Abstract
Pyriplatin (cis-diammine(pyridine)chloroplatinum(II) or cDPCP), a platinum-based antitumor drug candidate, is a cationic compound with anticancer properties in mice and is a substrate for the organic cation transporters, hOCT1 and hOCT2, which facilitate oxaliplatin uptake. Unlike cisplatin and oxaliplatin, which form DNA cross-links, pyriplatin binds DNA in a monofunctional manner. The antiproliferative effects of pyriplatin, as well as combintions of pyriplatin with known anticancer drugs (paclitaxel, gemcitabine, SN38, cisplatin or 5-fluorouracil), were evaluated in a panel of epithelial cancer cell lines, with direct comparison to cisplatin and oxaliplatin. The effects of pyriplatin on gene expression and platinum-DNA adduct formation were also investigated. Pyriplatin exhibited cytotoxic effects against human cell lines after 24 h (IC50: 171 – 443 μM), with maximum cytotoxicity in HOP-62 non-small cell lung cancer cells after 72 h (IC50: 24 μM). Pyriplatin caused a G2/M block of cell cycle similar to that induced by cisplatin and oxaliplatin. Apoptotic cell death was supported by Annexin-V analysis and detection of phosphorylated H2AX and Chk2. Treatment with pyriplatin caused an increase in CDKN1/p21 and decrease in ERCC1 mRNA expression. On a platinum-per-nucleotide basis, pyriplatin adducts resulted in less cytotoxicity than cisplatin- and oxaliplatin-DNA adducts. The mRNA levels of several genes implicated in drug transport and repair of DNA damage, including MSH2 and GSTP1, correlate with pyriplatin cellular activity in our panel of cell lines. Synergy was observed in combinations of pyriplatin with paclitaxel. Because it has a different spectrum of activitythan that of cisplatin or oxaliplatin, pyriplatin may be regarded as a lead compound for the development of other drug candidates with cytotoxicity profiles that differ from those of the drugs currently in use.
Keywords: cisplatin, pyriplatin, cytotoxicity, apoptosis, cell cycle, anticancer activity, DNA adducts
Introduction
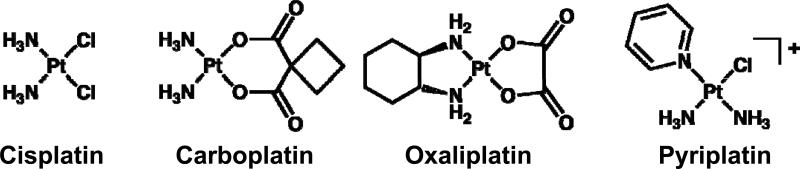
Three platinum compounds currently in use worldwide - cisplatin, carboplatin, and oxaliplatin (Figure 1) - have been developed with crucial support from the United States NCI, notably from screening of the NCI's 60-cell line panel (NCI60 screen) (1). This screen, together with the NCI COMPARE program, identified clear differences in activity profiles and mechanisms of action between platinum compounds, thus enabling the grouping of platinum compounds according to such characteristics (2). The cisplatin activity profile is similar to that of other diammineplatinum(II) compounds and to alkylating agents such as melphalan and camptothecin analogs. The oxaliplatin activity profile is similar to that of other platinum compounds containing the R,R-1,2-diaminocyclohexane ligand, including the platinum(IV) drug tetraplatin, and is also similar to those of acridines, organic compounds currently being developed as anticancer drugs (2).
Figure 1.
Structures of cisplatin, carboplatin, oxaliplatin, and pyriplatin.
Other classes of platinum compounds with activity different from cisplatin, oxaliplatin, or carboplatin have been defined on the basis of the NCI60 screen. The activity of the platinum-pyridines defines one group, into which some polyplatinum compounds including the clinically tested BBR3464 can be classified (3, 4). The platinum-silanes are another distinct group. Cells resistant to compounds from one group are commonly not cross-resistant to compounds from another. Similarly, because of the different mechanisms of action for each type of compound, it is possible for compounds of different groups to be used in combination with synergistic results, an example being the synergistic effect of combining cisplatin and oxaliplatin (2). The development of platinum compounds with mechanisms of action different from those of platinum-based drugs already on the market should facilitate identification of candidate compounds that are active in cancers for which cisplatin, carboplatin, or oxaliplatin are inactive. Unique mechanisms of action may derive from the mode of cellular uptake of the compound (5), preferential localization of the platinum compound to a specific body organ or cell organelle (6), manipulation of the cellular response to enhance cytotoxicity (7), or the prevention or retardation of drug inactivation by biotransformation, as occurs for platinum(IV) prodrugs (8-10).
Pyriplatin is a monofunctional, cationic platinum(II) compound that has previously shown antitumor activity in mice (11), and which forms only a single covalent bond with DNA, unlike cisplatin, carboplatin, or oxaliplatin, which bind in a bidentate manner. Besides this non-traditional structure when bound to DNA, there is also evidence for a unique cellular mode of pyriplatin uptake that differs from the uptake of cisplatin or oxaliplatin. Pyriplatin is an outstanding substrate for the organic cation transporters (OCT) 1 and 2 (12). The mechanism of RNA polymerase II inhibition by pyriplatin-DNA adducts is dramatically different from the inhibition seen with cisplatin-DNA adducts (13). The aim of the present study was to further characterize pyriplatin in vitro with direct comparison to cisplatin and oxaliplatin to gain insight into the mechanism of action and potential clinical applications for pyriplatin. We investigated cellular and molecular changes induced by pyriplatin in order to determine possible response biomarkers and predictive factors of pyriplatin activity. Effects of combination of pyriplatin with several chemotherapy drugs used in clinic were also studied.
Materials and Methods
Cell lines
All cell lines were obtained from the ATCC (Rockville, MD), and NCI cell collections. Cells were grown as monolayers in RPMI medium supplemented with 10% fetal calf serum (Invitrogen, Cergy-Pontoise, France), 2 mM glutamine, 100 units/ml penicillin and 100 μg/ml streptomycin. Cells were split twice a week using trypsin/EDTA (0.25%/0.02%; Invitrogen, Cergy-Pontoise, France) and seeded at a concentration of 2.5 × 104 cells/mL. All cell lines were tested regularly for Mycoplasma contamination by PCR using a Stratagene kit (La Jolla, CA).
Single agent evaluation
Pyriplatin was submitted to the National Cancer Institute (USA) for single agent, single dose testing in 2008. For evaluations performed in our laboratory, cells were seeded at 2 × 103 cells/well in 96-well plates and treated 24 h later with increasing concentrations of cisplatin, oxaliplatin, or pyriplatin. After 1, 2, 5, 24, or 72 h of incubation, the cells were washed and post-incubated in platinum-free medium for 72 h (after 1, 2, or 5 h) or 48 (after 24 or 72 h). Growth inhibition was then determined by the MTT assay (14). The resulting absorption at 560 nm of the control wells containing untreated cells was defined as 100% and the viability of treated samples was expressed as a percentage of the control. IC50 values were determined as platinum concentrations that reduced cell viability by 50%.
Cell cycle analysis
Exponentially growing cells were treated for 24 h with cisplatin, oxaliplatin, or pyriplatin at the IC50 concentrations (Table 1). At the end of treatment and following the 24, 48, or 72 h drug-washout period, the cells were counted, fixed in 70% cold ethanol, and kept at 4°C. The cells were washed with cold PBS and stained with 5 μg/mL propidium iodide in PBS and 12.5 μL/mL RNAse A. Flow cytometric cell cycle analysis was performed on a minimum of 2 × 104 cells per sample on a FACS Calibur instrument (Becton Dickinson, Sunnyvale, CA). A 488 nm laser and a dichroic mirror (570 nm) were used and fluorescence emission was detected using a filter for 620 ± 35 nm.
Table 1.
Potencies, expressed as IC50 concentrations for pyriplatin, cisplatin, and oxaliplatin, on cancer cell proliferation in the 10-cell line panel after a 24-h incubation period.
| Cell Line | Cancer Type | IC50 |
||
|---|---|---|---|---|
| Pyriplatin | Cisplatin | Oxaliplatin | ||
| HOP-92 | Non-Small Cell Lung | 171 ± 56 μM | 3.55 ± 3.2 μM | 2.70 ± 0.60 μM |
| HOP-62 | Non-Small Cell Lung | 190 ± 36 μM | 3.56 ± 1.4 μM | 6.86 ± 0.39 μM |
| IGROV1 | Ovarian | 230 ± 33 μM | 5.64 ± 1.3 μM | 8.08 ± 2.9 μM |
| COLO 205 | Colorectal | 266 ± 57 μM | 16.7 ± 7.2 μM | 2.84 ± 0.64 μM |
| HCT-116 | Colorectal | 281 ± 50 μM | 4.22 ± 2.5 μM | 1.10 ± 0.28 μM |
| OVCAR-3 | Ovarian | 328 ± 128 μM | 5.10 ± 3.0 μM | 1.24 ± 0.30 μM |
| MCF7 | Breast | 335 ± 104 μM | 15.6 ± 6.4 μM | 1.70 ± 0.54 μM |
| HCC-2998 | Colorectal | 381 ± 103 μM | 11.8 ± 4.0 μM | 7.27 ± 2.3 μM |
| MDA-435 | Breast /Melanoma | 401 + 156 μM | 5.81 + 4.0 μM | 12.7 + 5.6 μM |
| HT29 | Colorectal | 443 + 255 μM | 11.9 + 4.1 μM | 6.65 + 1.0 μM |
Data are means +/- SEM from three separate experiments, each performed in triplicate.
Evaluation of apoptosis
Cells were harvested after 24-h treatment with platinum compounds at IC50 concentrations and washed once with cold PBS, then pelleted and resuspended in 100 μL of a staining buffer containing Annexin V-FITC and 0.5 μg propidium iodide. Fluorescence analysis by flow cytometry was performed after 15-minute incubation in the dark and dilution of the sample to 500 μL.
Western blotting
To determine the protein level of several apoptotic markers, cells were treated for 24 h at the IC50 concentrations of pyriplatin, oxaliplatin, or cisplatin. The platinum-containing medium was removed and cells were lysed either immediately or 24, 48, or 72 h after removal of the platinum-containing medium. Protein concentration was quantified by the Bradford assay and extracts were analyzed on SDS-PAGE and transferred to PVDF membranes. Membranes were blocked, incubated with specific antibodies, and revealed by peroxidase-coupled secondary antibody using enzymatic chemiluminescence.
Combination evaluation
The antiproliferative effects of pyriplatin in combination with paclitaxel, gemcitabine, SN38, cisplatin, or 5-fluorouracil were investigated in the ovarian cancer line OVCAR-3 and the colon cancer line HT29. Combination studies were performed as described elsewhere (15, 16). Cells were seeded at 2 × 103 cells/well in 96-well plates and incubated for 24 h prior to treatment. The combination experiments were performed according to three different schedules. Cells were either treated with pyriplatin for 24 h followed by the combination drug for 24 h, treated with the combination drug for 24 h followed by pyriplatin for 24 h, or treated for 24 h with pyriplatin and the combination drug simultaneously. The concentrations of pyriplatin or the combination agent used ranged from the IC20 to IC60 concentrations. Antiproliferative effects were evaluated by the MTT assay and analyzed using the Chou and Talalay method which is based on the median-effect principle (17). A combination index (CI) of <1 indicates synergy, a value of 1 indicates additive effects, and a value >1 indicates antagonism. Data were analyzed using concentration-effect analysis CalcuSyn software (Biosoft, Cambridge, UK).
Measurement of platinum content
Cells were incubated for 2 or 24 h with 10 μM cisplatin, oxaliplatin, or pyriplatin. A time course of 2 h platinum exposure followed by a PBS wash and a 22-h incubation in platinum-free medium (2/22 schedule) was also evaluated. After trypsinization, cytosol and nuclei were separated in a hypotonic buffer. Cell lysis was performed in a buffer of 100 mM Tris (pH 7.5) 1 mM EDTA, 1 mM EGTA, 0.5 mM Na3VO4, 10 mM sodium β-glycerophosphate, 50 mM sodium fluoride, 5 mM sodium pyrophosphate and 1% Triton X-100. DNA was purified from nuclear extracts by phenol-chloroform extraction and ethanol precipitation. Cellular lysates, DNA samples and incubation medium were analyzed for platinum levels by inductively-coupled plasma mass spectrometry (ICP-MS).
Quantitative RT-PCR analysis
Cells were treated for 48-h with IC50 concentrations of drugs, followed by isolation of mRNA as described (18). Briefly, total RNA was reverse-transcribed before real-time quantitative PCR amplification using the ABI Prism 7900 Sequence Detection System (Perkin-Elmer Applied Biosystems). The transcripts of the gene coding for the TATA box-binding protein (TBP; a component of the DNA-binding protein complex TFIID) were used as the endogenous control RNA for normalization. Results were expressed as N-fold differences in target gene expression relative to the TBP gene. The mRNA expression of ERCC1, XPA, XPC, PARP1, XRCC1, RAD50, BRCA1, DNA-PK-cs, XRCC6, MSH2, MLH1, BCL2, PUMA, COX2, CDKN1A/p21, ABCB1, ABCC1, GSTP1 was evaluated in a panel of 10 cancer cell lines. Thermal cycling was performed with an initial denaturation step at 95 °C for 10 min, and 50 cycles of 15 s at 95 °C and 1 min at 65 °C. Experiments were performed in duplicate.
Statistical analysis
Statistical analyses were performed with Instat and Prism software (GraphPad, San Diego, USA). Results are expressed as the mean ± standard deviation of at least three experiments performed in duplicate. Means and standard deviations were compared using the Student's t-test (two-sided p value).
Results
Antiproliferative effects of single agent pyriplatin in a panel of human cancer cell lines
The potential anticancer activity of pyriplatin was tested at the National Cancer Institute using the NCI-60 tumor cell line panel screen. Results are shown in Supplementary Figure S1. Analysis of these results using the online COMPARE algorithm revealed that the antiproliferative profile of pyriplatin was not similar to those of cisplatin or oxaliplatin. The best correlation with a platinum compound in the NCI database was with “(carboxyphthalato) platinum” (NSC #S748451), with a correlation coefficient of 0.396. These data suggest that pyriplatin may have a mechanism of action that differs from classical platinum drugs.
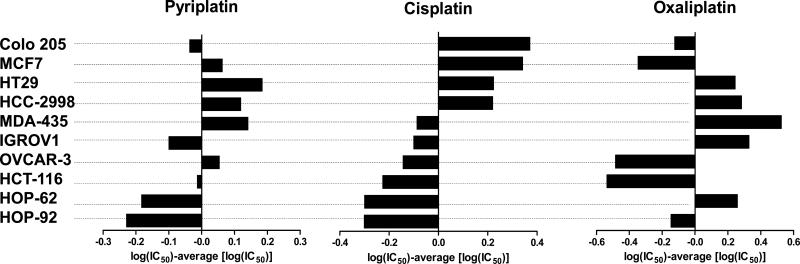
Pyriplatin was further evaluated in comparison with cisplatin and oxaliplatin using a well-characterized panel (19) of 10 cancer cell lines of different tissue origins (colorectal, breast, melanoma, ovarian, non-small cell lung). Cells were exposed for 24-h to pyriplatin (0.46-1000 μM), cisplatin (0.1-160 μM), or oxaliplatin (0.1-160 μM) and assessed for cytotoxicity by the MTT assay. Cell counting and the sulforhodamine B assay (data not shown) confirmed the results of the MTT-based antiproliferative assays. Table 1 shows the concentrations required to achieve 50% growth inhibition (IC50). The cytotoxicity profile for pyriplatin, shown in difference plots in Figure 2, was clearly different from those of both cisplatin and oxaliplatin.
Figure 2.
Difference plot of antiproliferative effects of pyriplatin, cisplatin, and oxaliplatin in a panel of cancer cell lines. The indicated values are calculated as follows: log (IC50 individual cell line) – mean (log IC50). Negative values indicate that the cell line is more sensitive than the average, whereas positive values indicate that the cell line is more resistant than the average.
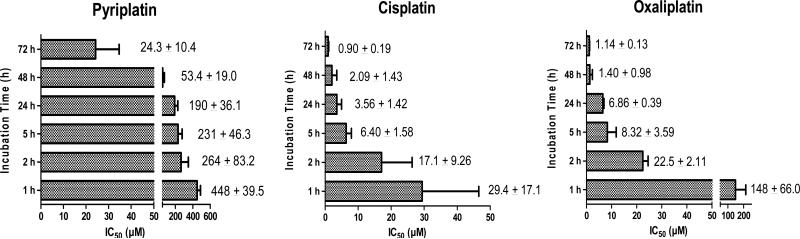
To study the effects of duration of pyriplatin exposure on cell proliferation, cells were treated for 1, 2, 5, 24, 48, or 72 h with pyriplatin, cisplatin or oxaliplatin, and then post-incubated for an additional 48- or 72-h period (as described above) in drug-free medium. Pyriplatin displayed dose- and time-dependent antiproliferative effects in HOP-62 cells, with 72 h being the most potent exposure duration (Figure 3). The IC50 of pyriplatin was only 15-fold higher than that of cisplatin at 1 and 2 h, but the difference between pyriplatin and cisplatin increased at 5 h (IC50 of pyriplatin was 36-fold that of cisplatin) and at 24 h (53-fold difference), suggesting that pyriplatin loses efficacy over time relative to cisplatin.
Figure 3.
Antiproliferative effects of pyriplatin, cisplatin, and oxaliplatin in HOP-62 cells over time. After 1, 2, 5, 24, or 72 h incubation, the cells were washed and post-incubated in platinum-free medium for 72 h (after 1, 2, or 5 h) or 48 (after 24 or 72 h). Cell viability was determined by the MTT assay. IC50 concentrations for different incubation times are shown as mean ± SD from at least three separate experiments.
Pyriplatin mechanism of action
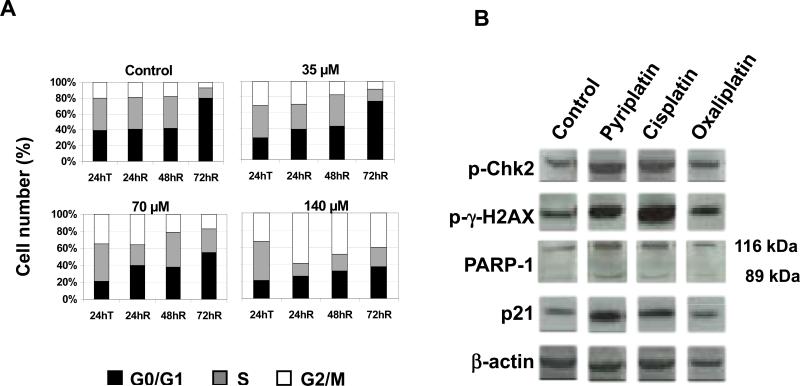
To investigate the mechanism of pyriplatin cytotoxicity, cell cycle analyses were performed in HCT-116 and MCF7 cell lines. All three platinum compounds caused dose-dependent progressive accumulation of cells in G2/M phase (see Figure 4A and Supplementary Figure S2). This block was apparent in both cell lines after a 24-h exposure to any of the three compounds, although oxaliplatin and pyriplatin induced only a slight G2/M block in MCF-7 cells. The cells were able to repair this block at 72 h after washout, except at high concentrations of platinum. In the case of pyriplatin in HCT-116 cells, the effect was reversible after treatment with 35 or 70 μM pyriplatin, but not after treatment with 140 μM.
Figure 4.
Pyriplatin-induced cell cycle changes. A) Cell cycle analysis in HCT-116 cells treated with increasing pyriplatin concentrations; T=immediately after drug exposure, R=after washout. B) Western blot of DNA damage and apoptosis-related signaling protein expression in HOP-62 cells after 24-h platinum treatment. β-actin was used as a loading control. Data are representative of 3 experiments.
In a next step, cells were stained with FITC-conjugated Annexin V for apoptosis detection and propidium iodide to detect necrosis prior to flow cytometric analysis. Annexin V binds to phosphatidylserine, a protein that is present in the cell membrane of apoptotic cells. Cells were treated for 24 h with IC50 platinum concentrations (Table 1). Apoptosis was determined immediately following 24 h drug exposure (24hT) and at 24 h (24hR), 48 h (48hR), and 72 h (72hR) after drug washout. At 24hT drug exposure, the percentage of apoptotic cells in pyriplatin, cisplatin, and oxaliplatin-treated cells was approximately twice the control values (Table 2). The percentage of apoptotic cells peaked at 24 h after drug washout for both cisplatin and pyriplatin-treated cells (24hR), and at 48h drug washout following oxaliplatin treatment (48hR). The detection of apoptosis 48 h after the start of exposure to cisplatin or 72 h after the beginning of incubation with oxaliplatin are in line with previously published results (20, 21). The peak of apoptosis in cells treated with pyriplatin corresponded to the maximal apoptotic response to cisplatin, suggesting that pyriplatin acts more quickly than oxaliplatin to induce cell death.
Table 2.
Apoptosis induction according to Annexin V staining in MCF7 cells treated with pyriplatin, cisplatin, or oxaliplatin followed by incubation in drug-free medium for 0, 24, 48, or 72 h. T=immediately after drug exposure, R=after wash-out. According to the experimental schedule, data are expressed as percentage of apoptotic cells.
| Control | Pyriplatin (25 μM) | Cisplatin (2 μM) | Oxaliplatin (0.4 μM) | |
|---|---|---|---|---|
| 24hT | 9.63% | 16.84% | 22.88% | 18.02% |
| 24hR | 21.40% | 51.70% | 48.70% | 21.94% |
| 48hR | 28.53% | 32.57% | 29.72% | 35.42% |
| 72hR | 22.26% | 16.66% | 13.98% | 16.76% |
To confirm apoptosis induction after pyriplatin exposure, its effects on the levels of H2AX and Chk2 phosphorylation, p21 expression, and poly (ADP-ribose) polymerase (PARP-1) cleavage were determined with Western blotting. As a downstream substrate of ATR, Chk2 is phosphorylated as part of the cellular response to cisplatin-induced DNA double-strand breaks (22). Phosphorylated H2AX (γ-H2AX) forms part of the repair complex that assembles at the site of DNA double-strand breaks and serves as a marker of DNA damage signaling (23). Cleavage of PARP-1 is observed in cells undergoing apoptosis (24) and produces two fragments of 89 and 24 kDa. Multiple roles of p21 and PARP-1 are also described in the context of DNA repair, regulation of cell cycle, apoptosis and gene transcription. In HOP-62 cells, the presence of DNA damage markers and apoptotic proteins was determined after treatment with pyriplatin, cisplatin, or oxaliplatin at IC50 concentrations. As shown in Figure 4B, γ-H2AX and Chk2 are detected following 24-h treatment with all three platinum compounds, and increases in p21 and slight PARP cleavage were seen after 24-hour pyriplatin exposure. Treatment with any of the three platinum compounds resulted in the appearance (Figure S3) of a band at 25 kDa corresponding to γ-H2AX, which was still present 72 hours after removal of the platinum-containing medium. The phosphorylation of these proteins suggests the formation of DNA double strand breaks and induction of apoptosis, a result that supports the apoptosis data obtained using the Annexin V assay.
Molecular determinants of pyriplatin activity
To evaluate platination of DNA after pyriplatin treatment, the platinum content was determined after exposure of HCT-116 cells to 10 μM cisplatin, oxaliplatin, or pyriplatin for 2, 6, 24, and 48 h. Data were also obtained from cells exposed to 2h treatment followed by a 22-h incubation in platinum-free medium. Table 3 and Figure S4 show platinum content on DNA purified from these cells, as measured by ICP-MS. After a 2-h exposure, pyriplatin induced 3.1- and 1.3-fold fewer DNA adducts than cisplatin and oxaliplatin, repectively. DNA platination induced by pyriplatin and oxaliplatin is decreased by 68-70% after the 22-h washout period, whereas cisplatin-treated cells showed higher levels of residual platinium bound to DNA. Specifically, levels decreased to only 40% after 22 h incubation in platinum-free medium. After 6 hof platinum exposure, DNA platination induced by pyriplatin was 2- and 3-fold less than DNA platination induced by oxaliplatin and cisplatin, respectively (data not shown). The differences in adduct formation increased at 24 h and again at 48 h, with cisplatin forming 4- and 25-fold more adducts than pyriplatin at 24 and 48 h, respectively, and oxaliplatin forming 6- and 48-fold more adducts.
Table 3.
DNA platinum content (ng Pt/mg DNA) after exposure to pyriplatin. HCT-116 cells were collected after 2-h platinum incubation, 2-h platinum incubation followed by 22-h in platinum-free medium, or 24-h platinum incubation. Extracted DNA was analyzed for platinum content by ICP-MS.
| 2 h platinum | 2 h platinum, 22 h washout | 24 h platinum | |
|---|---|---|---|
| pyriplatin | 2.44 | 0.74 | 3.71 |
| cisplatin | 7.49 | 4.48 | 15.31 |
| oxaliplatin | 3.07 | 1.00 | 23.81 |
After 2-h incubation with drug and 22 h without drug, cisplatin formed 6-fold more adducts and oxaliplatin formed 1.5-fold more adducts than pyriplatin. Compared with the 2-h data (i.e. minus the 22-h incubation without drug), the adduct levels and differences between pyriplatin and the other drugs are very similar, suggesting that no major adduct removal takes place by 24 h. Comparing the 2/22 schedule with the 24-h incubation, only slightly more pyriplatin-DNA adducts are observed at the 24-h mark, whereas significantly larger increases in both cisplatin-DNA and oxaliplatin-DNA adducts are observed. Pyriplatin may be inactivated in the cell at a greater rate than either cisplatin or oxaliplatin, rendering less pyriplatin available for binding over time.
The data indicate that pyriplatin forms fewer DNA adducts than oxaliplatin and cisplatin, which may play a role in the reduced cytotoxicity of pyriplatin relative to the two established drugs. On the other hand, although pyriplatin is 66- to over 200-fold less potent than cisplatin and oxaliplatin, respectively, at 24 h in HCT-116 cells, the difference in DNA adduct formation is not as stark, suggesting that each adduct of pyriplatin is less toxic than adducts of either cisplatin or oxaliplatin.
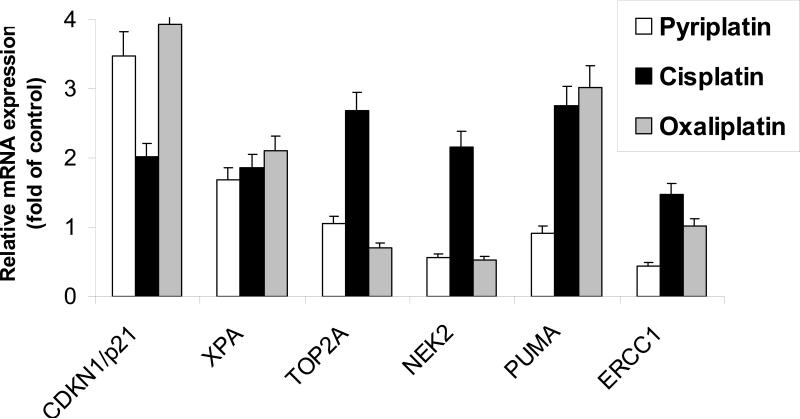
It was recently shown that exposure of colon cancer cells to oxaliplatin and cisplatin induced significant changes in expression of several genes implicated in drug transport, DNA repair, and cell cycle regulation (25). We compared the genetic effects induced by pyriplatin with those of oxaliplatin and cisplatin in the HCT-116 cell line. The mRNA levels encoded by selected genes involved in nucleotide excision repair (ERCC1, XPA, XPC), base excision repair (PARP1, XRCC1), homologous recombination (RAD50, BRCA1), mismatch repair (MSH2, MLH1), apoptosis (PUMA, CDKN1A/p21, COX2), transport (MDR1/ABCB1, ABCC1, GSTP1) and TOP2A, Ki67, and NEK2 were evaluated by RT-PCR after a 48-h exposure to pyriplatin, cisplatin, or oxaliplatin at IC50 concentrations. As shown in Figure 5 CDKN1/p21 mRNA expression was significantly induced following 48-h pyriplatin exposure (>3 fold). CDKN1/p21 mRNA levels were also induced after exposure to cisplatin and oxaliplatin. Pyriplatin also significantly decreased ERCC1 (2-fold decrease) expression. In contrast, exposure to cisplatin slightly increased the ERCC1 mRNA level (Figure 5), whereas exposure to oxaliplatin had little effect on ERCC1 mRNA. The mRNA levels of Ki67, ABCB1, XPA, TOP2A, MSH2, PUMA, RAD50, NEK2, and MLH1 was not significantly affected by pyriplatin exposure (Figure 5 and data not shown).
Figure 5.
Effects of pyriplatin, oxaliplatin, and cisplatin on gene expression. HCT116 cells were exposed to IC50 concentrations of pyriplatin, oxaliplatin and cisplatin for 48 hours. Relative mRNA gene expression of CDKN1/p21, XPA, TOP2A, NEK2, PUMA, ERCC1 was evaluated using quantitative RT-PCR. Data are representative of 3 experiments.
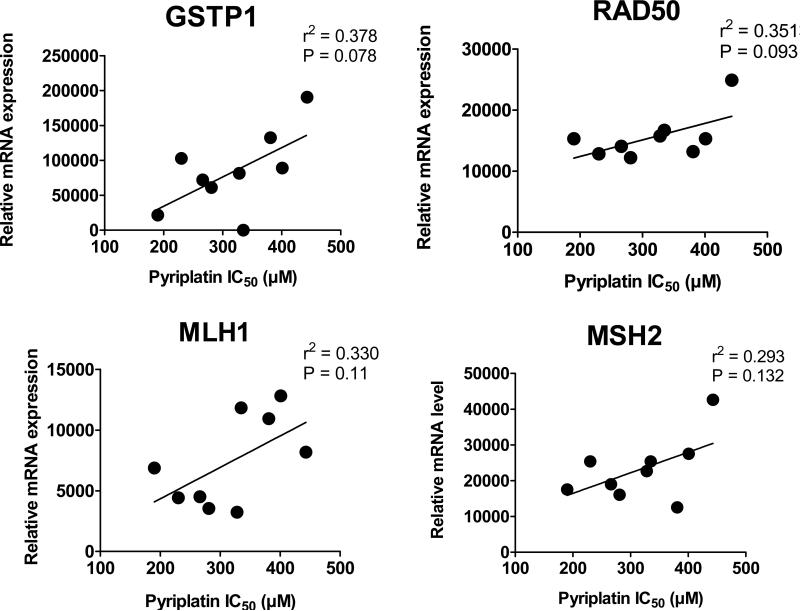
To study the potential predictive role of various genes associated with pyriplatin sensitivity, mRNA expression levels of 21 genes in the panel of 10 cell lines as measured by RT-PCR were plotted against pyriplatin IC50 values (Figure 6) to identify genes correlating closely with either resistance or sensitivity to pyriplatin. In this case, cells were not treated with platinum prior to RT-PCR analysis. Cells with high levels of RAD50 mRNA are more resistant to pyriplatin (r2 = 0.35), suggesting that double-strand breaks may play a role in the cellular consequences of pyriplatin-DNA lesions. Cells with high levels of mRNA coding for GSTP1 are also more resistant to pyriplatin (r2 = 0.38), indicating possible cellular inactivation of pyriplatin by modification with glutathione. Genetic factors GSTP1 and RAD50 are slightly correlated with sensitivity to pyriplatin and may serve as predictive factors of response.
Figure 6.
Correlation of pyriplatin cytotoxicity (IC50s) and mRNA expression levels of GSTP1, RAD50, MLH1 and MSH2 in a panel of 10 cancer cell lines.
Pyriplatin in combination with other anticancer agents
The effect of administering pyriplatin prior to, subsequently to, and simultaneously with five commonly used anticancer agents in the HT29 and OVCAR-3 cell lines was evaluated after 24-h exposure and interpreted using the Chou and Talalay method. Agents that had previously shown synergy in combination with platinum drugs were selected (15, 16), data were analyzed according to the method developed by Chou and Talalay, and combinatorial indices were calculated using the CalcuSyn program as described in the methods section. A CI of less than 1, indicating a synergistic effect, suggests that two drugs exert antiproliferative effects via separate mechanisms of action. Additive effects indicate that two drugs act via similar mechanisms of action.
A summary of pyriplatin-based combinations is shown in Table 4. Paclitaxel showed synergy (CI<1) when administered prior to pyriplatin in both cell lines. Similar to results for oxaliplatin and cisplatin (2), effects suggestive of synergy between pyriplatin and cisplatin were observed upon simultaneous addition of pyriplatin and cisplatin to cells (Table 4). Gemcitabine had antagonistic effects in both cell lines and with all schedules other than simultaneous exposure in HT29 cells.
Table 4.
Effects of pyriplatin in combination with various anticancer agents in HT29 and OVCAR-3 cancer cell lines. Data are presented as the median CI value and the 95% confidence interval. Schedule A: 24-h pyriplatin followed by the 24-h combination drug; Schedule B: 24-h combination drug followed by the 24-h pyriplatin; Schedule C: 24-h simultaneous exposure.
| Cell line | Schedule | Paclitaxel | Gemcitabine | SN38 | Cisplatin | 5-FU |
|---|---|---|---|---|---|---|
| HT29 | A | 0.87 (0.75-0.98) | 1.66 (0.70-2.76) | 0.90 (0.72-1.22) | 1.11 (0.89-1.23) | 0.94 (0.80-1.03) |
| B | 0.88 (0.69-1.22) | 1.09 (0.89-1.38) | 2.21 (1.42-3.31) | 0.96 (0.94-1.22) | ND | |
| C | 0.96 (0.55-1.21) | 0.86 (0.45-1.49) | 0.81 (0.62-1.34) | 0.84 (0.83-0.85) | 1.02 (0.87-1.20) | |
| OVCAR-3 | A | 0.89 (0.63-0.99) | 1.13 (0.69-1.91) | 1.08 (1.51-0.77) | 1.09 (0.96-1.19) | 0.99 (0.89-1.22) |
| B | 1.06 (0.63-1.55) | 1.31 (0.60-7.22) | 0.95 (0.94-0.97) | 0.84 (0.72-1.10) | ND | |
| C | 1.19 (0.87-1.38) | 1.21 (1.11-1.44) | 1.38 (1.07-1.99) | 0.79 (0.74-0.85) | 1.32 (1.22-1.55) | |
ND=not determined
Discussion
Platinum complexes are widely used in cancer therapy. The successful clinical applications of cisplatin, carboplatin, and oxaliplatin have inspired the synthesis and investigation of numerous platinum compounds as drug candidates. Of these compounds, those that show the most promise for clinical use have improved cytotoxicity, reduced side effects, or different mechanisms of action when compared with cisplatin, carboplatin, or oxaliplatin.
The cellular and molecular effects of the platinum derivative pyriplatin were investigated and directly compared to cisplatin and oxaliplatin. The pyriplatin cytotoxicity profile was demonstrated to be distinct from that of both cisplatin and oxaliplatin in a panel of 10 well-characterized cell lines and by the NCI single-dose screen. Although the IC50 values are 16 to 270 times higher for pyriplatin than for cisplatin or oxaliplatin, it is clear that the cell lines in which pyriplatin is most active (IGROV1, HOP-92, HOP-62, and COLO205, for example) are non-identical to those in which oxaliplatin is most active (HCT-116, OVCAR3, HOP-92, and MCF-7) or those in which cisplatin is most active (HCT-116, HOP-92, HOP-62, and OVCAR3), as shown in the difference plots in Figure 2 and in Table 1. The distinct cytotoxic profile of pyriplatin, coupled with a series of initial mechanistic studies, indicate that aspects of its mechanism of action are clearly different from those of either cisplatin or oxaliplatin. As is the case for cisplatin, the first cytotoxic effects of pyriplatin are seen as soon as one hour after the start of treatment. Relative to cisplatin and based on IC50 values in HOP-62 cells, pyriplatin is 15-fold less toxic than cisplatin after 1 or 2 h. This disparity increases at 5 and 24 h, possibly pointing to deactivation of pyriplatin over time by cellular and molecular mechanisms. IC50 values for pyriplatin decreased from 1 to 72 h, with efficacy peaking at an IC50 of 24 μM after 72 h in HOP-62 cells, the most sensitive cell line. At 1 and 2 h, pyriplatin activity contrasts with the relatively low activity of oxaliplatin, which must lose the oxalate prior to exerting cytotoxic action and is significantly less active after 1 or 2 h than after 5 h or longer.
Cell cycle studies indicate that, similar to cisplatin and oxaliplatin, pyriplatin induces a G2M block. Likewise, Annexin V staining and western blots showing activation of apoptosis-related proteins indicate that pyriplatin also displays an apoptotic mechanism of action, as occurs for cisplatin and oxaliplatin. The detection of apoptosis 48 h after initiating cisplatin exposure or 72 h after oxaliplatin exposure are in line with previously published results (20, 21). The peak of apoptosis in cells treated with pyriplatin corresponded to the maximal apoptotic response to cisplatin occurring 48 h after beginning treatment, suggesting that pyriplatin acts more quickly than oxaliplatin to induce cell death. An apoptotic response to pyriplatin was confirmed by the induction of H2AX phosphorylation.
When the effects of all three platinum compounds are compared at the same platinum concentration (10 μM), pyriplatin forms fewer DNA adducts than oxaliplatin or cisplatin after 24 h of treatment. However, after 2 h the levels of platinum per nucleotide are similar for pyriplatin and oxaliplatin. On the other hand, although pyriplatin is 66- to over 200-fold less potent than cisplatin and oxaliplatin respectively at 24 h in HCT-116 cells, the difference in DNA adduct formation is not as obvious, suggesting that pyriplatin adducts are less toxic than those of either cisplatin or oxaliplatin. Pyriplatin may be more susceptible to modification by cellular nucleophiles or inactivated more quickly than cisplatin or oxaliplatin, leaving less free pyriplatin available for binding DNA over longer times.
The cellular processing of platinum drugs involves a large number of cellular events that may play a role in the ultimate efficiency of these drugs: uptake and efflux, DNA adduct formation, recognition and repair of adducts, and signal transduction of DNA damage inducing cell death. In terms of molecular determinants of pyriplatin sensitivity, GSTP1, RAD50, MLH1, and MSH2 gene expression was higher in pyriplatin-resistant cell lines suggesting that MMR, homologous recombination, and detoxification systems participate in cellular response to pyriplatin. A slight correlation seen with GSTP1 expression may indicate possible cellular inactivation of pyriplatin by glutathione modification. Genetic factors GSTP1 and RAD50 may serve as predictive factors of pyriplatin response.
Previously it was shown (25) that exposure of colon cancer cells to cisplatin and oxaliplatin can induce expression of several genes implicated in drug transport, DNA repair, and cell cycle regulation. We compared the genetic effects induced by pyriplatin with those induced by cisplatin and oxaliplatin in HCT-116 cells. Significant increases in p21 expression were seen for all three platinum compounds whereas ERCC1 expression decreased in response to pyriplatin and increased in response to cisplatin exposure. The difference in ERCC1 expression, coupled with previous results showing that pyriplatin-DNA adducts evade repair by the nucleotide excision repair pathway as compared with repair of cisplatin-DNA adducts (26), supports the case that differential repair of cisplatin and pyriplatin adducts contribute to the different activity in our cell line panel.
The potential for use of pyriplatin in combination with other anti-cancer compounds was explored in terms of the antiproliferative potential of paclitaxel, gemcitabine, SN38, cisplatin, and 5-FU combinations. Interestingly, in both cell lines tested, pyriplatin was synergistic when administered simultaneously with cisplatin, as is seen with oxaliplatin (2). These results suggest a molecular mechanism of action distinct to that of cisplatin.
In conclusion, although pyriplatin is not likely to be developed due to its low cytotoxicity, it remains a promising lead compound for the generation of novel drug candidates with different cytotoxicity profiles from those of platinum drugs currently in use.
Supplementary Material
Supplementary data
Figure S1. NCI-60 10 μM pyriplatin single dose growth data.
Figure S2. Effect of increasing pyriplatin concentrations on the cell cycle in A) HCT-116 and B) MCF7 cells, by flow cytometry. T=immediately after drug exposure, R=after wash-out.
Figure S3. Level of γ-H2AX after 24-h treatment of HOP-62 cells with platinum followed by incubation in drug-free medium for 0, 24, 48, or 72 h. The α-tubulin was used as a loading control.
Figure S4. DNA platinum content after exposure to pyriplatin. HCT-116 cells were collected after 2-h platinum incubation, 2-h platinum incubation, followed by 22-h in platinum-free medium, and 24-h platinum incubation. Extracted DNA was analyzed for platinum content by ICP-MS.
Acknowledgements
We acknowledge Dr Sarah MacKenzie for assistance in preparation of this manuscript. Work in the lab of S.J. Lippard is supported by a grant from the US National Cancer Institute (CA34992). Data collected by ICP-MS was measured at the Salvatore Maugeri Foundation, Pavia, Italy.
Financial Support: Work in the lab of S.J. Lippard is supported by a grant from the US National Cancer Institute (CA034992).
Abbreviations
- cDPCP
cis-diammine(pyridine)chloroplatinum(II)
- ICP-MS
inductively coupled plasma mass spectrometry
- MMR
mismatch repair
- PARP
poly (ADP-ribose) polymerase
- TBP
TATA box-binding protein
References
- 1.Shoemaker RH. The NCI60 human tumour cell line anticancer drug screen. Nat Rev Cancer. 2006;6:813–23. doi: 10.1038/nrc1951. [DOI] [PubMed] [Google Scholar]
- 2.Rixe O, Ortuzar W, Alvarez M, Parker R, Reed E, Paull K, et al. Oxaliplatin, tetraplatin, cisplatin, and carboplatin: spectrum of activity in drug-resistant cell lines and in the cell lines of the National Cancer Institute's Anticancer Drug Screen panel. Biochem Pharmacol. 1996;52:1855–65. doi: 10.1016/s0006-2952(97)81490-6. [DOI] [PubMed] [Google Scholar]
- 3.Roberts JD, Peroutka J, Farrell N. Cellular pharmacology of polynuclear platinum anti-cancer agents. J Inorg Biochem. 1999;77:51–7. doi: 10.1016/s0162-0134(99)00147-6. [DOI] [PubMed] [Google Scholar]
- 4.Farrell N, Qu Y, Bierbach U, Valsecchi M, Menta E. Structure-activity relationships within di- and trinuclear platinum phase-I clinical anticancer agents. In: Lippert B, editor. Cisplatin: Chemistry and Biochemistry of a Leading Anticancer Drug. Wiley-VCH; Weinheim, Germany: 1999. pp. 479–96. [Google Scholar]
- 5.Hall MD, Okabe M, Shen D-W, Liang X-J, Gottesman MM. The role of cellular accumulation in determining sensitivity to platinum-based chemotherapy. Annu Rev Pharmacol Toxicol. 2008;48:495–535. doi: 10.1146/annurev.pharmtox.48.080907.180426. [DOI] [PubMed] [Google Scholar]
- 6.Molenaar CT,J, Heetebrij RJ, Tanke HJ, Reedijk J. New insights in the cellular processing of platinum antitumor compounds using fluorophore-labeled platinum complexes and digital fluorescence microscopy. J Biol Inorg Chem. 2000;5:655–65. doi: 10.1007/s007750000153. [DOI] [PubMed] [Google Scholar]
- 7.Barnes KR, Kutikov A, Lippard SJ. Synthesis, characterization, and cytotoxicity of a series of estrogen-tethered platinum(IV) complexes. Chem Biol. 2004;11:557–64. doi: 10.1016/j.chembiol.2004.03.024. [DOI] [PubMed] [Google Scholar]
- 8.Hall MD, Mellor HR, Callaghan R, Hambley TW. Basis for Design and Development of Platinum(IV) Anticancer Complexes. J Med Chem. 2007;50:3403–11. doi: 10.1021/jm070280u. [DOI] [PubMed] [Google Scholar]
- 9.Reisner E, Arion VB, Keppler BK, Pombeiro AJL. Electron-transfer activated metal-based anticancer drugs. Inorg Chim Acta. 2008;361:1569–83. [Google Scholar]
- 10.Mukhopadhyay S, Barnes CM, Haskel A, Short SM, Barnes KR, Lippard SJ. Conjugated platinum(IV)-peptide complexes for targeting angiogenic tumor vasculature. Bioconjug Chem. 2008;19:39–49. doi: 10.1021/bc070031k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hollis LS, Amundsen AR, Stern EW. Chemical and biological properties of a new series of cis-diammineplatinum(II) antitumor agents containing three nitrogen donors: cis-[Pt(NH3)2(N-donor) Cl]+. J Med Chem. 1989;32:128–36. doi: 10.1021/jm00121a024. [DOI] [PubMed] [Google Scholar]
- 12.Zhang S, Lovejoy KS, Shima JE, Lagpacan LL, Shu Y, Lapuk A, et al. Organic cation transporters are determinants of oxaliplatin cytotoxicity. Cancer Res. 2006;66:8847–57. doi: 10.1158/0008-5472.CAN-06-0769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang D, Zhu G, Huang X, Lippard SJ. X-ray structure and mechanism of RNA polymerase II stalled at an antineoplastic monofunctional platinum-DNA adduct. Proc Natl Acad Sci USA. 2010;107:9584–9. doi: 10.1073/pnas.1002565107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hansen MB, Nielsen SE, Berg K. Re-examination and further development of a precise and rapid dye method for measuring cell growth/cell kill. J Immunol Methods. 1989;119:203–10. doi: 10.1016/0022-1759(89)90397-9. [DOI] [PubMed] [Google Scholar]
- 15.Serova M, Calvo F, Lokiec F, Koeppel F, Poindessous V, Larsen AK, et al. Characterizations of irofulven cytotoxicity in combination with cisplatin and oxaliplatin in human colon, breast, and ovarian cancer cells. Cancer Chem Pharmacol. 2006;57:491–9. doi: 10.1007/s00280-005-0063-y. [DOI] [PubMed] [Google Scholar]
- 16.Raymond E, Buquet-Fagot C, Djelloul S, Mester J, Cvitkovic E, Allain P, et al. Antitumor activity of oxaliplatin in combination with 5-fluorouracil and the thymidylate synthase inhibitor AG337 in human colon, breast and ovarian cancers. Anti-Cancer Drugs. 1997;8:876–85. doi: 10.1097/00001813-199710000-00009. [DOI] [PubMed] [Google Scholar]
- 17.Chou TC, Talalay P. Quantitative analysis of dose-effect relationships: the combined effects of multiple drugs or enzyme inhibitors. Adv Enzyme Regul. 1984;22:27–55. doi: 10.1016/0065-2571(84)90007-4. [DOI] [PubMed] [Google Scholar]
- 18.Bieche I, Parfait B, Tozlu S, Lidereau R, Vidaud M. Quantitation of androgen receptor gene expression in sporadic breast tumors by real-time RT-PCR: evidence that MYC is an AR-regulated gene. Carcinogenesis. 2001;22:1521–6. doi: 10.1093/carcin/22.9.1521. [DOI] [PubMed] [Google Scholar]
- 19.Serova M, Galmarini CM, Ghoul A, Benhadji K, Green SR, Chiao J, et al. Antiproliferative effects of sapacitabine (CYC682), a novel 2'-deoxycytidine-derivative, in human cancer cells. Br J Cancer. 2007;97:628–36. doi: 10.1038/sj.bjc.6603896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gatti L, Supino R, Perego P, Pavesi R, Caserini C, Carenini N, et al. Apoptosis and growth arrest induced by platinum compounds in U2-OS cells reflect a specific DNA damage recognition associated with a different p53-mediated response. Cell Death Differ. 2002;9:1352–9. doi: 10.1038/sj.cdd.4401109. [DOI] [PubMed] [Google Scholar]
- 21.Sharma RI, Smith TA. Colorectal tumor cells treated with 5-FU, oxaliplatin, irinotecan, and cetuximab exhibit changes in 18F-FDG incorporation corresponding to hexokinase activity and glucose transport. J Nucl Med. 2008;49:1386–94. doi: 10.2967/jnumed.107.047886. [DOI] [PubMed] [Google Scholar]
- 22.Pabla N, Huang S, Mi Q-S, Daniel R, Dong Z. ATR-Chk2 Signaling in p53 Activation and DNA Damage Response during Cisplatin-induced Apoptosis. J Biol Chem. 2008;283:6572–83. doi: 10.1074/jbc.M707568200. [DOI] [PubMed] [Google Scholar]
- 23.Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem. 1998;273:5858–68. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- 24.Kaufmann SH, Desnoyers S, Ottaviano Y, Davidson NE, Poirier GG. Specific proteolytic cleavage of poly(ADP-ribose) polymerase: an early marker of chemotherapy-induced apoptosis. Cancer Res. 1993;53:3976–63. [PubMed] [Google Scholar]
- 25.Voland C, Bord A, Peleraux A, Penarier G, Carriere D, Galiegue S, et al. Repression of cell cycle-related proteins by oxaliplatin but not cisplatin in human colon cancer cells. Mol Cancer Ther. 2006;5:2149–57. doi: 10.1158/1535-7163.MCT-05-0212. [DOI] [PubMed] [Google Scholar]
- 26.Lovejoy KS, Todd RC, Zhang S, McCormick MS, D'Aquino JA, Reardon JT, et al. cis-diammine(pyridine)chloroplatinum(II), a monofunctional platinum(II) antitumor agent: uptake, structure, function, and prospects. Proc Natl Acad Sci USA. 2008;105:8902–7. doi: 10.1073/pnas.0803441105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data
Figure S1. NCI-60 10 μM pyriplatin single dose growth data.
Figure S2. Effect of increasing pyriplatin concentrations on the cell cycle in A) HCT-116 and B) MCF7 cells, by flow cytometry. T=immediately after drug exposure, R=after wash-out.
Figure S3. Level of γ-H2AX after 24-h treatment of HOP-62 cells with platinum followed by incubation in drug-free medium for 0, 24, 48, or 72 h. The α-tubulin was used as a loading control.
Figure S4. DNA platinum content after exposure to pyriplatin. HCT-116 cells were collected after 2-h platinum incubation, 2-h platinum incubation, followed by 22-h in platinum-free medium, and 24-h platinum incubation. Extracted DNA was analyzed for platinum content by ICP-MS.