Abstract
Approximately 47% of the genes of the Gram-positive bacterium Bacillus subtilis belong to paralogous gene families. The present studies were aimed at the functional analysis of the sip gene family of B. subtilis, consisting of five chromosomal genes, denoted sipS, sipT, sipU, sipV, and sipW. All five sip genes specify type I signal peptidases (SPases), which are actively involved in the processing of secretory preproteins. Interestingly, strains lacking as many as four of these SPases could be obtained. As shown with a temperature-sensitive SipS variant, only cells lacking both SipS and SipT were not viable, which may be caused by jamming of the secretion machinery with secretory preproteins. Thus, SipS and SipT are of major importance for protein secretion. This conclusion is underscored by the observation that only the transcription of the sipS and sipT genes is temporally controlled via the DegS–DegU regulatory system, in concert with the transcription of most genes for secretory preproteins. Notably, the newly identified SPase SipW is highly similar to SPases from archaea and the ER membrane of eukaryotes, suggesting that these enzymes form a subfamily of the type I SPases, which is conserved in the three domains of life.
Keywords: Bacillus subtilis, paralogous gene family, protein secretion, signal peptidase, leader peptidase
Since 1995, at least 12 microbial genomes have been completely sequenced and annotated (for a recent update, see the microbial database at http://www.tigr.org/tdb/mdb/mdb.html). A remarkable common property of these genomes is that many genes are present in multiple (paralogous) copies. A good example is the genome of Bacillus subtilis, which contains ∼4200 genes, 47% of which belong to paralogous gene families (Kunst et al. 1997). Close examination of the corresponding proteins of B. subtilis has indicated that many of these are involved in transcription regulation, and the transport of compounds into and out of the cell. Interestingly, most genes for components of the protein secretion machinery are present in only one copy, despite the fact that B. subtilis has a large capacity for protein secretion. The only known exception concerns the genes for type I signal peptidases (SPases; Tjalsma et al. 1997). These SPases remove amino-terminal signal peptides from secretory preproteins during, or shortly after their translocation across the cytoplasmic membrane, to release these proteins from the trans side of this membrane (for review, see Dalbey et al. 1997).
Homologous type I SPases have been identified in archaea, Gram-positive, and Gram-negative eubacteria, the inner membrane of yeast mitochondria, the thylakoid membrane of chloroplasts, and the endoplasmic reticular (ER) membranes of yeast and higher eukaryotes. Despite the fact that considerable similarities exist between the known type I SPases when amino acid sequences are compared pairwise, only few residues are strictly conserved in all known enzymes of this family (Dalbey et al. 1997). In particular, the type I SPases of eubacteria, mitochondria, and chloroplasts differ considerably from their homologs in archaea and the ER membrane, indicating that these enzymes belong to distinct subfamilies of SPases. Below, we have referred to these subfamilies as the P (prokaryotic)- and ER-type SPases, respectively. Most P-type SPases contain conserved serine and lysine residues that are essential for enzymatic activity, possibly by forming a catalytic dyad (Black 1993; Tschantz et al. 1993; van Dijl et al. 1995; Paetzel and Dalbey 1997). The putative active site serine residue of the P-type SPases is also conserved in the ER-type SPases. In contrast, the putative active site lysine residue of the P-type SPases is replaced with a histidine residue in the ER-type SPases (Dalbey and von Heijne 1992; van Dijl et al. 1992).
In many eubacteria, such as Escherichia coli (Blattner et al. 1997), Haemophilus influenzae (Fleischmann et al. 1995), Helicobacter pylori (Tomb et al. 1997), and Mycobacterium tuberculosis (Philipp et al. 1996), one P-type SPase seems to be sufficient for the processing of secretory pre-proteins. Similarly, the archaea Methanococcus jannaschii (Bult et al. 1996) and Methanobacterium thermoauthotrophicum (Smith et al 1997), and the yeast Saccharomyces cerevisiae (Goffeau et al. 1997) contain only one ER-type SPase. As shown for E. coli (Dalbey and Wickner 1985; van Dijl et al. 1988) and yeast (Böhni et al. 1988), these SPases are likely to be essential for cell viability. In contrast, two paralogous P-type SPases are found in Synechocystis PCC 6803 (Kaneko et al. 1996) and the inner membrane of yeast mitochondria (Behrens et al. 1991; Nunnari et al. 1993), whereas the presence of two paralogous ER-type SPases appears to be characteristic for most eukaryotic species (Dalbey et al. 1997). Finally, the largest numbers of paralogous SPases appear to be present in the archaeon Archaeoglobus fulgidus, which contains three genes for ER-type SPases (Klenk et al. 1997), and the Gram-positive bacterium B. subtilis in which seven sip genes for type I SPases have been identified. Five of the sip genes of B. subtilis (denoted sipS, sipT, sipU, sipV, and sipW) are located on the chromosome (van Dijl et al. 1992; Tjalsma et al. 1997); two additional sip genes (denoted sipP) are located on plasmids which were, thus far, only found in natto-producing strains of B. subtilis (Meijer et al. 1995).
Thus far, the presence of both P- and ER-type SPases in one organism was only evident for eukaryotes, which contain P-type SPases in their organelles and ER-type SPases in the ER (Dalbey et al. 1997). In this paper we document our surprising observation that one of the seven type I SPases of B. subtilis (ie. SipW) belongs to the ER-type subfamily of SPases. Thus, B. subtilis is the first organism known to contain SPases of the P- and the ER-type in one membrane. Furthermore, our studies, aimed at the functional analysis of the type I SPases of B. subtilis, show that SipS and SipT are the most important SPases of B. subtilis, whereas SipU, SipV, and SipW appear to have a minor role in precursor processing.
Results
SipW belongs to the subfamily of ER-type SPases
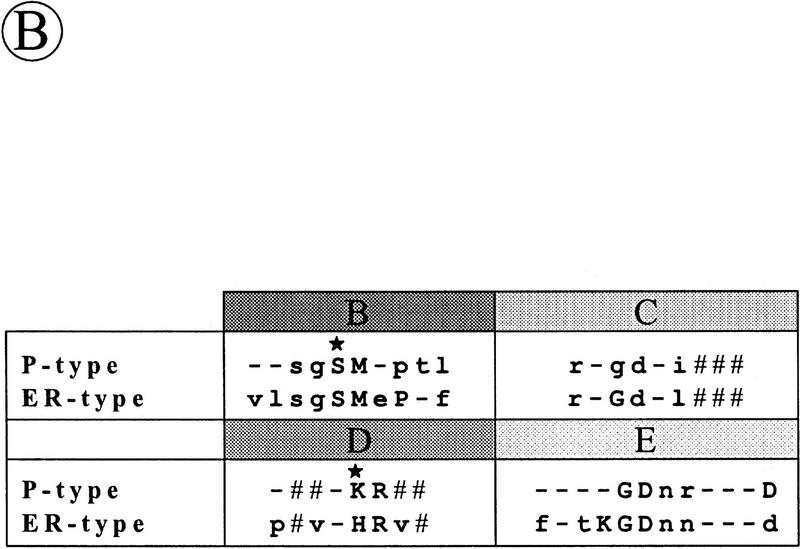
To make an inventory of the known type I SPases, database searches were performed with various P- and ER-type SPases. Unexpectedly, similarity searches with the ER-type SPase of M. jannaschii showed that the yqhE gene of B. subtilis (GenBank accession no. D84432) specifies a protein with a high degree of sequence similarity to the known ER-type SPases from archaea and eukaryotes. One of the SPases of A. fulgidus (GenBank locus AF1655) exhibited the highest degree of similarity with the deduced YqhE protein sequence (57% identical residues and conservative replacements in 131 residues). This observation strongly suggested that the yqhE gene specifies an ER-type SPase of B. subtilis. By analogy to other SPase-encoding genes of bacilli, we therefore renamed the yqhE gene sipW (Fig. 1A). As shown previously for the ER-type SPases of yeast (i.e., Sec11p) and Canis familiaris (i.e., SPC18 and SPC21; see Dalbey et al. 1997), SipW also shows amino acid sequence similarity with P-type SPases, but this similarity is mainly limited to the conserved domains B–E, which are present in all known SPases (Fig. 1B). Like the other known ER-type SPases, SipW contains a conserved histidine residue in domain D instead of the conserved lysine residue in domain D of the P-type SPases (Fig. 1A, B). Finally, SipW seems to contain one amino-terminal (AI), and one carboxy-terminal (AII) membrane anchor (Fig. 1C), as suggested previously for the P-type SPases of Rhodobacter capsulatus and yeast mitochondria (Dalbey et al. 1997).
Figure 1.
Conserved domains in ER-type SPases from eubacteria, archaea and eukaryotes. (A) Identification of conserved domains in the ER-type SPases from B. subtilis [SipW (Bsu); GenBank accession no. D84432], A. fulgidus [SPC21 (Afu), Sec11 (Afu) and Sip (Afu); GenBank loci AF1657, AF1791 and AF1655, respectively], M. thermoautotrophicum [Sip (Mth); GenBank locus MTH1448], S. cerevisiae [Sec11p (Sce); Böhni et al. 1988], C. familiaris [SPC18 (Cfa) and SPC21 (Cfa); Shelness and Blobel 1990; Greenburg et al. 1989], Rattus norvegicus [SPC18 (Rno); Signs and Jacquet 1994], M. jannaschii [Sip (Mja), GenBank locus MJU67481], and the partial amino acid sequence of a protein from Clostridium perfringens [SPC21 (Cpe); GenBank accession no. X86488]. The conserved domains B–E, which are present in all known type I SPases (van Dijl et al. 1992; Dalbey et al. 1997) are indicated. Because of limited similarities in the regions between conserved domains, only the four conserved domains B–E of Sip (Mja), Sip (Afu), Sec11 (Afu), and SPC21 (Cpe) are shown. Identical amino acids are indicated in bold when present in at least 6 of the 11 sequences shown. Putative transmembrane segments, indicated with A(nchor) I, or A(nchor) II, were predicted as described by Sipos and von Heijne (1993). (B) Consensus sequences of the conserved domains B–E of the P- and ER-type SPases. Strictly conserved residues are indicated by uppercase letters, conservative substitutions by lowercase letters, and conserved hydrophobic residues by a number sign (#). The serine and lysine residues that are critical for the activity of P-type SPases (Dalbey et al. 1997) are indicated (*). (C) Comparison of the predicted membrane topologies of SipS and SipW of B. subtilis. Membrane anchors (A, AI, AII) and the conserved domains B–E are indicated. (N) Amino terminus; (C) carboxyl terminus; (in) cytplasmic side of the membrane; (out) extracytoplasmic, cell wall-exposed side of the membrane.
The availability of the sequences of SipW of B. subtilis and recently identified SPases from archaea allowed the detailed comparison of the ER-type SPases, which is documented in Figure 1A. The comparison shows that these enzymes belong to a heterogeneous group of proteins. As documented previously for the P-type SPases, only the domains B–E are conserved in the ER-type SPases (Fig. 1A,B). A striking difference with the P-type SPases is that the domains B and C are separated by only one residue, and the domains D and E are separated by only 2–11 residues (Fig. 1A,C). The corresponding domains of the P-type SPases are separated by 19–42, and 23–118 residues, respectively (see Dalbey et al. 1997). Finally, the number of putative membrane spanning domains seems to vary in the ER-type SPases. All known enzymes of this subfamily have one amino-terminal membrane anchor (AI, Fig. 1A). In addition, some of these SPases, such as SipW of B. subtilis and SPC21 of A. fulgidus, seem to have a carboxy-terminal membrane anchor (AII, Fig. 1A,C), and a second SPase of A. fulgidus [Sip (Afu)] seems to have even three carboxy-terminal membrane anchors (data not shown).
Two quadruple SPase mutants of B. subtilis are viable
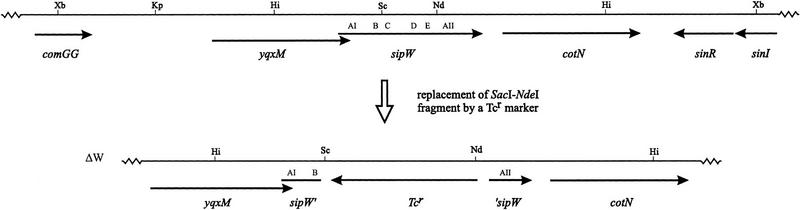
To determine whether SipW is required for growth or viability of the cell, an internal SacI–NdeI fragment of the chromosomal sipW gene, specifying the conserved domains C–E, was replaced with a tetracycline resistance (Tcr) marker (schematically shown in Fig. 2). As shown previously for strains lacking intact sipS, sipT, sipU, or sipV genes (Bolhuis et al. 1996; Tjalsma et al. 1997), the disruption of the sipW gene had no detectable effect on cell growth or viability (data not shown).
Figure 2.
Disruption of the B. subtilis sipW gene. Schematic presentation of the construction of B. subtilis 8G5 sipW–Tc (ΔW). The chromosomal sipW gene was disrupted with a Tcr marker by replacement recombination. To this purpose, B. subtilis 8G5 was transformed with the linearized plasmid pKW34dT, in which the 190-bp SacI–NdeI fragment (specifying the conserved domains C–E of SipW) was replaced with a Tcr marker. Only restriction sites relevant for the constructions are shown: (Hi) HindIII; (Kp) KpnI; (Nd) NdeI; (Sc) SacI; (Xb) XbaI. The relative positions of the comGG, yqxM, sipW, cotN, sinR, and sinI genes are indicated according to Kunst et al. (1997). Regions of sipW specifying the conserved domains B–E and the putative membrane anchors AI and AII are indicated. (sipW′), 3′-Truncated sipW gene; (‘sipW) 5′ truncated sipW gene.
To investigate which SPases are needed for growth and cell viability, various chromosomal sip gene deletions or disruptions were combined. The double, triple, and quadruple sip mutants obtained are listed in Table 1, which shows that most sip gene mutations could be combined, with one exception: It was not possible to construct strains lacking both sipS and sipT. Interestingly, the quadruple mutants jointly lacking sipS, sipU, sipV, and sipW (B. subtilis ΔSUVW), or sipT, sipU, sipV, and sipW (B. subtilis ΔTUVW) showed similar growth rates as the parental strain B. subtilis 8G5, irrespective of the growth medium used (data not shown). Thus, it seems that the secretory precursor processing machinery is functionally redundant, and that one SPase (i.e., SipS or SipT) is sufficient for growth and cell viability of B. subtilis.
Table 1.
Plasmids and bacterial strains
| Plasmids
|
Relevant
properties
|
Reference
|
|---|---|---|
| pGDL41 | encodes pre(A13i)–β-lactamase and SipS of B. subtilis; replicates in E. coli and B. subtilis; 8.1 kb; Apr; Kmr | van Dijl et al. (1992) |
| pGDL48 | lacks the sipS gene and contains a multiple cloning site; otherwise identical to pGDL41; 7.5 kb; Apr; Kmr | Meijer et al. (1995) |
| pMO | encodes pre(A13i)–β-lactamase, used for the site-directed mutagenesis of sipS | van Dijl et al. (1995) |
| pS-x | pMO derivatives carrying mutant sipS genes (x indicates the position and type of amino acid substitution in the corresponding mutant proteins); otherwise identical to pGDL41; 8.1 kb; Apr; Kmr | van Dijl et al. (1995) |
| pKTH10 | encodes the α-amylase (AmyQ) of B. amyloliquefaciens; 6.8 kb; Kmr | Palva (1982) |
| pCY66 | encodes the PSBT domain of the P. shermanii transcarboxylase 1.3 S subunit | Cronan (1990) |
| pBR322amyQ | encodes the α-amylase (AmyQ) of B. amyloliquefaciens | this paper |
| pBR322amyQ-BT | encodes the AmyQ–PSBT fusion protein | this paper |
| pKTH10-BT | as pKTH10, encodes the AmyQ–PSBT fusion protein; 7.0 kb | this paper |
| pX | vector for the integration of genes in the amyE locus of B.subtilis; integrated genes will be transcribed from the xylA promoter; carries the xylR gene; 7.5 kb; Apr; Cmr | Kim et al. (1996) |
| pXS | pX derivative; carries sipS downstream of the xylA promoter; 8.2 kb; Cmr | this paper |
| pXS-D146A | pX derivative; carries sipS–D146A downstream of the xylA promoter; 8.2 kb; Cmr | this paper |
| pLGW200 | integration vector for B. subtilis with a promoterless lacZ gene fused to the ribosome-binding site of the spoVG gene; 6.8 kb; Cmr | van Sinderen et al. (1990) |
| pLGW201 | pLGW200 derivative with a transcriptional sipW–lacZ fusion; 7.8 kb | this paper |
| pUK21 | cloning vector; 2.8 kb; Kmr | Vieria and Messing (1990) |
| pKW34dT | pUK21 derivative for the disruption of sipW; 6.6-kb Kmr; Tcr | this paper |
| Strains | ||
| E. coli | ||
| MC1061 | F; araD139; Δ(ara–leu)7696; Δ(lac)X74; galU; galK; hsdR2; mcrA; mcrBl; rspL | Wertmann et al. (1986) |
| B. subtilis | ||
| 8G5 | trpC2; tyr; his; nic; ura; rib; met; ade; lacks the sipP genes | Bron and Venema (1972) |
| 8G5 sipS (ΔS) | like 8G5; rib+; sipS | Bolhuis et al. (1996) |
| 8G5 sipT–Cm (ΔT) | like 8G5; sipT; Cmr | Tjalsma et al. (1997) |
| 8G5 sipT–Sp | like 8G5; sipT; Spr | this paper |
| 8G5 sipU | like 8G5; sipU | Tjalsma et al. (1997) |
| 8G5 sipV–Em | like 8G5; sipV; Emr | Tjalsma et al. (1997) |
| 8G5 sipV–Sp | like 8G5; sipV; Spr | this paper |
| 8G5 sipW–Tc (ΔW) | like 8G5; sipW; Tcr | this paper |
| ΔSU | like 8G5; rib+; sipS; sipU | this paper |
| ΔSV | like 8G5; rib+; sipS; sipV; Emr | this paper |
| ΔSW | like 8G5; rib+; sipS; sipW; Tcr | this paper |
| ΔSUV | like 8G5; rib+; sipS; sipU; sipV; Emr | this paper |
| ΔSUW | like 8G5; rib+; sipS; sipU; sipW; Tcr | this paper |
| ΔSVW | like 8G5; rib+; sipS; sipV; Emr; sipW; Tcr | this paper |
| ΔSUVW | like 8G5; rib+; sipS; sipU; sipV; Emr; sipW; Tcr | this paper |
| ΔTU | like 8G5; sipT; Cmr; sipU | this paper |
| ΔTV | like 8G5; sipT; Cmr; sipV; Emr | this paper |
| ΔTW | like 8G5; sipT; Cmr; sipW; Tcr | this paper |
| ΔTUV | like 8G5; sipT; Cmr; sipU; sipV; Emr | this paper |
| ΔTUW | like 8G5; sipT; Cmr; sipU; sipW; Tcr | this paper |
| ΔTUVW | like 8G5; sipT; Cmr; sipU; sipV; Spr; sipW; Tcr | this paper |
| ΔUV | like 8G5; sipU; sipV; Emr | this paper |
| ΔUW | like 8G5; sipU; sipW; Tcr | this paper |
| ΔUW | like 8G5; sipU; sipW; Tcr | this paper |
| ΔVW | like 8G5; sipV; Spr; sipW; Tcr | this paper |
| ΔST (pGDL41) | like 8G5; rib+; sipT; Cmr; contains pGDL41 | this paper |
| ΔST (pS-D146A) | like 8G5; rib+; sipT; Cmr; contains pS–D146A | this paper |
| ΔSTxS | like 8G5; rib+; sipT; Spr; PxylA–sipS; amyE; Cmr | this paper |
| ΔSTxS–D146A | like 8G5; rib+; sipT; Spr; PxylA–sipS–D146A; amyE; Cmr | this paper |
| 8G5::pGDE22 | 8G5 carrying pGDE22 (sipS–lacZ) in the chromosome; Cmr | Bolhuis et al., (1996) |
| 8G5::pLGT207 | 8G5 carrying pLGW207 (sipT–lacZ) in the chromosome; Cmr | Tjalsma et al. (1997) |
| 8G5::pLGU202 | 8G5 carrying pLGU202 (sipU–lacZ) in the chromosome; Cmr | Tjalsma et al. (1997) |
| 8G5::pLGV201 | 8G5 carrying pLGV201 (sipV–lacZ) in the chromosome; Cmr | Tjalsma et al. (1997) |
| 8G5::pLGW201 | 8G5 carrying pLGW201 (sipW–lacZ) in the chromosome; Cmr | this paper |
| 8G5 degS–degU | like 8G5; degS–degU; Kmr | Bolhuis et al. (1996) |
| 8G5 degU32(Hy) | like 8G5; degU32(Hy); Kmr | Bolhuis et al. (1996) |
| 8G5::pGDE22 | derivative of 8G5::pGDE22;degU32(Hy); Cmr; Kmr | |
| degU32(Hy) | Bolhuis et al. (1996) | |
| 8G5::pLGT207 | derivative of 8G5::pLGT207; degU32(Hy); Cmr; Kmr | |
| degU32(Hy) | this paper | |
| 8G5::pLGU202 | derivative of 8G5::pLGU202; degU32(Hy); Cmr; Kmr | |
| degU32(Hy) | this paper | |
| 8G5::pLGV201 | derivative of 8G5::pLGV201; degU32(Hy); Cmr; Kmr | |
| degU32(Hy) | this paper | |
| 8G5::pLGW201 | derivative of 8G5::pLGW201; degU32(Hy); Cmr; Kmr | |
| degU32(Hy) | this paper | |
Functional SipS is essential for viability of a B. subtilis strain lacking SipT
To evaluate the importance of SipS and SipT, a strain was constructed that produces a temperature-sensitive SipS mutant protein, but lacks wild-type SipS and SipT. To this purpose, we first selected temperature-sensitive variants from a large collection of previously constructed site-specific mutants of SipS (van Dijl et al. 1995). By use of a plate assay for SipS activity (van Dijl et al. 1992), five temperature-sensitive mutants were identified, which are active at 30°C, but not at 48°C (Table 2). These contain the L74A, Y81A, R84A, R84H, or D146A mutations, respectively. In a second step, B. subtilis 8G5 sipS, which lacks the chromosomal sipS gene (Bolhuis et al. 1996), was transformed with plasmids specifying the L74A, Y81A, R84A, R84H, or D146A SipS mutant proteins. In a third and final step, the sipT gene of the latter transformants was disrupted with a chloramphenicol resistance (Cmr) marker, as described previously (Tjalsma et al. 1997). The resulting mutant strains (ΔST sipSts) showed no growth defects at 30°C, 37°C, and 42°C (data not shown). However, after a temperature shift to 48°C during the exponential growth phase, all five ΔST sipSts strains stopped growing when they reached the transition phase between exponential and postexponential growth and, on continued incubation at 48°C, they even started to lyse. In contrast, the control strain ΔST (pGDL41), producing wild-type SipS, continued to grow in the postexponential growth phase (Fig. 3A). Because all ΔST sipSts mutant strains showed similar characteristics with respect to growth and cell viability, only one of these strains [i.e., ΔST (pS-D146A)] was selected for further analyses. As shown by Western blotting, the cellular levels of wild-type SipS were not affected by the temperature shift from 37°C to 48°C. In contrast, the cellular levels of SipS–D146A, which were already reduced a 37°C, were decreased to levels below detection at 48°C (Fig. 3B). In summary, these observations show that for growth and cell viability of B. subtilis, a functional SipS is required in the absence of SipT.
Table 2.
Identification of temperature-sensitive SipS mutants
| Plasmid
|
Temperature
|
|||
|---|---|---|---|---|
| 22°C
|
30°C
|
37°C
|
48°C
|
|
| pGDL48 (empty vector) | − | − | − | − |
| pGDL41 (sipS) | + | + | + | + |
| pS–L74A | + | + | +/− | − |
| pS–Y81A | − | + | − | − |
| pS–R84A | + | + | − | − |
| pS–R84H | + | + | − | − |
| pS–D146A | + | + | − | − |
A plate (halo)-assay for the processing of the hybrid precursor pre(A13i)–β-lactamase (van Dijl et al. 1992) was used to examine the temperature sensitivity of mutant SipS proteins. E coli MC1061 strains containing plasmids that specify wild-type SipS (pGDL41) or SipS mutant proteins (pS-x) were analyzed for halo formation at 22°C, 30°C, 37°C, and 48°C respectively. Halo formation is indicated with +.
Figure 3.
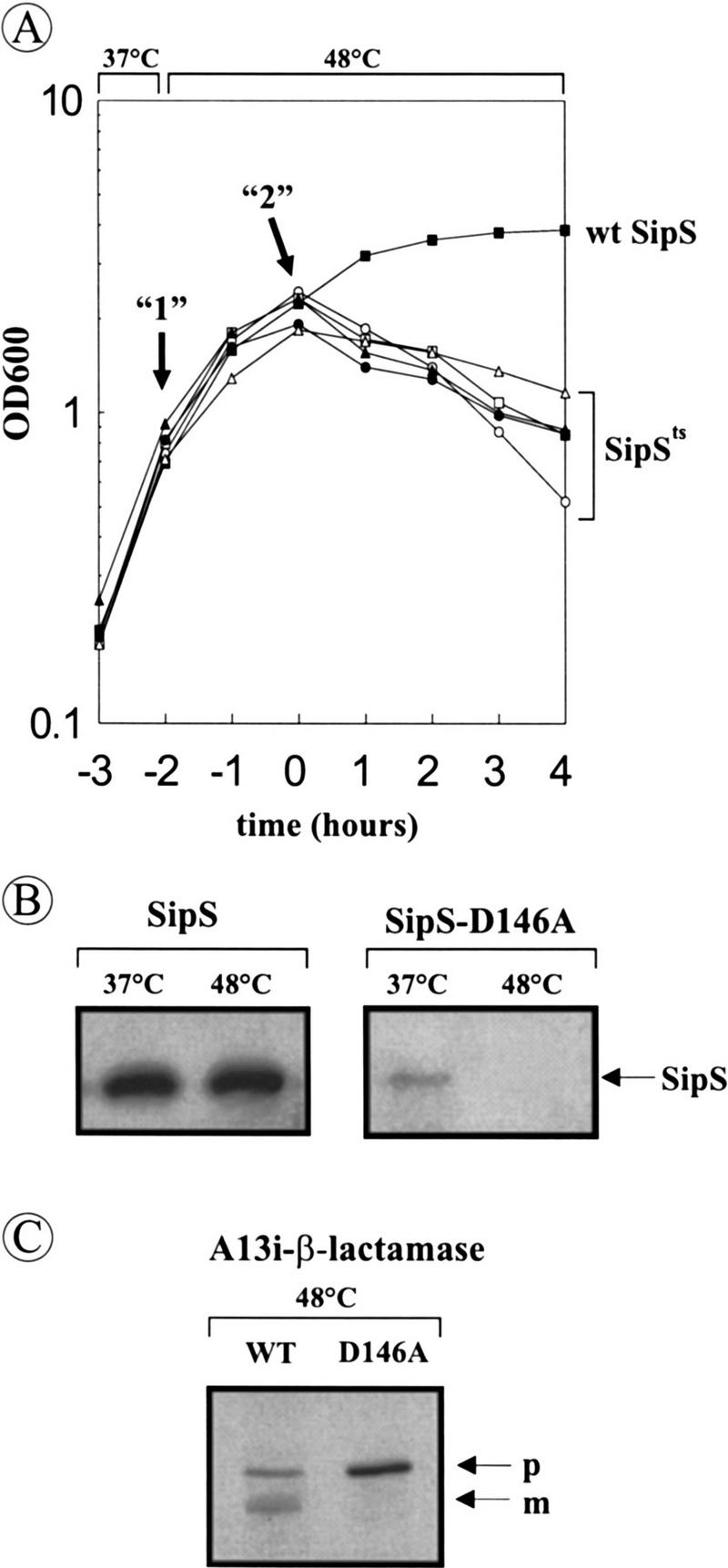
Temperature-sensitive growth of B. subtilis ΔST containing a sipSts gene. (A) Growth curves of B. subtilis strains that lack intact chromosomal copies of sipS and sipT but contain plasmids specifying the wild-type SipS or temperature-sensitive SipS mutant proteins. The names of the latter plasmids refer to the respective mutation in SipS. (○) B. subtilis ΔST (pS-L74A); ( ) B. subtilis ΔST (pS-Y81A); (•) B. subtilis ΔST (pS-R84A); (▴) B. subtilis ΔST (pS-R84H); (▵) B. subtilis ΔST (pS-D146A); (▪) B. subtilis ΔST (pGDL41; specifies wild-type SipS). Overnight cultures were diluted 100-fold in fresh TY medium and incubated at 37°C. When the cells reached an OD600 of ∼0.6, the temperature was shifted to 48°C. Samples were withdrawn shortly before the temperature shift (arrow “1”) and 2 hr after the temperature shift (arrow “2”). Zero time (t = 0) indicates the transition point between the exponential and postexponential growth phases. (B) The presence of wild-type SipS [B. subtilis ΔST (pGDL41)] and SipS–D146A [B. subtilis ΔST (pS-D146A)] in cells growing at 37°C (arrow “1” in A) and 48°C (arrow “2” in A) was analyzed by SDS-PAGE and Western blotting. The position of SipS/SipS–D146A is indicated. (C) Accumulation of pre(A13i)-β-lactamase in cells of B. subtilis ΔST (pGDL41; wild-type SipS) and ΔST(pS–D146A) at 48°C (arrow “2” in A) was analyzed by SDS-PAGE and Western blotting. (p) Precursor; (m) mature.
Accumulation of secretory preproteins in the absence of functional SipS and SipT
As a first approach to monitor the effects of the absence of functional SipS and SipT on the processing of secretory preproteins, we analyzed the processing of pre(A13i)-β-lactamase. This hybrid precursor is most efficiently processed by SipS and SipT (van Dijl et al. 1992; Tjalsma et al. 1997). As shown in Figure 3, B and C, the disappearance of the SipS–D146A mutant protein in B. subtilis ΔST (pS–D146A) at 48°C was paralleled by the accumulation of pre(A13i)-β-lactamase. In contrast, cells of B. subtilis ΔST (pGDL41; wild-type SipS) contained much smaller amounts of this precursor and, in addition, they contained the corresponding mature protein (Fig. 3C). These findings show that processing of pre(A13i)–β-lactamase is strongly affected in the absence of functional SipS and SipT.
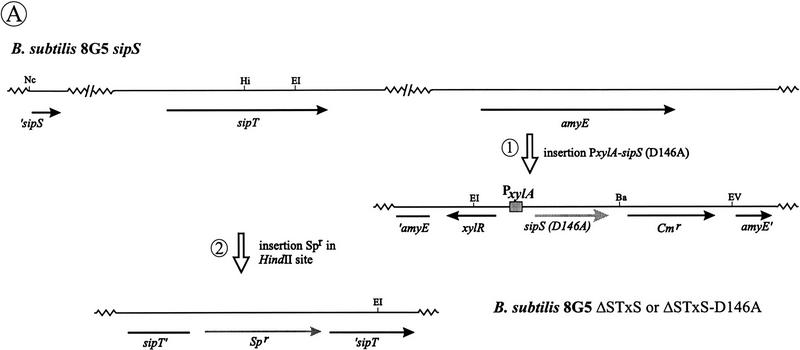
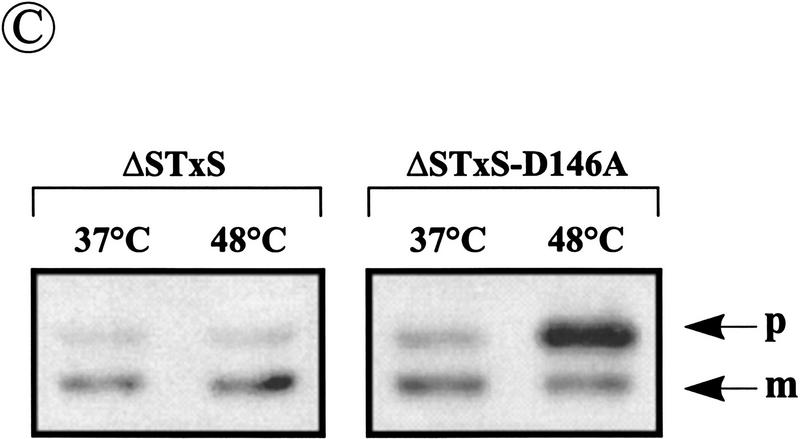
To investigate the effects of depletion of SipS in cells lacking SipT, two strains with a disrupted sipT gene were constructed in which the transcription of sipS or sipS–D146A is controlled by the xylose-inducible xylA promoter. In these strains, denoted ΔSTxS and ΔSTxS–D146A, respectively, the wild-type sipS gene was removed from its original locus and the xylA–sipS or xylA–sipS–D146A constructs were integrated into the amyE locus (Fig. 4A). Unexpectedly, even in the absence of xylose, both strains showed normal growth at 37°C and 42°C, suggesting that the xylA promoter was not completely repressed in the absence of xylose (data not shown). In fact, the inhibition of growth and subsequent cell lysis was only observed for B. subtilis ΔSTxS–D146A on incubation at 48°C in the absence of xylose (data not shown), similar to B. subtilis ΔST (pS–D146A) (Fig. 3A). As shown by Western blotting experiments with cells in the postexponential growth phase, the cellular levels of SipS or SipS–D146A in B. subtilis ΔSTxS and ΔSTxS–D146A, respectively, depended on the presence or absence of xylose. Compared with the parental strain B. subtilis 8G5, B. subtilis ΔSTxS contained slightly reduced levels of SipS in the absence of xylose, whereas SipS was about five-fold overproduced in the presence of xylose. In contrast, in B. subtilis ΔSTxS–D146A, only very low levels of SipS–D146A were detectable in the presence of xylose, whereas SipS–D146A was not detectable in the absence of xylose (Fig. 4B). The latter observation suggests that very small amounts of SipS–D146A are sufficient for growth and cell viability.
Figure 4.
Accumulation of pre-AmyQ in B. subtilis cells lacking functional SipS and SipT. (A) Schematic presentation of the construction of B. subtilis ΔSTxS and ΔSTxS–D146A. For this purpose, the sipS and sipS–D146A genes were amplified by PCR with the primers EX-1 (5′-ACTCTAGATGCCATTGAAATAGACC-3′) and EX-2 (5′-ATGGATCCTTGTTTAAAACTTGG-3′) with the plasmids pGDL41 and pS–D146A as templates. The amplified fragments were cleaved with XbaI and BamHI and ligated to the SpeI–BamHI sites of the plasmid pX (Kim et al. 1997), resulting in pXS and pXS–D146A, respectively. Next, both plasmids were linearized and used to transform B. subtilis 8G5 sipS. Thus, the amyE gene was replaced with the xylose-inducible sipS or sipS–D146A genes and a Cmr marker. The inactivation of the amyE gene was tested and confirmed by lack of halo formation during growth on plates containing 1% starch. Finally, the chromosomal sipT gene was disrupted with a Spr marker by homologous recombination, resulting in the strains ΔSTxS (sipS, sipT-Sp, PxylA–sipS, Cmr) and ΔSTxS–D146A (sipS, sipT–Sp, PxylA–sipS–D146A, Cmr). Only restriction sites relevant for the constructions are shown: (Ba) BamHI; (EI) EcoRI; (EV) EcoRV; (Hi) HindII; (Nc) NcoI. (′sipS) 5′-Truncated sipS gene (Bolhuis et al. 1996); (amyE′) 3′-truncated amyE gene; (′amyE) 5′-truncated amyE gene; (sipT′) 3′-truncated sipT gene; (′sipT) 5′-truncated sipT gene; (xylR) gene specifying the XylR repressor protein. (B) Detection of wild-type SipS or SipS–D146A in cells of B. subtilis 8G5 (WT), 8G5 sipS (ΔS), ΔSTxS and ΔSTxS–D146A. Cells were grown in TY medium at 37°C in the presence (+) or absence (−) of 1% xylose, and sample for SDS-PAGE and Western blotting were withdrawn 4 hr after the transition from exponential to postexponential growth. (C) Accumulation of pre-AmyQ in cells of B. subtilis ΔSTxS and ΔSTxS–D146A. Overnight cultures were diluted 100-fold in fresh TY medium without xylose and incubated at 37°C. When the cells reached an OD600 of ∼0.6, the temperature was shifted to 48°C. Samples withdrawn shortly before (t = −1; see Fig. 3A), and 30 min after the temperature shift (t = −0.5) were analyzed by SDS-PAGE and Western blotting. (p) Precursor; (m) mature.
To determine the effects of various levels of SipS (–D146A) activity on the accumulation of secretory precursor proteins in cells lacking SipT, B. subtilis ΔSTxS and ΔSTxS–D146A were transformed with plasmid pKTH10 that specifies the Bacillus amyloliquefaciens α-amylase AmyQ. The presence of pKTH10 results in the accumulation of pre-AmyQ, even in wild-type cells (Palva 1982; Kontinen and Sarvas 1988). As shown by Western blotting, cells of B. subtilis ΔSTxS–D146A, which were incubated at 48°C in the absence of xylose, accumulated strongly increased amounts of pre-AmyQ (Fig. 4C). In contrast, at 37°C, cells of the latter strain accumulated similar amounts of pre-AmyQ as B. subtilis ΔSTxS, producing close to wild-type levels of SipS (37°C and 48°C; Fig. 4C), or overproducing SipS (data not shown). Taken together, our observations show that the presence of SipS and SipT is required for efficient processing of pre-AmyQ, and that in cells lacking SipT, very small amounts of SipS, which are below the level of detection, are sufficient to prevent the accumulation of pre-AmyQ at high levels.
Impaired translocation of pre-AmyQ in cells lacking functional SipS and SipT
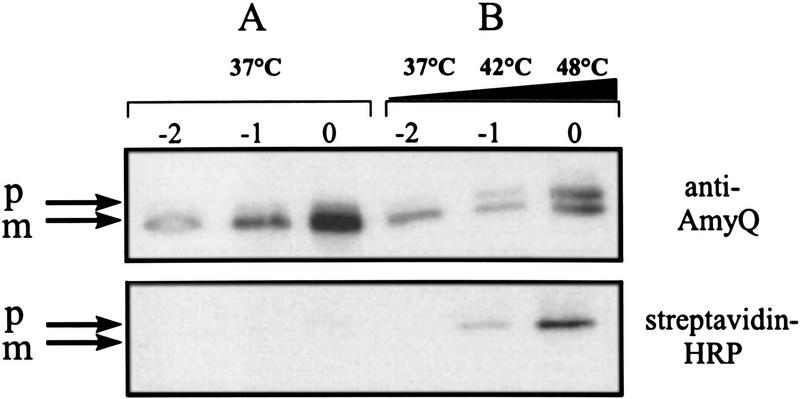
To investigate whether SipS and SipT depletion affects the translocation of pre-AmyQ, the sequences specifying the biotin-accepting domain of a transcarboxylase from Propionibacterium shermannii (PSBT; Jander et al. 1996) were fused to the 3′ end of the amyQ gene on pKTH10. The resulting plasmid pKTH10-BT was used to transform B. subtilis ΔSTxS–D146A and, subsequently, the accumulation of biotinylated (pre-)AmyQ–PSBT was determined. The rationale of this experiment is that pre-AmyQ–PSBT can only be biotinylated by the cytoplasmic biotin–ligase if the PSBT domain folds into its native three-dimensional structure in the cytoplasm. This will only happen if the translocation of pre-AmyQ–PSBT across the membrane is impaired. As shown in Figure 5A, processing of pre-AmyQ–PSBT occurred efficiently in cells of B. subtilis ΔSTxS–D146A growing at 37°C in the absence of xylose, as only small amounts of pre-AmyQ–PSBT were detectable. Under these conditions, mature nonbiotinylated AmyQ–PSBT was secreted into the growth medium (data not shown). Similar results were obtained with the parental strain 8G5, both at 37°C and 48°C (data not shown). In contrast, high levels of pre-AmyQ–PSBT accumulated in cells of B. subtilis ΔSTxS–D146A incubated in the absence of xylose at 42°C (permissive temperature for growth) or 48°C (nonpermissive temperature) (Fig. 5B, top panel). As shown with a streptavidin–horseradish peroxidase (HRP) conjugate, significant amounts of the accumulating pre-AmyQ–PSBT were biotinylated, showing that the translocation of these precursor molecules was impaired (Fig. 5B, bottom panel). Compared with cells of B. subtilis ΔSTxS–D146A, which were grown in the absence of xylose at 37°C, cells grown at 42°C, or 48°C accumulated significantly reduced levels of mature AmyQ–PSBT; although mature AmyQ–PSBT was produced in the absence of xylose at 42°C or 48°C, it was not detectably biotinylated (Fig 5A,B).
Figure 5.
Accumulation of biotinylated pre-AmyQ–PSBT in the absence of functional SipS and SipT. An overnight culture of B. subtilis ΔSTxS–D146A (pKTH10-BT) was diluted 100-fold in fresh TY medium without xylose and incubated at 37°C. When the culture reached an OD600 of ∼0.6, a sample (t = −2) was taken. Subsequently, the culture was divided in two equal moieties A and B. Culture A was kept for 2 hr at 37°C, and samples were taken at t = −1 and t = 0. Culture B was incubated for 1 hr at 42°C and, after a sample (t = −1) had been taken, incubation was continued for 1 hr at 48°C before a second sample (t = 0) was taken. Cells were collected by centrifugation and analyzed by SDS-PAGE and Western blotting. The presence of AmyQ–PSBT was visualized with specific antibodies. Biotinylated pre-AmyQ–PSBT was visualized with streptavidin–HRP. (p) Precursor; (m) mature.
Relative contributions of SipU, SipV, and SipW to the processing of pre-AmyQ
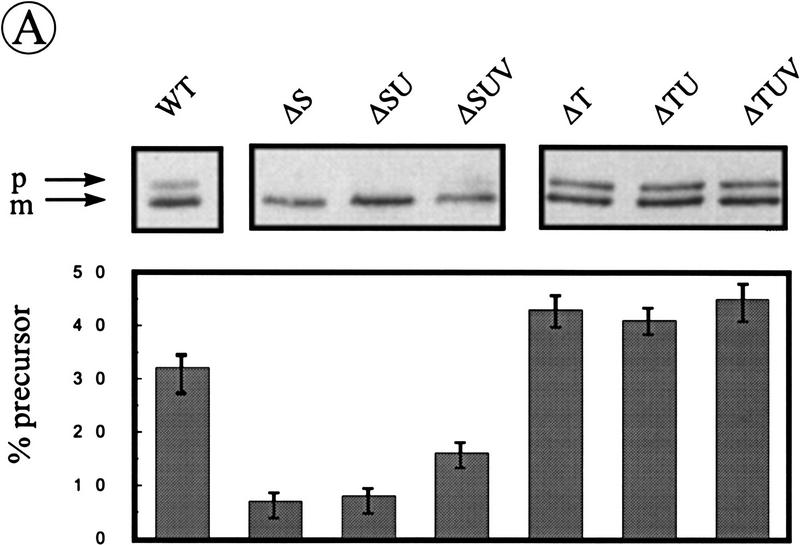
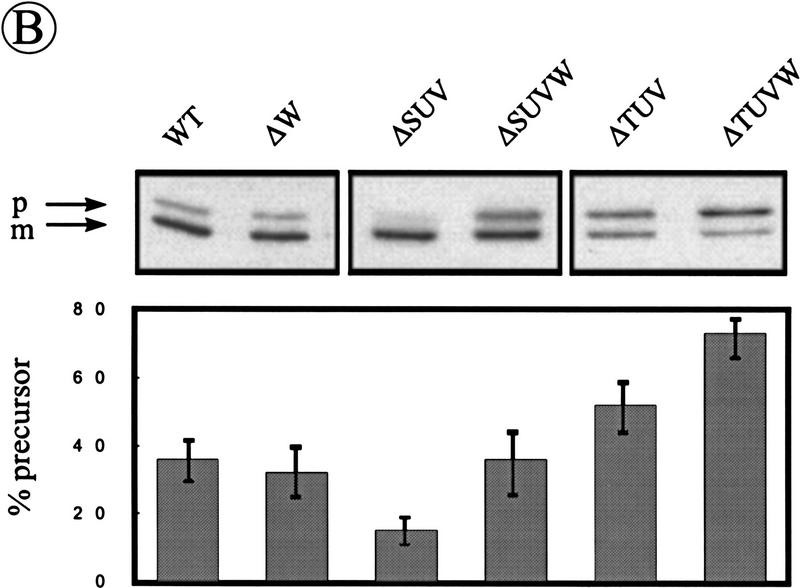
We have reported previously that the processing of pre-AmyQ is stimulated in the absence of SipS, and retarded in the absence of SipT, indicating that the production of SipS interferes with pre-AmyQ processing, and that this precursor is a preferred substrate of SipT (Tjalsma et al. 1997). To examine the relative contributions of SipU, SipV, and SipW to the processing of pre-AmyQ in the absence of SipS or SipT, Western blotting and pulse-chase labeling experiments were performed with strains lacking multiple SPases. The parental strain 8G5 was used as a control. As the results obtained by pulse-chase labeling confirmed those obtained by Western blotting, only the latter are documented. As shown in Figure 6A, cells of B. subtilis ΔS, or ΔSU, accumulated only very low levels of pre-AmyQ. Similar results were obtained with cells of B. subtilis ΔSV and ΔSW (data not shown). The level of pre–AmyQ was slightly increased in cells of B. subtilis ΔSUV, but remained lower than in the parental strain (Fig. 6A). The levels of pre-AmyQ accumulation in B. subtilis ΔSUW and ΔSVW were similar to those in B. subtilis ΔSUV (data not shown). Compared with B. subtilis ΔT, which accumulates more pre-AmyQ than the parental strain, cells of B. subtilis ΔTU and ΔTUV showed no additional accumulation of pre-AmyQ (Fig. 6A), and similar results were obtained with B. subtilis ΔTV, ΔTW, and ΔTUW (data not shown). The levels of pre-AmyQ in cells of B. subtilis ΔSUV or ΔTUV could only be increased further by disruption of the sipW gene, showing that SipW is actively involved in the processing of pre-AmyQ (Fig. 6B). In conclusion, these observations show that the accumulation of pre-AmyQ in cells lacking either SipS or SipT was significantly exacerbated only by the simultaneous removal of SipU, SipV, and SipW.
Figure 6.
Relative contribution of SipU, SipV, and SipW to pre-AmyQ processing in the absence of SipS or SipT. (A) The accumulation of pre-AmyQ in pKTH10-transformed cells of the parental strain B. subtilis 8G5 (WT) and in pKTH10-transformed cells lacking SipS (ΔS), SipS, and SipU (ΔSU), SipS, SipU, and SipV (ΔSUV), SipT (ΔT), SipT, and SipU (ΔTU), or SipT, SipU, and SipV (ΔTUV) was analyzed by SDS-PAGE and Western blotting. Samples were taken from overnight cultures in TY medium. (p) Precursor; (m) mature. Relative amounts of precursor and mature forms of AmyQ were determined by densitometer scanning of films. The average values of three independent experiments are shown, and the standard deviation is indicated by error bars. (B) The accumulation of pre-AmyQ in pKTH10-transformed cells of the parental strain B. subtilis 8G5 (WT), and pKTH10-transformed cells lacking SipW (ΔW), SipS, SipU, and SipV (ΔSUV), SipS, SipU, SipV, and SipW (ΔSUVW), SipT, SipU, and SipV (ΔTUV), or SipT, SipU, SipV, and SipW (ΔTUVW), was analyzed by SDS-PAGE and Western blotting. Samples were prepared from cells grown in TY medium until the early postexponential growth phase (t = 4). Relative amounts of precursor and mature forms of AmyQ are indicated as in A.
Transcription of only sipS and sipT is temporally regulated by the DegS–DegU system
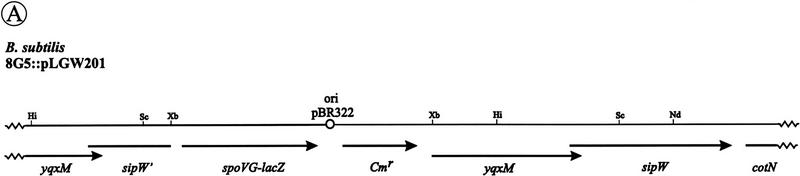
The transcription of the sipS and sipT genes is temporally controlled, whereas the sipU and sipV genes are constitutively transcribed (Bolhuis et al. 1996; Tjalsma et al. 1997). To analyze the transcription of the sipW gene, a transcriptional sipW–lacZ fusion was constructed, and introduced into the chromosome of B. subtilis 8G5 (schematically shown in Fig. 7A). Cells of the resulting strain were grown in TY or minimal medium, and samples withdrawn at hourly intervals were assayed for β-galactosidase activity. The results show that, irrespective of the growth medium used, sipW is constitutively transcribed at a low level (only the results for minimal medium are shown in Fig. 7B, ▴).
Figure 7.
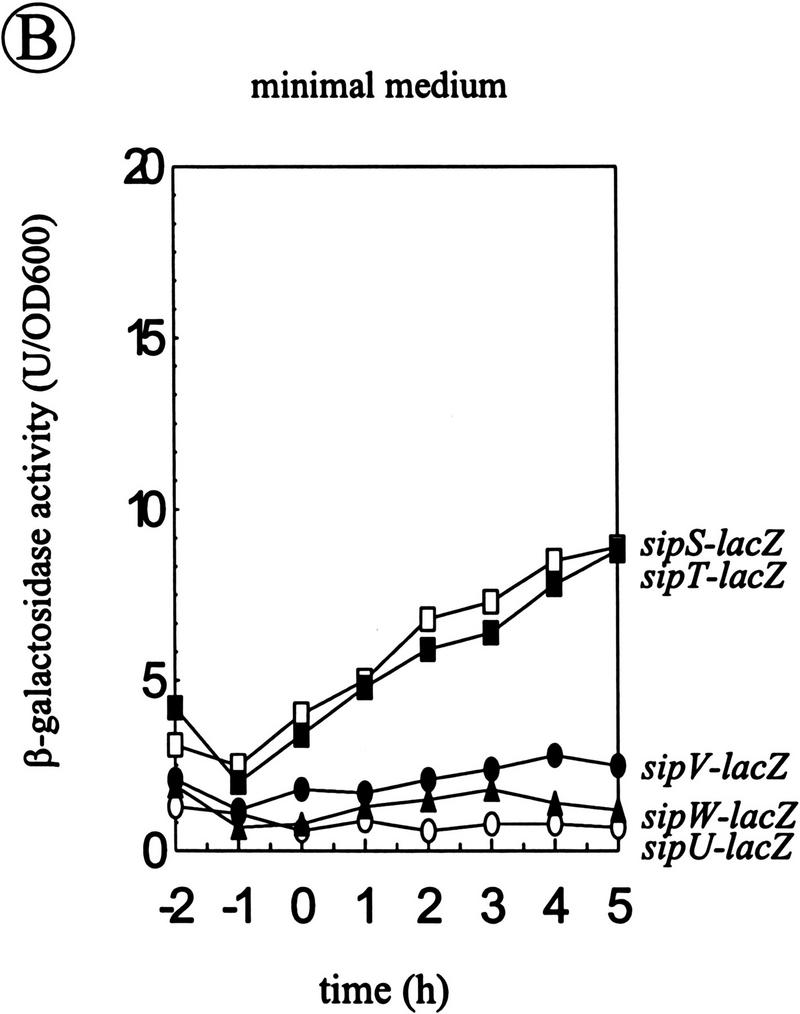
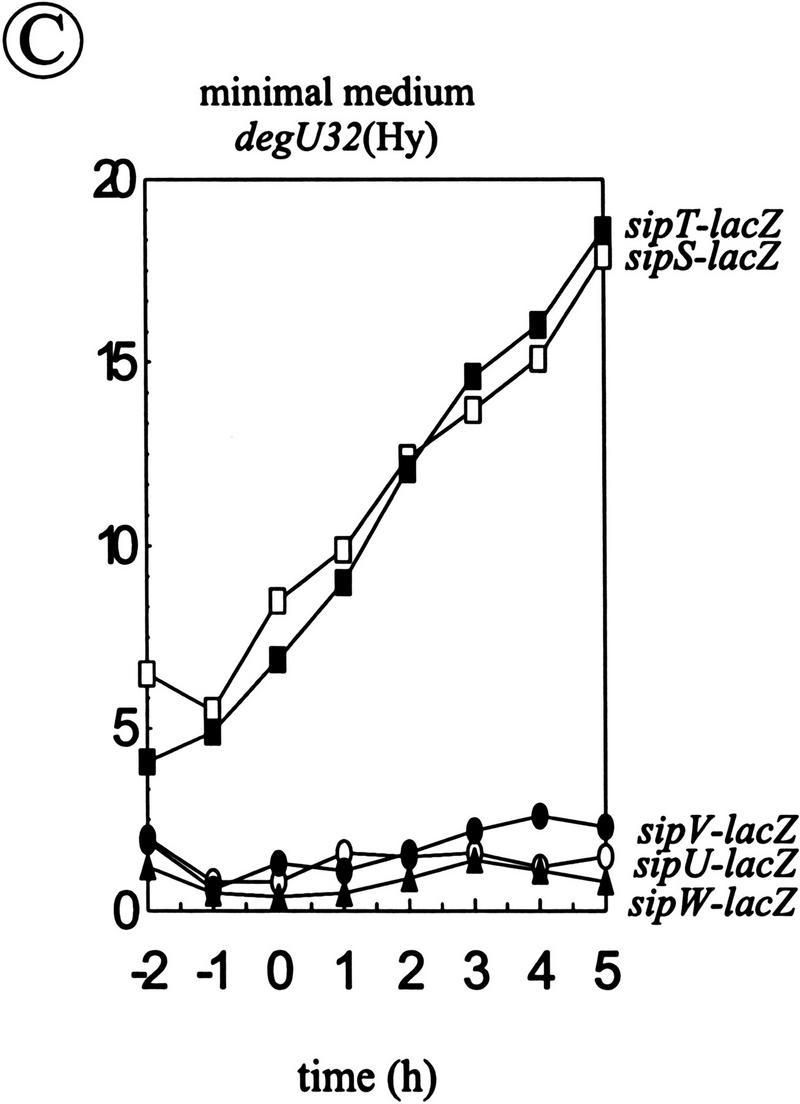
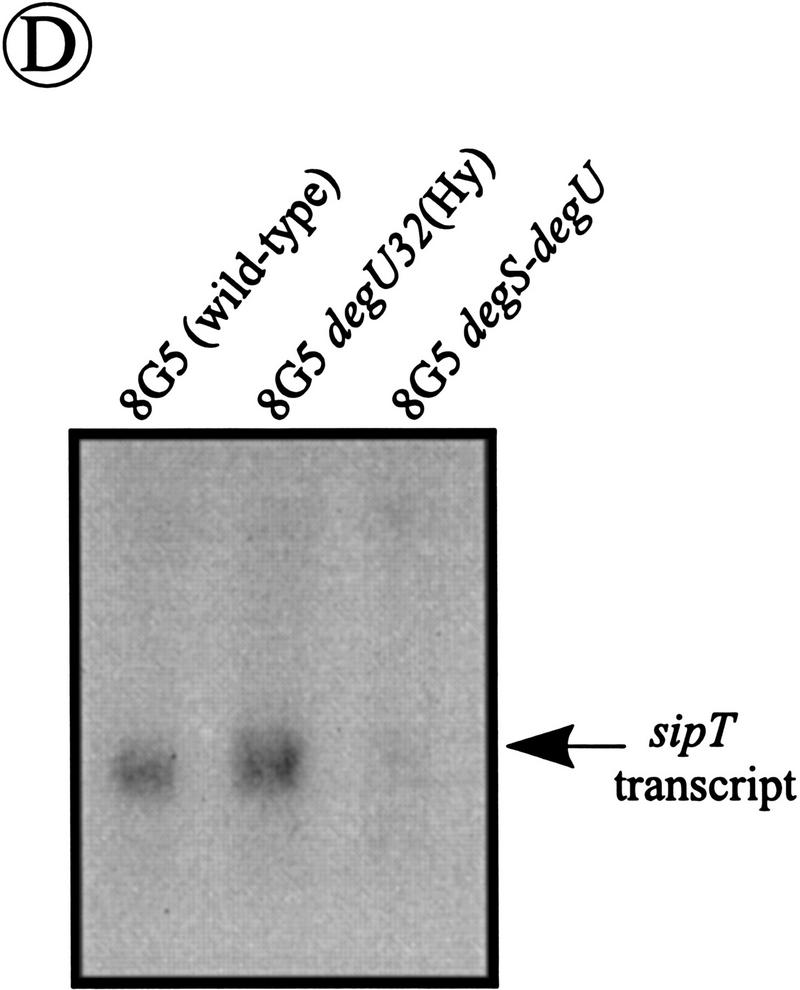
Analysis of the transcription of the sip genes of B. subtilis. (A) Schematic presentation of the sipW locus on the chromosome of B. subtilis 8G5::pLGW201. A transcriptional sipW–lacZ gene fusion was constructed with plasmid pLGW200 (van Sinderen et al. 1990), a chromosomal integration plasmid for B. subtilis containing a promoterless spoVG–lacZ gene fusion. For this purpose, a fragment comprising the yqxM gene and the 5′ end of sipW was amplified by PCR with the primers lbw-3 (5′-CATCTAGATACAGGAGGTAAGATATG-3′) and lbw-5 (5′-AGTTCTAGAGTATTTGCATCCTGC-3′). Next, the amplified fragment was cloned into the XbaI-site of the multiple cloning site upstream of the spoVG–lacZ gene fusion on pLGW200, resulting in pLGW201. Finally, the sipW–lacZ gene fusion was introduced in the chromosome of B. subtilis 8G5 by a Campbell-type integration of pLGW201, resulting in B. subtilis 8G5::pLGW201. In the latter strain, the transcription of lacZ is directed by the promoter(s) of sipW. Only restriction sites relevant for the constructions are shown: (Hi) HindIII; (Nd) NdeI; (Sc) SacI; (Xb) XbaI. (ori pBR322) Replication functions of pBR322. (B) Time courses of the expression of sip–lacZ gene fusions were determined in cells growing in minimal medium at 37°C. β-Galactosidase activities (in units per OD600) were determined for B. subtilis 8G5::pGDE22 ( ; sipS–lacZ), B. subtilis 8G5::pLGT207 (▪; sipT–lacZ), B. subtilis 8G5::pLGU202 (○; sipU–lacZ), B. subtilis 8G5::pLGV201 (•; sipV–lacZ), and B. subtilis 8G5::pLGW201 (▴; sipW–lacZ). Zero time (t = 0) indicates the transition point between the exponential and postexponential growth phases. (C) Effects of the degU32(Hy) mutation on the expression of sip–lacZ gene fusions were determined as in B. To this purpose, strains containing the sip–lacZ gene fusions indicated in B were transformed with chromosomal DNA of B. subtilis 8G5 degU32(Hy). (D) Detection of sipT-specific transcripts by Northern hybridization. Equal amounts of total RNA from cells of B. subtilis 8G5, 8G5 degU32(Hy), and 8G5 degU–degS in the transition state were loaded in each lane. A [32P]dCTP-labeled DNA fragment, amplified by PCR with the primers lbt-9 (5′-ATGAATTCAGCCCGGTTATCTCC-3′) and lbt-10 (5′-GGAAGTCGACATACGTACCTGGAATGG-3′), was used as a probe.
Because it was shown previously that the temporal control of sipS transcription is exerted by the DegS–DegU two-component regulatory system (Bolhuis et al. 1996), which is also required for the transcription of most genes for secreted degradative enzymes (for review, see Ferrari et al. 1993), we investigated whether this regulatory system is also involved in the transcription of the sipT, sipU, sipV, and sipW genes. To this purpose, the degU32(Hy) mutation, which leads to increased transcription of genes for degradative enzymes (Ferrari et al. 1993; Msadek et al. 1993) and sipS (Bolhuis et al. 1996), was introduced in strains carrying chromosomal sipS–lacZ, sipT–lacZ, sipU–lacZ, sipV–lacZ, or sipW–lacZ gene fusions. The resulting strains were grown in minimal medium and samples, withdrawn at hourly intervals, were assayed for β-galactosidase activity. The results show that only the transcription of sipS and sipT was stimulated in strains containing the degU32(Hy) mutation (Fig. 7B,C; indicated with and ▪ respectively). In contrast, the degU32(Hy) mutation had no effect on the transcription of sipU, sipV, and sipW (Fig. 7B,C; indicated with ○, •, ▴, respectively).
To verify the involvement of the DegS–DegU system in the transcription of the sipT gene, Northern hybridization experiments were performed with total RNA from the B. subtilis strains 8G5, 8G5 degU32(Hy), and 8G5 degS–degU. In contrast to sipS, for which two transcripts were detectable (Bolhuis et al. 1996), only one, monocistronic, sipT-specific transcript was detected. Compared with the parental strain 8G5, the amount of the sipT-specific transcript was about twofold increased in cells with the degU32(Hy) mutation, although it was hardly detectable in cells containing the degS–degU deletion (Fig. 7D). These findings show that the transcription of the sipT gene is controlled by the DegS–DegU system. Consistent with this conclusion, we identified the sequence: 5′-TATGATAAAGTA-3′ at a position 46 nucleotides upstream of the translational start of sipT. This sequence contains one mismatch and one base insertion compared with a consensus sequence (AMTGAAMAKWW in which K = G or T; M = A or C; W = A or T), which has been implicated in the DegU-dependent control of gene expression (Jacobs 1995). Taken together, these results show that the transcription of sipS and sipT is concerted with that of the genes for secreted degradative enzymes. In contrast, the sipU, sipV, and sipW genes are constitutively transcribed at a low level.
Discussion
Various microorganisms contain paralogous type I SPases. Interestingly, this seems to be a common feature of Gram-positive eubacteria in particular. For example, B. amyloliquefaciens (Hoang and Hofemeister 1995; Meijer et al. 1995), Staphylococcus aureus (Cregg et al. 1996) and Streptomyces lividans (S. Schacht and J. Anné, pers. comm.) contain at least two paralogous SPases of the P-type, whereas Deinococcus radiodurans seems to contain at least three of these enzymes (sequences are available via ftp://ftp.tigr.org/pub/data/d_radiodurans). Notably, we have identified one ER-type and six P-type SPases in B. subtilis. In the present studies, which were aimed at the functional analysis of the chromosomally encoded SPases of B. subtilis, we show that, in principle, the presence of one SPase (i.e., SipS or SipT) is sufficient for precursor processing, growth, and cell viability. Thus, the secretory precursor processing machinery of B. subtilis is functionally redundant, ensuring that this organism can avail of backup SPases. Under natural conditions, this may be of particular importance for the fitness of B. subtilis and other Gram-positive eubacteria, which can secrete large amounts of proteins into the medium as an adaptive response to changes in the environment.
Our results show that SipS and SipT play a major role in the processing of secretory preproteins and that, under the conditions tested, the three other chromosomally encoded SPases (SipU, SipV, and SipW) contribute only to a minor extent to this process. Interestingly, the transcription of sipS (Bolhuis et al. 1996) and sipT (this study) is temporally controlled via the DegS–DegU two-component regulatory system, in concert with the transcription of the genes for most secretory proteins. Thus, B. subtilis can increase its preprotein processing capacity under conditions of high-level protein secretion in the postexponential growth phase. Our observation that cells lacking SipS and SipT stop growing, and die in the transition phase between exponential and postexponential growth, indicates that the potential to increase the processing capacity is important for cell growth and viability. Nevertheless, the production of very small amounts of SipS–D146A, a temperature-sensitive variant of SipS, is sufficient for growth and cell viability. This suggests that the increased processing capacity provided by SipS and SipT relates to, as yet undefined, differences between the specificities of these two SPases and those of SipU, SipV, and/or SipW. This view is supported by the observation that SipU, SipV, and SipW cannot complement for the absence of SipS and SipT, even if the corresponding genes are placed on multi-copy plasmids (H. Tjalsma et al., unpubl.). In this respect, it is important to note that we have shown previously that SipS, SipT, SipU, and SipV have overlapping substrate specificities (Tjalsma et al. 1997). At present, we do not know the precise function of each of the SPases of B. subtilis, but it seems likely that SipS and SipT are required, in particular, for the processing of preproteins that are synthesized in the postexponential growth phase. In contrast, as the sipU, sipV, and sipW genes are transcribed constitutively, the corresponding SPases are most likely ivolved in the processing of preproteins which are synthesized during all growth phases.
The previously reported presence of SPase-encoding genes on endogenous plasmids (Meijer et al. 1995) seems to give certain strains of B. subtilis an additional possibility to increase their capacity for preprotein processing. The latter hypothesis is supported by our observation that the SPase SipP, specified by plasmid pTA1015, can complement for the absence of SipS and SipT (H. Tjalsma et al., unpubl.). Thus, it seems that SipS, SipT, and SipP have similar substrate specificities.
Dalbey and Wickner (1985) have shown that E. coli cells depleted of their type I SPase (i.e., leader peptidase) accumulate preproteins on the outer surface of the cytoplasmic membrane. Thus, processing by SPase was shown to be a prerequisite for the release of translocated proteins from the membrane. However, the question whether the accumulation of secretory preproteins in cells depleted of SPase would affect protein translocation across the membrane remained unresolved. Our observation that biotinylated pre-AmyQ–PSBT accumulated in B. subtilis cells lacking functional SipS and SipT shows that this is the case. Thus, the decreased viability of these cells may be due to impaired protein translocation across the membrane. However, the alternative explanation that certain proteins, which are essential for cell growth and viability, do not reach their correct cellular localization (e.g., the cell wall) cannot be excluded. Interestingly, no biotinylated mature AmyQ–PSBT was produced by cells lacking functional SipS and SipT, suggesting that biotinylated pre-AmyQ–PSBT is not translocated across the membrane of B. subtilis. As biotinylated proteins can be transported by the preprotein translocase of E. coli (Jander et al. 1996), it seems that the preprotein translocase of B. subtilis is more restrictive with respect to (partially) folded, biotinylated preproteins than that of E. coli.
An intriguing result of the present studies is that B. subtilis contains an ER-type SPase (SipW). Thus, B. subtilis is the first known eubacterium containing SPases of both the P- and the ER-type. This may not be a peculiarity of B. subtilis, as we identified the partial sequence of a SipW/SPC21-like protein from Clostridium perfringens by similarity searches in GenBank (accession no. X86488). Furthermore, the identification of SipW of B. subtilis has at least three interesting implications. First, the presence of ER-type SPases in all three domains of life suggests that the P-type SPases, which are only present in eubacteria and the organelles of eukaryotes, have evolved from the ER-type SPases. However, the alternative possibilities of convergent evolution of P- and ER-type SPases and/or horizontal transfer of ER-type SPases from archaea or eukaryotes to eubacteria cannot be excluded. Second, recent studies indicate that the P-type SPases make use of a serine–lysine catalytic dyad (see Dalbey et al. 1997). In contrast, the catalytic mechanism of the ER-type SPases is presently unclear, because the conserved lysine residue in domain D, which is invoked in the catalytic mechanism of the P-type SPases, is not conserved in the ER-type SPases; instead the ER-type SPases contain a conserved histidine residue at this position (Fig. 1B). This could mean that the ER-type SPases use the latter histidine residue for catalysis, for example in a serine-histidine-aspartic acid catalytic triad, like in the classical serine proteases. In addition to the conserved histidine residue in domain D, such a catalytic triad could involve the strictly conserved serine residue in domain B, which is essential for the activity of the P-type SPases, and the strictly conserved aspartic acid residue in domain E. Alternatively, the ER-type SPases could make use of a serine-lysine catalytic dyad involving the strictly conserved serine and lysine residues in domains B and E, respectively. Third, the ER-type SPase of yeast and higher eukaryotes are part of larger complexes, containing three additional subunits of which the SPC3 or SPC22/23 subunits, respectively, are essential for SPase activity (Newsome et al. 1992; Fang et al. 1997; Meyer and Hartmann 1997). None of these additional subunits is conserved in eubacteria or archaea (H. Tjalsma et al., unpubl.), suggesting that the ER-type SPases of the latter groups of organisms require no, or very different additional subunits for activity. The identification of the catalytic mechanism of the ER-type SPases, and the identification of their putative partner proteins in eubacteria and archaea, are major challenges for future research.
Finally, the present studies on the sip gene family of B. subtilis have important implications for projects on the functional analysis of the genomes of B. subtilis and other organisms, such as E. coli and yeast, because the presence of paralogous gene families is a common feature of these microbial genomes. As examplified by the lack of effect of the single disruption of the sipW gene, strategies based on single gene disruptions will probably be inadequate to assign functions to many multiplied genes with thus far unidentified functions.
Materials and methods
Plasmids, bacterial strains, and media
Table 1 lists the plasmids and bacterial strains used. TY medium (tryptone/yeast extract) contained Bacto-tryptone (1%), Bacto yeast extract (0.5%), and NaCl (1%). Minimal medium for B. subtilis contained: 100 mm potassium phosphate buffer at pH 7.0, glucose (1%), trisodium citrate (3 mm), MgSO4 (3 mm), casamino acids (0.1%), potassium glutamate (0.2%), tryptophan (20 μg/ml), histidine (20 μg/ml), methionine (20 μg/ml), tyrosine (20 μg/ml), adenine (20 μg/ml), uracil (20 μg/ml), nicotinic acid (0.4 μg/ml), riboflavin (0.4 μg/ml), and Fe–ammonium citrate (1.1 μg/ml). S7 media 1 and 3, used for labeling of B. subtilis proteins with [35S]methionine (Amersham), were prepared as described by van Dijl et al. (1990a, b). If required, medium for E. coli was supplemented with ampicillin (Ap, 50 μg/ml), erythromycin (Em, 100 μg/ml), or kanamycin (Km, 40 μg/ml); media for B. subtilis were supplemented with Cm (5 μg/ml), Em (2 μg/ml), Km (10 μg/ml), or spectinomycin (Sp, 100 μg/ml).
DNA and RNA techniques
Procedures for DNA purification, restriction, ligation, agarose gel electrophoresis, and transformation of E. coli were carried out as described by Sambrook et al. (1989). Enzymes were from Boehringer Mannheim. B. subtilis was transformed by growth in minimal medium until an OD600 of ±1, the subsequent addition of plasmid or chromosomal DNA to the culture, and continued incubation for at least 4 hr. Multiple sip mutants were constructed by transforming single mutants with chromosomal or linearized plasmid DNA containing a disrupted sip gene. To minimize the number of antibiotic resistance markers in the cells, triple sip-mutants were prepared from double sip-mutants containing no, or only one, antibiotic resistance marker. Correct integration of linearized DNA fragments, or plasmids in the chromosome of B. subtilis was verified by Southern hybridization. PCR was carried out with Vent DNA polymerase (New England Biolabs) as described by van Dijl et al. (1995). DNA and protein sequences were analyzed by use of version 6.7 of the PCGene Analysis Program (Intelligenetics Inc.). The BLAST algorithm (Altschul et al. 1997) was used for protein comparisons in GenBank. RNA was isolated with the RNeasy total RNA kit from Qiagen. RNA reference markers were from GIBCO BRL Life Technologies Inc. Northern hybridizations were performed as described by Bolhuis et al. (1996).
Plasmid pKTH10-BT was constructed in three steps. First, the amyQ gene was amplified by PCR with the primers amyQ5′ (5′-G GC CT AG GAT C CTA AGAGAG GGAGAGGA A ACA T GAT-3′) and amyQ3′ (5′-GGCCATGTCGACAGTACTGCATGCTTATCTAGATTTCTGAACATAAATGGAGAC-3′) using pKTH10 as a template. The amplified fragment was cleaved with BamHI and SalI, and cloned into the corresponding sites of pBR322, resulting in pBR322amyQ. Second, the sequences specifying the biotinylation domain (PSBT) of the P. shermanii transcarboxylase 1.3 S subunit were amplified by PCR with the primers PSBT1 (5′-GGCCATGAGCTCTCTAGAGTCGCCGGTAAGGCCGGAGAG-3′) and PSBT2 (5′-GAATTCGCATGCGCGTCAGCCGATCTTGATGAGACC-3′) using plasmid pCY66 (Cronan 1990) as a template. The amplified fragment was cleaved with XbaI and SphI, and cloned into the corresponding sites of pBR322amyQ, resulting in pBR322amyQ–BT. Third, the amyQ–PSBT gene fusion was amplified by PCR with the primers amyQ–BT5′ (5′-GGCCATAAGCTTTAATCAATCCGTGTTTGATGTTCC-3′) and amyQ–BT3′ (5′-GGCCATGGATCCGCGTCAGCCGATCTTGATGAGACC-3′), using pBR322amyQ–BT as a template. The amplified fragment was cleaved with HindIII and BamHI, and ligated into the corresponding sites of pKTH10, resulting in pKTH10–BT.
Plate assay for the processing of the (A13i)–β-lactamase precursor
The plate assay for the processing of the hybrid precursor pre(A13i)–β-lactamase was carried out as described by van Dijl et al. (1992).
Pulse-chase protein labeling, immunoprecipitation, SDS-PAGE, and fluorography
Pulse-chase labeling of B. subtilis, immunoprecipitation, SDS-PAGE, and fluorography were performed as described previously (van Dijl et al. 1991a, b). 14C-Methylated molecular weight markers were from Amersham International. Relative amounts of precursor and mature forms of secreted proteins were estimated by film scanning with an LKB ultroscan XL laser densitometer.
Western blot analysis
Western blotting was performed as described by Kyhse-Andersen (1984). After separation by SDS-PAGE, proteins were transferred to Immobilon–PVDF membranes (Millipore Corporation). To detect the α-amylase AmyQ, (A13i)-β-lactamase, or SipS, B. subtilis cells were separated from the growth medium by centrifugation (5 min, 12.000 rpm, room temperature), and samples for SDS-PAGE were prepared as described by van Dijl et al. (1991a). AmyQ, (A13i)–β-lactamase, or SipS were visualized with specific antibodies and HRP anti-rabbit IgG conjugates (Amersham International). A streptavidin-HRP conjugate (Amersham International) was used to detect biotinylated AmyQ–PSBT.
β-Galactosidase activity assay
Overnight cultures were diluted 100-fold in fresh medium and samples were taken at hourly intervals for OD600 readings and β-galactosidase activity determinations. The assay and the calculation of β-galactosidase units (expressed as units per OD600) were carried out as described by Miller et al. (1982).
Acknowledgments
We thank Dr. M. Sarvas for providing plasmid pKTH10 and specific sera against AmyQ, Dr. J. Cronan for providing plasmid pCY66, H. Mulder for preparing figures, and Dr. J. Jongbloed for useful discussions. H.T. was supported by Genencor International (Rijswijk, the Netherlands) and Gist brocades B.V. (Delft, the Netherlands), A.B., S.B., C.J.B., J.M.v.D., W.J.Q., W.S., and T.W. were supported by Biotechnology Grants (Bio2-CT93-0254 and Bio4-CT96-0097) from the European Union, M.L.v.R was supported by the Dutch Ministry of Economic Affairs through ABON (Associatie Biologische Onderzoeksscholen Nederland).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL J.M.VAN.DIJL@BIOL.RUG.NL; FAX 31503632348.
References
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrens M, Michaelis G, Pratje E. Mitochondrial inner membrane protease 1 of Saccharomyces cerevisiae shows sequence similarity to the Escherichia coli leader peptidase. Mol & Gen Genet. 1991;228:167–176. doi: 10.1007/BF00282462. [DOI] [PubMed] [Google Scholar]
- Black MT. Evidence that the catalytic activity of prokaryote leader peptidase depends upon the operation of a serine-lysine catalytic dyad. J Bacteriol. 1993;175:4957–4961. doi: 10.1128/jb.175.16.4957-4961.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blattner FR, Plunkett G, Bloch CA, Perna NT, Burland V, Riley M, Collado-Vides J, Glasner JD, Rode CK, Mayhew GF, et al. The complete genome sequence of Escherichia coliK-12. Science. 1997;277:1453–1462. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- Bolhuis A, Sorokin A, Azevedo V, Ehrlich SD, Braun PG, de Jong A, Venema G, Bron S, van Dijl JM. Bacillus subtilis can modulate its capacity and specificity for protein secretion by temporally expression of the sipSgene for signal peptidase I. Mol Microbiol. 1996;22:605–618. doi: 10.1046/j.1365-2958.1996.d01-4676.x. [DOI] [PubMed] [Google Scholar]
- Bron S, Venema G. Ultraviolet inactivation and excision repair in Bacillus subtilis.Construction and characterization of a transformable eightfold auxotrophic strain and two ultraviolet-sensitive derivatives. Mutat Res. 1972;15:1–10. doi: 10.1016/0027-5107(72)90086-3. [DOI] [PubMed] [Google Scholar]
- Böhni PC, Deshaies RJ, Schekman RW. Sec11 is required for signal peptide processing and yeast cell growth. J Cell Biol. 1988;106:1035–1042. doi: 10.1083/jcb.106.4.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bult CJ, White CO, Olsen GJ, Zhou L, Fleishmann RD, Sutton GG, Blake JA, Fitzgerald TM, Clayton RA, Gocayne JD, et al. Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science. 1996;273:1058–1073. doi: 10.1126/science.273.5278.1058. [DOI] [PubMed] [Google Scholar]
- Cregg KM, Wilding EI, Black MT. Molecular cloning and expression of the spsB gene encoding an essential type I signal peptidase from Staphylococcus aureus. J Bacteriol. 1996;178:5712–5718. doi: 10.1128/jb.178.19.5712-5718.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronan JE. Biotination of proteins in vivo. J Biol Chem. 1990;265:10327–10333. [PubMed] [Google Scholar]
- Dalbey RE, Wickner W. Leader peptidase catalyzes the release of exported proteins from the outer surface of the Escherichia coliplasma membrane. J Biol Chem. 1985;260:15925–15931. [PubMed] [Google Scholar]
- Dalbey RE, von Heijne G. Signal peptidases in prokaryotes and eukaryotes: A new protease family. Trends Biochem Sci. 1992;17:474–478. doi: 10.1016/0968-0004(92)90492-r. [DOI] [PubMed] [Google Scholar]
- Dalbey RE, Lively MO, Bron S, van Dijl JM. The chemistry and enzymology of the type I signal peptidases. Protein Sci. 1997;6:1129–1138. doi: 10.1002/pro.5560060601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang H, Mullins C, Green N. In addition to SEC11, a newly identified gene, SPC3, is essential for signal peptidase activity in the yeast endoplasmic reticulum. J Biol Chem. 1997;272:13152–13158. doi: 10.1074/jbc.272.20.13152. [DOI] [PubMed] [Google Scholar]
- Ferrari E, Jarnagin AS, Schmidt BF. In: Bacillus subtilis and other Gram-positive bacteria. Sonenshein AL, Hoch JA, Losick R, editors. Washington, D.C.: American Society for Microbiology; 1993. pp. 917–937. [Google Scholar]
- Fleischmann RD, Adams MD, White O, Clayton RA, Kirkness EF, Keslavage AR, Bult CJ, Tomb JF, Dougherty BA, Merrick JM, et al. Whole-genome random sequencing and assembly of Haemophilus influenzaeRd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- Goffeau A, Aert R, Agostini-Carbone ML, Ahmed A, Aigle M, Alberghina L, albermann K, Albers M, Aldea M, Alexandraki D, et al. The yeast genome directory. Nature. 1997;387:5–105. [Google Scholar]
- Greenburg G, Shelness GS, Blobel G. A subunit of mammalian signal pepsidase is homologous to yeast SEC11 protein. J Biol Chem. 1989;264:15762–15765. [PubMed] [Google Scholar]
- Hoang V, Hofemeister J. Bacillus amyloliquefacienspossesses a second type I signal peptidase with extensive sequence similarity to other Bacillus SPases. Biochim Biophys Acta. 1995;1269:64–68. doi: 10.1016/0167-4889(95)00101-w. [DOI] [PubMed] [Google Scholar]
- Jacobs MF. Expression of the subtilisin Carlsberg-encoding gene in Bacillus licheniformis and Bacillus subtilis. Gene. 1995;152:69–74. doi: 10.1016/0378-1119(94)00655-c. [DOI] [PubMed] [Google Scholar]
- Jander G, Cronan JE, Beckwith J. Biotinylation in vivo as a sensitive indicator of protein secretion and membrane protein insertion. J Bacteriol. 1996;178:3049–3058. doi: 10.1128/jb.178.11.3049-3058.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko T, Sato S, Kotani H, Tanaka A, Asamizu E, Nakamura Y, Miyajima N, Hirosawa M, Sugiura M, Sasamoto S, et al. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res. 1996;30:109–136. doi: 10.1093/dnares/3.3.109. [DOI] [PubMed] [Google Scholar]
- Kim L, Mogk A, Schumann W. A xylose-inducible Bacillus subtilisintegration vector and its application. Gene. 1996;181:71–76. doi: 10.1016/s0378-1119(96)00466-0. [DOI] [PubMed] [Google Scholar]
- Klenk HP, Clayton RA, Tomb JF, White O, Nelson KE, Ketchum KA, Dodson RJ, Gwinn M, Hickey EK, Peterson JD, et al. The complete genome sequence of the hyperthermophilic, sulphate-reducing archaeon Archaeoglobus fulgidus. Nature. 1997;390:364–370. doi: 10.1038/37052. [DOI] [PubMed] [Google Scholar]
- Kontinen VP, Sarvas M. Mutants of B. subtilisdefective in protein export. J Gen Microbiol. 1988;134:2333–2344. doi: 10.1099/00221287-134-8-2333. [DOI] [PubMed] [Google Scholar]
- Kunst F, Ogasawara N, Moszer I, Albertoni AM, Alloni G, Azevedo V, Bertero MG, Bessieres P, Bolotin A, Borchept S, et al. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature. 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- Kyhse-Andersen J. Electroblotting of multiple gels: A simple apparatus without buffer tank for rapid transfer of proteins from polyacrylamide to nitrocellulose. J Biochem Biophys Methods. 1984;10:203–209. doi: 10.1016/0165-022x(84)90040-x. [DOI] [PubMed] [Google Scholar]
- Meijer WJJ, de Jong A, Wisman GBA, Tjalsma H, Venema G, Bron S, van Dijl JM. The endogenous Bacillus subtilis (natto) plasmids pTA1015 and pTA1040 contain signal peptidase-encoding genes: Identification of a new structural module on cryptic plasmids. Mol Microbiol. 1995;17:621–631. doi: 10.1111/j.1365-2958.1995.mmi_17040621.x. [DOI] [PubMed] [Google Scholar]
- Meyer HA, Hartmann E. The yeast SPC22/23 homologue Spc3p is essential for signal peptidase activity. J Biol Chem. 1997;272:13159–13164. doi: 10.1074/jbc.272.20.13159. [DOI] [PubMed] [Google Scholar]
- Miller JH. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1982. [Google Scholar]
- Msadek T, Kunst F, Rapoport G. In: Bacillus subtilis and other Gram-positive bacteria. Sonenshein AL, Hoch JA, Losick R, editors. Washington, D.C.: American Society for Microbiology; 1993. pp. 729–745. [Google Scholar]
- Newsome AL, McLean JW, Lively MO. Molecular cloning of a cDNA encoding the glycoprotein of hen oviduct microsomal signal peptidase. Biochem J. 1992;282:447–452. doi: 10.1042/bj2820447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunnari J, Fox TD, Walter P. A mitochondrial protease with two catalytic subunits of nonoverlapping specificities. Science. 1993;262:1997–2004. doi: 10.1126/science.8266095. [DOI] [PubMed] [Google Scholar]
- Palva I. Molecular cloning of α-amylase gene from Bacillus amyloliquefaciens and its expression in Bacillus subtilis. Gene. 1982;19:81–87. doi: 10.1016/0378-1119(82)90191-3. [DOI] [PubMed] [Google Scholar]
- Paetzel M, Dalbey RE. Use of site-directed chemical modification to study an essential lysine in Escherichia coli leader peptidase. Trends Biochem Sci. 1997;22:28–31. doi: 10.1074/jbc.272.15.9994. [DOI] [PubMed] [Google Scholar]
- Philipp WJ, Poulet S, Eiglmeir K, Pascopella S, Balasubamanian V, Heym B, Bergh S, Bloom BR, Jacobs WR, Jr, Cole ST. An integrated map of the genome of the tubercle Bacillus, Mycobacterium tuberculosis H37Rv, and comparison with Mycobacterium leprae. Proc Natl Acad Sci. 1996;93:3132–3137. doi: 10.1073/pnas.93.7.3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: A laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Shelness GS, Blobel G. Two subunits of the canine signal peptidase complex are homologous to yeast Sec11 protein. J Biol Chem. 1990;265:9512–9519. [PubMed] [Google Scholar]
- Signs SA, Jacquet R. Induction of ethanol dependence increases signal peptidase mRNA levels in rat brain. Mol Cell Biochem. 1994;139:21–26. doi: 10.1007/BF00944199. [DOI] [PubMed] [Google Scholar]
- Sipos L, von Heijne G. Predicting the topology of eukaryotic membrane proteins. Eur J Biochem. 1993;213:1333–1340. doi: 10.1111/j.1432-1033.1993.tb17885.x. [DOI] [PubMed] [Google Scholar]
- Smith DR, Doucette-Stamm LA, Deloughery C, Lee H, Dubois J, Aldrege T, Bashirzadeh R, Blakely D, Cook R, Gilbert K, et al. Complete genome sequence of Methanobacterium thermoautotrophicumdeltaH: Functional analysis and comparative genomics. J Bacteriol. 1997;179:7135–7155. doi: 10.1128/jb.179.22.7135-7155.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tjalsma H, Noback MA, Bron S, Venema G, Yamane K, van Dijl JM. Bacillus subtiliscontains four closely related type I signal peptidases with overlapping substrate specificities: Constitutive and temporally controlled expression of different sip genes. J Biol Chem. 1997;272:25983–25992. doi: 10.1074/jbc.272.41.25983. [DOI] [PubMed] [Google Scholar]
- Tomb JF, White O, Kerlavage AR, Clayton RA, Sutton GG, Fleischmann RD, Ketchum KA, Klenk HP, Gill S, Dougherty BA, et al. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997;7:539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- Tschantz WR, Sung M, Delgado-Partin VM, Dalbey RE. A serine and a lysine residue implicated in the catalytic mechanism of the Escherichia colileader peptidase. J Biol Chem. 1993;268:27349–27354. [PubMed] [Google Scholar]
- van Dijl JM, de Jong A, Smith H, Bron S, Venema G. Synthesis and processing of Escherichia coli TEM-β-lactamase and Bacillus licheniformis α-amylase in Escherichia coli:The role of signal peptidase I. Mol & Gen Genet. 1988;214:55–61. doi: 10.1007/BF00340179. [DOI] [PubMed] [Google Scholar]
- van Dijl JM, de Jong A, Smith H, Bron S, Venema G. Signal peptidase I overproduction results in increased efficiencies of export and maturation of hybrid secretory proteins in Escherichia coli. Mol & Gen Genet. 1991a;227:40–48. doi: 10.1007/BF00260704. [DOI] [PubMed] [Google Scholar]
- ————— Non-functional expression of Escherichia coli signal peptidase I in Bacillus subtilis. J Gen Microbiol. 1991b;137:2073–2083. doi: 10.1099/00221287-137-9-2073. [DOI] [PubMed] [Google Scholar]
- van Dijl JM, de Jong A, Vehmaanperá J, Venema G, Bron S. Signal peptidase I of Bacillus subtilis:Patterns of conserved amino acids in prokaryotic and eukaryotic type I signal peptidases. EMBO J. 1992;11:2819–2828. doi: 10.1002/j.1460-2075.1992.tb05349.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dijl JM, de Jong A, Venema G, Bron S. Identification of the potential active site of the signal peptidase SipS of Bacillus subtilis. J Biol Chem. 1995;270:3611–3618. doi: 10.1074/jbc.270.8.3611. [DOI] [PubMed] [Google Scholar]
- van Sinderen D, Withoff S, Boels H, Venema G. Isolation and characterisation of comL, a transcriptional unit involved in competence development of Bacillus subtilis. Mol & Gen Genet. 1990;224:396–404. doi: 10.1007/BF00262434. [DOI] [PubMed] [Google Scholar]
- Vieria J, Messing J. New pUC-derived cloning vectors with different selectable markers and DNA replication origins. Gene. 1990;100:189–194. doi: 10.1016/0378-1119(91)90365-i. [DOI] [PubMed] [Google Scholar]
- Wertman KF, Wyman AR, Botstein D. Host/vector interactions which affect the viability of recombinant phage lambda clones. Gene. 1986;49:253–262. doi: 10.1016/0378-1119(86)90286-6. [DOI] [PubMed] [Google Scholar]