Abstract
Considerable attention has focused on the use of alternatives to the native ribose and phosphate backbone of small interfering RNAs for therapeutic applications of the RNA interference pathway. In this synopsis, we highlight the less common chemical modifications, namely those of the RNA nucleobases. Base modifications have the potential to lend insight into the mechanism of gene silencing and to lead to novel methods to overcome off-target effects that arise due to deleterious protein binding or mis-targeting of mRNA.
Upon the discovery of the RNA interference (RNAi) pathway,1,2 bioorganic chemists seized the opportunity to engineer chemically modified strands of RNA with altered and improved properties.3–5 Such synthetic modifications aid our understanding of the mechanism of RNA interference, and improve the potency and specificity of short interfering RNAs (siRNAs), the triggers of RNAi, for therapeutic applications. Numerous studies have illustrated the utility of sugar, backbone and bioconjugate modifications in improving the properties of siRNAs, such as increasing stability and cell permeability and decreasing immunostimulation.6–9 In this synopsis, we highlight recent work in chemical modifications of siRNA nucleobases.
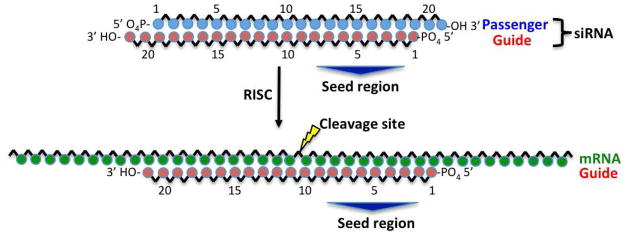
SiRNAs are 21 to 23-nucleotide RNA duplexes that engage the RNAi pathway. Upon cellular uptake, a complex of proteins binds the siRNA and loads one strand (termed the “guide strand”) into Argonaute 2 (Ago2), an RNase H-like endonuclease.10 The selection of the guide strand is based on the thermodynamic stability of the siRNA duplex ends, with the least stable end presenting the 5′-end of the guide strand.11,12 The other strand of the siRNA, termed the “passenger stand”, is then cleaved.13,14 Ago2 loaded with the guide strand is referred to as RISC, for RNA-induced silencing complex. RISC binds target messenger RNA (mRNA) that is Watson-Crick complementary to the guide strand and cleaves it, resulting in inhibition of expression of the corresponding gene product (termed “gene silencing”) (Figure 1).15,16 The interaction of RISC with mRNA is highly dependent on nucleotides 2 – 8 of the guide strand,17 which is termed the “seed region”. On its way to the RISC, the siRNA should avoid binding to certain proteins such as the Toll-like receptors (TLRs),18 the RNA-dependent protein kinase (PKR)19 and double-stranded RNA-specific adenosine deaminases (ADARs).20 Interaction with proteins other than RISC is a principle cause of off-target effects that reduce overall gene silencing activity, decrease target gene specificity, and trigger toxicity.21
Figure 1.
The guide strand of an siRNA directs cleavage of Watson-Crick complementary mRNA, mediated by the RNA-induced silencing complex (RISC). Cleavage of mRNA results in silencing of the corresponding gene.
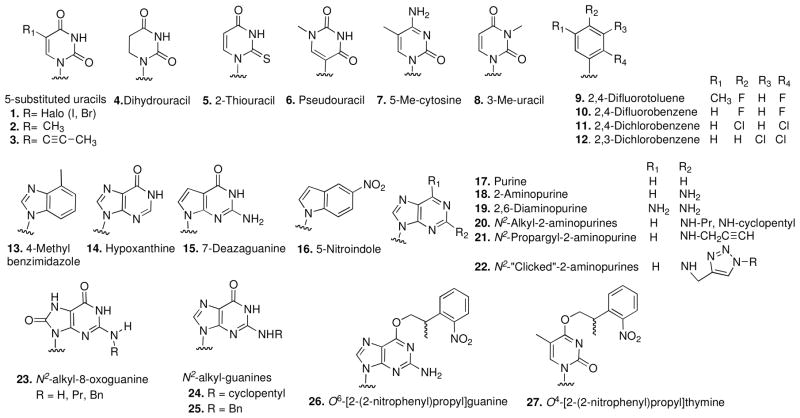
This review covers the nucleobase analogs 1–27, shown in Chart I. The chart is accompanied by Table I, which details the sites of incorporation of each modification (passenger or guide strand), the effect of each modification on the duplex melting temperature (Tm) relative to the unmodified siRNA and the effect of each modification on gene silencing activity (i.e. increasing, decreasing or maintaining the silencing activity of the unmodified siRNA).
Chart I.
Structures of siRNA base modifications.
Table 1.
Effects of Chemical Modification of siRNA Bases on Tm and RNAi Activity
| No. | Modification name | Strand | Δ Tm (°C) | RNAi activity | Ref |
|---|---|---|---|---|---|
| 1 | 5-Halouracil | G | NR | − | 3 |
| 2 | 5-Methyluracil | G | 0.2 to −0.7 | +/− | 22 |
| 3 | 5-Propynyluracil | G | 1.3 to 1.8 | − | 22 |
| 4 | Dihydrouracil | G, P | −3 to −5 | − to +/− | 23 |
| 5 | 2-Thiouracil | G, P | 0 to 2 | − to + | 23 |
| 6 | Pseudouracil | G, P | −1 to 1 | − to +/− | 23 |
| 7 | 5-Methylcytosine | G | 1 | +/− | 22, 24 |
| 8 | 3-Methyluracil | G | NR | --- | 3 |
| 9 | 2,4-Difluorotoluene | G, P | −5 to −12 | +/− to + | 25–27 |
| 10 | 2,4-Difluorobenzene | G, P | −4 to −10 | − to +/− | 28, 29 |
| 11 | 2,4-Dichlorobenzene | G | −4 | +/− | 29 |
| 12 | 2,3-Dichlorobenzene | G | −8 | − | 29 |
| 13 | 4-Methylbenzimidazole | G | −4 to −15 | − to +/− | 29 |
| 14 | Hypoxanthine | P | −4 | +/− | 25 |
| 15 | 7-Deazaguanine | G | NR | NR | 24 |
| 16 | 5-Nitroindole | P | −8 to −12 | +/− to + | 25 |
| 17 | Purine | P | −11 to −12 | +/− to + | 25 |
| 18 | 2-Aminopurine | P | −2 to −8 | +/− to + | 25 |
| 19 | 2,6-Diaminopurine | G | NR | − | 3 |
| 20 | N2-Propyl/N2-cyclopentyl-2-aminopurines | P | 0 | +/− | 30, 31 |
| 21 | N2-Propargyl-2-aminopurine | G, P | −4 to −5 | − to +/− | 31, 32 |
| 22 | N2-“Clicked”-2-aminopurines | G,P | −2 to −3 | --- to +/− | 31, 32 |
| 23 | N2-Alkyl-8-oxoguanine-(deoxyribose) | G | −3 to −13 | --- to + | 33, 34 |
| 24 | N2-Cyclopentyl-guanine | G, P | 0 | − to +/− | 30 |
| 25 | N2-Benzyl-guanine-(deoxyribose) | G | −3 | − to +/− | 35 |
| 26 | O6-[2-(2-nitrophenyl)propyl] guanine | G, P | NR | --- (Protected) +/− (Deprotected) |
36 |
| 27 | O4-[2-(2-nitrophenyl)propyl]thymine | G,P | NR | --- (Protected) +/−(Deprotected) |
36 |
P: Passenger strand; G: Guide strand; +: Increased silencing activity; −: Decreased silencing activity; ---: Severely decreased silencing activity/not tolerated; +/−: No significant change in silencing activity; NR: Not reported
Effects of thermal stability
Early work on siRNA chemical modifications suggested the importance of the thermal stability of the siRNA duplex, as measured by the melting temperature, Tm, on the gene silencing activity.3,37,38 However, there is no obvious correlation between the overall duplex Tm and the gene silencing activity of the siRNA. Rather, specific regions of the siRNA duplex have distinct tolerances towards stabilization and destabilization, resulting in position-specific changes of activity upon incorporation of chemical modifications that affect thermal stability.
In an initial study of siRNA chemical modifications, Chiu and Rana incorporated multiple 2,6-diaminopurines (19) at the 5′-end of an siRNA guide strand, replacing adenines.3 2,6-Diaminopurine uses its additional amine to form a third hydrogen bond with U,39 and thus increases the association of the base pair. The siRNA containing the stabilizing modifications displayed decreased RNAi activity. This observation is now supplemented by a large body of evidence that the 5′-end of the guide strand (i.e. the seed region) is particularly sensitive to changes in thermal stability, both positive and negative.40 More recently, Nawrot and coworkers23 reported that the activity of an siRNA could be increased by incorporating single 2-thiouracil (5) or pseudouracil bases (6) at the 3′ end of the guide stand in conjunction with a single dihydrouracil (4) base at the 3′ end of the passenger strand (i.e. opposite the seed region). 2-Thiouracil and pseudouracil favor a C3′-endo sugar pucker (the conformation preferred by an A-form RNA helix) and can increase the thermal stability by up to 2 °C.41–43 However, dihydrouracil, which favors a C2′-endo sugar pucker and lacks the base-stacking ability of aromatic heterocycles, decreases the thermal stability by 3 – 5 °C.44 Thus, the increase in activity of the siRNA was attributed to the enhanced thermodynamic asymmetry of the duplex ends, favoring the “opening” of the duplex at the 5′ end of the guide strand. As expected, incorporation of 2-thiouracil and pseudouracil at the 3′ end of the passenger strand was detrimental to activity.
In another study, Terrazas and Kool reported that small base modifications projecting into the RNA major groove (5-MeU, 2, and 5-MeC, 7) can improve siRNA thermal stability without impairing gene silencing, whereas bulkier groups (5-propynyl-U, 3) that increase thermal stability to a greater degree (~1.5 °C per modification) disrupted activity when incorporated at the 5′-end of the guide strand.22 This disruption could be due to close protein contacts around the seed region in addition to the selective thermal stabilization of the 5′-end of the guide strand. In examples such as this, further investigation is needed to distinguish the steric and thermal effects of modification.
Addepalli et al. investigated the introduction of the destabilizing modifications 2,4-difluorotoluene (9), hypoxanthine (14), 5-nitroindole (16), purine (17) and 2-aminopurine (18) at various positions within the passenger strand. The degree of thermal destabilization of the duplex varied between 1 and 12 °C. Their analysis revealed that the non-hydrogen-bonding nucleobase isosteres 2,4-difluorotoluene (9) and 5-nitroindole (16), or base-pair mismatches involving natural bases, can improve activity when incorporated at central locations in the passenger strand (i.e. nucleotides 9, 10, 11 and 12).25 However the effect was highly dependent on the specific position and type of modification. Interestingly, at positions 9, 11 and 12 the effect of the modifications on activity was fairly independent of the extent of thermal destabilization, however a strong correlation between thermal destabilization and activity was observed at position 10. Additional analysis, including replacement of the destabilizing base modifications with highly destabilizing abasic ribose modifications, confirmed that thermal destabilization is responsible for the activity enhancement. Their analysis also excluded the possibility that guide strand selection was biased by the presence of the chemical modifications in the passenger strand and also the possibility that the modifications inhibited the nucleolytic destruction of the passenger strand.
Effects of hydrogen-bonding and sterics
Crystallographic analyses of Argonaute proteins provide considerable insight into the mechanism of mRNA cleavage.26,45–47 However, other important factors, such as the necessity to maintain hydrogen bonds and steric effects, can be better understood by the introduction of nucleobase analogs within the guide strand. Principal to this analysis are the non-hydrogen-bonding nucleobase isosteres 9 – 13. Manoharan and coworkers showed that 2,4-difluorotoluene (9), which was introduced above in the context of passenger strand modifications, is an effective replacement for U at certain positions in the guide strand, such as position 7 and the 5′-end.27 However, when the modification was incorporated at position 10 of the guide stand, which is adjacent to the site of mRNA cleavage, the siRNA activity was reduced. Consistent with these observations, Kool and coworkers also showed that an siRNA containing either 2,4-difluorobenzene (10) or 2,4-dichlorobenzene (11) at position 7 of the guide strand displayed near-wild type activity, however the activity was dramatically decreased by the incorporation of 10 at positions 10 and 11 of the guide strand.28,29 High levels of activity were also observed for the incorporation of 10 at several other locations on the guide strand. Collectively, these results suggest that hydrogen bonding is not critical at several positions, most notably position 7, for effective cleavage of target mRNA, but critical at other locations, such as 10 and 11. Further evidence for the requirement of hydrogen-bonding at position 11 is provided by the fact that 3-methyluracil (8), which has a compromised Watson-Crick face due to the N-alkylation, is also not tolerated at this site.3
Incorporation of 10 or 11 at position 7 of the guide strand gave an siRNA with improved sequence selectivity for target mRNA in comparison to an siRNA containing the natural base (U) at this position.28,29 This target nucleotide preference was not due to the base-pairing selectivity of the RNAs alone, indicating that RISC enforces steric constraints on base-pairing interactions at this location. In contrast, the non-hydrogen-bonding base analogs 2,3-dichlorobenzene (12) and 4-methylbenzimidazole (13) showed preference for activity at position 7 when paired opposite U in the mRNA target, thus resembling the selectivity profile of A.29 Steric comparisons indicated that unlike 10 and 11, these two base analogs are closer in shape to A than U, thus consistent with their pairing preferences. This observation further supports the role of steric effects on mRNA selection and cleavage at certain locations within RISC.
Manoharan and coworkers also incorporated 9 in place of C, thereby forming 9:G base-pairs adjacent to the site of mRNA cleavage. This resulted in substantial loss of RNAi activity.26 Crystallographic analysis revealed a widening of the duplex at the 9:G base pairs, indicating that structural distortion of the duplex away from Watson-Crick geometry could be responsible for the decreased activity in this case. In addition, the incorporation of 9 at position 16 of the guide stand protected the siRNA duplex from endonuclease cleavage in human serum, however the silencing activity was halved.27
Minor groove modifications to prevent off-target effects
SiRNAs can stimulate the TLR-mediated innate immune response18 and can interact with off-target proteins such as PKR and ADAR, resulting in inefficient gene silencing20 and upregulation of off-target genes.48 The Beal laboratory has been pursuing minor-groove localized base modifications to prevent the interactions of siRNA with protein receptors other than the RISC. Recently, in a collaboration with Sirna Therapeutics, we demonstrated that N2-cyclopentylguanine (24) and N2-propyl or N2-cyclopentyl-2-aminopurines (20) could inhibit the immunostimulatory properties of a microRNA mimic (which can be considered as an siRNA).30 When this microRNA mimic was transfected into peripheral blood mononuclear cells (PBMCs), a single cyclopentyl modification at selected sites on the guide strand decreased production of the inflammatory cytokine tumor necrosis factor α (TNF-α) by at least 5-fold whilst maintaining the full RNAi activity of the unmodified RNA. The base-localized minor groove projections are likely preventing the activation of one or more of the RNA-sensing TLRs 3,49,50 7 and 8.51,52 These base modifications rival ribose modifications such as 2′-methoxy and 2′-fluoro at inhibiting immune stimulation.24,53–55 Unlike the minor-groove modifications, the major groove modification 7-deazaguanosine (15) did not inhibit immune stimulation when incorporated into an siRNA.24
In an earlier study, Puthenveetil et al. reported that incorporation of N2-benzyl-2′-deoxyguanosine (25) at specific positions in the passenger strand blocked activation of the RNA-dependent protein kinase (PKR) by siRNA.35 Since the double-stranded RNA binding motif present in PKR recognizes RNA through minor groove contacts,56,57 the minor groove localized benzyl substituent is poised to provide a steric block to protein binding. The Beal lab expanded on this idea by incorporating N2-propargyl-2-aminopurine (21) into siRNAs for subsequent conversion to bulky triazoles (22) via the copper(I)-catalyzed azide-alkyne cycloaddition (CuAAC) with azides.32 This initial report on synthetic RNA modification via the CuAAC reaction31 and the initial demonstration of RNAi with triazole-modified siRNAs32 was followed by recent reports from others presenting further CuAAC strategies for RNA modification.58–61 The propargyl-modified siRNAs and, surprisingly, the triazole-modified siRNAs showed minimal losses in activity when incorporated at certain locations in the passenger and guide strands. In fact, at one site in the guide strand where bulky triazole modifications were tolerated (position 14), a ribose 2′-methoxyethyl modification at this position has been reported to abolish activity.62 This highlights an advantage of base modifications over ribose modifications, in that large minor groove substituents projecting from the base can be introduced at critical sites where ribose modification at such sites appears to be detrimental to RNAi activity. The binding of PKR was reduced by both propargyl and triazole modifications, and the binding of the double-stranded RNA-specific adenosine deaminase 1 (ADAR1) was reduced by the bulky triazole modifications. These base modifications thus present a strategy for designing siRNAs that reduce off-target effects while retaining native RNAi activity.
Chemical Modification as a Tool to Switch on siRNA Activity
Introduction of modifications that prevent enzymatic degradation or off-target protein binding during delivery, but that can be removed when targeting mRNA in the RISC, could be highly advantageous. Ideally, the modified siRNA should be completely inactive in its delivery form, but fully active after triggering the removal of the blocking group. Two different designs have capitalized on this approach—one involving a photo-cleavable protecting group on an siRNA base,36,63–66 and the other using conformational switching to display or hide a steric blockage in the minor or major groove of siRNA.21,22
In the first of these examples, Mikat and Heckel introduced the 2-(2-nitrophenyl)propyl group (NPP group) on the nucleobases guanine (26) or thymine (27) at various positions of the guide and passenger strands and determined its effect on silencing activity.36 The modifications introduced on the bases near the mRNA cleavage site were capable of blocking the RNAi activity, but upon irradiation, the normal level of activity returned. Modifications introduced at other positions or even on backbone or terminal phosphates were incapable of achieving either complete inhibition of activity while protected or full reactivation upon deprotection.36,63–66 Overall, the photo-labile protective group was shown to be useful for turning on siRNA activity at a precise point in the RNAi mechanism.
A second example of switchable activity of chemically modified siRNAs was developed jointly in the Beal and Burrows laboratories. As discussed above, off-target effects due to deleterious siRNA-protein binding can be addressed through the introduction of minor groove modifications. We reasoned that the activity of these modified siRNAs could be improved by switching of the sterically interfering modifications from the minor groove to the major groove during RISC formation. In recent work, we showed that N2-alkyl-8-oxo-2′-deoxyguanosine analogues (alkyl = propyl, benzyl) (23), adopting either the syn or anti conformation depending upon their base-pairing partner, can be used as the switchable base to introduce a steric blockade to protein binding in one form (C opposite) vs. the other (A opposite) (see Figure 2). 33,34
Figure 2.

N2-alkyl-8-oxo-2′-deoxyguanosine can be used to switch an alkyl group from the minor to the major groove depending on the base opposite.
N2-Alkyl-8-oxo-2′-deoxyguanosine introduced in the guide strand can project the N2-alkyl group into the minor groove when in the anti conformation upon Watson-Crick pairing with C of the passenger strand during delivery. In this conformation, the steric blockage projecting into the minor groove reduces binding to off-target proteins such as PKR. The same modified nucleoside adopts the syn conformation on pairing with A in target mRNA which would then place the N2-alkyl modification in the major groove, where it is less likely to interfere with the target mRNA cleavage process. PKR binding and RNAi activity were found to be highly dependent on the guide strand position substituted with the N2-alkyl-8-oxo-2′-deoxyguanosines. For RNAi activity, introduction of these modifications was tolerated better at positions 11 and 16 of the guide strand than at position 4 (seed region).33 Particularly in the case of position 11 modifications, overall silencing efficiency improved compared to wild-type siRNA, suggesting avoidance of off-target protein binding could be improving efficacy. Furthermore, this case suggests that changes in base pair geometry can be tolerated at the cleavage site; in this case the guide:mRNA duplex is expected to make a Hoogsteen base pair (Figure 2) at the cleavage site. In most cases, siRNAs with multiple modifications led to reduced activity, although a propyl substituent at both positions 4 and 11 gave greater than expected activity. This study showed that a conformational switch of sterically blocking groups between the minor and major grooves of RNA could be an important new strategy for manipulating RNA-protein binding important to RNA interference.
Conclusion
The research efforts described here illustrate how chemical modification of the nucleobase components of siRNAs further our understanding of the RNAi mechanism and could be used to advance RNAi therapeutics. While unmodified siRNAs make poor drug candidates because they are prone to various types of off-target effects, are sensitive to nuclease degradation, and can have lack of specificity for the target mRNA, assembly of siRNAs with nucleoside analogs bearing structural changes to the sugars or the bases can improve these properties. As more siRNA-based drug candidates enter clinical trials, it is clear that organic chemists will be called upon to invent new ways to overcome the shortcomings of the natural siRNA structure. The next generation of siRNAs for therapeutic applications may indeed contain new nucleobases with enhanced base paring specificity, novel interactions within RISC and avoidance of immune receptors.
Acknowledgments
We thank NIGMS (R01 GM080784) for a grant supporting collaborative research on this topic in our laboratories.
Biography
 Hayden Peacock is a New Zealander who will soon complete his PhD degree in Peter Beal’s lab before heading to Andrew Hamilton’s group at Oxford University for postdoctoral studies. Arunkumar Kannan, originally from Thanjavur, India, defended his PhD thesis in April 2011 in the Burrows laboratory and will be starting a position at DPT Laboratories in San Antonio, TX. Part of the work described in this Synopsis has been a collaborative project between the Beal and Burrows labs at UC-Davis and the University of Utah, respectively. Both labs have a focus on base modification chemistry in nucleic acids, particularly RNA chemistry in the Beal group and DNA damage in the Burrows lab.
Hayden Peacock is a New Zealander who will soon complete his PhD degree in Peter Beal’s lab before heading to Andrew Hamilton’s group at Oxford University for postdoctoral studies. Arunkumar Kannan, originally from Thanjavur, India, defended his PhD thesis in April 2011 in the Burrows laboratory and will be starting a position at DPT Laboratories in San Antonio, TX. Part of the work described in this Synopsis has been a collaborative project between the Beal and Burrows labs at UC-Davis and the University of Utah, respectively. Both labs have a focus on base modification chemistry in nucleic acids, particularly RNA chemistry in the Beal group and DNA damage in the Burrows lab.
References
- 1.Hannon GJ. Nature. 2002;418:244. doi: 10.1038/418244a. [DOI] [PubMed] [Google Scholar]
- 2.McCaffrey AP, Meuse L, Pham TTT, Conklin DS, Hannon GJ, Kay MA. Nature. 2002;418:38. doi: 10.1038/418038a. [DOI] [PubMed] [Google Scholar]
- 3.Chiu YL, Rana TM. RNA. 2003;9:1034. doi: 10.1261/rna.5103703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Manoharan M. Curr Opin Chem Biol. 2004;8:570. doi: 10.1016/j.cbpa.2004.10.007. [DOI] [PubMed] [Google Scholar]
- 5.Parrish S, Fleenor J, Xu S, Mello C, Fire A. Mol Cell. 2000;6:1077. doi: 10.1016/s1097-2765(00)00106-4. [DOI] [PubMed] [Google Scholar]
- 6.Kurreck J. Angew Chem, Int Ed. 2009;48:1378. doi: 10.1002/anie.200802092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shukla S, Sumaria CS, Pradeepkumar PI. Chem Med Chem. 2010;5:328. doi: 10.1002/cmdc.200900444. [DOI] [PubMed] [Google Scholar]
- 8.Rozners E. Curr Org Chem. 2006;10:675. [Google Scholar]
- 9.Bramsen JB, Kjems J. Methods Mol Biol. 2011;721:77. doi: 10.1007/978-1-61779-037-9_5. [DOI] [PubMed] [Google Scholar]
- 10.MacRae IJ, Ma E, Zhou M, Robinson CV, Doudna JA. Proc Nat Acad Sci USA. 2008;105:512. doi: 10.1073/pnas.0710869105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khvorova A, Reynolds A, Jayasena SD. Cell. 2003;115:209. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- 12.Schwarz DS, Hutvagner G, Du T, Xu ZS, Aronin N, Zamore PD. Cell. 2003;115:199. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- 13.Matranga C, Tomari Y, Shin C, Bartel DP, Zamore PD. Cell. 2005;123:607. doi: 10.1016/j.cell.2005.08.044. [DOI] [PubMed] [Google Scholar]
- 14.Rand TA, Petersen S, Du FH, Wang XD. Cell. 2005;123:621. doi: 10.1016/j.cell.2005.10.020. [DOI] [PubMed] [Google Scholar]
- 15.Meister G, Landthaler M, Patkaniowska A, Dorsett Y, Teng G, Tuschl T. Mol Cell. 2004;15:185. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 16.Rand TA, Petersen S, Du F, Wang X. Cell. 2005;123:621. doi: 10.1016/j.cell.2005.10.020. [DOI] [PubMed] [Google Scholar]
- 17.Wang Y, Sheng G, Juranek S, Tuschl T, Patel D. J Nature. 2008;456:209. doi: 10.1038/nature07315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jackson AL, Linsley PS. Nat Rev Drug Discov. 2010;9:57. doi: 10.1038/nrd3010. [DOI] [PubMed] [Google Scholar]
- 19.Zhang Z, Weinschenk T, Guo K, Schluesener HJ. J Cell Biochem. 2006;97:1217. doi: 10.1002/jcb.20716. [DOI] [PubMed] [Google Scholar]
- 20.Yang W, Wang Q, Howell KL, Lee JT, Cho DSC, Murray JM, Nishikura K. J Biol Chem. 2005;280:3946. doi: 10.1074/jbc.M407876200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Robbins M, Judge A, Maclachlan I. Oligonucleotides. 2009;19:89. doi: 10.1089/oli.2009.0180. [DOI] [PubMed] [Google Scholar]
- 22.Terrazas M, Kool ET. Nucleic Acids Res. 2009;37:346. doi: 10.1093/nar/gkn958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sipa K, Sochacka E, Kazmierczak-Baranska J, Maszewska M, Janicka M, Nowak G, Nawrot B. RNA. 2007;13:1301. doi: 10.1261/rna.538907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eberle F, Giessler K, Deck C, Heeg K, Peter M, Richert C, Dalpke AH. J Immunol. 2008;180:3229. doi: 10.4049/jimmunol.180.5.3229. [DOI] [PubMed] [Google Scholar]
- 25.Addepalli H, Meena, Peng CG, Wang G, Fan Y, Charisse K, Jayaprakash KN, Rajeev KG, Pandey RK, Lavine G, Zhang L, Jahn-Hofmann K, Hadwiger P, Manoharan M, Maier MA. Nucleic Acids Res. 2010;38:7320. doi: 10.1093/nar/gkq568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li F, Pallan PS, Maier MA, Rajeev KG, Mathieu SL, Kreutz C, Fan Y, Sanghvi J, Micura R, Rozners E, Manoharan M, Egli M. Nucleic Acids Res. 2007;35:6424. doi: 10.1093/nar/gkm664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xia J, Noronha A, Toudjarska I, Li F, Akinc A, Braich R, Frank-Kamenetsky M, Rajeev KG, Egli M, Manoharan M. ACS Chem Biol. 2006;1:176. doi: 10.1021/cb600063p. [DOI] [PubMed] [Google Scholar]
- 28.Somoza A, Chelliserrykattil J, Kool ET. Angew Chem, Int Ed. 2006;45:4994. doi: 10.1002/anie.200601311. [DOI] [PubMed] [Google Scholar]
- 29.Somoza A, Silverman AP, Miller RM, Chelliserrykattil J, Kool ET. Chem –Eur J. 2008;14:7978. doi: 10.1002/chem.200800837. [DOI] [PubMed] [Google Scholar]
- 30.Peacock H, Fucini RV, Jayalath P, Ibarra-Soza JM, Haringsma HJ, Flanagan WM, Willingham A, Beal PA. J Am Chem Soc. 2011;133:9200. doi: 10.1021/ja202492e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peacock H, Maydanovych O, Beal PA. Org Lett. 2010;12:1044. doi: 10.1021/ol100019r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peacock H, Fostvedt E, Beal PAACS. Chem Biol. 2010;5:1115. doi: 10.1021/cb100245u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kannan A, Fostvedt E, Beal PA, Burrows CJ. J Am Chem Soc. 2011;133:6343. doi: 10.1021/ja2003878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kannan A, Burrows CJ. J Org Chem. 2010;76:720. doi: 10.1021/jo102187y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Puthenveetil S, Whitby L, Ren J, Kelnar K, Krebs JF, Beal PA. Nucleic Acids Res. 2006;34:4900. doi: 10.1093/nar/gkl464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mikat V, Heckel A. RNA. 2007;13:2341. doi: 10.1261/rna.753407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Prakash TP, Allerson CR, Dande P, Vickers TA, Sioufi N, Jarres R, Baker BF, Swayze EE, Griffey RH, Bhat B. J Med Chem. 2005;48:4247. doi: 10.1021/jm050044o. [DOI] [PubMed] [Google Scholar]
- 38.Watts JK, Choubdar N, Sadalapure K, Robert F, Wahba AS, Pelletier J, Mario Pinto B, Damha M. J Nucleic Acids Res. 2007;35:1441. doi: 10.1093/nar/gkl1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Saenger W. In: Principles of nucleic acid structure. Cantor CR, editor. Springer-Verlag; New York: 1984. p. 116. [Google Scholar]
- 40.Bramsen JB, Laursen MB, Nielsen AF, Hansen TB, Bus C, Langkjaer N, Babu BR, Hojland T, Abramov M, Van Aerschot A, Odadzic D, Smicius R, Haas J, Andree C, Barman J, Wenska M, Srivastava P, Zhou C, Honcharenko D, Hess S, Muller E, Bobkov GV, Mikhailov SN, Fava E, Meyer TF, Chattopadhyaya J, Zerial M, Engels JW, Herdewijn P, Wengel J, Kjems J. Nucleic Acids Res. 2009;37:2867. doi: 10.1093/nar/gkp106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Davis DR, Veltri CA, Nielsen L. J Biomol Struct Dyn. 1998;15:1121. doi: 10.1080/07391102.1998.10509006. [DOI] [PubMed] [Google Scholar]
- 42.Agris PF, Sierzputowskagracz H, Smith W, Malkiewicz A, Sochacka E, Nawrot B. J Am Chem Soc. 1992;114:2652. [Google Scholar]
- 43.Kumar RK, Davis DR. Nucleic Acids Res. 1997;25:1272. doi: 10.1093/nar/25.6.1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stuart JW, Basti MM, Smith WS, Forrest B, Guenther R, SierzputowskaGracz H, Nawrot B, Malkiewicz A, Agris PF. Nucleos Nucleot. 1996;15:1009. [Google Scholar]
- 45.Wang HW, Noland C, Siridechadilok B, Taylor DW, Ma E, Felderer K, Doudna JA, Nogales E. Nat Struct Mol Biol. 2009;16:1148. doi: 10.1038/nsmb.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang Y, Juranek S, Li H, Sheng G, Tuschl T, Patel DJ. Nature. 2008;456:921. doi: 10.1038/nature07666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang Y, Sheng G, Juranek S, Tuschl T, Patel DJ. Nature. 2008;456:209. doi: 10.1038/nature07315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Armstrong ME, Gantier M, Li L, Chung WY, McCann A, Baugh JA, Donnelly SC. J Immunol. 2008;180:7125. doi: 10.4049/jimmunol.180.11.7125. [DOI] [PubMed] [Google Scholar]
- 49.Kleinman M, Yamada K, Takeda A, Chandrasekaran V, Nozaki M, Baffi J, Albuquerque R, Yamasaki S, Itaya M, Pan Y. Nature. 2008;452:591. doi: 10.1038/nature06765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kariko K, Bhuyan P, Capodici J, Weissman D. J Immunol. 2004;172:6545. doi: 10.4049/jimmunol.172.11.6545. [DOI] [PubMed] [Google Scholar]
- 51.Sioud M. J Mol Biol. 2005;348:1079. doi: 10.1016/j.jmb.2005.03.013. [DOI] [PubMed] [Google Scholar]
- 52.Hornung V, Guenthner-Biller M, Bourquin C, Ablasser A, Schlee M, Uematsu S, Noronha A, Manoharan M, Akira S, De Fougerolles A, Endres S, Hartmann G. Nat Med. 2005;11:263. doi: 10.1038/nm1191. [DOI] [PubMed] [Google Scholar]
- 53.Sioud M. Eur J Immunol. 2006;36:1222. doi: 10.1002/eji.200535708. [DOI] [PubMed] [Google Scholar]
- 54.Hamm S, Latz E, Hangel D, Müller T, Yu P, Golenbock D, Sparwasser T, Wagner H, Bauer S. Immunobiol. 2010;215:559. doi: 10.1016/j.imbio.2009.09.003. [DOI] [PubMed] [Google Scholar]
- 55.Tluk S, Jurk M, Forsbach A, Weeratna R, Samulowitz U, Krieg A, Bauer S, Vollmer J. Int Immunol. 2009;21:607. doi: 10.1093/intimm/dxp030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ryter JM, Schultz SC. Embo J. 1998;17:7505. doi: 10.1093/emboj/17.24.7505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wu HH, Henras A, Chanfreau G, Feigon J. Proc Nat Acad Sci USA. 2004;101:8307. doi: 10.1073/pnas.0402627101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yamada T, Peng CG, Matsuda S, Addepalli H, Jayaprakash KN, Alam MR, Mills K, Maier MA, Charisse K, Sekine M, Manoharan M, Rajeev KG. J Org Chem. 2011;76:1198. doi: 10.1021/jo101761g. [DOI] [PubMed] [Google Scholar]
- 59.Jayaprakash KN, Peng CG, Butler D, Varghese JP, Maier MA, Rajeev KG, Manoharan M. Org Lett. 2010;12:5410. doi: 10.1021/ol102205j. [DOI] [PubMed] [Google Scholar]
- 60.van Delft P, Meeuwenoord NJ, Hoogendoorn S, Dinkelaar J, Overkleeft HS, van der Marel GA, Filippov DV. Org Lett. 2010;12:5486. doi: 10.1021/ol102357u. [DOI] [PubMed] [Google Scholar]
- 61.El-Sagheer AH, Brown T. Proc Nat Acad Sci USA. 2010;107:15329. doi: 10.1073/pnas.1006447107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lima WF, Wu H, Nichols JG, Sun H, Murray HM, Crooke ST. J Biol Chem. 2009;284:26017. doi: 10.1074/jbc.M109.010835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mayer G, Heckel A. Angew Chem, Int Ed. 2006;45:4900. doi: 10.1002/anie.200600387. [DOI] [PubMed] [Google Scholar]
- 64.Nguyen QN, Chavli RV, Marques JT, Conrad PG, II, Wang D, He W, Belisle BE, Zhang A, Pastor LM, Witney FR, Morris M, Heitz F, Divita G, Williams BRG, McMaster GK. BBA-Biomembranes. 2006;1758:394. doi: 10.1016/j.bbamem.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 65.Shah S, Jain PK, Kala A, Karunakaran D, Friedman SH. Nucleic Acids Res. 2009;37:4508. doi: 10.1093/nar/gkp415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shah S, Rangarajan S, Friedman SH. Angew Chem, Int Ed. 2005;44:1328. doi: 10.1002/anie.200461458. [DOI] [PubMed] [Google Scholar]