Abstract
Hematopoietic differentiation involves the commitment of multipotent progenitors to a given lineage, followed by the maturation of the committed cells. To study the transcriptional events controlling these processes, we have investigated the role of C/EBP proteins in lineage choice of multipotent hematopoietic progenitors (MEPs) transformed by the E26 virus. We found that forced expression of either the α or β isoforms of C/EBP in MEPs induced eosinophil differentiation and that in addition, C/EBPβ could induce myeloid differentiation. Conversely, dominant-negative versions of C/EBPβ inhibited myeloid differentiation. C/EBP-induced eosinophil differentiation could be separated into two distinct events, lineage commitment and maturation. Thus, eosinophils induced by transactivation-deficient C/EBPβ alleles were found to be blocked in their maturation, whereas those expressing wild-type C/EBP proteins were not. Likewise, a 1-day activation of a conditional C/EBPβ allele in multipotent progenitors led to the formation of immature eosinophils, whereas sustained activation produced mature eosinophils. These results show that C/EBP can induce both myeloid and eosinophil lineage commitment and that transactivation independent and dependent C/EBP functions are required during eosinophil lineage commitment and maturation, respectively.
Keywords: Hematopoietic differentiation, leukemia virus, multipotent progenitor, transcription factor
To understand how Myb–Ets multipotent hematopoietic progenitors (MEPs) choose to differentiate along a given lineage, it is necessary to identify transcription factors that have the ability to instruct multipotent cells to commit to specific differentiation programs. Candidate lineage-instructive transcription factors are those with restricted expression in the hematopoietic system. These include the myeloid/B-cell-specific factor PU.1 (Nerlov and Graf 1998) and C/EBP isoforms α and β, which are expressed in both myeloid cells and eosinophils (Scott et al. 1992; this paper).
In the hematopoietic system, the α, β, and δ C/EBP isoforms are all capable of activating the promoters of genes specifically expressed in the myeloid lineage in both mammalian and avian species (Ness et al. 1993; Oelgeschläger et al. 1996) and in chickens C/EBPα and C/EBPβ can activate the eosinophil-specific EOS47 gene (McNagny et al. 1998). Myeloid gene expression can be ectopically induced in quail fibroblasts through cooperation between C/EBPs and Myb (Burk et al. 1993; Ness et al. 1993). This observation, together with the fact that Myb is expressed in all immature cells of the hematopoietic system, and that C/EBPs are specifically expressed in both myeloid cells and eosinophils, has led to the suggestion that these two factors act in conjunction with PU.1 to specify the myeloid lineage (Tenen et al. 1997), and with GATA factors to determine the eosinophil lineage (Müller et al. 1995).
Hematopoietic lineage deficiencies have been reported for C/EBPα, C/EBPβ, and C/EBPε knockout mice. The phenotypes are at the level of maturation of myeloblasts into granulocytes, eosinophil maturation (C/EBPα, C/EBPε), and macrophage activation (C/EBPβ), respectively (Tanaka et al. 1995; Yamanaka et al. 1997; Zhang et al. 1997), but in neither case has lineage commitment been shown to be affected. However, because several C/EBP isoforms are expressed in hematopoietic cells (Scott et al. 1992), this may be explained, at least in part, by functional redundancy. Therefore, whereas it is clear that C/EBPs are important for myeloid- and eosinophil-specific gene expression and differentiation, their role in lineage commitment remains to be resolved.
To study this question, we have developed a system consisting of MEPs transformed by the Gag–Myb–Ets encoding E26 leukemia virus. These progenitors can be induced to differentiate along the myeloid, eosinophilic, thrombocytic, and erythroid lineages (Graf et al. 1992; Kraut et al. 1994; Frampton et al. 1995; Rossi et al. 1996a) and thus resemble normal multipotent progenitors of the CFU–GEMM type. Here we show that both the α and β isoforms of C/EBP promote eosinophil differentiation when constitutively expressed in MEPs. Commitment and maturation of eosinophils could be separated either by the sustained expression of alleles with impaired transactivating potential or by subjecting MEPs to short periods of activation of a conditional C/EBPβ allele; under the latter condition a fraction of the MEPs also differentiated along the myeloid lineage. Our results show that C/EBPs induce multipotent progenitors to acquire new fates, requiring both transactivation dependent and independent functions.
Results
Expression pattern of C/EBP isoforms in chicken hematopoietic cells
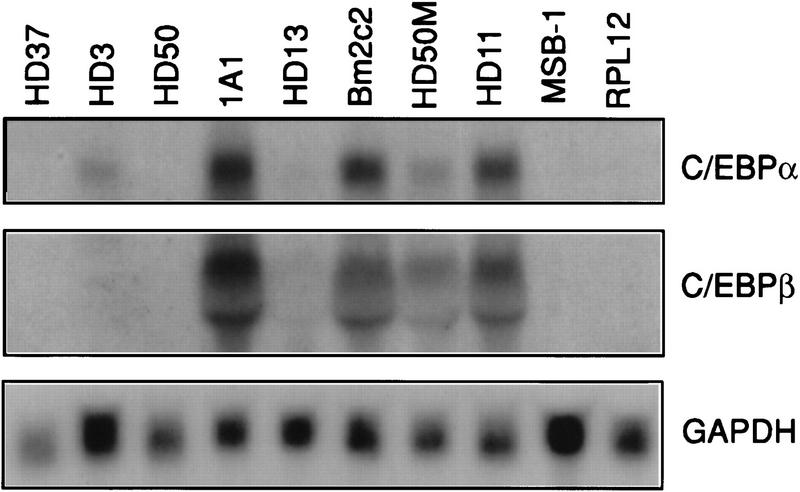
To determine the expression pattern of chicken C/EBPα and C/EBPβ (Calkhoven et al. 1992; Katz et al. 1993) in the hematopoietic system, we tested their expression in chicken cell lines of various lineages. As shown in Figure 1, the mRNAs for the two factors were both expressed in chicken myeloid and eosinophil cell lines but were absent from lymphoid, MEP, and erythroid cells (except for a weak expression of C/EBPα mRNA in HD3 erythroblasts). This pattern was confirmed at the protein level by gel-shift experiments with nuclear extracts from MEPs (HD50 cell line), eosinophils (1A1 cell line), and myeloid cells (HD50M cell line) showing the presence of specific complexes that could be supershifted by anti-C/EBPα and C/EBPβ antibodies in eosinophils and myeloid cells, but not in MEPs (data not shown).
Figure 1.
Expression of C/EBPα and C/EBPβ isoforms in chicken hematopoietic cell types. Twenty micrograms of total RNA extracted from HD3, HD37 (erythroblast), HD50 (MEP), 1A1 (eosinophil), HD13 (promyelocyte), Bm2c2 (monoblast), HD50M (myeloblast), HD11 (macrophage), RPL12 (B-cell), and MSB-1 (T-cell) were probed with chicken C/EBPα, C/EBPβ, and GAPDH cDNAs. Because of comigration with 18S RNA, the C/EBPβ signal appears as a doublet.
E26 viruses coexpressing either C/EBPα or C/EBPβ yield increased proportions of transformed colonies containing eosinophils
To address the role of C/EBP in the differentiation of MEPs, E26 proviral vectors were constructed that—in addition to the Gag–Myb–Ets oncoprotein—express either chicken C/EBPα (E26–C/EBPα), chicken C/EBPβ (E26–C/EBPβ), or a mutant form of C/EBPβ containing the point mutation Asp → Asn at amino acid number 63 (E26–βD63N). This mutation affects a region conserved between activating C/EBPs known to be involved in contacting the TBP and TFIIB basal transcription factors (Nerlov and Ziff 1995) and severely reduces its transactivation capacity on the myeloid-specific mim-1 and cMGF promoters (C. Nerlov and T. Graf, unpubl.). The C/EBP coding sequences were inserted into an E26 vector behind an IRES element placed downstream of the Gag–Myb–Ets coding region (Fig. 2A). To generate the corresponding viruses, the various constructs were transiently transfected into the Q2bn-packaging cell line. After 2 days, the virus-producing cells were cocultivated with cells from 2-day chicken blastoderm and the infected cells seeded in semisolid medium at 37°C for 2 weeks, at which time macroscopically visible hematopoietic colonies had developed. The cocultivation conditions used induce ∼50% of the E26–WT MEP clones to undergo myeloid differentiation (Graf et al. 1992). For each virus, between 14 and 30 transformed colonies were isolated, clonal populations expanded and phenotyped by indirect immunofluorescence and flow cytometry by use of lineage-specific cell surface markers (MEPs, MEP21; eosinophils, EOS47; myeloid cells, MYL51/2). Data from a representative subset of clones transformed by each virus are shown in Figure 2B. The myeloid cells detected in these assays are myeloblasts, as the Myb moiety of E26 blocks further differentiation along the myeloid lineage (Beug et al. 1984; Frampton et al. 1996). As can be seen from the distribution of the three types of antigen-positive cells, about half of the colonies transformed by E26–WT exhibited an MEP phenotype, the other half were myeloid with few, or no, eosinophils present. In contrast, most clones expressing C/EBPα or C/EBPβ, and to a lesser extent those transformed by E26–βD63N, contained EOS47-positive cells. The number of MEP21-positive cells remained relatively constant, whereas the abundance of MYL51/2-positive myeloid cells decreased in the order E26–βD63N, E26–C/EBPβ, and E26–C/EBPα; in the latter case they were essentially absent. Approximately 50%–60% of the clones transformed by E26–C/EBPα and E26–C/EBPβ were bilineage MEP21/EOS47, another 20% E26–C/EBPβ clones were trilineage. Interestingly, mixed MEP21/MYL51/2 and EOS47/MYL51/2 colonies were rare or absent. These results show that all three C/EBPs tested induce the differentiation of MEPs into eosinophils, with a concommitant decrease in myeloid cell formation.
Figure 2.
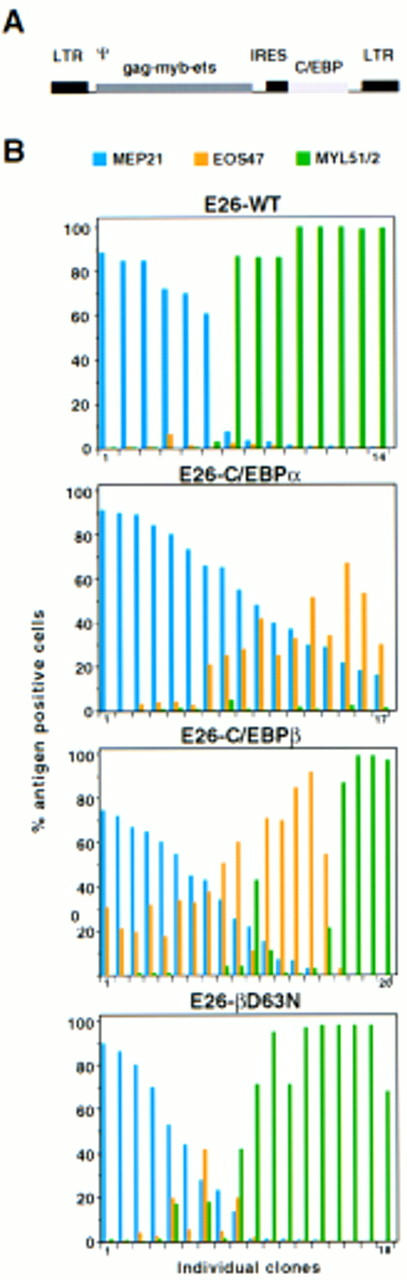
Effect in MEP cells of chicken C/EBP isoforms on myeloid and eosinophil differentiation. (A) Structure of recombinant E26-C/EBP proviruses. (LTR) Long terminal repeat; (Θ) packaging signal; (IRES) internal ribosomal entry site. (B) E26-WT, E26-C/EBPα, E26-C/EBPβ, and E26-βD63N viruses were generated and used to infect blastoderm cells by cocultivation with the Q2bn packaging cells. Transformed colonies were isolated 2 weeks later, expanded in liquid culture for ∼2 weeks more and phenotyped by FACS analysis. Histograms show the proportion of cells positive for MEP21 (MEP cells), EOS47 (eosinophils), and MYL51/2 (myeloblasts) in each single colony-derived population. For each virus, clones are ordered according to decreasing MEP21 expression (left to right).
C/EBP transactivation strength influences the choice between myeloid and eosinophil differentiation
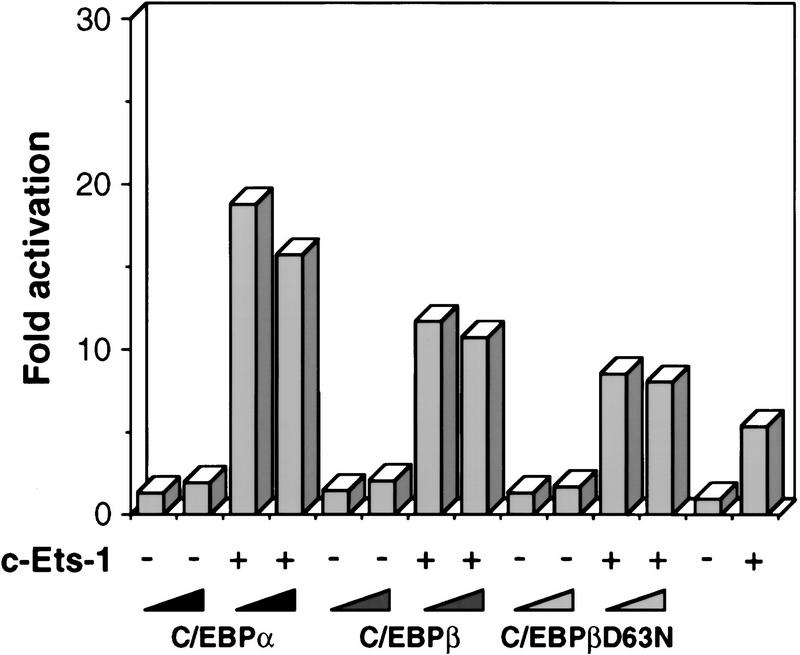
Next, we examined the transactivation capacities of the different chicken C/EBPs, using the promoter of the EOS47 gene. EOS47 is the earliest known eosinophil-specific marker (McNagny et al. 1996), and the regulation of this gene may thus reflect transcriptional events related to eosinophil lineage commitment. As C/EBP cooperates with Ets-1 on this promoter (McNagny et al. 1998), transient transfection experiments were performed in the presence and absence of Ets-1. As can be seen in Figure 3, the ability of the C/EBPs to transactivate the EOS47 promoter in the presence of Ets-1 decreased in the order C/EBPα, C/EBPβ, and βD63N. A similar hierachy was observed on the chicken mim-1 and cMGF promoters (C. Nerlov and T. Graf, unpubl.) and has been reported for the murine neutrophil elastase and human M-CSF receptor gene promoters (Oelgeschläger et al. 1996; Zhang et al. 1996). This order correlated with the increase in eosinophils, and, in particular, the decrease in myeloid cells observed in cultures transformed by the corresponding E26 constructs. Thus, these results suggest that a strong transactivation potential enhances the ability of C/EBPs to induce eosinophil differentiation at the expense of myeloid differentiation.
Figure 3.
Cooperative activation of the EOS47 promoter by Ets-1 and chicken C/EBPs. One microgram of reporter plasmid (EOS47/−152-LUC) and 250 ng internal control plasmid (pRSV–βGal) were cotransfected into Q2bn fibroblasts along with 100 ng expression vectors for c-Ets-1 (pCRNCM-c-Ets-1) and 10 ng and 30 ng CMV expression vectors for C/EBPα, C/EBPβ, and C/EBPβD63N (increasing left to right). Empty expression vector was added to achieve equivalent DNA concentrations in all transfections. Luciferase activity was determined 48 hr later and normalized to the β-galactosidase activity. The basal level obtained with pEOS/−152-LUC alone was arbitrarily assigned a value of 1. The values shown represent the average of three or more experiments with similar results.
Eosinophils induced by transactivation-deficient C/EBPβ are immature
To further characterize the EOS47-positive cells obtained with the various E26–C/EBP constructs, we determined their peroxidase expression, a marker for mature eosinophils in chickens (Brune and Spitznagel 1973; Daimon and Caxton-Martins 1977). To facilitate comparison, data are shown for clones containing ∼70% EOS47-positive cells (Fig. 4A). Cells transformed by E26–C/EBPα and E26–C/EBPβ expressed comparable levels of the EOS47 antigen and ∼30% of these were peroxidase positive. In contrast, E26–βD63N-transformed cells expressed somewhat lower levels of EOS47 antigen and were completely peroxidase negative. Similar observations were made with all (>10) E26–βD63N-transformed clones that contained significant numbers of EOS47-positive cells. The myeloid/eosinophil-specific mim-1 gene was expressed in the predominantly EOS47-positive clones, with few or no cells positive for myeloid surface antigens (data not shown). This further indicates that the E26–βD63N-transformed cells correspond to committed eosinophils, although it cannot be ruled out that they still have myeloid potential. To determine whether normal cells exist that exhibit a similar immature phenotype, we stained the bone marrow of a 2-week-old chick with the EOS47 monoclonal antibody and sorted the cells by FACS. The EOS47-positive cells (purity ∼95%) were then stained with peroxidase reagent, and counterstained with DAPI to visualize nuclei. All peroxidase-positive cells were contained in the EOS47-positive fraction (data not shown). However, as illustrated in Figure 4B, and as expected from earlier work (McNagny et al. 1992, 1996), the EOS47-positive fraction also contained cells that did not express peroxidase. The E26–βD63N transformed EOS47+/peroxidase− cells thus resemble immature normal eosinophils. This suggests that the C/EBPβD63N allele, although promoting commitment toward cells of the eosinophil lineage, prevents their maturation by arresting them at the pro-eosinophil stage.
Figure 4.
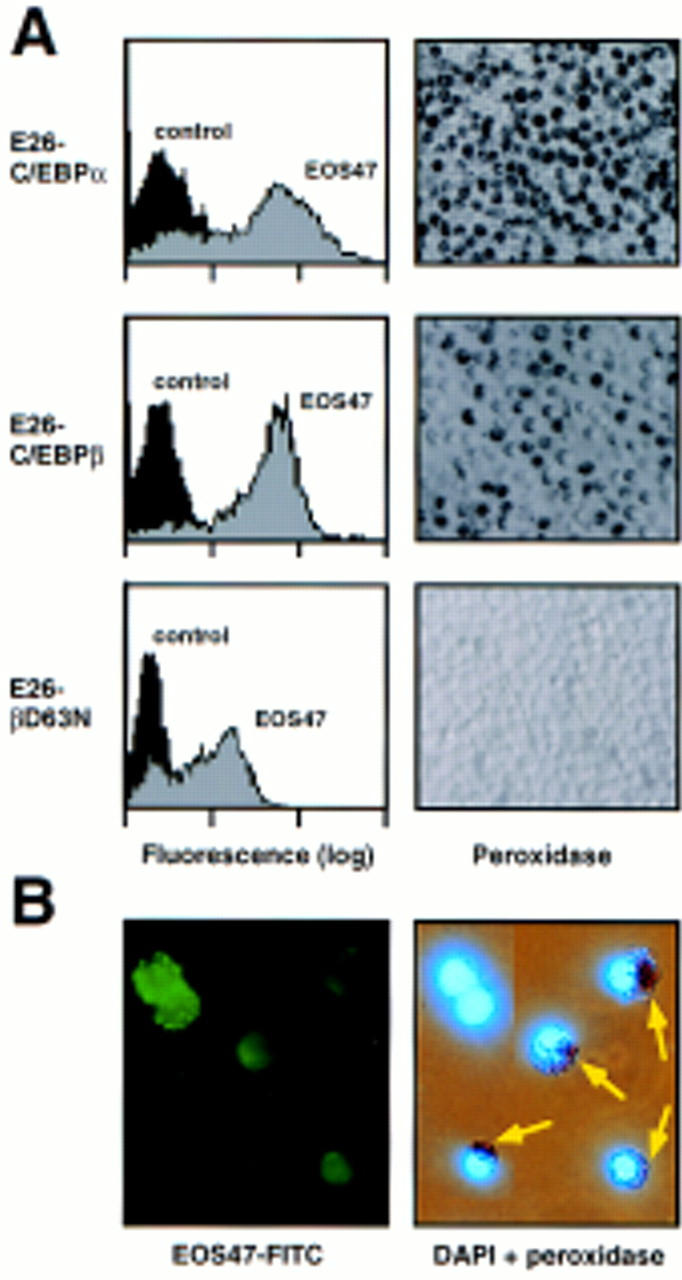
Characterization of eosinophils transformed by E26 viruses coexpressing various C/EBP forms. (A) FACScan profiles of EOS47 expression (left) and peroxidase staining (right) of representative eosinophil-rich clones transformed by E26–C/EBPα, E26–C/EBPβ, and E26–βD63N viruses. Peroxidase positive cells contain dark granules. (B) Immunofluorescence (left) and combined peroxidase and DAPI staining (right) of the same field of EOS47 FACS-sorted normal chick bone marrow cells. Peroxidase-positive cells are indicated by arrows.
Dominant-negative C/EBPβ alleles affect both eosinophil and myeloid cell differentiation
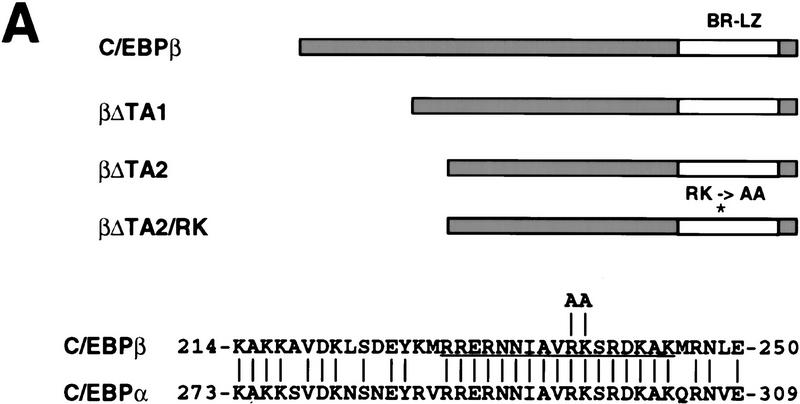
Next, we addressed two questions: To what extent is the induction of eosinophil commitment independent of the C/EBP transactivation function, and how does inhibition of C/EBP activity affect myeloid/eosinophil differentiation? For this purpose, we generated three types of dominant-negative C/EBPs (Fig. 5A), using the rat C/EBPβ protein whose transactivation domain is known to be confined to the amino-terminal part of the protein (Williams et al. 1995). The first mutant (amino acids 1–70; βΔTA1) lacked most, the second, all of the transactivation domain (amino acids 1–99; βΔTA2). Truncated proteins of this type are still able to heterodimerize with wild-type C/EBPs (C/EBP family members dimerize in all combinations tested; e.g., Cao et al. 1991; Ron and Habener 1992) and should thus increase the total level of C/EBP DNA-binding activity while reducing the level of transactivation via C/EBP sites. In the third mutant (βΔTA2/RK) we mutated, in addition, the basic region of the DNA-binding domain (R238 and K239 to alanines), rendering the corresponding homodimers as well as the heterodimers with C/EBPβ incapable of DNA binding (data not shown). Expression of this mutant in a C/EBP-expressing cell should, therefore, lead to a reduction in the overall levels of active C/EBP.
Figure 5.
Characterization of dominant negative mutants of C/EBPβ. (A) Structure of dominant-negative rat C/EBPβ mutants. (*) Position of the RK mutation. The sequences show a comparison between the basic regions (underlined) of rat C/EBPα and C/EBPβ, and the position of the substitutions introduced in the C/EBPβ/RK mutant (arginine-238 and lysine-239 into alanines). The numbers indicate the positions of the first and last amino acid shown for each protein. (B) Capacity of mutant C/EBPs to block the transactivation of the mim-1 promoter by C/EBPα. The ability of the βΔTA1, βΔTA2, and βΔTA2/RK alleles to inhibit C/EBP dependent transactivation was tested on the mim-1 promoter. One microgram of pMim-1ΔC–LUC was cotransfected with 0.25 μg of pRSVβGal, 0.5 μg pCMV–C/EBPα (chicken C/EBPα expressed from the CMV promoter). Two and five micrograms of CMV expression vector for the dominant-negative alleles were cotransfected as indicated. As a specificity control, 5 μg of a CMV expression vector for the I(16) protein (a dominant-negative inhibitor of the AML complex) was used. Relative transactivation values were calculated as in Fig. 3; the activity of pMim-1ΔC–LUC in the presence of C/EBPα was arbitrarily assigned a value of 1.
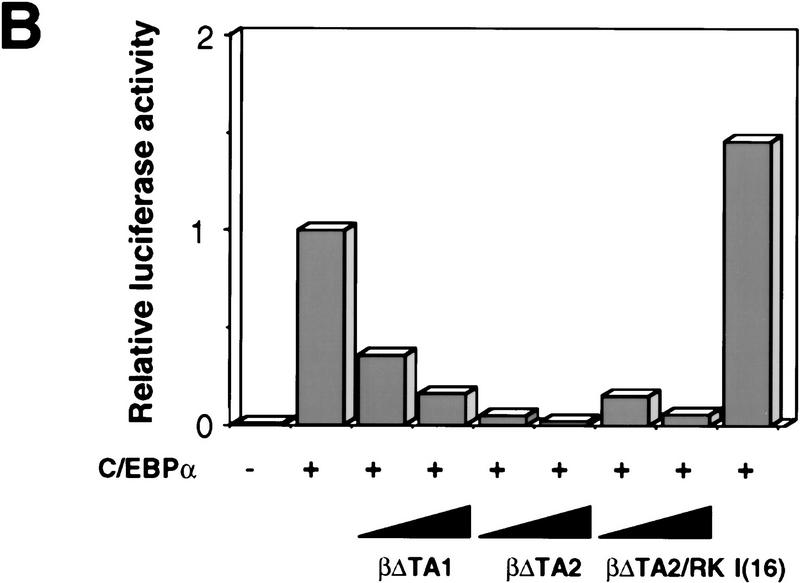
To test the ability of the mutants to inhibit C/EBP transactivation, we used chicken C/EBPα and the mim-1 promoter, as this activator/promoter combination provides a good dynamic range over which to assess the effect of dominant-negative proteins. The results in Figure 5B show that mim-1 promoter activation by C/EBPα was moderately inhibited by βΔTA1 and strongly by βΔTA2 and βΔTA2/RK. As a specificity control, we tested the I(16) dominant-negative allele of the core binding factor, derived from the inv(16) translocation (Liu et al. 1993). This protein did not inhibit mim-1 promoter activation by C/EBPα, nor did the C/EBPβ mutant proteins show a significant effect on the RSV promoter, used as internal control in these experiments (not shown).
To test their biological activity, the βΔTA1, βΔTA2, and βΔTA2/RK coding sequences were introduced into the E26 provirus as described in Figure 2. The recombinant viruses, as well as E26–WT were then used to infect 2-day chicken blastoderm, and transformed colonies were expanded and analyzed. The results in Figure 6 show a substantial increase in the proportion of EOS47-positive cells in clones transformed by E26–βΔTA1 and E26–βΔTA2, whereas myeloid cells were essentially absent from clones transformed by the E26–βΔTA2 and E26–βΔTA2/RK viruses. In addition, as for E26–WT, no significant numbers of eosinophils were observed in E26–βΔTA2/RK transformed clones. As expected from the results with the βD63N allele (Fig. 4A), the EOS47-positive cells transformed by E26–βΔTA1 and E26–βΔTA2 were negative for eosinophil peroxidase (data not shown). These results confirmed that transactivation-defective (but DNA-binding competent) C/EBP molecules can promote MEP commitment to the eosinophil lineage (without eosinophil maturation), and indicate that inhibition of C/EBP activity inhibits MEP differentitation along the myeloid lineage.
Figure 6.
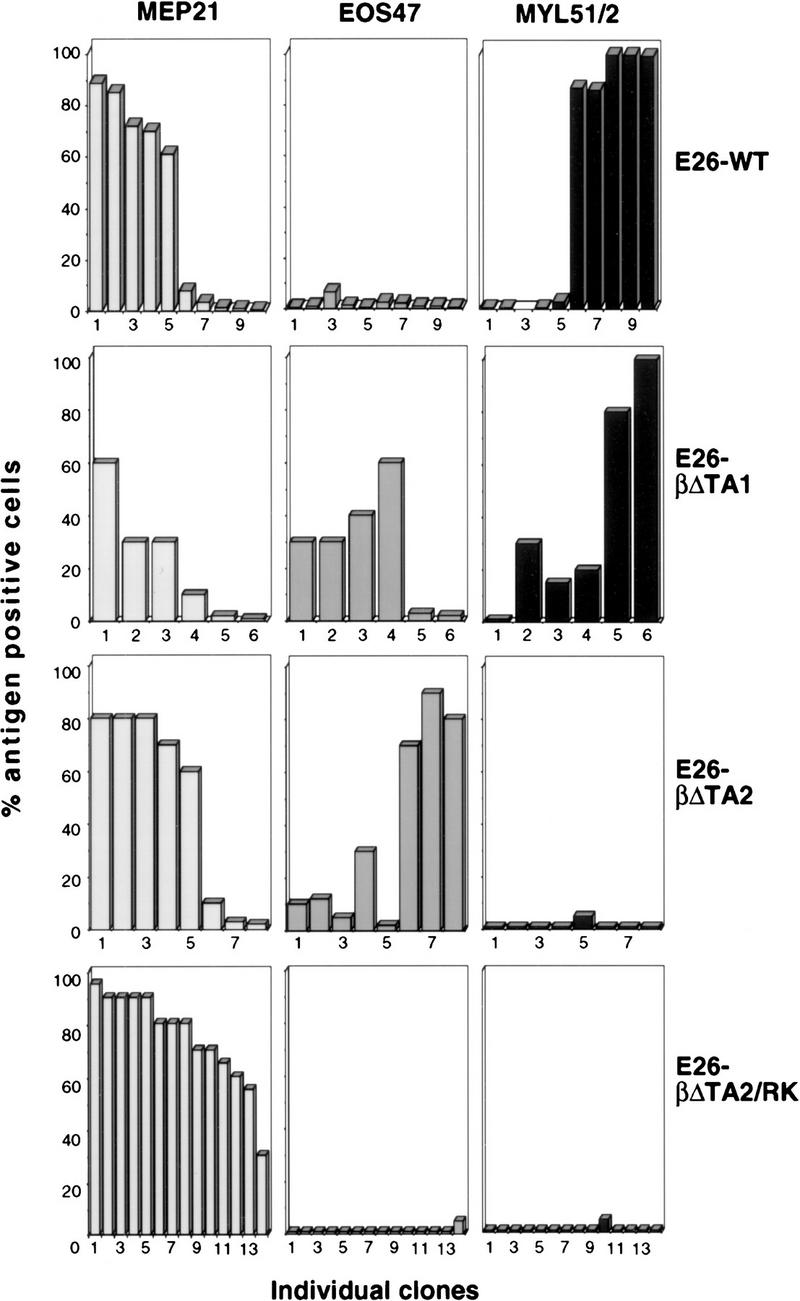
Effects of dominant-negative rat C/EBPβ mutants on MEP differentiation. Two-day blastoderm cells were infected by cocultivation with Q2bn cells transfected with E26–WT, E26–βΔTA1, E26–βΔTA2, and E26–βΔTA2/RK DNAs. Transformed colonies were expanded and phenotyped by FACS analysis. The percentages of MEP21-, EOS47-, and MYL51/2-positive cells for each clone analyzed is shown (the clone order is identical in each of the three columns).
Activation of a C/EBPβ–estrogen receptor fusion in MEPs induces myeloid lineage commitment and eosinophil maturation
To ensure that the observed increase in eosinophil differentiation was a direct effect of ectopic C/EBP expression (and not the result of selection or other indirect mechanisms) we made an E26 virus expressing the chicken C/EBPβ fused to the estrogen receptor (ER) hormone-binding domain (E26–βER; Fig. 7A). This C/EBPβ–ER fusion protein is transcriptionally active only in the presence of estrogen agonists (Müller et al. 1995). The virus was then used to generate clonal MEP populations after infection of chicken blastoderm. These E26–βER MEPs expressed a 85-kD C/EBPβ–ER fusion protein at a level similar to that of endogenous C/EBPβ in E26 transformed myeloblasts (Fig. 7B). They were then either treated with β-estradiol or mock treated, and their cell-surface antigen expression determined at different times thereafter. As shown in Figure 7C–E, for two clones of each type, a down-regulation of MEP21 antigen was observed in the β-estradiol treated E26–βER clones after 2 days, with <15% antigen-positive cells (with a >10-fold decrease in fluorescence intensity relative to the starting population; data not shown) remaining after 5 days. These changes were followed by the appearance of EOS47-positive cells in the hormone-treated samples (Fig. 7D) and, with some delay, of MYL51/2-positive cells (Fig. 7E). The up-regulation of the EOS47 antigen was paralleled by the appearance of cells staining for eosinophil peroxidase (data not shown). No significant changes in antigen expression or peroxidase staining were seen in control E26–WT-transformed MEPs with or without inducer (Fig. 7C–E), nor was any significant cell death associated with the observed change in phenotype (data not shown). We conclude that activation of C/EBPβ–ER in MEPs suppresses the MEP phenotype and induces their differentiation into either mature eosinophils or myeloblasts.
Figure 7.
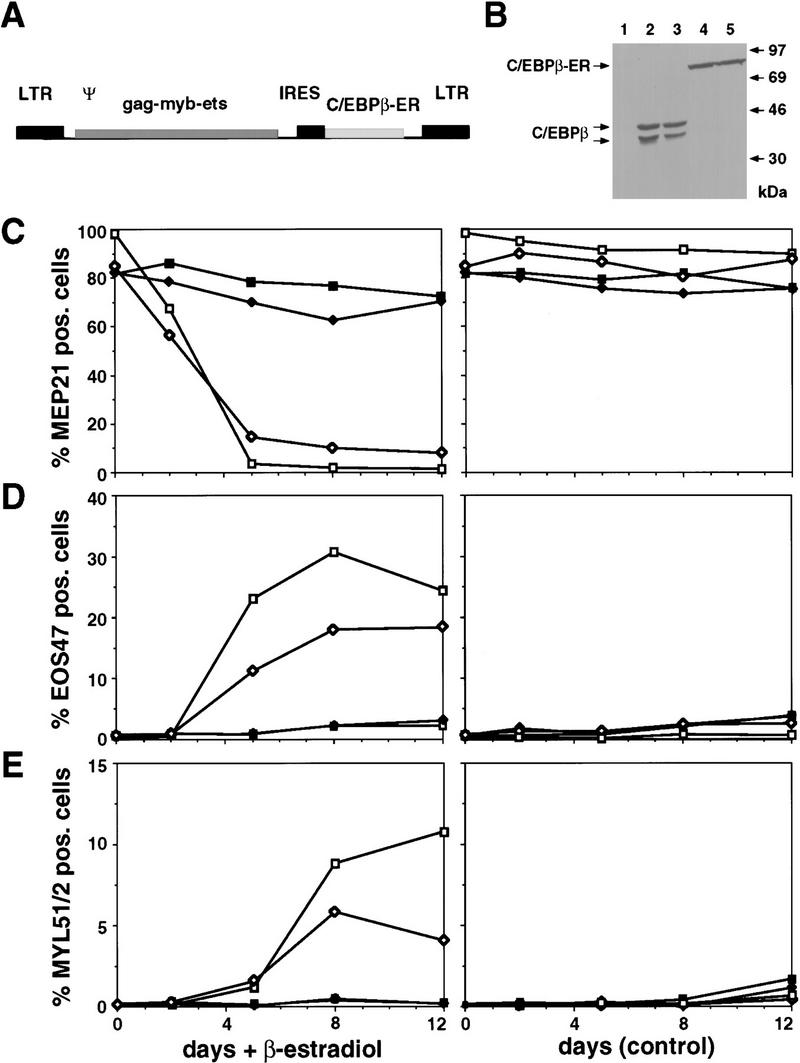
Effects of activating chicken C/EBPβ in MEPs transformed by E26–βER virus. (A) Structure of the E26–βER virus. (B) Western blot analysis of C/EBPβ–ER protein in E26–βER MEPs (βER cl.1 and cl.2; lanes 4,5) and of endogenous C/EBPβ in E26-WT-transformed myeloblasts (lanes 2,3). An E26–WT control MEP clone is shown for comparison (WT cl.1; lane 1). Western blotting was done with anti-chicken C/EBPβ antiserum on equal protein amounts from each clone. Bands corresponding to the endogenous C/EBPβ and exogenous C/EBPβ–ER are indicated. (C) Down-regulation of MEP21 expression by C/EBPβ–ER activation. E26–WT and E26–βER-transformed MEPs were treated with β-estradiol or mock treated, and their expression of MEP21 antigen, (D) EOS47 antigen, and (E) MYL51/2 antigen measured by FACS analysis after 2, 5, 8, and 12 days. Data from two representative clones of each type are shown. (▪) E26–WTcl.1; (♦) E26–WTcl.2; ( ) E26–βERcl.1; (⋄) E26–βERcl.2.
Transient induction of C/EBPβ separates eosinophil lineage commitment and terminal differentiation
To determine the time required for C/EBPβ to induce eosinophil and myeloid lineage commitment, we transiently activated C/EBPβ–ER in MEPs for increasing lengths of time and determined the effect on lineage commitment. This protocol involved exposing E26–βER and control E26–WT-transformed MEPs to β-estradiol for 1–4 days, removing the inducer by thorough washing and allowing differentiation to proceed for a total of 8 days (Fig. 8A). As can be seen from the experiment in Figure 8B–D, exposure of E26–βER MEPs to β-estradiol for 1 day led to significant eosinophil lineage commitment, as judged by the appearance of 10%–30% of the cells expressing EOS47 (but not MEP21) antigen. However, the cells obtained after this treatment were immature by morphology, had no granules, and were negative for eosinophil peroxidase (Fig. 8E, c,d) but could be distinguished from untreated E26–βER MEPs (panels a,b). In contrast, a 2-day pulse of β-estradiol yielded ∼80% of EOS47-positive cells, a high proportion of which contained cytoplasmic granules and were positive for eosinophil peroxidase (Fig. 8E, e,f). In addition, ∼5% of the cells expressed MYL51/2, a number that increased to 15%–20% after a 4-day pulse of β-estradiol. This showed that C/EBP-mediated eosinophil lineage commitment and maturation can be temporally separated, and that commitment to the eosinophil lineage induced by C/EBPβ–ER activation precedes commitment to the myeloid lineage.
Figure 8.
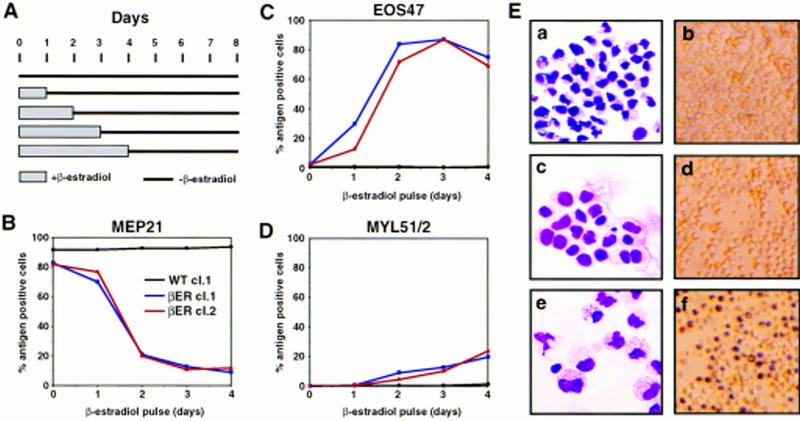
Effect of transient β-estradiol treatment on E26–βER-transformed MEP clones. (A) Experimental protocol. Cells were treated for 1–4 days with β-estradiol, thoroughly washed, and incubated in the absence of the hormone for a total of 8 days, at which time they were subjected to FACS analysis. Changes in MEP21 (B), EOS47 (C), and MYL51/2 (D) expression in MEP clones transformed by E26–WT and E26–βER after transient exposure to β-estradiol for 1–4 days (as indicated) were determined by antibody staining and FACS analysis at day 8 after initiation of the experiment. (Black line) E26–WTcl.1; (blue line) E26–βERcl.1; (red line) E26–βERcl.2. (E) May–Gruenwald–Giemsa (left) and peroxidase staining (right) of untreated E26–βER cl.1 cells (a,b), or cells treated for 1 day (c,d), or 2 days (e,f) with β-estradiol. Note that 30% of the cells in c and d are EOS47 positive.
Discussion
Induction of eosinophil and myeloid differentiation by C/EBP
In this report we have examined the effect of C/EBP on lineage choice by E26-transformed MEPs. Using recombinant E26 viruses, we found that both the α and β isoforms of C/EBP are able to induce eosinophil lineage commitment of MEPs. That this is an instructive property of C/EBP was shown by activation of a conditional C/EBPβ allele, which led to a synchronous change of phenotype in the entire cell population. These experiments also showed that C/EBPβ, in addition to inducing eosinophils, can induce myeloid lineage commitment, albeit with lower efficiency. The instructive capacity of C/EBP is further illustrated by the observation that a transient (2 day) C/EBPβ–ER activation was sufficient to induce >80% of MEPs to become eosinophils. That this effect is specific is indicated by the fact that we observed no expression of thrombocytic or erythoid markers after C/EBPβ–ER induction. In addition, the biological effect of C/EBPβ induction in MEPs contrasts with that of PU.1 in the same cells, in which myeloid differentiation was exclusively observed (Nerlov and Graf 1998; Fig. 9A). However, the finding that C/EBPβ protein is also capable of inducing myeloid differentiation suggests a partial functional overlap with PU.1.
Figure 9.
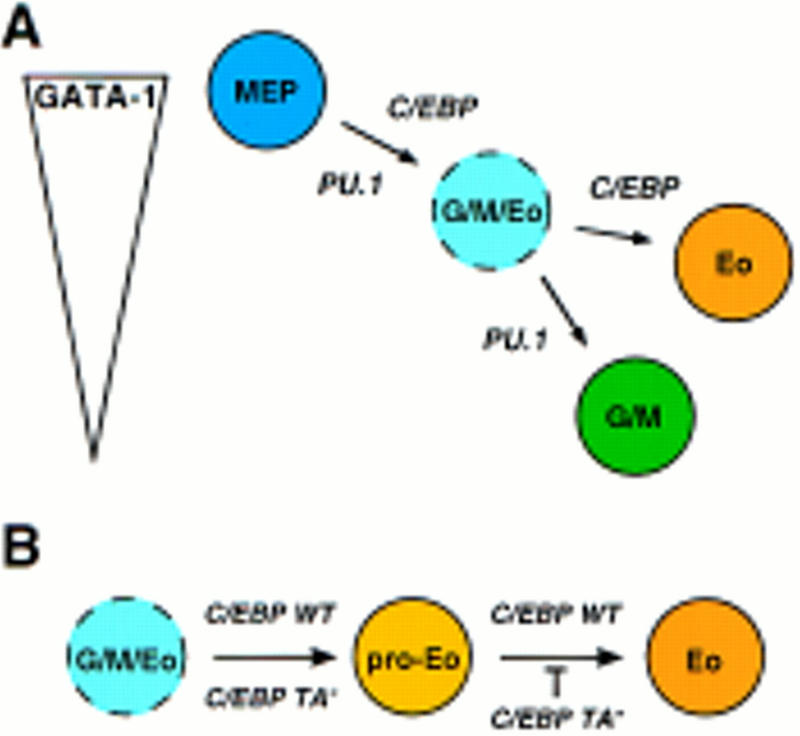
Model for the role of PU.1 and C/EBP in lineage commitment (A) and in eosinophil maturation (B). As explained in the text, C/EBPβ, although predominantly inducing eosinophil differentiation, may, in addition, be partially redundant with PU.1 with respect to induction of myeloid lineage commitment.
We have shown previously that eosinophil differentiation can be induced in an MEP cell line by activation of C/EBPβ–ER (Müller et al. 1995). However, in this case no myeloid cells were observed, possibly because of the massive cell death induced in the cell line after C/EBPβ–ER activation, an effect not observed in the primary clones used in the present study. Although this may simply illustrate the greater differentiation capacity of primary cells, an alternative explanation is that they express significantly lower levels of exogenous C/EBPβ than the cell line, levels that are comparable with that of the endogenous factor in myeloblasts.
Because the Gag–Myb–Ets oncoprotein contains the Ets-1 DNA-binding domain, it is possible that it, like c-Ets-1, cooperates with C/EBPs on the EOS47 promoter (and in eosinophil lineage induction). However, because the E26 virus transforms cells of most nonlymphoid lineages, it does not seem to participate directly in lineage specification. Furthermore, as the endogenous c-Ets-1 mRNA is expressed in MEPs (McNagny et al. 1998) the Gag–Myb–Ets oncoprotein is not likely to be required for the effect of C/EBP in MEPs.
Regulation of myeloid and eosinophil differentiation by C/EBP
All C/EBP forms tested, when constitutively expressed, promoted eosinophil lineage commitment of MEPs, with the exception of the βΔTA2/RK DNA-binding-deficient mutant. This suggests that, whereas the DNA-binding-domain is required for this function, the C/EBPβ-transactivation domain is, to a significant extent, dispensable. The effect of forced C/EBP expression on myeloblast formation is more complicated. On the one hand, in clones transformed by E26–C/EBP viruses, the proportion of myeloid cells relative to that observed in E26–WT-transformed clones decreased with increasing strength of the C/EBP transactivation potential. On the other hand, no myeloid cells were obtained after forced expression of deletion mutants that completely lacked the C/EBP transactivation domain. This suggests that myeloblast differentiation can be suppressed by C/EBP proteins that either exhibit strong or no transcriptional activity. As both C/EBPα and C/EBPβ are abundantly expressed in myeloid cells, neither is likely to adversely affect myeloid cell survival. We propose that C/EBPs with strong transactivation capacity divert MEP differentiation toward the eosinophil lineage, at the expense of spontaneously differentiating myeloid cells (see below; Fig. 9A).
A putative target of the dominant-negative C/EBP alleles is the cMGF receptor. This as yet unidentified receptor is required for growth of both normal and E26-transformed myeloid cells (Leutz et al. 1984). Expression of the mouse IL-6 and G-CSF receptor genes is dependent on C/EBPα (Zhang et al. 1997; P. Zhang and D. Tenen, pers. comm.), and mammalian IL-6 and G-CSF are homologous to cMGF structure and function (Leutz et al. 1989), raising the possibility that the corresponding receptors are regulated in a similar fashion.
C/EBPs act at two distinct steps during eosinophil differentiation.
We observed that short periods of C/EBPβ–ER activation in MEPs led to the formation of nongranulated immature eosinophils (EOS47 positive, peroxidase negative), whereas longer periods of activation yielded mature granulated eosinophils (EOS47 positive, peroxidase positive). In addition, as mentioned above, all DNA-binding-competent C/EBP proteins promoted eosinophil lineage commitment, whereas only those with an intact transactivation domain supported terminal differentiation. This indicates that C/EBP can regulate genes involved in eosinophil lineage commitment independently of its transactivation function. In contrast, C/EBP-mediated transactivation is required for the terminal differentiation of mature eosinophils, as these were only obtained with the wild-type forms of C/EBPα and C/EBPβ. The presence of EOS47-positive, peroxidase-negative cells in chicken bone marrow raises the possibility that a similar two-step mechanism of eosinophil differentiation also occurs during normal hematopoiesis. Our interpretation of the data is summarized in Figure 9B. Considering that both C/EBPα (Zhang et al. 1997) and C/EBPε (Yamanaka et al. 1997) have been shown to be required for eosinophil development in mice, the observation that interference with C/EBP function blocks eosinophil differentiation is consistent with similar roles for C/EBP proteins in mammalian and avian species. However, species differences may still exist, especially as C/EBP isoform sequences outside the highly conserved bZip region deviate considerably between mouse and chicken.
Role of GATA-1 in myeloid/eosinophil lineage choice
A surprising observation was that constitutive expression of potent C/EBP activators in MEPs not only promoted eosinophil differentiation, but led to partial (C/EBPβ) or almost complete (C/EBPα) inhibition of myeloid differentiation. Several lines of evidence suggest that GATA factors, in particular GATA-1, are involved in this phenomenon. First, forced expression of GATA-1 in myeloid cells (Kulessa et al. 1995; E. Querfurth, C. Nerlov, and T. Graf, unpubl.) and of C/EBP in MEPs (this paper) both lead to eosinophil formation. Therefore, these two factors are likely, on a hematopoietic background, to be sufficient to specify the eosinophil lineage, as other required factors would have to be ubiquitous among the three cell types. Second, the differentiation of MEPs into either myeloid cells or eosinophils requires the complete and partial down-regulation of GATA-1, respectively (Kulessa et al. 1995). The stabilization of GATA-1 expression at an intermediate level is therefore likely to be instrumental in the decision of a multipotent progenitor to choose the eosinophil over the myeloid lineage. In support of this notion is the finding that activation of a PU.1–ER fusion in MEPs leads to GATA-1 down-regulation followed by myeloid differentiation and that if this process is interrupted at a stage in which cells express an intermediate GATA-1 level, most cells commit to the eosinophil lineage (see Nerlov and Graf 1998). As summarized in Figure 9A, these results therefore indicate that myeloid/eosinophil differentiation proceeds via an MEP-derived intermediate (designated granulocyte/macrophage/eosinophil, or G/M/Eo, precursor), in which GATA-1 levels have been partially down-regulated, and which is directed by C/EBP towards an eosinophil fate (because C/EBP + intermediate GATA-1 specifies eosinophils) and by PU.1 toward a myeloid fate.
Materials and methods
Cell culture and FACS analysis
Infection of 2-day chicken blastoderm by cocultivation with Q2bn packaging cells transfected with E26 proviral DNA has been described (Rossi et al. 1996b). For analysis of bulk cultures, cells were plated in methylcellulose and single colonies picked and expanded. Cells were phenotyped by indirect immunoflourescence (IIF) on a Becton–Dickinson FACScan 2 weeks after infection (Graf et al. 1992). The MEP21, EOS47, and MYL51/2 monoclonal antibodies have been described (Kornfeld et al. 1983; Graf et al. 1992; McNagny et al. 1992). For analysis of individual MEP clones, colonies were picked and expanded, analyzed by IIF and clones positive for MEP21 and MEP26 antigen expression and negative for lineage-specific markers selected. For induction, cells were cultured in the presence of 0.5 μm β-estradiol. In pulse-chase experiments, cells were washed three times in medium after the indicated period of β-estradiol treatment and replated in the absence of inducer. Phenotypes were analyzed by IIF as above. Peroxidase activity was detected as described (Graf et al. 1992), and morphological analyses were performed on cytospins stained with May Gruenwald Giemsa (Diff-Quik, Harleco).
The origins of the cell lines used as sources of RNA have been described previously: HD3 erythroblasts (Beug et al. 1982); HD37 erythroblasts (Metz and Graf 1991); HD11 macrophages (Beug et al. 1979); HD13 granulocytes (Golay et al. 1988); HD50 MEPs and HD50M myeloblasts (Graf et al. 1992); 1A1 eosinophils (Kulessa et al. 1995); MSB-1 T cells (Akiyama and Kato 1974); RP-12 B cells (Siegfried and Olson 1972); Bm2c2 monoblasts (Moscovici et al. 1983).
All cells were grown in blastoderm medium (Graf et al. 1992) composed of DMEM supplemented with 10% fetal calf serum, 2.5% chicken serum, 0.15% NaHCO3, 56 μg/ml conalbumin, 80 μm of 2-mercaptoethanol, 0.9 mg/ml insulin, and the standard complement of antibiotics at 37°C in 5% CO2. Medium for HD50M and HD11 cells was supplemented with ∼10 U/ml of recombinant chicken myelomonocytic growth factor (cMGF; Leutz et al. 1989).
RNA extraction and Northern blotting
Total cellular RNA was prepared according to Chomczynski and Sacchi (1987). After electrophoresis through a 1.2% formaldehyde–agarose gel, RNA was transferred to Duralose (Stratagene) by capillary blotting and probed with chicken C/EBPα (Calkhoven et al. 1992), C/EBPβ (Katz et al. 1993), and GAPDH cDNA (Dugaiczyk 1983) labeled by random priming.
Transfection and Western blot analyses
Transient transfection into Q2bn fibroblasts using the EOS47/−152-LUC (containing bp −152 to +157 of the EOS47 gene, relative to the transcription start site, driving a firefly luciferase gene; McNagny et al. 1998), the mim-1 promoter construct pMim-1-ΔC–LUC (Ness et al. 1989), and pRSV–βGal internal control plasmid was done as described (Frampton et al. 1996). Cellular extracts were made and Western blotting performed as described (Frampton et al. 1996), with anti-chicken C/EBPβ antiserum (formerly known as anti-NF-M antiserum; Katz et al. 1993; kindly provided by Dr. A. Leutz) to detect C/EBPβ proteins. This antiserum was used at a 1:3000 dilution.
DNA constructs
Proviral constructs were based on an E26 provirus containing the EMC virus IRES element (Frampton et al. 1996). This construct we refer to as E26–WT. The chicken C/EBPα, C/EBPβ, and C/EBPβD63N cDNAs (Calkhoven et al. 1992; Katz et al. 1993) were inserted into E26–WT as NcoI–XbaI fragments derived from pcDNAI-based expression vectors, in all cases using the first NcoI site of the C/EBP coding sequence. The C/EBPβ–ER fusion (Müller et al. 1995) was inserted in the same way. Dominant-negative C/EBP alleles were expressed as fusions with a leader peptide containing the 9E10 Myc epitope tag (MEQKLISEEDLNGA), also from the NcoI site of the EMCV IRES. The βΔTA2 alleles contained the rat C/EBPβ amino acids 100–297 (Descombes et al. 1990), the βΔTA1 allele amino acids 71–297. The RK mutant additionally contained the double amino acid substitution R238A, K239A. Mutations in the C/EBPβ coding sequences were introduced as described (Nerlov et al. 1991), except for the chicken C/EBPβ D63N mutant, which was a random mutation generated by PCR amplification. For transfection analyses, the C/EBP alleles described above were expressed from the CMV promoter of the pcDNAI vector (Invitrogen), as was the I(16) (or CBFβ–SMMHC) protein, derived from the pCL1 expression plasmid (Hajra et al. 1995).
Cell sorting and immunofluorescence analyses
Bone marrow cells from a 10-day-old chick were dispersed into a single cell suspension and stained with the EOS47 mAb or isotype-matched control antibody followed by goat anti-mouse IgG antibodies coupled to FITC. Cells were then sorted with a FACS Vantage (Becton Dickinson) cytometer. Sorted cells were reanalyzed to determine the purity of the populations (for the EOS47 positive cells it was ∼95%) and dispensed onto glass slides by use of a cytospin apparatus (Shandon), air dried, and stained for granular peroxidase (an exclusive marker of chicken eosinophil granules; Brune and Spitznagel 1973). The slides were then fixed with methanol, DAPI stained and observed by fluorescence microscopy.
Acknowledgments
We thank Drs. Steve McKnight and Achim Leutz for providing C/EBP cDNAs and antibodies, Dr. Francis Collins for the I(16)/CBFβ–SMMHC cDNA, Dr. Daniel Tenen for sharing unpublished data, and Servane Leillard for assistance in preparing the report. C.N. thanks Dr. Jon Frampton for help with primary cell cultures. C.N. is a fellow of the Danish Medical Research Council and K.M.M. was supported by National Research Service Award Fellowship no. F32 HL0736 from the National Heart, Lung, and Blood Institute, National Institutes of Health. This work was partially funded by the Deutsche Forschungs Gemeinschaft (SFB 229).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL graf@embl-heidelberg.de; FAX 49 6221 387 516.
References
- Akiyama Y, Kato S. Two cell lines from lymphomas of Marek’s desease. Biken J. 1974;17:105–116. [PubMed] [Google Scholar]
- Beug H, von Kirchbach A, Döderlein G, Conscience J-F, Graf T. Chicken hematopoietic cells transformed by seven strains of defective avian leukemia viruses display three distinct phenotypes of differentiation. Cell. 1979;18:375–390. doi: 10.1016/0092-8674(79)90057-6. [DOI] [PubMed] [Google Scholar]
- Beug, H., G. Doederlein, C. Freudenstein, and T. Graf. 1982. Erythroblast cell lines transformed by a temperature sensitive mutant of avian erythroblastosis virus: A model system to study erythroid differentiation in vitro. J. Cell. Physiol. (Suppl.) 1: 195–207. [DOI] [PubMed]
- Beug H, Leutz A, Kahn P, Graf T. Ts mutants of E26 leukemia virus allow transformed myeloblasts, but not erythroblasts or fibroblasts, to differentiate at the nonpermissive temperature. Cell. 1984;39:579–588. doi: 10.1016/0092-8674(84)90465-3. [DOI] [PubMed] [Google Scholar]
- Brune K, Spitznagel JK. Peroxidaseless chicken leukocytes: Isolation and characterization of antibacterial granules. J Infect Dis. 1973;127:84–94. doi: 10.1093/infdis/127.1.84. [DOI] [PubMed] [Google Scholar]
- Burk O, Mink S, Ringwald M, Klempnauer K-H. Synergistic activation of the chicken mim-1gene by v-Myb and C/EBP transcription factors. EMBO J. 1993;12:2027–2038. doi: 10.1002/j.1460-2075.1993.tb05852.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calkhoven CF, Geert AB, Wijnholds J. C/EBP, a chicken transcription factor of the leucine-zipper C/EBP family. Nucleic Acids Res. 1992;20:4093. doi: 10.1093/nar/20.15.4093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Z, Umek RM, McKnight SL. Regulated expression of three C/EBP isoforms during adipose conversion of 3T3-L1 cells. Genes & Dev. 1991;5:1538–1552. doi: 10.1101/gad.5.9.1538. [DOI] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N. Single step isolation of RNA by acid guanidinium isothiocyanate method. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Daimon T, Caxton-Martins A. Electron microscopic and enzyme cytochemical studies on granules of mature chicken granular leukocytes. J Anat. 1977;123:553–563. [PMC free article] [PubMed] [Google Scholar]
- Descombes P, Chojkier M, Lichtsteiner S, Falvey E, Schibler U. LAP, a novel member of the C/EBP gene family, encodes a liver-enriched transcriptional activator protein. Genes & Dev. 1990;4:1541–1550. doi: 10.1101/gad.4.9.1541. [DOI] [PubMed] [Google Scholar]
- Dugaiczyk A. Cloning and sequencing of a deoxyribonucleic acid copy of glyceraldehyde-3-phosphate dehydrogenase messenger ribonucleic acid isolated from chicken muscle. Biochemistry. 1983;22:1605–1613. doi: 10.1021/bi00276a013. [DOI] [PubMed] [Google Scholar]
- Frampton J, McNagny KM, Sieweke M, Philip A, Smith G, Graf T. v-Myb DNA binding is required to block thrombocytic differentiation of Myb-Ets-transformed multipotent hematopoitic progenitors. EMBO J. 1995;14:2866–2875. doi: 10.1002/j.1460-2075.1995.tb07286.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frampton J, Ramqvist T, Graf T. v-Myb of E26 leukemia virus up-regulates bcl-2and suppresses apoptosis in myeloid cells. Genes & Dev. 1996;10:2720–2731. doi: 10.1101/gad.10.21.2720. [DOI] [PubMed] [Google Scholar]
- Golay J, Introna M, Graf T. A single point mutation in the v-etsoncogene affects both erythroid and myelomoncytic cell differentiation. Cell. 1988;55:1147–1158. doi: 10.1016/0092-8674(88)90259-0. [DOI] [PubMed] [Google Scholar]
- Graf T, McNagny KM, Brady G, Frampton J. Chicken “Erythroid” cells transformed by the Gag-Myb-Ets encoding E26 leukemia virus are multipotent. Cell. 1992;70:201–213. doi: 10.1016/0092-8674(92)90096-u. [DOI] [PubMed] [Google Scholar]
- Hajra A, Liu PP, Wang Q, Kelley CA, Stacy T, Adelstein RS, Speck NA, Collins FS. The leukemic core binding factor β-smooth muscle myosin heavy chain (CBFβ-SMMHC) chimeric protein requires both CBFβ and myosin heavy chain domains for transformation of NIH3T3 cells. Proc Natl Acad Sci. 1995;92:1926–1930. doi: 10.1073/pnas.92.6.1926. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Katz S, Kowenz-Leutz E, Müller C, Meese K, Ness SA, Leutz A. The NF-M transcription factor is related to C/EBPβ and plays a role in signal transduction, differentiation and leukemogenesis of avian myelomonocytic cells. EMBO J. 1993;12:1321–1332. doi: 10.1002/j.1460-2075.1993.tb05777.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornfeld S, Beug H, Doederlein G, Graf T. Detection of avian hematopoietic cell surface antigens with monoclonal antibodies to myeloid cells. Exp Cell Res. 1983;143:383–394. doi: 10.1016/0014-4827(83)90065-4. [DOI] [PubMed] [Google Scholar]
- Kraut N, Frampton J, McNagny KM, Graf T. A functional Ets DNA-binding domain is required to maintain multipotency of hematopoietic progenitors transformed by Myb–Ets. Genes & Dev. 1994;8:33–44. doi: 10.1101/gad.8.1.33. [DOI] [PubMed] [Google Scholar]
- Kulessa H, Frampton J, Graf T. GATA-1 reprograms avian myelomonocytic cell lines into eosinophils, thromboblasts and erythroblasts. Genes & Dev. 1995;9:1250–1262. doi: 10.1101/gad.9.10.1250. [DOI] [PubMed] [Google Scholar]
- Leutz A, Beug H, Graf T. Purification and characterization of cMGF, a novel chicken myelomonocytic growth factor. EMBO J. 1984;3:3191–3197. doi: 10.1002/j.1460-2075.1984.tb02278.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leutz A, Damm K, Sterneck E, Kowenz E, Ness S, Frank R, Gausepohl H, Pan Y-C E, Smart J, Hayman M, Graf T. Molecular cloning of the chicken myelomonocytic growth factor (cMGF) reveals relationship to interleukin 6 and granulocyte colony stimulating factor. EMBO J. 1989;8:175–181. doi: 10.1002/j.1460-2075.1989.tb03362.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Tarle SA, Hajra A, Claxton DF, Marlton P, Freedman M, Siciliano M, Collins FS. Fusion between transcription factor CBFβ/PEBP2β and a myosin heavy chain in acute myeloid leukemia. Science. 1993;261:1041–1044. doi: 10.1126/science.8351518. [DOI] [PubMed] [Google Scholar]
- McNagny KM, Lim F, Grieser S, Graf T. Cell surface proteins of chicken hematopoietic progenitors, thrombocytes and eosinophils detected by novel monoclonal antibodies. Leukemia. 1992;6:975–984. [PubMed] [Google Scholar]
- McNagny KM, Rossi F, Smith G, Graf T. The eosinophil-specific cell surface antegen, EOS47, is a chicken homologue of the oncofetal antigen melanotransferrin. Blood. 1996;87:1343–1352. [PubMed] [Google Scholar]
- McNagny KM, Sieweke M, Döderlein G, Graf T, Nerlov C. Regulation of eosinophil-specific gene expression by a C/EBP-Ets complex and GATA-1. EMBO J. 1998;17:3669–3680. doi: 10.1093/emboj/17.13.3669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metz T, Graf T. v-myb and v-etstransform chicken erythroid cells and cooperate both in trans and in cis to induce distinct differentiation phenotypes. Genes & Dev. 1991;5:369–380. doi: 10.1101/gad.5.3.369. [DOI] [PubMed] [Google Scholar]
- Moscovici MG, Jurdic P, Samarut J, Gazzolo L, Mura CV, Moscovici C. Characteristics of the hemopoietic target cells for the avian leukemia virus E26. Virology. 1983;129:65–78. doi: 10.1016/0042-6822(83)90396-3. [DOI] [PubMed] [Google Scholar]
- Müller C, Kowenz-Leutz E, Grieser-Ade S, Graf T, Leutz A. NF-M (chicken C/EBPβ) induces eosinophilic differentiation and apoptosis in a hematopoietic progenitor cell line. EMBO J. 1995;14:6127–6135. doi: 10.1002/j.1460-2075.1995.tb00303.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nerlov, C. and T. Graf. 1998. PU.1 induces myeloid linkage commitment in multipotent hematopoetic progenitors. Genes & Dev. (this issue). [DOI] [PMC free article] [PubMed]
- Nerlov C, Ziff EB. CCAAT/enhancer binding protein-α amino acid motifs with dual TBP and TFIIB binding ability mediate transcriptional activation in both yeast and mammalian cells. EMBO J. 1995;14:4318–4328. doi: 10.1002/j.1460-2075.1995.tb00106.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nerlov C, Rørth P, Blasi F, Johnsen M. Essential AP-1 and PEA3 binding elements in the human urokinase enhancer display cell type-specific activity. Oncogene. 1991;6:1583–1592. [PubMed] [Google Scholar]
- Ness SA, Marknell Å, Graf T. The v-myb oncogene product binds to and activates the promyelocyte-specific mim-1gene. Cell. 1989;59:1115–1125. doi: 10.1016/0092-8674(89)90767-8. [DOI] [PubMed] [Google Scholar]
- Ness SA, Kowenz-Leutz E, Casini T, Graf T, Leutz A. Myb and NF-M: Combinatorial activators of myeloid genes in heterologous cell types. Genes & Dev. 1993;7:749–759. doi: 10.1101/gad.7.5.749. [DOI] [PubMed] [Google Scholar]
- Oelgeschläger M, Nuchprayoon I, Lüscher B, Friedman AD. C/EBP, c-Myb, and PU.1 cooperate to regulate the neutrophil elastase promoter. Mol Cell Biol. 1996;16:4717–4725. doi: 10.1128/mcb.16.9.4717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ron D, Habener JF. CHOP, a novel developmentally regulated nuclear protein that dimerizes with transcription factors C/EBP and LAP and functions as a dominant-negative inhibitor of gene transcription. Genes & Dev. 1992;6:439–453. doi: 10.1101/gad.6.3.439. [DOI] [PubMed] [Google Scholar]
- Rossi F, McNagny KM, Logie C, Stewart AF, Graf T. Excision of Ets by an inducible site-specific recombinase causes differentiation of Myb-Ets-transformed hematopoietic progenitors. Curr Biol. 1996a;6:866–872. doi: 10.1016/s0960-9822(02)00610-3. [DOI] [PubMed] [Google Scholar]
- Rossi F, McNagny KM, Smith G, Frampton J, Graf T. Lineage commitment of transformed haematopoietic progenitors is determined by the level of PKC activity. EMBO J. 1996b;15:1984–1901. [PMC free article] [PubMed] [Google Scholar]
- Scott LM, Civin CI, Rorth P, Friedman AD. A novel temporal expression pattern of three C/EBP family members in differentiating myelomonocytic cells. Blood. 1992;80:1725–1735. [PubMed] [Google Scholar]
- Siegfried LM, Olson C. Characteristics of avian transmissible lymphoid tumor cells maintained in culture. J Natl Cancer Inst. 1972;48:791–795. [PubMed] [Google Scholar]
- Tanaka T, Akira S, Yoshida K, Umemoto M, Yoneda Y, Shirafuji N, Fujiwara H, Suematsu S, Yoshida N, Kishimoto T. Targeted disruption of the NF-IL6 gene discloses its essential role in bacteria killing and tumor cytotoxicity by macrophages. Cell. 1995;80:353–361. doi: 10.1016/0092-8674(95)90418-2. [DOI] [PubMed] [Google Scholar]
- Tenen DG, Hromas R, Licht JD, Zhang D-E. Transcription factors, normal myeloid development, and leukemia. Blood. 1997;90:489–519. [PubMed] [Google Scholar]
- Williams SC, Baer M, Dilner AJ, Johnson PF. CRP2 (C/EBPβ) contains a bipartite regulatory domain that controls transcriptional activation, DNA binding and cell specificity. EMBO J. 1995;14:3170–3183. doi: 10.1002/j.1460-2075.1995.tb07319.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka R, Barlow C, Lekstrom-Himes J, Castilla LH, Liu PP, Eckhaus M, Decker T, Wynshaw-Boris A, Xanthopoulos KG. Impaired granulopoiesis, myelodysplasia, and early lethality in CCAAT/enhancer binding protein ε-deficient mice. Proc Natl Acad Sci. 1997;94:13187–13192. doi: 10.1073/pnas.94.24.13187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D-E, Hetherington CJ, Meyers S, Rhoades KL, Larson CJ, Chen H-M, Hiebert SW, Tenen DG. CCAAT enhancer-binding protein (C/EBP) and AML1 (CBFa2) synergistically activate the macrophage colony-stimulating factor receptor promoter. Mol Cell Biol. 1996;16:1231–1240. doi: 10.1128/mcb.16.3.1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D-E, Zhang P, Wang N-D, Hetherington CJ, Darlington GJ, Tenen D. Absence of granulocyte colony-stimulating factor signalling and neutrophil development in CCAAT enhancer binding protein α-deficient mice. Proc Natl Acad Sci. 1997;94:569–574. doi: 10.1073/pnas.94.2.569. [DOI] [PMC free article] [PubMed] [Google Scholar]