Abstract
E-cadherin is an important tumor suppressor gene whose expression is lost when cells acquire a metastatic phenotype. We analyzed the role of E-cadherin mis-splicing as a mechanism of its downregulation by analyzing a mis-spliced E-cadherin transcript that lacks exon 11 of this gene. This results in a frame shift and a premature termination codon which targets this transcript for degradation. Tumor tissues including breast (20%, n=9)), prostate (30%, n=9) and Head and Neck (H&N) (75%, n=8) cancer, express the exon 11 skipped transcripts (versus non-malignant controls) and its levels inversely correlate with E-cadherin expression. This is a novel mechanism of E-cadherin downregulation by mis-splicing in tumor cells which is observed in highly prevalent human tumors. In the H&N cancer model, non-tumorigenic keratinocytes express exon 11 skipped splice product 2–6 fold lower than the H&N tumor cell lines. Mechanistic studies reveal that SFRS2 (SC35), a splicing factor as one of the regulators that increases mis-splicing and downregulates E-cadherin expression. Furthermore, this splicing factor was found to be over expressed in five out of seven H&N cell lines and primary H&N tumors. Also, methylation of E-cadherin gene acts as a regulator of this aberrant splicing process. In two H&N cell lines, wild type transcript expression increased 16–25 folds while the percentage of exon 11 skipped transcripts in both the cell lines decreased 5–30 folds when cells were treated with a hypomethylating agent, azacytidine. Our findings reveal that promoter methylation and an upregulated splicing factor (SFRS2) are involved in the E-cadherin mis-splicing in tumors.
Keywords: E-cadherin, gene splicing, epigenetics
Introduction
Presence of a premature termination codon mutation (PTC) results in an in-frame stop codon in RNA. These RNA transcripts are degraded by pathway called the nonsense mediated decay pathway (NMD) that is active in all mammalian cells (1–3). Theoretically, the NMD prevents expression of truncated proteins that could act as dominant negatives and have adverse effects. PTC in a RNA transcript could be due to a point mutation or a frame shift and the rapid degradation of PTC-containing RNAs can be a mechanism by which loss-of-function of potential tumor suppressor genes can occur. Alternative splicing affects more than 60% of the human genes (4) and cancer cells are known to have different splicing patterns as compared to their normal counterparts (5). Cancer cells are also associated with alternative splicing of several tumor suppressors genes, such as BRCA1/2, APC, WT1, mdm2 and ATM (6–10) with subsequent NMD-mediated degradation and loss of expression.
Changes in splicing patterns in tumor cells could be secondary to aberrant expression of splice factors affecting the splicing patterns of a number of genes. Recent reports on aberrant expression of SR proteins (serine-arginine rich), SFRS1 and SFRS2 splicing factors exemplify their potential role in transformation (11,12). Splicing factors are also regulated by oncogenic signaling pathways that affect activity of SR proteins (13).
Methylation affects transcription of the gene (14) which in turn is intricately linked to splicing as both processes occur simultaneously (15–16). As splicing factors are also known to associate with the transcriptional machinery (17), the nature of the transcriptional complex assembled for the transcription of a particular gene influences the splicing factors available for splicing which in turn affects the splicing patterns. Methylation changes can also occur at the lysine residues of the histones that are able to modify the chromatin structure which are now recognized as important regulator of alternative splicing (18,19) and in fact define the exon-intron boundaries (20). The epigenetic events associated with methylation could thereby potentially affect splicing with change in transcriptional rates and histone modifications.
We have previously reported an E-cadherin exon 11 skipped, alternatively spliced transcript that is subject to NMD degradation in chronic lymphocytic leukemia cells (CLL) (21). This tumor suppressor gene has a critical role in maintaining cell-cell adhesion and cell-matrix interactions (reviewed in 22, 23). As these functions are important for the epithelial membrane integrity, mis-splicing of this transcript was analyzed in epithelial cancers. We report the presence of this exon 11 skipped transcript in primary human cancer tissues and further characterize its regulation by splicing factors and methylation status of the E-cadherin gene.
Materials and Methods
Primary human tissues and TissueScan arrays
Primary human H&N cancer specimens (8 matched pairs) and matched normal controls were obtained from Cooperative human tissue network. Tissue RNA was isolated by grinding frozen tissues in RLT buffer (Qiagen RNA kit, Valencia, CA) followed by the kit isolation protocol. TissueScan qPCR arrays for breast, prostate and other cancers were purchased from Origene Rockville, MD. Each tissue type has nine individual tumor cDNA and three tissue matched normals. Both primary human tissues and Tissue scan arrays are analyzed by light microscopy to confirm the quality and the nature of the tissues.
Transcript specific real time PCR
E-cadherin expression in cells was analyzed by a real time PCR. The 5’ primer GGATGTGCTGGATGTGAATG localizes to exon 10 of the E-cadherin gene. The 3’ primer CACATCAGACAGGATCAGCAGAA localizes to the exon 12. The taqman probe 10–11 (TAACATATCGGATTTGGAGAGAC) determining the wild type E-cadherin transcript level, binds to the junction of exon 10-exon 11. The expression level of the skipped or aberrant transcript (transcript lacking exon 11) is determined by the same set of PCR primers as mentioned above with a taqman probe 10–12 (CAGAAAATAACGTTCTCCAGTTG) binding the exon 10-exon 12 junction. Real time PCR for actin expression was used as a control and relative expression of transcripts was determined by comparative Ct method of Pfall (24) that compares transcript levels in different samples using a co-amplified internal control.
Cell Culture and Microarray Analysis
The H&N cancer cell lines UMSCC1, 2, 12, 14,22b were obtained from University of Michigan, Ann Arbor, and MI. Cal27 was obtained from ATCC. Telomerase reverse transcriptase immortalized keratinocytes (tert-kert) and normal human oral keratinocytes (NHOK) cells were obtained from Dr. David Wong’s lab (UCLA Dental School) and cultured as previously described (25). No further authentication of the cell lines was done by the authors. Emetine treatment was performed at concentration of 100µg/ml for 8 hours (10). In some groups actinomycin D was added at a concentration of 2µg/ml, together with emetine (100µg/ml) (26). For microarray analysis cRNA was obtained as per standard protocols and hybridized to the Affymetrix chip 133A (Affymetrix, Santa Clara, CA). The NTEI (nonsense transcript enrichment index) (26) score is the ratio of upregulation of the signal intensity on emetine treatment of HNSCC cells of a particular probe set, divided by the ratio of upregulation of signal intensity of non-malignant tert-kert cells for the same probe set.
Minigene constructs
Exon 10, 11 and 15 of the E-cadherin gene along with their flanking intronic regions were cloned into a minigene construct pDup (gift from Dr. Black, UCLA, ref 27). The exon 10 is 245 base pairs and a DNA fragment from position −325 from 5’ exon-intron junction to + 338 from the 3’ exon-intron junction was amplified by PCR from normal human PBMC DNA and cloned in the pDup vector at the Apa1 and BglII site. Similarly, for exon 11 (146 bp) a fragment −623 to +544 and for exon 15, a fragment from −477 to +518 was cloned. The constructs were transiently transfected in SCC12 cells and cells were analyzed by RT-PCR analysis. The primers for amplification are GACACCATGCATGGTGCACC and GCAGCTCACTCAGTGTGGCA which bind to β-globin exon E1 and E3 respectively. To quantify the amount of correctly or aberrantly spliced transcripts a real time PCR was performed with taqman probes specifically binding the junction of β-globin E1-exon 11 or β-globin E1-β-globin E3. Cells were transfected with 1µg of plasmid along with 5 µg of bluescript DNA and Lipofectamine 2000 (Invitrogen, Carlsbad, CA). Forty eight hours after transfection, cells were analyzed by real time RT-PCR.
Analysis of splicing factors and SiRNA knockdown
H&N cell lines were analyzed for expression of splicing factors SFRS1,2,5, and 6 with real time PCR (Applied Biosystems, CA). The SCC12 and SCC1 lines transfected with 10nM SiRNA and 1 µg of the minigene exon 11 construct SiRNA against SFRS2 (Santa Cruz Biotechnology, CA). Western blot analysis for SFRS2 expression was performed with Santa Cruz Biotechnology antibody, E-16. 48 hours after transfection the cells were analyzed for SRFS2 expression and quantitative transcript specific real time PCR analysis for E-cadherin splicing.
DNA Hypomethylation
SCC1 and 22b cell lines were treated with 5-aza-deoxycytidine (5µM) for 96 hours and cells analyzed for E-cadherin RNA expression by transcript specific real time PCR. In parallel, DNA was isolated and bisulfite modified methylation specific PCR performed as described (28) along with western blot analysis for E-cadherin.
Results
Wild type and Exon 11 skipped transcript
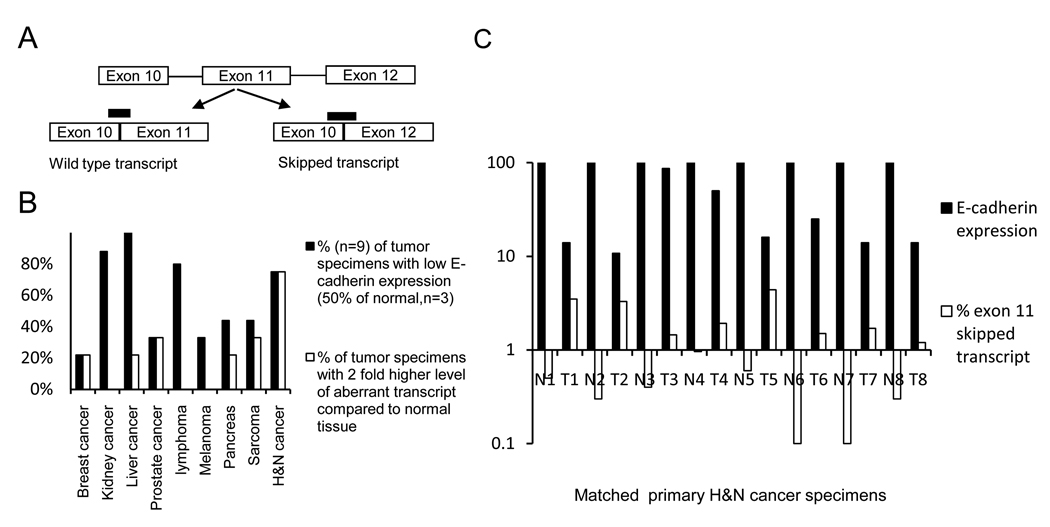
To ascertain if exon 11 skipped transcripts are present in solid tumors we screened eight pairs of H&N cancer primary tissues (tumor and matched normal). For lymphomas, melanomas, sarcomas, breast, kidney, liver, pancreas prostate cancers, a tissue scan PCR array with nine tumor cDNAs and three normal tissue cDNAs was analyzed. A transcript specific real time PCR was performed on all tissues which distinguishes normal wild type transcripts from the exon 11 skipped transcripts (figure 1A).
Figure 1. Aberrant transcripts in primary human tumor tissues.
A, Schematic showing the location of the probes to identify the wild type and aberrant transcript (exon 11 skipped). B, Different types of human cancer cDNA analyzed by real time PCR analysis for wild type and aberrant E-cadherin transcript (n=9 for each tumor, n=3 for matched organ specific normal tissue). Black bars represent % of tumor cDNAs with E-cadherin expression less than 50% of matched normal tissue, open bars represent the percentage of tumor cDNAs in which the percentage of aberrant transcript is at least two-fold greater than the aberrant transcript in matched normal tissue. For the H&N cancer group the data shows, relative levels of total E-cadherin expression in 8 pairs of matched normal (N) and tumor tissues (T), relative to actin and adjusted to the matched normal control. C, H&N cancer data from eight matched groups is shown with wild type and percentage of exon 11 skipped transcript in each matched pair (log scale).
Figure 1B shows percent of each tumor type with E-cadherin RNA expression, which is decreased, compared to corresponding control tissue (at least 50% expression decrease vs. non-malignant control tissue, black bars). To estimate the relative abundance of the aberrant transcript as compared to the wild type in tissues, the difference between the delta Ct of the transcript specific PCR reactions was calculated by using the cycle difference in the two transcripts to calculate the relative abundance, e.g. with 8 cycle difference between the two transcripts the exon 11 skipped transcript is 0.4% of the wild type. The percent of each tumor type with at least a two-fold higher level of exon 11 skipped transcript compared to corresponding normal tissue is shown in figure 1B (open bars). As shown in figure 1B, tumor types with the highest incidence of loss of normal E-cadherin RNA expression were kidney cancer (90%), liver cancer (100%), lymphoma (80%) and H&N cancer (75%). Although the sample size in this screening is relatively small, these data are consistent with previous literature (29–31) that also identified loss of E-cadherin expression as being frequent in these cancers. Of these four tumor types, the one with the highest frequency of increased expression of exon 11 skipped transcripts was H&N cancer (open bar, 7 of 9 specimens) and this tumor was chosen for further analysis.
Fig 1C depicts the results of the transcript specific PCR assay for each H&N cancer specimen. Relative amounts of wild type RNA are shown in closed bars and % of exon 11 skipped transcripts in open bars (log scale). As can be seen there is an inverse correlation between the relative amount of normal E-cadherin RNA expression and exon 11 skipped RNA (p=0.08, correlative coefficient). It should be noted that this is an underestimation of the exon 11 skipped transcripts as the NMD pathway had been active in these cells, which would have caused constant rapid degradation of this transcript and not the wild type.
Microarray analysis with inhibition of NMD pathway
An unbiased screen for PTC codons in five H&N cancer cell lines (UMSCC1, 2, 12, 14,22b) also supported the frequency of a PTC bearing transcript that was caused by exon 11 skipping. In this screen, the NMD pathway is paralyzed by a short exposure to emetine as previously described (26) and microarray RNA profiling is performed with or without NMD paralysis. Immortalized non-transformed tert-kert cells were used as control cell and compared to five H&N cell lines (GEO accession number GSE29788). A more than 5 fold mean emetine induced upregulation was observed in 199 of 23000 probes. The initial “hits” (significant upregulations by emetine) were also analyzed for RNA signal intensity versus tert keratinocytes in the absence of emetine. Our reasoning was that those with downregulated expression were most likely to represent loss-of-function tumor suppressors. The E-cadherin gene was identified as one of the top 100 maximally downregulated genes when comparing H&N tumor lines to tert keratinocytes and its RNA signal was significantly increased upon NMD paralysis with emetine (supplementary data).
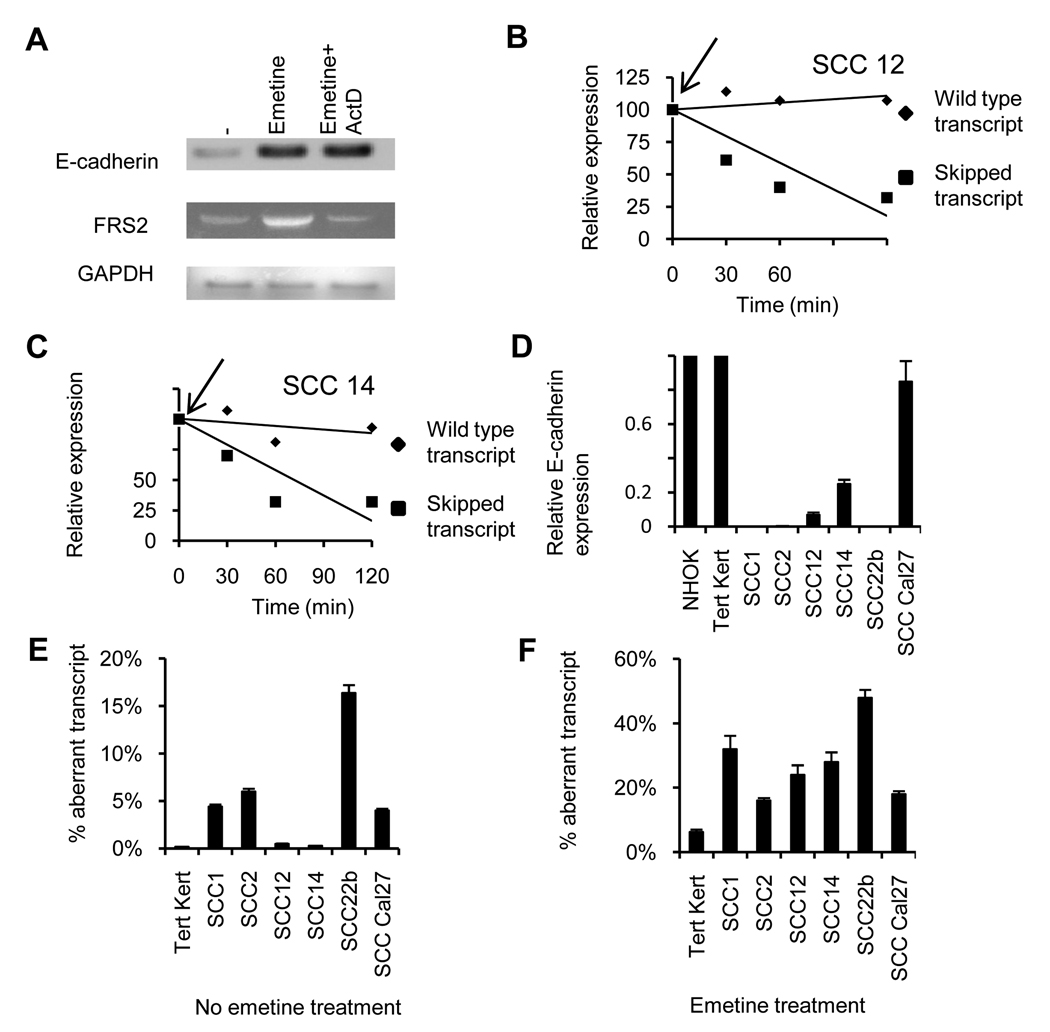
To confirm these findings, we treated the H&N line UMSCC14A with emetine to paralyze the NMD. As emetine exposure also results in upregulation of a number of stress response genes via stress induced transcription, actinomycin D was added to block the upregulation of stress-induced transcription (32). Following exposure to emetine or emetine+actinomycin D, RT-PCR analysis was performed (figure 2A). The primers for this PCR bind to the 3’ end of the E-cadherin gene and a stronger band was observed in the emetine treated cells. Importantly, addition of actinomycin D did not prevent this upregulation indicating the emetine-induced upregulation is not due to a stress response that activates transcription. Figure 2A shows a contrasting example of the FRS2 (fibroblast growth factor receptor substrate 2) gene for which the emetine induced upregulation was prevented by addition actinomycin D. Upon sequencing the entire cDNA of FRS2 no PTC mutations were identified. In contrast, sequencing demonstrated that, in addition to the wild type E-cadherin transcript, a smaller transcript was identified which lacked exon 11 and resulted in a frame-shift and a PTC located in exon 12 as previously described (21).
Figure 2. Analysis of the aberrant transcript.
A, RT-PCR analysis of untreated UMSCC12 cells treated with emetine or emetine plus actinomycin-D and analyzed for expression of E-cadherin RNA. With the same cDNA preparation FRS2 and GAPDH were amplified. B & C UMSCC14 and UMSCC12 cells were treated with actinomycin-D, and transcript specific PCR performed at different time-points. D, Relative expression of E-cadherin in Tert keratinocytes, NHOK and H&N cell lines relative to expression in Tert-kert cells. E, Percentage exon 11 skipped transcript calculated by the cycle difference between the two transcripts with transcript specific real time PCR of untreated (E) and emetine treated, (F) cells.
Half-life of the exon 11 skipped transcript
To further support the notion that the PTC-containing RNA is rapidly degraded by the NMD, we studied the turnover of the aberrant transcript versus wild type transcripts. UMSCC14 and UMSCC12 H&N cells were treated with actinomycin D (2 µg/ml) to block transcription and then at different time points, the two transcripts were determined by real time PCR. In both cell lines (figure 2B,C), the wild type E-cadherin transcript is stable over the 2 hour time course while the signal from the exon 11 skipped transcript is rapidly lost (t1/2 approx 60 min) indicating, as expected, its short half-life in H&N lines. With this short half-life of the exon 11 skipped transcript, our assays on tissues and cells with active NMD pathway, underestimate the amount of this transcript.
Quantification of wild type and exon 11 skipped transcript
To determine the relative abundance of this aberrant transcript, the H&N cell lines were analyzed by real time PCR analysis using the transcript-specific PCR approach shown in figure 1. Figure 2D shows the relative (adjusted to actin) E-cadherin expression in the cell lines compared to two normal non-transformed counterparts, normal human oral keratinocytes (NHOK) and Tert Kert cells. Except for the Cal27 cell line, all the other cell lines have lower E-cadherin levels as compared to the two normal non-transformed control cells. Also the relative abundance of exon 11 skipped transcript in the tumorigenic cell lines is higher than in the Tert-Kert cells (4–24 fold higher in 4 of the 6 H&N cancer cell lines, figure 2E). A similar experiment was performed on the H&N cancer cell lines and Tert-Kert cells treated with emetine (figure 2F) which the percentage of aberrant E-cadherin transcripts is 3–8 fold higher in all the H&N cell lines than Tert Kert cells. Thus, exon 11 skipping occurs at a low level in non-transformed oral epithelial cells but is markedly increased in malignant H&N epithelial tissues.
Minigene constructs
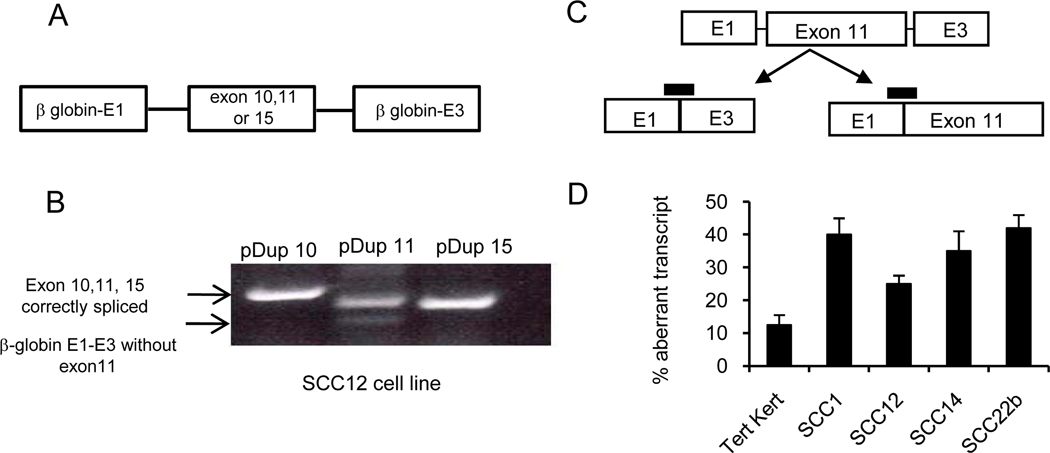
Exon 11 along with its intronic regions were amplified and sequenced from UMSCC12 and UMSCC22b cells and no mutations were identified. This was not unexpected since as described above, the aberrant splicing is also seen but quantitatively less in normal cells. To further study the mechanism of aberrant splicing, and to investigate why it is significantly increased in malignant tissue, exon 11 along with its intronic regions were cloned into the pDup minigene construct (27). As controls, we also cloned E-cadherin exon 10 and 15. The cloned exons are flanked by beta-globin exons E1 and E3 (figure 3A). Transfection of the plasmid constructs into UMSCC12 cells and RT-PCR analysis reveals that there is a correctly spliced product (upper band) in cells transfected with exon 10 and 15 pDup constructs. In the case of pDup exon 11 construct, an additional smaller band is seen which lacked exon 11 on sequencing. A transcript specific real time PCR strategy similar to the above described E-cadherin wild type and aberrant transcripts (figure 2) was developed to quantify improperly spliced E1–E3, and the correctly spliced E1-exon11-E3 transcripts in the minigene construct (figure 3C). Tert-kert cells and H&N cell lines were transiently transfected with the exon 11 minigene construct and 48hrs later a real time RT-PCR analysis was performed. Figure 3D demonstrates H&N cell lines with a 2–3 fold increase in aberrant splicing versus non-transformed tert-keratinocytes providing additional evidence that the E-cadherin exon 11 is improperly spliced in H&N cell lines.
Figure 3. Minigene experiments show similar splicing patterns.
A. Schematic showing the minigene constructs with E-cadherin exon 10, 11, and 15. B, Transient transfection and RT-PCR analysis (DNA gel) of UMSCC12 cells transfected with various minigene constructs. Primers bind to β globin exon E1 and E3. Upper arrow indicates the correct expected size and the lower arrow indicates a transcript lacking exon 11. C, Schematic for the location of probes for transcript specific PCR. D, Real time PCR data showing the percentage aberrant transcript in Tert-kert and H&N cell lines (mean±SD).
Role of splicing factors in aberrant splicing
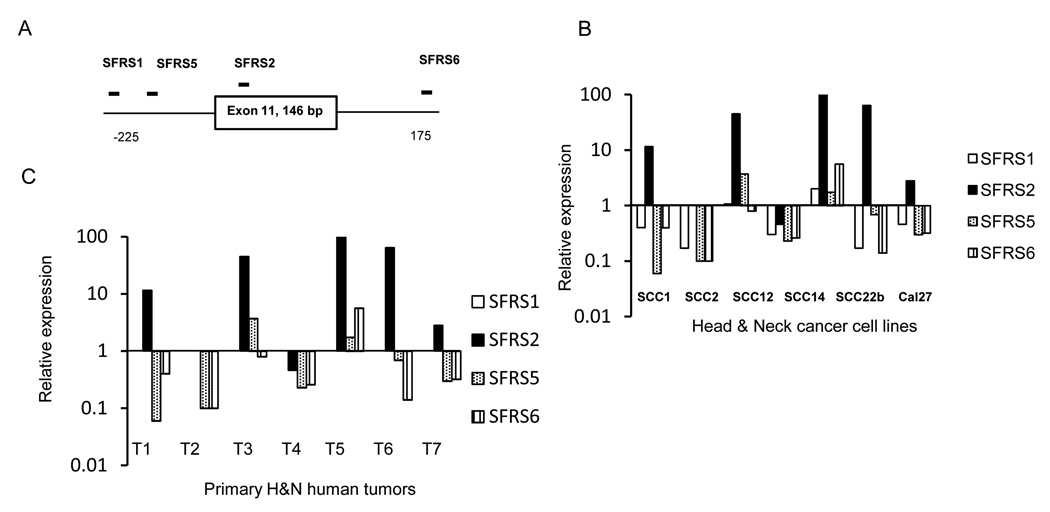
Since we did not identify mutations in exon 11 and the intronic region in two H&N cell lines, we analyzed splicing factors as potential modulators of exon 11 skipping. We analyzed the exon 11 sequence with the Exonic splicing enhancer (ESE) finder software program (33) to determine the putative binding sites of known splicing factors and observed binding sites for the SFRS1,2,5 and 6 splice factors (34,35) in exon 11 and its neighboring intronic region (−225 to +175 bp, figure 4). The expression of these splicing factors in H&N lines relative to Tert-Kert is shown in figure 4B. A consistent finding was the upregulation of SFRS2 splicing factor which formed the rationale for further analysis. Similar analysis in Figure 4C shows expression of splicing factors in primary H&N tumors relative to the matched normal tissue. In five out of seven pairs analyzed, SFRS2 is also upregulated in tumor tissues as compared to the other splicing factors which corroborates the H&N cell line data.
Figure 4. Role of splicing factors in aberrant splicing.
A. Schematic with predicted binding sites for splicing factors around E-cadherin exon 11. B. Real time PCR data for expression of SFRS1,2,5 and 6 splicing factors in cell lines relative to Tert-Kert cells. C, Expression of splicing factors by real time PCR in primary human H&N tumors relative to matched normal tissue (7 matched pairs).
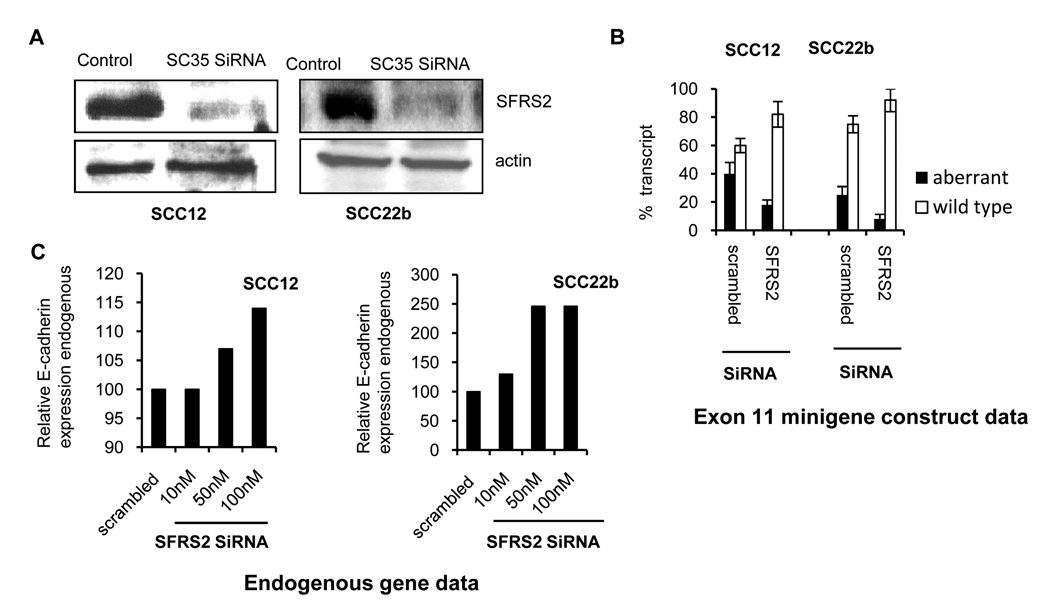
This was further analyzed with the minigene exon 11 construct (figure 3A) which was co-transfected with SiRNA (10nM) targeting the splice factor SFRS2 (12) in UMSCC12 and UMSCC22b cells. A scrambled SiRNA control transfected along with the minigene construct was used as the control to determine the relative ratio of the correctly spliced E1-11-E3 transcript. Western blot analysis (figure 5A) confirmed successful knockdown of SFRS2 in UMSCC12 and UMSCC22b cells. Figure 5B shows the data from three transfections (mean±SD). In UMSCC12 cells, knockdown of SFRS2 resulted in decrease in aberrant transcript from 40±8% to 18±3.6% (mean±SD, two-tailed p=0.012) and in UMSCC22b cells the aberrant transcript decreases from 25±6% to 8± 3.4% (mean±SD, two-tailed p=0.013). Both cell lines show that inactivation of SFRS2 splicing factor results in an increase in the correctly spliced exon 11 minigene transcript and decrease in the aberrant transcript. To determine the effect of SiRNA knockdown on the endogenous E-cadherin gene splicing, these two cell lines were transiently transfected with varying amounts (10,50 and 100nm) of SFRS2 SiRNA (figure 5C) and analyzed for RNA transcripts. We observe an increase in wild type E-cadherin RNA when cells were transfected with increasing amounts of SiRNA. The increase in UMSCC12 cells is marginal while in UMSCC22b cells there is a 2.5 fold increase in wild type transcripts.
Figure 5. Effect of SiRNA mediated inactivation of SFRS2 on splicing.
A. Western blot analysis for UMSCC12 and UMSCC22b cells transfected with SFRS2 SiRNA or scrambled control sequence. B. Bar diagram with percentage of aberrantly spliced transcript in cells transfected with exon 11 minigene construct and SFRS2 SiRNA as determined by transcript specific real time PCR in UMSCC12 and UMSCC22b. Control cells were transfected with the exon 11 minigene construct and scrambled SiRNA. Data are mean ± SD, n=3. C. E-cadherin expression by real time PCR analysis in two cell lines transfected with varying amounts of SFRS2 SiRNA.
Methylation status and splicing
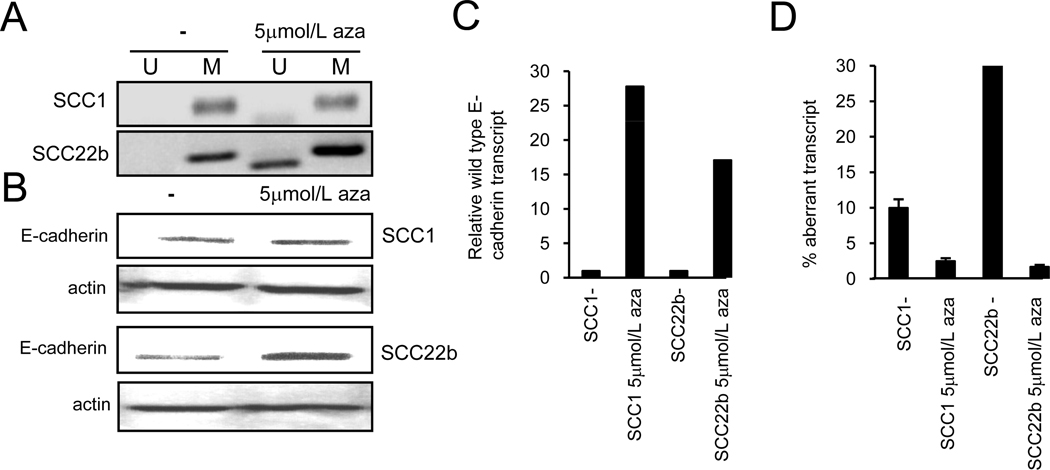
Promoter methylation of the E-cadherin gene downregulates its expression (22,23,28) that can be reversed by hypomethylating agents like azacytidine which are also being used in the clinical setting as well (reviewed in 36). Also as transcription and splicing occur simultaneously in the cell and inclusion of certain exons is dependent upon the transcriptional state of the gene, we investigated whether changes in E-cadherin transcription by hypomethylation could affect exon 11 splicing. Two H&N cancer cell lines UMSCC1 and 22b were treated with 5-aza-deoxycytidine (5µm for 4 days) and analyzed for DNA hypomethylation, E-cadherin expression and quantification of the two transcripts. Figure 6A shows the methylation specific PCR performed on bisulfite treated DNA from untreated and azacytidine treated cells (28). As expected azacytidine treatment results in the amplification of a band with primers specific for the non-methylated DNA which is not seen in DNA from un-treated cells. Identically treated cells also show increase in E-cadherin expression by western blot analysis in the two cell lines (figure 6B). Figures 6C,D describe the RNA transcript analysis with data, UMSCC1 and UMSCC22b cells upon azacytidine treatment show an increase in the wild type E-cadherin transcript (16–28 fold) with a corresponding decrease in the exon 11 skipped transcript decreases relative to the wild type transcript (5–30 fold decrease). As the overall transcription increases secondary to promoter hypomethylation, it is expected that both the transcripts would increase, and potentially increase to the same extent. However the results point towards a preferential increase in the wild type transcript in the azacytidine treated cells. The data shows that the exon 11 skipped transcript can be clearly modulated by the hypomethylation process and is not a random phenomenon.
Figure 6. Hypomethylating agent and aberrant splicing.
A, Methylation specific PCR for UMSCC1 and UMSCC22b cell lines with and without azacytidine treatment. B, Western blot analysis for UMSCC1 and UMSCC22b showing E-cadherin expression and actin control with azacytidine treatment. C, Expression of wild type E-cadherin transcripts relative to the untreated cells and adjusted to actin, along with % aberrant transcript after azacytidine treatment.
Discussion
We initiated this study to determine if a previously identified E-cadherin RNA splicing abnormality that results in loss of E-cadherin expression in chronic lymphocytic leukemia cells (21) could also be found in solid tumors. Since majority of human cancers are epithelial in origin and E-cadherin has a well documented role in cell-cell adhesion and junctional complexes, we thought it important to see if exon 11 skipping could play a role in loss of expression. The results demonstrate an increase in exon 11 skipped transcript in several solid tumor types along with decreased total E-cadherin expression. Because the frequency of exon 11 skipping was highest in H&N tumors, we focused on this tumor for subsequent studies. Even though the exon 11 skipped transcript can be amplified from normal control tissues, it is more abundantly expressed in H&N tumors and cell lines that have a lower expression of E-cadherin.
The differential splicing or changes in alternative splicing patterns of exon 11 between normal and tumor cells could be secondary to differential expression of splicing-factors. We chose to study the SFRS2 splicing factor because it has a putative binding site in the E-cadherin exon 11 and is over-expressed in H&N cell lines and primary tumor tissues as compared to the other splicing factors (figure 4, 5). This splicing factor is reported to have a role in the alternative splicing of the tumor suppressor gene KLF6 (12). To determine if the over-expression of this factor plays a role in aberrant splicing, this was knocked down in two cell lines and changes in minigene construct splicing pattern were observed (figure 5) along with a dose dependent increase in wild type E-cadherin expression when different amounts of SFRS2 SiRNA were used. The ability of SFRS2 siRNA to significantly enhance exon 11 incorporated transcripts suggests the upregulation of SFRS2 in H&N cell lines may be involved in aberrant exon 11 splicing. In addition, the ability of knockdown to significantly increase expression of the wild type transcript in the UMSCC22b cell line, suggests that the enhanced aberrant splicing plays a role in loss-of-expression of this tumor suppressor gene rather than just representing an epi-phenomenon. However, it is certainly possible that additional splicing factors not studied also play a role in the observed aberrant splicing.
Our studies with azacytidine address two related processes, gene transcription and splicing. Our data shows that azacytidine treatment results in hypomethylation of the E-cadherin promoter and an increase in transcription (figure 6) along with changes the splicing pattern of the cells with a preferential increase in the wild type transcript. Conversely, methylation of this gene that is frequently observed in human malignancies can potentially decrease E-cadherin expression with both transcriptional repression and an increase in exon 11 mis-splicing. There are two possible explanations for methylation status to result in splicing changes. Firstly, hypomethylation changes the nature of the transcriptional complex formed at the promoter which then recruits additional factors which have a dual role as transcriptional factors and also affect splicing of exons (37–40). In the fibronectin gene model, the C-terminal domain (CTD) is required for interaction with inhibitory splicing factors such as SRp20 to promote exon skipping (17). Secondly, methylation could influence the lysine residues of the histones (reviewed in 14, 18, 36, 41) and the resulting chromatin changes increase exon 11 mis-splicing which is reversed by azacytidine treatment. Studies are ongoing to further elucidate the relative contribution of these two processes in exon 11 skipping.
In summary, H&N cancer cell lines, primary tissues and a number of other malignancies harbor an aberrantly spliced E-cadherin transcript which is present in much larger amounts as compared to the normal cells. As expression of E-cadherin gene is frequently lost in tumor cells, mis-splicing of this exon is an interesting model to study potential role of genetic and epigenetic events which affect splicing in cancer cells. It is also plausible that transcription factors involved in epithelial to mesenchymal transformation, e.g. snail and zeb (42,43) change the splicing milieu of the cells as they downregulate E-cadherin expression. The findings have a potential for clinical implications as epigenetic events can be reversed for re-expressing this tumor suppressor gene.
Supplementary Material
Acknowledgements
We thank Dr. Douglas Black HHMI UCLA for useful suggestions and discussions and Dr. Eri Srivatsan and Gwen Jordaan for help with cell culture work. Microarray experiments were performed at the UCLA Microarray core facilities.
Financial support: SS is a recipient of a grant from Flight Attendants Medical Research Institute (FAMRI), ASCO Foundation Young Investigator Award and a VA Merit award. AL is supported by grant CA 96920 and CA111448, and research funds from the Veteran's Administration. XZ is supported by K22DE014847, RO1CA139596, RO3CA135992, and a grant from Prevent Cancer Foundation.
Footnotes
Conflict of Interest: The authors have no conflict of interest.
References
- 1.Chang YF, Imam JS, Wilkinson MF. The nonsense-mediated decay RNA surveillance pathway. Annu Rev Biochem. 2007;76:51–74. doi: 10.1146/annurev.biochem.76.050106.093909. [DOI] [PubMed] [Google Scholar]
- 2.Culbertson MR. RNA surveillance. Unforeseen consequences for gene expression, inherited genetic disorders and cancer. Trends Genet. 1999;15:74–80. doi: 10.1016/s0168-9525(98)01658-8. [DOI] [PubMed] [Google Scholar]
- 3.Frischmeyer PA, van Hoof A, O'Donnell K, Guerrerio AL, Parker R, Dietz HC. An mRNA surveillance mechanism that eliminates transcripts lacking termination codons. Science. 2002;295:2258–2261. doi: 10.1126/science.1067338. [DOI] [PubMed] [Google Scholar]
- 4.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 5.Venables JP. Unbalanced alternative splicing and its significance in cancer. Bioessays. 2006;28:378–386. doi: 10.1002/bies.20390. [DOI] [PubMed] [Google Scholar]
- 6.Couch FJ, Weber BL. Mutations and polymorphisms in the familial early-onset breast cancer (BRCA1) gene. Breast Cancer Information Core. Hum Mutat. 1996;8:8–18. doi: 10.1002/humu.1380080102. [DOI] [PubMed] [Google Scholar]
- 7.El-Bchiri J, Buhard O, Penard-Lacronique V, Thomas G, Hamelin R, Duval A. Differential nonsense mediated decay of mutated mRNAs in mismatch repair deficient colorectal cancers. Hum Mol Genet. 2005;14:2435–2442. doi: 10.1093/hmg/ddi245. [DOI] [PubMed] [Google Scholar]
- 8.Abbas S, Erpelinck-Verschueren CA, Goudswaard CS, Lowenberg B, Valk PJ. Mutant Wilms' tumor 1 (WT1) mRNA with premature termination codons in acute myeloid leukemia (AML) is sensitive to nonsense-mediated RNA decay (NMD) Leukemia. 2009;24:660–663. doi: 10.1038/leu.2009.265. [DOI] [PubMed] [Google Scholar]
- 9.Bartel F, Taubert H, Harris LC. Alternative and aberrant splicing of MDM2 mRNA in human cancer. Cancer Cell. 2002;2:9–15. doi: 10.1016/s1535-6108(02)00091-0. [DOI] [PubMed] [Google Scholar]
- 10.Gilad S, Khosravi R, Shkedy D, Uziel T, Ziv Y, Savitsky K, et al. Predominance of null mutations in ataxia-telangiectasia. Hum Mol Genet. 1996;5:433–439. doi: 10.1093/hmg/5.4.433. [DOI] [PubMed] [Google Scholar]
- 11.Karni R, de Stanchina E, Lowe SW, Sinha R, Mu D, Krainer AR. The gene encoding the splicing factor SF2/ASF is a proto-oncogene. Nat Struct Mol Biol. 2007;14:185–193. doi: 10.1038/nsmb1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shi J, Hu Z, Pabon K, Scotto KW. Caffeine regulates alternative splicing in a subset of cancer-associated genes: a role for SC35. Mol Cell Biol. 2008;28:883–895. doi: 10.1128/MCB.01345-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eisenreich A, Malz R, Pepke W, Ayral Y, Poller W, Schultheiss HP, et al. Role of the phosphatidylinositol 3-kinase/protein kinase B pathway in regulating alternative splicing of tissue factor mRNA in human endothelial cells. Circ J. 2009;73:1746–1752. doi: 10.1253/circj.cj-99-0225. [DOI] [PubMed] [Google Scholar]
- 14.Sharma S, Kelly TK, Jones PA. Epigenetics in cancer. Carcinogenesis. 2010;31:27–36. doi: 10.1093/carcin/bgp220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kornblihtt AR. Chromatin, transcript elongation and alternative splicing. Nat Struct Mol Biol. 2006;13:5–7. doi: 10.1038/nsmb0106-5. [DOI] [PubMed] [Google Scholar]
- 16.Kornblihtt AR. Coupling transcription and alternative splicing. Adv Exp Med Biol. 2007;623:175–189. doi: 10.1007/978-0-387-77374-2_11. [DOI] [PubMed] [Google Scholar]
- 17.de la Mata M, Kornblihtt AR. RNA polymerase II C-terminal domain mediates regulation of alternative splicing by SRp20. Nat Struct Mol Biol. 2006;13:973–980. doi: 10.1038/nsmb1155. [DOI] [PubMed] [Google Scholar]
- 18.Schwartz S, Ast G. Chromatin density and splicing destiny: on the cross-talk between chromatin structure and splicing. EMBO J. 2010;29:1629–1636. doi: 10.1038/emboj.2010.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luco RF, Pan Q, Tominaga K, Blencowe BJ, Pereira-Smith OM, Misteli T. Regulation of alternative splicing by histone modifications. Science. 2010;327:996–1000. doi: 10.1126/science.1184208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schwartz S, Meshorer E, Ast G. Chromatin organization marks exon-intron structure. Nat Struct Mol Biol. 2009;16:990–995. doi: 10.1038/nsmb.1659. [DOI] [PubMed] [Google Scholar]
- 21.Sharma S, Lichtenstein A. Aberrant splicing of the E-cadherin transcript is a novel mechanism of gene silencing in chronic lymphocytic leukemia cells. Blood. 2009;114:4179–4185. doi: 10.1182/blood-2009-03-206482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jeanes A, Gottardi CJ, Yap AS. Cadherins and cancer: how does cadherin dysfunction promote tumor progression? Oncogene. 2008;27:6920–6929. doi: 10.1038/onc.2008.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pecina-Slaus N. Tumor suppressor gene E-cadherin and its role in normal and malignant cells. Cancer Cell Int. 2003;3:17. doi: 10.1186/1475-2867-3-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Park NH, Min BM, Li SL, Huang MZ, Cherick HM, Doniger J. Immortalization of normal human oral keratinocytes with type 16 human papillomavirus. Carcinogenesis. 1991;12:1627–1631. doi: 10.1093/carcin/12.9.1627. [DOI] [PubMed] [Google Scholar]
- 26.Noensie EN, Dietz HC. A strategy for disease gene identification through nonsense-mediated mRNA decay inhibition. Nat Biotechnol. 2001;19:434–439. doi: 10.1038/88099. [DOI] [PubMed] [Google Scholar]
- 27.Modafferi EF, Black DL. A complex intronic splicing enhancer from the c-src pre-mRNA activates inclusion of a heterologous exon. Mol Cell Biol. 1997;17:6537–6545. doi: 10.1128/mcb.17.11.6537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakayama S, Sasaki A, Mese H, Alcalde RE, Tsuji T, Matsumura T. The E-cadherin gene is silenced by CpG methylation in human oral squamous cell carcinomas. Int J Cancer. 2001;93:667–673. doi: 10.1002/ijc.1386. [DOI] [PubMed] [Google Scholar]
- 29.Gervais ML, Henry PC, Saravanan A, Burry TN, Gallie BL, Jewett MA, et al. Nuclear E-cadherin and VHL immunoreactivity are prognostic indicators of clear-cell renal cell carcinoma. Lab Invest. 2007;87:1252–1264. doi: 10.1038/labinvest.3700684. [DOI] [PubMed] [Google Scholar]
- 30.Endo K, Ueda T, Ueyama J, Ohta T, Terada T. Immunoreactive E-cadherin, alpha-catenin, beta-catenin, and gamma-catenin proteins in hepatocellular carcinoma: relationships with tumor grade, clinicopathologic parameters, and patients' survival. Hum Pathol. 2000;31:558–565. doi: 10.1053/hp.2000.6683. [DOI] [PubMed] [Google Scholar]
- 31.Kurtz KA, Hoffman HT, Zimmerman MB, Robinson RA. Decreased E-cadherin but not beta-catenin expression is associated with vascular invasion and decreased survival in head and neck squamous carcinomas. Otolaryngol Head Neck Surg. 2006;134:142–146. doi: 10.1016/j.otohns.2005.08.026. [DOI] [PubMed] [Google Scholar]
- 32.Ionov Y, Nowak N, Perucho M, Markowitz S, Cowell JK. Manipulation of nonsense mediated decay identifies gene mutations in colon cancer Cells with microsatellite instability. Oncogene. 2004;23:639–645. doi: 10.1038/sj.onc.1207178. [DOI] [PubMed] [Google Scholar]
- 33.Cartegni L, Wang J, Zhu Z, Zhang MQ, Krainer AR. ESEfinder: A web resource to identify exonic splicing enhancers. Nucleic Acids Res. 2003;31:3568–3571. doi: 10.1093/nar/gkg616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ghosh G, Adams JA. Phosphorylation mechanism and structure of serine-arginine protein kinases. FEBS J. 2011;278:587–597. doi: 10.1111/j.1742-4658.2010.07992.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lin S, Fu XD. SR proteins and related factors in alternative splicing. Adv Exp Med Biol. 2007;623:107–122. doi: 10.1007/978-0-387-77374-2_7. [DOI] [PubMed] [Google Scholar]
- 36.Yang X, Lay F, Han H, Jones PA. Targeting DNA methylation for epigenetic therapy. Trends Pharmacol Sci. 2010;31:536–546. doi: 10.1016/j.tips.2010.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Luco RF, Allo M, Schor IE, Kornblihtt AR, Misteli T. Epigenetics in alternative pre-mRNA splicing. Cell. 2011;144:16–26. doi: 10.1016/j.cell.2010.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Munoz MJ, de la Mata M, Kornblihtt AR. The carboxy terminal domain of RNA polymerase II and alternative splicing. Trends Biochem Sci. 2010;35:497–504. doi: 10.1016/j.tibs.2010.03.010. [DOI] [PubMed] [Google Scholar]
- 39.Kornblihtt AR. Promoter usage and alternative splicing. Curr Opin Cell Biol. 2005;17:262–268. doi: 10.1016/j.ceb.2005.04.014. [DOI] [PubMed] [Google Scholar]
- 40.Perales R, Bentley D. "Cotranscriptionality": the transcription elongation complex as a nexus for nuclear transactions. Mol Cell. 2009;36:178–191. doi: 10.1016/j.molcel.2009.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kolasinska-Zwierz P, Down T, Latorre I, Liu T, Liu XS, Ahringer J. Differential chromatin marking of introns and expressed exons by H3K36me3. Nat Genet. 2009;41:376–381. doi: 10.1038/ng.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Peinado H, Olmeda D, Cano A. Snail, Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype? Nat Rev Cancer. 2007;7:415–428. doi: 10.1038/nrc2131. [DOI] [PubMed] [Google Scholar]
- 43.Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.