Abstract
Brucella spp. infect hosts primarily by adhering and penetrating mucosal surfaces; however the initial molecular phenomena of this host:pathogen interaction remain poorly understood. Using cDNA microarray analysis, we characterized the transcriptional profile of the intracellular pathogen Brucella melitensis at 4 h (adaptational period) and 12 h (replicative phase) following HeLa cells infection. The intracellular pathogen transcriptome was determined using initially enriched and then amplified B. melitensis RNA from total RNA of B. melitensis-infected HeLa cells. Analysis of microarray results identified 161 and 115 pathogen genes differentially expressed at 4 and 12 h p.i., respectively. In concordance with phenotypic studies, most of the genes expressed were involved in pathogen growth and metabolism, and were down-regulated at the earliest time point (78%), but up-regulated at 12 h p.i. (75%). Further characterization of specific genes identified in this study will elucidate biological processes and pathways to help understand how both host and Brucella interact during the early infectious process to the eventual benefit of the pathogen and to the detriment of the naïve host.
Keywords: Brucella melitensis, Gene expression, Microarray, Bacterial pathogenesis
1. Introduction
Brucella spp. are small aerobes non-motile Gram negative coccobacilli that are facultative intracellular pathogens responsible for zoonotic infections. The brucellae infect hosts primarily by penetrating the natural mucosa [1]. The mucosal surface of the alimentary tract is the principal route of entry for B. melitensis and B. abortus, while the mucosa of the genital tract is a major route for B. ovis, B. suis and B. canis penetration [2]. Conjunctivae and intranasal mucosa are also permeable to Brucella [3,4,5]. In vivo, Brucella quickly translocate through the epithelium layer and are endocytosed by mucosal macrophages. Virulent Brucella have the ability to survive and replicate inside phagocytic cells for long periods of time, where they evade the host immunity and later re-emerge and systemically disseminate throughout of the body [6].
Due to the need to understand the pathogenesis of brucellosis in greater depth, research has focused on identifying virulence factor-encoded genes involved in the intracellular replication and survival of Brucella in professional mononuclear phagocytic cells. However, in spite of the transient nature of the encounter, the interaction between Brucella and the epithelial cells is crucial to the outcome of the infection. The current understanding about the molecular mechanisms and factors employed by Brucella to adhere and migrate through the epithelium is limited. Among individual gene products involved in adhesion, invasion and survival of Brucella in non-professional phagocytic cells, only a few Brucella genes have been identified, such as the two-component regulatory system, BvrR/BvrS. This gene system is critical for regulating the structure of outer membrane components necessary for penetration, vacuole maturation and intracellular trafficking [7,8]. BvrR/BvrS Brucella mutants had reduced invasiveness in HeLa cells when compared to wild type strain [9]. Also, an adhesin called SP41 (for Surface Protein 41kDa) associated with adhesion and invasion of non-professional phagocytic cells has been characterized. A wild-type strain of B. suis was 40 to 50 times more invasive in epithelial cell lines than the mutant strain, indicating that invasion was affected in the absence of this surface protein [10]. The presence of antibodies against SP41 in 70% of human with acute brucellosis indicates strong immune recognition and some role during initial stage of in vivo infection. More recently, the hypothetical protein encoded by BMEI0216 gene was found to be involved in B. melitensis internalization to non-professional phagocytic cells, but not in macrophages [11]. Additionally, 5 metabolic-process defective B. abortus mutants had reduced abilities to internalize and/or replicate intracellularly in HeLa cells as compared to the wild type strain [12] as well as the type IV secretion system (T4SS) mutant strains [13,14,15].
A variety of techniques are employed to identify differences in global gene expression levels between identical cells subjected to different stimuli or between different cellular phenotypes under the same conditions in a single experiment. Microarrays have features that have made them the most widely used method for profiling mRNA expression. To identify novel Brucella adhesion and internalization-encoded genes in non-professional phagocytic cells, we used Brucella cDNA microarrys to compare the differences in gene expression between the most and the least invasive growth phase of B. melitensis cultures [16]. Among genes differentially expressed between these two conditions, several transcriptional regulator genes and genes encoding cell envelope and outer membrane components were detected. Here, we analyze the transcriptome of intracellular Brucella following infection of HeLa cells to identify initial strategies employed by the pathogen to survive, replicate and invade susceptible hosts. The hybridization of enriched and amplified B. melitensis RNA from total RNA of B. melitensis-infected HeLa cells to a custom B. melitensis microarray revealed a broadly down-regulated expression profile at 4 h post-infection (p.i.), which transitions to up-regulated transcriptional activity at 12 h p.i. The analysis of microarray results indicates that the pathogen undergoes an adaptation period during the first 4 h p.i. that is subsequently overcome, facilitating Brucella to replicate intracellularly. The results presented here are expected to generate new hypotheses regarding the initial molecular pathogenesis of Brucella.
2. Results
2.1. Global array data
In concordance with previous publications [17, 13] our experiments have shown that the intracellular replication of B. melitensis in non-phagocytic cells begins after an initial adaptation period (data not shown). To understand the intracellular behavior of Brucella at the molecular level, we studied its transcriptional profile at the adaptation and the replicative phases (i.e. 4 and 12 h p.i.). RNA samples used in the experiment were of good to excellent quality (RIN –RNA Integrative Number- > 7.0; 28S/18S ratio ≥ 1.6, OD260/280 ≥ 1.75, OD260/230 > 1.6). On average, the enriched and amplified B. melitensis RNA samples generated readable signals in 86% and 87% for the B. melitensis microarray probes, at 4 and 12 h p.i., respectively, while the reference sample (B. melitensis gDNA) generated readable signal intensities in more than 95% of the genes on the microarray (SNR > 3SD above background).
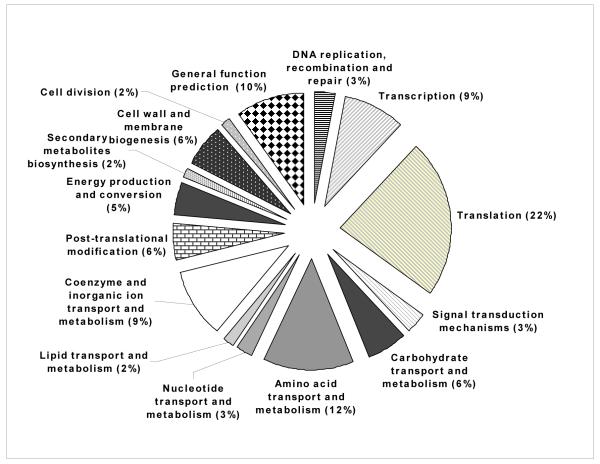
Microarray analysis revealed that the vast majority (126 genes, 78%) of the 161 genes differentially expressed (fold-change > 2 and P < 0.05), were down-regulated at 4h p.i. (Supplemental Table S1). The greatest number of transcriptional changes at this time post-infection occurred in genes whose products are associated with transcription, translation, coenzyme and inorganic ion transport and metabolism and carbohydrate and amino acids transport and metabolism (Fig. 1A). The relative changes in gene expression ranged from a 142.5-fold induction of the narG gene (BMEII0949) to a 60.9-fold down-regulation (0.01643) of the BMEI0299 locus (hypothetical protein).
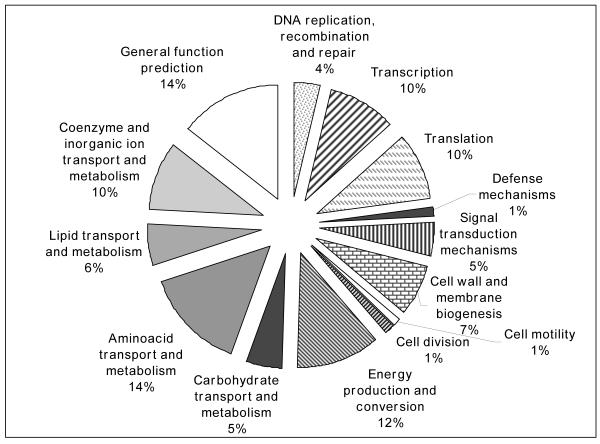
Figure 1.
Proportional representation of functional categories differentially expressed at 4 (A) and 12h (B) p.i. in the HeLa host cell as compared to extracellular control conditions. Detailed information is presented in Supplemental tables 1 (Fig A) and 2 (Fig B).
Contrarily, 115 genes were differentially expressed by microarray analysis at 12 h p.i., and the majority of them (86 genes, 75%) were up-regulated (Supplemental Table S2). The greatest number of transcriptional changes at this time post-infection occurred in genes whose products are associated with DNA replication, transcription, transport and intermediate metabolism, as well as energy production and conversion (Fig. 1B). narG (BMEII0949) was also the most highly expressed gene (103.3) and the most down-regulated gene (−13.5 fold, 0.0739) was ptsP (BMEI0190), whose product is involved in regulation of carbon and nitrogen utilization.
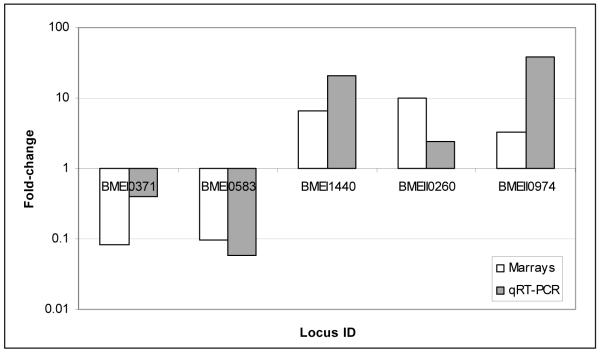
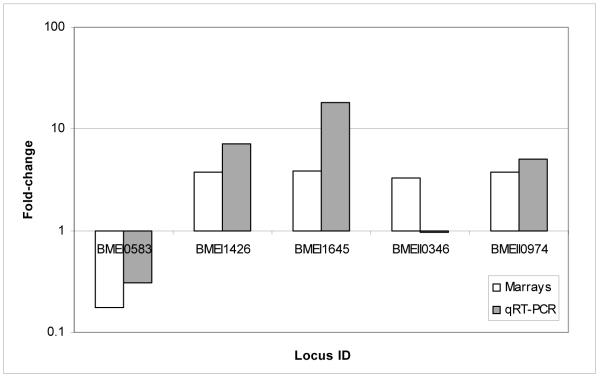
To confirm the microarray results, we randomly selected 10 differentially expressed genes (5 from each time point, i.e. 4 and 12h p.i.) and conducted qRT-PCR. Quantitative RT-PCR results confirmed 90% of the Brucella genes tested to be greater than 2.0-fold up- or down-regulated and in the same direction as was determined by microarray analysis (Fig. 2A and B).
Figure 2. Validation of Brucella melitensis microarray results by quantitative Real Time - PCR.
A. Five of 5 ORFs (100%) tested at 4 h p.i had fold-change greater than 2-fold and in the same direction than microarray. B. Four of 5 ORFs (80%) tested at 12 h p.i. had fold-change greater than 2-fold and in the same direction by both methodologies.
2.2. The lack of intracellular B. melitensis replication at 4 h p.i. correlates with stress response at the molecular level
In agreement with our kinetic studies of B. melitensis intracellular replication in HeLa cells that indicated almost no bacterial replication in the first 4 h p.i., cell division genes ftsQ (BMEI0583) and ftsA (BMEI0584) were down-regulated, as well as genes involved in DNA replication, transcription and translation, transport and intermediate metabolism, and cell envelope, biogenesis and outer membrane activities. Ultrastructural studies in HeLa cells indicate that virulent Brucella are located inside the stressful environment of autophagic vesicles at 4 h p.i. [18], and data from our microarrays agree with this finding. For example, the observed down-regulation of ribosomal protein genes and RNA polymerase (BMEI0750) are indications of amino acid starvation, consistent with the poor nutritional intra-vacuolar microenvironment of Brucella [19]. In addition, ppx (BMEII0598) that encodes an exopolyphosphatase that is a major regulator of bacterial adjustment to stress [20] was significantly up-regulated based on our microarray analysis results.
The up-regulation of the czcD gene (BMEI1438) and the catalytic subunits of the denitrifying reductase genes (BMEII0949, BMEII0974 and BMEII0998) are also consistent with growth inside autophagic vesicles. Host cells deliver divalent cations from the cytosol to the phagosome compartment to kill invaders that are taken up through constitutively expressed transporters. CzcD is an integral membrane protein and part of the Co/Zn/Cd efflux system component that reduces the intracellular concentration of toxic heavy metals via active cation efflux to the extracellular medium. Conversely, the low oxygen level inside the phagosome requires the pathogen to adapt from aerobic metabolism to microaerobic or anaerobic metabolism to survive. Analysis of our microarray results revealed up-regulation of narG (BMEII0949), norB (BMEII0998) and nosZ (BMEII0974) genes, which encode catalytic subunits of enzymes involved in electron transport during nitrate respiration, allowing Brucella to survive under low-oxygen conditions [21]. In addition to its role in de-nitrification, the protein encoded by norB reduces nitric oxide (NO) to nitrous oxide (N2O). This response may decrease the presence of intravacuolar NO, an important host cell defense element in the autophagic vacuole, thereby increasing Brucella’s intracellular survival. Also down-regulated at 4 h p.i. were B. melitensis genes that encode an iron uptake protein (BMEI0375 and BMEII0844). Iron is an essential cofactor in various biosynthetic and bioenergetic pathways and is also important for bacterial growth. The down-regulation of BMEI0375 and BMEII0844 suggests further confirmation of the slow growth by B. melitensis during this initial intracellular phase. Overall, these results suggest that after internalization, B. melitensis encounters a hostile environment obligating the bacteria to regulate their metabolism to survive.
2.3. Gene expression profile indicates that Brucella have an active intracellular life style at 12 h p.i
The molecular information, combined with the intracellular replication of B. melitensis observed in infected HeLa cells, collectively indicate that by 12 h p.i. the bacteria have adapted to the intracellular environment and are actively replicating. Only 3 transcripts for ribosomal proteins (BMEI0202, BMEI0759 and BMEI0823) exhibited decreased expression at 12 h p.i., compared to 17 at the earlier time point. Also at 12 h p.i., no translation factors or tRNA synthetases were observed to be down-regulated which is indicative of translation re-activation and bacterial division.
Microarray data at 12 h p.i. indicated that there were seven transcriptional regulators with enhanced expression in intracellular Brucella. Consistent with our data, two of them (BMEI0169 and BMEI0320) were previously identified as necessary for intracellular B. melitensis survival and replication, both in vivo and in cell culture models [22]. Another regulator with enhanced expression at 12 h p.i. was nikR. In Helicobacter pylori, the product of this gene transcriptionally represses the expression of a nickel transport system (encoded by nik operon –nikABCDE-) but induces urease expression by binding to the ureA promoter [23]. In our study, coincidently, no gene from nik operon was observed differentially expressed, but ureA (BMEI0649) was highly up-regulated (10.57); thus nikR not only may transcriptionally represses nik operon but only activates urease expression in Brucella. We speculate that urease may be used to hydrolyze urea to produce ammonia and neutralizes acidic pH in the Brucella-intravacuolar compartment, perhaps necessary at this stage of the infection. Experimental evidence indicates that urease does not likely play a role in cell models of intracellular survival of Brucella, but it is necessary for intestinal infection in mice [24,25].
There were 4 differentially expressed genes with possible involvement in Brucella intracellular survival: acrB, motD, phoQ, and ftsQ. AcrB (acriflavin-resistance protein B) is a component of the efflux pumps that protect the organism from antibiotics and other substances produced by the host. The up-regulation of this transcript suggests the protein might be important in Brucella pathogenesis, as it is in Salmonella [26]. The up-regulation of this transcript and other uncharacterized transport systems, may have possible implications for Brucella protection from deleterious host and environmental factor effects. The role of these up-regulated defense-encoded genes in Brucella intracellular survival and how they interact with the host counterpart also warrant further study. The motD gene (or fliK) was up-regulated and encodes a regulator of flagellar hook length in the alpha subgroup of the Proteobacteria [27] that is essential for proper formation of flagellum filaments bundles. Previous work demonstrated the requirement of flagellum expression for persistence of Brucella in a mouse model of infection [28]. Also up-regulated was phoQ (BMEI1336) that in Salmonella encodes one of the two components of the regulatory system PhoP/PhoQ, which regulates the expression of virulence factors necessary for survival inside macrophages, and defensin and acid resistance [29]. To date, no studies describing the function of phoQ or its product in Brucella genus have been published. Similar to the observations at 4 h p.i., ftsQ transcription was down-regulated, though to a lesser degree (−5.63 vs −10.25), possibly due to the intracellular replication of Brucella.
One iron transport system gene (frpB, BMEII0105) was up-regulated at 12 h, consistent with the requirement for iron during Brucella replication. Genes encoding transporters of carbohydrates, lipids and amino acids, as well as metabolic genes were also up-regulated. Genes encoding different components of amino acid ABC-transport systems were the most extensively expressed (BMEI1627, BMEI1728, BMEII0070, BMEII0196, BMEII0631) suggesting either the need for amino acids in protein synthesis during this active growth period or possibly the use of amino acids as carbon sources. Together, these results indicate a reactivation of Brucella gene expression at 12 h p.i. (replicative period) compared with the earlier adaptation period.
Analysis of our microarray data also revealed modification in the expression of genes encoding a group of proteins with unknown, predicted or moderately known functions at 4 and 12 h p.i., compared to the control samples (Supplemental Tables 1 and 2). These novel findings may have implications for Brucella virulence or intracellular survival and thus warrant further study.
3. Discussion
A general overview of our molecular analysis correlates well with phenotypic studies. In agreement with a lack of B. melitensis intacellular replication in HeLa cells at 4 h p.i., our microarray analysis reveals an adaptive-associated Brucella transcriptome. In the past, only one study has been published describing intracellular Brucella gene expression during the first 4 h p.i. [30]. Using a differential fluorescence induction approach, the authors identified only 34 B. abortus 2308-ORFs differentially activated within RAW264.7 macrophages 4 h after infection. Of these 34 ORF, only 9 were identified based on similarity with other bacterial sequences in GenBank. In agreement with our results, the Brucella genes identified were involved in adaptation to intracellular environmental conditions. Similar results were obtained by Lamontagne et al. who did a proteomic analysis of virulent intracellular B. abortus following infection of macrophages [31]. In their study, the authors reported that early after infection (3 h p.i.) virulent B. abortus reduce most of its metabolic functions, including transcriptional and translational synthetic metabolism, central carbon metabolism and synthesis of components for outer membrane. Also in concordance with our study, proteomic analysis strongly suggested that Brucella switch to a low oxygen tension type of respiration and has a low capacity to capture iron at early time p.i. Together, the previously published data and data from our experiments strongly suggest that even in two different cell types, Brucella undergo adaptation in the first 4 h p.i. In concordance, an adaptive period was also identified in two other intracellular pathogens, Shigella flexneri and Chlamydia trachomatis, during the initial infection process of Hela cells [32,33]. Collectively, these results indicate that an adaptive period occurs at the initial phases of the infectious process which appears to be necessary and crucial for successful persistence of intracellular pathogens.
Two Brucella key elements for invasion and intracellular survival are the Bvr two-component system (BvrR/BvrS) and the type IV secretion system (T4SS) [9,13]. BvrR/BvrS is a regulatory system that modulates the expression of outer membrane proteins necessary for cell penetration [8,9]. The ChvI/ChvG (encoded by BMEI2036 and BMEI2035, respectively) represents the B. melitensis homolog of the B. abortus two-component regulatory system BvrR/BvrS. Contrary to expectations, but in concordance with a previous proteomic study of intramacrophage B. abortus [31], chvI expression was down-regulated early after infection, and later reverted to pre-infection levels. Possibly, the lower expression level of the two-component regulatory system that occurs in in vitro cultures does not turn off gene expression as was observed with bvrR/S- mutants, yet is somehow necessary to facilitate Brucella penetration and survival into host cells. A functional type IV secretion system (T4SS), encoded by the virB operon, is essential for B. melitensis to complete autophagic vacuole maturation process to establish an intracellular replication niche in HeLa cells [14,15]. Previous studies demonstrated that B. suis virB operon is induced in macrophages within 3 h p.i. [34]. However, its expression was not always detected among those genes or peptides expressed in intracellular B. abortus early after infection [30,31]. In our study, analysis of the microarray results did not identify virB genes differentially expressed in B. melitensis inside HeLa cells, compared to the inoculum. Previously, we found that several genes from the B. melitensis virB operon were already up-regulated in the inoculum growth under our laboratory conditions [16]. Perhaps, the expression level reached by the virB operon in in vitro cultures was appropriately maintained for normal B. melitensis intracellular trafficking. Another possibility is that the role of these genes was not analyzable under our experimental conditions.
A different scenario was observed at 12 h p.i. The pathogen transcriptional profile was clearly up-regulated. Genes whose products are involved in growth and metabolism (DNA replication, transcription and translation, cell wall and membrane biogenesis, energy production and conversion and carbohydrate, lipid and amino acid metabolism) were mainly up-regulated, in agreement with an active intracellular Brucella replication. At this time point of the in vitro infection of non-phagocytic cells, virulent Brucella are delivered to the perinuclear endoplasmic reticulum where actual bacterial multiplication occurs [18]. This has been interpreted as an opportunity for Brucella to take advantage of the metabolites synthesized or translocated by the host cell to this compartment to obtain nutritional requirements for growth. For instance, Legionella pneumophila, another intracellular pathogen that impairs phago-lysosomal fusion and replicates inside macrophages similar to Brucella, scavenges host proteins and amino acids for nutrients [35]. Our microarray data indicate a reactivation of transcription of Brucella genes involved in translational processes, compared with 4 h p.i., while it has been demonstrated that de novo host protein synthesis is not required during intracellular Brucella replication in non-phagocytic cells [36]. These results suggest that Brucella is actively using host amino acid availability for synthesizing their own proteins, or possibly utilize them as a carbon source.
A previous proteomic study demonstrated that the peptides initially differentially expressed, gradually recover to their original expression levels over the Brucella infection timecourse [31] while in our study, the transcriptome reverts from an under expression to a over expression. We speculate that this difference may be that Brucella establish a long lasting relationship in macrophages but a transient relationship in non-phagocytic cells.
In summary, we analyzed the transcriptional profile of intracellular Brucella melitensis during early infection time points in epithelial cells. We conclude that there is a down-regulation of the pathogen transcriptome at the first 4 h p.i. that is reversed at the later time point (12 h p.i.) in concordance with Brucella intracellular replication. These data provide specific genes and biological processes to further elucidate how Brucella survive and replicate inside epithelial cells to the eventual benefit of the pathogen and to the detriment of the naïve host. The integrated results facilitate the development of new hypotheses regarding the initial molecular pathogenesis of Brucella.
4. Materials and Methods
4.1. Bacterial strain, media and culture conditions
Saturated culture of a frozen glycerol stock of smooth virulent Brucella melitensis 16M Biotype 1 (ATCC 23456) (American Type Culture Collection, Manassas, VA) was sub-cultured into cell culture media [F12K medium (ATCC) supplemented with 10% heat-inactivated fetal bovine serum (HI-FBS) (ATCC)] and incubated with the lid loose and shaking (200 rpm) at 37°C with 5% CO2 until the late-log growth phase (OD600= 0.4) was reached [16].
4.2. HeLa cell infection
HeLa S3 cell line (ATCC; CCL-2.2), between passages 8 and 15, was grown in F12K medium containing 10% HI-FBS at 37°C with 5% CO2. Twenty-four hours prior to infection, cells were suspended and cultured in 24 well plates (Corning, Corning, NY) at a concentration of 1×105 cells/well and placed in the incubator. Before infection, cells from 2 wells were detached and counted. For infections, the medium was replaced with a bacterial inoculum grown in cell culture media (F12K with 10% HI-FBS) at a multiplicity of infection (MOI) of 1,000 bacteria per HeLa cell. Bacteria were centrifuged onto the cells at 800X g for 10 min followed by 30 min of incubation at 37°C. Then, cells were washed once with phosphate buffer solution (PBS) to remove extracellular bacteria and re-incubated with F12K media supplemented with 100 μg ml−1 of gentamicin solution (Sigma, St. Louis, MO) for 1 hour. After antibiotic treatment, infected cultures were washed 3 times with PBS, and re-incubated in 1 ml of fresh cell culture medium.
4.3. Sample isolation, preparation and slide hybridization
Sample isolation, labeling and hybridization procedures were repeated from our previous experiments [16,37]. Microarrays containing all B. melitensis 16M ORFs were designed at the Pathogen Expression Core (Dr. S.A. Johnston’s Laboratory at Arizona State University) as previously reported [16]. Briefly, total RNA from 4 different infected cell cultures (biological replicates) was extracted at 4 and 12 h p.i. (n = 8) by TRI-Reagent® (Ambion, Austin, TX) according to manufacturer’s instructions. B. melitensis total RNA was initially enriched and then amplified (E&A) from 50 μg of total RNA from B. melitensis-infected HeLa cells at 4 and 12 h p.i. Ten μg of E&A B. melitensis RNA were reverse transcribed overnight to amino-allyl cDNA using 1.5 μg of B. melitensis genomic directed primers (BmGDPs) [37], 0.6 μl 50X dNTPs (Invitrogen) / aa-dUTP (Ambion) mix (2:3 aa-dUTP:dTTP) and 400U Superscript III (Invitrogen). Experimental samples (i.e., E&A B. melitensis RNA and total RNA from B. melitensis - infected Hela cells) were labeled with Cy3-ester (Amersham Pharmacia Biosciences) with one hour incubation in the dark and dye incorporation was calculated by NanoDrop® ND-1000 (NanoDrop). Dried, labeled cDNA samples were resuspended in nuclease–free water (Ambion) and mixed with 0.5 μg of Cy5-labeled B. melitensis gDNA to the final volume of 35 μl. Following incubation at 45°C, 35 μl of 2X formamide-based hybridization buffer [50% formamide; 10X SSC; 0.2% SDS] was mixed with each sample and applied to a custom 3.2K B. melitensis oligo-array. Slides were hybridized at 45°C for 20 h in a dark, humid chamber (Corning) and washed for 10 min at 45°C with low stringency buffer [1X SSC, 0.2% SDS] followed by two 5-min washes in a higher stringency buffer [0.1X SSC, 0.2% SDS and 0.1X SSC] at room temperature with agitation. Slides were dried and immediately scanned.
4.4. Data acquisition and microarray data analysis
Microarrays were scanned using a commercial laser scanner (GenePix 4100; Axon Instruments Inc., Foster City, CA). The genes represented on the arrays were adjusted for background and normalized to internal controls using image analysis software (GenePixPro 6.0; Axon Instruments Inc.). Genes with fluorescent signal values below background were disregarded in all analyses. Pathogen arrays were normalized against B. melitensis genomic DNA, as previously described [38]. The intracellular B. melitensis gene expression was compared to the gene expression of the inoculum (i.e. in vitro-grown cultures of B. melitensis at late-log phase of growth) (n = 2). Data were analyzed using GeneSifter (VizX Labs, Seattle, WA). The signal values of every gene (triplicate spots in 4 arrays = 12 spots) for each experiment (i.e. 4 and 12 h) were averaged, the fold-change calculated, and Student’s t test performed. At each time point, genes determined to be expressed at statistically significant levels (fold-change > 2 and P value < 0.05) using pathogen arrays hybridized with probes generated from B. melitensis-infected cells were subtracted from the final list of differentially expressed genes. Microarray data have been deposited in Gene Expression Omnibus (GEO) database at NCBI [Accession # GSE14704].
4.5. Validation of microarray results
Five randomly selected genes from B. melitensis with differential expression at 4 and 12 h p.i. (n = 10) by microarray results were analyzed by quantitative RT-PCR (qRT-PCR). Two micrograms of RNA were reverse transcribed using TaqMan® Reverse Transcription Reagents (Applied Biosystems, Foster City, CA). For relative quantitation of target cDNA, samples were analyzed in individual tubes in SmartCycler II (Cepheid, Sunnyvale, CA). One SmartMix bead (Cepheid) was used for 2 - 25 μl PCR reactions along with 20 ng of cDNA, 0.2X SYBR Green I dye (Invitrogen) and 0.3 μM forward and reverse primers (Sigma Genosys) designed using Primer Express Software v2.0 (Applied Biosystems) (Table 1). For each gene tested, the individual calculated threshold cycles (Ct) were averaged among each condition and normalized to the Ct of the gyrA gene, from the same cDNA samples before calculating the fold change using the ΔΔCt method [39]. For each primer pair, a negative control (water) and an RNA sample without reverse transcriptase (to determine genomic DNA contamination) were included as controls during cDNA quantitation. Pathogen array data were considered valid if the fold-change of each gene tested by qRT-PCR was > 2.0 and in the same direction as determined by microarray analysis.
TABLE 1.
Primers for Real Time – PCR tested genes on B. melitensis
| Locus ID | Gene name | Forward primers (5′-3′) | Reverse primers (5′-3′) |
|---|---|---|---|
| BMEI0371 | RNA polymerase S70 | AGGCATGGGCCAAGCA | AGATCAAGCGTGCCATATTGC |
| BMEI0583 | Cell division protein FtsQ | TCAAGGGTTTTGTGGACCAGAT | TGTTTTTCCCGATCAAGCTTCT |
| BMEI0884 | Gyrase A | AAGGCCTCGATGATCGAGAAG | ACGAGGTCTGCAAAGGCGTATA |
| BMEI1426 | Putative undecaprenyl-phophate alpha | TGCACTTATCATCGCAATCAATG | GAACAGGGCAAAACCGAGAA |
| BMEI1440 | Thio:disulfide interchange protein DsbA | CGAAATTGGCCGGTTTTACA | CCCGACATCTCCTCAAACGA |
| BMEI1645 | Acriflavin resistance protein B | CTGATCCGCCAGGAACTCA | CACCTGAACCGGCAATCG |
| BMEII0260 | GTP-binding protein LepA | AGGGCTATGCCTCGTTCGA | ATATGTTGCGGGATCAGTTCCT |
| BMEII0346 | Transcriptional regulator, AsnC family | GATCGCGAGATTCTGGCTATTC | TCGCCCGGATGATATTGCT |
| BMEII0974 | Nitrous-oxide reductase | TCAGTTGCCGAACCAGCATA | GGCGACCTTCATCGTTTCAC |
Supplementary Material
Highlights.
- cDNA microarray revealed different pathogen expression profile at 4 and 12 h p.i.
- An adaptive-associated Brucella transcriptome was detected at 4 h p.i.
- Conversely, a replicate-associated transcriptional activity was observed at 12 h p.i.
- Molecular analysis correlates well with phenotypic studies.
Acknowledgments
We thank Dr. Thomas A. Ficht for providing the B. melitensis 16M strain and Drs. Renée M. Tsolis and Sara D. Lawhon for critical reading of the manuscript. We are grateful to the Western Regional Center of Excellence (WRCE) Pathogen Expression Core (Dr. Mitchell McGee, Dr. Rhonda Friedberg and Dr. Stephen A. Johnston, A.S.U.) for developing and printing the B. melitensis cDNA microarrays. This study was supported by U.S. Department of Homeland Security – National Center of Excellence for Foreign Animal and Zoonotic Disease (FAZD) Defense grant ONR-N00014-04-1-0 and a NIH grant 2U54AI057156-06. C.A.R. was sponsored by Fulbright-INTA scholarship from Argentina. This work was part of the C.A.R. doctoral dissertation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Enright FM. The pathogenesis and pathobiology of Brucella infection in domestic animals. In: Nielsen K, Duncan JR, editors. Animal brucellosis. CRC Press; Boca Raton, Florida: 1990. pp. 301–20. [Google Scholar]
- [2].Olsen SC, Thoen CO, Cheville NF. Brucella. In: Gyles CL, Prescott JF, Songer JG, Thoen CO, editors. Pathogenesis of bacterial infections in animals. Third ed Blackwell Publishing Ltd.; Ames, Iowa: 2004. pp. 309–19. [Google Scholar]
- [3].Plant JW, Eamens GJ, Seaman JT. Serological, bacteriological and pathological changes in rams following different routes of exposure to Brucella ovis. Aust Vet J. 1986;63(12):409–12. doi: 10.1111/j.1751-0813.1986.tb15919.x. [DOI] [PubMed] [Google Scholar]
- [4].Meador VP, Deyoe BL. Intracellular localization of Brucella abortus in bovine placenta. Vet Pathol. 1989;26(6):513–5. doi: 10.1177/030098588902600609. [DOI] [PubMed] [Google Scholar]
- [5].Mense MG, van de Verg LL, Bhattacharjee AK, Garrett JL, Hart JA, et al. Bacteriologic and histologic features in mice after intranasal inoculation of Brucella melitensis. Am J Vet Res. 2001;62(3):398–405. doi: 10.2460/ajvr.2001.62.398. [DOI] [PubMed] [Google Scholar]
- [6].Roop RM, II, Bellaire BH, Valderas MW, Cardelli JA. Adaptation of the brucellae to their intracellular niche. Mol Microbiol. 2004;52(3):621–30. doi: 10.1111/j.1365-2958.2004.04017.x. [DOI] [PubMed] [Google Scholar]
- [7].Verri C Guzman, Manterola L, Sola-Landa A, Parra A, Cloeckaert A, Garin J, et al. The two-component system BvrR/BvrS essential for Brucella abortus virulence regulates the expression of outer membrane proteins with counterparts in members of the Rhizobiaceae. Proc Natl Acad Sci USA. 2002;99(19):12375–80. doi: 10.1073/pnas.192439399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].López-Goni I, Guzmán-Verri C, Manterola L, Sola-Landa A, Moriyon I, Moreno E. Regulation of Brucella virulence by the two-component system BvrR/BvrS. Vet Microbiol. 2002;90(1-4):329–39. doi: 10.1016/s0378-1135(02)00218-3. [DOI] [PubMed] [Google Scholar]
- [9].Sola-Landa A, Pizarro-Cerdá J, Grilló MJ, Moreno E, Moriyón I, et al. A two-component regulatory system playing a critical role in plant pathogens and endosymbionts is present in Brucella abortus and controls cell invasion and virulence. Mol Microbiol. 1998;29(1):125–38. doi: 10.1046/j.1365-2958.1998.00913.x. [DOI] [PubMed] [Google Scholar]
- [10].Castañeda-Roldán EI, Ouahrani-Bettache S, Saldana Z, Avelino-Flores F, Rendón MA, et al. Characterization of SP41, a surface protein of Brucella associated with adherence and invasion of host epithelial cells. Cell Microbiol. 2006;8(12):1877–87. doi: 10.1111/j.1462-5822.2006.00754.x. [DOI] [PubMed] [Google Scholar]
- [11].Hernández-Castro R, Verdugo-Rodriguez A, Puente JL, Suarez-Guemes F. The BMEI0216 gene of Brucella melitensis is required for internalization in HeLa cells. Microb Pathog. 2008;44(1):28–33. doi: 10.1016/j.micpath.2007.08.008. [DOI] [PubMed] [Google Scholar]
- [12].Kim S, Watarai M, Kondo Y, Erdenebaatar J, Makino S, Shirahata T. Isolation and characterization of mini-Tn5Km2 insertion mutants of Brucella abortus deficient in internalization and intracellular growth in HeLa cells. Infect Immun. 2003;71(6):3020–7. doi: 10.1128/IAI.71.6.3020-3027.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Sieira R, Comerci DJ, Sánchez DO, Ugalde RA. A homologue of an operon required for DNA transfer in Agrobacterium is required in Brucella abortus for virulence and intracellular multiplication. J Bacteriol. 2000;182(17):4849–55. doi: 10.1128/jb.182.17.4849-4855.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Comerci DJ, Martínez-Lorenzo MJ, Sieira R, Gorvel JP, Ugalde RA. Essential role of the VirB machinery in the maturation of the Brucella abortus-containing vacuole. Cell Microbiol. 2001;3(3):159–68. doi: 10.1046/j.1462-5822.2001.00102.x. [DOI] [PubMed] [Google Scholar]
- [15].Delrue RM, Martínez-Lorenzo MJ, Lestrate P, Danese I, Bielarz V, et al. Identification of Brucella spp. genes involved in intracellular trafficking. Cell Microbiol. 2001;3(7):487–97. doi: 10.1046/j.1462-5822.2001.00131.x. [DOI] [PubMed] [Google Scholar]
- [16].Rossetti CA, Galindo CL, Lawhon S, Garner H, Adams LG. Brucella melitensis global gene expression study provides novel information on growth phase-specific gene regulation with potential insights for understanding Brucella:host interaction. BMC Microbiol. 2009;9:81. doi: 10.1186/1471-2180-9-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].O’Callaghan D, Cazevieille C, Allardet-Servent A, Boschiroli ML, Bourg G, Foulongne V, et al. A homologue of the Agrobacterium tumefaciens virB and Bordetella pertussis Ptl type IV secretion systems is esssential for intracellular survival of Brucella suis. Mol Microbiol. 1999;33(6):1210–20. doi: 10.1046/j.1365-2958.1999.01569.x. [DOI] [PubMed] [Google Scholar]
- [18].Pizarro-Cerdá J, Méresse S, Parton RG, van der Goot G, Sola-Landa A, et al. Brucella abortus transits through the autophagic pathway and replicates in the endoplasmic reticulum of non-professional phagocytes. Infect Immun. 1998;66(12):5711–24. doi: 10.1128/iai.66.12.5711-5724.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Kohler S, Foulongne V, Ouahrani-Bettache S, Bourg G, Teyssier J, et al. The analysis of the intramacrophagic virulome of Brucella suis deciphers the environment encountered by the pathogen inside the macrophage host cell. Proc Natl Acad Sci USA. 2002;99(24):15711–6. doi: 10.1073/pnas.232454299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Keasling JD, Bertsch L, Kornberg A. Guanosine pentaphosphate phosphohydrolase of Escherichia coli is a long-chain exopolyphosphatase. Proc Natl Acad Sci USA. 1993;90(15):7029–33. doi: 10.1073/pnas.90.15.7029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Haine V, Dozot M, Dornand J, Letesson JJ, De Bolle X. NnrA is required for full virulence and regulates several Brucella melitensis denitrification genes. J Bacteriol. 2006;188(4):1615–9. doi: 10.1128/JB.188.4.1615-1619.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Haine V, Sinon A, Van Steen F, Rousseau S, Dozot M, et al. Systematic targeted mutagenesis of Brucella melitensis 16M reveals a major role for GntR regulators in the control of virulence. Infect Immun. 2005;73(9):5578–86. doi: 10.1128/IAI.73.9.5578-5586.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Ernst FD, Kuipers EJ, Heijens A, Sarwari R, Stoof J, et al. The nickel-responsive regulator NikR controls activation and repression of gene transcription in Helicobacter pylori. Infect Immun. 2005;73(11):7252–8. doi: 10.1128/IAI.73.11.7252-7258.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Bandara AB, Contreras A, Contreras-Rodriguez A, Martins AM, Dobrean V, et al. Brucella suis urease encoded by ure1 but not ure2 is necessary for intestinal infection of BALB/C mice. BMC Microbiol. 2007;7:57. doi: 10.1186/1471-2180-7-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Sangari FJ, Seoane A, Rodriguez MC, Aguero J, Lobo JM García. Characterization of the urease operon of Brucella abortus and assessment of its role in virulence of the bacterium. Infect Immun. 2007;75(2):774–80. doi: 10.1128/IAI.01244-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Buckley AM, Webber MA, Cooles S, Randall LP, La Ragione RM, et al. The AcrAB-TolC efflux system of Salmonella enterica serovar Typhimurium plays a role in pathogenesis. Cell Microbiol. 2006;8(5):847–56. doi: 10.1111/j.1462-5822.2005.00671.x. [DOI] [PubMed] [Google Scholar]
- [27].Eggenhofer E, Rachel R, Haslbeck M, Scharf B. MotD of Sinorhizobium meliloti and related alpha proteobacteria is the flagellar-hook-length regulator and therefore reassigned as FliK. J Bacteriol. 2006;188(6):2144–53. doi: 10.1128/JB.188.6.2144-2153.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Fretin D, Faucommier A, Kohler S, Halling SM, Léonard S, et al. The sheathed flagellum of Brucella melitensis is involved in persistence in a murine model of infection. Cell Microbiol. 2005;7(5):687–98. doi: 10.1111/j.1462-5822.2005.00502.x. [DOI] [PubMed] [Google Scholar]
- [29].Miller SI. PhoP/PhoQ: macrophage-specific modulators of Salmonella virulence? Mol Microbiol. 1991;5(9):2073–78. doi: 10.1111/j.1365-2958.1991.tb02135.x. [DOI] [PubMed] [Google Scholar]
- [30].Eskra L, Canavessi A, Carey M, Splitter G. Brucella abortus genes identified following constitutive growth and macrophage infection. Infect Immun. 2001;69(12):7736–42. doi: 10.1128/IAI.69.12.7736-7742.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Lamontagne J, Forest A, Marazzo E, Denis F, Butler H, et al. Intracellular adaptation of Brucella abortus. J Proteome Res. 2009;8(3):1594–609. doi: 10.1021/pr800978p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Lucchini S, Liu H, Jin Q, Hinton JCD, Yu J. Transcriptional adaptation of Shigella flexneri during infection of macrophages and epithelial cells: insights into the strategies of a cytosolic bacterial pathogen. Infect Immun. 2005;73(1):88–102. doi: 10.1128/IAI.73.1.88-102.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Belland RJ, Zhong G, Crane DD, Hogan D, Sturdevant D, et al. Genomic transcriptional profiling of the developmental cycle of Chlamydia trachomatis. Proc Natl Acad Sci USA. 2003;100(14):8478–83. doi: 10.1073/pnas.1331135100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Boschiroli ML, Ouahrani-Bettache S, Foulongne V, Michaux-Charachon S, Bourg G, et al. The Brucella suis virB operon is induced intracellularly in macrophages. Proc Natl Acad Sci USA. 2002;99(3):1544–9. doi: 10.1073/pnas.032514299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Bruggemann H, Hagman A, Jules M, Sismeiro O, Dillies MA, et al. Virulence strategies for infecting phagocytes deduced from the in vivo transcriptional program of Legionella pheumophila. Cell Microbiol. 2006;8(8):1228–40. doi: 10.1111/j.1462-5822.2006.00703.x. [DOI] [PubMed] [Google Scholar]
- [36].Detilleux PG, Deyoe BL, Cheville NF. Effect of endocytic and metabolic inhibitors on the internalization and intracellular growth of Brucella abortus in Vero cells. Am J Vet Res. 1991;52(10):1658–64. [PubMed] [Google Scholar]
- [37].Rossetti CA, Galindo CL, Garner HR, Adams LG. Selective amplification of Brucella melitensis mRNA from a mixed host-pathogen total RNA. BMC Res Notes. 2010;3:244. doi: 10.1186/1756-0500-3-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Talaat AM, Howard ST, Hale W, IV, Lyons R, Garner H, et al. Genomic DNA standards for gene expression profiling in Mycobacterium tuberculosis. Nucleic Acids Res. 2002;30:e104. doi: 10.1093/nar/gnf103. (109 pages) [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.