Abstract
Pancreatic cancer is an especially deadly form of cancer with a survival rate <2%. Pancreatic cancers respond poorly to existing chemotherapeutic agents and radiation, and progress for the treatment of pancreatic cancer remains elusive. To address this unmet medical need, a better understanding of critical pathways and molecular mechanisms involved in pancreatic tumor development, progression and resistance to traditional therapy is therefore critical. Reduction-oxidation (redox) signaling systems are emerging as important targets in pancreatic cancer. AP endonuclease1/ Redox effector factor 1 (APE1/Ref-1) is upregulated in human pancreatic cancer cells and modulation of its redox activity blocks the proliferation and migration of pancreatic cancer cells as well as pancreatic cancer-associated endothelial cells (PCECs) in vitro. Modulation of APE1/Ref-1 using a specific inhibitor of APE1/Ref-1’s redox function, E3330 leads to a decrease in transcription factor activity for NFκB, AP-1, and HIF1 in vitro. This study aims to further establish the redox signaling protein APE1/Ref-1 as a molecular target in pancreatic cancer. Here, we show that inhibition of APE1/Ref-1 via E3330 results in tumor growth inhibition in cell lines as well as pancreatic cancer xenograft models in mice. Pharmacokinetic (PK) studies also demonstrate that E3330 attains >10 μM blood concentrations and is detectable in tumor xenografts. Through inhibition of APE1/Ref-1, the activity of NFκB, AP-1, and HIF1α which are key transcriptional regulators involved in survival, invasion and metastasis is blocked. These data indicate that E3330, inhibitor of APE1/Ref-1, has potential in pancreatic cancer and clinical investigation of APE1/Ref-1 molecular target is warranted.
Keywords: Pancreatic cancer, animal models of cancer, new targets, xenograft models, cellular responses to anticancer drugs, cellular pharmacology, pharmacokinetics and pharmacodynamics, agents with other mechanisms of action
INTRODUCTION
Pancreatic cancer is a particularly insidious form of cancer with the worst 5-year survival rate of any cancer at <2%.(1) There is no early detection method for pancreatic cancer, which often displays only non-specific symptoms such as abdominal pain, weight loss, and vomiting, until the cancer is well-advanced.(2) Pancreatic cancers are hypoxic tumors that respond poorly to existing chemotherapeutic agents and radiation.(3) NFκB and HIF-1α have been identified as leading drivers of cell growth in pancreatic cancer; both are under APE1/Ref-1 redox signaling control which is the focus of our studies.(4-6)
Cellular response to base DNA damage is a highly regulated and complex biological process. APE1/Ref-1 is a vital protein in this process and acts as a master regulator of the DNA damage response by contributing to the maintenance of the genome. APE1/Ref-1 is a dual function protein involved in base excision repair (BER) pathways of DNA lesions, as the major apurinic/apyrimidinic endonuclease, and in eukaryotic transcriptional regulation of gene expression as a reduction-oxidation (redox) factor. APE1/Ref-1 can stimulate DNA binding activity of numerous transcription factors that are involved in cancer promotion and progression such as HIF-1α, NFκB, AP-1, p53 and others. (6-13) The functional regions of APE1/Ref-1, redox and DNA repair, are completely independent in their function; i.e., mutations of the cysteine at position 65 removes the redox function but does not affect the DNA repair function, and vice versa. (14) While the DNA repair active site of APE1/Ref-1 is delineated, (15) the redox region is less obvious. Recently the mechanism of APE1/Ref-1 redox structure-function is being elucidated.(16-18) APE1/Ref-1 redox inhibitor, E3330 recognizes an alternate, redox active conformation of APE1/Ref-1, and potentially inhibits its redox activity by inducing disulfide bond formation within APE1/Ref-1.(16)
To investigate the role of redox regulation by APE1/Ref-1 in pancreatic cancer, we utilized E3330, a highly selective inhibitor of APE1/Ref-1 redox function. Originally discovered in a search for NFκB inhibitors, E3330 was used in liver inflammation and hepatitis, but never investigated for its therapeutic potential in cancer.(19, 20) We previously demonstrated that APE1/Ref-1 is upregulated in human pancreatic cancer cells and modulation of its redox activity blocks the proliferation and migration of pancreatic cancer cells (21, 22) as well as pancreatic cancer-associated endothelial cells (PCECs) in vitro.(23) Here we study the effects of redox signaling through APE1/Ref-1 in animal models of pancreatic cancer. The effectiveness of E3330 in vivo is demonstrated with good pharmacokinetic (PK) and pharmacodynamic properties (PD) as well as tumor growth reduction. Our in vitro data supports the in vivo results showing that blocking the redox activity of APE1/Ref-1 inhibits the proliferation and adhesion of pancreatic cancer cell lines, arrests cell cycle progression, and decreases the transcriptional activation of three major transcription factors known to be important in pancreatic cancer progression, survival and metastasis (NFκB, HIF-1α and AP-1(24)). In conclusion, this is the first specific APE1/Ref-1 redox inhibitor that demonstrates in vivo effectiveness and further work on this and other related compounds will allow the development of “first in class” and “first in human” agents for the treatment of pancreatic cancer.
MATERIALS AND METHODS
Cell lines
Panc-1 and PaCa-2 were purchased from and authenticated by ATCC (Manassas, VA). Both were maintained at 37°C in 5% CO2 and grown in DMEM (Invitrogen; Carlsbad, CA) with 10% Cosmic Calf Serum (Hyclone; Logan, UT).
Proliferation of PaCa-2 and Panc-1 cells
The proliferative capacity of PaCa-2 and Panc-1 cells was assessed using Trypan Blue (0.4%; Invitrogen; Carlsbad, CA) and the xCELLigence system (25, 26), and E3330 was synthesized as previously described.(16, 26) For Trypan Blue assays, PaCa-2 and Panc-1 cells were treated with E3330 for 72- and 48-h, respectively. Effects of E3330 on the growth were determined using the xCELLigence DP System (Roche Applied Science, Indianapolis IN).(25) Following background reading, cells (PaCa-2: 3,000 and Panc-1: 4,000 cells/well) were plated in 16-well plates in 100 μl volume. After adding cells to the wells, plates were kept at room temperature for 30 minutes after which they were inserted into the cradles. Cells were allowed to grow for 18 – 22 h before E3330 was added. Continuous impedance measurements were then monitored over 72 h. Assays were performed in triplicate.
Adhesion assay
For the adhesion assays,(27) serum-free media containing increasing concentrations of E3330 was added to each well in triplicate. A background reading was taken and then PaCa-2 or Panc-1 cells were added in complete media. Plates were kept at room temperature for 15 min and inserted into the cradle. Continuous impedance measurements were taken every 15 – 30 min over 30-h.
Western blot analysis
Cells were harvested, lysed in RIPA buffer (Santa Cruz Biotechnology; Santa Cruz, CA), and protein was quantified and electrophoresed on a 12% SDS-polyacrylamide gel. The following antibodies were used: APE1/Ref-1 (Novus Biologicals; Littleton, CO), PARP-1 (Cell Signaling Technology Inc.; Danvers, MA), tubulin (Sigma Aldrich), p21 and p27 (Santa Cruz Biotechnology).(22)
Apoptosis Assays via Alexa Fluor 488-Conjugated Annexin-V (Annexin-V) / Propidium Iodide Staining
Apoptosis was assayed 48-h after E3330 treatment. Cells were trypsinized, pelleted, washed in ice-cold PBS and resuspended in 1x Binding Buffer. Apoptosis was analyzed using the Alexa Fluor® 488 Annexin-V Vybrant® Apoptosis Assay Kit in combination with propidium iodide (PI) (Molecular Probes; Eugene, OR) as previously described (9).
Cell cycle staining via BrdU
To stain the cells for DNA content and analyze the movement of cells through G0/G1, S and G2/M, cells were plated, allowed to attach overnight, and treated with E3330 in serum free media for 24 hrs. Following treatment, cells were processed according to the manufacturer’s directions (BD Pharmingen; San Diego, CA). (9)
Transient Luciferase Reporter Assays
Panc-1 and PaCa-2 cells were co-transfected with constructs containing luciferase driven by NFκB, AP-1, or HIF1 (pLuc-MCS with the NFκB, AP-1, or HIF1 responsive promoter; PathDetect cis-Reporting Systems, Stratagene, La Jolla, CA(5)) and a Renilla luciferase control reporter vector pRL-TK (Promega Corp., Madison, WI) in a 20:1 ratio by using lipofectamine TM 2000 (Invitrogen Life Technologies, Carlsbad, CA). After transfection for 16-h, cells were treated with E3330 in serum free media for 24-h (for HIF1 drug added immediately before placement in hypoxic chamber). Firefly and Renilla luciferase activities were assayed by using the Dual Luciferase Reporter Assay System (Promega Corp.) with Renilla luciferase activity for normalization in a 96-well microtiter plate luminometer (Thermolabs Systems, Franklin, MA). Each time, a “media only” control was included, and the final data is expressed as the fold change of the relative luciferase units (RLU) compared to the vehicle control, DMSO. DMSO concentration was equivalent in all samples and the final concentration was less than 0.25%. All of the transfection experiments were performed in triplicate and repeated at least 3 times in independent experiments.
Stable Cell lines for Reporter assays
Using lentiviral transcriptional reporter vectors, pGreenFire-NFκB, pGreenFire-HIF1, and pGreenFire-mCMV (negative control), from System Biosciences Inc. (Mountainview, CA), we transfected HEK293-T cells (Lonza, Inc. Allendale, NJ) with the constructs and collected the media containing the viral supernatants. To generate stably expressing reporter cell lines, we infected pancreatic cancer cells with the viral supernatant and selected the cells with puromycin (5 ng/mL) for 5 days. For the experiments inducing NFκB activity via TNFα, Panc-1 and PaCa-2 were treated with E3330 for 24-h in serum free media and then treated with TNFα for 6-h. Cells were then assayed for luciferase activity as described above except the normalization was done using MTS-based assay. For the MTS assay (11, 22), 7.5×104 cells per 96-well were plated in parallel with the cells for luciferase assay, allowed to adhere overnight, and then treated with E3330 and TNFα as above. Alongside the luciferase assay, MTS reagent was added to the 96-well plates for 2-h. Values were standardized to wells containing media alone and then used to account for effects of drug treatment on the cells.
qRT-PCR Reactions
qRT-PCR was used to measure the mRNA expression levels of HIF1 downstream target, CA-IX (carbonic anhydrase-IX) gene. PaCa-2 and Panc-1 were treated with increasing amounts of E3330 in the presence or absence of hypoxia (1% O2) for 24-h, then total RNA was extracted from cells using the Qiagen RNeasy Mini kit (Valencia, CA) according to the manufacturer’s instructions(28).The extracted RNA was quantified by a Qubit fluorometer (Invitrogen Corp, Carlsbad, CA). First-strand cDNA was obtained from RNA using random hexamers and MultiScribe reverse transcriptase (Applied Biosystems, Foster City, CA). Quantitative PCR was performed using Taqman Gene Expression assays and Universal PCR master mix (Applied Biosystems) in a 7900HT Sequence detection system (Applied Biosystems). The relative quantitative mRNA level was determined using the comparative Ct method using 18S rRNA as the reference gene(28-30).The primers for CA-IX and 18S are commercially available (Applied Biosystems). Experiments were performed in triplicate for each sample.
PK Studies in NOD/SCID mice
NOD/SCID mice were administered 25mg/kg E3330 i.p. in DMSO(9%)/sterile saline for initial PK studies. To determine PK parameters, following administration of E3330, blood was collected via tail vein at multiple timepoints between 0.5 – 36-h following administration. We collected two samples (~25 μL) per mouse with each sample collected at least 6-h apart. Following half-life determination, a dosing regimen for the tumor studies was established in mice that consisted of two doses of E3330 each day, approximately 8-h apart for 14 days.
E3330 was quantified in plasma and solid tissue using an internal standard (bromomethylcoumarin, BMC), liquid-liquid extraction with ethyl acetate and HPLC-MS/MS (Agilent HPLC, Applied Biosystems API 4000). The HPLC was run in isocratic mode using acetonitrile:5mM ammonium acetate (60:40, v/v). The API 4000 was run in negative mode for E3330 (Q1/Q3: 377/333) and positive mode for BMC (Q1/Q3, 301/220). The lower limit of quantification was 1 ng/mL using 10 μL of blood or plasma. For E3330 in solid tissue, tissue was weighed, reconstituted with phosphate buffer (100 mM NaPO4, pH = 7.4) to up to two mL of total volume, and homogenized by a hand held homogenizer. An aliquot of the homogenate was used for the extraction. Tissue concentration is expressed in ng/g tissue.
Pharmacokinetic parameters for E3330 including area under the curve (AUC), area under the moment curve (AUMC), and t½ were estimated using noncompartmental methods with Excel®. The maximum plasma concentration (Cmax) and time of Cmax (tmax) were obtained from the data. The AUC from zero to infinity (AUC0-∞) was estimated from the AUC0-t (time zero to the last quantifiable concentration Clast) and the AUC from Clast to infinity, Clast/kel, where kel is the rate constant of elimination. The AUMC0-∞ was estimated by an analogous manner. The systemic clearance (Cl/F, where F = bioavailability) of E3330 was calculated from the dose and AUC0-∞. The apparent volume of distribution (Vdss) was estimated by the following equation: (dosage/AUC0-∞) x (AUMC0-∞/AUC0-∞)
Animals
All animal studies were conducted under the guidelines of the National Institutes of Health and were approved by the Institutional Animal Care and Use Committee of Indiana University School of Medicine and Johns Hopkins School of Medicine. Animals were maintained under pathogen-free conditions and a 12 h light-dark cycle.
For the patient-derived xenografts, 6-week-old female nu/nu athymic mice (Harlan) were utilized while PaCa-2 cells were grown as ectopic xenografts in NOD/scid mice. NOD/scid (NOD.CB17-Prkdcscid/J) mice were obtained from the In Vivo Therapeutics Core of the Indiana University Simon Cancer Center.
Establishment of ectopic xenografts from PaCa-2 cells
PaCa-2 cells (2.5×106) in 0.2 ml of DMEM media were implanted subcutaneously (s.c.) into the right flanks of NOD/SCID mice. E3330 was dissolved in 4% CremophorEL:EtOH (1:1) / saline solution or methylcelluose (0.5%, Sigma St. Louis, MO). When tumor volumes were greater than 100mm3, E3330 was administered twice daily, 8 hours apart, at 25 mg/kg for 10 - 12 days (5 days on and 2 days off schedule). Tumors were measured biweekly and followed for ~6 weeks. Tumor volumes were monitored by caliper measurement [tumor volume = length x (perpendicular width)2 x 0.5] and the average tumor volume in mm3 for each treatment group was plotted. Treatment corresponds to Days 1 – 15. Average tumor volume ± SE for the vehicle- and E3330-treated PaCa-2 xenografts (n=7) was analyzed by statistical analysis.
Ectopic xenograft established from patient-derived cells
Fresh pancreatic cancer specimens resected from patients at the time of surgery, with informed written patient consent, were implanted s.c. into the flanks of 6-week-old female nu/nu athymic mice (Harlan). The patients had not undergone chemotherapy or radiation therapy before surgery. Grafted tumors were subsequently transplanted from mouse to mouse and maintained as a live PancXenoBank according to an IRB approved protocol. Panc253 xenograft was used for the study. When tumors reached a volume of ~200 mm3, mice were individually identified and randomly assigned to treatment groups, with 5–6 mice (8-10 evaluable tumors) in each group: 1) control; 2) E3330, administered twice daily, 8 hours apart, at 25 mg/kg for a total of 10 doses (5 days on 2 days off, 5 days on and 2 days off schedule).
Statistics
For the PaCa-2 xenograft data, all data points for vehicle and E3330-treated mice were analyzed. Continuous variables were summarized by typical parameters such as mean, standard deviation and range and compared using two-sample T test (if the normality assumption holds) or Wilcoxon rank-sum test (if the normality assumption does not hold). Normality of distribution was determined using the Kolmogorov-Smirnov goodness-of-fit test. Categorical data were summarized by frequency and percentage and analyzed using the Chi-square or Fishers exact test, as appropriate. A linear mixed-effects model was fit to the repeatedly collected tumor size data. The fixed effects include experimental group, and distinct linear, quadratic and cubic effects of time for each of the two groups. The random effects include intercept and linear effect of time (31). The model was estimated using SAS 9.2 (SAS Institute, Inc., Cary, NC). A p-value less than 0.05 was considered statistically significant.
RESULTS
APE1/Ref-1 redox inhibitor, E3330 inhibits the proliferation and ability of pancreatic cancer cell lines to adhere but does not induce apoptosis
We tested the effect of APE1/Ref-1 redox inhibition on the growth of pancreatic cancer cells using a trypan blue assay. E3330 was found to effectively slow the growth rate of cells in a dose-dependent manner (Figure 1A), with an ED50 of 135 and 87 μM for PaCa-2 and Panc-1, respectively. The ability of E3330 to inhibit proliferation and/or reduce survival of PaCa-2 and Panc-1 cells was further characterized using the xCELLigence DP system (Roche, Indianapolis IN). This system measures a dimensionless parameter called Cell Index (CI), which integrates information on cell viability, number, morphology, and adhesion.(27) In this assay, E3330 dramatically reduced CI of PaCa-2 and Panc-1 cells efficiently and in a dose-dependent manner, suggesting that E3330 inhibits growth and proliferation of pancreatic cancer cells as well as affects morphology and adhesion (Figure 1B). The ED50 of E3330 was reduced by 2-fold using the xCELLigence system (43 and 54 μM for PaCa-2 and Panc-1 cells), therefore we used the xCELLigence system to monitor the adhesion of PaCa-2 and Panc-1 cells. Adhesion of the cells was blocked at doses greater than 33.75μM (Figure 1D). The CI value was reduced by 50% in both cell lines at 67.5 μM between 8 – 12 h. Doses that blocked proliferation also had a dramatic effect on the cells’ ability to adhere to the plate. The sensitivity of the xCELLigence system demonstrates the effects of APE1/Ref-1 redox inhibition on proliferation, adherence, and morphology.
Figure 1.
Inhibition of APE1/Ref-1 redox activity blocks the proliferation and adhesion of pancreatic cancer cells without inducing apoptosis. A, Cells were counted in triplicate using Trypan Blue after 48-h (Panc-1) or 72-h (PaCa-2) of E3330 exposure. Day 0 refers to the cell number 24-h after plating. xCELLigence DP system was used to determine cell growth (B) and adhesion (D) after E3330 treatment. Assays were performed in triplicate, three individual times with a representative experiment shown. C, Histogram plots (The x-axis represents Annexin-488 staining and the y-axis represents PI staining, Top) and Western blot (Bottom) probed with PARP-1 after treatment with increasing concentrations of E3330 for 48 hours (FL, full length).
To investigate whether the cells were undergoing apoptosis following treatment with E3330, we utilized two methods: Annexin / PI staining and PARP-1 cleavage (Figure 1C). Both assays demonstrated similar results showing that although we had a reduction in proliferation, there was not a corresponding increase in apoptosis. These data are consistent with the idea that inhibition of the redox function of APE1/Ref-1 affects the cells’ proliferative capacity while the DNA repair function is critical for cell survival.(4)
Inhibition of the redox function of APE1/Ref-1 via E3330 arrests cell cycle progression
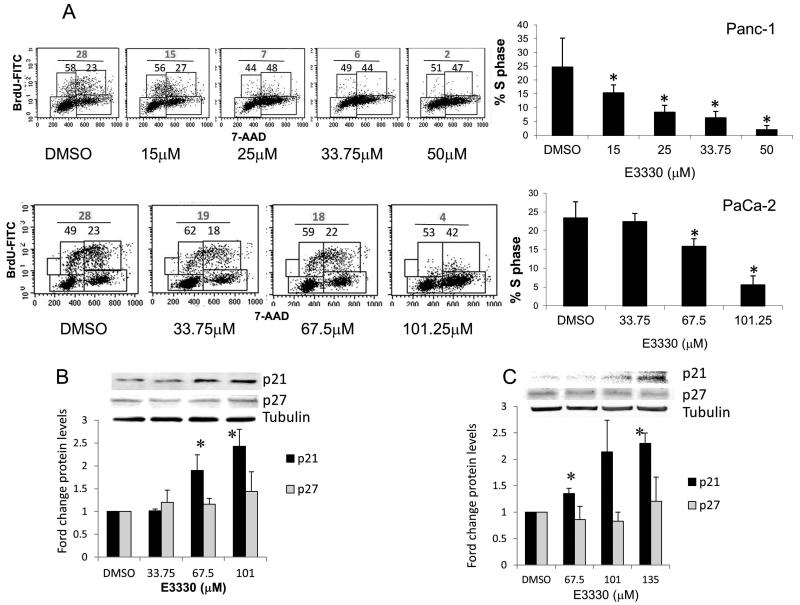
Using BrdU incorporation assays, the percentage of PaCa-2 and Panc-1 cells in S phase is significantly decreased following treatment with E3330 (Figure 2A). DMSO, the vehicle control, does not differ significantly from the media (data not shown). Furthermore, we synchronized PaCa-2 cells and monitored re-entry into cell cycle following treatment with E3330. At 8 h after adding serum-containing media, 60% less cells have entered S phase (Supp. Fig. 1), demonstrating that progression out of G1 is slower in the PaCa-2 cells when the redox function of APE1/Ref-1 is inhibited.
Figure 2.
Inhibition of APE1/Ref-1 redox activity alters cell cycle profile and levels of p21 protein. A, Representative dot plots of BrdU assays in Panc-1 (Top) and PaCa-2 cells (Bottom) treated with E3330. The x-axis represents 7-AAD staining and the y-axis represents BrdU-FITC staining. Western blots of cell-cycle proteins, p21, p27, and tubulin as loading control in PaCa-2 (B) and Panc-1 (C). Bar graphs demonstrate quantitation of ≥ 3 independent experiments with average +/− SE. * p < 0.05 using Student’s t-test, comparing DMSO vs. E3330.
We quantitated the levels of endogenous cell cycle inhibitors, p21 and p27 (Figure 2B, C). While we observe no change in p27 levels, the levels of p21 increase 2 - 2.5 fold in cells that have been treated with E3330. The effects on cell cycle progression and levels of p21 and p27 are very similar to our recent report of the effects of APE1/Ref-1 siRNA on PaCa-2 and Panc-1 cells.(22) However, the increase in apoptosis that we observe when APE1/Ref-1 protein levels are decreased by siRNA is not observed when we inhibit the redox function of APE1/Ref-1. These data suggest that the redox function of APE1/Ref-1 affects both cell cycle progression and the time the cells take to traverse the cell cycle in pancreatic cancer cells.
Inhibition of APE1/Ref-1 via E3330 results in a decrease in transcriptional activity for three known targets of APE1/Ref-1 redox activity
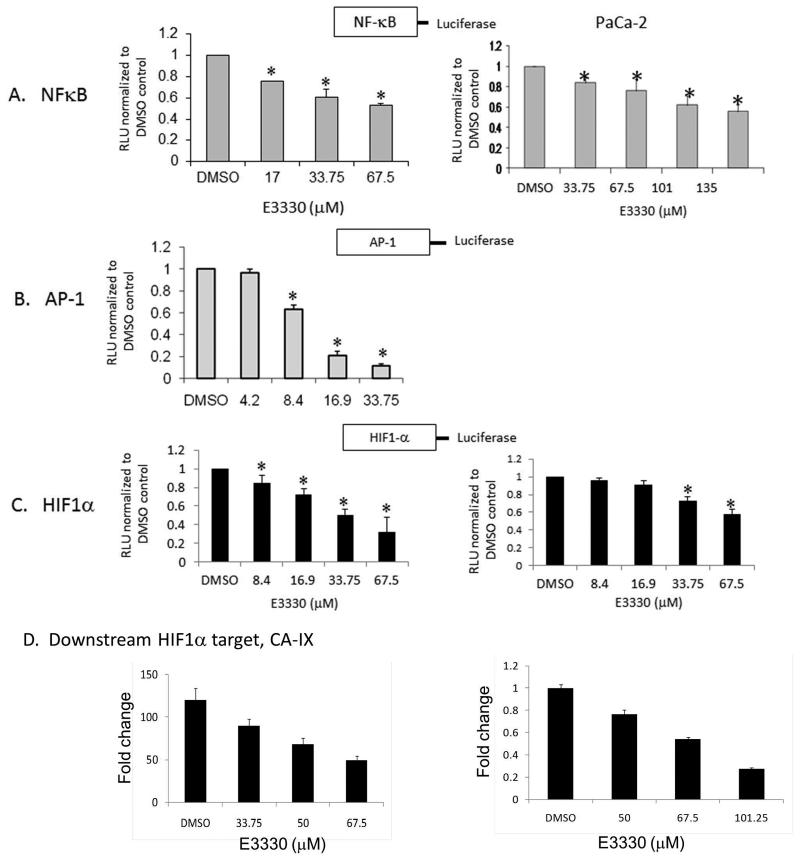
To further characterize the redox function of APE1/Ref-1 on pancreatic cancer cell signaling, we utilized transient luciferase assays. In these experiments, the luciferase gene expression was driven by NFκB, HIF1, or AP-1 and normalized to Renilla gene expression for transfection efficiency. Inhibition of APE1/Ref-1 via E3330 results in a dose-dependent reduction in NFκB, HIF1, and AP-1 activity in vitro (Figure 3, A-C). HIF1 activity was induced by exposure to hypoxia for 24 hrs prior to luciferase assay. The basal levels of AP-1 activity in PaCa-2 are ~20-fold lower than the basal activity in Panc-1 cells and we were unable to detect a significant decrease in activity with the low levels of basal activity in PaCa-2 cells (data not shown). The doses of E3330 required to inhibit 50% of promoter activity were ~67.5μM for NFκB and HIF1 and ~15μM for AP-1. To further demonstrate that inhibition of APE1/Ref-1 blocks downstream signaling of HIF1, we treated Panc-1 and PaCa-2 cells with E3330 for 24-h. We then analyzed HIF1 target, CA-IX(32) mRNA levels using qRT-PCR assay. As predicted, there was a significant dose-dependent decrease in HIF1 target, CA-IX mRNA in pancreatic cancer cells following APE1/Ref-1 inhibition (Fig. 3D). Because inhibition of mRNA for CA-IX serves as a biomarker for HIF1 activity(32),we hypothesize that inhibition of APE1/Ref-1’s redox function by E3330 could disable the tumors’ ability to respond to hypoxic conditions which is known to contribute to the chemotherapeutic resistance of these tumors.
Figure 3.
E3330 inhibits the activity of NFκB, HIF1, and AP-1 in a cell-based reporter assay. A – C, Panc-1 and PaCa-2 cells were transfected with NFκB, HIF1, or AP-1–Luc construct and cotransfected with a Renilla vector, pRL-TK. After 16-h, cells were treated with E3330 for 24-h, and Firefly and Renilla luciferase activities were assayed using Renilla luciferase activity for normalization. All transfection experiments were performed in triplicate and repeated at least 4 times in independent experiments. Data are expressed as Relative Luciferase Units (RLU) normalized to DMSO showing the mean ± SE. Student’s t tests were performed, and * p < 0.05 level comparing DMSO to E3330. D) qPCR analysis of CA-IX expression following E3330 treatment and hypoxia. Expression in Panc-1 cells was normalized to DMSO in normoxia (left). However, basal levels of CA-IX were undetectable in PaCa-2 cells in normoxia, therefore the CA-IX expression was normalized to DMSO under hypoxia (right). Shown is one representative experiment. Samples were run in triplicate and experiment performed in both cell lines in duplicate.
Blockade of APE1/Ref-1 redox activity diminishes the pancreatic cancer cells ability to induce HIF1α and NFκB
We generated PaCa-2 and Panc-1 lines that stably express NFκB-driven and HIF1–driven luciferase/GFP using lentiviral constructs, pGreenFire (pGF) from System Biosciences Inc. As a negative control, stable cell lines were generated that contain the luciferase/GFP cassette but do not have the NFκB or HIF1 response element (pGF-mCMV). Inhibition of the redox function of APE1/Ref-1 blocks the ability of PaCa-2 and Panc-1 cells to induce NFκB through TNFα (Figure 4A, B) as well as HIF1 activity (Figure 4C). PaCa-2 and Panc-1 cells that express pGF-mCMV did not show detectable luciferase activity above baseline and treatment with TNFα or hypoxia did not induce expression of luciferase activity (data not shown).
Figure 4.
E3330 inhibits NFκB and HIF1 transactivation in stable luciferase-expressing cell lines. A, PaCa-2 cells pGF-NFκB or pGF-mCMV were treated with E3330 for 24-h, followed by TNFα (50ng/mL, 6-h). Green cells indicate induction of NFκB which is blocked by treatment with E3330. B, NFκB activity of cells induced with TNFα, M = media control which shows basal NFκB activity. C, HIF1 activity of cells following E3330 treatment. Firefly luciferase activity was assayed as above and normalized to MTS assay data and then to vehicle control. Experiments were performed in triplicate and repeated at least 3 times in independent experiments. Student’s t tests were performed, and * p < 0.05 level comparing DMSO to E3330.
Inhibition of APE1/Ref-1 redox activity delays the growth of pancreatic cancer xenografts in vivo and exhibits favorable pharmacokinetic parameters
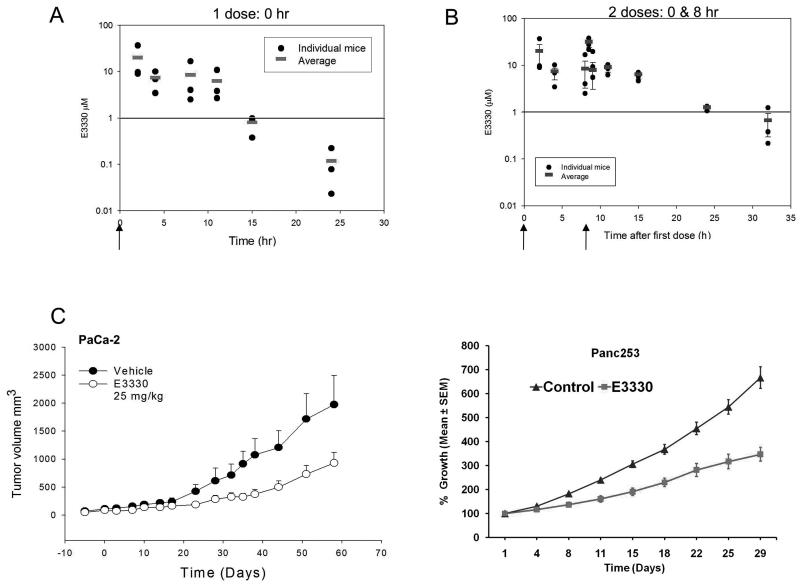
The estimated half-life (t ½) of E3330 was 3.7hrs. (Figure 5A). From this, a dosing regimen was established in mice that consisted of two doses of E3330 each day, approximately 8-h apart. As shown in Figure 5B, two dosages of E3330 maintained the concentrations of E3330 ≥1 μM for over 24-h. In formulation studies, E3330 was dissolved in 4% CremophorEL: EtOH (1:1) in saline or 0.5% methylcellulose. Both vehicles were similar in their PK levels in plasma and tumor tissue (Supplemental Fig. 2) and replaced the less desirable vehicle, DMSO. Concentrations of E3330 attained in the blood of mice are within the required range for target inhibition from in vitro studies suggesting that E3330 has favorable properties in vivo.
Figure 5.
E3330 pharmacokinetic profile and analysis of tumor growth rate in PaCa-2 and patient-derived xenografts. A, Serum concentrations of E3330 following one dose at 25 mg/kg, ip in 9% DMSO; B, Serum concentrations of E3330 (25 mg/kg, ip, 9% DMSO) following dosing at 0-h and 8-h as indicated by the arrows. C, Tumor growth delay following treatment with E3330 in PaCa-2 xenografts (left) and patient-derived tissue, Panc253 (right).
Next we evaluated the anti-tumor efficacy of E3330 using both patient-derived tumors and xenografts from established cell line, PaCa-2. In contrast to vehicle-control tumors, both patient-derived and PaCa-2 xenografts demonstrated a significant tumor growth delay (Figure 5C). All animals treated with E3330 had detectable drug in the tumor tissue (40 -7500 ng/g tissue). E3330 was detectable in pancreas, liver, lung, kidney, heart, and brain, but was not overly toxic to the animals as measured by weight loss and bone marrow cellularity (data not shown and Supplemental Fig. 2A, B). Furthermore, Ki67 staining is reduced in E3330-treated tumors at Day 7 and Day 11 supporting the notion that APE1/Ref-1 redox activity is blocking proliferation of the tumor cells (Supplemental Fig. 2C). Another patient-derived xenograft was utilized and did not show the dramatic growth delay that we observe in Figure 5C. However, tumor tissue analysis indicated that levels of E3330 in the unresponsive tumors were 10-fold less than in the Panc 253 responsive tumors: 2500ng/g (~6.6μM) versus 250ng/g (~0.7μM), data not shown. These results indicate that if we can efficiently deliver APE1/Ref-1 redox inhibitor, tumor growth is strongly inhibited.
DISCUSSION
Although today’s standard of care for pancreatic cancer strives for a cure, debulking surgery along with chemotherapy and/or radiation is almost always palliative rather than curative, with few cases of long-term regression.(33) Researchers agree that pancreatic cancer defies most of what we have come to know about other types of cancer; therefore, a different therapeutic approach is needed.(1, 33, 34) Blocking a single step in a pathway or a single pathway has very limited clinical utility in the face of the tumors’ cumulative defects. Jones et al. found that pancreatic cancers contain a core set of 12 cellular signaling pathways and processes, each of which was altered in 67 to 100% of the tumors analyzed.(24) Novel targets that modulate multiple pathways may offer the most promise for clinical utility against this dreaded disease. Transcription factors including NFκB, AP-1, and HIF1 are key in the regulation of multiple signals in pancreatic cancer which provides strong evidence for investigating the effects of targeting APE1/Ref-1 to kill pancreatic cancer cells. In this report, we show that inhibition of APE1/Ref-1 reduces the proliferation of ectopic pancreatic tumors in mice using both established pancreatic cancer cell lines and primary human pancreatic tumors. Inhibition of APE1/Ref-1 redox activity led to cell cycle arrest and a reduction in NFκB, AP-1, and HIF1 activity, key regulators of pathways that are involved in the progression, maintenance, invasive and metastatic potential of pancreatic cancer.(5, 35-37) Furthermore, we show down-regulation of HIF1 target, CA-IX and surmise that APE1/Ref-1 inhibition may be able to sensitize these tumors to therapy by disabling their response to the hypoxic environment in which they are growing.
We recently demonstrated that human adenocarcinoma and peri-pancreatic metastases have a significant increase in APE1/Ref-1 expression in adenocarcinoma as compared to normal pancreas tissue.(22) The experiments described here and our previous work showing both in vitro and in vivo data support APE1/Ref-1 as a viable target in this deadly disease.(21, 22) We utilized the redox-specific APE1/Ref-1 inhibitor, E3330, which recognizes an alternate, redox active conformation of APE1/Ref-1, and potentially inhibits its redox activity by inducing disulfide bond formation.(16, 17) Biochemical studies using radiolabeled E3330 and proteins renatured on membrane blots demonstrated that 14C-labeled E3330 very selectively bound to both recombinant APE1/Ref-1 and purified APE1/Ref-1 from cell nuclear extracts.(20) While E3330 blocks Ref-1’s redox function, it has no effect on APE1/Ref-1 endonuclease activity or BER activity of an AP site.(5) These studies formally demonstrate the specificity of E3330 for APE1/Ref-1’s redox activity, without impacting on its DNA repair function. The PK/PD studies (Figure 5) demonstrate that tumors with significant levels of E3330 have their rate of growth significantly reduced. We are currently investigating additional delivery methodology such as nanoparticle technology to ensure efficient delivery of APE1/Ref-1 inhibitor as well as novel compounds with sub-micromolar inhibition of APE1/Ref-1 redox activity. Another potential mechanism for the different amount of E3330 in these two patient-derived tumors is that the drug is pumped out of the cells. For example, there are several ATP-binding cassette transporters in the MDR/MRP gene family. Given the chemical structure of E3330, we predict that it is not a substrate for P-gp (MDR1, ABCB1) since this efflux transporter typically binds cationic, lipophilic structures. However, E3330 could be a substrate for MRP (ABCC1) or BCRP (ABCG2), as these transporters are known to bind anionic substrates including glutathione conjugates.(38) However, it should be noted that MRP is not always associated with extracellular transport, but may concentrate substrates into intracellular vesicles, thus protecting potential targets from drug. This is one possibility for the tumors with low levels of E3330 detected. However, further research is warranted.
The mechanism of action of E3330 is through the blockage of the transcriptional regulation of APE1/Ref-1 therefore, it blocks activity of NFκB, AP-1, and HIF1 as shown in Fig. 3, 4. These data provide fundamental evidence that E3330 inhibits three known transcriptional targets of APE1/Ref-1 in pancreatic cancer cells. Pancreatic tumors are characterized as one of the most hypoxic tumors that clinicians encounter.(3, 35) Our previous work and the data here demonstrate that blockade of APE1/Ref-1 redox signaling does affect the activity of HIF1 in pancreatic cancer cells. (21, 23) This supports our hypothesis that inhibition of APE1/Ref-1 could disable the tumors’ ability to respond to hypoxic conditions which contributes to chemotherapeutic resistance. Using in vitro luciferase-based assays, E3330 can inhibit the activity of these three important transcription factors in pancreatic cancer. Using a lentiviral system, we created stable cell lines with a construct containing an NFκB or HIF1 response element which drives GFP and luciferase. In PaCa-2 and Panc-1 cells that express luciferase driven by the NFκB or HIF1 promoter, E3330 treatment results in a reduction in activity as shown by reduction of GFP positivity and luciferase activity (Figure 4 and data not shown). Inhibition of NFκB (with and without induction by TNFα) and HIF1 activity is observed both in transient and stable luciferase assays. Decreases in the activity of downstream transcription factors provide important support for APE1/Ref-1 as a target in pancreatic cancer. We can now use these stable cell lines in vivo as markers of NFκB, AP-1, and HIF1 and dissect which of these transcription factors are critical for the survival and spread of pancreatic cancer.
Inhibition of APE1/Ref-1 via E3330 inhibits the growth of pancreatic tumor xenografts, both the cell line as well as xenograft models, and we plan to extend our studies to orthotopic models. In addition to evaluating E3330 as a single agent, combination of E3330 with Gemcitabine will also be tested to know whether E3330 potentiates Gemcitabine sensitivity. A reduction in APE1/Ref-1 protein expression does indeed sensitize pancreatic cells in vitro to gemcitabine(39) suggesting that combination therapy might be useful in treating this disease. Due to the decrease in proliferative capacity and the decrease in cells in S phase (Figure 1, 2), careful selection of the agent(s) that are chosen for combination therapy as well as the dosing schedules should be chosen carefully. The studies here provide pre-clinical validation for a novel therapeutic strategy for this largely incurable cancer.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to acknowledge the IVT Core in Indiana University Simon Cancer Center, specifically Jayne Silver, for expertise in xenograft studies. We are grateful to Dr. Shanbao Cai for his assistance in the bone marrow cellularity studies shown in the supplemental data.
Financial Support: Financial support for this work was provided by the National Institutes of Health NCI CA121168, CA114571, and CA121168S1 (MRK), CA122298 (MLF), CA138798 (KEP), CA134767, CA113669 and CA134292 (AM), by the American Cancer Society RSG 0924401TBG (MI), the Riley Children’s Foundation (MRK) and the Ralph W. and Grace M. Showalter Research Trust Fund (MLF).
Abbreviations List
- AP
Apurinic/apyrimidinic
- AP-1
Activator protein-1
- APE1/Ref-1
AP endonuclease1/Redox factor-1
- AUC
area under the curve
- AUMC
area under the moment curve
- ATP
adenosine-5′-triphosphate
- BER
base excision repair
- CA-IX
carbonic anhydrase-IX
- Cmax
maximum plasma concentration
- CI
cell index
- HIF1α
hypoxia-inducible factor 1, alpha subunit
- mCMV
mouse cytomegalovirus
- NFkB
Nuclear Factor kappa b
- PCEC
pancreatic cancer-associated endothelial cells
- pGF
pGreenFire
- PD
pharmacodynamic properties
- PK
pharmacokinetic
- PI
propidium iodide
- Redox
reduction-oxidation
- Ref-1
Redox effector factor 1
- RLU
relative luciferase units
- tmax
time of Cmax
- Vdss
volume of distribution
Footnotes
Declaration of interest: In addition the authors declare that they have no conflict of interest except for Dr. Mark R. Kelley who is also a consultant for Apex Therapeutics, a company that has licensed IP from his work
References
- 1.Moore MJ. Chemotherapy in pancreatic carcinoma. Current Oncology. 2003;5:s24–s6. [Google Scholar]
- 2.Tanase CP, Neagu M, Albulescu R, Hinescu ME. Advances in pancreatic cancer detection. Adv Clin Chem. 2010;51:145–80. doi: 10.1016/s0065-2423(10)51006-0. [DOI] [PubMed] [Google Scholar]
- 3.Duffy JP, Eibl G, Reber HA, Hines OJ. Influence of hypoxia and neoangiogenesis on the growth of pancreatic cancer. Mol Cancer. 2003;2:12. doi: 10.1186/1476-4598-2-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tell G, Quadrifoglio F, Tiribelli C, Kelley MR. The Many Functions of APE1/Ref-1: Not Only a DNA Repair Enzyme. Antioxid Redox Signal. 2009;11:601–20. doi: 10.1089/ars.2008.2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Luo M, Delaplane S, Jiang A, Reed A, He Y, Fishel M, et al. Role of the multifunctional DNA repair and redox signaling protein Ape1/Ref-1 in cancer and endothelial cells: small-molecule inhibition of the redox function of Ape1. Antioxid Redox Signal. 2008;10:1853–67. doi: 10.1089/ars.2008.2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bapat A, Fishel M, Kelley MR. Going Ape as an Approach to Cancer Therapeutics. Antioxid Redox Signal. 2009;11:651–68. doi: 10.1089/ars.2008.2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kelley MR, Fishel ML. DNA repair proteins as molecular targets for cancer therapeutics. Anticancer Agents Med Chem. 2008;8:417–25. doi: 10.2174/187152008784220294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Georgiadis MM, Luo M, Gaur RK, Delaplane S, Li X, Kelley MR. Evolution of the redox function in mammalian apurinic/apyrimidinic endonuclease. Mutat Res. 2008;643:54–63. doi: 10.1016/j.mrfmmm.2008.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fishel ML, He Y, Reed AM, Chin-Sinex H, Hutchins GD, Mendonca MS, et al. Knockdown of the DNA repair and redox signaling protein Ape1/Ref-1 blocks ovarian cancer cell and tumor growth. DNA Repair (Amst) 2008;7:177–86. doi: 10.1016/j.dnarep.2007.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fishel ML, Kelley MR. The DNA base excision repair protein Ape1/Ref-1 as a therapeutic and chemopreventive target. Mol Aspects Med. 2007;28:375–95. doi: 10.1016/j.mam.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 11.Fishel ML, He Y, Smith ML, Kelley MR. Manipulation of base excision repair to sensitize ovarian cancer cells to alkylating agent temozolomide. Clin Cancer Res. 2007;13:260–7. doi: 10.1158/1078-0432.CCR-06-1920. [DOI] [PubMed] [Google Scholar]
- 12.Tell G, Damante G, Caldwell D, Kelley MR. The intracellular localization of APE1/Ref-1: more than a passive phenomenon? Antioxid Redox Signal. 2005;7:367–84. doi: 10.1089/ars.2005.7.367. [DOI] [PubMed] [Google Scholar]
- 13.Evans AR, Limp-Foster M, Kelley MR. Going APE over ref-1. Mutation Research. 2000;461:83–108. doi: 10.1016/s0921-8777(00)00046-x. [DOI] [PubMed] [Google Scholar]
- 14.McNeill DR, Wilson DM., 3rd A Dominant-Negative Form of the Major Human Abasic Endonuclease Enhances Cellular Sensitivity to Laboratory and Clinical DNA-Damaging Agents. Mol Cancer Res. 2007;5:61–70. doi: 10.1158/1541-7786.MCR-06-0329. [DOI] [PubMed] [Google Scholar]
- 15.Gorman MA, Morera S, Rothwell DG, de la Fortelle E, Mol CD, Tainer JA, et al. The crystal structure of the human DNA repair endonuclease HAP1 suggests the recognition of extra-helical deoxyribose at DNA abasic sites. Embo J. 1997;16:6548–58. doi: 10.1093/emboj/16.21.6548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Su DG, Delaplane S, Luo M, Rempel DL, Vu B, Kelley MR, et al. Interactions of APE1 with a redox inhibitor: Evidence for an alternate conformation of the enzyme. Biochemistry. 2011;50:82–92. doi: 10.1021/bi101248s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kelley MR, Luo M, Reed A, Su D, Delaplane S, Borch R, et al. Functional analysis of novel analogs of E3330 that block the redox signaling activity of the multifunctional AP endonuclease/redox signaling enzyme APE1/Ref-1. Antioxid Redox Signal. 2011;14:1387–401. doi: 10.1089/ars.2010.3410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luo M, He H, Kelley MR, Georgiadis M. Redox Regulation of DNA Repair: Implications for Human Health and Cancer Therapeutic Development. Antioxid Redox Signal. 2010;12:1247–69. doi: 10.1089/ars.2009.2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hiramoto M, Shimizu N, Sugimoto K, Tang J, Kawakami Y, Ito M, et al. Nuclear targeted suppression of NF-kappa B activity by the novel quinone derivative E3330. J Immunol. 1998;160:810–9. [PubMed] [Google Scholar]
- 20.Shimizu N, Sugimoto K, Tang J, Nishi T, Sato I, Hiramoto M, et al. High-performance affinity beads for identifying drug receptors. Nat Biotechnol. 2000;18:877–81. doi: 10.1038/78496. [DOI] [PubMed] [Google Scholar]
- 21.Zou GM, Maitra A. Small-molecule inhibitor of the AP endonuclease 1/REF-1 E3330 inhibits pancreatic cancer cell growth and migration. Mol Cancer Ther. 2008;7:2012–21. doi: 10.1158/1535-7163.MCT-08-0113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiang Y, Zhou S, Sandusky GE, Kelley MR, Fishel ML. Reduced expression of DNA repair and redox signaling protein APE1/Ref-1 impairs human pancreatic cancer cell survival, proliferation, and cell cycle progression. Cancer Investigation. 2010;28:885–95. doi: 10.3109/07357907.2010.512816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zou GM, Karikari C, Kabe Y, Handa H, Anders RA, Maitra A. The Ape-1/Ref-1 redox antagonist E3330 inhibits the growth of tumor endothelium and endothelial progenitor cells: therapeutic implications in tumor angiogenesis. J Cell Physiol. 2009;219:209–18. doi: 10.1002/jcp.21666. [DOI] [PubMed] [Google Scholar]
- 24.Jones S, Zhang X, Parsons DW, Lin JC, Leary RJ, Angenendt P, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science (New York, NY) 2008;321:1801–6. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bapat A, Glass LS, Luo M, Fishel ML, Long EC, Georgiadis MM, et al. Novel small molecule inhibitor of Ape1 endonuclease blocks proliferation and reduces viability of glioblastoma cells. J Pharmacol Exp Ther. 2010;334:988–98. doi: 10.1124/jpet.110.169128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fishel ML, Colvin ES, Luo M, Kelley MR, Robertson KA. Inhibition of the Redox Function of APE1/Ref-1 in Myeloid Leukemia Cell Lines Results in a Hypersensitive Response to Retinoic Acid-induced Differentiation and Apoptosis. Exp Hematol. 2010;38:1178–88. doi: 10.1016/j.exphem.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Glamann J, Hansen AJ. Dynamic detection of natural killer cell-mediated cytotoxicity and cell adhesion by electrical impedance measurements. Assay Drug Dev Technol. 2006;4:555–63. doi: 10.1089/adt.2006.4.555. [DOI] [PubMed] [Google Scholar]
- 28.Jiang Y, Guo C, Fishel ML, Wang Z-Y, Vasko MR, Kelley MR. Role of APE1 in differentiated neuroblastoma SH-SY5Y cells in response to oxidative stress: Use of APE1 small molecule inhibitors to delineate APE1 functions. DNA Repair. 2009;8:1273–82. doi: 10.1016/j.dnarep.2009.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 30.Fishel ML, Rabik CA, Bleibel WK, Li X, Moschel RC, Dolan ME. Role of GADD34 in modulation of cisplatin cytotoxicity. Biochem Pharmacol. 2006;71:239–47. doi: 10.1016/j.bcp.2005.10.039. [DOI] [PubMed] [Google Scholar]
- 31.Laird NM, Ware JH. Random-effects models for longitudinal data. Biometrics. 1982;38:963–74. [PubMed] [Google Scholar]
- 32.Kaluz S, Kaluzova M, Liao SY, Lerman M, Stanbridge EJ. Transcriptional control of the tumor- and hypoxia-marker carbonic anhydrase 9: A one transcription factor (HIF-1) show? Biochim Biophys Acta. 2009;1795:162–72. doi: 10.1016/j.bbcan.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Burris HA., 3rd Recent updates on the role of chemotherapy in pancreatic cancer. Semin Oncol. 2005;32:1–3. doi: 10.1053/j.seminoncol.2005.06.022. [DOI] [PubMed] [Google Scholar]
- 34.Bria E, Milella M, Gelibter A, Cuppone F, Pino MS, Ruggeri EM, et al. Gemcitabine-based combinations for inoperable pancreatic cancer: have we made real progress? A meta-analysis of 20 phase 3 trials. Cancer. 2007;110:525–33. doi: 10.1002/cncr.22809. [DOI] [PubMed] [Google Scholar]
- 35.Garcea G, Doucas H, Steward WP, Dennison AR, Berry DP. Hypoxia and angiogenesis in pancreatic cancer. ANZ J Surg. 2006;76:830–42. doi: 10.1111/j.1445-2197.2006.03872.x. [DOI] [PubMed] [Google Scholar]
- 36.Fujioka S, Niu J, Schmidt C, Sclabas GM, Peng B, Uwagawa T, et al. NF-kappaB and AP-1 connection: mechanism of NF-kappaB-dependent regulation of AP-1 activity. Mol Cell Biol. 2004;24:7806–19. doi: 10.1128/MCB.24.17.7806-7819.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Greten FR, Weber CK, Greten TF, Schneider G, Wagner M, Adler G, et al. Stat3 and NF-kappaB activation prevents apoptosis in pancreatic carcinogenesis. Gastroenterology. 2002;123:2052–63. doi: 10.1053/gast.2002.37075. [DOI] [PubMed] [Google Scholar]
- 38.Franke RM, Gardner ER, Sparreboom A. Pharmacogenetics of drug transporters. Curr Pharm Des. 2010;16:220–30. doi: 10.2174/138161210790112683. [DOI] [PubMed] [Google Scholar]
- 39.Lau JP, Weatherdon KL, Skalski V, Hedley DW. Effects of gemcitabine on APE/ref-1 endonuclease activity in pancreatic cancer cells, and the therapeutic potential of antisense oligonucleotides. British Journal of Cancer. 2004;91:1166–73. doi: 10.1038/sj.bjc.6602080. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.