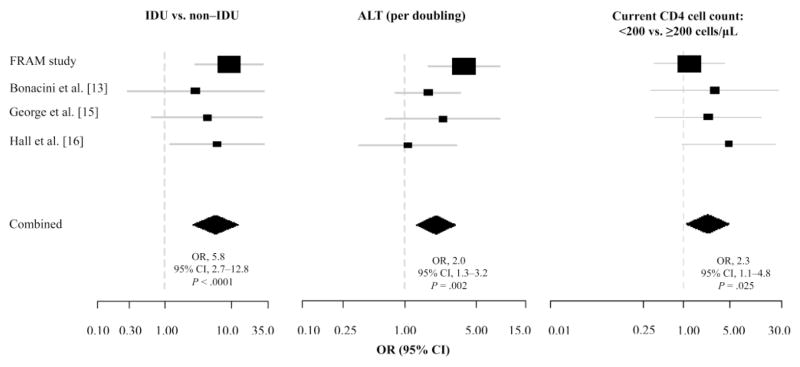
Multivariate analysis, which included history of IDU, ALT level, current CD4 cell count <200 cells/μL, and sex for each of the 4 cohorts and all of the cohorts combined, is shown in figure 1. For the combined data, a history of IDU (OR, 5.8; 95% CI, 2.7–12.8; P < .0001), increasing ALT level (OR, 2.0 per doubling; 95% CI, 1.3–3.2; P =.002), and a current CD4 cell count <200 cells/μL (OR, 2.3; 95% CI, 1.1–4.8; P =.025) were all independently associated with HCV RNA positivity among anti-HCV–negative patients.
Figure 1.
Multivariate logistic regression analysis of factors associated with testing hepatitis C virus (HCV) RNA positive among HCV antibody (anti-HCV)–negative subjects (by study and combined). Model controls for study were gender, injection drug use, alanine aminotransferase (ALT) level, and current CD4 cell count. Box size is proportional to study size. Location of box and whiskers denote estimate of OR and 95% CI, respectively. Diamond represents the OR and 95% CI for the combined studies.
Through multivariate analysis of each cohort, a history of IDU was associated with an increased odds of testing HCV RNA positive among anti-HCV–negative participants, but the association reached significance only in the FRAM (OR, 9.3; 95% CI, 2.9–29.5; P = .0002) and the Hall and colleagues cohorts (OR, 6.1; 95% CI, 1.2–31.0; P = .029). Similarly, an increasing ALT level and a CD4 cell count <200 cells/μL were associated with testing HCV RNA positive in each of the cohorts. However, for the association between ALT level and testing HCV RNA positive, statistical significance was reached only in the FRAM cohort (OR, 3.8 per doubling; 95% CI, 1.7–8.4; P = .001), and for CD4 cell count <200 cells/μL, the association did not reach significance in any of the individual cohorts.
Finally, among the 93 anti-HCV–negative subjects with a history of IDU combined from all 4 cohorts, the pooled prevalence of testing HCV RNA positive was 15% (95% CI, 8.5%–24%), but the pooled prevalence was highest at 24% (95% CI, 13%–39%) among the subgroup of those with a history of IDU who had either a CD4 cell count <200 cells/μL or an ALT greater than the upper limit of normal.
DISCUSSION
In the largest study to date, to our knowledge, to examine the prevalence and predictors of seronegative HCV infection, we found a pooled prevalence of seronegative HCV infection among anti-HCV–negative subjects of 3.2%. Although studies of HIV-infected [13] and HIV-uninfected [24, 25] individuals suggest that the HCV EIA 3.0 is a more sensitive assay, our findings of a low prevalence of seronegative HCV infection among anti-HCV–negative subjects suggest that the HCV EIA 2.0, which remains the standard clinical HCV screening assay used at most large hospital laboratories and at most of our institutions, is a reasonable screening tool to use for HIV-infected populations.
The FRAM cohort, which is representative of HIV-infected patients in medical care in the United States, demonstrated the lowest prevalence of seronegative HCV infection in anti-HCV–negative subjects among the previously reported studies using the HCV EIA 2.0. It should be noted that our study used commercially available HCV RNA tests to detect HCV infection. Some studies have reported HCV infection among anti-HCV–negative patients even in the absence of detectable HCV viremia in serum when more sensitive noncommercial methods to detect HCV RNA have been used for whole blood [15] and liver tissue [10] specimens. However, the 2% prevalence of seronegative HCV infection in the FRAM cohort remains higher than reported in HIV-uninfected individuals. Among US blood donors, seronegative HCV infection has been estimated to occur, on average, in 1 in 250,000 donors with the majority of cases being a result of acute HCV infection [25].
Although the prevalence of seronegative HCV infection appears low among HIV-infected subjects, we identified subgroups of anti-HCV–negative subjects who tested HCV RNA positive; particularly among persons with an elevated ALT level, a history of IDU, and a CD4 cell count <200 cells/μL. Our finding that an elevated ALT level was associated with being HCV RNA positive is not surprising, because an elevated ALT level may reflect HCV-related liver damage. Given the high pretest probability of HCV infection in those with a history of IDU, the finding of an association between a history of IDU and being HCV RNA positive is also expected. Finally, we establish that immunosuppression is associated with testing HCV RNA positive among anti-HCV–negative individuals. When we examined the predictors for testing anti-HCV negative among the HCV RNA–positive subjects to understand the limitations of standard HCV screening in HIV infection, we confirmed that immunosuppression is associated with testing anti-HCV negative among HCV RNA–positive individuals. It is noteworthy that for anti-HCV–negative, HIV-infected subjects with a history of IDU and either a CD4 cell count <200 cells/μL or an abnormal ALT level, we found a pooled prevalence of 24% for testing HCV RNA positive. Therefore, HCV RNA testing may be necessary for such individuals to exclude HCV infection.
A limitation of our study was that, in the FRAM cohort, we could not rule out acute HCV infection in 7 cases, although the ALT levels in these patients were not suggestive of acute HCV infection (range, 16–92 mg/dL). On the other hand, the anti-HCV–negative but HCV RNA–positive patients described by George et al. [15] and the majority of those described by Hall et al. [16] and Bonacini et al. [13] appeared to have chronic seronegative HCV infection, shown through longitudinal HCV EIA and HCV RNA testing of collected serum samples. Because of the nature of our study, HCV RNA testing was neither systematic nor of optimal sensitivity. The bDNA assay, which has a lower sensitivity to detect low HCV RNA levels, was used for the FRAM and the Hall et al. [16] cohorts. However, we do not believe that the use of the bDNA assay substantially underestimated the prevalence of seronegative HCV infection in these 2 cohorts. HIV-HCV–coinfected patients have been shown to have higher HCV RNA levels than HCV-monoinfected patients; therefore, low HCV RNA levels would be unexpected [26]. Finally, although the characteristics of the cohorts included in the study were not completely comparable, we found that the ORs for the most significant factors associated with testing HCV RNA positive were in a similar direction for all 4 cohorts, which provided reassurance regarding study comparability.
In summary, the low overall prevalence of seronegative HCV infection among those who were anti-HCV negative suggests that the HCV EIA 2.0 assay is a sufficiently sensitive screening tool to determine antibody status. Our findings suggest that HCV RNA testing should be performed in anti-HCV–negative, HIV-infected patients, especially those with a history of IDU and either a CD4 cell count <200 cells/μL or an abnormal ALT level.
Acknowledgments
We would like to thank the FRAM study investigators.
Financial support. The FRAM study was supported by the National Institutes of Health (NIH; RO1-DK57508, HL74814, HL 53359, and AI 027767) and the NIH General Clinical Research Center (M01-RR00036, RR00051, RR00052, RR00054, RR00083, RR0636, and RR00865). P.C.T. was supported by the National Institute of Allergy and Infectious Diseases (K23 AI 66943-01). The REACH Cohort was supported by the NIH (MH54907). D.R.B. received support from the National Institute on Alcohol Abuse and Alcoholism (AA015287).
Footnotes
Potential conflicts of interest. E.T.O. has been conducting research funded by Abbott, Bristol-Myers Squibb, Bavarian Nordic, Tibotec, GlaxoSmithKline, Merck, and Pfizer; has been on the speakers’ bureau for GlaxoSmithKline, Bristol-Myers Squibb, Tibotec, and Gilead; and has been a consultant for GlaxoSmithKline, Abbott, Roche, and Gilead. All other authors: no conflicts.
References
- 1.Bica I, McGovern B, Dhar R, et al. Increasing mortality due to end-stage liver disease in patients with human immunodeficiency virus infection. Clin Infect Dis. 2001;32:492–7. doi: 10.1086/318501. [DOI] [PubMed] [Google Scholar]
- 2.Martin-Carbonero L, Soriano V, Valencia E, Garcia-Samaniego J, Lopez M, Gonzalez-Lahoz J. Increasing impact of chronic viral hepatitis on hospital admissions and mortality among HIV-infected patients. AIDS Res Hum Retroviruses. 2001;17:1467–71. doi: 10.1089/08892220152644160. [DOI] [PubMed] [Google Scholar]
- 3.Backus LI, Phillips BR, Boothroyd DB, et al. Effects of hepatitis C coinfection on survival in veterans with HIV treated with highly active antiretroviral therapy. J Acqui Immune Def Syndr. 2005;39:613–9. [PubMed] [Google Scholar]
- 4.Sherman KE, Rouster SD, Chung RT, Rajicic N. Hepatitis C Virus prevalence among patients infected with human immunodeficiency virus: a cross-sectional analysis of the US adult AIDS Clinical Trials Group. Clin Infect Dis. 2002;34:831–7. doi: 10.1086/339042. [DOI] [PubMed] [Google Scholar]
- 5.Sulkowski MS, Mast EE, Seeff LB, Thomas DL. Hepatitis C virus infection as an opportunistic disease in persons infected with human immunodeficiency virus. Clin Infect Dis. 2000;30(Suppl 1):S77–84. doi: 10.1086/313842. [DOI] [PubMed] [Google Scholar]
- 6.Sulkowski M, Brinkley-Laughton S, Thomas D. HCV and HIV co-infection: prevalence, genotype distribution and severity of liver disease in an urban HIV clinic [abstract 204] Hepatology. 2000;32:212A. [Google Scholar]
- 7.2001 USPHS/IDSA guidelines for the prevention of opportunistic infections in persons infected with human immunodeficiency virus. HIV Clin Trials. 2001;2:493–554. doi: 10.1310/AQML-UABK-5LLB-E615. [DOI] [PubMed] [Google Scholar]
- 8.Tien PC. Management and treatment of hepatitis C virus infection in HIV-infected adults: recommendations from the Veterans Affairs Hepatitis C Resource Center Program and National Hepatitis C Program Office. Am J Gastroenterol. 2005;100:2338–54. doi: 10.1111/j.1572-0241.2005.00222.x. [DOI] [PubMed] [Google Scholar]
- 9.Arrojo IP, Pareja MO, Orta MD, et al. Detection of a healthy carrier of HCV with no evidence of antibodies for over four years. Transfusion. 2003;43:953–7. doi: 10.1046/j.1537-2995.2003.00445.x. [DOI] [PubMed] [Google Scholar]
- 10.Castillo I, Pardo M, Bartolome J, et al. Occult hepatitis C virus infection in patients in whom the etiology of persistently abnormal results of liver-function tests is unknown. J Infect Dis. 2004;189:7–14. doi: 10.1086/380202. [DOI] [PubMed] [Google Scholar]
- 11.Schmidt WN, Wu P, Cederna J, Mitros FA, LaBrecque DR, Stapleton JT. Surreptitious hepatitis C virus (HCV) infection detected in the majority of patients with cryptogenic chronic hepatitis and negative HCV antibody tests. J Infect Dis. 1997;176:27–33. doi: 10.1086/514033. [DOI] [PubMed] [Google Scholar]
- 12.Stapleton JT, Klinzman D, Schmidt WN, et al. Prospective comparison of whole-blood- and plasma-based hepatitis C virus RNA detection systems: improved detection using whole blood as the source of viral RNA. J Clin Microbiol. 1999;37:484–9. doi: 10.1128/jcm.37.3.484-489.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bonacini M, Lin HJ, Hollinger FB. Effect of coexisting HIV-1 infection on the diagnosis and evaluation of hepatitis C virus. J Acquir Immune Defic Syndr. 2001;26:340–4. doi: 10.1097/00126334-200104010-00008. [DOI] [PubMed] [Google Scholar]
- 14.Cribier B, Rey D, Schmitt C, Lang JM, Kirn A, Stoll-Keller F. High hepatitis C viraemia and impaired antibody response in patients coinfected with HIV. AIDS. 1995;9:1131–6. doi: 10.1097/00002030-199510000-00003. [DOI] [PubMed] [Google Scholar]
- 15.George SL, Gebhardt J, Klinzman D, et al. Hepatitis C virus viremia in HIV-infected individuals with negative HCV antibody tests. J Acquir Immune Defic Syndr. 2002;31:154–62. doi: 10.1097/00126334-200210010-00005. [DOI] [PubMed] [Google Scholar]
- 16.Hall CS, Charlebois ED, Hahn JA, Moss AR, Bangsberg DR. Hepatitis C virus infection in San Francisco’s HIV-infected urban poor. J Gen Intern Med. 2004;19:357–65. doi: 10.1111/j.1525-1497.2004.30613.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marcellin P, Martinot-Peignoux M, Elias A, et al. Hepatitis C virus (HCV) viremia in human immunodeficiency virus-seronegative and -seropositive patients with indeterminate HCV recombinant immunoblot assay. J Infect Dis. 1994;170:433–5. doi: 10.1093/infdis/170.2.433. [DOI] [PubMed] [Google Scholar]
- 18.Bassett SE, Brasky KM, Lanford RE. Analysis of hepatitis C virus-inoculated chimpanzees reveals unexpected clinical profiles. J Virol. 1998;72:2589–99. doi: 10.1128/jvi.72.4.2589-2599.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sorbi D, Shen D, Lake-Bakaar G. Influence of HIV disease on serum anti-HCV antibody titers: a study of intravenous drug users. J Acquir Immune Defic Syndr Hum Retrovirol. 1996;13:295–6. doi: 10.1097/00042560-199611010-00014. [DOI] [PubMed] [Google Scholar]
- 20.Thio CL, Nolt KR, Astemborski J, Vlahov D, Nelson KE, Thomas DL. Screening for hepatitis C virus in human immunodeficiency virus-infected individuals. J Clin Microbiol. 2000;38:575–7. doi: 10.1128/jcm.38.2.575-577.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tien PC, Benson C, Zolopa AR, Sidney S, Osmond D, Grunfeld C. The study of fat redistribution and metabolic change in HIV infection (FRAM): methods, design, and sample characteristics. Am J Epidemiol. 2006;163:860–9. doi: 10.1093/aje/kwj111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hosmer DJ, Lemeshow S. Applied logistic regression. 2. New York: John Wiley & Sons; 2000. [Google Scholar]
- 23.SAS/STAT user’s guide, version 9.1. Cary, NC: SAS Institute; 2004. [Google Scholar]
- 24.Galel SA, Strong DM, Tegtmeier GE, et al. Comparative yield of HCV RNA testing in blood donors screened by 2.0 versus 3. 0 antibody assays. Transfusion. 2002;42:1507–13. doi: 10.1046/j.1537-2995.2002.00236.x. [DOI] [PubMed] [Google Scholar]
- 25.Stramer SL, Glynn SA, Kleinman SH, et al. Detection of HIV-1 and HCV infections among antibody-negative blood donors by nucleic acid-amplification testing. N Engl J Med. 2004;351:760–8. doi: 10.1056/NEJMoa040085. [DOI] [PubMed] [Google Scholar]
- 26.Thomas DL, Astemborski J, Vlahov D, et al. Determinants of the quantity of hepatitis C virus RNA. J Infect Dis. 2000;181:844–51. doi: 10.1086/315314. [DOI] [PubMed] [Google Scholar]