Abstract
Homologous recombination in Trypanosoma brucei is used for moving variant surface glycoprotein (VSG) genes into expression sites during immune evasion by antigenic variation. A major route for such VSG switching is gene conversion reactions in which RAD51, a universally conserved recombinase, catalyses homology-directed strand exchange. In any eukaryote, RAD51-directed strand exchange in vivo is mediated by further factors, including RAD51-related proteins termed Rad51 paralogues. These appear to be ubiquitously conserved, although their detailed roles in recombination remain unclear. In T. brucei, four putative RAD51 paralogue genes have been identified by sequence homology. Here we show that all four RAD51 paralogues act in DNA repair, recombination and RAD51 subnuclear dynamics, though not equivalently, while mutation of only one RAD51 paralogue gene significantly impedes VSG switching. We also show that the T. brucei RAD51 paralogues interact, and that the complexes they form may explain the distinct phenotypes of the mutants as well as observed expression interdependency. Finally, we document the Rad51 paralogues that are encoded by a wide range of protists, demonstrating that the Rad51 paralogue repertoire in T. brucei is unusually large among microbial eukaryotes and that one member of the protein family corresponds with a key, conserved eukaryotic Rad51 paralogue.
Introduction
Homologous recombination (HR) is of central importance in the maintenance and transmission of the genome. The key factor in this process, termed RAD51 in eukaryotes, RecA in bacteria and RadA in archaea, appears to be universally conserved (Story et al., 1993; Brendel et al., 1997; Lin et al., 2006; Haldenby et al., 2009). HR is one of a number of DNA repair pathways that can reverse genotoxic damage, and acts in particular on DNA double-strand breaks (DSBs) (San Filippo et al., 2008; Kass and Jasin, 2010) and the restart of blocked or collapsed replication forks (Petermann and Helleday, 2010). Despite the core functions of HR, some organisms have co-opted the process for rearrangements targeted to a defined locus or loci, such as in mating type gene switching in fungi (Fraser and Heitman, 2003; Klar, 2007). Several pathogenic microbes, including the bacteria Neisseria gonorrhoeae and Borellia sp. and the eukaryotes Babesia bovis and Anaplasma marginale, use HR for antigenic variation, the changing of surface antigens to evade host immunity (Deitsch et al., 2009; Morrison et al., 2009). African trypanosomes, including Trypanosoma brucei, are protistan parasites of mammals belonging to the eukaryotic order Kinetoplastida and are responsible for devastating disease in humans and livestock in sub-Saharan Africa. African trypanosomes proliferate extracellularly in the bloodstream and tissue fluids of mammals and undergo antigenic variation of their major surface antigen, the variant surface glycoprotein (VSG) (Barry and McCulloch, 2001; Horn and Barry, 2005; Taylor and Rudenko, 2006; Morrison et al., 2009).
The success of antigenic variation in African trypanosomes relies upon each cell expressing one antigenic form of VSG at a time, and switching this to an antigenically distinct VSG. Such VSG switching, which is not induced by the host immune response, allows some of the infecting population to evade elimination by antibody-mediated killing, prolonging the infection and enhancing transmission to a new mammal, via the tsetse fly. Singular VSG expression in the mammal is achieved because a VSG gene must be present in a bloodstream VSG expression site (BES) for transcription. However, multiple BESs are found in the genome, and therefore a switch in the expressed VSG can be enacted by inactivating the fully transcribed BES and activating full transcription from one of the silent BESs (Vanhamme et al., 2001; Borst, 2002; Pays, 2005). The mechanisms of, and trigger for, this transcriptional, or in situ, switching reaction are not yet understood, but it appears to be a co-ordinated process (Chaves et al., 1999; Ulbert et al., 2002) and some factors have been described whose mutation elevates transcriptional switching frequency (Landeira et al., 2009) or alters its dynamics (Figueiredo et al., 2008). In addition to the VSGs in the 20 or so BES, the T. brucei genome contains > 1600 VSG genes (Berriman et al., 2005; Marcello and Barry, 2007a). Most are concentrated in arrays in the subtelomeres of T. brucei megabase chromosomes (of which there are 11 diploid copies) (Melville et al., 1998), where ∼ 85% are pseudogenes. Large numbers of VSGs are also found in minichromosomes and intermediate chromosomes, chromosome classes specific to African trypanosomes among the Kinetoplastida (Ersfeld et al., 1999; Wickstead et al., 2004). Such VSGs are transcriptionally silent unless activated by recombination into the BESs. The most commonly observed recombination reactions in VSG switching are gene conversions where a silent VSG is copied and displaces the VSG residing in the BES (Hoeijmakers et al., 1980; Pays et al., 1981; Robinson et al., 1999). VSG gene conversion can encompass more sequence that the VSG ORF: copies of silent array VSGs normally extend to upstream VSG-associated 70 bp repeats (Liu et al., 1983), while much greater amounts of sequence can be copied when the silent VSG is present in a BES (Laurent et al., 1983; Pays et al., 1983; McCulloch et al., 1997; Kim and Cross, 2010); in some cases the entire transcription unit is duplicated (Hertz-Fowler et al., 2008). Downstream, gene conversion can terminate in the VSG ORF or 3′ UTR, but can encompass the telomere if the silent VSG is at the chromosome end (de Lange et al., 1983). Other recombination reactions also act in VSG switching. Cross-over, or reciprocal, recombination reactions where telomeric VSGs are exchanged between chromosomes are found infrequently (Pays et al., 1985; Rudenko et al., 1996). Segmental gene conversions in which novel VSG‘mosaic’ genes are constructed from silent VSG ORFs also occur (Longacre and Eisen, 1986; Roth et al., 1986; Kamper and Barbet, 1992). Although mosaic VSG switching has been considered rare, it predominates late in T. brucei infections (Marcello and Barry, 2007a) and allows the parasite to use the huge numbers of VSG pseudogenes to construct intact VSGs. This is likely to be a key contributing reaction for the function of antigenic variation in trypanosome survival (Barbet and Kamper, 1993; Marcello and Barry, 2007b; Morrison et al., 2009).
Genetic analysis has uncovered a number of factors that direct T. brucei VSG switching by recombination. Mutation of RAD51, the catalytic recombinase in eukaryotic HR, or BRCA2, a mediator of RAD51 nucleoprotein filament formation, impairs VSG switching frequency (McCulloch and Barry, 1999; Hartley and McCulloch, 2008). More recently, mutation of TOPO3α, an orthologue of the Saccharomyces cerevisiae Top3 component of the Sgs1-Top3-Rmi1 complex (Chu and Hickson, 2009), was shown to elevate VSG switching frequency (Kim and Cross, 2010). These data suggest VSG switching, at least of intact VSG genes, is largely driven by an HR reaction in which strand exchange is mediated by RAD51 and recombination intermediates that lead to cross-overs are suppressed. This reaction mechanism is consistent with the suggestion that VSG switching may be promoted by DSBs in the 70 bp repeats of the actively transcribed BES (Boothroyd et al., 2009). Nevertheless, a number of findings may indicate VSG switching is a specific adaptation of HR. The Mre11-Rad50-Xrs2/Nbs1 complex is critical in sensing and processing DSBs during eukaryotic HR (Rupnik et al., 2010). T. brucei MRE11 mutants are impaired in DNA damage repair and recombination (Robinson et al., 2002; Tan et al., 2002), but show no discernible defect in VSG switching (Robinson et al., 2002). Similarly, mismatch repair is an important regulator of HR efficiency in eukaryotes (Jiricny, 2006), including T. brucei, but does not detectably influence VSG switching (Bell and McCulloch, 2003; Bell et al., 2004). Other observations suggest that VSG switching may exploit more than one HR pathway: although mutation of RAD51 or BRCA2 impairs VSG switching, VSG gene conversions can still be detected (McCulloch and Barry, 1999; Hartley and McCulloch, 2008); and ablation of the putative VSG switch-initiating 70 bp repeats from the active BES does not detectably impair VSG switching (McCulloch et al., 1997; Boothroyd et al., 2009).
Rad51-directed strand exchange in eukaryotic HR is regulated by a number of factors, including Brca2, Rad52, Rad59 and Rad54 (San Filippo et al., 2008). Among such HR ‘mediators’, the functions of Rad51 paralogues (Thacker, 2005) are least well understood. Rad51 paralogues are found in all eukaryotes (Lin et al., 2006) and are Rad51-like proteins that display much greater sequence divergence from Rad51 than Dmc1, another widespread Rad51-like protein that acts in meiosis (San Filippo et al., 2008). Rad51 paralogues are not specific to eukaryotes, as RadA-related proteins are found in archaea (Haldenby et al., 2009) and bacteria encode Sms/RadA, a RecA-like protein implicated in HR (Beam et al., 2002). Two Rad51 paralogues, Rad55 and Rad57, are found in S. cerevisiae and have been shown to heterodimerize and interact with Rad51 (Hays et al., 1995; Johnson and Symington, 1995), mediating Rad51 nucleoprotein filament formation or stability (Sung, 1997; Fortin and Symington, 2002). Mutants of Rad55 or Rad57 are sensitive to DNA damage and display reduced HR (Hays et al., 1995; Sugawara et al., 1995). In vertebrates five Rad51 paralogues are found, XRCC2, XRCC3, Rad51B (Rad51L1), Rad51C (Rad51L2) and Rad51D (Rad51L3), forming at least two complexes (Schild et al., 2000; Masson et al., 2001a,b; Liu et al., 2002). Although Rad51 paralogue mutants are lethal during mouse embryogenesis (Shu et al., 1999; Deans et al., 2000; Pittman and Schimenti, 2000; Kuznetsov et al., 2009), mutations of each gene in cultured cells demonstrate roles in DNA damage repair (Liu et al., 1998; Lio et al., 2004; Daboussi et al., 2005), chromosome stability (Cui et al., 1999; Smiraldo et al., 2005), HR (Johnson et al., 1999; Pierce et al., 1999; Brenneman et al., 2000; Drexler et al., 2004; Hatanaka et al., 2005) and Rad51 localization in subnuclear foci after DNA damage (Bishop et al., 1998; Takata et al., 2000; O'Regan et al., 2001; Takata et al., 2001; Tarsounas et al., 2004a; van Veelen et al., 2005). Like S. cerevisiae Rad55-Rad57, in vitro assays suggest some vertebrate Rad51 paralogues mediate Rad51 HR (Sigurdsson et al., 2001; Lio et al., 2003; Shim et al., 2004). Arabidopsis thaliana also encodes five Rad51 paralogues, with roles in DNA damage repair and meiosis (Bleuyard and White, 2004; Bleuyard et al., 2005; Osakabe et al., 2005), while four are found in Drosophila melanogaster (Ghabrial et al., 1998; Abdu et al., 2003; Radford and Sekelsky, 2004). The reasons that these multicellular organisms require an expanded repertoire of Rad51 paralogues is unclear, since Caenorhabditis elegans possesses a single such protein, Rfs1 (Ward et al., 2007). No work to date has looked beyond mammals and yeast to ask if Rad51 paralogues interact in functionally or structurally conserved complexes.
Previously, we described four T. brucei genes encoding putative RAD51 paralogues, as well as a DMC1 orthologue (Proudfoot and McCulloch, 2005; 2006;). Only two of the Rad51 paralogues (RAD51-3 and RAD51-5) were examined genetically and shown to have roles in T. brucei DNA repair and recombination. Surprisingly, only one of the two factors (RAD51-3) appeared to act in VSG switching (Proudfoot and McCulloch, 2005). To understand if each of the four genes indeed encode Rad51 paralogues, we describe here the consequences of mutating them individually. This confirms that all four genes contribute to DNA repair and recombination, although we show that their roles are not equivalent, suggesting distinct functions. In addition, we asked if the T. brucei RAD51 paralogues interact and if the complexes they form are comparable with those described in mammals and yeast. Finally, we examined the Rad51 paralogues that can be identified in the completed genome sequences of a wide range of parasitic and non-parasitic protists, microbial organisms that contribute much of the diversity of the eukaryotic kingdom, but where analysis of HR is limited (Bhattacharyya et al., 2005; Lopez-Casamichana et al., 2008; Lopez-Camarillo et al., 2009). In so doing, we show that the repertoire of Rad51 paralogues in T. brucei and related kinetoplastids is uniquely large among protsists, matched only by Naegleria gruberi, and detail a key member of the Rad51 paralogue family.
Results
Kinetoplastids contain an unusually large repertoire of RAD51 paralogues
In previous work, we described the RAD51 paralogues in T. brucei and the related kinetoplastid parasites T. cruzi and Leishmania major (Proudfoot and McCulloch, 2005); a summary of the domain organization and size of the T. brucei (Tb) proteins relative to TbRAD51, TbDMC1 and to Escherichia coli RecA is shown in Fig. 1, illustrating the divergence of the RAD51 paralogues from the catalytic recombinases. Since then, the genomes of a larger number of protists have been sequenced, including non-parasitic organisms (Fritz-Laylin et al., 2010). To ask what RAD51 paralogues are encoded by protists, we searched for RAD51 paralogues in a number of these genomes (Table 1). To do this, we used two strategies. First, we used blast searches with full-length TbRAD51 and TbRAD51-3 sequences, as well as with S. cerevisiae RAD51 and RAD57, and iterative searches within each genome using the RAD51-like proteins identified, manually searching for Walker A and B box motifs within a ‘RecA fold’, the conserved core of RAD51/RecA/RadA-like factors in eukaryotes, bacteria (Brendel et al., 1997) and archaea (Haldenby et al., 2009). Second, an alignment was extracted of the 40 most diverse sequences contributing to the Position Specific Scoring Matrix for the Conserved Domain Database (Marchler-Bauer et al., 2009) domain type cd01393 (RecA-like family including prokaryotic RecA, eukaryotic Rad51 and DCM1, and archeal RabA and RabB homologues). This alignment was then used to generate a Hidden Markov model using the hmmbuild program of HMMER2.3.2 (Eddy, 1998). Following calibration using the hmmcalibrate option this profile was used to search (hmmsearch, per-sequence Eval cutoff: ≤ 10) the UniprotKB peptide sequence database (current at January 2008). Virtually all the factors identified by the manual blast and annotation approach were found by this latter search (with Expectation values < 10−30), which additionally identified proteins clearly not Rad51/RecA-related, such as ABC multidrug resistance proteins and AAA+ ATPase factors (members of the P-loop containing nucleoside triphosphate hydrolase superfamily, Pfam CL0023) (Finn et al., 2006), all of which were discarded.
Fig. 1.
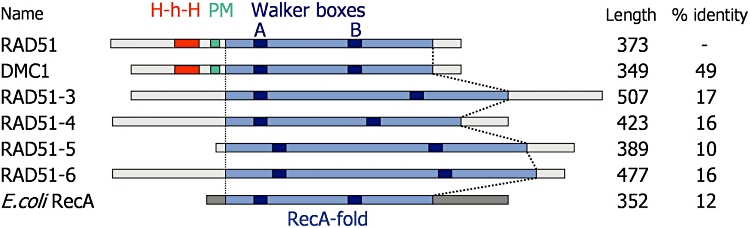
RAD51, DMC1 and RAD51 paralogues in T. brucei. Primary structures of T. brucei RAD51-related proteins relative to each other and to Escherichia coli RecA are shown; the proteins' lengths (amino acids) and sequence identity with T. brucei RAD51 are indicated. A conserved core (RecA-fold; light blue) is present in all proteins, containing Walker A and B motifs (dark blue). All the T. brucei proteins, except RAD51-5, contain N-terminal extensions that are not conserved with RecA; in this region RAD51 and DMC1 encode helix–hairpin–helix (H–h–H) and polymerization (PM) motifs not found in the RAD51 paralogues. A C-terminal extension in RecA is conserved with other bacteria RecA proteins, but is not conserved with the RAD51-related proteins.
Table 1.
A comparison of the presence (+), absence (−) or number of detectable RAD51, DMC1, RAD51 paralogue (Rad51par) and RecA proteins encoded by the genomes of a range of eukaryotic species, which are grouped into taxa and into five supergroups
| Supergroupa | Taxon | Organism | RAD51 | DMC1 | RAD51par | RecAb |
|---|---|---|---|---|---|---|
| Excavata | Parabasalida | T. vaginalis | + | + | 2 | − |
| Diplomonadida | G. intestinalis | − | 2 | − | − | |
| Euglenozoa | T. brucei | + | + | 4 | − | |
| T. vivax | + | + | 4 | − | ||
| T. congolense | + | + | 4 | − | ||
| T. cruzi | + | + | 4 | − | ||
| L. major | + | + | 3 | − | ||
| L. braziliensis | + | + | 3 | − | ||
| L. infantum | + | + | 3 | − | ||
| Heterolobosea | N. gruberi | + | + | 3 | − | |
| Chromalveolata | Stramenopile | T. pseudonana | + | − | 2 | − |
| Apicomplexa | C. parvum | + | + | 2 | − | |
| T. annulata | + | + | 1 | − | ||
| P. falciparum | + | + | 1 | − | ||
| T. gondii | + | + | 1 | − | ||
| Ciliophora | T. thermophila | + | + | 1 | − | |
| Plantae | Viridiplantae | O. sativa | + | + | 5 | 3 |
| A. thaliana | + | + | 5 | 3 | ||
| Amoebozoa | Entamoebidae | E. histolytica | + | + | 1 | − |
| Mycetozoa | D. discoideum | + | − | 5 | 1 | |
| Opisthokonta | Microsporidia | E. cuniculi | + | − | 1 | − |
| Fungi | S. pombe | + | + | 4 | − | |
| S. cerevisiae | + | + | 2 | − | ||
| U. maydis | + | − | 1 | − | ||
| Metazoa | C. elegans | + | − | 1 | − | |
| D. melanogaster | + | − | 4 | − | ||
| H. sapiens | + | + | 5 | − | ||
| G. gallus | + | + | 5 | − |
The supergroup Excavata is often split into two supergroups, one containing Euglenozoan, Heterolobosean and Jackobid (not shown) organisms, and the other containing Parabasalid, Diplomonad and Oxymonad (not shown) organisms.
Where characterized, eukaryotic RecA proteins are targeted to the mitochondrion or chloroplast.
We consider that the combined output of these two bioinformatic approaches provides a comprehensive listing of canonical eukaryotic RAD51 paralogues in the genomes analysed (accession numbers or genome identifiers are provided in a supplementary file). None of the putative RAD51 paralogues showed evidence for helix–hairpin–helix domains, which were conserved in the N-termini of all the RAD51 and DMC1 orthologues (data not shown), illustrating their divergence from the recombinases. It is striking that the kinetoplastids possess among the largest repertoire of RAD51 paralogues among protists, and indeed among single-celled eukaryotes (Table 1). Each Trypanosoma species examined contains four syntenic RAD51-like genes that encode recognizable orthologues of RAD51-3, RAD51-4, RAD51-5 and RAD51-6 in T. brucei, while each Leishmania species encodes three (all lacking RAD51-5; see Discussion). Only in multicellular eukaryotes, or in the social amoeba Dictyostelium discoideum, were greater numbers of RAD51 paralogues identified. Three putative Rad51 paralogues were found in N. gruberi, the free-living protist whose genome has been sequenced that is most closely related to the Euglenozoa, which includes T. brucei (Fritz-Laylin et al., 2010). A further protein (ID 70092) was distantly related to Rad51, but appeared not to possess Walker A and B motifs, and did not therefore match our criteria for Rad51 paralogues.
All four T. brucei RAD51 paralogues act in nuclear DNA repair and recombination
To date, the functions of only RAD51-3 and RAD51-5 have been examined among the RAD51 paralogues of any kinetoplastid (Proudfoot and McCulloch, 2005). To ask if RAD51-4 and RAD51-6 provide related functions, we made homozygous mutants of each gene in bloodstream stage T. brucei. To do this, and to allow comparison between all mutants, we followed the same gene disruption strategy adopted previously for RAD51-3, RAD51-5 and RAD51, replacing the central core of the ORFs (including both Walker boxes) with antibiotic resistance cassettes (Fig. S1). T. brucei is diploid in the housekeeping core of the megabase chromosomes, and therefore two rounds of transformation were used for each gene, sequentially generating heterozygous (+/−) and homozygous (−/−) mutants that were selected via blasticidin and then puromycin resistance. Southern blotting and RT-PCR analysis showed that the gene disruptions had occurred as expected and that intact RAD51-4 or RAD51-6 genes and mRNAs were absent from the −/− clones (Fig. S1). To ensure reproducibility, two +/− mutants were generated independently for each gene, from which two independent −/− mutants were derived. In addition, we re-expressed RAD51-4 or RAD51-6 in the −/− mutants by targeting (via phleomycin selection) the intact ORF to the constitutively transcribed tubulin array [such re-expressers are designated −/−/+, and were verified by Southern blotting (data not shown) and RT-PCR; Fig. S1].
In common with T. brucei rad51-3−/− and rad51-5−/− mutants, rad51-4−/− and rad51-6−/− mutants were viable but displayed growth impairment relative to wild-type (wt) or +/− mutants. The population numbers of wt cells doubled in vitro on average in 6.69 (± 0.01) h, and +/− and −/−/+ cells grew at the same rate (data not shown). In contrast, rad51-6−/− mutants doubled in 9.52 (± 0.79) h, a growth impairment very similar to that measured in parallel for rad51-3−/− (9.80 ± 0.89 h) and rad51−/− cells (9.99 ± 1.0 h). rad51-5−/− and rad51-4−/− mutants showed population doubling times of 8.37 (± 0.31) h and 7.87 (± 0.73) h, which suggests somewhat less growth impairment than the other mutants, though still significantly different from wt (P-values each < 0.001; unpaired, two-tailed Student's t-test).
To ask if RAD51-4 and RAD51-6 act in T. brucei DNA repair, we assayed the sensitivity of the mutants to exogenous DNA damage. First, we measured the survival of the cells in increasing concentrations of phleomycin, a glycopeptide antibiotic member of the bleomycin family that produces single-strand and double-strand breaks in DNA (Giloni et al., 1981). To do this, the IC50s of wt, +/− and −/− cells were determined over a wide range of phleomycin concentrations using Alamar Blue (Raz et al., 1997) as an indicator of metabolic function (Fig. 2A). In addition, we verified these findings using a clonal survival assay we have described previously (Conway et al., 2002a;Robinson et al., 2002; Proudfoot and McCulloch, 2005; 2006;), employing a more restricted range of phleomycin concentrations (Fig. S3). The rad51-6−/− mutants were approximately fourfold to sixfold more sensitive to phleomycin than the wt or RAD51-6+/− cells, consistent with an impairment in repair of such damage. rad51-4−/− mutants also displayed greater sensitivity to phleomycin damage than wt cells or their heterozygous antecedents, but to a lesser extant (approximately twofold to fourfold). To ask how these phenotypes of increased sensitivity compare with the other RAD51 paralogue mutants, and with RAD51 mutants, we measured the IC50s of all T. brucei−/− mutants (Fig. 2B), showing that each differed significantly from wt cells (P-values < 0.05 in all cases; unpaired, two-tailed Student's t-tests). These data also show that whereas rad51-3−/−, rad51-5−/− and rad51-6−/− mutants showed essentially equivalent increased sensitivity to phleomycin compared with each other and with the rad51−/− mutant, rad51-4−/− cells were less sensitive (P-values < 0.05, unpaired, two-tailed Student's t-tests). We next determined the sensitivity of the mutants to methyl methanesulphonate (MMS), an SN2 alkylating agent, using the same assays (Figs S2 and S3 for IC50s and clonal survival respectively). We found again that both rad51-4−/− and rad51-6−/− mutants were more sensitive to MMS than wt or +/− cells, and also demonstrated that re-expressing the protein was able to complement this phenotype. In addition, the rad51-4−/− mutants were the least sensitive of the mutants to MMS damage, although this was not as pronounced as seen for phleomycin. Taken together, these data indicate that all the RAD51 paralogues contribute to T. brucei DNA repair.
Fig. 2.
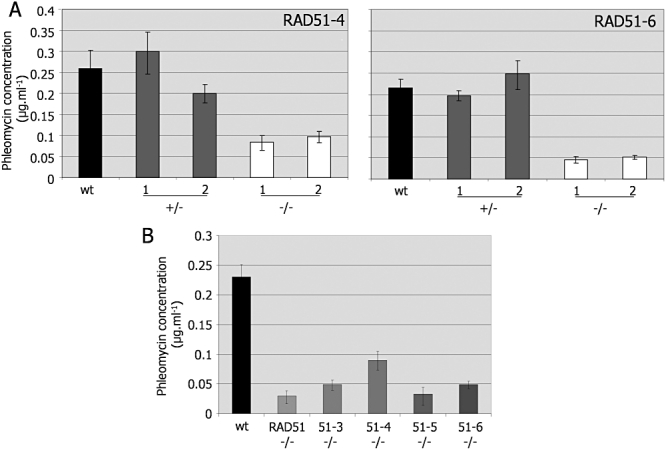
DNA damage sensitivity in T. brucei RAD51 and RAD51 paralogue mutants. A. The concentration of phleomycin that caused 50% growth inhibition (IC50) of wild-type (wt) cells is compared with two (1, 2) independent heterozygous (+/−) and homozygous (−/−) mutants of RAD51-4 and RAD51-6. B. Comparison of the phleomycin IC50s of wt, rad51−/−, rad51-3−/−, rad51-4−/−, rad51-5−/− and rad51-6−/− cells. Values are averages from at least three experiments; bars indicate standard deviation.
To ask if the above repair functions are due to the action of the RAD51 paralogues in T. brucei recombination, we measured the efficiency with which the mutants generate antibiotic-resistant transformants following electroporation with the construct tub-BLE-tub, which confers phleomycin resistance following HR-mediated integration into the tubulin locus. In a number of previous studies we have demonstrated the validity of this approach to measure T. brucei recombination frequency (Conway et al., 2002b; Robinson et al., 2002; Bell and McCulloch, 2003; Proudfoot and McCulloch, 2005; Barnes and McCulloch, 2007; Hartley and McCulloch, 2008), and that it is primarily a measure of HR efficiency as non-HR has never been observed in this organism (Burton et al., 2007; Glover et al., 2008). Figure 3A compares the transformation efficiency of the rad51-4−/− and rad51-6−/− mutants with +/− antecedents and with wt cells, and shows significant impairment in each homozygous mutant (P-values < 0.05 in all cases where the −/− cells were compared with wt or +/− cells by paired, two-tailed Student's t-tests). To compare the contribution to recombination of RAD51-4 and RAD51-6 with that of the other RAD51 paralogues and with RAD51 itself, the transformation efficiency of each −/− mutant was compared with each other and with wt cells. Because the effectiveness of phleomycin selection depends on oxygen and metal ion concentrations (Claussen and Long, 1999), transformation rates can vary between experiments. For this reason, we compared the transformation of the mutants in a single experiment using cells grown and selected after transformation in a single batch of medium, and using single preparations of DNA construct and phleomycin stock (Fig. 3B). These data show that each RAD51 paralogue −/− mutant is impaired in transformation, reflecting a reduced capacity for HR. None of the mutants, with the possible exception of rad51-6−/−, was as compromised in transformation as rad51−/− cells, perhaps consistent with supporting roles in the recombination reaction.
Fig. 3.
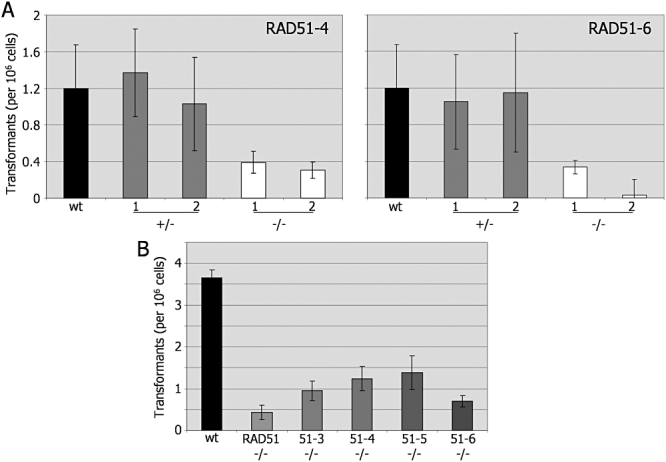
Homologous recombination in T. brucei RAD51 and RAD51 paralogue mutants. A. The number of transformants generated (per 106 cells put on antibiotic selection) when the construct tub-BLE-tub was electroporated into wild-type (wt) T. brucei, or into two independent heterozygous (+/− 1, 2) or homozygous (−/− 1, 2) mutants of RAD51-4 or RAD51-6, is shown. B. A comparison of the transformation frequency of wt, rad51−/−, rad51-3−/−, rad51-4−/−, rad51-5−/− and rad51-6−/− cells. Values are averages from at least three experiments; bars indicate standard deviation.
T. brucei RAD51 paralogues act non-equivalently in RAD51 foci formation
To ask if all the T. brucei RAD51 paralogues contribute to the function of RAD51 during DNA repair, we next examined the capacity of the mutants to support the relocalization of RAD51 to subnuclear foci following DNA damage. Previously, we have shown in T. brucei that RAD51 can be detected in discrete foci in the parasite cell's nucleus following phleomycin treatment (Proudfoot and McCulloch, 2005; Hartley and McCulloch, 2008) or the generation of a site-specific DSB by the I-SceI meganuclease (Glover et al., 2008). The formation of such foci appears to be a conserved response to various forms of genotoxic damage in eukaryotes and bacteria (Kidane and Graumann, 2005; Renzette et al., 2005), and the structures are thought to represent the concentration of a number of repair enzymes at sites of damage (Lisby and Rothstein, 2009). The formation or stabilization of RAD51 foci is, furthermore, dependent on a number of factors in eukaryotes, of which RAD51-3, RAD51-5 and BRCA2 have been shown to play a role in T. brucei after phleomycin treatment (Proudfoot and McCulloch, 2005; Hartley and McCulloch, 2008).
Figure 4A compares the number of RAD51 subnuclear foci detected by indirect immunofluorescence in RAD51-4 and RAD51-6 mutants after growth of the cells for 18 h in 1 µg ml−1 phleomycin. In the absence of phleomycin treatment, only a very small proportion (∼ 1–4%) of wt cells, or the +/− or −/− mutants, displayed RAD51 foci (data not shown) (Proudfoot and McCulloch, 2005; Hartley and McCulloch, 2008). In contrast, foci were seen in ∼ 80% of wt cells grown in phleomycin, with ∼ 50% displaying one to two foci and smaller numbers with ≥ 3 (an example of a wt cell with four foci is shown in Fig. 4B, and Fig. S4 shows examples of the range of foci formed). Although the protocol we have adopted here for RAD51 immunofluorescence is distinct from previous analyses (Proudfoot and McCulloch, 2005; Hartley and McCulloch, 2008), the DNA damage response in wt cells was very comparable. RAD51-4+/− and RAD51-6+/− cells also readily formed RAD51 foci, consistent with the lack of increased sensitivity to phleomycin-induced damage (Fig. 2). In contrast, rad51-6−/− cells were compromised in their ability to form RAD51 foci: in only 13–17% of the cells were RAD51 foci detectable, and this was primarily (10–11%) cells with a single focus (Fig. 4B). Because the rad51-6−/− mutants are more sensitive to phleomycin damage than wt or RAD51-6+/− cells, we also treated the cells with 0.25 µg ml−1 phleomycin. In these conditions, fewer foci were detected in wt (68% displayed foci, and 43% had only one focus) and rad51-6−/− cells (4–5% displayed one to three foci), showing that the impairment in RAD51 focal accumulation reflects the absence of RAD51-6 rather than abrogation of the DNA damage response due to increased cell death.
Fig. 4.
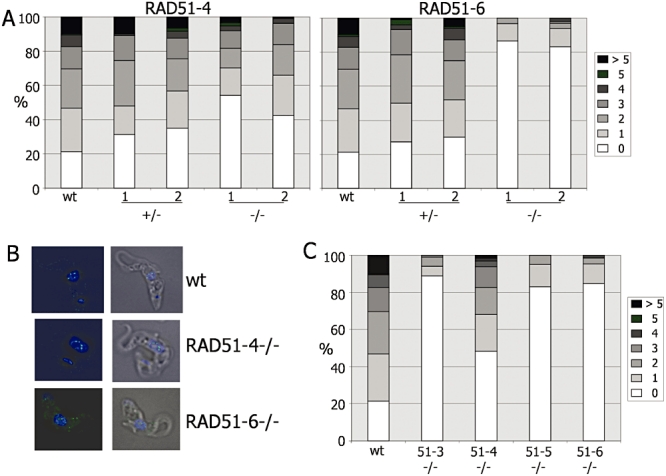
RAD51 subnuclear foci formation in T. brucei bloodstream form RAD51 paralogue mutants. A. Quantification of RAD51 foci formation in wild-type (wt) cells compared with two independent heterozygous (+/− 1, 2) or homozygous (−/− 1, 2) mutants of RAD51-4 or RAD51-6 after growth for 18 h in 1.0 µg ml−1 phleomycin; the number of subnuclear RAD51 foci in individual cells is shown as a percentage of the population (counting > 100 cells). B. Examples of RAD51 localization in wt, rad51-4−/−and rad51-6−/− heterozygous cells after 18 h growth in 1.0 µg ml−1 phleomycin. Cells were visualized by differential interference contrast (DIC), DNA was stained with DAPI, and RAD51 was visualized using polyclonal anti-RAD51 antiserum and SFX-conjugated goat-derived anti-rabbit secondary. Merged DAPI and RAD51 (left), and DAPI, RAD51 and DIC (right), images are shown. C. Quantification of RAD51 foci in wt, rad51−/−, rad51-3−/−, rad51-4−/−, rad51-5−/− and rad51-6−/− cells.
Figure 4A–C show that rad51-4−/− mutants are also compromised in their ability to form RAD51 foci, but that this is less marked than in other RAD51 paralogue mutants. After 18 h growth in 1 µg ml−1 phleomycin ∼ 45–58% of rad51-4−/− cells contained RAD51 foci, fewer than in wt (∼ 80%) or in RAD51-4+/− cells (66–68%), but considerably more than observed for the rad51-6−/− mutants. To ensure that this was not simply an effect seen at this concentration of drug, we analysed RAD51 foci after growth for 18 h in 0.25, 0.5 and 2.0 µg ml−1 phleomycin. The proportion of rad51-4−/− cells that contained RAD51 foci increased in parallel with increasing phleomycin, but was consistently lower than in wt cells (data not shown). To compare the contribution of the four RAD51 paralogues to RAD51 foci formation, each homozygous mutant was grown in 1 µg ml−1 phleomycin for 18 h (Fig. 4C), using a single preparation of medium and drug to avoid variation (see above). This confirmed our previous report that rad51-3−/− and rad51-5−/− mutants are impaired in RAD51 foci formation (Proudfoot and McCulloch, 2005), and highlights that rad51-4−/− mutants have markedly the least impairment in this process. Indeed, it is only in rad51-4−/− mutants that clearly defined RAD51 foci could be seen (see Fig. 4B and Fig. S5 for examples); in all the other mutants (Fig. 4C and Figs S5 and S6) punctate staining was more pronounced throughout the cells and may have been incorrectly scored as foci if it overlapped the nucleus. Finally, western blot analysis showed that RAD51 was expressed in each RAD51 paralogue −/− mutant (Fig. S7), demonstrating that impairment in RAD51 foci formation is not a consequence of loss of the recombinase.
T. brucei RAD51 paralogues act non-equivalently in antigenic variation
Given the roles of RAD51-4 and RAD51-6 in the repair of induced DNA damage and in HR, and their distinct contributions to RAD51 subnuclear focal accumulation after phleomycin treatment, we next asked if the factors act in T. brucei antigenic variation. As in previous studies, where we demonstrated a role for RAD51, RAD51-3 and BRCA2 in VSG switching (McCulloch and Barry, 1999; Proudfoot and McCulloch, 2005; Barnes and McCulloch, 2007; Hartley and McCulloch, 2008), RAD51-4 and RAD51-6 mutants were generated in the Lister 427-derived transgenic strain 3174, which allows VSG switching frequency and pathways to be measured from a number of switched variants that arise after a fixed number of generations growth in vitro (Rudenko et al., 1996; McCulloch et al., 1997) from a starting population that exclusively expresses a bloodstream expression site (BES1) containing VSG 427-2 (also called VSG221, or MITat1.2) (Hertz-Fowler et al., 2008). The switch frequencies determined for the RAD51-4 and RAD51-6 mutants are shown in Fig. 5A, comparing the −/− mutants with wt and +/− cells, as well as with paralogue re-expresser cells (−/−/+) generated in one of the −/− mutants. In each case, the data indicate a trend of lower numbers of switched variants recovered in the −/− mutants. However, although the switch frequencies were significantly different when comparing each independent −/− line with its +/− antecedent (each P-value < 0.02; unpaired, two-tailed Student's t-tests), significant differences were not observed uniformly relative to the wt or −/−/+ cells.
Fig. 5.
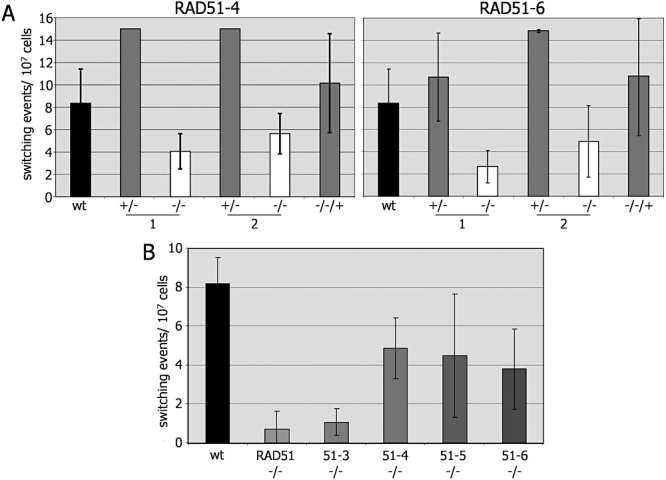
Effect of RAD51 or RAD51 paralogue mutation on the frequency of T. brucei VSG switching. A. Analysis of the effect if RAD51-4 or RAD51-6 mutation on VSG switching. The frequency with which VSG switch variants arose is shown in wild-type T. brucei cells (strain 3174.2; wt), two independent heterozygous (+/− 1, 2) or homozygous mutants (−/− 1, 2), and in −/− cells re-expressing the mutated gene (−/−/+). B. A comparison of VSG switching frequency in wt, rad51−/−, rad51-3−/−, rad51-4−/−, rad51-5−/− and rad51-6−/− cells. Values are the means of at least three independent experiments, and bars indicate standard deviation.
To ask about the contribution of each RAD51 paralogue to antigenic variation, the switch frequencies of all the −/− mutants were compared with each other, and with rad51−/− mutants and wt cells. To do this, the switch frequencies of the two independent −/− mutants generated for each RAD51 paralogue (rad51-4−/− and rad51-6−/− from this study; rad51-3−/− and rad51-5−/− from Proudfoot and McCulloch, 2005) were considered to be equivalent and switch frequencies averaged from all experiments performed (> 6 in each case). The switch frequency of the rad51−/− mutants was derived from three independent −/− mutants (nine experiments) (McCulloch and Barry, 1999), and the wt from 14 experiments (McCulloch and Barry, 1999; Proudfoot and McCulloch, 2005; Hartley and McCulloch, 2008). Taken together, these data show that rad51−/− and rad51-3−/− mutants show significantly impaired ability to undergo VSG switching, with average switch frequencies (0.7 × 10−7 and 1.1 × 10−7 switches per cell respectively) ∼ 12- and ∼ 8-fold reduced relative to wt (8.2 × 10−7 switches per cell). In contrast, rad51-4−/− and rad51-6−/− mutants were comparable with rad51-5−/− mutants in generating VSG switched variants approximately fourfold to fivefold more frequently than the rad51−/− or rad51-3−/− mutants and displaying, at best, only a minor impairment relative to wt cells. It seems likely therefore that RAD51-4, RAD51-6 and RAD51-5 either play no role in VSG switching, or that each individually provides a small contribution that the assay used here is too insensitive to detect. To ask further if RAD51-4 or RAD51-6 act in VSG switching, we assayed the relative contribution of recombination and in situ switching reactions, as described previously (McCulloch et al., 1997). No clear differences could be discerned in the rad51-4−/− or rad51-6−/− mutants relative to wt or +/− cells (Fig. S8). This result is perhaps not surprising, since in this T. brucei strain rad51−/−, rad51-3−/− and brca2−/− mutants, despite switching VSG less efficiently, display the same broad profiles of switching as wt cells (predominantly using either transcriptional switching or gene conversions from the silent BES, and using gene conversions limited to the region encompassing the VSG and upstream 70 bp repeats less frequently) (McCulloch et al., 1997; McCulloch and Barry, 1999; Proudfoot and McCulloch, 2005; Hartley and McCulloch, 2008). Nevertheless, this appears to rule out a major role for RAD51-4 or RAD51-6 in reaction pathway choice during VSG switching.
Analysis of T. brucei RAD51 paralogue interactions
The above genetic analyses indicate distinct functions of the T. brucei RAD51 paralogues in some aspects of DNA repair and immune evasion. To ask if this is because each paralogue acts as an isolated protein, or whether they form a complex, or complexes, we next sought to examine interactions between the proteins. The ORF of each RAD51 paralogue, and of RAD51, was cloned into the vectors pHybLex/Zeo and pYesTrp2 (Invitrogen), allowing yeast two-hybrid analysis by expressing the proteins as N-terminal fusions with the LexA DNA binding domain or with the B42 activating domain respectively. All pairwise combinations of pHybLex/Zeo-RAD51/paralogue and YesTrp2-RAD51/paralogue constructs were co-transformed into S. cerevisiae L40 cells and expression of each fusion protein confirmed by western analysis in two clonal co-transformants (data not shown). As a control, each of the pHybLex/Zeo-RAD51/paralogue vectors was also co-transformed with the pYesTrp2 vector alone. Interactions between the proteins were assayed via activation of a β-galactosidase reporter gene, by quantifying enzymatic activity in co-transformant lysates (Fig. 6A). The results were additionally verified by activation of a HIS3 reporter gene, by testing for growth of the co-transformants on agar plates lacking histidine and containing 15 mM 3′ aminotriazole (data not shown). Only in those cells coexpressing LexA-RAD51 and B42-RAD51, or expressing LexA-RAD51-3 and B42-RAD51-4 or B42-RAD51-6, was there evidence for β-galactosidase activity above background (LexA fusion alone). This suggests that RAD51 is capable of self-interaction, but does not interact with the RAD51 paralogues in these conditions, and that RAD51-3 can interact with RAD51-4 and RAD51-6. The RAD51-3–RAD51-6 interaction appeared to be directional, as it was not detected in the co-transformants expressing LexA-RAD51-6 and B42-RAD51-3. No interactions between RAD51-5 and any other protein were detected. Finally, it is not clear from these data if RAD51-4 and RAD51-6 can interact, since the LexA–RAD51-4 fusion protein caused activation of β-galactosidase when expressed alone (∼ 3000–6000 units; 15- to 30-fold higher than the background level seen for all other proteins; data not shown). To attempt to dissect this, we expressed the N-terminal 151-amino-acid residues of RAD51-4 as a LexA fusion, as well as the C-terminal 330-amino-acid residues, based on previous work (Miller et al., 2004) that predicted distinct N- and C-terminal domains in human RAD51 paralogues that can modulate interactions. However, even the truncated RAD51-4 proteins caused self-activation of the reporter genes, precluding interaction analysis (data not shown).
Fig. 6.
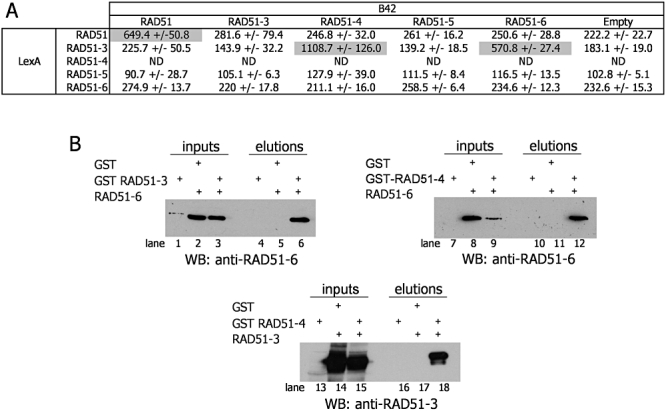
Interactions among T. brucei RAD51 paralogues and with RAD51. A. Yeast two-hybrid analysis of interactions. β-Galactosidase activity is shown for yeast cells carrying plasmids that express RAD51, RAD51-3, RAD51-4, RAD51-5 or RAD51-6 fused with a LexA DNA binding domain (LexA), in combination with plasmids expressing any of the proteins fused with a B42 transcriptional activation domain (B42); activity is also shown for control yeast cells carrying the LexA fusion-expressing plasmids and the B42 vector without a T. brucei gene (Empty). Values are the means of three independent experiments for two independent co-transformant yeast clones; deviation in activity around the mean is indicated (ND, not determined). B. Interactions between a GST–RAD51-3 fusion coexpressed in E. coli with native RAD51-6, or with a GST–RAD51-4 fusion coexpressed with native RAD51-3 or RAD51-6. Western blots (anti-RAD51 paralogue antiserum used is indicated by WB) are shown of the native proteins in cell extracts before purification (input), and after GST pull down of the GST fusion proteins from the cell extract (eluate) using glutathione beads. Controls are shown with GST fusion proteins expressed on their own, and with native RAD51 paralogues expressed with GST alone. Expression of the GST fusions, or of GST, by western analysis is not shown.
To validate and extend the above yeast two-hybrid analysis, we next assayed for interactions between the RAD51 paralogues when expressed in E. coli. To facilitate this analysis, each of the T. brucei RAD51 paralogues was expressed in E. coli with 10 histidine residues fused to the C-terminus, allowing each protein to be purified and polyclonal antisera to be raised; western analysis with the recombinant proteins showed that each antiserum was specific to the cognate, purified RAD51 paralogue (Fig. S9). To analyse interactions, we coexpressed combinations of native (from the plasmid pRSF1b; Novagen) and N-terminal glutathione S-transferase (GST) fusions (from the plasmid pGEX-4T3; GE life sciences) of the RAD51 paralogues. Glutathione beads were used to recover the GST fusion proteins from E. coli extracts, and western analysis with the RAD51 paralogue antiserum was then used to determine if a coexpressed, native RAD51 paralogue was also recovered, indicating stable interaction. As controls, the native RAD51 paralogues were coexpressed with GST alone and subjected to the same purification, and GST–RAD51 paralogue fusions were expressed on their own and recovered in the same way (to control for antibody cross-reactivity). The results of this analysis are shown in Fig. 6B. This confirms the yeast two-hybrid data that showed RAD51-3–RAD51-4 and RAD51-3–RAD51-6 interaction (Fig. 6B, lanes 6 and 12 respectively). In addition, this approach revealed interaction between RAD51-4 and RAD51-6 (Fig. 6B, lane 18). Unfortunately, we were unable to assess whether or not RAD51-5 interacts with any of the other RAD51 paralogues, as GST–RAD51-5 fusions were insoluble and native RAD51-5 was not detected using the available antiserum when coexpressed with GST fusions of the other RAD51 paralogues. Given that it is possible that the RAD51 paralogues are able to bind DNA, we were concerned this may give artefactual co-purification during the centrifugation of the extracts. We therefore repeated the analysis and treated the extracts with DNAseI prior to GST purification, with the same results (data not shown), consistent with protein-mediated interactions.
Having found evidence for interactions between the T. brucei RAD51 paralogues in heterologous systems, we now used the antisera against the proteins to attempt to ask if they form complexes in vivo. Western analysis of wt and RAD51 paralogue −/− mutants with each antiserum showed that only RAD51-3 and RAD51-4 could be detected in whole cell T. brucei extracts, even after affinity purification of each antiserum using cognate RAD51 paralogue protein purified from E. coli as a GST fusion (Fig. S9). Nevertheless, we performed immunoprecipitation with each antisera from wt and −/−T. brucei cells. Figure 7A shows that when RAD51-3 was immunoprecipitated from wt, but not rad51-3−/− cells, both RAD51-3 and RAD51-4 can be detected in the precipitated material, consistent with interaction. This was confirmed by immunoprecipitation of RAD51-4, which precipitated RAD51-3 also. In both of these immunoprecipitations, RAD51-5 and RAD51-6 could not be detected with the available antisera (data not shown). Immunoprecipitation with antiserum against RAD51-5 or RAD51-6 did not detect the cognate protein, nor any of the other RAD51 paralogues, including RAD51-3 or RAD51-4, so we suspect that these antisera are inadequate for this experimental analysis. Finally, we probed for RAD51 (readily detected in cell extracts; Fig. S7) in immunoprecipitates generated by the RAD51 paralogue antisera, and performed immunoprecipitation with anti-RAD51 antiserum and probed for the RAD51 paralogues in the immunoprecipitates, but found no evidence for interaction (data not shown).
Fig. 7.
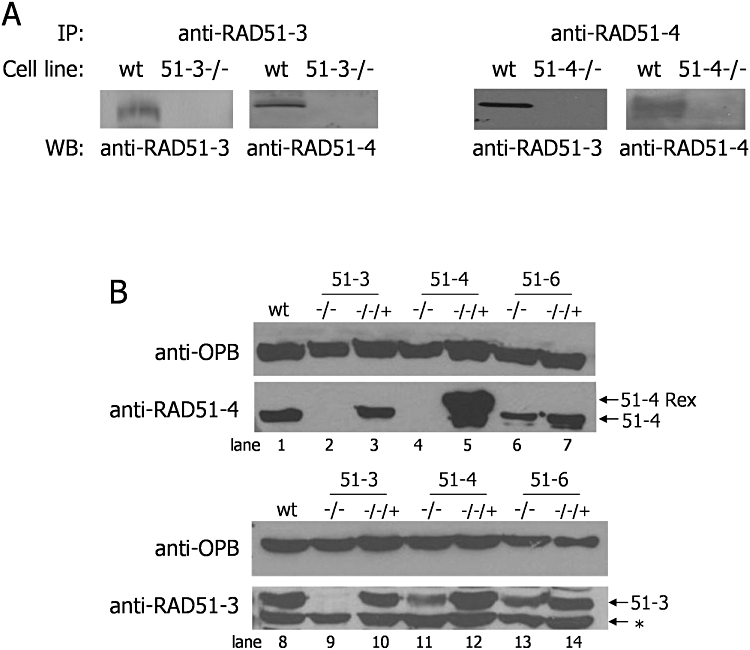
Interactions and expression interdependency of T. brucei RAD51 paralogues in vivo. A. RAD51-3 and RAD51-4 interact in vivo. Affinity-purified polyclonal antiserum against RAD51-3 or RAD51-4 was used to immunoprecipitate (IP) cognate protein from wild-type (wt) and rad51-3−/− or rad51-4−/− cells respectively. The presence or absence of RAD51-3 and RAD51-4 in all cases is shown by Western blots (WB) of the precipitated proteins, after SDS-PAGE, using anti-RAD51-3 and anti-RAD51-4 antiserum. B. Western blots are shown of whole cell extracts, after SDS-PAGE, from wt cells and from homozygous mutants (−/−) and re-expressers of RAD51-3, RAD51-4 and RAD51-6. Expression of RAD51-3 (51-3) or RAD51-4 (51-4) was assayed using affinity-purified polyclonal antiserum; a cross-reacting band is indicated in the anti-RAD51-3 blot (*), and a larger band seen only in the RAD51-4 re-expresser cell probed with anti-RAD51-4 antiserum is indicated (51-4Rex). As a loading control, the blots were stripped and re-probed with anti-OPB antiserum (gift, J. Mottram).
Expression levels of the RAD51 paralogues show interdependency
To probe potential interactions between the RAD51 paralogues further, we next asked if stable expression of the proteins is influenced by the presence or absence of the other factors. To do this, we concentrated on RAD51-3, RAD51-4 and RAD51-6, since we have evidence for interactions between the proteins in vivo and in heterologous systems. RAD51-3 and RAD51-4 protein levels were examined in Western blots of whole cell extracts from wt cells and compared with −/− mutants of each of the three genes and in re-expresser cell lines (−/−/+) (Fig. 7B). RAD51-4 was, as expected, undetectable in rad51-4−/− cells (lane 4). In addition, the protein was also absent in rad51-3−/− cells (lane 2). The same absence of RAD51-4 was not seen in rad51-6−/− cells (lane 6), although slightly lowered levels were apparent. Despite that fact that RAD51-4 abundance was strongly dependent on expression of RAD51-3, the opposite was not as clearly seen, despite considerable evidence that these proteins interact: RAD51-3 protein was detectable in rad51-4−/− cells (lane 11), though at lower levels than in wt or −/−/+RAD51-4 cells. In addition, there was little evidence for alteration of RAD51-3 levels in rad51-6−/− cells (lane 13). Taken together, these data are consistent with the observations of interactions among these three T. brucei RAD51 paralogues. It suggests, furthermore, that the nature of the interaction between RAD51-3 and RAD51-4 renders RAD51-4 unstable or reduces gene expression in the absence of RAD51-3, while putative RAD51-3–RAD51-6 and RAD51-4–RAD51-6 interactions do not have as pronounced an effect.
Discussion
In this study we have examined the functions of, and interactions among, RAD51 paralogues in the kinetoplastid parasite T. brucei, providing insight into the factors that contribute to DNA repair, recombination and antigenic variation in this important pathogen. In addition, this work provides an evolutionary perspective on the conservation of these recombination mediators (San Filippo et al., 2008), since functional analysis of recombination has been most extensive in fungi, vertebrates, plants and the nematode C. elegans, but less extensive in protists, which contribute a great deal of the evolutionary diversity of eukaryotes (Yoon et al., 2008). Within the RAD51 paralogues of T. brucei we show that one, RAD51-3, corresponds with the earliest emerging Rad51 paralogues in eukaryotes.
Variability in RAD51 paralogue numbers in eukaryotes
Four RAD51 paralogues are found in African and American Trypanosoma species, as defined by identification of Walker A and B boxes within recognizable RecA-fold homology (Brendel et al., 1997). These proteins are present in addition to RAD51 and its meiotic cousin DMC1 (McCulloch and Barry, 1999; Proudfoot and McCulloch, 2006), and represent a larger number of RAD51 paralogues identified by sequence homology than in any other single-celled eukaryote thus far examined (excluding potentially highly diverged Rad51 paralogues recently described in S. pombe) (Martin et al., 2006). The work described here validates these bioinformatic predictions for T. brucei, showing that mutants of each predicted RAD51 paralogue are phenotypically impaired in repair of induced DNA damage, recombination of transformed DNA into the nucleus and relocalization of RAD51 to subnuclear foci following phleomycin treatment. What feature(s) of Trypanosoma biology may have selected for this expanded repertoire of nuclear repair factors is unclear. For instance, it appears not to reflect genome size, as Trichomonas vaginalis encodes only two Rad51 paralogues, despite having a genome (∼ 160 Mb) (Carlton et al., 2007) around fivefold larger than T. brucei, while C. elegans (∼ 100 Mb genome) has only one paralogue, Rfs-1 (Ward et al., 2007).
In this study we have identified a broader range of eukaryotic RAD51 paralogues than has been documented to date (Table 1). To ask if these provide insight into the evolutionary origins of these proteins, we performed phylogenetic analyses that included bacterial RecA and archaeal RadA and RadB representatives (data not shown). In common with previous such analysis (Brendel et al., 1997; Lin et al., 2006; Haldenby et al., 2009), eukaryotic Rad51 and Dmc1 proteins (including all characterized protozoan Rad51 and Dmc1 orthologues; Table 1) grouped consistently with archaeal RadA, and bacterial RecA proteins grouped with organeller RecA-like proteins in eukaryotes, in each case suggesting common origins. The most complete analysis of eukaryotic Rad51 paralogue evolution to date is by Lin et al. (2006), who reported phylogenetic relationships primarily between vertebrates, plants and sea urchins, and found that the five Rad51 paralogues present in each grouped consistently, suggesting they were present in a common ancestor. Establishing the evolutionary relationships of the protistan Rad51 paralogues, including from T. brucei, was difficult as they did not consistently map to the well-supported groupings reported by Lin et al. This reflects the considerable sequence divergence of these proteins from each other and from the catalytic recombinases (Rad51, Dmc1, RadA and RecA). However, among protists that possess only one Rad51 paralogue (Table 1), these grouped most readily with Rad51C in some (e.g. E. histolytica) and XRCC3 in others (e.g. P. falciparum and T. gondii), both in phylogenetic analysis and as revealed by the ‘best’ hits in blast searches (data not shown). This seems consistent with the suggestion that these are the most ancient of the Rad51 paralogues (Lin et al., 2006). Perhaps surprisingly, given that mammalian Rad51C and XRCC3 interact (see below) (Masson et al., 2001a), in protists with two Rad51 paralogues, such as T. vaginalis and C. parvum, only one was identifiable as Rad51C-like or XRCC3-like, and the other was not easily assigned. T. brucei, T. cruzi and Leishmania sp., despite possessing more RAD51 paralogues, were consistent with this: in each, RAD51-3 most consistently grouped with Rad51C, and the other Rad51 paralogues had less certain orthology (Fig. 8; data not shown). Taken together, these data build upon the evolutionary picture of Lin et al. (Lin et al., 2006). Rad51C and/or XRCC3 appear to be the central, earliest emerging Rad51 paralogue(s), related to archaeal RadB, and in many protists these are the sole, extant orthologues. In other protists, further Rad51 paralogues are found. It is possible that gene duplication and diversification beyond the core Rad51C and/or XRCC3 Rad51 paralogue(s) occurred independently in different eukaryote lineages. It is alternatively possible that the overall smaller number of Rad51 paralogues found in most protists and fungi relative to animals and plants reflects gene loss from an initial repertoire of five Rad51 paralogues (Lin et al., 2006). Many sequenced protists are parasites and may therefore have undergone genome reduction similar to that seen in Encephalitozoon cuniculi (Gill and Fast, 2007), an intracellular microsporidian parasite related to fungi, though possessing only one RAD51 paralogue and lacking DMC1 (Table 1). Indeed, whole-genome comparisons between N. gruberi, a free-living protist that shares ancestry with kinetoplastids, suggest that parasitism has resulted in the loss of > 2000 conserved eukaryotic gene families in T. brucei (Fritz-Laylin et al., 2010). In this evolutionary context, the finding that Trypanosoma RAD51 paralogue numbers are potentially greater than in N. gruberi (Table 1) is striking. Irrespective of how the Rad51 paralogues evolved, the variability in their number and the extent of their sequence divergence appears to suggest considerable functional diversity, which we show is reflected in diversity in Rad51 paralogue interactions found in yeast, humans and T. brucei (see below). Moreover, it is possible that similar diversification has occurred during archaeal evolution, where variable numbers of RadA paralogues, including some distinct from RadB, have been described in a number of species (Haldenby et al., 2009).
Fig. 8.
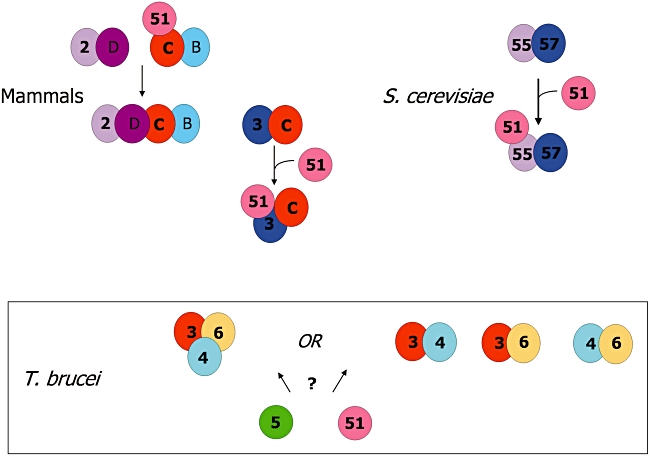
A comparison of RAD51 paralogue complexes, and their interaction with RAD51, in mammals, S. cerevisiae and T. brucei. The two principle Rad51 paralogue complexes found in mammals, XRCC2-Rad51D-Rad51C-Rad51B (2-D-C-B) and XRCC3-Rad51C (3-C), are shown as well as two further complexes (2-D, C-B). In S. cerevisiae, Rad55 (55) and Rad57 (57) are the only Rad51 paralogues, and form a stable complex. Experimentally determined interactions between the mammalian and yeast Rad51 paralogue complexes and Rad51 (51) are shown. In T. brucei, three Rad51 paralogues, RAD51-3 (3), RAD51-4 (4) and RAD51-6 (6), interact and experiments indicate either a stable trimer or distinct dimeric complexes; interactions with RAD51 or with a fourth Rad51 paralogue, RAD51-5 (5), have not been detected. Proteins are coloured to indicate potential orthology in the three organisms: Rad51 is orthologous in all (pink); Rad57 is considered orthologous with XRCC3 (dark blue) (Lin et al., 2006); and we propose that T. brucei RAD51-3 is orthologous with Rad51C (red).
Interactions among T. brucei RAD51 paralogues
The work presented here indicates that at least three of the T. brucei RAD51 paralogues do not function in isolation. Instead, and in common with Rad51 paralogues from yeast and mammals, they are found as a complex or complexes. Yeast two-hybrid analysis demonstrated RAD51-3 interaction with both RAD51-4 and RAD51-6. These interactions were confirmed by in vitro GST pull down analysis, which additionally revealed interaction between RAD51-4 and RAD51-6. Finally, co-immunoprecipitation of RAD51-3 and RAD51-4 from T. brucei cell extracts confirmed interaction in vivo. Taken together, these data are compatible with these T. brucei RAD51 paralogues forming a single trimeric complex. Alternatively, interaction between the RAD51 paralogues may be more dynamic, with one or more dimeric sub-complexes in existence; Fig. 8 provides an extreme example, with three distinct dimeric sub-complexes, although we cannot discount there being one predominant dimer that associates with a third RAD51 paralogue in some circumstances. A number of observations suggest that a stable, trimeric complex is unlikely. First, we find that the contributions of RAD51-3, RAD51-4 and RAD51-6 to DNA repair are not equivalent, since rad51-4−/− mutants are significantly less sensitive to phleomycin-induced DNA damage than any other T. brucei RAD51 paralogue mutant. Second, and consistent with the above, rad51-4−/− mutants were markedly different from other T. brucei RAD51 paralogue mutants in that RAD51 foci numbers increased strongly following phleomycin-induced DNA damage, indicating a lesser role in either the formation or stabilization of these structures. Finally, we document variable levels of protein expression interdependency among the T. brucei RAD51 paralogues. Strikingly, RAD51-4 is not detectably expressed in a rad51-3−/− mutant, whereas RAD51-3 can be detected in a rad51-4−/− mutant. We are currently unable to assess RAD51-6 expression in these mutants, but find that in rad51-6−/− mutants RAD51-3 and RAD51-4 are both detectably expressed. These data are comparable with studies in human cells, where RNAi depletion of Rad51C resulted in similar depletion of XRCC3, but had less effect, if any, on the levels of Rad51D, Rad51B or RAD51, despite evidence that they also interact with Rad51C (Lio et al., 2004). Such expression interdependency presumably reflects co-stabilization of proteins in a heterodimer. The fact that T. brucei RAD51-4 stability is dependent on the presence of RAD51-3, but not vice versa, suggests that RAD51-3 is the central component of T. brucei RAD51 paralogue interactions. This appears telling, as it mirrors the centrality of mammalian Rad51C in two different complexes (XRCC2-Rad51D-Rad51C-Rad51B and XRCC3-Rad51C; Fig. 8), and is compatible with the proposed evolutionary relationship between T. brucei RAD51-3 and mammalian and plant Rad51C (see above).
Further work will be required to determine the topology of interactions between T. brucei RAD51-3, RAD51-4 and RAD51-6. We also have not yet been able to determine if T. brucei RAD51 interacts with any, or all, of these factors. This is possible (Fig. 8), as Rad51 has been documented to interact with mammalian Rad51C in the Rad51C-Rad51B sub-complex (Schild et al., 2000; Masson et al., 2001b; Lio et al., 2003) and with XRCC3 in the Rad51C-XRCC3 complex (Liu et al., 2002), while S. cerevisiae Rad51 binds Rad55 in the Rad55-Rad57 complex (Hays et al., 1995; Johnson and Symington, 1995; Sung, 1997). Equally importantly, it is unclear whether or not RAD51-5 interacts with the putative T. brucei RAD51 paralogue complexes. Yeast two-hybrid analysis failed to reveal any interactions between this factor and the other T. brucei RAD51 paralogues, or with RAD51, while technical details hampered our attempts to assay such interactions in vitro or in vivo. RAD51-5 is the smallest, most diverged protein in the T. brucei RAD51 paralogue family (Fig. 1). Homology searches of Leishmania species revealed only three RAD51 paralogues, with RAD51-5 not identified (Table 1). However, synteny analysis identified a gene on L. major chromosome 33 that is positionally conserved with T. brucei RAD51-5 (chromosome 10; Fig. S10). Though encoding a protein with only distant homology with T. brucei RAD51-5 (∼ 14% amino acid identity), this appears to have arisen from a RAD51 paralogue, retaining a Walker B motif but lacking a Walker A motif or wider RecA-fold homology (Fig. S11). This may suggest recent, Leishmania-specific neofunctionalization of RAD51-5, perhaps consistent with a distinct role for RAD51-5 in kinetoplastid repair and recombination, which may have ceased to be important in the biology of Leishmania parasites.
Comparing the interaction data for mammalian Rad51 paralogues with the data we present here, and with the stable heterodimer found in S. cerevisiae, fails to reveal an evolutionarily conserved pattern. In mammals two primary complexes are found, XRCC2-Rad51D-Rad51C-Rad51B and XRCC3-Rad51C, and in addition two sub-complexes of the former, XRCC2-Rad51D and Rad51C-Rad51B (Masson et al., 2001a,b; Liu et al., 2002; Miller et al., 2002; Wiese et al., 2002). It has been argued that S. cerevisiae Rad57 is orthologous with XRCC3, and Rad55 with XRCC2 (Lin et al., 2006). The fact that the yeast proteins form a stable heterodimer (Sung, 1997) is then surprising, as no evidence has found direct interaction between XRCC2-XRCC3 in mammals, and they are not common to a complex (Fig. 8). The work here represents the only analysis of Rad51 paralogue interactions to date beyond yeast and mammals. We have argued that T. brucei RAD51-3 is Rad51C-like, but cannot confidently predict the orthology of RAD51-4, RAD51-5 or RAD51-6. We suggest that this reflects flexibility in Rad51 paralogue interactions in the eukarya. Although Rad51C or XRCC3-like factors appear central to Rad51 paralogue complexes when they form, the interacting factors are less conserved. Moreover, it appears that Rad51 paralogue complexes are needed only in some organisms, since a number of protists (Table 1) and examples of microsporidia, fungi and nematodes are found that appear to utilize only a single Rad51 paralogue (Table 1) (Kojic et al., 2006; Ward et al., 2007). This further underlines the flexibility in Rad51 paralogue evolution.
Functions of T. brucei RAD51 paralogues?
The relatively broad phenotypic analysis of the T. brucei RAD51 paralogue mutants described here allows us only some insight into potential functions of these factors. Each contributes to the repair of phleomycin- and MMS-induced DNA damage, to the genomic recombination of transformed DNA constructs and to the relocalization of RAD51 into discrete foci following phleomycin damage. Although these contributions are not equivalent, since RAD51-4 appears less important in DNA repair and RAD51 foci dynamics, these data argue that each factor acts in RAD51-dependent HR. In any eukaryotic organism, the detailed functions of Rad51 paralogues are still being determined (San Filippo et al., 2008). In S. cerevisiae, mutation of Rad55 affects the recruitment of Rad51 to DNA damage (Sugawara et al., 2003), while the Rad55-Rad57 heterodimer promotes Rad51 strand exchange in vitro (Sung, 1997), consistent with mediating Rad51 function. In addition, Rad55 is phosphorylated in response to DNA damage, providing a checkpoint function that may be more important in stalled replication restart that in repair of an induced break (Herzberg et al., 2006). In mammals, each Rad51 paralogue complex contributes to Rad51-mediated repair, since mutations of any Rad51 paralogue results in increased DNA damage sensitivity (Takata et al., 2001) and attenuates Rad51 foci formation (van Veelen et al., 2005), similar to what we describe for T. brucei. Further work has begun to detail functions of the mammalian Rad51 paralogue complexes, consistent with mediating Rad51 function: Rad51B-Rad51C binds DNA and can promote Rad51 strand exchange (Sigurdsson et al., 2001); Rad51D-XRCC2 binds DNA (Kurumizaka et al., 2002); and both XRCC2-Rad51D-Rad51C-Rad51B and XRCC3-Rad51C bind to DNA junction structures in preference to intact DNA (Masson et al., 2001b; Yokoyama et al., 2004; Compton et al., 2010). However, increasing evidence also points to functions for Rad51 paralogues and their complexes beyond simply aiding Rad51 strand exchange. Mammalian Rad51C-XRCC3 has been associated with Holliday junction resolution (Liu et al., 2007), a late stage in recombination that may explain longer gene conversion tracts in mutants (Brenneman et al., 2002). Rad51C in mammals has been implicated in checkpoint signalling following DNA damage (Badie et al., 2009), in translocation of Rad51 to the nucleus (Gildemeister et al., 2009) and in mitosis (Renglin et al., 2007). Finally, mammalian Rad51D has been implicated in telomere maintenance (Tarsounas et al., 2004b), which may be a conserved function of the sole C. elegans Rad51 paralogue, RFS-1 (Yanowitz, 2008), which is considered to be Rad51D-like (Lin et al., 2006) and to play specific roles in recombination at replication blocks, rather than in general break repair (Ward et al., 2007). Given this, if the T. brucei RAD51 paralogues do indeed form a number of complexes, they may provide redundant functions in RAD51 strand exchange, or they may provide specific functions, perhaps in particular genomic contexts or in response to different DNA lesions.
In contrast to the equivalent contributions of T. brucei RAD51-3, RAD51-5 and RAD51-6 to the repair of phleomycin damage, the relative contributions of the proteins to the efficiency of VSG switching is distinct. Here, we find clear evidence for RAD51-3 acting to promote the process but, at best, only a minor contribution of RAD51-5 (Proudfoot and McCulloch, 2005), RAD51-6 and RAD51-4. Two explanations are possible. First, it is possible that the three RAD51 paralogue complexes we propose exist and each contributes to VSG switching. We have shown that rad51-3−/− mutants ablate both RAD51-3 and RAD51-4 expression, whereas RAD51-3 is expressed in rad51-4−/− and rad51-6−/− mutants, and RAD51-4 is expressed in rad51-6−/− cells. Thus, the greater effect of the rad51-3−/− mutation in VSG switching might arise because all complexes are functionally ablated, whereas RAD51-3–RAD51-6 and RAD51-3–RAD51-4 complexes remain in rad51-4−/− and rad51-6−/− cells respectively. If correct, this could either indicate redundancy in a specific step (such as RAD51-directed strand exchange), or that the complexes contribute to different recombination pathways that can fuel VSG switching. A complication in this scenario, and indeed in eukaryotic recombination in general, is BRCA2. This factor is also known to promote Rad51 strand exchange, most likely by co-ordinating the formation and disassembly of the recombinase nucleoprotein filaments (Lord and Ashworth, 2007; Jensen et al., 2010). In T. brucei, BRCA2 also promotes VSG switching, since brca2−/− mutants are defective to an extent comparable with rad51−/− and rad51-3−/− mutants (Hartley and McCulloch, 2008). To what extent BRCA2 function overlaps with, or even competes with, that of Rad51 paralogues is unknown. The second explanation is that RAD51-3 alone provides a VSG switching-specific recombination function. What this might be is unclear, but perhaps RAD51-3 acts on VSG switch-initiating lesions (Barry and McCulloch, 2009; Boothroyd et al., 2009), recombination intermediates (Kim and Cross, 2010) or links the process to cell cycle progression. Alternatively, recent work has shown that mammalian Rad51 paralogues can interact with other DNA repair mechanisms and cellular processes (Wilson et al., 2008; Rajesh et al., 2009), and perhaps this is the key to understanding T. brucei VSG switching.
Experimental procedures
T. brucei strains, growth, transformation and VSG switching
Trypanosoma brucei bloodstream cells of strain 3174, a derivative of MITat1.2a (McCulloch et al., 1997), were used throughout and grown at 37°C in HMI-9 medium (Hirumi and Hirumi, 1989). Cell concentration was determined with a haemocytometer (bright-line, Sigma). Electroporation of DNA constructs was conducted as described previously (Conway et al., 2002b; Bell and McCulloch, 2003). For both RAD51-4 and RAD51-6 mutants, transformants were selected with 0.5 µg ml−1 puromycin or 2.5 µg ml−1 blasticidin. Recombination was assayed by transformation with 5 µg of tub-BLE-tub (pRM450) digested with NotI and ApaI, and transformants selected with 2 µg ml−1 phleomycin, as described previously (Proudfoot and McCulloch, 2005). VSG switching frequency was determined as described previously (Proudfoot and McCulloch, 2005; Hartley and McCulloch, 2008). The type of VSG switching reactions used in the cells was determined by a combination of PCR and antibiotic resistance, as described previously (Proudfoot and McCulloch, 2005; Hartley and McCulloch, 2008), and no clear differences in pathway usage were observed in the mutants relative to wt (data not shown).
Generation and analysis of RAD51-4 and RAD51-6 mutants
Constructs for gene disruption employed cassettes for puromycin N-acetyl transferase (puromycin) or blasticidin S deaminase (blasticidin) expression described previously (Hartley and McCulloch, 2008). These were flanked by 5′ and 3′ sequences derived from the RAD51-4 or RAD51-4 ORFs that had been PCR-amplified (primers available on request). For RAD51-4, the 5′ flank was 290 bp beginning at the predicted start codon; the 3′ flank was 402 bp and ended at the stop codon. For RAD51-6, the 5′ flank was also 290 bp, beginning at the start codon, while the 3′ flank was 354 bp, ending at the codon prior to the stop. All gene disruption constructs were digested with XbaI and XhoI before transformation. Re-expression constructs for RAD51-4 and RAD51-6 was generated by PCR-amplifying the genes' ORFs with Herculase polymerase and cloning them into EcoRV-digested construct pRM481. Drug-resistant transformants were analysed by probing Southern blots of restriction digested genomic DNA separated on 0.8% agarose gels, and using DNA fragments labelled using α-32P. For RT-PCR, total RNA was prepared from T. brucei using an RNeasy kit (Qiagen) and cDNA was synthesized with random hexamers and Superscript reverse transcriptase (Life Technologies). Primers to assay for gene expression are available on request.
Assaying DNA damage sensitivity
Sensitivity of the cells to both MMS and phleomycin was assessed by the metabolic reduction of Alamar blue (Raz et al., 1997). Here, cells in mid-logarithmic growth were plated at a density of 1 × 105 cells ml−1 in a 96-well dish (200 µl well−1) in HMI-9 medium containing doubling dilution concentrations of drug. After 48 h growth, 20 µl of Alamar blue (from a 12.5 mg ml−1 stock of resazurin, Sigma) was added to each well and the plates incubated in the dark for 24 h, when the fluorescence in each well was determined fluorometrically (PerkinElmer LS55) at 530 nm excitation wavelength and 590 nm emission wavelength. In addition, the sensitivity of the cell lines to the agents was analysed by a clonal survival assay that has been described before (Conway et al., 2002a; Robinson et al., 2002; Proudfoot and McCulloch, 2005; 2006;) (data not shown).
RAD51 immunolocalization
RAD51 localization was performed by a modified technique to that described previously (Proudfoot and McCulloch, 2005; Hartley and McCulloch, 2008). Cells were grown to a density of approximately 1 × 106 cells per millilitre, and pelleted at 600 g for 1 min. The supernatant was removed, and the cells were resuspended in 100 µl PBS (Sigma). 1 ml of 1% (v/v) formaldehyde/PBS was added and the solution inverted several times and then stored at 4°C for 1–24 h. The formaldehyde-fixed cells were then centrifuged at 600 g for 1 min at 4°C. The pellet was washed twice in 1 ml chilled PBS, followed by addition of 1% Triton X-100 (Sigma) to a final concentration of 0.1%. This was incubated at room temperature for 10 min, then glycine (Sigma) was added to a final concentration of 0.1 M and incubated at room temperature 10 min. The cells were pelleted and resuspended in 200 µl of PBS and spread evenly onto a glass slide (Menzel-glaser) and allowed to dry for 2–18 h. The slides were next washed with 1 × PBS for 5 min in Coplin jars and then blocked with TB buffer (0.1% Triton X-100, 0.1% BSA in PBS) for 15 min. This was removed and rabbit polyclonal anti-RAD51 antiserum, diluted 1:500 in TB buffer, was added and left for 55 min. The slides were then washed three times in PBS, 5 min each time, and secondary antiserum (goat anti-rabbit conjugated with SFX; fluorescein, succinimidyl ester, Molecular Probes), diluted in TB buffer, was added for 45 min in the dark. The slides were then washed as before and counter stained with two drops of VectorShield with 4,6-diamidino-2-phenylindole (DAPI; Vector Labs). A coverslip was then placed on the slides and sealed with nail polish. Once dried, the slides were visualized using an Axioskop 2 microscope (Zeiss) and Openlab 5.00 software (Improvision). The images were prepared using PhotoShop 6 (Adobe).
Yeast two-hybrid analysis
Saccharomyces cerevisiae L40 strain was grown on YPD media [10 g of yeast extract (Melford), 20 g of bactopeptone (Melford) and 20 g of dextrose (Fisher) in 1 L at pH 5.8, supplemented with adenine at 0.01%; Sigma] at 30°C. If plates were required, agar was added per manufacturer's instructions (Melford). Minimal media (YC-W; Clontech) was used for the growth of cells containing pYesTrp2-Prey constructs, while selection with 300 µg ml−1 zeocin (Invitrogen) was used for cells containing pHybLex/Zeo-Bait constructs. β-Galactosidase was assayed by a kit by Pierce, following manufacturer's instructions. Briefly, two independent co-transformant clones were grown as liquid cultures in YC-W with 300 µg ml−1 zeocin at 30°C for 48 h. Cell density was quantified by measuring the optical density (OD) at 620 nm. Cell lysis solution and o-nitrophenyl-β-D-galactopyranoside (ONPG) was added to the cells. β-Galactosidase-mediated conversion of ONPG to o-nitrophenol was quantified by measuring absorbance at 420 nm using a 96-well plate reader (Wallace Envision, 2102 Multi-label reader). β-Galactosidase activity was calculated using the following equation: (1000 × Absorbance 420 nm) ÷ (t × V × OD 620 nm), where t is the assay reaction time in minutes and V is the volume of culture used in the assay in ml. β-Galactosidase activity of the cells was also assayed by a filter lift procedure (data not shown), and interactions assayed via histidine auxotrophy (data not shown); details of these approaches can be supplied.
Purification of RAD51 and RAD51 paralogues, and antiserum generation
RAD51 and RAD51 paralogues were purified both as histidine (His)-tagged variants and as GST-tagged variants. The genes were C-terminally tagged with 10 × His by cloning the full-length ORFs into pET51b after PCR amplification; N-terminal GST fusions were generated by cloning in pGex-4T3 (Amersham). All proteins were expressed after transforming the plasmids into E. coli Rosetta 2 cells (Novagen). RAD51-His expression was induced with 1 mM IPTG for 2–4 h at 37°C, yielding soluble protein. All His-tagged RAD51 paralogues were expressed in the same way, but were purified from inclusion bodies. Purification was performed using a 5 ml Histrap column connected to an AKTAprime (both GE Healthcare). After induction, cells were harvested by centrifugation (5200 g, 20 min) and resuspend in 10–20 ml binding buffer (20 mM Na phosphate pH 7.5, 0.5 M NaCl, 20 mM imidazole, 1 mM DTT for soluble protein; supplemented with 8 M Urea for insoluble protein). For soluble protein, the cells were then lysed by sonication on an ice/water mix (10 cycles of 20 s on/30 s off, amplitude 18) and centrifuged (20 000 g, 4°C, 25 min). For insoluble protein, the cells were lysed using a dounce homogenizer and centrifuged at room temperature (20 000 g, 25 min). In either case, supernatant was then applied to the column after filtering (0.2 µm syringe filter for soluble, vacuum filter for insoluble). Proteins were recovered from the column in elution buffer (20 mM Na phosphate pH 7.5, 0.5 M NaCl, 400 mM imidazole, 1 mM DTT for soluble protein; supplemented with 8 M Urea for insoluble protein), after a wash in 30% elution buffer. Proteins were then dialysed (4°C, 3 times, using 30 volumes for 90 min per step) in non-denaturing buffer (for RAD51, RAD51-3, RAD51-4 and RAD51-6, 20 mM Tris-Cl pH 7.5, 0.5 M NaCl, 1 mM DTT, 50% glycerol; for RAD51-5, same buffer but with 0.2 M NaCl), centrifuged (20 000 g, 4°C, 15 min), snap-frozen on dry ice and stored at −80°C before being sent for antibody production. The conditions for expression and purification of the GST fusion proteins are described for GST pull downs (below). Proteins were purified from cell extracts using 1 ml GST trap HP columns connected to an AKTAprime (both GE Healthcare) following the manufacturers' instructions and using the binding and elution buffers described below. It was not possible to express GST–RAD51-5 solubly, and therefore untagged protein was purified from inclusion bodies using anion and cation exchange (data not shown). Polyclonal antiserum were raised (Scottish Blood Transfusion Board, Edinburgh) against the His-tagged proteins in lop-eared rabbits (RAD51 and RAD51-4), sheep (RAD51-3), rats (RAD51-6) or chicken (RAD51-5). For anti-RAD51-5 antiserum IgY was purified from the egg yolk using a chicken IgY purification kit (Pierce), according to manufacturer's instructions. Where necessary, the antisera were affinity-purified using GST fusion proteins. Here, the GST fusion proteins were eluted in 50 mM Na phosphate pH 8.0, 10 mM reduced glutathione, 200 mM NaCl, 1 mM DTT. Using a buffer exchange column (Bio-Rad) they were then moved to coupling buffer (4 M Guanidine HCl, 100 mM Na phosphate pH 6.4, 0.05% Na-azide), protein concentration determined (Bio-Rad microassay) and coupled to beads (Aminolink resin; Pierce) to form an affinity column. For affinity purification, the resin was washed with 5 ml 0.05% sodium azide/PBS (Sigma) and antiserum added. Approximately 100 µl of 0.05% sodium azide/PBS was added and allowed to flow through, after which the column was closed at the bottom, 500 µl of 0.05% sodium azide/PBS added, the top capped and incubated at room temperature for 1 h. Next, the column was washed with 12 ml of PBS and antiserum eluted (in 0.5 ml fractions) by addition of 4 ml of 100 mM glycine-HCl (pH 2.7). Fractions were neutralized by addition of 50 µl of 1 M Tris-HCl (pH 7.5) and protein concentrations determined (Bradford assay, Bio-Rad) after dilution with 40 volumes oH2O. The buffer of fractions to be used was exchanged into PBS using a Bio-Rad Econo-Pac 10DG column, following manufacturer's instructions.
GST pull down
GST fusions were expressed from pGex-4T3 (Amersham) and untagged proteins were expressed from pRSF1b (Merck); in all cases full-length T. brucei RAD51 or RAD51 paralogue ORFs were cloned by PCR (primers available on request). Normally, Rosetta E. coli cells were transformed the GST fusion and untagged protein expression vectors to allow coexpression and GST pull down. In cases where coexpression was not seen, the GST fusion and untagged proteins were expressed individually and GST pull down performed by combining individual cells extracts. In either case, a 50 ml culture of E. coli cells carrying the appropriate overexpression vectors, maintained by selection with antibiotics, were grown at 20°C until an OD of ∼ 0.5, at which time protein expression was induced with 1 mM IPTG overnight. Cells were then harvested by centrifugation (3750 g, 20 min, 4°C), resuspended in wash/binding buffer (PBS, containing 1 mM DTT, 0.1% NP40, 0.1% DNaseI and protease inhibitors; Roche) and stored at −80°C. The resuspensions were allowed to thaw at room temperature and were then lysed by sonication (4 cycles: 10 s on, 20 s off) and centrifuged (20 000 g, 20 min, 4°C). Samples of the supernatant were retained as input. To perform the GST pull down, glutathione sepharose beads (100 µl per cell extract) were washed twice with 5 ml wash/binding buffer and then resuspended in 10 volumes of the same buffer, forming a bead-slurry. Supernatant from the E. coli extract, or extracts, was added to the slurry and incubated at 4°C with rotation for 1 h. The beads were then harvested by centrifugation (500 g, 2 min, 4°C) and washed four times with 5 ml of wash/binding buffer. After the final wash, 0.5 ml of elution buffer (50 mM Tris-Cl pH 8.0, 200 mM NaCl, 20 mM glutathione, 0.1% NP40, 1 mM DTT) was added to the beads and incubated at 4°C with rotation for up to 30 min. Proteins were eluted from the beads by centrifugation at 500 g for 2 min (4°C) and the supernatant (eluate) added to protein loading buffer. After running on an SDS-PAGE gel, proteins were visualized with unpurified anti-RAD51 paralogue antiserum (see above) or with anti-GST antiserum (Novagen).
Acknowledgments
This work was supported by grants from the Medical Research Council and the Wellcome Trust. We thank Chris Irons for his experimental input, Marko Prorocic for his comments, and Lillian Fritz-Laylin and Michael Ginger for discussions regarding Naegleria.
Supporting information
Additional supporting information may be found in the online version of this article.
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Abdu U, Gonzalez-Reyes A, Ghabrial A, Schupbach T. The Drosophila spn-D gene encodes a RAD51C-like protein that is required exclusively during meiosis. Genetics. 2003;165:197–204. doi: 10.1093/genetics/165.1.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badie S, Liao C, Thanasoula M, Barber P, Hill MA, Tarsounas M. RAD51C facilitates checkpoint signaling by promoting CHK2 phosphorylation. J Cell Biol. 2009;185:587–600. doi: 10.1083/jcb.200811079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbet AF, Kamper SM. The importance of mosaic genes to trypanosome survival. Parasitol Today. 1993;9:63–66. doi: 10.1016/0169-4758(93)90039-i. [DOI] [PubMed] [Google Scholar]
- Barnes RL, McCulloch R. Trypanosoma brucei homologous recombination is dependent on substrate length and homology, though displays a differential dependence on mismatch repair as substrate length decreases. Nucleic Acids Res. 2007;35:3478–3493. doi: 10.1093/nar/gkm249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry D, McCulloch R. Molecular microbiology: a key event in survival. Nature. 2009;459:172–173. doi: 10.1038/459172a. [DOI] [PubMed] [Google Scholar]
- Barry JD, McCulloch R. Antigenic variation in trypanosomes: enhanced phenotypic variation in a eukaryotic parasite. Adv Parasitol. 2001;49:1–70. doi: 10.1016/s0065-308x(01)49037-3. [DOI] [PubMed] [Google Scholar]
- Beam CE, Saveson CJ, Lovett ST. Role for radA/sms in recombination intermediate processing in Escherichia coli. J Bacteriol. 2002;184:6836–6844. doi: 10.1128/JB.184.24.6836-6844.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell JS, McCulloch R. Mismatch repair regulates homologous recombination, but has little influence on antigenic variation, in Trypanosoma brucei. J Biol Chem. 2003;278:45182–45188. doi: 10.1074/jbc.M308123200. [DOI] [PubMed] [Google Scholar]
- Bell JS, Harvey TI, Sims AM, McCulloch R. Characterization of components of the mismatch repair machinery in Trypanosoma brucei. Mol Microbiol. 2004;51:159–173. doi: 10.1046/j.1365-2958.2003.03804.x. [DOI] [PubMed] [Google Scholar]
- Berriman M, Ghedin E, Hertz-Fowler C, Blandin G, Renauld H, Bartholomeu DC, et al. The genome of the African trypanosome Trypanosoma brucei. Science. 2005;309:416–422. doi: 10.1126/science.1112642. [DOI] [PubMed] [Google Scholar]
- Bhattacharyya MK, Bhattacharyya nee DS, Jayabalasingham B, Kumar N. Characterization of kinetics of DNA strand-exchange and ATP hydrolysis activities of recombinant PfRad51, a Plasmodium falciparum recombinase. Mol Biochem Parasitol. 2005;139:33–39. doi: 10.1016/j.molbiopara.2004.09.007. [DOI] [PubMed] [Google Scholar]
- Bishop DK, Ear U, Bhattacharyya A, Calderone C, Beckett M, Weichselbaum RR, Shinohara A. Xrcc3 is required for assembly of Rad51 complexes in vivo. J Biol Chem. 1998;273:21482–21488. doi: 10.1074/jbc.273.34.21482. [DOI] [PubMed] [Google Scholar]
- Bleuyard JY, White CI. The Arabidopsis homologue of Xrcc3 plays an essential role in meiosis. EMBO J. 2004;23:439–449. doi: 10.1038/sj.emboj.7600055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleuyard JY, Gallego ME, Savigny F, White CI. Differing requirements for the Arabidopsis Rad51 paralogs in meiosis and DNA repair. Plant J. 2005;41:533–545. doi: 10.1111/j.1365-313X.2004.02318.x. [DOI] [PubMed] [Google Scholar]
- Boothroyd CE, Dreesen O, Leonova T, Ly KI, Figueiredo LM, Cross GA, Papavasiliou FN. A yeast-endonuclease-generated DNA break induces antigenic switching in Trypanosoma brucei. Nature. 2009;459:278–281. doi: 10.1038/nature07982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borst P. Antigenic variation and allelic exclusion. Cell. 2002;109:5–8. doi: 10.1016/s0092-8674(02)00711-0. [DOI] [PubMed] [Google Scholar]
- Brendel V, Brocchieri L, Sandler SJ, Clark AJ, Karlin S. Evolutionary comparisons of RecA-like proteins across all major kingdoms of living organisms. J Mol Evol. 1997;44:528–541. doi: 10.1007/pl00006177. [DOI] [PubMed] [Google Scholar]
- Brenneman MA, Weiss AE, Nickoloff JA, Chen DJ. XRCC3 is required for efficient repair of chromosome breaks by homologous recombination. Mutat Res. 2000;459:89–97. doi: 10.1016/s0921-8777(00)00002-1. [DOI] [PubMed] [Google Scholar]
- Brenneman MA, Wagener BM, Miller CA, Allen C, Nickoloff JA. XRCC3 controls the fidelity of homologous recombination: roles for XRCC3 in late stages of recombination. Mol Cell. 2002;10:387–395. doi: 10.1016/s1097-2765(02)00595-6. [DOI] [PubMed] [Google Scholar]
- Burton P, McBride DJ, Wilkes JM, Barry JD, McCulloch R. Ku heterodimer-independent end joining in Trypanosoma brucei cell extracts relies upon sequence microhomology. Eukaryot Cell. 2007;6:1773–1781. doi: 10.1128/EC.00212-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlton JM, Hirt RP, Silva JC, Delcher AL, Schatz M, Zhao Q, et al. Draft genome sequence of the sexually transmitted pathogen Trichomonas vaginalis. Science. 2007;315:207–212. doi: 10.1126/science.1132894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaves I, Rudenko G, Dirks-Mulder A, Cross M, Borst P. Control of variant surface glycoprotein gene-expression sites in Trypanosoma brucei. EMBO J. 1999;18:4846–4855. doi: 10.1093/emboj/18.17.4846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu WK, Hickson ID. RecQ helicases: multifunctional genome caretakers. Nat Rev Cancer. 2009;9:644–654. doi: 10.1038/nrc2682. [DOI] [PubMed] [Google Scholar]
- Claussen CA, Long EC. Nucleic acid recognition by metal complexes of bleomycin. Chem Rev. 1999;99:2797–2816. doi: 10.1021/cr980449z. [DOI] [PubMed] [Google Scholar]
- Compton SA, Ozgur S, Griffith JD. Ring-shaped Rad51 paralog protein complexes bind Holliday junctions and replication forks as visualized by electron microscopy. J Biol Chem. 2010;285:13349–13356. doi: 10.1074/jbc.M109.074286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway C, McCulloch R, Ginger ML, Robinson NP, Browitt A, Barry JD. Ku is important for telomere maintenance, but not for differential expression of telomeric VSG genes, in African trypanosomes. J Biol Chem. 2002a;277:21269–21277. doi: 10.1074/jbc.M200550200. [DOI] [PubMed] [Google Scholar]
- Conway C, Proudfoot C, Burton P, Barry JD, McCulloch R. Two pathways of homologous recombination in Trypanosoma brucei. Mol Microbiol. 2002b;45:1687–1700. doi: 10.1046/j.1365-2958.2002.03122.x. [DOI] [PubMed] [Google Scholar]
- Cui X, Brenneman M, Meyne J, Oshimura M, Goodwin EH, Chen DJ. The XRCC2 and XRCC3 repair genes are required for chromosome stability in mammalian cells. Mutat Res. 1999;434:75–88. doi: 10.1016/s0921-8777(99)00010-5. [DOI] [PubMed] [Google Scholar]
- Daboussi F, Thacker J, Lopez BS. Genetic interactions between RAD51 and its paralogues for centrosome fragmentation and ploidy control, independently of the sensitivity to genotoxic stresses. Oncogene. 2005;24:3691–3696. doi: 10.1038/sj.onc.1208438. [DOI] [PubMed] [Google Scholar]
- Deans B, Griffin CS, Maconochie M, Thacker J. Xrcc2 is required for genetic stability, embryonic neurogenesis and viability in mice. EMBO J. 2000;19:6675–6685. doi: 10.1093/emboj/19.24.6675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deitsch KW, Lukehart SA, Stringer JR. Common strategies for antigenic variation by bacterial, fungal and protozoan pathogens. Nat Rev Microbiol. 2009;7:493–503. doi: 10.1038/nrmicro2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drexler GA, Rogge S, Beisker W, Eckardt-Schupp F, Zdzienicka MZ, Fritz E. Spontaneous homologous recombination is decreased in Rad51C-deficient hamster cells. DNA Repair (Amst) 2004;3:1335–1343. doi: 10.1016/j.dnarep.2004.05.002. [DOI] [PubMed] [Google Scholar]
- Eddy SR. Profile hidden Markov models. Bioinformatics. 1998;14:755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- Ersfeld K, Melville SE, Gull K. Nuclear and genome organization of Trypanosoma brucei. Parasitol Today. 1999;15:58–63. doi: 10.1016/s0169-4758(98)01378-7. [DOI] [PubMed] [Google Scholar]
- Figueiredo LM, Janzen CJ, Cross GA. A histone methyltransferase modulates antigenic variation in African trypanosomes. PLoS Biol. 2008;6:e161. doi: 10.1371/journal.pbio.0060161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Mistry J, Schuster-Bockler B, Griffiths-Jones S, Hollich V, Lassmann T, et al. Pfam: clans, web tools and services. Nucleic Acids Res. 2006;34:D247–D251. doi: 10.1093/nar/gkj149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortin GS, Symington LS. Mutations in yeast Rad51 that partially bypass the requirement for Rad55 and Rad57 in DNA repair by increasing the stability of Rad51-DNA complexes. EMBO J. 2002;21:3160–3170. doi: 10.1093/emboj/cdf293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser JA, Heitman J. Fungal mating-type loci. Curr Biol. 2003;13:R792–R795. doi: 10.1016/j.cub.2003.09.046. [DOI] [PubMed] [Google Scholar]
- Fritz-Laylin LK, Prochnik SE, Ginger ML, Dacks JB, Carpenter ML, Field MC, et al. The genome of Naegleria gruberi illuminates early eukaryotic versatility. Cell. 2010;140:631–642. doi: 10.1016/j.cell.2010.01.032. [DOI] [PubMed] [Google Scholar]
- Ghabrial A, Ray RP, Schupbach T. okra and spindle-B encode components of the RAD52 DNA repair pathway and affect meiosis and patterning in Drosophila oogenesis. Genes Dev. 1998;12:2711–2723. doi: 10.1101/gad.12.17.2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gildemeister OS, Sage JM, Knight KL. Cellular redistribution of Rad51 in response to DNA damage: a novel role for Rad51C. J Biol Chem. 2009;284:31945–31952. doi: 10.1074/jbc.M109.024646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill EE, Fast NM. Stripped-down DNA repair in a highly reduced parasite. BMC Mol Biol. 2007;8:24. doi: 10.1186/1471-2199-8-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giloni L, Takeshita M, Johnson F, Iden C, Grollman AP. Bleomycin-induced strand-scission of DNA. Mechanism of deoxyribose cleavage. J Biol Chem. 1981;256:8608–8615. [PubMed] [Google Scholar]
- Glover L, McCulloch R, Horn D. Sequence homology and microhomology dominate chromosomal double-strand break repair in African trypanosomes. Nucleic Acids Res. 2008;36:2608–2618. doi: 10.1093/nar/gkn104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldenby S, White MF, Allers T. RecA family proteins in archaea: RadA and its cousins. Biochem Soc Trans. 2009;37:102–107. doi: 10.1042/BST0370102. [DOI] [PubMed] [Google Scholar]
- Hartley CL, McCulloch R. Trypanosoma brucei BRCA2 acts in antigenic variation and has undergone a recent expansion in BRC repeat number that is important during homologous recombination. Mol Microbiol. 2008;68:1237–1251. doi: 10.1111/j.1365-2958.2008.06230.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatanaka A, Yamazoe M, Sale JE, Takata M, Yamamoto K, Kitao H, et al. Similar effects of Brca2 truncation and Rad51 paralog deficiency on immunoglobulin V gene diversification in DT40 cells support an early role for Rad51 paralogs in homologous recombination. Mol Cell Biol. 2005;25:1124–1134. doi: 10.1128/MCB.25.3.1124-1134.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hays SL, Firmenich AA, Berg P. Complex formation in yeast double-strand break repair: participation of Rad51, Rad52, Rad55, and Rad57 proteins. Proc Natl Acad Sci USA. 1995;92:6925–6929. doi: 10.1073/pnas.92.15.6925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertz-Fowler C, Figueiredo LM, Quail MA, Becker M, Jackson A, Bason N, et al. Telomeric expression sites are highly conserved in Trypanosoma brucei. PLoS ONE. 2008;3:e3527. doi: 10.1371/journal.pone.0003527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzberg K, Bashkirov VI, Rolfsmeier M, Haghnazari E, McDonald WH, Anderson S, et al. Phosphorylation of Rad55 on serines 2, 8, and 14 is required for efficient homologous recombination in the recovery of stalled replication forks. Mol Cell Biol. 2006;26:8396–8409. doi: 10.1128/MCB.01317-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirumi H, Hirumi K. Continuous cultivation of Trypanosoma brucei blood stream forms in a medium containing a low concentration of serum protein without feeder cell layers. J Parasitol. 1989;75:985–989. [PubMed] [Google Scholar]
- Hoeijmakers JH, Frasch AC, Bernards A, Borst P, Cross GA. Novel expression-linked copies of the genes for variant surface antigens in trypanosomes. Nature. 1980;284:78–80. doi: 10.1038/284078a0. [DOI] [PubMed] [Google Scholar]
- Horn D, Barry JD. The central roles of telomeres and subtelomeres in antigenic variation in African trypanosomes. Chromosome Res. 2005;13:525–533. doi: 10.1007/s10577-005-0991-8. [DOI] [PubMed] [Google Scholar]
- Jensen RB, Carreira A, Kowalczykowski SC. Purified human BRCA2 stimulates RAD51-mediated recombination. Nature. 2010;467:678–683. doi: 10.1038/nature09399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiricny J. The multifaceted mismatch-repair system. Nat Rev Mol Cell Biol. 2006;7:335–346. doi: 10.1038/nrm1907. [DOI] [PubMed] [Google Scholar]
- Johnson RD, Symington LS. Functional differences and interactions among the putative RecA homologs Rad51, Rad55, and Rad57. Mol Cell Biol. 1995;15:4843–4850. doi: 10.1128/mcb.15.9.4843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson RD, Liu N, Jasin M. Mammalian XRCC2 promotes the repair of DNA double-strand breaks by homologous recombination. Nature. 1999;401:397–399. doi: 10.1038/43932. [DOI] [PubMed] [Google Scholar]
- Kamper SM, Barbet AF. Surface epitope variation via mosaic gene formation is potential key to long-term survival of Trypanosoma brucei. Mol Biochem Parasitol. 1992;53:33–44. doi: 10.1016/0166-6851(92)90004-4. [DOI] [PubMed] [Google Scholar]
- Kass EM, Jasin M. Collaboration and competition between DNA double-strand break repair pathways. FEBS Lett. 2010;584:3703–3708. doi: 10.1016/j.febslet.2010.07.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidane D, Graumann PL. Dynamic formation of RecA filaments at DNA double strand break repair centers in live cells. J Cell Biol. 2005;170:357–366. doi: 10.1083/jcb.200412090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HS, Cross GA. TOPO3alpha influences antigenic variation by monitoring expression-site-associated VSG switching in Trypanosoma brucei. PLoS Pathog. 2010;6:e1000992. doi: 10.1371/journal.ppat.1000992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klar AJ. Lessons learned from studies of fission yeast mating-type switching and silencing. Annu Rev Genet. 2007;41:213–236. doi: 10.1146/annurev.genet.39.073103.094316. [DOI] [PubMed] [Google Scholar]
- Kojic M, Zhou Q, Lisby M, Holloman WK. Rec2 interplay with both Brh2 and Rad51 balances recombinational repair in Ustilago maydis. Mol Cell Biol. 2006;26:678–688. doi: 10.1128/MCB.26.2.678-688.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurumizaka H, Ikawa S, Nakada M, Enomoto R, Kagawa W, Kinebuchi T, et al. Homologous pairing and ring and filament structure formation activities of the human Xrcc2*Rad51D complex. J Biol Chem. 2002;277:14315–14320. doi: 10.1074/jbc.M105719200. [DOI] [PubMed] [Google Scholar]
- Kuznetsov SG, Haines DC, Martin BK, Sharan SK. Loss of Rad51c leads to embryonic lethality and modulation of Trp53-dependent tumorigenesis in mice. Cancer Res. 2009;69:863–872. doi: 10.1158/0008-5472.CAN-08-3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landeira D, Bart JM, Van Tyne D, Navarro M. Cohesin regulates VSG monoallelic expression in trypanosomes. J Cell Biol. 2009;186:243–254. doi: 10.1083/jcb.200902119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lange T, Kooter JM, Michels PA, Borst P. Telomere conversion in trypanosomes. Nucleic Acids Res. 1983;11:8149–8165. doi: 10.1093/nar/11.23.8149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurent M, Pays E, Magnus E, Van Meirvenne N, Matthyssens G, Williams RO, Steinert M. DNA rearrangements linked to expression of a predominant surface antigen gene of trypanosomes. Nature. 1983;302:263–266. doi: 10.1038/302263a0. [DOI] [PubMed] [Google Scholar]
- Lin Z, Kong H, Nei M, Ma H. Origins and evolution of the recA/RAD51 gene family: evidence for ancient gene duplication and endosymbiotic gene transfer. Proc Natl Acad Sci USA. 2006;103:10328–10333. doi: 10.1073/pnas.0604232103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lio YC, Mazin AV, Kowalczykowski SC, Chen DJ. Complex formation by the human Rad51B and Rad51C DNA repair proteins and their activities in vitro. J Biol Chem. 2003;278:2469–2478. doi: 10.1074/jbc.M211038200. [DOI] [PubMed] [Google Scholar]
- Lio YC, Schild D, Brenneman MA, Redpath JL, Chen DJ. Human Rad51C deficiency destabilizes XRCC3, impairs recombination, and radiosensitizes S/G2-phase cells. J Biol Chem. 2004;279:42313–42320. doi: 10.1074/jbc.M405212200. [DOI] [PubMed] [Google Scholar]
- Lisby M, Rothstein R. Choreography of recombination proteins during the DNA damage response. DNA Repair (Amst) 2009;8:1068–1076. doi: 10.1016/j.dnarep.2009.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu AY, Van der Ploeg LH, Rijsewijk FA, Borst P. The transposition unit of variant surface glycoprotein gene 118 of Trypanosoma brucei. Presence of repeated elements at its border and absence of promoter-associated sequences. J Mol Biol. 1983;167:57–75. doi: 10.1016/s0022-2836(83)80034-5. [DOI] [PubMed] [Google Scholar]
- Liu N, Lamerdin JE, Tebbs RS, Schild D, Tucker JD, Shen MR, et al. XRCC2 and XRCC3, new human Rad51-family members, promote chromosome stability and protect against DNA cross-links and other damages. Mol Cell. 1998;1:783–793. doi: 10.1016/s1097-2765(00)80078-7. [DOI] [PubMed] [Google Scholar]
- Liu N, Schild D, Thelen MP, Thompson LH. Involvement of Rad51C in two distinct protein complexes of Rad51 paralogs in human cells. Nucleic Acids Res. 2002;30:1009–1015. doi: 10.1093/nar/30.4.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Tarsounas M, O'Regan P, West SC. Role of RAD51C and XRCC3 in genetic recombination and DNA repair. J Biol Chem. 2007;282:1973–1979. doi: 10.1074/jbc.M609066200. [DOI] [PubMed] [Google Scholar]
- Longacre S, Eisen H. Expression of whole and hybrid genes in Trypanosoma equiperdum antigenic variation. EMBO J. 1986;5:1057–1063. doi: 10.1002/j.1460-2075.1986.tb04322.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Camarillo C, Lopez-Casamichana M, Weber C, Guillen N, Orozco E, Marchat LA. DNA repair mechanisms in eukaryotes: special focus in Entamoeba histolytica and related protozoan parasites. Infect Genet Evol. 2009;9:1051–1056. doi: 10.1016/j.meegid.2009.06.024. [DOI] [PubMed] [Google Scholar]
- Lopez-Casamichana M, Orozco E, Marchat LA, Lopez-Camarillo C. Transcriptional profile of the homologous recombination machinery and characterization of the EhRAD51 recombinase in response to DNA damage in Entamoeba histolytica. BMC Mol Biol. 2008;9:35. doi: 10.1186/1471-2199-9-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord CJ, Ashworth A. RAD51, BRCA2 and DNA repair: a partial resolution. Nat Struct Mol Biol. 2007;14:461–462. doi: 10.1038/nsmb0607-461. [DOI] [PubMed] [Google Scholar]
- McCulloch R, Barry JD. A role for RAD51 and homologous recombination in Trypanosoma brucei antigenic variation. Genes Dev. 1999;13:2875–2888. doi: 10.1101/gad.13.21.2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCulloch R, Rudenko G, Borst P. Gene conversions mediating antigenic variation in Trypanosoma brucei can occur in variant surface glycoprotein expression sites lacking 70- base-pair repeat sequences. Mol Cell Biol. 1997;17:833–843. doi: 10.1128/mcb.17.2.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcello L, Barry JD. Analysis of the VSG gene silent archive in Trypanosoma brucei reveals that mosaic gene expression is prominent in antigenic variation and is favored by archive substructure. Genome Res. 2007a;17:1344–1352. doi: 10.1101/gr.6421207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcello L, Barry JD. From silent genes to noisy populations-dialogue between the genotype and phenotypes of antigenic variation. J Eukaryot Microbiol. 2007b;54:14–17. doi: 10.1111/j.1550-7408.2006.00227.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, et al. CDD: specific functional annotation with the Conserved Domain Database. Nucleic Acids Res. 2009;37:D205–D210. doi: 10.1093/nar/gkn845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin V, Chahwan C, Gao H, Blais V, Wohlschlegel J, Yates JR, et al. Sws1 is a conserved regulator of homologous recombination in eukaryotic cells. EMBO J. 2006;25:2564–2574. doi: 10.1038/sj.emboj.7601141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masson JY, Stasiak AZ, Stasiak A, Benson FE, West SC. Complex formation by the human RAD51C and XRCC3 recombination repair proteins. Proc Natl Acad Sci USA. 2001a;98:8440–8446. doi: 10.1073/pnas.111005698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masson JY, Tarsounas MC, Stasiak AZ, Stasiak A, Shah R, McIlwraith MJ, et al. Identification and purification of two distinct complexes containing the five RAD51 paralogs. Genes Dev. 2001b;15:3296–3307. doi: 10.1101/gad.947001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melville SE, Leech V, Gerrard CS, Tait A, Blackwell JM. The molecular karyotype of the megabase chromosomes of Trypanosoma brucei and the assignment of chromosome markers. Mol Biochem Parasitol. 1998;94:155–173. doi: 10.1016/s0166-6851(98)00054-1. [DOI] [PubMed] [Google Scholar]
- Miller KA, Yoshikawa DM, McConnell IR, Clark R, Schild D, Albala JS. RAD51C interacts with RAD51B and is central to a larger protein complex in vivo exclusive of RAD51. J Biol Chem. 2002;277:8406–8411. doi: 10.1074/jbc.M108306200. [DOI] [PubMed] [Google Scholar]
- Miller KA, Sawicka D, Barsky D, Albala JS. Domain mapping of the Rad51 paralog protein complexes. Nucleic Acids Res. 2004;32:169–178. doi: 10.1093/nar/gkg925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison LJ, Marcello L, McCulloch R. Antigenic variation in the African trypanosome: molecular mechanisms and phenotypic complexity. Cell Microbiol. 2009;11:1724–1734. doi: 10.1111/j.1462-5822.2009.01383.x. [DOI] [PubMed] [Google Scholar]
- O'Regan P, Wilson C, Townsend S, Thacker J. XRCC2 is a nuclear RAD51-like protein required for damage-dependent RAD51 focus formation without the need for ATP binding. J Biol Chem. 2001;276:22148–22153. doi: 10.1074/jbc.M102396200. [DOI] [PubMed] [Google Scholar]
- Osakabe K, Abe K, Yamanouchi H, Takyuu T, Yoshioka T, Ito Y, et al. Arabidopsis Rad51B is important for double-strand DNA breaks repair in somatic cells. Plant Mol Biol. 2005;57:819–833. doi: 10.1007/s11103-005-2187-1. [DOI] [PubMed] [Google Scholar]
- Pays E. Regulation of antigen gene expression in Trypanosoma brucei. Trends Parasitol. 2005;21:517–520. doi: 10.1016/j.pt.2005.08.016. [DOI] [PubMed] [Google Scholar]
- Pays E, Van Meirvenne N, Le Ray D, Steinert M. Gene duplication and transposition linked to antigenic variation in Trypanosoma brucei. Proc Natl Acad Sci USA. 1981;78:2673–2677. doi: 10.1073/pnas.78.5.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pays E, Van Assel S, Laurent M, Dero B, Michiels F, Kronenberger P, et al. At least two transposed sequences are associated in the expression site of a surface antigen gene in different trypanosome clones. Cell. 1983;34:359–369. doi: 10.1016/0092-8674(83)90370-7. [DOI] [PubMed] [Google Scholar]
- Pays E, Guyaux M, Aerts D, Van Meirvenne N, Steinert M. Telomeric reciprocal recombination as a possible mechanism for antigenic variation in trypanosomes. Nature. 1985;316:562–564. doi: 10.1038/316562a0. [DOI] [PubMed] [Google Scholar]
- Petermann E, Helleday T. Pathways of mammalian replication fork restart. Nat Rev Mol Cell Biol. 2010;11:683–687. doi: 10.1038/nrm2974. [DOI] [PubMed] [Google Scholar]
- Pierce AJ, Johnson RD, Thompson LH, Jasin M. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev. 1999;13:2633–2638. doi: 10.1101/gad.13.20.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittman DL, Schimenti JC. Midgestation lethality in mice deficient for the RecA-related gene, Rad51d/Rad51l3. Genesis. 2000;26:167–173. doi: 10.1002/(sici)1526-968x(200003)26:3<167::aid-gene1>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- Proudfoot C, McCulloch R. Distinct roles for two RAD51-related genes in Trypanosoma brucei antigenic variation. Nucleic Acids Res. 2005;33:6906–6919. doi: 10.1093/nar/gki996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudfoot C, McCulloch R. Trypanosoma brucei DMC1 does not act in DNA recombination, repair or antigenic variation in bloodstream stage cells. Mol Biochem Parasitol. 2006;145:245–253. doi: 10.1016/j.molbiopara.2005.10.007. [DOI] [PubMed] [Google Scholar]
- Radford SJ, Sekelsky JJ. Taking Drosophila Rad51 for a SPiN. Nat Struct Mol Biol. 2004;11:9–10. doi: 10.1038/nsmb0104-9. [DOI] [PubMed] [Google Scholar]
- Rajesh C, Gruver AM, Basrur V, Pittman DL. The interaction profile of homologous recombination repair proteins RAD51C, RAD51D and XRCC2 as determined by proteomic analysis. Proteomics. 2009;9:4071–4086. doi: 10.1002/pmic.200800977. [DOI] [PubMed] [Google Scholar]
- Raz B, Iten M, Grether-Buhler Y, Kaminsky R, Brun R. The Alamar Blue assay to determine drug sensitivity of African trypanosomes (T.b. rhodesiense and T.b. gambiense) in vitro. Acta Trop. 1997;68:139–147. doi: 10.1016/s0001-706x(97)00079-x. [DOI] [PubMed] [Google Scholar]
- Renglin LA, Schultz N, Saleh-Gohari N, Helleday T. RAD51C (RAD51L2) is involved in maintaining centrosome number in mitosis. Cytogenet Genome Res. 2007;116:38–45. doi: 10.1159/000097416. [DOI] [PubMed] [Google Scholar]
- Renzette N, Gumlaw N, Nordman JT, Krieger M, Yeh SP, Long E, et al. Localization of RecA in Escherichia coli K-12 using RecA-GFP. Mol Microbiol. 2005;57:1074–1085. doi: 10.1111/j.1365-2958.2005.04755.x. [DOI] [PubMed] [Google Scholar]
- Robinson NP, Burman N, Melville SE, Barry JD. Predominance of duplicative VSG gene conversion in antigenic variation in African trypanosomes. Mol Cell Biol. 1999;19:5839–5846. doi: 10.1128/mcb.19.9.5839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson NP, McCulloch R, Conway C, Browitt A, Barry JD. Inactivation of Mre11 does not affect VSG gene duplication mediated by homologous recombination in Trypanosoma brucei. J Biol Chem. 2002;277:26185–26193. doi: 10.1074/jbc.M203205200. [DOI] [PubMed] [Google Scholar]
- Roth CW, Longacre S, Raibaud A, Baltz T, Eisen H. The use of incomplete genes for the construction of a Trypanosoma equiperdum variant surface glycoprotein gene. EMBO J. 1986;5:1065–1070. doi: 10.1002/j.1460-2075.1986.tb04323.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudenko G, McCulloch R, Dirks-Mulder A, Borst P. Telomere exchange can be an important mechanism of variant surface glycoprotein gene switching in Trypanosoma brucei. Mol Biochem Parasitol. 1996;80:65–75. doi: 10.1016/0166-6851(96)02669-2. [DOI] [PubMed] [Google Scholar]
- Rupnik A, Lowndes NF, Grenon M. MRN and the race to the break. Chromosoma. 2010;119:115–135. doi: 10.1007/s00412-009-0242-4. [DOI] [PubMed] [Google Scholar]
- San Filippo J, Sung P, Klein H. Mechanism of eukaryotic homologous recombination. Annu Rev Biochem. 2008;77:229–257. doi: 10.1146/annurev.biochem.77.061306.125255. [DOI] [PubMed] [Google Scholar]
- Schild D, Lio YC, Collins DW, Tsomondo T, Chen DJ. Evidence for simultaneous protein interactions between human Rad51 paralogs. J Biol Chem. 2000;275:16443–16449. doi: 10.1074/jbc.M001473200. [DOI] [PubMed] [Google Scholar]
- Shim KS, Schmutte C, Tombline G, Heinen CD, Fishel R. hXRCC2 enhances ADP/ATP processing and strand exchange by hRAD51. J Biol Chem. 2004;279:30385–30394. doi: 10.1074/jbc.M306066200. [DOI] [PubMed] [Google Scholar]
- Shu Z, Smith S, Wang L, Rice MC, Kmiec EB. Disruption of muREC2/RAD51L1 in mice results in early embryonic lethality which can Be partially rescued in a p53(-/-) background. Mol Cell Biol. 1999;19:8686–8693. doi: 10.1128/mcb.19.12.8686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigurdsson S, Van Komen S, Bussen W, Schild D, Albala JS, Sung P. Mediator function of the human Rad51B-Rad51C complex in Rad51/RPA-catalyzed DNA strand exchange. Genes Dev. 2001;15:3308–3318. doi: 10.1101/gad.935501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smiraldo PG, Gruver AM, Osborn JC, Pittman DL. Extensive chromosomal instability in Rad51d-deficient mouse cells. Cancer Res. 2005;65:2089–2096. doi: 10.1158/0008-5472.CAN-04-2079. [DOI] [PubMed] [Google Scholar]
- Story RM, Bishop DK, Kleckner N, Steitz TA. Structural relationship of bacterial RecA proteins to recombination proteins from bacteriophage T4 and yeast. Science. 1993;259:1892–1896. doi: 10.1126/science.8456313. [DOI] [PubMed] [Google Scholar]
- Sugawara N, Ivanov EL, Fishman-Lobell J, Ray BL, Wu X, Haber JE. DNA structure-dependent requirements for yeast RAD genes in gene conversion. Nature. 1995;373:84–86. doi: 10.1038/373084a0. [DOI] [PubMed] [Google Scholar]
- Sugawara N, Wang X, Haber JE. In vivo roles of Rad52, Rad54, and Rad55 proteins in Rad51-mediated recombination. Mol Cell. 2003;12:209–219. doi: 10.1016/s1097-2765(03)00269-7. [DOI] [PubMed] [Google Scholar]
- Sung P. Yeast Rad55 and Rad57 proteins form a heterodimer that functions with replication protein A to promote DNA strand exchange by Rad51 recombinase. Genes Dev. 1997;11:1111–1121. doi: 10.1101/gad.11.9.1111. [DOI] [PubMed] [Google Scholar]
- Takata M, Sasaki MS, Sonoda E, Fukushima T, Morrison C, Albala JS, et al. The Rad51 paralog Rad51B promotes homologous recombinational repair. Mol Cell Biol. 2000;20:6476–6482. doi: 10.1128/mcb.20.17.6476-6482.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takata M, Sasaki MS, Tachiiri S, Fukushima T, Sonoda E, Schild D, et al. Chromosome instability and defective recombinational repair in knockout mutants of the five Rad51 paralogs. Mol Cell Biol. 2001;21:2858–2866. doi: 10.1128/MCB.21.8.2858-2866.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan KS, Leal ST, Cross GA. Trypanosoma brucei MRE11 is non-essential but influences growth, homologous recombination and DNA double-strand break repair. Mol Biochem Parasitol. 2002;125:11–21. doi: 10.1016/s0166-6851(02)00165-2. [DOI] [PubMed] [Google Scholar]
- Tarsounas M, Davies AA, West SC. RAD51 localization and activation following DNA damage. Philos Trans R Soc Lond B Biol Sci. 2004a;359:87–93. doi: 10.1098/rstb.2003.1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarsounas M, Munoz P, Claas A, Smiraldo PG, Pittman DL, Blasco MA, West SC. Telomere maintenance requires the RAD51D recombination/repair protein. Cell. 2004b;117:337–347. doi: 10.1016/s0092-8674(04)00337-x. [DOI] [PubMed] [Google Scholar]
- Taylor JE, Rudenko G. Switching trypanosome coats: what's in the wardrobe? Trends Genet. 2006;22:614–620. doi: 10.1016/j.tig.2006.08.003. [DOI] [PubMed] [Google Scholar]
- Thacker J. The RAD51 gene family, genetic instability and cancer. Cancer Lett. 2005;219:125–135. doi: 10.1016/j.canlet.2004.08.018. [DOI] [PubMed] [Google Scholar]
- Ulbert S, Chaves I, Borst P. Expression site activation in Trypanosoma brucei with three marked variant surface glycoprotein gene expression sites. Mol Biochem Parasitol. 2002;120:225–235. doi: 10.1016/s0166-6851(02)00003-8. [DOI] [PubMed] [Google Scholar]
- Vanhamme L, Pays E, McCulloch R, Barry JD. An update on antigenic variation in African trypanosomes. Trends Parasitol. 2001;17:338–343. doi: 10.1016/s1471-4922(01)01922-5. [DOI] [PubMed] [Google Scholar]
- van Veelen LR, Essers J, van de Rakt MW, Odijk H, Pastink A, Zdzienicka MZ, et al. Ionizing radiation-induced foci formation of mammalian Rad51 and Rad54 depends on the Rad51 paralogs, but not on Rad52. Mutat Res. 2005;574:34–49. doi: 10.1016/j.mrfmmm.2005.01.020. [DOI] [PubMed] [Google Scholar]
- Ward JD, Barber LJ, Petalcorin MI, Yanowitz J, Boulton SJ. Replication blocking lesions present a unique substrate for homologous recombination. EMBO J. 2007;26:3384–3396. doi: 10.1038/sj.emboj.7601766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickstead B, Ersfeld K, Gull K. The small chromosomes of Trypanosoma brucei involved in antigenic variation are constructed around repetitive palindromes. Genome Res. 2004;14:1014–1024. doi: 10.1101/gr.2227704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiese C, Collins DW, Albala JS, Thompson LH, Kronenberg A, Schild D. Interactions involving the Rad51 paralogs Rad51C and XRCC3 in human cells. Nucleic Acids Res. 2002;30:1001–1008. doi: 10.1093/nar/30.4.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson JB, Yamamoto K, Marriott AS, Hussain S, Sung P, Hoatlin ME, et al. FANCG promotes formation of a newly identified protein complex containing BRCA2, FANCD2 and XRCC3. Oncogene. 2008;27:3641–3652. doi: 10.1038/sj.onc.1211034. [DOI] [PubMed] [Google Scholar]
- Yanowitz JL. Genome integrity is regulated by the Caenorhabditis elegans Rad51D homolog rfs-1. Genetics. 2008;179:249–262. doi: 10.1534/genetics.107.076877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama H, Sarai N, Kagawa W, Enomoto R, Shibata T, Kurumizaka H, Yokoyama S. Preferential binding to branched DNA strands and strand-annealing activity of the human Rad51B, Rad51C, Rad51D and Xrcc2 protein complex. Nucleic Acids Res. 2004;32:2556–2565. doi: 10.1093/nar/gkh578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon HS, Grant J, Tekle YI, Wu M, Chaon BC, Cole JC, et al. Broadly sampled multigene trees of eukaryotes. BMC Evol Biol. 2008;8:14. doi: 10.1186/1471-2148-8-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


