Abstract
Autophagy is a homeostatic, catabolic degradation process whereby cellular proteins and organelles are engulfed into autophagosomes, digested in lysosomes and recycled to sustain cellular metabolism. Autophagy has dual roles in cancer, acting as both a tumor suppressor by preventing the accumulation of damaged proteins and organelles and as a mechanism of cell survival that can promote the growth of established tumors. Tumor cells activate autophagy in response to cellular stress including hypoxia and increased metabolic demands related to rapid cell proliferation. Autophagy-related stress tolerance can enable cell survival by maintaining energy production that can lead to tumor growth and therapeutic resistance, as shown in preclinical models where the inhibition of autophagy can restore chemosensitivity and enhance tumor cell death. These results established autophagy as a therapeutic target and have led to multiple early phase clinical trials in humans evaluating autophagy inhibition using hydroxychloroquine in combination with chemotherapy or targeted agents. Targeting autophagy in cancer provides new opportunities for drug development since more potent and specific inhibitors of autophagy are needed. The role of autophagy and its regulation in cancer cells continues to emerge and studies aim to define optimal strategies to modulate autophagy for therapeutic advantage.
Introduction
Macroautophagy (hereafter referred to as autophagy) is a homeostatic and evolutionarily conserved process that degrades cellular organelles and proteins and maintains cellular biosynthesis during nutrient deprivation or metabolic stress (1). Autophagy begins with the formation of double membrane vesicles, known as autophagosomes, that engulf cytoplasmic constituents. The autophagosomes then fuse with lysosomes where the sequestered contents undergo degradation and recycling (1). Autophagy is important in all cells for the removal of damaged or long-lived proteins and organelles. Autophagy defects are associated with susceptibility to metabolic stress, genomic damage, and tumorigenesis in mice indicating a role for autophagy in tumor suppression (2). Monoallelic loss of the essential autophagy gene, Beclin 1, has been found in 40 to 75% of human breast, prostate and ovarian cancers (2), suggesting that autophagy may play a role in preventing these tumors. While autophagy is a mechanism of tumor suppression, it confers stress tolerance that enables tumor cell survival under adverse conditions (1). Stress in tumor cells is compounded by the high metabolic demand associated with rapid cell proliferation. Autophagy is induced in tumor cells within hypoxic tumor regions (3). Stress-induced autophagy in tumor cells can lead to treatment resistance and tumor dormancy with eventual tumor regrowth and progression (4). In preclinical models, inhibition of pro-survival autophagy by genetic or pharmacological means can kill tumor cells and trigger apoptotic cell death (1, 5–9). Furthermore, autophagy inhibitors given in combination with chemotherapy suppressed tumor growth and triggered cell death to a greater extent than did chemotherapy alone both in vitro and in vivo (Table 1). These data indicate that pro-survival autophagy may represent a major impediment to successful cancer therapy and thus, represents a novel therapeutic target. However, autophagy has been referred to as a double-edge sword since in certain cellular contexts, excessive or sustained tumor cell autophagy may be pro-death, particularly in apoptosis-defective cells (10). Understanding the role of autophagy in cancer treatment is critical since many anti-cancer therapies have been shown to activate autophagy, although the consequences of this autophagy activation in this context are unclear. The complex role of autophagy in cancer continues to emerge and elucidating the mechanisms by which autophagy influences tumorigenesis as well as treatment response are critical. Analysis of autophagic signaling may identify novel therapeutic targets for modulation and therapeutic advantage. In this review, the multiple roles of autophagy in tumor biology will be outlined including its emergence as a therapeutic target for both cancer prevention and therapy.
Table 1.
Pre-clinical and ongoing clinical studies using autophagy inhibitors, chloroquine and hydroxychloroquine in cancer treatment
| Tumor Type | Development Status | Therapeutic Combination |
|---|---|---|
| Colorectal cancer | In vitro, in vivo Phase II |
CQ + bortezomib (63) CQ + vorinostat (56) HCQ + XELOX + bevacizumab |
| Gastrointestinal stromal tumor | In vitro, in vivo | CQ + imatinib (75) |
| Prostate cancer | In vitro, in vivo | CQ + Src kinase inhibitors (57) |
| Vulvar cancer | In vitro | CQ + cetuximab (34) |
| Chronic myelogenous leukemia | In vitro Phase II |
CQ + vorinostat (73) HCQ + imatinib |
| Lymphoma | In vivo | CQ + cyclophosphamide (27) |
| Pancreatic cancer | Phase II Phase I/II |
HCQ only HCQ + gemcitabine |
| Prostate cancer | Phase II | HCQ + docetaxel |
| Lung cancer | Phase II | HCQ + erlotinib |
| Glioblastoma multiforme | Phase I/II | HCQ + temozolomide + radiation |
| Multiple myeloma | Phase I/II | HCQ + bortezomib |
| Renal cell carcinoma | Phase I | HCQ only |
| Breast cancer | Phase II | HCQ only |
| Chronic lymphocytic leukemia | Phase II | HCQ only |
| Advanced solid tumor | Phase I Phase I Phase I Phase I |
HCQ + sirolimus or vorinostat HCQ + temsirolimus HCQ + sunitinib HCQ + temozolomide |
Regulation of Autophagy
Autophagy involves the formation of the autophagosome that assembles around and encapsulate damaged organelles or cellular debris and then fuses with the lysosome to degrade its contents (11). The initiation of autophagy is controlled by the ULK1 (human homolog of ATG1) kinase complex (consists of ULK1, Atg13, and Atg17) that integrates stress signals from the mTOR complex 1 (mTORC1) (12, 13). When mTORC1 kinase activity is inhibited, autophagosome formation can occur and involves vacuolar sorting protein 34 (Vps34), a class III phosphoinositide 3-kinase (PI3K), that forms a complex with Beclin 1 (11). The production of PtIns3P by the Beclin 1/Vps34 complex is essential for the recruitment other autophagy-related gene (Atg) products that are critical for autophagosome formation (11). During the initiation phase, formation of Atg5-Atg12-Atg16 complex promotes the recruitment and conversion of cytosolic-associated protein light chain 3 (LC3-I) to the membrane-bound, lipidated form LC3-II(14). LC3 is conjugated to PE and incorporated into the membrane by an Atg7- and Atg3-dependent activation and transfer cascade that follows cleavage of LC3 by the cysteine protease Atg4 (15). Upon completion of autophagosome formation and with the exception of a proportion of LC3-II bound to the luminal membrane, the Atg proteins are then recycled in the cytosol (11). LC3-II remains on mature autophagosomes until after fusion with lysosomes and is commonly used to monitor autophagy. LC3-II also binds to the adaptor protein p62/sequestosome1 (SQSTM 1) that is involved in trafficking proteins to the proteasome and serves to facilitate the autophagic degradation of ubiquitinated protein aggregates (16). p62/SQSTM 1 is normally degraded during autophagy and accumulates when autophagy is impaired, as has also been shown in autophagy-deficient mice (17). The late events in autophagy involve the final maturation and fusion of autophagosomes with lysosomes to form an autolysosome, a step requiring small Rab GTPases, and lysosome-associated membrane protein 2 (Lamp2) (18, 19).
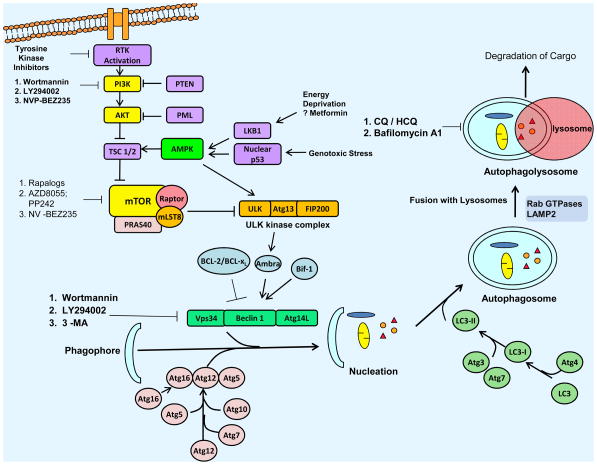
A major regulator of autophagy is the mammalian target of rapamycin (mTOR) pathway that consists of two distinct signaling complexes, known as mTORC1 and mTORC2 (11). mTOR is activated downstream of PI3K-AKT, a pathway commonly dysregulated in human cancer (20) (Figure 1). Activation of mTOR can also occur due to loss of tumor suppressors (LKB1, PML, PTEN, TSC1/2) or through gain-of-function mutations in receptor tyrosine kinases (21). Cellular stress leads to downregulation of mTOR1 activity that triggers autophagy (11) and in this regard, mTOR inhibitors including rapamycin have been shown to induce autophagy in tumor cells (20). mTOR negatively regulates autophagy by causing phosphorylation of Atg13 that reduces its interaction with ULK1 and inhibits formation of a trimeric complex required for autophagosome formation (12). A decrease in intracellular energy also results in activation of adenosine monophosphate kinase (AMPK) that is a central metabolic sensor with important functions in regulating lipid and glucose metabolism. Activation of AMPK serves to repress mTOR and initiate autophagy (12). A recent study found that AMPK can directly phosphorylate ULK1 that is required for mitochondrial homeostasis and cell survival during starvation (22). Autophagy can also be induced by hypoxia which is regulated through various mechanisms including HIF (hypoxia-inducible factor) (21).
Figure 1. Overview of the Autophagy Pathway.
The initiation of autophagy is controlled by the ULK1 kinase complex that integrates stress signals from mTORC1. When mTORC1 kinase activity is inhibited, autophagosome formation can occur from the phagophore and involves vacuolar sorting protein 34 (Vps34), a class III phosphoinositide 3-kinase (PI3K), that forms a complex with Beclin 1. Beclin 1 interacts with factors (Ambra, Bif1, Bcl-2) that modulate its binding to Vps34 whose lipid kinase activity is essential for autophagy. In addition to these two complexes, autophagosome formation requires the participation of two ubiquitin-like protein (Atg12 and LC3) conjugation systems that are essential for the formation of the phagophore. In addition, the LC3 system is required for autophagosome transport and maturation. Mature autophagosomes fuse their external membranes with those from lysosomes to degrade their cargo, and recycle essential biomolecules. Autophagy can be inhibited by drugs that target early or late stages in the pathway. Chloroquine (CQ) and hydroxychloroquine (HCQ) inhibit autophagy at a late stage by blocking lysosomal acidification resulting in an inability to digest its engulfed cargo.
Autophagy can be potently induced by the unfolded protein response (UPR), a component of the endoplasmic reticulum (ER) stress pathway. The binding of misfolded proteins to the ER chaperone Bip/GRP78 leads to release of three ER membrane-associated proteins, PKR-like eIF2α kinase (PERK), activating transcription factor-6 (ATF6), and inositol-requiring enzyme 1 (IRE1) (23). While PERK and ATF6 are autophagy inducers, IRE1 negatively regulates autophagy. Other factors that link cellular stress with autophagy is the transcription factor NF-kappa β and its upstream regulators IKK complex and TAK1 that integrate diverse stress signals including starvation or ER stress with the autophagy pathway (24).
The tumor suppressor p53 protein can modulate autophagy depending upon its cellular localization. Nuclear p53 acts as a transcription factor that transactivates several autophagy inducers including DRAM1 and Sestrin2 to activate autophagy (25), whereas cytoplasmic p53 inhibits autophagy by an unknown mechanism. Inducers of autophagy can stimulate proteosome-mediated degradation of p53 (26). Recently, some novel regulators of autophagy have been found. Ataxia-telangiectasia mutated (ATM) is a cellular damage sensor that coordinates the cell cycle with DNA damage-response checkpoints and DNA repair, and engages the TSC2/mTORC1 signaling axis to regulate autophagy (27). Additionally, the high mobility group box 1 (HMGB1) is an immune modulator and regulator of stress-induced autophagy that directly interacts with Beclin 1 (28).
Role of Autophagy in Cancer
Autophagy in tumor suppression
Autophagy is a homeostatic mechanism that when disrupted, can promote and accelerate tumorigenesis. Autophagy functions as a tumor suppression mechanism by removing damaged organelles/proteins and limiting cell growth and genomic instability (17). Beclin 1 is a protein required for autophagy induction and Beclin 1+/− mice were shown to be tumor-prone indicating that Beclin 1 is a haploinsufficient tumor suppressor gene (2). In contrast, excessive stimulation of autophagy due to Beclin 1 overexpression can inhibit tumor development (29). A potential molecular link between defective autophagy and tumorigenesis involves the accumulation of p62/SQSTM 1 protein aggregates, damaged mitochondria, and misfolded proteins that lead to the production of reactive oxygen species (ROS) to cause DNA damage that can lead to genomic instability (17). Knockdown of p62/SQSTM 1 in autophagy defective cells prevented ROS and the DNA damage response (17). The relationship between defective autophagy and p62/SQSTM 1 accumulation with tumorigenesis was also shown in p62/SQSTM 1−/− mice were protected from Ras-induced lung carcinomas compared to wild-type animals (30). Autophagy may also protect against tumorigenesis through limiting necrosis and chronic inflammation that are associated with the release of pro-inflammatory HMGB1 (28). Together, these findings establish a role for autophagy as a mechanism of tumor suppression.
Autophagy in Established Tumors
Evidence indicates that the predominant role of autophagy in cancer cells is to confer stress tolerance that can maintain tumor cell survival (1). Knockdown of essential autophagy genes in tumor cells has been shown to confer or potentiate the induction of cell death (7). Cancer cells have high metabolic demands due to increased cellular proliferation and in vivo models, exposure to metabolic stress impairs survival in autophagy-deficient in contrast to autophagy-proficient cells (17). Cytotoxic and metabolic stress, including hypoxia or nutrient deprivation, can activate autophagy for recycling of ATP and to maintain cellular biosynthesis and survival. Autophagy is induced in hypoxic tumor cells from regions that are distal to blood vessels and can be enhanced by HIF-1α expression, and HIF-1α can also increase expression of angiogenic factors such as vascular endothelial growth factor (VEGF), platelet derived growth factor (PDGF), and nitric oxide synthase (NOS) (21). Increased basal levels of autophagy were detected in human pancreatic cancer cell lines and tumor specimens, and were shown to enable tumor cell growth by maintaining cellular energy production. Inhibition of autophagy in these cells led to tumor regression and extended survival in pancreatic cancer xenografts and genetic mouse models (31). In cancer cells surviving chemotherapy and/or radiation, activation of autophagy may enable a state of dormancy in residual cancer cells that may contribute to tumor recurrence and progression (4). Inhibition of autophagy in tumor cells has been shown to enhance the efficacy of anti-cancer drugs (Table 1), supporting its role in cytoprotection, as discussed below.
Recent data indicate that human cancer cell lines bearing activating mutations in H-ras or K-ras have high basal levels of autophagy even in the presence of abundant nutrients (32). In a subset of these cells, suppression of essential autophagy proteins was shown to inhibit cell growth, indicating that autophagy maintains tumor cell survival and suggesting that blocking autophagy in tumors ‘addicted to’ autophagy, such as Ras- driven cancers, may be an effective treatment approach.
Autophagy as a Mechanism of Cell Death
In contrast to the cytoprotective function of autophagy that is supported by abundant evidence, induction of autophagic cell death has also been proposed as a mechanism of cell death given that features of autophagy have been observed in dying cells. In cancer cells, autophagy accompanied by non-apoptotic cell death has been described (33, 34). Prolonged stress and sustained autophagy may eventually lead to cell death when protein and organelle turnover overwhelm the capacity of the cell. Induction of autophagic cell death by anti-cancer drugs may occur depending upon the cell type and genetic background. In VHL-deficient renal cell carcinoma cells, a novel small molecule (STF-62247) was shown to promote cell death through induction of autophagy (35). However, in vivo evidence is limited and whether induction of autophagic death in tumor cells can be achieved for cancer therapy remains unknown.
Autophagy and Cell Senescence
Autophagy and senescence represent two distinct cellular responses to stress that can serve as tumor suppressors. Recently, autophagy was shown to mediate Ras oncogene-induced senescence (36). Cellular senescence represents a state of cell cycle arrest maintained by the expression of cell cycle inhibitors (p16Ink4a, p21Cip1, p27Kip1) in metabolically viable cells (37). The senescence phenotype can be induced by oncogenes, DNA damaging drugs or oxidative stress, and their ability to induce senescence is enhanced by functional p53 and Rb tumor suppressor genes (37). Senescence has been suggested as a mechanism for autophagy-mediated tumor dormancy (4, 38). Conversely, the inhibition of autophagy in tumor cells was shown to delay the senescence phenotype (36). A subset of autophagy-related genes (ULK1, ULK3) are up-regulated during senescence and overexpression of ULK3 was shown to induce autophagy and senescence (36).
Autophagy Modulation for Cancer Therapy
Autophagy inducers
Conventional cytotoxic drugs and irradiation have been shown to induce autophagy (5, 39–43). Other anti-cancer drugs that can induce autophagy include the BCR-ABL tyrosine kinase inhibitor imatinib (44), the anti-EGFR (epidermal growth factor receptor) cetuximab (45), proteosome inhibitors (46), TRAIL (TNF-related apoptosis–inducing ligand) (47), and the HDAC inhibitors vorinostat (SAHA; suberoylanilide hydroamic acid) and OSU-HDAC42 (48). Arsenic trioxide was shown to induce autophagy in leukemia and glioma cells that was regulated by the mitochondrial stress sensor, BNIP3, in malignant glioma (33, 49). Furthermore, agents with diverse mechanisms of action have also been shown to induce autophagy in tumor cells including tamoxifen (50), cyclooxygenase inhibitors (51) and the protease inhibitor, nelfinavir (52). While the consequence of promoting autophagy in tumor cells is incompletely understood and may depend upon multiple factors including extent of induction, duration, and cellular context. Excessive or sustained autophagy has the potential to induce tumor cell death (35), however, the relevance of this finding to the in vivo situation is unknown. mTOR is a central coordinator of cell growth that is involved in both protein translation and autophagy. Rapamycin is a naturally occurring allosteric mTOR inhibitor and its analogs temsirolimus (CCI-779), everolimus (RAD-001), and deforolimus (AP-23573) selectively target mTORC1 to stimulate autophagy. With the exception of renal cell and neuroendocrine carcinomas and lymphoma, rapamycin and its analogs (rapalogs) have had limited success in the clinical setting (53). Rapamycin or rapalogs do not inhibit mTORC2 and are unable to abrogate the S6K-IRS1-mediated negative feedback loop that can result in rebound AKT activation (54). These limitations led to the development of ATP-competitive inhibitors of both mTORC1 and mTORC2 (e.g. Torin1, PP242, AZD8055, WYE132) and the dual PI3K-mTOR inhibitor NVP-BEZ235. In preclinical studies, dual inhibitors of mTORC1 and mTORC2 demonstrate anti-tumor activity (55–57) and have been shown to be more potent inducers of autophagy (58, 59). Of note, the dual PI3K-mTOR inhibitor, PI-103 was shown to induce autophagy in glioma cells and inhibitors of autophagy cooperated with PI-103 to induce apoptosis. . Furthermore, the PI3K-mTOR inhibitor NVP-BEZ235, which is in clinical trials, synergized with chloroquine to induce apoptosis in glioma xenografts (60). These data indicate that induction of pro-survival autophagy by these dual inhibitors may antagonize their anti-tumor effects.
The anti-diabetic, biguanide drug metformin has been shown to inhibit mTOR signaling through its upstream mediator, AMPK and has a cytostatic effect in certain cancer cell types (61). While metformin was shown to induce autophagy in colon cancer cells (62), it inhibited 2-deoxyglucose-induced autophagy, decreased Beclin 1 expression, and triggered a switch from cell survival to cell death in prostate cancer cells (61). This finding is inconsistent with the ability of metformin to activate AMPK that is expected to induce autophagy. Other autophagy stimulators include the selective serotonin reuptake inhibitor, fluoxetine, the norepinephrine reuptake inhibitor, maprotiline (63), and the anti-epileptic drug valproic (64). Using cell-based screening assays, the anti-hypertensive drugs verapamil, minoxidil and clonidine were found to induce autophagy through an mTOR-independent pathway involving calpain (65).
Autophagy inhibitors
Multiple studies have shown that genetic knockdown of autophagy-related genes (Atgs) or pharmacological inhibition of autophagy can effectively enhance tumor cell death induced by diverse anti-cancer drugs in preclinical models (7–9)(Table 1). Inhibition of autophagy in preclinical models improves response to alkylating agents in tumor cells (5). In apoptosis-defective leukemic and colon cancer cell lines, inhibition of autophagy was shown to sensitize resistant cells to TRAIL-mediated apoptosis (47). Furthermore, inhibition of autophagy enhanced apoptosis induction by cetuximab, an antibody against the epidermal growth factor receptor (EGFR) (45).
Pharmacological inhibitors of autophagy can be broadly classified as early versus late stage inhibitors of the pathway. Early stage inhibitors include 3-methyadenine (3-MA), wortmannin, and LY294002 that target the class III PI3K (Vps34) and interfere with its recruitment to the membranes. Late stage inhibitors include the anti-malarial drugs chloroquine (CQ) or hydroxychloroquine (HCQ), bafilomycin A1, and monensin. Bafilomycin A1 is a specific inhibitor of vacuolar-ATPase (66) while monensin and CQ/HCQ are lysosomotropic drugs that prevent acidification of lysosomes whose digesting hydrolases depend on low pH. Autophagosomes and lysosomes move along microtubules and microtubule disrupting agents (taxanes, nocodazole, colchicine, vinca alkaloids) inhibit fusion of autophagosomes to lysosomes. Other inhibitors of autophagy that block autophagosome degradation include the tricyclic antidepressant drug, clomipramine, and anti-schistome agent, lucanthone (67, 68).
The ability of autophagy inhibition to enhance chemosensitivity and tumor regression has been confirmed in animal models. In a Myc-induced murine lymphoma model, inhibition of autophagy by CQ was shown to enhance cyclophosphamide-induced tumor cell death to a similar extent as did shRNA knockdown of Atg5, and delayed the time-to-tumor recurrence (5). In a colon cancer xenograft model, the addition of CQ to vorinostat was shown to significantly reduce tumor burden and to increase apoptosis (69). Similarly, CQ enhanced the therapeutic efficacy of the Src inhibitor, saracatinib, in a prostate cancer xenograft mouse model (70). Saracatinib decreased tumor growth by 26% compared to control-treated mice, and CQ plus saracatinib further inhibited tumor growth by 64% (70). This combination also led to at least a 2-fold increase in the number of apoptotic tumor cells in the group treated with saracatinib plus CQ, suggesting that suppression of autophagy drives cells into apoptosis (70). Autophagy inhibition by 3-MA increased apoptosis induction by 5-fluorouracil (5-FU) in association with tumor regression in colon cancer xenografts (42). These data indicate that autophagy inhibition can enhance the anti-tumor efficacy of chemotherapeutic agents that utilize diverse cellular mechanisms. Of the known autophagy inhibitors, only CQ/HCQ have been evaluated in humans given their common usage as anti-malarial drugs and in autoimmune disorders. These drugs cross the blood-brain barrier, and HCQ is preferred to CQ in humans given its more favorable side effect profile (71). Based upon preclinical data, several phase I/II trials are ongoing that evaluate the combination of HCQ with cytotoxic drugs in a variety of tumor types (Table 1). Challenges include the long half-life of HCQ and the need for micromolar concentrations to inhibit autophagy that may limit its efficacy in human studies. A recently reported Phase I trial of HCQ in combination with adjuvant temozolamide and radiation in patients with glioblastoma found that the maximum tolerated dose (MTD) of HCQ was 600 mg a day and this dose achieved concentrations of HCQ required for autophagy inhibition in preclinical studies (72). In this trial, dose-dependent autophagy inhibition was observed as indicated by increases in autophagic vesicles (by electron microscopy) with confirmatory elevations in LC3-II detected in peripheral blood mononuclear cells (PBMCs) (72). In a phase I trial of 2-deoxyglucose, an agent that blocks glucose metabolism, autophagy occurred in association with a reduction in p62/SQSTM1 in PBMCs (73). These biomarker data suggest the potential for evaluating autophagy modulation during therapy and to correlate with treatment outcome.
Intracellular proteins are degraded within lysosomes during autophagy and by the ubiquitin-proteosome pathway (74). Given the primary role of these pathways in protein degradation, it has been postulated that their combined blockade may lead to ER stress-induced cytotoxicity through the accumulation of unfolded protein aggregates that can activate autophagy through JNK (75) or PERK/eIF2α (76). The combination of bortezomib and CQ was shown to suppress tumor growth to a greater extent than did either drug alone in colon cancer xenografts (6). Phase I/II clinical trials evaluating this combination are ongoing in patients with relapsed/refractory multiple myeloma.
Interplay of Autophagy and Apoptosis
When autophagy is inhibited, apoptosis is promoted in cancer cells with intact apoptotic signaling (10). Disabled apoptosis is a frequent occurrence in human cancers and tumors under stress generally die by other cell death mechanisms. Autophagy may be an alternative mode of cell death in apoptosis-resistant cells (10, 50); however, the conditions under which autophagy can function as a primary cell death mechanism remain to be defined. Cross-talk between autophagy and apoptosis exists at many levels since both pathways share mediators ranging from the core machinery to upstream regulators (10). Recent finding suggest a link between autophagy and extrinsic apoptotic pathway that is mediated by p62/SQSTM 1 (77). p62/SQSTM 1 was shown to bind caspase-8 to enable its aggregation and activation that enhanced TRAIL-mediated apoptosis (77). TRAIL is cytokine with activity against multiple tumor types where it induces apoptosis. TRAIL or death receptor agonists are currently being evaluated in phase I/II studies.
Anti-apoptotic Bcl-2 family proteins are overexpressed in multiple tumor types where they contribute to both intrinsic and acquired treatment resistance. While Bcl-2 family proteins function to inhibit apoptosis, data indicate that they can also inhibit autophagy (78). The mechanism of this effect is due to the fact that Bcl-2/Bcl-xL proteins can bind to and disrupt the autophagic function of Beclin 1 which contains a BH3 domain (78) (Figure 1). Small molecule antagonists of Bcl-2/Bcl-xL, known asBH3 mimetics (ABT-737/263, obatoclax) can competitively disrupt the Beclin 1-Bcl-2/Bcl-xL interaction to trigger autophagy (78). These data suggest that inhibition of cytoprotective autophagy by BH3 mimetics may further enhance apoptosis. In this regard, the selective cyclooxygenase-2 inhibitor, celecoxib, was shown to induce apoptosis that was enhanced by the BH3 mimetic, ABT-737, and was further augmented by inhibition of autophagy in colon cancer cells (51).
Targeting Autophagy for Cancer Prevention
Since autophagy plays a role is tumor suppression, the induction of autophagy may be an important strategy for cancer prevention. PI3K-AKT-mTOR signaling is frequently dysregulated in human tumors and the inhibition of mTOR signaling can induce autophagy. In accordance with this observation, treatment with the mTOR inhibitor rapamycin was associated with a 90% reduction in carcinogen-induced lung tumors in a murine model (79). In another study, inhibition of mTOR signaling by metformin attenuated tumorigenesis in the same tumor model (80). Furthermore, continuous, low dose rapamycin treatment in APCMin/+ mice with enhanced AKT-mTOR signaling was shown to markedly inhibit intestinal neoplasia (81). Defective autophagy has been linked to colonic tumor formation through a mechanism involving the aberrant activation of Wnt signaling from impaired degradation of Dishevelled (Dvl) by autophagy (82). Other pharmacological activators of the autophagy may also be of potential benefit for cancer chemoprevention and further studies are awaited.
Conclusions
Autophagy serves a dual role as a mechanism of tumor suppression and as an adaptive stress response in tumor cells to maintain their survival in the setting of increased metabolic demands, a hypoxic microenvironment, or cancer therapy. Maintenance of cell survival by autophagy can promote the growth of established tumors. Abundant preclinical evidence indicates that stress-induced autophagy in tumor cells is predominantly cytoprotective and that inhibition of autophagy can enhance tumor cell death by diverse anti-cancer therapies. These data establish autophagy as a novel therapeutic target whose modulation presents new opportunities for cancer treatment. While several drugs can inhibit autophagy, most lack specificity and anti-tumor activity. Chloroquine is the most widely tested in preclinical models and multiple ongoing phase I and II clinical trials are evaluating hydroxychloroquine alone or in combination with cytotoxic chemotherapy or targeted agents, mostly in patients with solid tumors. Targeting autophagy in cancer provides new opportunities for drug development since more potent and specific inhibitors of autophagy are clearly needed. High-throughput screening of chemical libraries to identify small molecule inhibitors of autophagy is ongoing. Biomarkers to measure autophagy modulation during treatment should be an important component of drug development efforts. While important strides have been made, several key questions remain unanswered and include how autophagy is regulated in tumor cells, its interplay with apoptosis, and the specific mechanism by which autophagy confers treatment resistance. An increased understanding of autophagy in cancer is important for its optimal exploitation for therapeutic advantage. Given the role of autophagy in tumor suppression, activation of autophagy may be an important strategy for cancer chemoprevention.
Acknowledgments
This manuscript is supported, in part, by NCI K05 CA142885-01 to FAS
References
- 1.Degenhardt K, Mathew R, Beaudoin B, Bray K, Anderson D, Chen G, et al. Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer Cell. 2006;10:51–64. doi: 10.1016/j.ccr.2006.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Qu X, Yu J, Bhagat G, Furuya N, Hibshoosh H, Troxel A, et al. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J Clin Invest. 2003;112:1809–20. doi: 10.1172/JCI20039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mazure NM, Pouyssegur J. Hypoxia-induced autophagy: cell death or cell survival? Curr Opin Cell Biol. 2010;22:177–80. doi: 10.1016/j.ceb.2009.11.015. [DOI] [PubMed] [Google Scholar]
- 4.Lu Z, Luo RZ, Lu Y, Zhang X, Yu Q, Khare S, et al. The tumor suppressor gene ARHI regulates autophagy and tumor dormancy in human ovarian cancer cells. J Clin Invest. 2008;118:3917–29. doi: 10.1172/JCI35512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Amaravadi RK, Yu D, Lum JJ, Bui T, Christophorou MA, Evan GI, et al. Autophagy inhibition enhances therapy-induced apoptosis in a Myc-induced model of lymphoma. J Clin Invest. 2007;117:326–36. doi: 10.1172/JCI28833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ding WX, Ni HM, Gao W, Chen X, Kang JH, Stolz DB, et al. Oncogenic transformation confers a selective susceptibility to the combined suppression of the proteasome and autophagy. Mol Cancer Ther. 2009;8:2036–45. doi: 10.1158/1535-7163.MCT-08-1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.White E, DiPaola RS. The double-edged sword of autophagy modulation in cancer. Clin Cancer Res. 2009;15:5308–16. doi: 10.1158/1078-0432.CCR-07-5023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Katayama M, Kawaguchi T, Berger MS, Pieper RO. DNA damaging agent-induced autophagy produces a cytoprotective adenosine triphosphate surge in malignant glioma cells. Cell Death Differ. 2007;14:548–58. doi: 10.1038/sj.cdd.4402030. [DOI] [PubMed] [Google Scholar]
- 9.Carew JS, Nawrocki ST, Kahue CN, Zhang H, Yang C, Chung L, et al. Targeting autophagy augments the anticancer activity of the histone deacetylase inhibitor SAHA to overcome Bcr-Abl-mediated drug resistance. Blood. 2007;110:313–22. doi: 10.1182/blood-2006-10-050260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maiuri MC, Zalckvar E, Kimchi A, Kroemer G. Self-eating and self-killing: crosstalk between autophagy and apoptosis. Nat Rev Mol Cell Biol. 2007;8:741–52. doi: 10.1038/nrm2239. [DOI] [PubMed] [Google Scholar]
- 11.Pattingre S, Espert L, Biard-Piechaczyk M, Codogno P. Regulation of macroautophagy by mTOR and Beclin 1 complexes. Biochimie. 2008;90:313–23. doi: 10.1016/j.biochi.2007.08.014. [DOI] [PubMed] [Google Scholar]
- 12.Jung CH, Jun CB, Ro SH, Kim YM, Otto NM, Cao J, et al. ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol Biol Cell. 2009;20:1992–2003. doi: 10.1091/mbc.E08-12-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mizushima N. The role of the Atg1/ULK1 complex in autophagy regulation. Curr Opin Cell Biol. 2010;22:132–9. doi: 10.1016/j.ceb.2009.12.004. [DOI] [PubMed] [Google Scholar]
- 14.Matsushita M, Suzuki NN, Obara K, Fujioka Y, Ohsumi Y, Inagaki F. Structure of Atg5.Atg16, a complex essential for autophagy. J Biol Chem. 2007;282:6763–72. doi: 10.1074/jbc.M609876200. [DOI] [PubMed] [Google Scholar]
- 15.Satoo K, Noda NN, Kumeta H, Fujioka Y, Mizushima N, Ohsumi Y, et al. The structure of Atg4B-LC3 complex reveals the mechanism of LC3 processing and delipidation during autophagy. EMBO J. 2009;28:1341–50. doi: 10.1038/emboj.2009.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282:24131–45. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 17.Mathew R, Karp CM, Beaudoin B, Vuong N, Chen G, Chen HY, et al. Autophagy suppresses tumorigenesis through elimination of p62. Cell. 2009;137:1062–75. doi: 10.1016/j.cell.2009.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jager S, Bucci C, Tanida I, Ueno T, Kominami E, Saftig P, et al. Role for Rab7 in maturation of late autophagic vacuoles. J Cell Sci. 2004;117:4837–48. doi: 10.1242/jcs.01370. [DOI] [PubMed] [Google Scholar]
- 19.Gonzalez-Polo RA, Boya P, Pauleau AL, Jalil A, Larochette N, Souquere S, et al. The apoptosis/autophagy paradox: autophagic vacuolization before apoptotic death. J Cell Sci. 2005;118:3091–102. doi: 10.1242/jcs.02447. [DOI] [PubMed] [Google Scholar]
- 20.Takeuchi H, Kondo Y, Fujiwara K, Kanzawa T, Aoki H, Mills GB, et al. Synergistic augmentation of rapamycin-induced autophagy in malignant glioma cells by phosphatidylinositol 3-kinase/protein kinase B inhibitors. Cancer Res. 2005;65:3336–46. doi: 10.1158/0008-5472.CAN-04-3640. [DOI] [PubMed] [Google Scholar]
- 21.Semenza GL. HIF-1: upstream and downstream of cancer metabolism. Curr Opin Genet Dev. 2010;20:51–6. doi: 10.1016/j.gde.2009.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Egan DF, Shackelford DB, Mihaylova MM, Gelino S, Kohnz RA, Mair W, et al. Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science. 2011;331:456–61. doi: 10.1126/science.1196371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shen J, Chen X, Hendershot L, Prywes R. ER stress regulation of ATF6 localization by dissociation of BiP/GRP78 binding and unmasking of Golgi localization signals. Dev Cell. 2002;3:99–111. doi: 10.1016/s1534-5807(02)00203-4. [DOI] [PubMed] [Google Scholar]
- 24.Criollo A, Senovilla L, Authier H, Maiuri MC, Morselli E, Vitale I, et al. The IKK complex contributes to the induction of autophagy. EMBO J. 2010;29:619–31. doi: 10.1038/emboj.2009.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maiuri MC, Malik SA, Morselli E, Kepp O, Criollo A, Mouchel PL, et al. Stimulation of autophagy by the p53 target gene Sestrin2. Cell Cycle. 2009;8:1571–6. doi: 10.4161/cc.8.10.8498. [DOI] [PubMed] [Google Scholar]
- 26.Tasdemir E, Maiuri MC, Galluzzi L, Vitale I, Djavaheri–Mergny M, D’Amelio M, et al. Regulation of autophagy by cytoplasmic p53. Nat Cell Biol. 2008;10:676–87. doi: 10.1038/ncb1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alexander A, Cai SL, Kim J, Nanez A, Sahin M, MacLean KH, et al. ATM signals to TSC2 in the cytoplasm to regulate mTORC1 in response to ROS. Proc Natl Acad Sci U S A. 2010;107:4153–8. doi: 10.1073/pnas.0913860107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tang D, Kang R, Livesey KM, Cheh CW, Farkas A, Loughran P, et al. Endogenous HMGB1 regulates autophagy. J Cell Biol. 2010;190:881–92. doi: 10.1083/jcb.200911078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liang XH, Jackson S, Seaman M, Brown K, Kempkes B, Hibshoosh H, et al. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402:672–6. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- 30.Duran A, Linares JF, Galvez AS, Wikenheiser K, Flores JM, Diaz-Meco MT, et al. The signaling adaptor p62 is an important NF-kappaB mediator in tumorigenesis. Cancer Cell. 2008;13:343–54. doi: 10.1016/j.ccr.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 31.Yang S, Wang X, Contino G, Liesa M, Sahin E, Ying H, et al. Pancreatic cancers require autophagy for tumor growth. Genes Dev. 2011;25:717–29. doi: 10.1101/gad.2016111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guo JY, Chen HY, Mathew R, Fan J, Strohecker AM, Karsli-Uzunbas G, et al. Activated Ras requires autophagy to maintain oxidative metabolism and tumorigenesis. Genes Dev. 2011;25:460–70. doi: 10.1101/gad.2016311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kanzawa T, Zhang L, Xiao L, Germano IM, Kondo Y, Kondo S. Arsenic trioxide induces autophagic cell death in malignant glioma cells by upregulation of mitochondrial cell death protein BNIP3. Oncogene. 2005;24:980–91. doi: 10.1038/sj.onc.1208095. [DOI] [PubMed] [Google Scholar]
- 34.Shao Y, Gao Z, Marks PA, Jiang X. Apoptotic and autophagic cell death induced by histone deacetylase inhibitors. Proc Natl Acad Sci U S A. 2004;101:18030–5. doi: 10.1073/pnas.0408345102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Turcotte S, Chan DA, Sutphin PD, Hay MP, Denny WA, Giaccia AJ. A molecule targeting VHL-deficient renal cell carcinoma that induces autophagy. Cancer Cell. 2008;14:90–102. doi: 10.1016/j.ccr.2008.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Young AR, Narita M, Ferreira M, Kirschner K, Sadaie M, Darot JF, et al. Autophagy mediates the mitotic senescence transition. Genes Dev. 2009;23:798–803. doi: 10.1101/gad.519709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ewald JA, Desotelle JA, Wilding G, Jarrard DF. Therapy-induced senescence in cancer. J Natl Cancer Inst. 2010;102:1536–46. doi: 10.1093/jnci/djq364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gewirtz DA. Autophagy, senescence and tumor dormancy in cancer therapy. Autophagy. 2009;5:1232–4. doi: 10.4161/auto.5.8.9896. [DOI] [PubMed] [Google Scholar]
- 39.Kanzawa T, Germano IM, Komata T, Ito H, Kondo Y, Kondo S. Role of autophagy in temozolomide-induced cytotoxicity for malignant glioma cells. Cell Death Differ. 2004;11:448–57. doi: 10.1038/sj.cdd.4401359. [DOI] [PubMed] [Google Scholar]
- 40.Abedin MJ, Wang D, McDonnell MA, Lehmann U, Kelekar A. Autophagy delays apoptotic death in breast cancer cells following DNA damage. Cell Death Differ. 2007;14:500–10. doi: 10.1038/sj.cdd.4402039. [DOI] [PubMed] [Google Scholar]
- 41.Mukubou H, Tsujimura T, Sasaki R, Ku Y. The role of autophagy in the treatment of pancreatic cancer with gemcitabine and ionizing radiation. Int J Oncol. 2010;37:821–8. doi: 10.3892/ijo_00000732. [DOI] [PubMed] [Google Scholar]
- 42.Li J, Hou N, Faried A, Tsutsumi S, Kuwano H. Inhibition of autophagy augments 5-fluorouracil chemotherapy in human colon cancer in vitro and in vivo model. Eur J Cancer. 2010;46:1900–9. doi: 10.1016/j.ejca.2010.02.021. [DOI] [PubMed] [Google Scholar]
- 43.Paglin S, Hollister T, Delohery T, Hackett N, McMahill M, Sphicas E, et al. A novel response of cancer cells to radiation involves autophagy and formation of acidic vesicles. Cancer Res. 2001;61:439–44. [PubMed] [Google Scholar]
- 44.Ertmer A, Huber V, Gilch S, Yoshimori T, Erfle V, Duyster J, et al. The anticancer drug imatinib induces cellular autophagy. Leukemia. 2007;21:936–42. doi: 10.1038/sj.leu.2404606. [DOI] [PubMed] [Google Scholar]
- 45.Li X, Fan Z. The epidermal growth factor receptor antibody cetuximab induces autophagy in cancer cells by downregulating HIF-1alpha and Bcl-2 and activating the beclin 1/hVps34 complex. Cancer Res. 2010;70:5942–52. doi: 10.1158/0008-5472.CAN-10-0157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhu K, Dunner K, Jr, McConkey DJ. Proteasome inhibitors activate autophagy as a cytoprotective response in human prostate cancer cells. Oncogene. 2010;29:451–62. doi: 10.1038/onc.2009.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Han J, Hou W, Goldstein LA, Lu C, Stolz DB, Yin XM, et al. Involvement of protective autophagy in TRAIL resistance of apoptosis-defective tumor cells. J Biol Chem. 2008;283:19665–77. doi: 10.1074/jbc.M710169200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu YL, Yang PM, Shun CT, Wu MS, Weng JR, Chen CC. Autophagy potentiates the anti-cancer effects of the histone deacetylase inhibitors in hepatocellular carcinoma. Autophagy. 2010;6:1057–65. doi: 10.4161/auto.6.8.13365. [DOI] [PubMed] [Google Scholar]
- 49.Goussetis DJ, Altman JK, Glaser H, McNeer JL, Tallman MS, Platanias LC. Autophagy is a critical mechanism for the induction of the antileukemic effects of arsenic trioxide. J Biol Chem. 2010 doi: 10.1074/jbc.M109.090530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bursch W, Ellinger A, Kienzl H, Torok L, Pandey S, Sikorska M, et al. Active cell death induced by the anti-estrogens tamoxifen and ICI 164 384 in human mammary carcinoma cells (MCF-7) in culture: the role of autophagy. Carcinogenesis. 1996;17:1595–607. doi: 10.1093/carcin/17.8.1595. [DOI] [PubMed] [Google Scholar]
- 51.Huang S, Sinicrope FA. Celecoxib-induced apoptosis is enhanced by ABT-737 and by inhibition of autophagy in human colorectal cancer cells. Autophagy. 2010;6:256–69. doi: 10.4161/auto.6.2.11124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gills JJ, Lopiccolo J, Dennis PA. Nelfinavir, a new anti-cancer drug with pleiotropic effects and many paths to autophagy. Autophagy. 2008;4:107–9. doi: 10.4161/auto.5224. [DOI] [PubMed] [Google Scholar]
- 53.Meric-Bernstam F, Gonzalez-Angulo AM. Targeting the mTOR signaling network for cancer therapy. J Clin Oncol. 2009;27:2278–87. doi: 10.1200/JCO.2008.20.0766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.O’Reilly KE, Rojo F, She QB, Solit D, Mills GB, Smith D, et al. mTOR inhibition induces upstream receptor tyrosine kinase signaling and activates Akt. Cancer Res. 2006;66:1500–8. doi: 10.1158/0008-5472.CAN-05-2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yu K, Shi C, Toral-Barza L, Lucas J, Shor B, Kim JE, et al. Beyond rapalog therapy: preclinical pharmacology and antitumor activity of WYE-125132, an ATP-competitive and specific inhibitor of mTORC1 and mTORC2. Cancer Res. 2010;70:621–31. doi: 10.1158/0008-5472.CAN-09-2340. [DOI] [PubMed] [Google Scholar]
- 56.Chresta CM, Davies BR, Hickson I, Harding T, Cosulich S, Critchlow SE, et al. AZD8055 is a potent, selective, and orally bioavailable ATP-competitive mammalian target of rapamycin kinase inhibitor with in vitro and in vivo antitumor activity. Cancer Res. 2010;70:288–98. doi: 10.1158/0008-5472.CAN-09-1751. [DOI] [PubMed] [Google Scholar]
- 57.Feldman ME, Apsel B, Uotila A, Loewith R, Knight ZA, Ruggero D, et al. Active-site inhibitors of mTOR target rapamycin-resistant outputs of mTORC1 and mTORC2. PLoS Biol. 2009;7:e38. doi: 10.1371/journal.pbio.1000038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Thoreen CC, Kang SA, Chang JW, Liu Q, Zhang J, Gao Y, et al. An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. J Biol Chem. 2009;284:8023–32. doi: 10.1074/jbc.M900301200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Janes MR, Limon JJ, So L, Chen J, Lim RJ, Chavez MA, et al. Effective and selective targeting of leukemia cells using a TORC1/2 kinase inhibitor. Nat Med. 16:205–13. doi: 10.1038/nm.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fan QW, Cheng C, Hackett C, Feldman M, Houseman BT, Nicolaides T, et al. Akt and autophagy cooperate to promote survival of drug-resistant glioma. Sci Signal. 2010;3:ra81. doi: 10.1126/scisignal.2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ben Sahra I, Laurent K, Giuliano S, Larbret F, Ponzio G, Gounon P, et al. Targeting cancer cell metabolism: the combination of metformin and 2-deoxyglucose induces p53-dependent apoptosis in prostate cancer cells. Cancer Res. 2010;70:2465–75. doi: 10.1158/0008-5472.CAN-09-2782. [DOI] [PubMed] [Google Scholar]
- 62.Buzzai M, Jones RG, Amaravadi RK, Lum JJ, DeBerardinis RJ, Zhao F, et al. Systemic treatment with the antidiabetic drug metformin selectively impairs p53-deficient tumor cell growth. Cancer Res. 2007;67:6745–52. doi: 10.1158/0008-5472.CAN-06-4447. [DOI] [PubMed] [Google Scholar]
- 63.Cloonan SM, Williams DC. The antidepressants maprotiline and fluoxetine induce Type II autophagic cell death in drug-resistant Burkitt’s lymphoma. Int J Cancer. 2010 doi: 10.1002/ijc.25477. [DOI] [PubMed] [Google Scholar]
- 64.Fu J, Shao CJ, Chen FR, Ng HK, Chen ZP. Autophagy induced by valproic acid is associated with oxidative stress in glioma cell lines. Neuro Oncol. 2010;12:328–40. doi: 10.1093/neuonc/nop005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Williams A, Sarkar S, Cuddon P, Ttofi EK, Saiki S, Siddiqi FH, et al. Novel targets for Huntington’s disease in an mTOR-independent autophagy pathway. Nat Chem Biol. 2008;4:295–305. doi: 10.1038/nchembio.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shacka JJ, Klocke BJ, Roth KA. Autophagy, bafilomycin and cell death: the “a-B-cs” of plecomacrolide-induced neuroprotection. Autophagy. 2006;2:228–30. doi: 10.4161/auto.2703. [DOI] [PubMed] [Google Scholar]
- 67.Rossi M, Munarriz ER, Bartesaghi S, Milanese M, Dinsdale D, Guerra-Martin MA, et al. Desmethylclomipramine induces the accumulation of autophagy markers by blocking autophagic flux. J Cell Sci. 2009;122:3330–9. doi: 10.1242/jcs.048181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Carew JS, Espitia CM, Esquivel JA, 2nd, Mahalingam D, Kelly KR, Reddy G, et al. Lucanthone: A novel inhibitor of autophagy that induces cathepsin D-mediated apoptosis. J Biol Chem. 2010 doi: 10.1074/jbc.M110.151324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Carew JS, Medina EC, Esquivel JA, 2nd, Mahalingam D, Swords R, Kelly K, et al. Autophagy inhibition enhances vorinostat-induced apoptosis via ubiquitinated protein accumulation. J Cell Mol Med. 2009 doi: 10.1111/j.1582-4934.2009.00832.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wu Z, Chang PC, Yang JC, Chu CY, Wang LY, Chen NT, et al. Autophagy Blockade Sensitizes Prostate Cancer Cells towards Src Family Kinase Inhibitors. Genes Cancer. 2010;1:40–9. doi: 10.1177/1947601909358324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ruiz-Irastorza G, Ramos-Casals M, Brito-Zeron P, Khamashta MA. Clinical efficacy and side effects of antimalarials in systemic lupus erythematosus: a systematic review. Ann Rheum Dis. 2010;69:20–8. doi: 10.1136/ard.2008.101766. [DOI] [PubMed] [Google Scholar]
- 72.Rosenfeld MR, GSA, Brem S, Mikkelsen T, Wang D, Piao S, Davis LE, O’Dwyer PJ, Amaravadi RK. Pharmacokinetic analysis and pharmacodynamic evidence of autophagy inhibition in patients with newly diagnosed glioblastoma treated on a phase I trial of hydroxychloroquine in combination with adjuvant temozolomide and radiation (ABTC 0603) J Clin Oncol. 2010;28 [Google Scholar]
- 73.Stein M, Lin H, Jeyamohan C, Dvorzhinski D, Gounder M, Bray K, et al. Targeting tumor metabolism with 2-deoxyglucose in patients with castrate-resistant prostate cancer and advanced malignancies. Prostate. 2010;70:1388–94. doi: 10.1002/pros.21172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Korolchuk VI, Mansilla A, Menzies FM, Rubinsztein DC. Autophagy inhibition compromises degradation of ubiquitin-proteasome pathway substrates. Mol Cell. 2009;33:517–27. doi: 10.1016/j.molcel.2009.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ogata M, Hino S, Saito A, Morikawa K, Kondo S, Kanemoto S, et al. Autophagy is activated for cell survival after endoplasmic reticulum stress. Mol Cell Biol. 2006;26:9220–31. doi: 10.1128/MCB.01453-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kouroku Y, Fujita E, Tanida I, Ueno T, Isoai A, Kumagai H, et al. ER stress (PERK/eIF2alpha phosphorylation) mediates the polyglutamine-induced LC3 conversion, an essential step for autophagy formation. Cell Death Differ. 2007;14:230–9. doi: 10.1038/sj.cdd.4401984. [DOI] [PubMed] [Google Scholar]
- 77.Jin Z, Li Y, Pitti R, Lawrence D, Pham VC, Lill JR, et al. Cullin3-based polyubiquitination and p62-dependent aggregation of caspase-8 mediate extrinsic apoptosis signaling. Cell. 2009;137:721–35. doi: 10.1016/j.cell.2009.03.015. [DOI] [PubMed] [Google Scholar]
- 78.Levine B, Sinha S, Kroemer G. Bcl-2 family members: dual regulators of apoptosis and autophagy. Autophagy. 2008;4:600–6. doi: 10.4161/auto.6260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Granville CA, Warfel N, Tsurutani J, Hollander MC, Robertson M, Fox SD, et al. Identification of a highly effective rapamycin schedule that markedly reduces the size, multiplicity, and phenotypic progression of tobacco carcinogen-induced murine lung tumors. Clin Cancer Res. 2007;13:2281–9. doi: 10.1158/1078-0432.CCR-06-2570. [DOI] [PubMed] [Google Scholar]
- 80.Memmott RM, Mercado JR, Maier CR, Kawabata S, Fox SD, Dennis PA. Metformin prevents tobacco carcinogen-induced lung tumorigenesis. Cancer prevention research (Philadelphia, Pa. 3:1066–76. doi: 10.1158/1940-6207.CAPR-10-0055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Koehl GE, Spitzner M, Ousingsawat J, Schreiber R, Geissler EK, Kunzelmann K. Rapamycin inhibits oncogenic intestinal ion channels and neoplasia in APC(Min/+) mice. Oncogene. 29:1553–60. doi: 10.1038/onc.2009.435. [DOI] [PubMed] [Google Scholar]
- 82.Gao C, Cao W, Bao L, Zuo W, Xie G, Cai T, et al. Autophagy negatively regulates Wnt signalling by promoting Dishevelled degradation. Nat Cell Biol. 2010;12:781–90. doi: 10.1038/ncb2082. [DOI] [PubMed] [Google Scholar]