Abstract
The aim of research on infectious diseases is their prevention, and brucellosis and salmonellosis as such are classic examples of worldwide zoonoses for application of a systems biology approach for enhanced rational vaccine development. When used optimally, vaccines prevent disease manifestations, reduce transmission of disease, decrease the need for pharmaceutical intervention, and improve the health and welfare of animals, as well as indirectly protecting against zoonotic diseases of people. Advances in the last decade or so using comprehensive systems biology approaches linking genomics, proteomics, bioinformatics, and biotechnology with immunology, pathogenesis and vaccine formulation and delivery are expected to enable enhanced approaches to vaccine development. The goal of this paper is to evaluate the role of computational systems biology analysis of host:pathogen interactions (the interactome) as a tool for enhanced rational design of vaccines. Systems biology is bringing a new, more robust approach to veterinary vaccine design based upon a deeper understanding of the host-pathogen interactions and its impact on the host's molecular network of the immune system. A computational systems biology method was utilized to create interactome models of the host responses to Brucella melitensis (BMEL), Mycobacterium avium paratuberculosis (MAP), Salmonella enterica Typhimurium (STM), and a Salmonella mutant (isogenic ΔsipA, sopABDE2) and linked to the basis for rational development of vaccines for brucellosis and salmonellosis as reviewed by Adams and Ficht (Adams et al. 2009; Ficht et al. 2009). A bovine ligated ileal loop biological model was established to capture the host gene expression response at multiple time points post infection. New methods based on Dynamic Bayesian Network (DBN) machine learning were employed to conduct a comparative pathogenicity analysis of 219 signaling and metabolic pathways and 1620 Gene Ontology (GO) categories that defined the host's biosignatures to each infectious condition. Through this DBN computational approach, the method identified significantly perturbed pathways and GO category groups of genes that define the pathogenicity signatures of the infectious agent. Our preliminary results provide deeper understanding of the overall complexity of host innate immune response as well as the identification of host gene perturbations that defines a unique host temporal biosignature response to each pathogen. The application of advanced computational methods for developing interactome models based on DBNs has proven to be instrumental in elucidating novel host responses and improved functional biological insight into the host defensive mechanisms. Evaluating the unique differences in pathway and GO perturbations across pathogen conditions allowed the identification of plausible host-pathogen interaction mechanisms. Accordingly, a systems biology approach to study molecular pathway gene expression profiles of host cellular responses to microbial pathogens holds great promise as a methodology to identify, model and predict the overall dynamics of the host-pathogen interactome. Thus, we propose that such an approach has immediate application to the rational design of brucellosis and salmonellosis vaccines.
Introduction
Some of the veterinary vaccines licensed for controlling infectious disease of domestic animal species today are still based on empirical technology that was introduced by Edward Jenner, using live vaccines in 1796, and later Louis Pasteur, using killed whole organism vaccines. Indeed, Jenner derived the term “vaccine” from his use of the less innocuous zoonotic cowpox virus (Latin variolae vaccinae, adapted from vaccinus, from vacca cow) to provide protection against smallpox. Much of veterinary vaccinology is driven by the realities that exist in raising production animals or working in veterinary practice, where making a living depends on keeping the animals healthy. Livestock production is an industry where vaccines are like insurance policies – protection from events that one hopes never happen (Adams et al. 2009). For example, the USDA recognizes these varying levels of protection in the way that they allow label claims: 1) “aids in disease control”, 2) “for the prevention of disease”, and 3) “for the prevention of infection”. Additionally there may be indirect protection, or herd immunity, that results from vaccination of sufficient numbers of animals in a given population resulting in the reduction of the ability of a disease to transmit through the vaccinated individuals. The perception that vaccines provide sterilizing immunity, where the disease agent does not establish an infection, while widely held, is generally unfounded and largely unrealistic. In the last 15 years, genomics, proteomics, bioinformatics, biotechnology, immunology, pathogenesis and vaccine formulation and delivery have dramatically enabled novel approaches to vaccine development. When used optimally, vaccines prevent disease manifestations, reduce transmission of disease, decrease the need for pharmaceutical intervention, and improve the health and welfare of animals, as well as indirectly protecting against zoonotic diseases of people. The challenge in developing an optimal vaccination program is in dealing with the great diversity that exists within the animal world, and as such there probably is no single optimal program for all situations. While there is no single strategy to optimizing vaccination programs for animals, nonetheless, a solid understanding of the animal's innate and environmental risk factors as well as the variables such as stress, will enable the development of tailored vaccination schedules that best meet the needs of the animal. The use of vaccines in animal health is not restricted to the protection of morbidity and mortality of the animal hosts themselves, but they are regularly employed as key elements in public health programs. When appropriate biopreparedness, management modeling strategies and contingency plans of the future are linked with 1) protective DIVA vaccines against zoonoses, 2) effective predictive modeling, and 3) deployable implementation policies, control and prevention of serious zoonotic diseases of man and animals will become more achievable at local, state and national levels.
Systems biology is bringing a new, more robust approach to vaccine design that is based upon understanding the molecular network of the immune system of two interacting systems. With this approach, it is within the realm of possibility to develop more effective vaccines supported by a fuller understanding of the complexities of the host-pathogen interactions (interactome) as a product of the innovations of the past 15 years. On the other hand, the massive crush of data now being generated to enhance our understanding of the host-pathogen interactions may not have as much utility as expected unless more dynamic biologically sound models are developed and validated to comprehend and apply to vaccine design. The complexity of host-pathogen interactions across multiple species of hosts and pathogens requires a system level understanding of the entire hierarchy of biological interactions and dynamics. A systems biology approach can provide systematic insights into the dynamic/temporal difference in gene regulation, interaction, and function, and thereby deliver an improved understanding and more comprehensive hypotheses of the underlying mechanisms (Musser and DeLeo 2005; Franke, Müller et al. 2008). The ability to consolidate complex data and knowledge into plausible interactome models is essential to promote the effective discovery of key points of interaction. Accordingly, a systems biology approach to study molecular pathway gene expression profiles of host cellular responses to microbial pathogens holds great promise as a methodology to identify, model and predict the overall dynamics of the host-pathogen interactome. It is believed that such an approach will be essential for the rational design of both animal and human vaccines when incorporated into a method of incremental refinement of these models so that new knowledge can be accrued and utilized for future vaccine developments. What is even more challenging in animal vaccine development is the spectrum of animal hosts in which vaccines must perform effectively. Interactome models can be employed to assess the viability of vaccines in other species and help reduce unnecessary animal experiments. Accordingly, the systems biology approach to study molecular pathway gene expression profiles of host cellular responses to microbial pathogens holds great promise as a methodology to identify, model and predict the overall dynamics of the host-pathogen interactome to facilitate the rational design of brucellosis and salmonellosis vaccines.
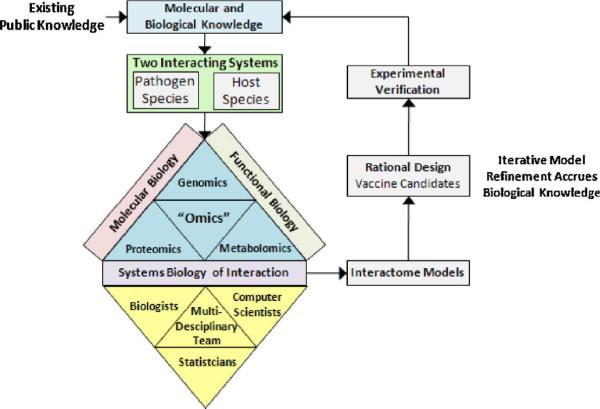
As researchers hypothesize and deduce the sequences and structures of pathogenic proteins and develop detailed knowledge of their regulatory roles in the host, they can rationally design vaccines with defined components in order to maximize effectiveness and minimize safety concerns. Computational capabilities are emerging for creating host-pathogen interactome models. Such models, utilizing data at the genomic, transcriptomic, proteomic, metabolomics, etc. levels, can be used to learn and understand the underlying mechanisms and points of interaction governing the host innate and adaptive responses to pathogens and their vaccines. Such models are envisioned to play an increasingly integral part in the vaccine and immunotherapeutic development process, with incremental model improvements accruing as new biological knowledge is collected from translational in vivo and ex vivo efficacy and safety studies (non-clinical through clinical trials). An exciting prospect of such incremental modeling is the role these models can play in a forward -looking vaccine rational design strategy. Figure 1 illustrates the strategy of employing a vaccine-immunotherapeutic development methodology referred to as incremental systems biology interactome modeling. Multiple elements must come together to implement such a methodology. Prior biological knowledge (molecular and functional biology) must be current for both the host and pathogen biological systems. Often such knowledge is minimal for many of the veterinary animal species and extra steps of obtaining latest genome and proteome annotations and interaction predictions are necessary and labor intensive. The role of the computer scientist, statistician, and biologist is integral to the successful development, refinement and verification of such models. The interactome model cannot just be a list of possible interaction prediction, but must be part of a dynamic model in which the relationships governing the host immune response can be captured, interpreted and refined. The interactome model becomes a tool that can be interrogated and employed in simulation to help guide vaccine development and/or immunotherapeutic drug candidate selections. Experimental verification will always be a necessary element, and as such experiments are conducted, the resulting biological information should be retained and employed as new biological knowledge for creating the next refined interactome model.
Figure 1.
Methods
Gene Expression Data Acquisition
An established in vivo perinatal calf ligated ileal loop model, in conjunction with custom bovine microarrays, was used to study the early temporal changes in the host response to a previously optimized dosage of 1 × 109 colony forming units of STM, STM mutant, BMEL, or MAP at four common sampling time-points post infection (0.5, 1, 2, and 4 hours post-infection) conducted under protocols approved by the Texas A&M University Institutional Animal Use and Care Committee. Gene expression and phenotyping data were collected at the College of Veterinary Medicine, Texas A&M University, following a surgical and sample collection methodology described elsewhere (Santos, Zhang et al. 2002; Zhang, Santos et al. 2002; Khare, Nunes et al. 2009; Santos, Raffatellu et al. 2009). For each pathogen condition, under approved BSL2/BSL3 conditions, there were four biological replicate non-survival surgeries for each pathogen performed on 3-week old male Salmonella, Brucella and Mycobacterium-free Holstein calves. Host RNA (from each host-pathogen surgery) was collected and co-hybridized in quadruplicate against bovine reference RNA to 13K custom bovine arrays (fabricated by W. M. Keck Center, University of Illinois at Urbana-Champaign) to allow for cross-comparison between experimental conditions. These custom microarrays consist of 70-mer oligonucleotides representing 13,258 unique oligos with 12,220 cattle open reading frames (ORFs). A detailed description of the design and development of the microarray has been published elsewhere (Loor, Everts et al. 2007). Time matched RNA from non-infected ligated ileal loops were used as healthy state controls. Proteomics analysis of the STM-Host samples was provided by Pacific Northwest National Labs (PNNL) utilizing an approach referred to as the accurate mass and time (AMT) tag approach. The microarray annotation was updated to the latest bovine Unigene Build #95.
The microarrays were scanned using a GenePix scanner. The spots representing genes on the arrays were adjusted for background and normalized to internal controls using GenePixPro image analysis software. Spots with fluorescent signal values below background were disregarded in all analyses. Samples were normalized against the bovine reference RNA signals across array and within each array (across duplicate spots). Pairwise comparisons and Student's t test with Benjamini-Hochberg correction were performed using GeneSifter and Spotfire DecisionSite. Note, the statistical analysis results are not provided herein, because this paper is focused on interactome analysis and modeling. However, these normalized data were employed in the interactome analysis and modeling as described next.
Interactome Systems Biology Analysis and Modeling
Interactome analysis and modeling were completed using the Seralogix (Seralogix, LLC, Austin, TX http://www.seralogix.com/) suite of software with an integrated platform enabling a systems biology computational pipeline for multi-conditional analysis and modeling, termed the BioSignature Discovery System (BioSignatureDS™). Its core tools are based on Dynamic Bayesian Networks (DBN) (Murphy 2002), an advanced form of machine learning and pattern recognition. The platform enables comprehensive cross-comparisons of the genomic/proteomic data to identify key pathway/GO perturbations and underlying mechanistic regulatory points. BioSignatureDS™ is used to identify groups and individual genes that capture the perturbation in a pathway or biological process over time. This technique is named Dynamic Bayesian Gene Group Activation (DBGGA™). DBGGA utilizes the normalized microarray data, as described above, as its data input. A subset of the complete array of genes that map to the analyzed pathways and gene ontology groups are used in the analysis.
DBGGA creates a Dynamic Bayesian network (DBN) model for each pathway based on KEGG (KEGG; Kanehisa and Goto 2000) while a naïve Bayesian classifier (Friedman, Geiger et al. 1997) type network model is created for each GO category. Each network model is trained using the gene expression data from the control condition. The other experimental condition expression data are then used as evidence to test the goodness-of-fit of this data against the trained DBN control model. Goodness-of-fit is determined by Bayesian likelihood ratio tests that are subsequently transformed to a z-score test statistic (Bayesian z-score) to permit comparison of scores across all pathways and GO categories (i.e., biological processes). The DBGGA computational method scores and rank groups of interrelated genes within a given pathway or gene ontology category across all time points in lieu of just one gene in a single time point (such as used in traditional t-tests), and thus determines the differences and commonalities between experimental and control conditions. DBGGA can also determine which genes are the significant sources of the perturbation. Such genes are designated as “candidate mechanistic genes”, a term we coined to describe those genes within a pathway that individually contributed significantly to the overall pathway Bayesian z-scores divergence, and thus are considered mechanistic candidates that may play key roles in governing the host's immune response. Only those genes that are associated with a given pathway or GO category were examined using the DBGGA modeling approach.
BioSignatureDS™ utilizes the significantly perturbed pathways, GO categories and the “candidate mechanistic genes” as building blocks to construct a system level interactome network model of the disease/condition. Encapsulating global time-course patterns and multi-conditional behaviors of a large group of genes/proteins, the systems level model has great discriminating power even when the effects of individual genes are small. Thus, the disease models are useful for more efficient comparative modeling, pattern recognition (diagnostics) and simulations (prognostics). Further, proteomic data are effectively integrated into the models as an overlay to individual pathway and system-level models to both confirm the presence of proteins for their encoding genes as well as to visualize the temporal patterns of protein abundance.
Results
To evaluate the potential for computational systems biology analysis of host:pathogen interactions (the interactome) to be used as a tool for enhanced rational design of vaccines, each host/pathogen interaction condition was modeled and scored 219 known metabolic and signaling pathways and 1620 biological processes (gene groups associated with Gene Ontology (GO) terms) at four time points. DBGGA was employed to identify the perturbations between pathogen conditions for pathways, GO categories, and genes. The DBGGA method generates Bayesian log likelihood scores that are normalized and transformed to a z-score equivalent (the Bayesian z-score) so that all pathways and GO categories across all host/pathogen conditions can be equivalently compared and assessed for significance.
DBGGA Gene Ontology Analysis
A DBGGA analysis was conducted for gene GO categories. For each pathogen condition, 1620 biological process GO categories were scored. Each condition produced its own unique set of highly scored GO functions, but for comparison purposes, we chose a small subset of highly perturbed categories to illustrate the different temporal responses as shown in Figure 2(a). Figure 2 (b) illustrates the comparative analysis of gene scores for just the phagocytosis GO term category. The gene ontology group scores show a very diverse pattern over time. As can be seen, the induction or suppression of groups of genes allows us to identify specific biological process groups that define the pathogenicity biosignatures of each pathogen. Individual gene patterns within the groups can be further compared as show in Figure 2(b). For example the gene encoding ADORA1 (adenosine A1 receptor) is up modulated in BMEL for at all four time points, but is not significantly expressed in MAP or STM. Comparative pathogenicity can provide important insights into the mechanistic differences and guiding the research biologists to identify unique and common mechanisms that may be new targets for immunotherapeutic drugs or indicators of immunogenicity of novel vaccine candidates. Such comparative modeling could also be utilized to compare the effectiveness of vaccine candidates across multiple species.
Figure 2.
DBGGA Pathway Analysis
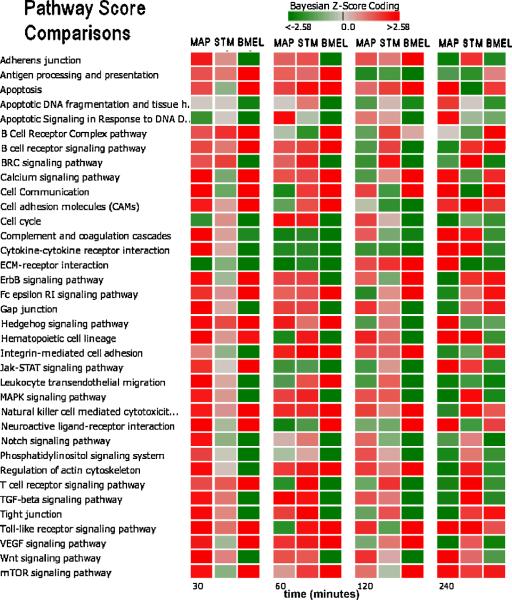
Of the 219 signaling/metabolic pathways scored, we focused on a subset of immune response related pathways as listed in Figure 3. This figure shows a heat map comparison of pathway Bayesian z-scores between pathogen conditions over time post infection. There were considerable differences between the host response profiles. MAP had strong early (30 minute) induction of the majority of its pathways and appeared to reverse to a more suppressive state by 240 minutes. STM's early response indicated mild perturbations at 30 minutes that increased over time until several pathways were strongly induced by 240 minutes. BMEL was more strongly suppressive for the majority of pathways over time. At early times (30, 60 minutes) there were a few commonly induced pathways: Antigen Processing and Presentation, B Cell Receptor Signaling, Fc epsilon RI Signaling, Hedgehog Signaling, and Natural Killer Cell Mediated Cytotoxicity. In contrast, only ECM-receptor Interaction, Apoptotic Signaling and Apoptotic DNA Fragmentation had similar suppressions at 30 and 60 minutes. Interestingly, there was no single pathway at later times (120, 240 minutes) with similar perturbed states, implying that the host defenses have divergent biosignatures against the various virulent mechanisms presented by the pathogens.
Figure 3.
Significant divergent responses between conditions were observed for MAPK Signaling and Regulation of Actin Cytoskeleton, for example. For MAP, the MAPK pathway reversed from induced to suppressed, while STM increased induction and BMEL maintained a suppressed state. The MAPK Signaling Pathway has been implicated in bacterial pathogenesis for a number of pathogens such as Salmonella enterica serovar Typhimurium (Hobbie, Chen et al. 1997), Yersinia spp. (Ruckdeschel, Harb et al. 1998), Listeria monocytogenes (Tang, Sutherland et al. 1998), and Mycobacterium spp. (Schorey and Cooper 2003). For the MAP condition, the Regulation of Actin Cytoskeleton pathway was induced within the first 30 minutes and became suppressed after 240 minutes, while for STM, this pathway became more strongly induced over the course of 240 minutes. The BMEL condition had a biphasic response from suppressed to induced and back to being suppressed over the 240 minute time course. The manipulation of the Actin Cytoskeleton pathway by STM to invade host cells has been well established (Guiney and Lesnick 2005), but not as well documented for MAP or BMEL.
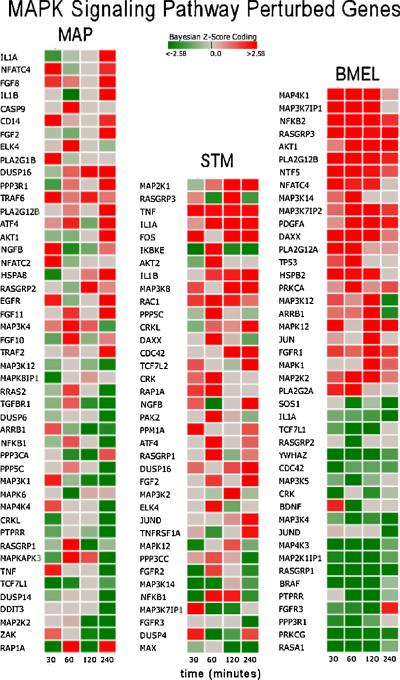
The MAPK pathway was selected as a potential candidate gene for more detailed discussion with regard to gene perturbations, mechanistic interpretations, and gene knockout simulation. Figure 4 is a heat map of significantly perturbed genes for the MAPK pathway by pathogen condition. In this figure, the genes are sorted in order of highest up modulation to lowest down modulation, and for a gene to be included in this figure, a Bayesian z-score>|2.24| at any one time point was required. The Bayesian z-score > |2.24| reflects 99% confidence in the data. It is easy to observe that the perturbed genes and their expression patterns are quite different between conditions. Surprisingly, of the 171 measured genes in this pathway, only two genes in Figure 4 were found to be commonly perturbed across all three pathogen conditions: 1) IL1A, which encodes interleukin 1 protein involved in various immune responses, inflammatory processes, and hematopoiesis; and 2) RASGRP1, which encodes a protein characterized by the presence of a Ras superfamily guanine nucleotide exchange factor (GEF) domain that activates the Erk/MAP kinase cascade and regulates T-cell and B-cell development, homeostasis and differentiation. The perturbation of IL1A and RASGRP1 is consistent with genes involved in immune response, but the expression patterns for these two genes vary significantly between pathogens.
Figure 4.
Simply comparing and contrasting the expression patterns of perturbed genes was inadequate for deciphering the MAPK pathway response dynamics. Clearly, the uniqueness of the MAPK pathway responses suggested that very different invasion/evasion mechanisms have evolved for each pathogen. More sophisticated methods are needed to identify potential points of host response disruptions. This is done by interrogating the trained DBN model for the MAPK Pathway for genes that exceed threshold Bayesian z-scores>|2.24| (“mechanistic genes”) and gene-gene network relationships (arcs). For example, Figure 5 shows the visualization of the MAPK pathway network. The network can be employed to visualize several key features that would otherwise be difficult to discern by looking at spreadsheet lists of genes. For example the state of gene modulation is distinguished by color-coded nodes. The state of upstream and downstream genes can be easily identified. Various threshold levels can be modified to identify significantly perturbed genes (annotated with orange circles, Figure 5). The strength of correlation between gene pairs is indicated by the color and thickness of the arcs connecting the genes.
Figure 5.
In Table 1, we show a list of 20 specific gene-to-gene relations associated with the MAPK pathway (Figure 3) having strong positive and/or negative arc weight correlations. We normalized the DBN arc weights to allow equivalent comparison to other pathway gene-gene relations. In this arc weight table, a few significant relationships are numbered in the table and on the network (Figure 3) with an encircled number and arrow pointing to the corresponding arc. It is hypothesized that virulence factors from each pathogen can have different disruptive influences on the host's MAPK Signaling Pathway and that such disruption can be used to identify pathogenic mechanisms unique to each pathogen, thus providing a rationale for development of deletion mutants of the corresponding pathogen virulence factors as potential vaccine candidates. For example, the relation arc TRAF2->FLNA had a strong positive weight (correlated) for STM-Host (0.241) and for the BMEL-Host (0.204) while MAP-Host had a large negative weight (anti-correlated) of −0.17.
Table 1.
Arc weight correlation table showing highly correlated (positive weights) or anti-correlated gene-gene relationships (negative weights) learned from the model training.
| Arc label | Gene start | Normalized arc weight | Gene end | Relation type |
|---|---|---|---|---|
| 1 | SOS1 | 0.372 | GRB2 | Binding |
| 2 | FGF1 | 0.306 | FGFR2 | Activation |
| 3 | IL1B | 0.271 | IL1R2 | Activation |
| 4 | TRAF2 | 0.241 | FLNA | Binding |
| FGF1 | 0.234 | FGFR4 | Activation | |
| MAPK4 | 0.204 | PLA2G12B | Activation | |
| MAPK8IP1 | 0.202 | MAP4K1 | Binding | |
| DUSP4 | 0.173 | MAPK3 | Inhibition | |
| MAPK4 | 0.185 | MAP2K1IP1 | Binding | |
| DUSP4 | 0.178 | MAPK6 | Inhibition | |
| MAPK6 | 0.176 | RPS6KA3 | Phosphorylation | |
| MAPK12 | 0.173 | TP53 | Phosphorylation | |
| MAP3K7IP1 | 0.092 | MAPK12 | Phosphorylation | |
| CASP9 | −0.008 | PAK1 | Phosphorylation | |
| CASP7 | −0.123 | MAP4K1 | Phosphorylation | |
| RASGRP4 | −0.175 | RRAS2 | Activation | |
| MAPK12 | −0.184 | RPS6KA5 | Phosphorylation | |
| MAPK1 | −0.191 | YWHAZ | Activation | |
| 5 | DUSP4 | −0.199 | MAPK4 | Inhibition |
| 6 | PRKACB | −0.209 | RAP1B | Activation |
The arc label column with numbers 1–6 correspond to the arcs labeled in Fig. 4. Arcs define a relationship between a starting gene (parent) and an ending gene (child).
The reversal of gene-to-gene arc weight of MAP-Host may indicate a disruption of either TRAF or FLNA gene by virulent factors of the pathogens, identifying potential novel points of interaction. TRAF2 encodes a protein that is a member of the TNF receptor associated factor (TRAF) protein family. This protein is required for TNF-alpha-mediated activation of MAPK8/JNK and NF-κB. It has a binding relationship with the filamin-A protein encoded by the FLNA gene. FLNA participates in the anchoring of membrane proteins to the actin cytoskeleton. This type of interaction analysis can be done for every pathway and used to identify novel differences between pathogen conditions. The visualization of mechanistic genes and arc weight enables an efficient identification of the differences between pathogenic influences.
Interrogating the Influence of Genes by Interactome Knockout Simulations
More detailed interrogation of the BMEL-Host model found that the gene, MAPK1, was significantly upregulated in the BMEL condition while not in MAP or STM. Further, it was observed that MAPK1 had a number of interactions with other genes within the MAPK Signaling pathway model that showed either very strong anti-correlated relationship such as the MAPK1->YWHAZ or strongly correlated relationship such as MAPK1->MAPT, while in MAP and STM host interactome models, the influence of MAPK1 was negligible. Specifically for the BMEL condition, we found that MAPK1 has a series of direct relationships with highly significant correlations as listed in Table 2. This could imply that MAPK1 has a unique role in BMEL pathogenesis during early host cell invasion. Mitogen-activated protein kinase 1 (MAPK1 or ERK 1/2) controls many biological functions (Johnson and Lapadat 2002). The MAPK signaling cascade, represented by 3 well characterized subfamilies of MAPKs (ERK1/2, JNK and p38), has been implicated in bacterial internalization (Tang, Sutherland et al. 1998) and intracellular survival and replication (Hobbie, Chen et al. 1997; Palmer, Hobbie et al. 1998; Schorey and Cooper 2003). Jimenez-Bagues et al. (Bagués, Gross et al. 2005) demonstrated the importance that the integrity of the MEK - MAPK - ERK 1/2 pathway has on the elimination of rough Brucella suis in macrophages. To identify the importance of this MAPK signaling pathway in BMEL invasion and intracellular survival in HeLa cells, a siRNA molecule (ID1449) was used to knock-down MAPK1 expression. Our results confirmed that the internalization of BMEL decreased more than 60% when the gene was knocked-down with the siRNA molecule as shown in Figure 6.
Table 2.
MAPK1 gene interactions pairs that have significant correlated or anit-correlated relationships determined by interrogating the MAPK Signaling pathway model.
| Gene start | Normalized correlation weight | Gene end | Gene end description | Relation |
|---|---|---|---|---|
| MAPK1 | 0.132 | PLA2G12A | Phospholipase A2, group XIIA | Activation |
| MAPK1 | −0.13 | PLA2G1B | Phospholipase A2, group IB (pancreas) | Activation |
| MAPK1 | 0.137 | PLA2G12B | Phospholipase A2, group XIIB | Activation |
| MAPK1 | −0.167 | YWHAZ | Tyrosine3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide | Activation |
| MAPK1 | 0.134 | PLA2G4A | Phospholipase A2, group IVA (cytosolic, calcium-dependent) | Activation |
| MAPK1 | −0.176 | MKNK1 | MAP kinase interacting serine/threonine kinase 1 | Phosphorylation |
| MAPK1 | 0.137 | MAPT | Microtubule-associated protein tau | Activation |
| MAPK1 | −0.122 | MAP2K1IP1 | Mitogen-activated protein kinase kinase 1 interacting protein 1 | Binding |
| MAPK1 | −0.114 | MKNK2 | MAP kinase interacting serine/threonine kinase 2 | Phosphorylation |
Figure 6.
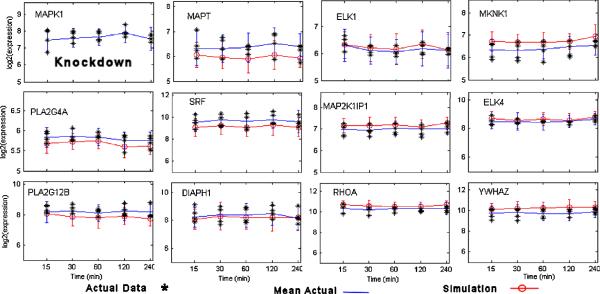
To gain better insight regarding the influence of MAPK1 on other genes, we employed the interactome model to simulate the MAPK1 knockdown in both the MAPK Signaling and Regulation of Actin Cytoskeleton models. The simulation identified a set of genes that were heavily influenced by MAPK1 as shown in the gene expression plots of Figure 7 in which the simulation data is plotted in comparison to the actual data used to train the interactome model. Interestingly, the simulation identified genes that were both in correlation with the reduced MAPK1 knockdown expression as well as several that had an increase in expression (anti-correlated). Either set of correlated or anti-correlated genes could be considered important contributors to the observed internalized reduction of BMEL in the HeLa host cells. For example, a correlated gene, SRF (serum response factor), is known to be involved in actin filament organization, regulation of cell adhesion, negative regulation of cell migration, negative regulation of cell proliferation, and regulation of transcription. Another correlated gene, MAPT (Microtubule-associated protein tau) is associated with regulation of microtubule depolymerization. The anti-correlated gene, YWHAZ (14-3-3 protein zeta/delta), is known to be involved in the biological processes of anti-apoptosis, histamine secretion by mast cell and signal transduction. The anti-correlated gene RHOA (Transforming protein RhoA) is associated with actin cytoskeleton organization, regulation of I-kappaB kinase/NF-kappaB cascade, and cell adhesion. This type of analysis is an integral part of the “incremental systems biology interactome modeling” process and introduced here as preliminary illustration as to how simulation/inferencing of the interactome model can be employed to guide next phases of in vitro and in vivo experimentation.
Figure 7.
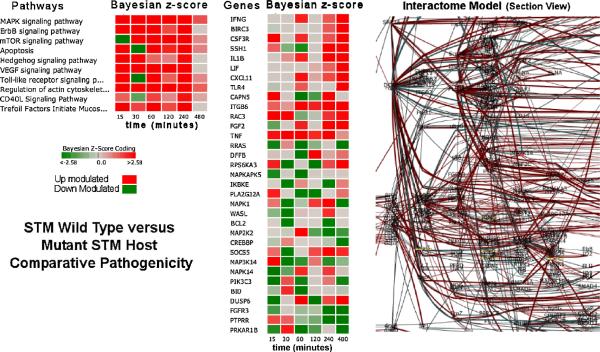
Biological System Model Generation for Comparative Pathogenicity Analysis
Comparative pathogenicity is a method by which the host response between different pathogens or pathogen vaccine candidates can be utilized to elicit unique and/or common biomarkers of immunogenicity. Utilizing BioSignatureDS™, the significantly perturbed pathways and gene groups from DBGGA were integrated to construct a plausible system level model of the STM wild type (WT) condition versus an isogenic ΔsipA, sopABDE2 mutant. The system model encompasses whole time-course patterns and multi-conditional behaviors of larger groups of genes and proteins than utilized only in the pathways. The system model expands the relationship of genes across related pathways and can be used for more efficient comparative modeling, pattern recognition and simulation supporting “what-if” type of analyses as previously described at the pathway level. The system model is constructed from a method based on merging of pathways with known gene/protein relationships and produces a trained and optimized network model similar to the MAPK signaling pathway network shown in Figure 5. Following this procedure, a system model was constructed from 10 selected pathways (listed in Figure 8) showing significant perturbation between the host infected with STM WT and STM mutant. The resulting model has a common network structure that is trained using the host response data for the WT, mutant and control conditions and was comprised of 930 genes and over 1500 gene-to-gene relations. By interrogating the model, we identified a number of significantly differentially expressed genes (|Bayesian z-score| >= 2.24) as shown in the center heatmap of Figure 8. From this heatmap, the difference in the STM mutant shown as green in the heatmap are a subset of genes which were found to be very highly up regulated in the STM mutant compared to the wild type and could be considered candidate genes governing the effective immune response of the host. These genes also form the basis of a biosignature that can be correlated to immunogenicity for more rational vaccine development. As expected for the WT, we found increased expression of genes associated with immune response such as those encoding IFNG, TNF, TLR4, and as well as genes associated with signaling and regulation of the actin cytoskeleton.
Figure 8.
Discussion and Conclusions
Advances in the last decade or so using comprehensive systems biology approaches linking genomics, proteomics, bioinformatics, and biotechnology with immunology, pathogenesis and vaccine formulation and delivery have dramatically enabled modern approaches to vaccine development. Systems biology is bringing a new, more robust approach to veterinary vaccine design based upon a deeper understanding of the host-pathogen interactions and their impact on the host's molecular network of the immune system. A computational systems biology method was utilized to create interactome models of the host responses to Brucella melitensis (BMEL), Mycobacterium avium paratuberculosis (MAP), Salmonella enterica Typhimurium (STM), and a Salmonella mutant (isogenic ΔsipA, sopABDE2). A bovine ligated ileal loop biological model was established to capture the host gene expression response at multiple time points post infection. New methods based on Dynamic Bayesian Network (DBN) machine learning were employed to conduct a comparative pathogenicity analysis of 219 signaling and metabolic pathways and 1620 Gene Ontology (GO) categories that defined the host's biosignatures to each infectious condition. Through this DBN computational approach, the method identified significantly perturbed pathways and GO category groups of genes that define the pathogenicity signatures of the infectious agent. Our preliminary results provide deeper understanding of the overall complexity of host innate immune response as well as the identification of host gene perturbations that defines unique host temporal biosignature responses to each pathogen. The application of advanced computational methods for developing interactome models based on DBNs has proven to be instrumental in elucidating novel host responses and improved functional biological insight into the host defensive mechanisms. Evaluating the unique differences in pathway and GO perturbations across pathogen conditions facilitated the identification of plausible host-pathogen interaction mechanisms. Accordingly, a systems biology approach to study molecular pathway gene expression profiles of host cellular responses to microbial pathogens holds great promise as a methodology to identify, model and predict the overall dynamics of the host-pathogen interactome. Thus, we propose that such an approach will have direct application to the rational design of brucellosis, salmonellosis and other vaccines for zoonotic diseases where application of veterinary vaccines directly impact disease transmission in animal populations and indirectly transmission of zoonoses to human populations.
Vaccination of reservoir species, including domestic animals, when efficacious vaccines are available, offer significant advantages to combating zoonoses. When appropriate biopreparedness, management strategies and contingency plans are linked with 1) protective rationally designed vaccines against zoonoses, 2) effective predictive disease modeling and 3) deployable field implementation policies, control and prevention of serious zoonotic diseases of man and animals become more achievable.
Acknowledgement
The animal studies were supported by U.S. Department of Homeland Security – National Center of Excellence for Foreign Animal and Zoonotic Disease (FAZD) Defense grant ONR-N00014-04-1-0. The project described was supported by Grant Number U54 AI057156, AI040124, AI044170, AI079173 and AI076246 from NIAID/NIH. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the RCE Programs Office, NIAID, or NIH. SDL was supported by AI060933. The computational analysis was supported in part by the National Institutes of Allergies and Infectious Diseases SBIR grants 2R44AI058362-02 and R43AI084223-01.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Pacific Northwest National Laboratory (Mass Spec) http://www.pnl.gov/biology/programs/msd/measurements.stm.
- Adams LG, Babiuk Lorne, McGavin David, Nordgren Robert. Vaccines for Biodefense and Emerging Diseases. Academic Press; Amsterdam: 2009. Chapter 16. Special Issues Around Veterinary Vaccines; pp. 226–254. A. D. T. Barrett, Stanberry, Lawrence R. [Google Scholar]
- Bagués M. P. J. d., Gross A, et al. Regulation of the mitogen-activated protein kinases by Brucella spp. expressing a smooth and rough phenotype: Relationship to pathogen invasiveness. Infection and Immunity. 2005;73:3178–3183. doi: 10.1128/IAI.73.5.3178-3183.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ficht TA, Adams L. Garry. Vaccines for Biodefense and Emerging and Neglected Diseases. Academic Press; Amsterdam: 2009. Chapter 42. Brucellosis; pp. 808–829. A. D. T. Barrett, Stanberry, Lawrence R. [Google Scholar]
- Franke R, Müller M, et al. Host-pathogen systems biology: Logical modelling of hepatocyte growth factor and Helicobacter pylori induced c-Met signal transduction. BMC Systems Biology. 2008;2(4) doi: 10.1186/1752-0509-2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman N, Geiger D, et al. Bayesian Network Classifiers. Machine Learning. 1997;29:131–161. [Google Scholar]
- Guiney DG, Lesnick M. Targeting of the actin cytoskeleton during infection by Salmonella strains. Clinical Immunology. 2005;114:248–255. doi: 10.1016/j.clim.2004.07.014. [DOI] [PubMed] [Google Scholar]
- Hobbie S, Chen LM, et al. Involvement of mitogen-activated protein kinase pathways in the nuclear responses and cytokine production induced by Salmonella typhimurium in cultured intestinal epithelial cells. J. Immunol. 1997;159:5550–5559. [PubMed] [Google Scholar]
- Johnson JL, Lapadat R. Mitogen-activated protein kinase pathways mediated by ERK, JNK, and p38 protein kinases. Science. 2002;298:1911–1912. doi: 10.1126/science.1072682. [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Goto S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Research. 2000;28(1):27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KEGG Kyoto Encyclopedia of Genes and Genomes. www.kegg.com.
- Khare S, Nunes JS, et al. Early phase morphological lesions and transcriptional responses of bovine ileum infected with Mycobacterium avium subsp. paratuberculosis. Vet Pathol. 2009;46(4):717–728. doi: 10.1354/vp.08-VP-0187-G-FL. [DOI] [PubMed] [Google Scholar]
- Loor JJ, Everts RE, et al. Nutrition-induced ketosis alters metabolic and signaling gene networks in liver of periparturient dairy cows. Physiol Genomics. 2007;32(1):105–116. doi: 10.1152/physiolgenomics.00188.2007. [DOI] [PubMed] [Google Scholar]
- Murphy K. Dynamic Bayesian Networks: Representation, Inference and Learning. UC Berkeley: 2002. [Google Scholar]
- Musser JM, DeLeo FR. Toward a genome-wide systems biology analysis of host-pathogen interactions in group A Streptococcus. Am J Pathol. 2005;167(6):1461–1472. doi: 10.1016/S0002-9440(10)61232-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer LE, Hobbie S, et al. YopJ of Yersinia pseudotuberculosis is required for the inhibition of macrophage TNF-alpha production and downregulation of the MAP kinases p38 and JNK. Molecular Microbiology. 1998;27:953–965. doi: 10.1046/j.1365-2958.1998.00740.x. [DOI] [PubMed] [Google Scholar]
- Ruckdeschel K, Harb S, et al. Yersinia enterocolitica impairs activation of transcription factor NF-kB: involvement in the induction of programmed cell death and in the suppression of the macrophage tumor necrosis factor production. J. Exp. Med. 1998;187:1069–1079. doi: 10.1084/jem.187.7.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos RL, Raffatellu M, et al. Life in the Inflammed Intestine, Salmonella Style. Trends in Microbiology. 2009;(17):498–506. doi: 10.1016/j.tim.2009.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos RL, Zhang S, et al. Morphologic and molecular characterization of S. typhimurium infection in neonatal calves. Veterinary Pathology. 2002;(39):200–215. doi: 10.1354/vp.39-2-200. [DOI] [PubMed] [Google Scholar]
- Schorey JS, Cooper AM. Macrophage signalling upon mycobacterial infection: the MAP kinases lead the way. Cell. Microbiol. 2003;5:133–142. doi: 10.1046/j.1462-5822.2003.00263.x. [DOI] [PubMed] [Google Scholar]
- Tang P, Sutherland CL, et al. Listeria monocytogenes invasion of epithelial cells requires the MEK-1/ERK-2 mitogen-activated protein kinase pathway. Infect. Immun. 1998;66:1106–1112. doi: 10.1128/iai.66.3.1106-1112.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang P, Sutherland CL, et al. Listeria monocytogenes invasion of epithelial cells requires the MEK-1/ERK-2 mitogenactivated protein kinase pathway. Infection and Immunity. 1998;66:1106–1112. doi: 10.1128/iai.66.3.1106-1112.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Santos RL, et al. The Salmonella enterica serotype typhimurium effector proteins SipA, SopA, SopB, SopD, and SopE2 act in concert to induce diarrhea in calves. Infect Immun. 2002;70:3843–3855. doi: 10.1128/IAI.70.7.3843-3855.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]