Abstract
Rem2 is a member of the Rad/Rem/Rem2/Gem/Kir (RGK) subfamily of small Ras-like GTPases that was identified as an important mediator of synapse development. We performed a comprehensive, loss-of-function analysis of Rem2 function in cultured hippocampal neurons using RNAi to substantially decrease Rem2 protein levels. We found that knockdown of Rem2 decreases the density and maturity of dendritic spines, the primary site of excitatory synapses onto pyramidal neurons in the hippocampus. Knockdown of Rem2 also alters the gross morphology of dendritic arborizations, increasing the number of dendritic branches without altering total neurite length. Thus, Rem2 functions to inhibit dendritic branching and promote the development of dendritic spines and excitatory synapses. Interestingly, binding to the calcium binding protein calmodulin (CaM) is required for Rem2 regulation of dendritic branching. However, this interaction is completely dispensable for synapse development. Overall, our results suggest that Rem2 regulates dendritic branching and synapse development via distinct and overlapping signal transduction pathways.
Keywords: synapse, dendrite, spine, GTPase, Rem2
Introduction
The development of the mature nervous system involves a variety of complex and dynamic events that contribute to shaping the eventual structure and connectivity of individual neurons. For example, the development of synapses, the sites of cell-cell communication between neurons, is a multistep process that begins with the guidance of the axon of a presynaptic neuron towards a target site on a postsynaptic neuron (Plachez and Richards 2005). When the pre- and postsynaptic neurons make contact, proteins including synaptic vesicle proteins, scaffolding proteins, and receptors are recruited to the target site where a functional synapse forms (McAllister 2007). Ultimately, synapses in the maturing nervous system undergo experience-dependent synaptic refinement where some synapses are strengthened and maintained while others are eliminated (Maffei & Turrigiano 2008; Flavell & Greenberg 2008; Holtmaat & Svoboda 2009). The majority of excitatory synapses in the mammalian central nervous system form onto actin-rich structures known as dendritic spines (Nimchinsky et al. 2002; Tada & Sheng 2006). A number of signaling proteins, both intracellular and transmembrane, have been implicated in various steps of synapse and dendritic spine development (Shapiro et al. 2007; Saneyoshi et al. 2010). However, a complete understanding of the molecular mechanisms underlying this important developmental process remains unknown.
In addition, each type of neuron in the central nervous system has a distinctive dendritic arborization, which describes the length, branching, and overall shape of the dendrites, where the majority of synaptic inputs occur (Ramon y Cajal 1911; Jan & Jan 2010). The dendritic arborization influences the specific function of each neuron in part by ensuring that the proper synaptic connections are made (Elston 2000). Importantly, disruption or misregulation of development of the dendritic arbor and dendritic spines is a major contributing factor in a variety of neurodevelopmental disorders, including mental retardation and autism spectrum disorders (Newey et al. 2005; Pardo & Eberhart 2007; Jan & Jan 2010). Therefore, achieving a full understanding of the proteins involved in these developmental processes is an important first step to uncovering the underlying pathology of these disorders.
Previously, an RNAi-based screen for molecules essential for synapse development identified the small GTPase Rem2 as an important mediator of both excitatory and inhibitory synapse development (Paradis et al. 2007). RNAi-mediated decrease of Rem2 expression results in decreased excitatory synapse density, as assessed by immunostaining against synapsin I, a synaptic vesicle-associated protein that marks presynaptic sites, and PSD-95, a scaffolding protein that is highly enriched at the postsynaptic side of excitatory synapses. Neurons in which Rem2 expression has been decreased by RNAi also display decreased AMPA receptor-mediated miniature excitatory postsynaptic current (mEPSC) frequency and amplitude, and fewer AMPA receptor-containing synapses as assessed by immunostaining (Paradis et al. 2007). Taken together, this data indicates the Rem2 mediates the development of functional, glutamatergic synapses in the mammalian hippocampus.
Rem2 is a member of the Rad/Rem/Rem2/Gem/Kir (RGK) protein family, a Ras-related subfamily of small GTPases, and is the only member of this family that is highly expressed in the central nervous system (Finlin et al. 2000). RGK proteins have been characterized as regulators of calcium current and cytoskeletal structure via overexpression studies in a variety of cell types (Beguin et al. 2005b, 2006; Finlin et al. 2003; Piddini et al. 2001). However, a role for RGK proteins in these biological processes using a loss-of-function approach in these cell types has not been reported. Overexpression of Rem2 results in a strong inhibition of calcium current through L-type and N-type high-voltage activated calcium channels in heterologous cells and sympathetic neurons, respectively (Finlin et al. 2005; Chen et al. 2005). Overexpression studies have also shown that Rem2 and other RGK proteins induce neurite-like outgrowths in endothelial cells, at least in part by antagonizing the activity of the Rho GTPase-associated kinase ROKβ (Piddini et al. 2001; Beguin et al. 2007). Interestingly, Rem2 has also been recently implicated as a survival factor and regulator of the cell cycle in human embryonic stem cells (hESCs) (Edel et al. 2010). Based on a loss-of-function analysis, we have discovered a previously unknown role for the Rem2 GTPase in regulating dendritic arborization and spine development.
It is well-established that small GTPases play a major role in the regulation of both dendritic morphology and dendritic spine development, as exemplified by studies of the Rho family of GTPases, which include RhoA, Rac1, and Cdc42 (Li et al. 2000; McNair et al. 2010; Talens-Visconti et al. 2010; Fu et al. 2007; Randazzo et al. 2005; Sakane et al. 2010). These two proteins appear to play opposing roles in regulating dendritic arbor complexity, and interestingly, dendritic spine development as well; RhoA acts to inhibit, and Rac1 and Cdc42 to promote, dendritic complexity and spine development (Nakayama et al. 2000; Tashiro et al. 2000). Additionally, many proteins involved in the regulation of GTPases, including GTPase activating proteins (GAPs) and guanine nucleotide exchange factors (GEFs), are implicated in spine and dendrite morphology (Tolias et al. 2005; Penzes et al. 2003; Xie et al. 2007). As many of these GTPases and associated proteins have opposing actions on dendritic and spine morphology, it is not well understood how neurons integrate information from these different signaling pathways to arrive at the appropriate dendritic arborization and spine density during development.
The calcium/calmodulin-dependent protein kinase (CaMK) family is another group of proteins that have been implicated in regulating the development of dendritic arbors and spines (Fink et al. 2003; Redmond et al. 2002). Increases in intracellular calcium levels are sensed by the calcium binding protein calmodulin (CaM) which, upon calcium binding, activates CaMKs. CaMKs play a crucial role in the initiation of signal transduction pathways that are activated in response to rises in intracellular calcium (Soderling 1999). Several CaMK family members have been shown to modulate dendritic morphology and spine development, although at present the identity of the specific CaMK that mediates these effects remains somewhat controversial (Fink et al. 2003; Redmond et al. 2002). Interestingly, Rem2 also binds to CaM, leading to changes in Rem2 subcellular localization and filopodial dynamics in non-neuronal cells (Beguin et al. 2005a). A biological role for the interaction of Rem2 with CaM in neurons has not been previously reported.
While a variety of molecules have been shown to simultaneously regulate dendritic arborization and spine development, previous studies have not been able to dissociate the different signaling pathways through which these processes are regulated. Using an RNAi-based approach to significantly decrease the expression of the Rem2 protein, we found that Rem2 is required for dendritic spine development and proper dendritic arborization. Further, we demonstrated that the ability of Rem2 to mediate changes in dendritic morphology is dependent on Rem2 binding to CaM, while the effects of Rem2 on synapse development are independent of this interaction. This indicates that Rem2 mediates these biological processes via separable signal transduction pathways. Thus, regulation of Rem2 represents an important avenue into dissociating and defining the complex signaling events underlying the development of dendritic spines and the dendritic arbor.
Materials and Methods
293T cell culture, antibody generation, and Western blotting
We generated a full length Rem2-GST fusion protein and purified this protein from bacterial lysate by binding to glutathione sepharose beads. Rabbits were initially injected with 100μg Rem2-GST and boosted with 500μg Rem2-GST approximately every 2 months at Cocalico Biologicals Custom Antibody Services (Reamstown, PA). Sera were assessed for immunoreactivity to Rem2 by Western blotting of myc-tagged Rem2 from HEK 293T cell lysate.
HEK 293T cells grown in 12 well tissue culture plates were transfected by the calcium phosphate method (Xia et al. 1996) with 200 ng GFP, 100 ng myc-CMV Rem2, and 1 μg shRNA per well against either Rem2 or Rad2 (as a negative control). For Western blotting, the HEK 293T cells were lysed in 3× Sample Buffer (SDS, bromphenol blue, 1M Tris pH 6.8, glycerol) and proteins were separated by electroporesis on a 10% SDS/PAGE gel. The proteins were transferred to a nitrocellulose membrane which was then blocked in a 5% milk/1× TBST solution for one hour, then incubated overnight at 4°C in primary antibody (anti-Rem2 (1:100) and anti-β-actin (Abcam; 1:1000)). The following day, the membrane was washed two times for 20 minutes in 1× TBST, then incubated in appropriate secondary antibody (Licor; 1:5000) for 2 hours at room temperature. Following two 20-minute washes in 1× TBST, the blot was developed using the Odyssey Western Blotting system (Licor).
Neuronal cell culture and transfection
Neurons were cultured at low density on an astrocyte feeder layer. Astrocytes were isolated from P0 rat cortex by plating dissociated cells at low density in DMEM+10%FBS on uncoated 10 cm tissue culture dishes. The media was exchanged one day after plating to remove cells that failed to adhere. The glia were trypsinized once confluent and plated on 12 mm glass coverslips which had been coated overnight at 37°C with poly-D-lysine (20 μg/ml) and laminin (3.4 μg/ml) in 24 well plates. Dissociated hippocampal neurons from E18 rats were plated at a density of approximately 80,000/well onto the confluent glia and grown in Neurobasal media with B27 supplement (Invitrogen). AraC (Sigma) was added to a final concentration of 5 μM 24 hrs after plating.
At 4DIV, neurons were transfected by the calcium phosphate method (Xia et al. 1996) with 500 ng pCMV-GFP plasmid/well. For control conditions, neurons were also transfected with empty pSuper vector at 33ng/well and 100ng/well. For RNAi and rescue conditions, neurons were also transfected with pSuper-shRNA plasmids containing an shRNA against Rem2 (see Paradis et al. 2007) at 33ng/well. For overexpression and rescue conditions, neurons were also transfected with the RNAi-resistant Rem2 cDNA or, in the case of the structure/function experiments, the relevant RNAi-resistant Rem2 mutant cDNA at 100 ng/well.
The relatively low transfection efficiency (less than 10%, our unpublished observations) of the calcium phosphate method (Xia et al. 1996) allows for a transfected neuron to develop in the context of a network of otherwise unaffected cells. As GFP signal alone was used to identify transfected neurons for imaging experiments, we sought to determine the efficiency of cotransfection of three cDNAs simultaneously as is used periodically in this study (for example “rescue condition” experiments as outlined above). We cotransfected pCMV-RFP (500 μg), pCMV-myc-Rem2 (100μg), and a vector expressing both GFP and an shRNA against Rem2 (100μg) (note that this vector was utilized to visualize GFP expression in this control experiment only and not in any of the Rem2 RNAi experiments as its efficacy as a vehicle for shRNA delivery has not yet been verified) into neurons at 4DIV. Neurons were fixed and stained with mouse anti-myc primary antibody (1:500; Santa Cruz Biotechnology) and anti-mouse Cy5 secondary antibody (1:500; Jackson ImmunoResearch Laboratories) at 7 DIV to visualize the myc-Rem2 signal; GFP and RFP signals were apparent without antibody stain. Approximately 30 GFP-expressing neurons (blinded to the RFP and Cy5 (myc) channels) and 30 RFP-expressing neurons (blinded to the GFP and Cy5 (myc) channels) were selected for imaging, and the percentage of neurons that expressed all three proteins was calculated. We found that the efficiency of simultaneous cotransfection of three different plasmids via the calcium phosphate method was over 90% (Supplementary Figure 2), independent of whether GFP or RFP was used to identify the transfected cells. This result confirms the validity of simultaneous cotransfection of three cDNAs and of using GFP alone as a marker for transfected neurons.
Immunocytochemistry
As synapse development in primary hippocampal culture occurs over a period of approximately two weeks (Rao et al. 1998; Ziv & Smith 1996), neurons were fixed and stained for synaptic markers at 14 DIV. Neuronal media was replaced with 1× PBS and neurons were then fixed with 4% paraformaldehyde/4% sucrose for 8 minutes at room temperature. Coverslips were then washed three times with 1× PBS for 5 minutes each and incubated overnight at 4°C in a humidified chamber with primary antibody.
For the Rem2 localization experiments, the following primary antibodies were used: rabbit anti-Rem2 (1:500) and mouse anti-MAP2 (1:1000; Sigma). For the nuclear localization experiments, primary antibodies were rabbit anti-Rem2 (1:500) and mouse anti-NeuN (1:100; Millipore). For all other experiments, the primary antibodies were: mouse anti-PSD-95 (1:500; ABR) and rabbit anti-synapsin I (1:1000; Millipore). All antibody dilutions were prepared in 1× GDB (0.1% gelatin, 0.3% TritonX-100, 4.2% 0.4 M phosphate buffer, 9% 5 M NaCl).
After overnight incubation, coverslips were washed three times with 1× PBS for 5 minutes each and then incubated with appropriate Cy3- and Cy5-conjugated secondary antibodies (1:500 each; Jackson ImmunoResearch Laboratories) in 1× GDB for 2 hours at room temperature. Coverslips were then washed three times with 1× PBS for 10 minutes each, dipped in dH2O, and mounted on glass slides with Aquamount (Lerner Laboratories).
Quantification of synapse density and staining intensity
Image acquisition and quantification were performed in a blinded manner. 12-bit images of neurons were acquired on an Olympus Fluoview300 confocal microscope using a 60× objective. Within each experiment, images were acquired with identical settings for laser power, detector gain, and amplifier offset. Note that for the Rem2 localization experiments, separate settings and coverslips were used for control vs. RNAi and control vs. overexpression conditions to insure that the immunofluorescence intensity was kept within the linear range of the detector. Images were acquired as a z-stack (5-15 optical sections, 0.5 μm step size) and maximum intensity projections were created from the stacks.
For PSD-95/synapsin I experiments, synapse density was quantified as the overlap of GFP, anti-PSD-95, and anti-synapsin I staining using MetaMorph image analysis software. For each experiment, the threshold for the PSD-95 and synapsin I channels was determined visually using an image of a control neuron. The threshold was chosen such that all punctate structures would be included in the analysis. This threshold was then applied across all images within the experiment. The threshold for GFP was determined independently for each image. A binary mask including all pixels above the threshold was created for all channels for each image. The cell body was then manually deleted from the GFP mask. The “logical and” function was used to determine regions of triple co-localization at least 1 pixel in size. To calculate synapse density, this number was divided by the area of the neuron as measured using the GFP mask minus the cell body. Approximately 15-20 images from at least two separate coverslips were acquired and analyzed for each condition within an experiment, for a total of three experiments.
To determine the number of PSD-95/Synapsin puncta overlapping with GFP per unit length of dendrite, the above procedure was repeated after reblinding all coverslips for each experiment, and the analysis was limited to a 300 μm stretch of dendrite.
Synapse density values within each experiment were normalized, to account for the variation in antibody staining and neuronal density from experiment to experiment, before combining data from separate experiments. Within an experiment, the average synapse density value was obtained for the control and for experimental conditions. The normalized value of each experiment is the experimental average value divided by the control average value. See Paradis et al. 2007 for details of this conversion. Statistical analysis was performed comparing each experimental condition to control on the combined raw data from all experiments using SPSS Software to run a two-way between-effect ANOVA (factors were transfection group, and date of experiment), followed by the Tukey posthoc test for significance. Error bars denote standard error.
For Rem2/MAP2 and Rem2/NeuN experiments, staining intensity was quantified using the MetaMorph image analysis software. For the Rem2/MAP2 experiments, the soma and an approximately 25 μm stretch of dendrite were carefully traced using the GFP channel as a guide, and these regions were then copied to the Rem2 and MAP2 channels, where the average pixel intensity of these regions was calculated. For the Rem2/NeuN experiments, the region of NeuN staining in a GFP-transfected neuron was carefully traced, and then copied to the Rem2 channel, where the average pixel intensity of the nuclear staining was calculated. Then, the soma of a GFP-transfected neuron, was carefully traced and copied to the Rem2 channel, and the average pixel intensity of the Rem2 staining was calculated. The value of the nuclear Rem2 staining intensity was then subtracted from this value to yield the somatic cytoplasmic Rem2 staining intensity. Approximately 15-20 images from at least two separate coverslips were acquired and analyzed for each condition within an experiment, for a total of three experiments. The staining intensity for each condition was calculated by averaging the intensities for each image across all experiments, and significant differences between conditions were determined via a two-sample t-test
Spine analysis
Image acquisition and quantification were performed in a blinded manner. Since 14 DIV neurons do not contain many mature spines, neurons were cultured for additional days (Rao et al. 1998; Ziv & Smith 1996). Specifically, neurons were cultured and transfected as described above at 6 DIV, then fixed and stained for PSD-95/synapsin I at 17 DIV. To measure spine density, three dendritic segments totaling approximately 150 μm of dendritic length per neuron were carefully traced, and the number of spines was counted manually using the Cell Counter plugin for ImageJ (NIH). Between eight and ten transfected neurons were quantified per experiment, and three independent experiments were utilized. For quantification of spine morphology, images were analyzed by manually tracing both the spine length and width at widest point for approximately 1000 spines per condition. For both spine density and morphology experiments, statistical significance was calculated by using SPSS Software to run a two-way between-effect ANOVA (factors were transfection group, and date of experiment), followed by the Tukey posthoc test for significance.
Sholl analysis and morphology measurements
Image acquisition and quantification were performed in a blinded manner. 12-bit images were acquired on an Olympus Fluoview300 confocal microscope using a 20× oil objective. All images were acquired with identical settings for laser power, detector gain, and amplifier offset. Images were acquired as a z-stack (5-10 optical sections, 1.0 μm step size). Maximum intensity projections were created from the images and analyzed using MetaMorph image analysis software and ImageJ (NIH). For each experiment, 10-20 images from two separate coverslips were acquired and analyzed for each condition, and each condition was analyzed in three independent experiments.
For Sholl analysis, a series of 11 concentric circles of increasing radii (10 μm intervals) was drawn around the center of the cell body, and the number of dendrite crossings at each circle was counted (Sholl 1953). The number of dendrite crossings at each radius for each condition was calculated by averaging the number of crossings from every image. For measurements of neurite length and number, the NeuronJ plugin for ImageJ (NIH) was used to manually trace all projections for each image and indicate those that were primary, secondary, or higher-order. The plugin automatically and simultaneously recorded the number, total length, and mean length for these conditions, and these values were compared between experimental conditions and the control condition. Significance was determined using SPSS Software to run a two-way repeated measures ANOVA (factors were transfection group, and circle radius) followed by the Tukey posthoc test for significance.
Generation of additional Rem2 constructs
The following Rem2 constructs were generated using the QuikChange Site Directed Mutagenesis Kit (Stratagene). The forward primers used are listed, with the introduced mutations indicated in lower case.
| Rem2 RNAi-resistant | GACACCTATGAGAGgCGaATtATGGTGGACAAAGAA |
| Rem2 L317G-calmodulin binding | GCTAAGCGCTTCggCGCCAACCTGGTGC |
| Rem2 S69A/S334A-14-3-3 binding | CCAGACGAAGAGGAgcCATGCCCGTGCCC GCAACGCTCCAGGgCATGTCACGACCTC |
| Rem2 S129N-GTP binding | CGTGGGCAAGAaCACTCTAGCGGGCAC |
| Rem2 R236A/L263A-Ca channel binding | GTGACCTGGCCgcCTCCCGGGAGGTATC GTCGGCCGCTgcCCACCACAACACTCG |
Statistical analyses
See Supplementary Table 1 for statistical test used, F values, degrees of freedom, and p values for all experiments.
Results
Rem2 is widely expressed in neurons
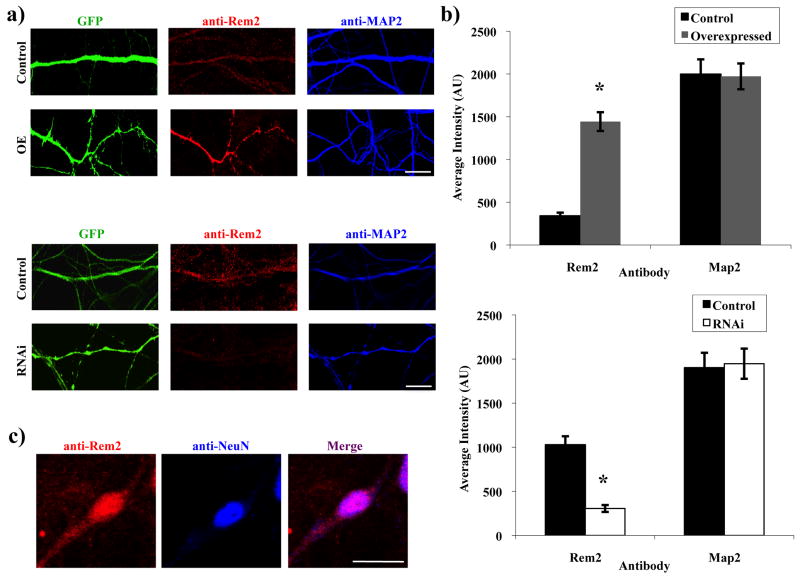
To examine the subcellular localization of endogenous Rem2 in neurons, we developed an antibody against Rem2 and verified the specificity of this antibody by Western blot (data not shown) and on fixed hippocampal neurons using immunocytochemistry. To increase or decrease the level of Rem2 expression, hippocampal neurons were cultured and transfected with plasmids expressing either Rem2 cDNA or an shRNA targeting Rem2 at 4 DIV. The cultures were fixed and stained at 14 DIV with antibodies against Rem2 and the microtubule-associated protein MAP2 as a control. The average intensity of the Rem2 antibody staining was increased in the overexpressed condition and decreased in the RNAi condition, compared to control neurons transfected with GFP alone. As expected, MAP2 staining intensity was not altered between conditions (Figure 1a,b).
Figure 1.
Rem2 is widely expressed in primary hippocampal neurons. (a) Representative images of a stretch of dendrite immunostained for Rem2 (red) and MAP2 (blue) from a neuron transfected with either a GFP-expressing plasmid plus: (top rows) a control plasmid (“Control”), or a Rem2 RNAi-resistant cDNA (“OE”); (bottom rows) a control plasmid (“Control”), or a shRNA plasmid targeting Rem2 (“RNAi”). (b) Quantification of Rem2 and MAP2 staining intensity in Control and overexpressed conditions. (c) Representative images of a Control neuron immunostained for Rem2 and NeuN. Asterisks indicate p<0.01 compared to control conditions by two-sample t-test. Scale bars indicate (a) 5 μm and (b) 50 μm. n > 50 neurons/condition for all experiments. (See Supplementary Table 1 for detailed statistical information for the data presented in this and subsequent figures.)
We found that Rem2 was widely expressed in neurons, with variable degrees of immunostaining visible throughout the soma and dendritic and axonal projections. This suggests that the sequestering of Rem2 at specific sites where synapses will form is not necessarily required for its involvement in that process, and also implies that Rem2 may be a component of other cellular pathways as well. Interestingly, RGK proteins also lack the canonical C-terminal CAAX domain, which serves as a prenylation signal targeting other GTPases to the plasma membrane (Kelly 2005). It has been previously noted that Rem2 is also present in the nucleus (Mahalakshmi et al. 2007). We also found that Rem2 colocalizes with the neuronal nuclear stain NeuN, confirming that it is present in the nucleus in cultured hippocampal neurons (Figure 1c).
Rem2 is important for the development and morphology of dendritic spines
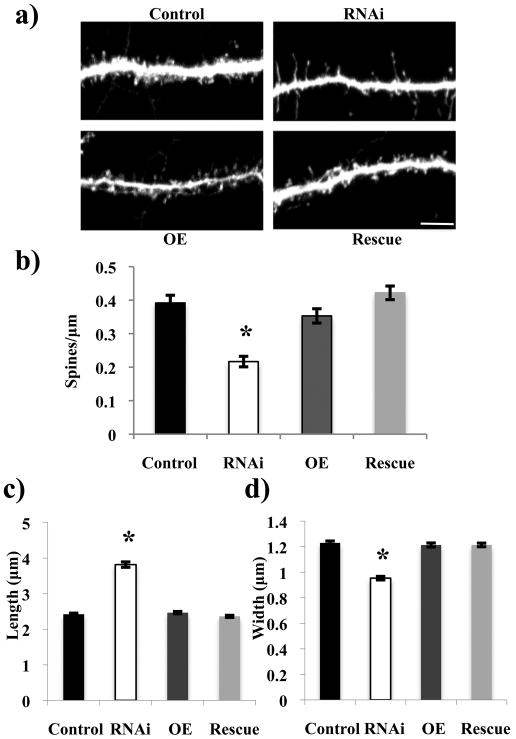
As a result of the known role of Rem2 in excitatory synapse development, as assessed by immunostaining and electrophysiology (Paradis et al. 2007), we sought to further characterize the role of Rem2 in this process by determining whether Rem2 might also be an important regulator of the development or morphology of dendritic spines as well. To pursue this question, we cotransfected hippocampal neurons at 6 DIV with GFP and an shRNA construct targeting Rem2 and fixed these neurons at 17 DIV. This experimental time course was chosen as it envelops a period of robust dendritic spine morphogenesis in cultured neurons; by the end point of 17 DIV, pyramidal neurons have a considerable density of dendritic spines. The dendritic spine density of these neurons was assayed by counting the number of spines present along three independent stretches of dendrite totaling 150 micrometers in length per neuron for at least 25 neurons per condition. Neurons in which Rem2 levels were reduced by RNAi had fewer dendritic spines per micron compared to control conditions (Figure 2a,b). Traditionally, mature spines are considered “mushroom-shaped,” with a short length and wider head (Bourne & Harris 2007). We measured the length and width of dendritic spines on neurons in which Rem2 levels were reduced by RNAi versus control neurons to observe any differences in spine morphology as a result of the shRNA-mediated decrease in Rem2. We found that those spines present on the neurons in the RNAi condition were increased in length and decreased in width compared to those in the control condition (Figure 2a,c-d), indicating a less mature dendritic spine structure in the absence of Rem2. Thus, we conclude that Rem2 plays a role both in the development and subsequent maturation of dendritic spines.
Figure 2.
Rem2 regulates the development and morphological maturation of dendritic spines. (a) Representative images of a stretch of dendrite from a neuron that has been transfected with either a GFP-expressing plasmid plus (clockwise from top left) a control plasmid (“Control”), a shRNA plasmid targeting Rem2 (“RNAi”), a shRNA plasmid targeting Rem2 plus a Rem2 RNAi-resistant cDNA (“Rescue”), or a Rem2 RNAi-resistant cDNA alone (“OE”). (b) Quantification of dendritic spine density for each condition. n > 20 neurons/condition. (c) Quantification of dendritic spine length and (d) dendritic spine head width for each condition. Asterisks indicate p<0.001 compared to control condition by two-way between-subjects ANOVA with Tukey posthoc test. n > 1000 spines/condition. Scale bar indicates 5 μm.
To confirm that the decrease in spine density and change in spine morphology were specifically due to a decrease in Rem2 levels, we took a rescue approach. First, we engineered a Rem2 cDNA that was resistant to knockdown by the shRNA by introducing silent mutations into the Rem2 cDNA at the site recognized by the shRNA. This disrupts the ability of the shRNA to recognize Rem2 mRNA, without altering the coding sequence of the protein. Next, we confirmed that this construct was indeed RNAi-resistant in HEK 293T cells by coexpressing either wild-type myc-tagged Rem2 cDNA or RNAi-resistant Rem2 cDNA along with the Rem2 shRNA or an shRNA targeting the related gene Rad2 as a control (Supplementary Figure 1a). The Rem2 shRNA significantly decreased wild-type Rem2 protein expression on a Western blot, while the level of RNAi-resistant Rem2 protein was not affected by co-transfection with the Rem2 shRNA (Supplementary Figure 1a).
Next, we sought to determine if the RNAi-resistant construct was able to rescue the previously reported decrease in excitatory synapse density in primary hippocampal culture as assayed by PSD-95/synapsin immunostaining (Supplementary Figure 1b,c). Neurons were transfected at 4 DIV with GFP alone (“Control”), an shRNA against Rem2 (“RNAi”), RNAi-resistant Rem2 cDNA (“OE”), or both the shRNA and RNAi-resistant Rem2 cDNA (“Rescue”). These neurons were fixed after 14 DIV and co-stained using antibodies against the synaptic markers PSD-95 (postsynaptic) and Synapsin I (presynaptic) to identify excitatory synapses. The synapse density of each neuron was calculated as the number of overlapping GFP/PSD-95/Synapsin-1 puncta, divided by the total GFP area (excluding the soma). Although slightly different from the experimental time course noted above to assay dendritic spine density, this experimental time course again corresponds to a robust period of synaptogenesis in cultured neurons and was chosen to more closely match previously reported experimental paradigms (Paradis et al. 2007). Importantly, we confirmed that over 90% of the GFP-expressing neurons in which three cDNAs were simultaneously transfected (e.g. GFP, shRNA targeting Rem2, and Rem2 RNAi-resistant cDNA) expressed all three cDNAs (see Methods and Supplemental Figure 3).
We found that while the excitatory synapse density of neurons in the Rem2 RNAi condition was decreased relative to Control neurons, this effect was rescued for neurons in the Rescue condition to an excitatory synapse density that was not significantly different from Control neurons (Supplementary Figure 1b,c). This result supports our conclusion that the RNAi-resistant cDNA is, in fact, resistant to knock down by the Rem2 shRNA and further validates our conclusion that Rem2 is required for excitatory synapse development (Paradis et al. 2007). In addition, co-transfection of the RNAi-resistant Rem2 cDNA with the shRNA targeting Rem2 (“Rescue”) completely rescued the decrease in dendritic spine density and change in dendritic spine morphology observed with the Rem2 RNAi condition (Figure 2), confirming the results of the PSD-95/synapsin immunostaining experiments. Based on these results, we conclude that the decrease in number and altered morphology of dendritic spines in the absence of Rem2 is due specifically to knockdown of Rem2 and not to other “off-target” effects of the Rem2 shRNA. Further, we conclude that Rem2 normally functions to promote the development and maturation of dendritic spines, the structures on which excitatory synapses form.
Interestingly, we did not observe a concomitant increase in excitatory synapse density, as assessed both by quantifying PSD-95/synapsin immunostaining and by measuring dendritic spine density, in response to Rem2 overexpression (“OE,” Figures 2 and Supplementary Figure 1) with the transfection conditions used in these experiments. In addition, we were unable to observe a change in excitatory synapse density by further increasing amounts of Rem2 cDNA transfected into neurons (data not shown). This result is perhaps unsurprising, as RGK family members are known to exhibit very slow kinetics of GTP hydrolysis, rendering them constantly GTP-bound and therefore constitutively active (Cohen et al. 1994; Finlin et al. 2000). Thus, one possibility is that the introduction of additional Rem2 protein is unable to substantially increase overall Rem2 activity levels with regards to regulating excitatory synapse development in neurons.
Rem2 affects dendritic morphology by limiting branching while maintaining the length of neurites
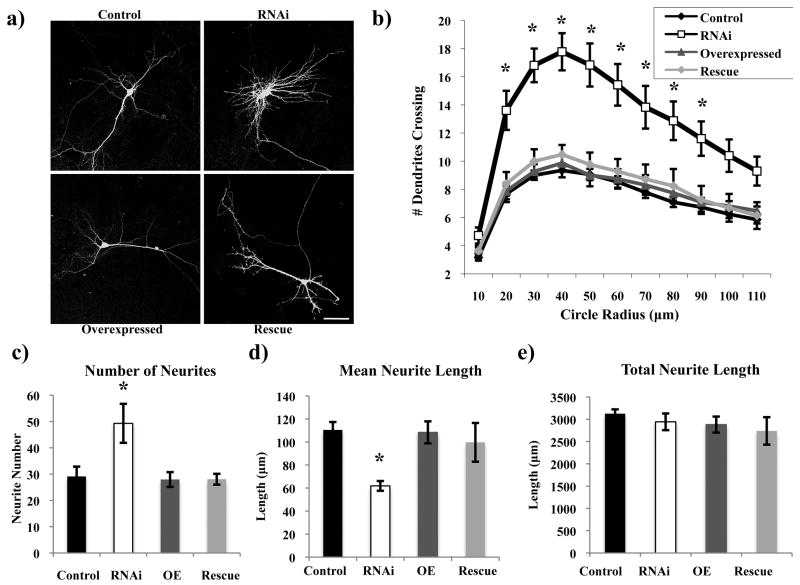
In the course of assaying the effects of Rem2 RNAi on spine density and morphology, it became clear that the neurons in the Rem2 RNAi condition exhibited a strongly altered dendritic morphology phenotype. Overall, we observed that neurons in which the level of the Rem2 protein was decreased by RNAi displayed a more complex dendritic arborization, with more highly branched but shorter dendrites, compared to control neurons (Figure 3a). To quantify dendritic complexity, we performed a Sholl analysis (Sholl 1953), which consists of superimposing on an image of a neuron a series of concentric circles of increasing diameter, centered at the soma, and quantifying the number of dendrites that intersect these circles. The Rem2 RNAi neurons showed a pronounced increase in branching at distances close to the soma, particularly from 20 to 60 micrometers, compared to control neurons (Figure 3b).
Figure 3.
Rem2 inhibits branching but maintains the total length of the dendritic arborization. (a) Representative images of the morphology of a (clockwise from top left) Control, RNAi, Rescue, and OE neuron. Total neurite lengths are as follows (in μm): Control= 2997.16, RNAi= 3057.23, OE= 2882.6, Rescue= 2776.21. (b) Quantification of dendritic branching for each condition via Sholl analysis. Asterisks indicate p<0.001 compared to control condition by two-way repeated measures ANOVA with Tukey posthoc test. n > 40 neurons/condition. (c) Quantification of the number of neurites for each condition. Asterisk indicates p<0.001 compared to control condition by two-way between-subjects ANOVA with Tukey posthoc test. (d) Quantification of the mean and (e) total neurite length for each condition. Asterisks indicate p<0.05 compared to control condition by two-way between-subjects ANOVA with Tukey posthoc test. n > 800 neurites/condition. Scale bar indicates 50 μm.
In addition, we utilized the NeuronJ plugin for ImageJ (NIH) to trace the projections of these neurons and quantified the total number, mean length, and total length of neurites. We found that the shRNA-mediated knockdown of Rem2 caused an increase in the number of neurites, but a decrease in the mean length of each neurite, compared to control neurons. Thus, the summed total length of all neurites was not affected (Figure 3c-e). These results were similar for both primary and higher-order neurites, and the phenotype was rescued upon co-transfection with the RNAi-resistant Rem2 cDNA. Again, we did not observe a change in dendritic branching or length as a result of Rem2 overexpression under these transfection conditions. Based on these studies, we conclude that normal function of Rem2 is to limit dendritic branching, without affecting the length of overall dendritic projections.
Rem2 structure/function analysis reveals distinct pathways for synapse development and dendritic morphology
Although little is known about the signal transduction pathways upstream or downstream of Rem2, several protein-binding domains on Rem2 have been identified. These include a GTP-binding domain, and amino acid residues that are responsible for Rem2 binding to calmodulin, 14-3-3 proteins, and the β subunit of high-voltage-activated calcium channels (Beguin et al. 2005a, 2007). We chose to examine a role for Rem2 binding to these proteins in excitatory synapse development and dendritic morphology by generating mutant constructs of Rem2 that were both resistant to RNAi and in which key amino acid residues were mutated to render Rem2 unable to bind to these various proteins. We then asked whether these constructs were able to rescue the Rem2 RNAi-based decrease in excitatory synapse density and increase in dendritic branching.
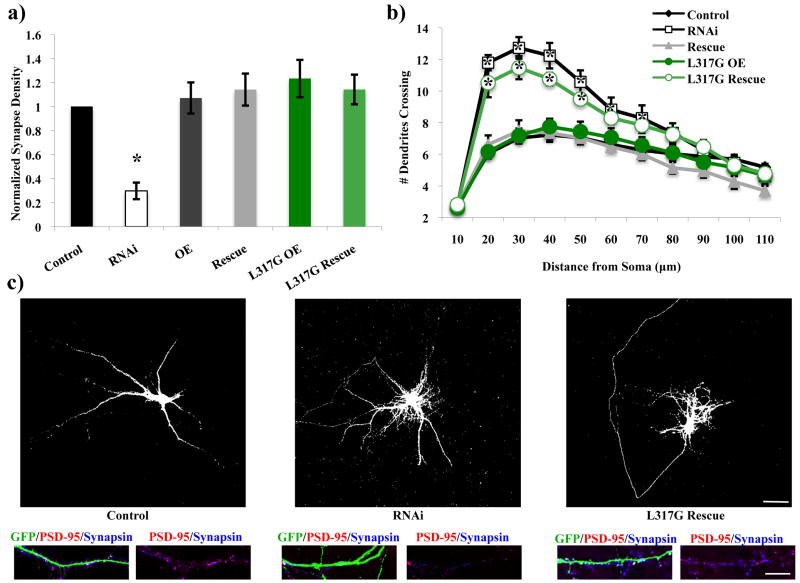
We found that a mutant construct of Rem2 that was unable to bind calmodulin (L317G) was able to rescue the Rem2 RNAi-mediated decrease in excitatory synapse density as assessed by PSD-95/synapsin immunostaining, but failed to rescue the increase in dendritic branching of these neurons as assessed by Sholl analysis (Figure 4a-c,Supplementary Figure 3). Based on these results, we conclude that Rem2 affects excitatory synapse density and dendritic branching via two distinct pathways, one of which requires calmodulin (i.e. dendritic branching) and one of which does not (i.e. excitatory synapse development). To verify that the PSD-95/synapsin immunostaining result was not due to a Rem2-dependent alteration of PSD-95 expression in the transfected neuron, we assessed the intensity of the PSD-95 puncta in the transfected neurons for each transfection condition (Figure 4), and found that these values did not differ between conditions (Supplementary Figure 5b,c). This analysis confirms that the observed changes in synapse density are not due to altered postsynaptic PSD-95 expression, and supports our conclusion that Rem2 regulates dendritic branching and synapse development by distinct molecular mechanisms.
Figure 4.
Rem2 affects excitatory synapse development and dendritic branching via distinct pathways. L317G= an RNAi-resistant cDNA encoding a Rem2 protein that does not bind calmodulin. (a) Quantification of excitatory synapse density as PSD-95/synapsin overlapping puncta for Control, RNAi, OE, Rescue, L317G OE, and L317G Rescue conditions. Asterisk indicates p<0.001 compared to control condition by two-way between-subjects ANOVA with Tukey posthoc test. (b) Quantification of dendritic branching for all conditions via Sholl analysis. Asterisks indicate p<0.05 compared to control condition by two-way repeated measures ANOVA with Tukey posthoc test. (c) Representative images of neurons from Control, RNAi, and L317G Rescue conditions. Top image is GFP signal to depict the dendritic arbor. Bottom images are a stretch of dendrite from above neuron to depict synaptic staining (see Supplementary Figure 3 for separated fluorescence channels for all experiments with synaptic staining). Scale bars indicate 50 μm (neuron images) or 5 μm (stretch of dendrite images). n > 40 neurons/condition for all experiments.
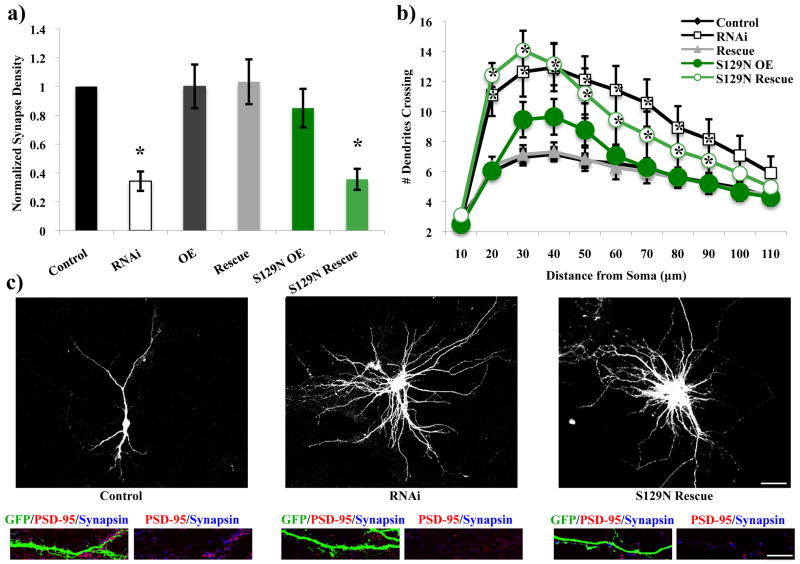
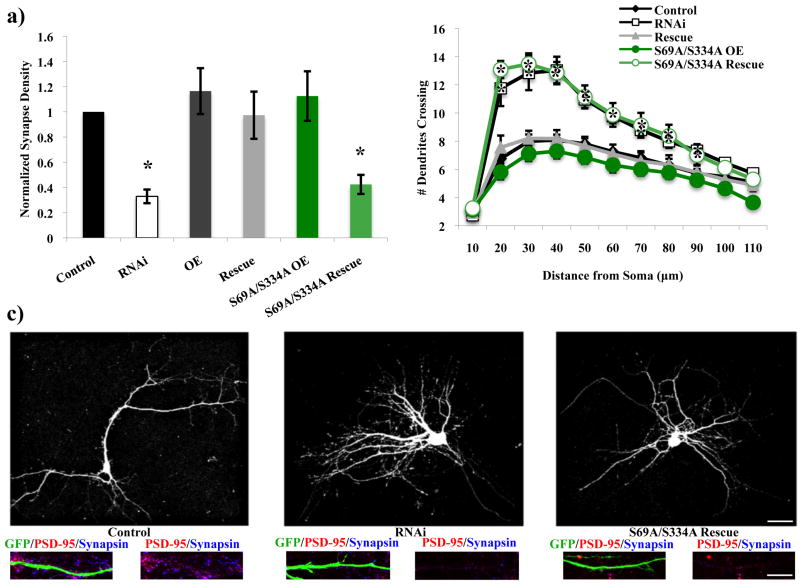
In contrast, mutant constructs that were unable to bind GTP (S129N) or were unable to bind 14-3-3 proteins (S69A/S334A) failed to rescue either the decrease in excitatory synapse density or the increase in dendritic branching caused by RNAi of Rem2 (Figure 5a-c, Figure 6a-c, Supplementary Figure 3). 14-3-3 proteins are implicated in nuclear trafficking and proteins that are 14-3-3-bound are frequently found sequestered outside the nucleus (Morrison 2009; van Heusden 2009). In addition, interference with the ability of Rem2 to bind 14-3-3 proteins has been shown to disrupt its normal function and subcellular localization when overexpressed in certain heterologous cell types (Beguin et al. 2005a). We quantified Rem2 immunoreactivity in the nucleus (as indicated by colocalization between anti-Rem2 and anti-NeuN immunostaining) and somatic cytoplasm of neurons that had been transfected with a Rem2 shRNA and either the Rem2 RNAi-resistant S69A/S334A cDNA or a wild type RNAi-resistant Rem2 cDNA. We found an increase in nuclear Rem2 immunostaining in the Rem2 S69A/S334A transfection condition compared to the wild-type Rem2 transfection condition (Supplementary Figure 4a). This result, combined with the Rem2 S69A/S334A failure to rescue either the excitatory synapse density or dendritic branching phenotypes of Rem2 RNAi, suggests that nuclear Rem2 is not an important regulator of these biological processes. Overall, our results indicate that Rem2 must bind to GTP and 14-3-3 proteins in order to regulate excitatory synapse development and dendritic arborization.
Figure 5.
The ability of Rem2 to bind GTP is essential for excitatory synapse development and proper dendritic arborization. S129N= an RNAi-resistant cDNA encoding a Rem2 protein that does not bind GTP. (a) Quantification of excitatory synapse density as PSD-95/synapsin overlapping puncta for Control, RNAi, OE, Rescue, S129N OE, and S129N Rescue conditions. Asterisks indicate p<0.001 compared to control condition by two-way between-subjects ANOVA with Tukey posthoc test. (b) Quantification of dendritic branching for all conditions via Sholl analysis. Asterisks indicate p<0.05 compared to control condition by two-way repeated measures ANOVA with Tukey posthoc test. (c) Representative images of neurons from Control, RNAi, and S129N Rescue conditions. Top image is GFP signal to depict the dendritic arbor. Bottom images are a stretch of dendrite from above neuron to depict synaptic staining. Scale bars indicate 50 μm (neuron images) or 5 μm (stretch of dendrite images). n > 40 neurons/condition for all experiments.
Figure 6.
The ability of Rem2 to bind 14-3-3 proteins is essential for excitatory synapse development and proper dendritic arborization. S69A/S334A= an RNAi-resistant cDNA encoding a Rem2 protein that does not bind 14-3-3 proteins. (a) Quantification of excitatory synapse density as PSD-95/synapsin overlapping puncta for Control, RNAi, OE, Rescue, S69A/S334A OE, and S69A/S334A Rescue conditions. Asterisks indicate p<0.05 compared to control condition by two-way between-subjects ANOVA with Tukey posthoc test. (b) Quantification of dendritic branching for all conditions via Sholl analysis. Asterisks indicate p<0.05 compared to control condition by two-way repeated measures ANOVA with Tukey posthoc test. (c) Representative images of neurons from Control, RNAi, and S69A/S334A Rescue conditions. Top image is GFP signal to depict the dendritic arbor. Bottom images are a stretch of dendrite from above neuron to depict synaptic staining. Scale bars indicate 50 μm (neuron images) or 5 μm (stretch of dendrite images). n > 40 neurons/condition for all experiments.
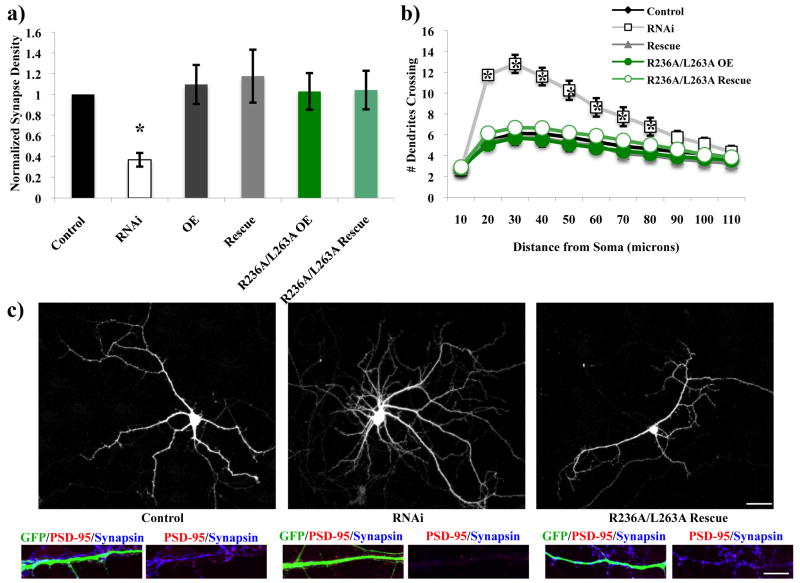
Rem2 binds to the β subunit of calcium channels, which is required to traffic the channels to the plasma membrane (Beguin et al. 2007), and overexpression of Rem2 inhibits the influx of calcium into cells (Finlin et al. 2005; Chen et al. 2005). However, it remains controversial whether the inhibition of calcium current is actually due to decreased channel trafficking to the membrane, or alternatively if the binding of Rem2 to the β subunit of channels already present at the membrane leads to their subsequent inactivation (Chen et al. 2005). Interestingly, a mutant construct of Rem2 that was unable to bind the β-subunit of calcium channels (R236A/L263A) rescued both the excitatory synapse density and dendritic morphology phenotypes (Figure 7; Supplementary Figure 3). Our results demonstrate that Rem2 binding to the β-subunit of calcium channels is not required for Rem2 to promote excitatory synapse development or restrict dendritic branching. Importantly, these results suggest that the role of Rem2 in regulating excitatory synapse development and dendritic morphology is independent of changes in calcium that might occur in the cell specifically in response to decreases in Rem2 protein levels.
Figure 7.
The roles of Rem2 in excitatory synapse development and dendritic branching are independent of Rem2 binding to calcium channels. R236A/L263A= an RNAi-resistant cDNA encoding a Rem2 protein that does not bind the β subunit of calcium channels. (a) Quantification of excitatory synapse density as PSD-95/synapsin overlapping puncta for Control, RNAi, OE, Rescue, R236A/L263A OE, and R236A/L263A Rescue conditions. Asterisks indicate p<0.05 compared to control condition by two-way between-subjects ANOVA with Tukey posthoc test. (b) Quantification of dendritic branching for all conditions via Sholl analysis. Asterisks indicate p<0.01 compared to control condition by two-way repeated measures ANOVA with Tukey posthoc test. (c) Representative images of neurons from Control, RNAi, and R236A/L263A Rescue conditions. Top image is GFP signal to depict the dendritic arbor. Bottom images are a stretch of dendrite from above neuron to depict synaptic staining. Scale bars indicate 50 μm (neuron images) or 5 μm (stretch of dendrite images). n > 40 neurons/condition for all experiments.
Discussion
Using an RNAi-based approach to significantly decrease levels of Rem2 protein in cultured hippocampal neurons, we demonstrate that Rem2 is required for normal dendritic spine development. Further, we also find that Rem2 regulates neuronal morphology by inhibiting dendritic branching while promoting the lengthening of individual dendrites. In addition, we show that Rem2 is widely expressed in neurons and can be found throughout the soma, dendrites, and nucleus. Further, our structure/function analysis of Rem2 demonstrates that binding to CaM is required for Rem2 regulation of dendritic branching but not excitatory synapse development. Importantly, this suggests that Rem2 regulates these two processes by dissociable signal transduction pathways, making Rem2 the first GTPase to our knowledge for which these biological processes have been distinguished. We also demonstrate that binding to GTP and 14-3-3 proteins is required for Rem2 regulation of both dendritic branching and excitatory synapse development, while binding to the β subunit of calcium channels is dispensable for both functions. Taken together, this data suggests that Rem2 regulates dendritic branching and excitatory synapse development via distinct and overlapping signal transduction pathways.
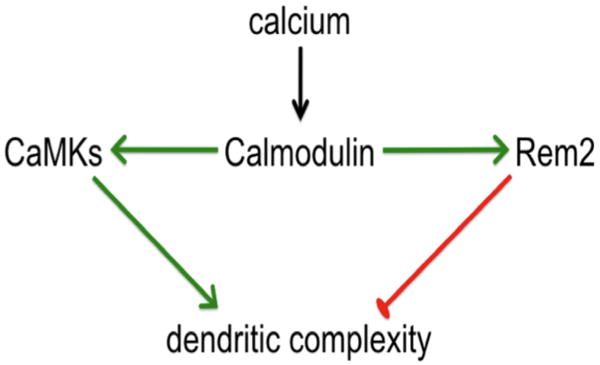
The formation of the dendritic arbor of a neuron is a dynamic process involving both the addition of branches and lengthening of individual dendrites. In addition, changes in dendritic arborization can be regulated by changes in the activity level of individual neurons. For example, the dendritic arbor of Xenopus laevis tectal neurons increases dramatically in response to light stimulation after a period of deprivation (Sin et al. 2002). Further, activation of CaMK family members, which are themselves responsive to neuronal activity, has been shown to regulate dendritic arborization (Fink et al. 2003; Redmond et al. 2002). A key finding of our structure/function analysis of Rem2 is that the ability of Rem2 to inhibit dendritic branching is dependent on an interaction with CaM. Interestingly, it has been shown that the ability of another RGK family member, Rem, to bind to CaM is calcium dependent (Correll et al. 2008c). Therefore, one model that is consistent with our results is that Rem2 acts to constrain dendritic branching in response to increased neuronal activity by binding to and sequestering calcium-bound CaM away from CaMK family members (Figure 8). In response to decreased activation of CaMK family members, dendritic arborization does not proceed as robustly. Thus, in the case where Rem2 levels are decreased by RNAi, dendritic branching is increased due to increased availability of calcium-bound CaM that activates CaMK family members.
Figure 8.
Model depicting a putative interaction between Rem2 and CaMK signaling pathways. Rem2 limits dendritic complexity by sequestering calcium-bound CaM from CaMKs, thus limiting CaMK-dependent dendritic outgrowth.
Although the overall effect of neuronal activity appears to be to promote dendritic branching (Chen & Ghosh 2005; Van Aelst & Cline 2004), it is apparent that some molecules function to constrain dendritic arborization. For example, while Rac and Cdc42 act to promote branching of dendrites, RhoA acts antagonistically to inhibit lengthening of dendrites (Li et al. 2000; Linseman & Louck 2008). Interestingly, recent studies of human embryonic stem cells suggest Rem2 antagonizes Rho signaling to regulate stem cell apoptosis (Edel et al. 2010). Our results suggest that Rem2 may in general behave similarly to RhoA as an inhibitor of overall dendritic arborization, but may also act antagonistically towards the Rho signaling cascade with regards to dendritic length. However, Rem2 has the additional function of being able to modulate dendritic arborization in response to neuronal activity through an interaction with CaM.
Dendritic spines are actin-rich structures that rely on precise regulation of the cytoskeleton in order to form and mature (Tada & Sheng 2006). In addition, similar to their role in the regulation of dendritic arborization, Rho family GTPases have been extensively implicated in the regulation of dendritic spine development (Nakayama et al. 2000; Tashiro et al. 2000; Govek et al. 2005). Our current study extends our previous finding of a role for Rem2 in excitatory synapse development, as assayed by immunostaining for excitatory synaptic markers and electrophysiology (Paradis et al. 2007), by demonstrating that Rem2 regulates the density of dendritic spines as well. This suggests that at least part of the function of Rem2 is to regulate the development of the dendritic spines onto which the AMPA receptor-containing synapses will form. Indeed, it has been shown in vivo that the outgrowth of cortical dendritic spines occurs prior to the formation of active synapses onto those spines (Knott et al. 2006). However, since we also observe a decrease in excitatory synapse density as assayed by immunostaining at DIV 14, when the neurons still lack appreciable dendritic spines and the majority of excitatory synapses are found on the dendritic shaft, it is likely that Rem2 plays multiple roles in excitatory synapse development. For example, Rem2 could signal to recruit proteins such as glutamate receptors to a newly-formed synaptic contact onto a dendritic shaft early in development, while also promoting the morphogenesis of dendritic spines by remodeling the cytoskeleton later in development. Synapse development refers to the processes of synapse formation, maturation, and elimination. Our experiments to date have not addressed which specific step(s) in synapse development requires the function of Rem2.
Interestingly, Rem2 modulates the cytoskeleton through a poorly understood mechanism in a variety of cell types (Correll et al. 2008b). Rem2 binds the Rho kinase ROKβ (Beguin et al. 2007), suggesting that Rem2 is involved in the Rho/Rac signaling cascade. However, a biological function for this interaction remains to be established. When activated by Rho GTPases, Rho kinases initiate a signaling cascade that ultimately leads to the phosphorylation of cofilin. This results in the disassembly of actin filaments and the rearrangement of the actin cytoskeleton that in turn limits cytoskeletal-dependent outgrowth (Luo 2000; Schmandke et al. 2007). In neurons, the ability to signal through Rho kinases has been shown to be essential for proper dendritic spine development and dendritic arborization (Nakayama et al. 2000; Zhou et al 2009). In addition, the expression of the RGK family member Gem has been shown to inhibit the ability of ROKβ to phosphorylate its downstream targets in non-neuronal cells (Ward & Kelly 2006). Thus, Rem2 may function similarly in neurons to mediate its effects by preventing the depolymerization of actin filaments by antagonizing one or multiple steps along this Rho kinase-mediated pathway.
We have demonstrated that Rem2 plays an important role in excitatory synapse development, the development and maturation of dendritic spines, and that it limits branching while promoting lengthening of individual dendrites. Rem2 and other RGK family members are regulated at the transcriptional level in response to a variety of extracellular stimuli (Maguire et al. 1994; Laville et al. 1996; Finlin & Andres 1997; Paradis et al. 2007). It is known that synapse number, spine morphology, and dendritic arborization can be altered in response to changes in neuronal activity (Chen & Ghosh 2005; Flavell & Greenberg 2008). However, the molecular mechanisms by which extracellular signals are transduced to cause morphological changes in neurons are entirely unclear. It is possible that Rem2 represents a novel link between the ability of neurons to sense changes in the environment and respond by altering their morphology as needed.
Supplementary Material
Supplementary Figure 1: Validation of the RNAi-resistant Rem2 cDNA. (a) Western blot with anti-Rem2 antibody of HEK 293T cell lysates. anti-β-actin serves as a loading control. Developed using the Odyssey IR System. (b) Quantification of excitatory synapse density as PSD-95/synapsin overlapping puncta for Control, RNAi, OE, and Rescue conditions. Asterisk indicates p<0.05 from control by two-way between-subjects ANOVA with Tukey posthoc test. (C) Representative images of a stretch of dendrite of a neuron from the Control, RNAi, OE, and Rescue conditions. Scale bar indicates 50 μm. n > 50 neurons/condition.
Supplementary Figure 2: Simultaneous cotransfection of three different plasmids into the same neuron via the calcium phosphate method. (a) Representative images of neurons cotransfected with myc-Rem2-, RFP- and GFP-expressing plasmids, and subsequently stained for myc. (b) Quantification of the percentage of cells triple cotransfected (as detected by RFP, GFP, and anti-myc expression), double cotransfected (as detected by RFP and GFP, RFP and anti-myc, or GFP and anti-myc), or single transfected (as detected by RFP or GFP expression alone). Scale bar indicates 50 μm. n = 60 neurons (30 selected by GFP expression, 30 selected by RFP expression).
Supplementary Figure 3: The same images from (a) Supplementary Figure 1, (b) Figure 4, (c) Figure 5, (d) Figure 6, and (e) Figure 7, separated to show individual fluorescence channels. Note that synaptic immunostaining associated with the processes of non-transfected neurons is visible in some images (for example, images extracted from Figure 5). Scale bars indicate 5 μm. n > 40 neurons/condition for all experiments.
Supplementary Figure 4: Rem2 that cannot bind 14-3-3 proteins is more abundant in the nucleus than is wild-type Rem2. (a) Quantification of nuclear anti-Rem2 staining for Control, RNAi, Rescue, and S69A/S334A Rescue conditions. (b) Quantification of somatic, cytoplasmic anti-Rem2 staining intensity for each condition. (c) Representative images of neurons in each condition immunostained for anti-Rem2 and anti-NeuN. Asterisks indicate p<0.01 from control, unless otherwise indicated, by two-sample t-test. Scale bar indicates 5 μm. n > 30 neurons/condition.
Supplementary Figure 5: Further analysis of Rem2 L317G (no CaM binding) mutant. (a) Synapsin I puncta density onto GFP- positive neurons transfected with the indicated constructs (compare to Figure 4). Rem2 RNAi in the postsynaptic neuron significantly decreases the density of synapsin puncta, and L317G (Rem2 that cannot bind CaM) rescues this effect (* = p<0.01). (b) The intensity of PSD-95 immunostaining in the transfected neuron overlapping with Synapsin I immunostaining does not change significantly between conditions; thus, we conclude that L317G (Rem2 that cannot bind CaM) does not alter the postsynaptic expression of PSD-95. (c) The intensity of total PSD-95 immunostaining in the transfected neuron does not change significantly between conditions; thus, we conclude that L317G (Rem2 that cannot bind CaM) does not alter the postsynaptic expression of PSD-95.
Acknowledgments
This work was supported by the Richard and Susan Smith Family Foundation, Chestnut Hill, MA, by an NIH grant R01NS065856 to S.P., and by an NIH Grant P30NS45713 for Core Facilities for Neurobiology at Brandeis University. We thank Calla Olson for help with antibody generation, Dr. Don Katz for advice and assistance with statistical analyses, and Dr. Gina Turrigiano for helpful advice and suggestions and for critical reading of the manuscript.
References
- Beguin P, Mahalakshmi RN, et al. Nuclear sequestration of beta-subunits by Rad and Rem is controlled by 14-3-3 and calmodulin and reveals a novel mechanism for Ca2+ channel regulation. J Mol Biol. 2006;355(1):34–46. doi: 10.1016/j.jmb.2005.10.013. [DOI] [PubMed] [Google Scholar]
- Beguin P, Mahalakshmi RN, et al. Roles of 14-3-3 and calmodulin binding in subcellular localization and function of the small G-protein Rem2. Biochem J. 2005;390(Pt 1):67–75. doi: 10.1042/BJ20050414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beguin P, Mahalakshmi RN, et al. 14-3-3 and calmodulin control subcellular distribution of Kir/Gem and its regulation of cell shape and calcium channel activity. J Cell Sci. 2005;118(Pt 9):1923–1934. doi: 10.1242/jcs.02321. [DOI] [PubMed] [Google Scholar]
- Beguin P, Ng YJ, et al. RGK small GTP-binding proteins interact with the nucleotide kinase domain of Ca2+-channel beta-subunits via an uncommon effector binding domain. J Biol Chem. 2007;282(15):11509–11520. doi: 10.1074/jbc.M606423200. [DOI] [PubMed] [Google Scholar]
- Bourne J, Harris KM. Do thin spines learn to be mushroom spines that remember? Curr Opin Neurobiol. 2007;17(3):381–386. doi: 10.1016/j.conb.2007.04.009. [DOI] [PubMed] [Google Scholar]
- Chen H, Puhl HL, 3rd, et al. Expression of Rem2, an RGK family small GTPase, reduces N-type calcium current without affecting channel surface density. J Neurosci. 2005;25(42):9762–9772. doi: 10.1523/JNEUROSCI.3111-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Ghosh A. Regulation of dendritic development by neuronal activity. J Neurobiol. 2005;64(1):4–10. doi: 10.1002/neu.20150. [DOI] [PubMed] [Google Scholar]
- Correll RN, Botzet GJ, et al. Analysis of the Rem2 - voltage dependant calcium channel beta subunit interaction and Rem2 interaction with phosphorylated phosphatidylinositide lipids. Cell Signal. 2008;20(2):400–408. doi: 10.1016/j.cellsig.2007.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correll RN, Pang C, et al. The RGK family of GTP-binding proteins: regulators of voltage-dependent calcium channels and cytoskeleton remodeling. Cell Signal. 2008;20(2):292–300. doi: 10.1016/j.cellsig.2007.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correll RN, Pang C, et al. Calmodulin binding is dispensable for Rem-mediated Ca2+ channel inhibition. Mol Cell Biochem. 2008;310(1-2):103–110. doi: 10.1007/s11010-007-9670-8. [DOI] [PubMed] [Google Scholar]
- Edel MJ, Boue S, et al. Rem2 GTPase controls proliferation and apoptosis of neurons during embryo development. Cell Cycle. 2010;9(17) doi: 10.4161/cc.9.17.12719. [DOI] [PubMed] [Google Scholar]
- Elston GN. Pyramidal cells of the frontal lobe: all the more spinous to think with. J Neurosci. 2000;20(18) doi: 10.1523/JNEUROSCI.20-18-j0002.2000. RC95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink CC, Bayer KU, et al. Selective regulation of neurite extension and synapse formation by the beta but not the alpha isoform of CaMKII. Neuron. 2003;39(2):283–297. doi: 10.1016/s0896-6273(03)00428-8. [DOI] [PubMed] [Google Scholar]
- Finlin BS, Andres DA. Rem is a new member of the Rad- and Gem/Kir Ras-related GTP-binding protein family repressed by lipopolysaccharide stimulation. J Biol Chem. 1997;272(35):21982–21988. doi: 10.1074/jbc.272.35.21982. [DOI] [PubMed] [Google Scholar]
- Finlin BS, Crump SM, et al. Regulation of voltage-gated calcium channel activity by the Rem and Rad GTPases. Proc Natl Acad Sci U S A. 2003;100(24):14469–14474. doi: 10.1073/pnas.2437756100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finlin BS, Mosley AL, et al. Regulation of L-type Ca2+ channel activity and insulin secretion by the Rem2 GTPase. J Biol Chem. 2005;280(51):41864–41871. doi: 10.1074/jbc.M414261200. [DOI] [PubMed] [Google Scholar]
- Finlin BS, Shao H, et al. Rem2, a new member of the Rem/Rad/Gem/Kir family of Ras-related GTPases. Biochem J. 2000;347(Pt 1):223–231. [PMC free article] [PubMed] [Google Scholar]
- Flavell SW, Greenberg ME. Signaling mechanisms linking neuronal activity to gene expression and plasticity of the nervous system. Annu Rev Neurosci. 2008;31:563–590. doi: 10.1146/annurev.neuro.31.060407.125631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Z, Lee SH, et al. Differential roles of Rap1 and Rap2 small GTPases in neurite retraction and synapse elimination in hippocampal spiny neurons. J Neurochem. 2007;100(1):118–131. doi: 10.1111/j.1471-4159.2006.04195.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govek EE, Newey SE, et al. The role of the Rho GTPases in neuronal development. Genes Dev. 2005;19(1):1–49. doi: 10.1101/gad.1256405. [DOI] [PubMed] [Google Scholar]
- Holtmaat A, Svoboda K. Experience-dependent structural synaptic plasticity in the mammalian brain. Nat Rev Neurosci. 2009;10(9):647–658. doi: 10.1038/nrn2699. [DOI] [PubMed] [Google Scholar]
- Jan YN, Jan LY. Branching out: mechanisms of dendritic arborization. Nat Rev Neurosci. 2010;11(5):316–328. doi: 10.1038/nrn2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly K. The RGK family: a regulatory tail of small GTP-binding proteins. Trends Cell Biol. 2005;15(12):640–643. doi: 10.1016/j.tcb.2005.10.002. [DOI] [PubMed] [Google Scholar]
- Knott GW, Holtmaat A, et al. Spine growth precedes synapse formation in the adult neocortex in vivo. Nat Neurosci. 2006;9(9):1117–1124. doi: 10.1038/nn1747. [DOI] [PubMed] [Google Scholar]
- Laville M, Auboeuf D, et al. Acute regulation by insulin of phosphatidylinositol-3-kinase, Rad, Glut 4, and lipoprotein lipase mRNA levels in human muscle. J Clin Invest. 1996;98(1):43–49. doi: 10.1172/JCI118775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Van Aelst L, et al. Rho GTPases regulate distinct aspects of dendritic arbor growth in Xenopus central neurons in vivo. Nat Neurosci. 2000;3(3):217–225. doi: 10.1038/72920. [DOI] [PubMed] [Google Scholar]
- Linseman DA, Loucks FA. Diverse roles of Rho family GTPases in neuronal development, survival, and death. Front Biosci. 2008;13:657–676. doi: 10.2741/2710. [DOI] [PubMed] [Google Scholar]
- Luo L. Rho GTPases in neuronal morphogenesis. Nat Rev Neurosci. 2000;1(3):173–180. doi: 10.1038/35044547. [DOI] [PubMed] [Google Scholar]
- Maffei A, Turrigiano G. The age of plasticity: developmental regulation of synaptic plasticity in neocortical microcircuits. Prog Brain Res. 2008;169:211–223. doi: 10.1016/S0079-6123(07)00012-X. [DOI] [PubMed] [Google Scholar]
- Maguire J, Santoro T, et al. Gem: an induced, immediate early protein belonging to the Ras family. Science. 1994;265(5169):241–244. doi: 10.1126/science.7912851. [DOI] [PubMed] [Google Scholar]
- Mahalakshmi RN, Ng MY, et al. Nuclear localization of endogenous RGK proteins and modulation of cell shape remodeling by regulated nuclear transport. Traffic. 2007;8(9):1164–1178. doi: 10.1111/j.1600-0854.2007.00599.x. [DOI] [PubMed] [Google Scholar]
- McAllister AK. Dynamic aspects of CNS synapse formation. Annu Rev Neurosci. 2007;30:425–450. doi: 10.1146/annurev.neuro.29.051605.112830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNair K, Spike R, et al. A role for RhoB in synaptic plasticity and the regulation of neuronal morphology. J Neurosci. 2010;30(9):3508–3517. doi: 10.1523/JNEUROSCI.5386-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison DK. The 14-3-3 proteins: integrators of diverse signaling cues that impact cell fate and cancer development. Trends Cell Biol. 2009;19(1):16–23. doi: 10.1016/j.tcb.2008.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama AY, Harms MB, et al. Small GTPases Rac and Rho in the maintenance of dendritic spines and branches in hippocampal pyramidal neurons. J Neurosci. 2000;20(14):5329–5338. doi: 10.1523/JNEUROSCI.20-14-05329.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newey SE, Velamoor V, et al. Rho GTPases, dendritic structure, and mental retardation. J Neurobiol. 2005;64(1):58–74. doi: 10.1002/neu.20153. [DOI] [PubMed] [Google Scholar]
- Nimchinsky EA, Sabatini BL, et al. Structure and function of dendritic spines. Annu Rev Physiol. 2002;64:313–353. doi: 10.1146/annurev.physiol.64.081501.160008. [DOI] [PubMed] [Google Scholar]
- Paradis S, Harrar DB, et al. An RNAi-based approach identifies molecules required for glutamatergic and GABAergic synapse development. Neuron. 2007;53(2):217–232. doi: 10.1016/j.neuron.2006.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardo CA, Eberhart CG. The neurobiology of autism. Brain Pathol. 2007;17(4):434–447. doi: 10.1111/j.1750-3639.2007.00102.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penzes P, Beeser A, et al. Rapid induction of dendritic spine morphogenesis by trans-synaptic ephrinB-EphB receptor activation of the Rho-GEF kalirin. Neuron. 2003;37(2):263–274. doi: 10.1016/s0896-6273(02)01168-6. [DOI] [PubMed] [Google Scholar]
- Piddini E, Schmid JA, et al. The Ras-like GTPase Gem is involved in cell shape remodelling and interacts with the novel kinesin-like protein KIF9. EMBO J. 2001;20(15):4076–4087. doi: 10.1093/emboj/20.15.4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plachez C, Richards LJ. Mechanisms of axon guidance in the developing nervous system. Curr Top Dev Biol. 2005;69:267–346. doi: 10.1016/S0070-2153(05)69010-2. [DOI] [PubMed] [Google Scholar]
- Ramon y Cajal S. In: Histology of the Nervous System of Man and Vertebrate. Swanson N, Swanson LW, translators. Oxford University Press; New York: 1911. 1995. [Google Scholar]
- Randazzo PA, Inoue H, et al. Arf GAPs as regulators of the actin cytoskeleton. Biol Cell. 2007;99(10):583–600. doi: 10.1042/bc20070034. [DOI] [PubMed] [Google Scholar]
- Rao A, Kim E, et al. Heterogeneity in the molecular composition of excitatory postsynaptic sites during development of hippocampal neurons in culture. J Neurosci. 1998;18(4):1217–1229. doi: 10.1523/JNEUROSCI.18-04-01217.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redmond L, Kashani AH, et al. Calcium regulation of dendritic growth via CaM kinase IV and CREB-mediated transcription. Neuron. 2002;34(6):999–1010. doi: 10.1016/s0896-6273(02)00737-7. [DOI] [PubMed] [Google Scholar]
- Sakane A, Honda K, et al. Rab13 regulates neurite outgrowth in PC12 cells through its effector protein, JRAB/MICAL-L2. Mol Cell Biol. 2010;30(4):1077–1087. doi: 10.1128/MCB.01067-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saneyoshi T, Fortin DA, et al. Regulation of spine and synapse formation by activity-dependent intracellular signaling pathways. Curr Opin Neurobiol. 2010;20(1):108–115. doi: 10.1016/j.conb.2009.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmandke A, Strittmatter SM. ROCK and Rho: biochemistry and neuronal functions of Rho-associated protein kinases. Neuroscientist. 2007;13(5):454–469. doi: 10.1177/1073858407303611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro L, Love J, et al. Adhesion molecules in the nervous system: structural insights into function and diversity. Annu Rev Neurosci. 2007;30:451–474. doi: 10.1146/annurev.neuro.29.051605.113034. [DOI] [PubMed] [Google Scholar]
- Sholl DA. Dendritic organization in the neurons of the visual and motor cortices of the cat. J Anat. 1953;87(4):387–406. [PMC free article] [PubMed] [Google Scholar]
- Sin WC, Haas K, et al. Dendrite growth increased by visual activity requires NMDA receptor and Rho GTPases. Nature. 2002;419(6906):475–480. doi: 10.1038/nature00987. [DOI] [PubMed] [Google Scholar]
- Soderling TR. The Ca-calmodulin-dependent protein kinase cascade. Trends Biochem Sci. 1999;24(6):232–236. doi: 10.1016/s0968-0004(99)01383-3. [DOI] [PubMed] [Google Scholar]
- Tada T, Sheng M. Molecular mechanisms of dendritic spine morphogenesis. Curr Opin Neurobiol. 2006;16(1):95–101. doi: 10.1016/j.conb.2005.12.001. [DOI] [PubMed] [Google Scholar]
- Talens-Visconti R, Peris B, et al. RhoE stimulates neurite-like outgrowth in PC12 cells through inhibition of the RhoA/ROCK-I signalling. J Neurochem. 2010;112(4):1074–1087. doi: 10.1111/j.1471-4159.2009.06526.x. [DOI] [PubMed] [Google Scholar]
- Tashiro A, Minden A, et al. Regulation of dendritic spine morphology by the rho family of small GTPases: antagonistic roles of Rac and Rho. Cereb Cortex. 2000;10(10):927–938. doi: 10.1093/cercor/10.10.927. [DOI] [PubMed] [Google Scholar]
- Tolias KF, Bikoff JB, et al. The Rac1-GEF Tiam1 couples the NMDA receptor to the activity-dependent development of dendritic arbors and spines. Neuron. 2005;45(4):525–538. doi: 10.1016/j.neuron.2005.01.024. [DOI] [PubMed] [Google Scholar]
- Van Aelst L, Cline HT. Rho GTPases and activity-dependent dendrite development. Curr Opin Neurobiol. 2004;14(3):297–304. doi: 10.1016/j.conb.2004.05.012. [DOI] [PubMed] [Google Scholar]
- van Heusden GP. 14-3-3 Proteins: insights from genome-wide studies in yeast. Genomics. 2009;94(5):287–293. doi: 10.1016/j.ygeno.2009.07.004. [DOI] [PubMed] [Google Scholar]
- Ward Y, Kelly K. Gem protein signaling and regulation. Methods Enzymol. 2006;407:468–483. doi: 10.1016/S0076-6879(05)07038-2. [DOI] [PubMed] [Google Scholar]
- Xia Z, Dudek H, et al. Calcium influx via the NMDA receptor induces immediate early gene transcription by a MAP kinase/ERK-dependent mechanism. J Neurosci. 1996;16(17):5425–5436. doi: 10.1523/JNEUROSCI.16-17-05425.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Srivastava DP, et al. Kalirin-7 controls activity-dependent structural and functional plasticity of dendritic spines. Neuron. 2007;56(4):640–656. doi: 10.1016/j.neuron.2007.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Meng Y, et al. A critical role of Rho-kinase ROCK2 in the regulation of spine and synaptic function. Neuropharmacology. 2009;56(1):81–89. doi: 10.1016/j.neuropharm.2008.07.031. [DOI] [PubMed] [Google Scholar]
- Ziv NE, Smith SJ. Evidence for a role of dendritic filopodia in synaptogenesis and spine formation. Neuron. 1996;17(1):91–102. doi: 10.1016/s0896-6273(00)80283-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1: Validation of the RNAi-resistant Rem2 cDNA. (a) Western blot with anti-Rem2 antibody of HEK 293T cell lysates. anti-β-actin serves as a loading control. Developed using the Odyssey IR System. (b) Quantification of excitatory synapse density as PSD-95/synapsin overlapping puncta for Control, RNAi, OE, and Rescue conditions. Asterisk indicates p<0.05 from control by two-way between-subjects ANOVA with Tukey posthoc test. (C) Representative images of a stretch of dendrite of a neuron from the Control, RNAi, OE, and Rescue conditions. Scale bar indicates 50 μm. n > 50 neurons/condition.
Supplementary Figure 2: Simultaneous cotransfection of three different plasmids into the same neuron via the calcium phosphate method. (a) Representative images of neurons cotransfected with myc-Rem2-, RFP- and GFP-expressing plasmids, and subsequently stained for myc. (b) Quantification of the percentage of cells triple cotransfected (as detected by RFP, GFP, and anti-myc expression), double cotransfected (as detected by RFP and GFP, RFP and anti-myc, or GFP and anti-myc), or single transfected (as detected by RFP or GFP expression alone). Scale bar indicates 50 μm. n = 60 neurons (30 selected by GFP expression, 30 selected by RFP expression).
Supplementary Figure 3: The same images from (a) Supplementary Figure 1, (b) Figure 4, (c) Figure 5, (d) Figure 6, and (e) Figure 7, separated to show individual fluorescence channels. Note that synaptic immunostaining associated with the processes of non-transfected neurons is visible in some images (for example, images extracted from Figure 5). Scale bars indicate 5 μm. n > 40 neurons/condition for all experiments.
Supplementary Figure 4: Rem2 that cannot bind 14-3-3 proteins is more abundant in the nucleus than is wild-type Rem2. (a) Quantification of nuclear anti-Rem2 staining for Control, RNAi, Rescue, and S69A/S334A Rescue conditions. (b) Quantification of somatic, cytoplasmic anti-Rem2 staining intensity for each condition. (c) Representative images of neurons in each condition immunostained for anti-Rem2 and anti-NeuN. Asterisks indicate p<0.01 from control, unless otherwise indicated, by two-sample t-test. Scale bar indicates 5 μm. n > 30 neurons/condition.
Supplementary Figure 5: Further analysis of Rem2 L317G (no CaM binding) mutant. (a) Synapsin I puncta density onto GFP- positive neurons transfected with the indicated constructs (compare to Figure 4). Rem2 RNAi in the postsynaptic neuron significantly decreases the density of synapsin puncta, and L317G (Rem2 that cannot bind CaM) rescues this effect (* = p<0.01). (b) The intensity of PSD-95 immunostaining in the transfected neuron overlapping with Synapsin I immunostaining does not change significantly between conditions; thus, we conclude that L317G (Rem2 that cannot bind CaM) does not alter the postsynaptic expression of PSD-95. (c) The intensity of total PSD-95 immunostaining in the transfected neuron does not change significantly between conditions; thus, we conclude that L317G (Rem2 that cannot bind CaM) does not alter the postsynaptic expression of PSD-95.