Abstract
Recent epidemiological and immunological studies provide evidence for an association between Epstein–Barr virus infection and multiple sclerosis, suggesting a role of Epstein–Barr virus infection in disease induction and pathogenesis. A key question in this context is whether Epstein–Barr virus-infected B lymphocytes are present within the central nervous system and the lesions of patients with multiple sclerosis. Previous studies on this topic provided highly controversial results, showing Epstein–Barr virus reactivity in B cells in the vast majority of multiple sclerosis cases and lesions, or only exceptional Epstein–Barr virus-positive B cells in rare cases. In an attempt to explain the reasons for these divergent results, a workshop was organized under the umbrella of the European Union FP6 NeuroproMiSe project, the outcome of which is presented here. This report summarizes the current knowledge of Epstein–Barr virus biology and shows that Epstein–Barr virus infection is highly complex. There are still major controversies, how to unequivocally identify Epstein–Barr virus infection in pathological tissues, particularly in situations other than Epstein–Barr virus-driven lymphomas or acute Epstein–Barr virus infections. It further highlights that unequivocal proof of Epstein–Barr virus infection in multiple sclerosis lesions is still lacking, due to issues related to the sensitivity and specificity of the detection methods.
Keywords: multiple sclerosis, Epstein–Barr virus, EBV biology, EBV detection in tissue
Introduction
Multiple sclerosis is a chronic inflammatory disease of the CNS that leads to demyelination and a variable degree of neurodegeneration (Lassmann et al., 2007; Frischer et al., 2009). However, little is currently known regarding the mechanisms that drive the chronic inflammatory process. Autoimmunity against CNS antigens is one possible explanation and this concept is, in part, supported by experimental data and by the detection of autoimmune reactions in some patients with multiple sclerosis (Hohlfeld and Wekerle, 2004). Another possible explanation could be that a persistent infection in the CNS or elsewhere in the body triggers an immunopathological response either directly or through autoimmunity. One of the potential candidates for a persistent infection, which has received increasing attention during the last years, is the Epstein–Barr virus (EBV) (Lünemann and Münz, 2009; Salvetti et al., 2009; Ascherio and Munger, 2010; Maghzi et al., 2010). Epidemiological evidence strongly supports an association of EBV infection with multiple sclerosis (Handel et al., 2010; Levin et al., 2010) and higher levels of EBV antibodies [in particular those specific for Epstein–Barr nuclear antigen 1 (EBNA1)] have been consistently found in the serum of patients with multiple sclerosis compared with control individuals (Ascherio and Munger, 2010). Evidence is also growing that patients with multiple sclerosis have a higher EBV-specific cellular immune response, both at the level of CD4 and CD8 T cells (Höllsberg et al., 2003; Lünemann et al., 2006, 2008; Jilek et al., 2008; Jaquiery et al., 2010). While enhanced immune reactivity to EBV in multiple sclerosis is indicative of altered virus–host interactions, it is not clear yet whether EBV has a direct role in multiple sclerosis development or acts as an activator of the underlying disease process (Salvetti et al., 2009; Pender, 2011). EBV employs numerous mechanisms to perturb the immune system, including molecular mimicry, induction of heat shock proteins and superantigens, and immortalization of autoreactive B cells. In favour of molecular mimicry, earlier studies showed that a myelin basic protein-specific T cell clone derived from a patient with multiple sclerosis cross-reacts with an EBV DNA polymerase peptide (Ufret-Vincenty et al., 1998; Lang et al., 2002). EBV-specific T cells cross-reacting with myelin basic protein were found in the CSF of a patient with multiple sclerosis (Holmoy et al., 2004) and it has been shown that EBNA1-specific CD4+ T cells from some patients with multiple sclerosis partially cross-react with myelin antigens (Lünemann et al., 2008). Alternatively or in addition, chronic persistence of EBV-infected cells in the CNS, possibly associated with lytic reactivation, could directly drive an immunopathological response causing tissue injury in the patients, a scenario supported by the finding that EBV-specific CD8 T cells accumulate in the CSF of patients with multiple sclerosis (Jaquiery et al., 2010). Given the importance of elucidating the link between altered immune reactivity to EBV and brain pathology in multiple sclerosis, the topic of this report is to critically review the available evidence for the presence of EBV in the multiple sclerosis brain.
EBV infects a very high percentage of humans and persists in a latent form in the B cells of the host (Evans, 1989; Thorley-Lawson, 2001). Persistent intrathecal B cell activation is the hallmark of multiple sclerosis (Obermeier et al., 2008; Owens et al., 2009; Lovato et al., 2011) and B cells and plasma cells are part of the inflammatory infiltrates in multiple sclerosis white matter lesions and meninges (Esiri, 1977; Prineas and Wright, 1978; Serafini et al., 2004, 2007; Magliozzi et al., 2007, 2010; Frischer et al., 2009; Lovato et al., 2011). In patients with multiple sclerosis with progressive disease and a very severe inflammatory and neurodegenerative pathology, large B cell aggregates have been described in the meninges. These structures show some features of lymphoid B cell follicles with germinal centres (Serafini et al., 2004, 2007; Aloisi and Pujol-Borrell, 2006; Magliozzi et al., 2007, 2010). Given the consistency of the serological data and the increasing literature on altered T cell responses to EBV in patients with multiple sclerosis, it has been hypothesized that, in the presence of a susceptible genetic background, EBV infection may not be controlled properly, that circulating infected B cells might enter the CNS and that intracerebral activation of the infection might sustain the chronic inflammatory process leading to multiple sclerosis (Pender, 2003, 2011).
Support for this concept came from recent studies (Serafini et al., 2007, 2010) which—using immunohistochemical, in situ hybridization and reverse transcriptase-polymerase chain reaction techniques in post-mortem brain tissue—found evidence for dysregulated EBV infection in the multiple sclerosis brain. These authors described a high frequency of EBV-infected cells among B cells infiltrating white matter lesions and meninges in multiple sclerosis, but not in other inflammatory diseases of the CNS (Serafini et al., 2007). EBV was detected in 95% of the tissue samples analysed (21 out of 22) and a highly significant correlation was found between EBV-infected cells and the total number of CD20+ B cells. The data also suggested an interaction between cytotoxic CD8+ T cells and EBV-infected B cells (Serafini et al., 2007). However, subsequent studies using similar technologies were unable to detect EBV in the brain tissue of the same and other cohorts of patients with multiple sclerosis (Willis et al., 2009; Peferoen et al., 2010; Sargsyan et al., 2010; Torkildsen et al., 2010). These divergent results may have several explanations. Although basically similar technologies have been used, several methodological differences may lead to differences in the sensitivity of the assays. This may be a particular issue when dealing with autopsy brain samples with suboptimal tissue preservation and subjected to different processing procedures. In addition, there is a wide spectrum of pathological features of multiple sclerosis lesions, depending upon the type and severity of the disease as well as the stage of the lesions (Lucchinetti et al., 2000; Lassmann et al., 2007).
In an attempt to identify the reasons underlying these discrepant results, a 2-day workshop promoted by the European Union FP6 Integrated Project ‘NeuroproMiSe’ was organized at the Centre for Brain Research of the Medical University of Vienna in July 2010. By bringing together all research groups that have addressed the issue of EBV infection in the multiple sclerosis brain, the workshop aimed to provide an update on EBV biology, discuss the individual experiences regarding the search of EBV in pathological tissues and define future research directions.
Virology and biology of the Epstein–Barr virus infection
Basic features of the biology of Epstein–Barr virus infection
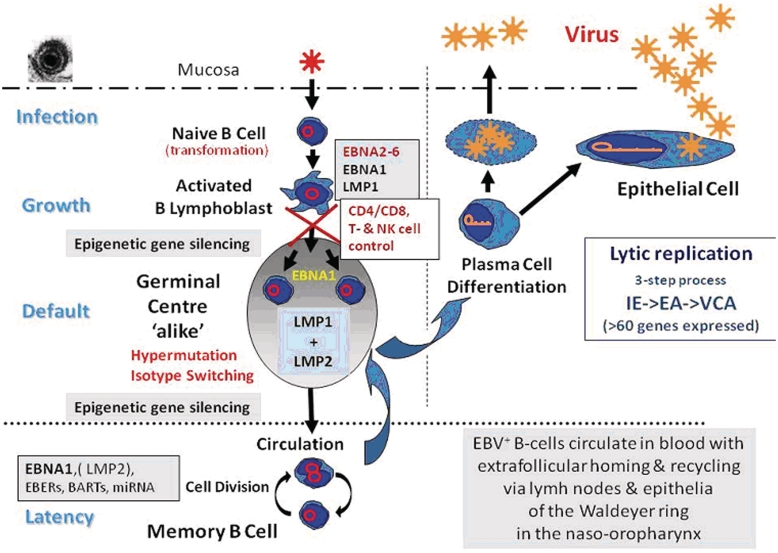
The currently favoured model of the EBV life cycle is schematically depicted in Fig. 1 and Table 1. Primary EBV infection occurs via the oral route leading to infection of submucosal B cells in oropharyngeal lymphoid tissues (Hutt-Fletcher, 2007). Upon infection, EBV transforms these cells into actively growing immunoglobulin-producing B cell blasts through a limited number of essential viral gene products [EBNA1–6, latent membrane proteins 1, 2a and 2b (LMP1, 2a, 2b) and non-coding small RNAs, EBER1, 2 and several microRNAs] (Middeldorp et al., 2003; Young and Rickinson, 2004; Swaminathan, 2008; Chaganti et al., 2009). This type of EBV infection (latency-III or growth programme) is also seen in vitro in lymphoblastoid cell lines and does not usually involve virus production (Thorley-Lawson, 2001).
Figure 1.
Schematic presentation of EBV infection and persistence in vivo. EA = early antigen; IE = immediate early; VCA = viral capsid antigen.
Table 1.
Latency types of EBV-infected cells and their gene expression repertoire
| Latency type | Gene repertoire | Associated diseases |
|---|---|---|
| Type -0/-1 (true latency) | EBER1, 2 and BARTs (microRNA), EBNA1 | Peripheral blood (memory) B cells, Burkitt's lymphoma, dividing EBV+ B cells |
| Type -2 (default) | EBER1, 2 and BARTs (microRNA), EBNA1, LMP1, LMP2A, 2Band BARF1a | Hodgkin disease, B cell non-Hodgkin lymphoma, T-/NK-cell non-Hodgkin lymphoma and nasopharyngeal carcinomaa |
| Type -3 (growth) | EBER1, 2 and BARTs/BHRF1 (microRNA) EBNA1, EBNA2, EBNA3A, 3B, 3C, EBNA-LP, LMP1, LMP2A, 2B | Lymphoblastoid cell lines, post-transplant lymphoproliferative disorders, AIDS-related lymphomas and infectious mononucleosis |
| Others | EBER1, 2 and BARTs (microRNA) EBNA1, LMP2a and BARF1a | Gastric carcinomaa |
a BARF1 is only expressed in epithelial malignancies (e.g. nasopharyngeal carcinoma and gastric carcinoma).
Genes indicated in bold highlight those important for stage-specific differentiation.
These EBV-infected B-blasts trigger a robust T cell response that can comprise nearly 60% of all T cells, and are subsequently largely eliminated (Hislop et al., 2007, 2010). The cytokines produced during this process cause the clinical syndrome of infectious mononucleosis. Subsequently, EBV establishes a lifelong persistent infection of memory B cells, a process accompanied by epigenetic silencing of most viral coding genes, but leaving expression of non-coding small RNAs unaffected (Thorley-Lawson and Allday, 2008; Paschos et al., 2009). By shutting down the expression of most EBV-encoded antigens, EBV-infected memory B cells become invisible to the immune system and circulate permanently in the blood in low but stable numbers (∼1/105 B cells) for the lifetime of the host (Latency-0/-I or true latency programme; Khan et al., 1996). When proliferating, these B cells may switch on EBNA1, which is crucial for maintenance of the viral episome in dividing cells (Hochberg et al., 2004). T cell control of EBV remains at high levels for life (∼1% of all T cells are permanently reactive to EBV antigens) and provides an effective control against re-emergence of EBV-driven B-blasts.
How EBV enters the memory B cell pool is not entirely clear. Three possibilities are discussed, that may not be mutually exclusive: EBV may directly infect memory B cells during primary infection (Kurth et al., 2000; Chaganti et al., 2009). Alternatively, EBV may infect naïve B cells, which differentiate into memory B cells by passage through germinal centre reactions exploiting physiological B cell differentiation pathways (Niedobitek et al., 1992; Khan, 2006). Finally, EBV may itself drive naïve B cell differentiation towards memory B cells by substituting signals that are normally provided to antigen-selected B cells in germinal centres through CD40 and immunoglobulin (Ig) receptor by constitutive signalling through its proteins LMP1 and LMP2, respectively. In support of this concept, EBV+ B cells expressing activation-induced cytidine deaminase (AID), a protein required for immunoglobulin class switch as well as for somatic hypermutation, have been detected outside germinal centres in infectious mononucleosis tonsils (Tobollik et al., 2006). Intriguingly, in this scenario, it would become possible for B cells expressing autoreactive antibodies to enter the memory B cell compartment.
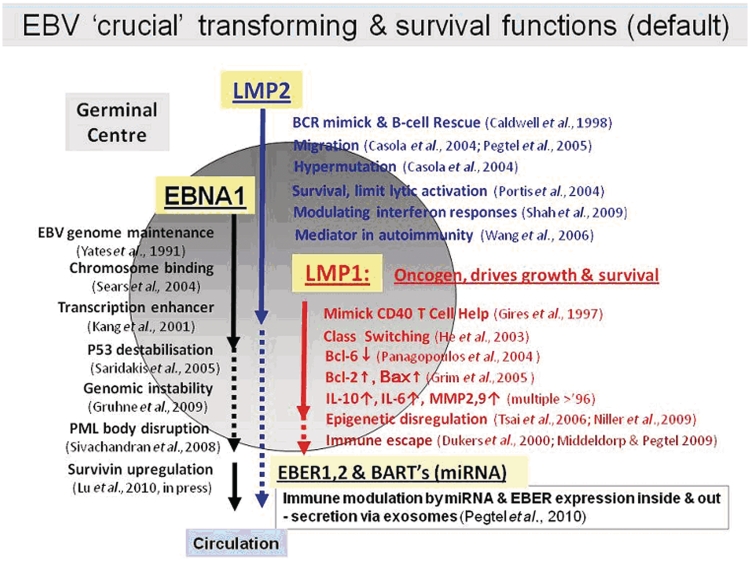
Circulating EBV-infected memory B cells preferentially home to lymphoid tissues in the head and neck region but may be detectable anywhere in the organism where B cells accumulate. Upon re-entry into lymphoid tissues, they may express the LMPs and EBNA1, and undergo limited activation and proliferation mimicking the germinal centre process (Latency-II or default programme) (Laichalk et al., 2002; Roughan et al., 2010). In this setting, potentially transforming viral gene products are allowed to be expressed together and to drive a limited expansion of EBV-infected B cells (Fig. 2; Middeldorp et al., 2003). LMP2a may provide additional survival signals and prevent B cell differentiation into plasma cells. Importantly, no EBNA2-6 re-expression is found in these cells, reflecting tight epigenetic promoter silencing. This process of EBV latent reactivation into the default programme may be triggered particularly by local inflammatory cytokines (Kis et al., 2006, 2010), but is kept under tight control by EBV-specific T cells in the healthy host (Hislop et al., 2005). Thus, latently EBV-infected B cells may be attracted to sites of inflammation, e.g. driven by TNF, IL6, CXCL13, to become locally activated into proliferation. Interestingly, subtle physiological influences, including stress hormones, can trigger EBV reactivation from latency causing temporary enhanced viral gene expression and triggering aberrant immune responses (Glaser et al., 2005).
Figure 2.
EBV gene expression mimicking the migration and germinal centre selection process. miRNA = microRNA.
Pathological conditions involving aberrant B cell triggering, such as severe chronic inflammation or infection, with temporary immune imbalance may be responsible for altering the balance between EBV and its host and set the stage for more serious complications (Thorley-Lawson, 2001; Middeldorp et al., 2003; Young and Rickinson, 2004). By providing an array of survival signals and manipulating host gene regulation, EBV influences and overrides a variety of molecular control pathways that are crucial for normal B cell function. These influences are exerted through a limited number of EBV gene products (Fig. 2), which provide advantage to EBV and B cell survival, but carry the inherent risk of oncogenic host cell transformation or allow the survival of normally forbidden (e.g. autoimmune) clones.
When triggered to become plasma cells, EBV+ B cells express the full coding content of the viral genome to allow virus replication, which is accompanied by virus-driven anti-apoptotic signalling and immune evasion strategies ensuring cell survival for the production of viral progeny (Laichalk and Thorley-Lawson, 2005; Ressing et al., 2008). Viral replication is initiated by the ZEBRA protein (also known as BZLF1 or Zta), a nuclear transcription factor driving viral and host gene expression and mediating evasion from innate immune responses (Miller et al., 2007). The ZEBRA protein is under scrutiny by T cells limiting the number and lifespan of virus producing cells (Hislop et al., 2007). Upon ZEBRA expression, a cascade of lytic cycle gene expression is induced involving distinct sets of interdependent immediate early genes (transcription factors and regulators), early genes (including enzymes involved in nucleotide metabolism, viral DNA replication, immune evasion, apoptosis resistance, etc.) and late genes (virus structural proteins and immune evasins). Besides full virus replication, EBV may also express a limited ‘abortive’ form of lytic infection, where gene expression is limited to Zta and a few early viral proteins. Interestingly, the switch to lytic replication is accompanied by a stop in the expression of small EBER RNAs, whereas other latent products (i.e. LMP1) are upregulated (Gilligan et al., 1990; Webster-Cyriaque et al., 2000). It is unclear if virus replication in plasma cells results in the liberation of infectious virus particles in quantities sufficient to allow spread of the virus to other individuals. It is considered likely that additional amplification of virus production in oropharyngeal epithelial cells is required (Hadinoto et al., 2009).
Recently, it was found that viral latency products (i.e. LMP1, LMP2, small RNAs) can be released from EBV-infected cells in exosomes, and can manipulate cells in the microenvironment and even systemically support inflammation and immune escape (Dukers et al., 2000; Flanagan et al., 2003; Houali et al., 2007; Iwakiri et al., 2009; Pegtel et al., 2010). This may lead to the accumulation of virus-related products in non-infected cells (Pegtel et al., 2010). If this phenomenon interferes with diagnostic techniques used for the in situ detection of EBV is presently unknown.
Disruption of the host–virus immune balance may lead to aberrant behaviour of EBV-infected cells resulting in several distinct benign and malignant disease syndromes. Although infrequent, neurological disease has been associated with acute and chronic EBV infection with detectable virus in the CNS, and it is well established that EBV-positive diffuse large B cell lymphomas may develop in the brains of HIV-infected individuals. Therefore, there may be a link between EBV and neurological disease, whether this is acute, chronic or malignant.
In summary, current evidence suggests that EBV has hijacked B cell biology for its own survival through a limited number of viral gene products, while remaining largely invisible to the immune system. In healthy individuals, EBV persists in memory B cells that preferentially home to lymphoid tissues in the head and neck region, but which can also travel to areas of inflammation. Here EBV carrying B cells may become activated and can multiply upon temporary or local loss of immune control. When activated, EBV+ B cells may secrete cytokines and viral components thus possibly contributing to inflammation as well as immune escape. These events may take place in inflammatory regions of the CNS of patients with multiple sclerosis.
Subcellular localization of different Epstein–Barr virus encoded RNAs or proteins and their detection in tissue sections
In order to assess the possible contribution of EBV to any neoplastic or non-neoplastic disease, in situ detection of viral genomes or gene products is of pivotal importance. For example, EBV genomes may be detectable in DNA extracts from human tumours by polymerase chain reaction suggesting an EBV association. In situ analysis of such cases, however, may reveal occasional EBV-positive lymphocytes admixed with the EBV-negative tumour cells as the source of the polymerase chain reaction signal.
In general, EBV gene products have a defined subcellular localization relating to their function and can be detected equally well in (fixed) cell lines in vitro as well as in (tumour) tissues ex vivo. The basic features of EBV biology, as described above, should be considered when applying defined EBV-specific reagents to conditions with suspected EBV involvement, aiming to understand or explain possible EBV-driven pathogenic events. Therefore, appropriate negative (e.g. reactive lymph node) and positive tissues (EBV+ Hodgkin lymphoma, nasopharyngeal carcinoma or infectious mononucleosis tonsil) should always be processed in parallel with the tissue to be examined, in addition to negative reagent controls.
In situ detection of EBV infection should follow a step-wise, hierarchical procedure. First, the presence of EBV infection must be established. Ideally, this is done by EBV DNA in situ hybridization since this technique detects viral genomes and is independent of viral gene expression. Sensitive methods, suitable for the detection of single viral copies, have been described in certain model systems, although this technique may require the use of radiolabelled probes (Niedobitek and Herbst, 2006). In practice, EBV DNA in situ hybridization has been successfully used to detect EBV in certain tumours. However, it is not sufficiently sensitive or robust to definitely exclude the presence of EBV in case of negative results.
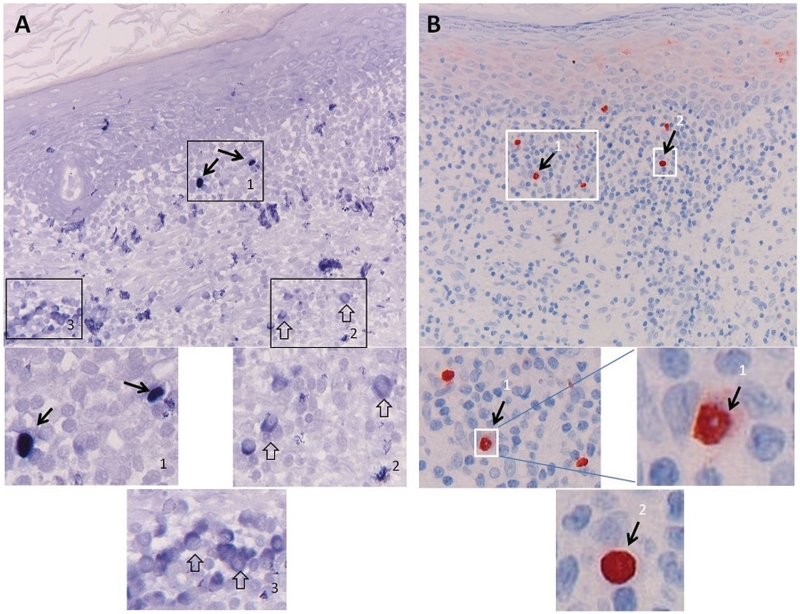
The non-coding small RNAs, called EBERs, which are abundantly expressed in all known forms of EBV latency in vivo and in vitro and serve as gold standard for detecting latent EBV infection in situ (Khan et al., 1992; Hamilton-Dutoit and Pallesen, 1994), are considered to reside exclusively or at least predominantly in the nucleus. Cytoplasmic staining is, therefore, generally considered as non-specific (Gulley et al., 2002; Gulley and Tang, 2008). An exception to this rule is cells undergoing mitosis, which may show diffuse cytoplasmic labelling as a consequence of the loss of the nuclear membrane. Recent studies have, however, shown a function for EBERs in the cytoplasm, and secretion of EBERs via cytoplasmic vesicles or as La-EBER protein–RNA complexes may be physiological, thus weakening this strict diagnostic criterion (Iwakiri et al., 2009). However, generally RNA in situ hybridization reveals pure nuclear staining in EBV-associated tumours (Fig. 3B). EBERs are estimated to be present at >1 million copies per cell and should be easily detected when RNA quality is preserved. The advantage of EBER staining by RNA in situ hybridization is that even sporadic EBV+ B cells can be clearly visualized in a pathological tissue. However, detection of EBERs requires well-controlled staining procedures in order to avoid false positive (over staining) or negative results (due to RNA degradation). For proper detection of EBERs by RNA in situ hybridization, precautions should be taken to preserve tissue and RNA integrity (proper tissue fixation and use of RNase-free cutting knife and buffers, preferably in a clean cross-flow cabinet), and RNA integrity should be checked in each sample by performing RNA in situ hybridization for a house-keeping gene, such as the U6 cellular RNA (Niedobitek et al., 2000). Several (commercial) methods may be valid (Gulley et al., 2002, 2008). The frequently used alkaline phosphatase-based detection of RNA in situ hybridization signals with BCIP/NBT as substrate/chromogen may provide very strong signals, but is prone to non-specific background staining, which may make interpretation difficult (Fig. 3A).
Figure 3.
EBER peptide nucleic acid staining pitfalls: comparison of detection by alkaline phosphatase and horseradish peroxidise. EBER expression was studied in an inflammatory epithelial lesion using the identical EBER peptide nucleic acid (PNA) probe labelled with FITC (DAKO Kit) applied under identical standardized tissue preparation and hybridization conditions (as described in the DAKO EBER PNA procedure) with subsequent detection (A) according to the manufacturer's protocol with standard rabbit anti-FITC labelled with alkaline phosphatase and BCIP/NBT developing reagent, or (B) using a modified protocol involving rabbit anti-FITC and Streptavidin–horseradish peroxidase (DAKO) detection and diaminobenzidene as developing reagent. Although both methods allowed detection of EBER-positive nuclei (A1 and B1) as appropriate sign of EBV presence in the tissue (black arrows), the alkaline phosphatase detection generally gave a problematic background staining especially in the cytoplasm of non-EBV-infected lymphoid cells (open arrow in A2 and A3). These cells may be plasma cells as judged by the enlarged cytoplasm. Such cytoplasmic staining is regularly observed in infiltrating lymphoid cells with plasma cell appearance in otherwise EBV negative inflammatory tissues. By using the alternative protocol (B), no such cytoplasmic staining was revealed and generally a very sharp nuclear boundary was produced (B1, B2), with occasionally less precise lining (single cell in B1), suggesting of EBER leakage into the cytoplasm. This should be considered when interpreting published data on EBV involvement in multiple sclerosis with commercial detection kits (Serafini et al., 2007, 2010; Peferoen et al., 2010).
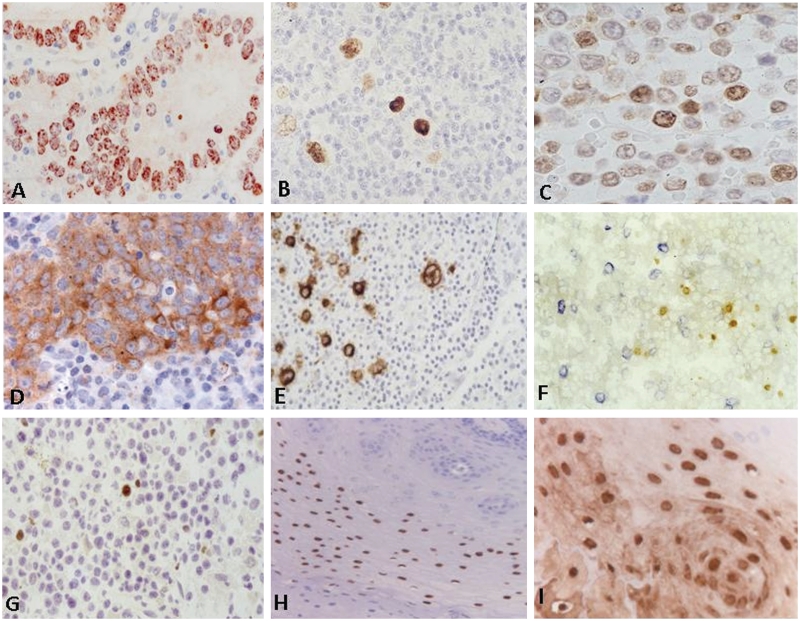
The EBNA1 protein, which is also expressed in all forms of EBV latency, is largely confined to the nucleus as revealed by immunohistochemistry (Fig. 4A–C). Multiple anti-EBNA1 reagents are available, some of which have potential false reactivity due to epitope sharing. Thus, one rat monoclonal antibody, designated 2B4, has also been reported to produce nuclear staining of some EBV-negative cells in the original publications (Grasser et al., 1994; Murray et al., 1996). Nevertheless, this antibody has been repeatedly used for the detection of EBV infection in human tumours leading to reports describing the detection of EBV in unexpected situations, e.g. breast and prostate cancer (Bonnet et al., 1999; Grinstein et al., 2002; Preciado et al., 2005). More recently, it was shown that this antibody, like the 1H4 antibody, cross-reacts with MAGE-4 (Hennard et al., 2006), at least partially explaining unusual results observed by some groups (Murray et al., 2003). Other antibodies, while more specific, often produce only weak staining. Thus, EBNA1 immunohistochemistry, on its own, has to be considered unreliable for the detection of EBV infection.
Figure 4.
Detection of EBV antigen expression in human tissues. A variety of EBV antigens can be detected by a characteristic staining pattern using defined monoclonal antibody reagents. EBNA1 expression (detected by mouse monoclonal antibody OT1x) is characterized by a (sometimes punctuated) nuclear staining as revealed in gastric cancer (A), Hodgkin's lymphoma (B) and B cell lymphoproliferative lesions (C). LMP1 expression (detected by mouse monoclonal antibody OT21C) is characterized by a homogeneous cytoplasmic and membraneous staining as revealed in nasopharyngeal carcinoma (D), Hodgkin's lymphoma (E) and blastoid cells in post-transplant lymphoproliferative disease (F, LMP1 stained in blue). Note that the parallel expression of nuclear EBNA2 (detected by rat monoclonal antibody R3) in smaller cells in the same lesion (F, EBNA2 stained in brown), suggesting a heterogeneous proliferative process (Brink et al., 1997). Zebra protein expression (detected by the mouse monoclonal antibody BZ1) is strictly nuclear as revealed by staining of post-transplant lymphoproliferative disease (G) or oral hairy leukoplakia (H). The structural capsid protein VCA-p18 can be abundantly detected in oral hairy leukoplakia as revealed by rat monoclonal antibody OT15E (I).
Nevertheless, once EBV infection has been established by EBER specific in situ hybridization, immunohistochemistry is useful for characterizing the type of EBV latency and to identify cells entering into the lytic cycle of EBV infection. Antibodies suitable for the detection of EBNA2 and LMP1 have been widely used to study EBV latency in various settings (Fig. 4D and F; Herbst et al., 1991; Niedobitek et al., 1992, 1995, 1997). The immunohistochemical detection of LMP2A expression is also possible but is technically more challenging and is also prone to false positive staining, e.g. of mantle zone lymphocytes (Niedobitek et al., 1997; Heussinger et al., 2004).
The switch from latent to lytic infection is triggered by the BZLF1 protein, also known as ZEBRA or Zta, and immunohistochemistry using a BZLF1-specific monoclonal antibody has proved useful for the in situ detection of lytic EBV infection (Herrmann et al., 2002; Frangou et al., 2005). ZEBRA protein can be readily detected by the BZ1 monoclonal antibody that is commercially available and gives a strong nuclear signal in cells undergoing lytic infection (Fig. 4G and H). Additional reagents directed against early or late viral proteins are useful to establish to what extent virus replication is complete or remains abortive.
For detection of full lytic EBV replication, it is necessary to demonstrate additional early and late viral gene products. Preferred early antigens comprise the BMRF1 (pol-accessory protein) and BALF2 (major DNA binding protein) products (Zeng et al., 1997; Zhang et al., 1998; Herrmann et al., 2002; Frangou et al., 2005). An additional early marker is the viral bcl2 homologue BHRF1 (Nicholls et al., 2001). The detection of true late antigens associating with productive replication is possible by using monoclonal antibodies to viral capsid components [BFRF3 (VCA-18), BdRF1 (VCA-p40), BcLF1 (VCA-p160)] or the gp350/220 envelope protein (BLLF1) (Zhang et al., 1998; Hermann et al., 2002; Frangou et al., 2005). The detection of lytic gene expression can be most clearly demonstrated in AIDS-associated oral hairy leukoplakia (Fig. 3I), which is the only pathological condition associated with full EBV replication (Webster-Cyriaque et al., 2000). Most of the early and lytic markers have a nuclear and cytoplasmic localization, whereas the gp350/220 has a cytoplasmic membrane localization. Virion structural antigens tend to spread into the micro-environment (Fig. 3I). Although only few studies have directly addressed co-expression of EBV proteins in situ, it is generally believed that full lytic replication is a continuous process once triggered beyond the stage of early antigen expression. Some evidence suggest that cells may undergo abortive infection when expressing the BZLF1 (Zebra) protein only, but this largely relies on in vitro data.
During uncomplicated virus persistence, the in situ detection of EBV latent genes and even more so of viral lytic gene expression has proven extremely difficult (Frangou et al., 2005; Hudnall et al., 2005; Chen and Hudnall, 2006). However under various pathological conditions, EBV gene expression can be detected in defined tissues as outlined above. In subclinical situations, such as stress and inflammation, EBV reactivation occurs, which may temporarily lead to increased EBV production, as reflected by elevated EBV DNA levels in blood and saliva as well as elevated antibody responses (Glaser et al., 2005; Gulley, 2008).
In summary, on the basis of currently available evidence, it is recommended that investigators use EBER-specific in situ hybridization for the detection of EBV infection followed by immunohistochemistry to identify the type of virus latency and the switch from latency to lytic infection.
In recent years, multiple approaches using polymerase chain reaction technology have been used to detect EBV genomes (DNA load) in tissue fluids, in particular blood, plasma, saliva and CSF. In general, during uncomplicated virus persistence, EBV DNA can easily be detected in saliva, but is rare in internal compartments (Stevens et al., 2002; Gulley, 2008; Hadinoto et al., 2009). In fact, the blood compartment contains usually <10 infected B cells per millilitre.
In general, in the immunocompetent host, fluctuations in anti-EBV immune responses reflect the host–virus relationship, being aberrant in (sub)clinical situations involving EBV (Glaser et al., 2005; de Sanjose et al., 2007; Stevens et al., 2007). Interestingly, although not detailed here, there is a consistency in studies reporting abnormalities in the immune response to a particular EBV protein EBNA1, but not to other EBV proteins or other viruses, in patients with multiple sclerosis (Lünemann et al., 2008; Sundstrom et al., 2009; Jafari et al., 2010). This appears to involve both T cell and B cell responses to defined regions (epitopes) in the middle to C-terminus of EBNA1, and is linked to an increased risk of developing multiple sclerosis. Importantly, it has been reported that certain EBNA1-specific CD4+ T cell clones cross-react with myelin to produce interferon gamma upon stimulation (Lünemann et al., 2008). How and if this anti-EBNA1 immune response reflects or contributes to inflammation in multiple sclerosis remains to be determined, but aberrant EBNA1 reactivity has been suggested as surrogate marker for multiple sclerosis risk (Lünemann and Munz, 2009; Ascherio and Munger, 2010, Simon et al., 2010).
Presence of Epstein–Barr virus in the multiple sclerosis brain
Polymerase chain reaction-based evidence
Several groups have attempted to identify EBV infection in the multiple sclerosis brain by polymerase chain reaction methodology. Summarizing the experience of all laboratories, there is good agreement that with polymerase chain reaction technology it is easily possible to detect infection in EBV+ tumour control tissue or in lymphoblastoid cell lines. However, EBV infection in whole multiple sclerosis brain sections or single cells isolated from the CSF was very rarely detected (Morré et al., 2001; Willis et al., 2009; Peferoen et al., 2010; Sargsyan et al., 2010). A positive result was obtained only by using pre-amplification polymerase chain reaction techniques (Sargsyan et al., 2010; Serafini et al., 2010) and/or by selectively isolating regions with dense B cell infiltrates using laser capture microdissection (Serafini et al., 2010).
Willis et al. (2009) performed real-time polymerase chain reaction for the detection of genomic EBV as well as real-time polymerase chain reaction for the abundant EBV-encoded RNA. A first set of 17 brain samples from five cases with multiple sclerosis, which contained white matter lesions, were completely negative with both techniques despite a robust signal for CD20 messenger RNA and the presence of CD20+ B cells, confirmed by immunohistochemistry. In a second set, the investigation focused on 12 tissue samples, three of which contained B cell aggregates in the brain parenchyma and four of which contained a loose B cell infiltrate in the meninges. EBV RNA was seen in two out of 12 cases and one of these also contained EBV DNA. The conclusion was that, if present at all, only very few EBV+ cells were contained in the samples. Similarly, a negative result was obtained with real-time polymerase chain reaction by Torkildsen et al. (2010) for EBNA1, EBNA2, LMP1 and LMP2 messenger RNA. Morré et al. (2001) and Sargsyan et al. (2010) did not detect EBER1 transcripts in single B lymphocytes and plasma cells derived from multiple sclerosis CSF, nor EBV-specific transcripts (EBER1, EBNA2, LMP1 and BFRF-1) in 15 multiple sclerosis lesions from 14 patients, five of them being reported previously by Serafini et al. (2007) to have a high load of EBV-infected B cells. However, in a second study Sargsyan et al. (2010) evaluated an additional group of paraffin-embedded multiple sclerosis plaques and positive and negative control lymphoma tissue pre-screened for EBV DNA by nested real-time polymerase chain reaction and confirmed the presence of EBV in rare multiple sclerosis tissue samples. In contrast to EBV+ lymphoma tissue, EBER1-positive multiple sclerosis plaque contained no additional EBV-specific transcripts such as EBNA2, LMP1 and BFRF-1, consistent with latency stage 0 of EBV infection. In a recent report, Serafini et al. (2010) were unable to detect LMP2A and EBNA1 transcripts by quantitative real-time polymerase chain reaction in multiple sclerosis whole brain sections. However, selective pre-amplification of complementary DNAs allowed detection of a low polymerase chain reaction signal for LMP2A and EBNA1 in two and three of four brain samples, respectively. Moreover, these authors showed high levels of transcripts for LMP2A and EBNA1 relative to the house-keeping gene GAPDH and to the B cell-associated gene CD19 in all inflammatory cell infiltrates that were isolated from the meninges and white matter lesions (both active and chronic active) with laser capture microdissection (Serafini et al., 2010). According to Serafini et al. (2010), these results are consistent with the observation that, even in the most infiltrated multiple sclerosis brain samples, total and EBV-infected B cells represent only a very minor proportion of the total population of CNS resident and inflammatory cells and are highly concentrated in very tiny areas, like the perivascular spaces of post-capillary venules and the leptomeningeal space. Besides aspects related to B cell frequency and localization, the possibility that the levels of viral transcripts in EBV-infected B cells surviving in a chronically inflamed environment are much lower than in EBV+ tumour cells could help explain why selection of B cell-enriched areas and increased polymerase chain reaction sensitivity are so crucial for successful detection of EBV in the multiple sclerosis brain.
Evidence based on in situ hybridization for EBER and immunocytochemistry
The largest discrepancies between the groups related to the morphological detection of EBV-infected cells in tissue sections. Serafini et al. (2007, 2010) report that a very high percentage of CNS infiltrating B cells are positive for EBER by in situ hybridization and for the EBV latent proteins LMP1 and LMP2A by immunohistochemistry. This abundant positive staining is in contrast to the completely negative findings obtained by Willis et al. (2009) and H. Lassmann (unpublished results). Largely negative results have also been obtained by the Dutch group (Peferoen et al., 2010). In the latter study, >632 tissue blocks from 94 patients with multiple sclerosis were screened for the presence of B cells. From the B cell-rich group (16 cases) comprising 60 blocks, 1–2 blocks from each patient (28 in total) were analysed for the presence of EBV using EBER in situ hybridization. In addition, eight blocks from other cases with multiple sclerosis in the Dutch cohort in which B cells were observed in the meninges were also screened. The only positive signal for EBER was seen in a few cells within a single lymphocyte-rich region in a single block of a single multiple sclerosis case. On the contrary, U. Meier, J. Tzartos and G. Khan (London) discussed unpublished results, reporting the presence of EBER-expressing cells in preselected IFN-α over-expressing active multiple sclerosis lesions, but surprisingly also in acute stroke lesions. In contrast to the above-mentioned studies, these authors used a non-commercial set of EBER1 and -2 probes in an in situ hybridization protocol, which they have established to be capable of detecting single EBV-infected lymphocytes (Khan et al., 1992). A problem of this particular study is that the morphology of cells with nuclear EBER signal within the lesions was not always typical for B cells, particularly in stroke lesions and double staining for EBER expression in B cells has not been performed.
Similar discrepancies exist regarding immunocytochemical detection of EBV antigens in multiple sclerosis brains and lesions. There was full agreement that the antibodies used for detection of EBV antigens in multiple sclerosis lesions are suitable to detect staining of EBV-infected cells in infected control tissue. In multiple sclerosis brain tissue, however, the results show major discrepancies. Several groups report completely negative results with antibodies against latent antigens (LMP1 or EBNA2; Willis et al., 2009; Peferoen et al., 2010; Torkildsen et al., 2010) with the exception of expression of antigens related to lytic infection in a single lesion of a single case (Peferoen et al., 2010). In contrast, Serafini et al. (2010) observed that 40–80% of all CD20+ B cells in the multiple sclerosis brain expressed LMP2A on their membrane. Most B cells also showed staining using an LMP1-specific reagent, while EBNA2+ cells were rarely observed, suggesting expression of the default programme. The same group also found that a substantial proportion of plasma cells in the inflamed meninges, ectopic B cell follicles and perivascular cuffs of active white matter lesions (30–55%), but not in chronic active lesions, also expressed BFRF1, an antigen associated with the early phase of lytic infection (Serafini et al., 2007). Such abundance of plasma cells is not confirmed by others and the specificity of the anti-BFRF1 antibody is not confirmed (J. Middeldorp, unpublished results). The Serafini et al. (2007) study further describes widespread expression of LMP1 and BFRF1 in B cell-rich perivascular cuffs of two patients who died from fulminate acute multiple sclerosis. However, this early study did not analyse the expression lytic switch ZEBRA protein, for which a well-reactive reagent (BZ-1 antibody) is available. More recently, the presence of EBV lytically infected plasma cells in the most inflamed multiple sclerosis brains was confirmed using the BZ-1 antibody specific for Zebra (B. Serafini, unpublished results). However, the prevalence of EBV-infected B cells and their frequent (re-)activation into default or lytic cycle in specific reactive B cell-rich regions of the brain in patients with multiple sclerosis, as described by the Serafini et al. (2007, 2010), remains to be confirmed.
Interpretation of the polymerase chain reaction and morphological data
The profound discrepancies between the results of the different groups are currently difficult to explain. Thus, most of the discussion during the meeting was devoted to this topic. No consensus, acceptable to all participants, could be reached.
The view, shared by the majority (H.L., G.N., J.M.M., K.O.C., J.B., W.B., C.S., S.A., P.v.V., U.C.M.) was that EBV infection in the multiple sclerosis brain is, when present at all, restricted to a very low number of B cells. The majority group also agreed that evidence from inflammatory control brain tissue is at present insufficient to judge whether it is multiple sclerosis specific or just reflects an accumulation of EBV-infected B cells in B cell-rich inflammatory human brain lesions as a bystander phenomenon. Conversely, based on the immunohistochemical and EBER in situ hybridization data obtained in 12 cases with other inflammatory neurological diseases including primary vasculitis, viral encephalitis, mycotic and bacterial meningoencephalitis and one case with neuromyelitis optica, (Serafini et al., 2007, 2010 and unpublished results), these authors concluded that intracerebral enrichment in EBV-infected B cells is characteristic of and specific for the multiple sclerosis brain only.
The discrepancies between the results of the different studies were felt by the majority to be mainly due to technical issues and interpretation of immunocytochemistry and in situ hybridization, but not due to differences in case/lesion selection or the preservation of brain tissue samples. These conclusions are based on the following considerations: First, evidence from attempts to detect EBV infection in the multiple sclerosis brains by (real time) polymerase chain reaction suggests that EBV-related DNA or RNA in multiple sclerosis brains and lesions are rare and most likely restricted to a small number of cells. Secondly, the chance of its detection appears to increase with enrichment of B cells within the tissue sample and, thus, is highest when B cell infiltrates are selectively isolated by laser microdissection. This, however, requires independent confirmation. Whether EBV-infected B cells within the multiple sclerosis brain are disease-specific or just reflect B cell infiltration and activation in the inflamed brain remains unclear, since proper inflammatory controls with comparable B cell infiltration and activation in the brain (such specimens are very rare) have so far not been included in the polymerase chain reaction studies. Altogether, this suggests that highly sensitive techniques have to be used to obtain positive results with polymerase chain reaction technology. This notion apparently contrasts with the reported high percentage of EBV-infected B cells as detected by EBER in situ hybridization and immunohistochemistry in cases with multiple sclerosis (Serafini et al., 2007, 2010).
Quantitative studies on the composition of inflammatory infiltrates in multiple sclerosis agree that the numbers of B lymphocytes within the lesions are highly variable between cases. Thus, one potential explanation for the divergent results could be that the pathological spectrum of the cases, in particular regarding B cell infiltrates, differed between the studies. This, however, is not likely for several reasons. First, all studies took care to include cases with substantial B cell infiltrates and larger B cell aggregates and this is well documented in the respective publications. Secondly, several groups (Rome, Boston/Göttingen, Amsterdam) have used in part tissue blocks from the same patients derived from a single tissue bank (UK Multiple Sclerosis Tissue Bank). EBER in situ hybridization has even been performed on consecutive frozen or paraffin sections from the same blocks with divergent results between different centres (Rome, Amsterdam and Vienna). Providing the same cases from the same tissue bank excludes differences in pre-mortem conditions, autolysis time, fixation and handling of the tissue blocks may explain the divergent results, although it does not exclude differences in tissue processing and storage in the individual laboratories.
Thus, technical issues related to the sensitivity of EBV detection in tissue sections became the centre of interest. One aspect, which seems to be important, is the type of material is used. The positive findings have been obtained in Rome by using to a large extent cryopreserved tissue fixed with formaldehyde (either prior or after freezing and cutting) but also a smaller number of paraffin samples obtained from Vienna and Amsterdam. In the negative studies by Willis et al. (2009), Sargsyan et al. (2010) and Peferoen et al. (2010), both frozen- and paraffin-embedded tissue has been used in parallel. Although the EBER in situ hybridization as well as the immunocytochemistry for EBV in principle works in both types of tissue sections, higher sensitivity results may be reached in frozen sections, but possibly at the expense of a higher chance of non-specific reactions. Yet, it is unlikely that this explains the divergent results, since in the studies by Willis et al. (2009) and Peferoen et al. (2010), tissue material from cases with multiple sclerosis that were rated positive in the study by Serafini et al. (2007), were obtained from the UK Multiple Sclerosis Tissue Bank under identical conditions of tissue preservation.
Thus, issues related to the specificity of the in situ hybridization and immunohistochemical reactions have to be considered, which are of particular relevance when dealing with plasma cells, with their very high cytoplasmic content of RNA and immunoglobulin. In situ hybridization for EBER has been performed in the studies by Serafini et al. (2007) with the methodology that was used by J.M.M. to stain the tumour tissues shown in Fig. 3A. A more detailed inspection of the original Fig. 2 of this article (Serafini et al., 2007) at high magnification reveals that the majority of EBER-positive cells show an unstained central portion, surrounded by a darkly stained halo. Such a staining pattern is highly suggestive of cytoplasmic reactivity. As discussed above, this subcellular distribution of reactivity is unexpected for the EBERs in latently EBV-infected cells. EBERs may be exported from the nuclei into cytoplasmic exosomes (Pegtel et al., 2010), but this in general is rare and quantitatively minor compared with nuclear EBER expression in B lymphocytes. In diagnostic pathology exclusively cytoplasmic EBER reactivity is considered non-specific, unrelated to EBV infection. An exception to this are cells undergoing mitosis, where breakdown of nuclear membrane leads to leaking of EBERs into the cytoplasm. Also the immunohistochemical results were not regarded as fully convincing due to a rather low signal to background ratio (see Serafini et al., 2010, Fig. 1 in the respective study). In addition, the very high number of cells, expressing early lytic EBV antigens in relation to those expressing latent antigens or just B cell markers, was seen as rather unusual for EBV-infected tissue, since in other situations, such as infectious mononucleosis or post-transplant lymphoproliferative disorders, the proportion of EBV-infected lymphocytes entering into the lytic cycle is extremely small. Finally, one antibody specific for BFRF1, which showed a highly positive result for the group based in Rome, was also evaluated by the Amsterdam group and revealed negative results in their multiple sclerosis samples and false positive results in control lymphoid tissue.
A fundamentally different interpretation was provided by the group from Rome (F.A., B.S.) as discussed in detail by Aloisi et al. (2010). In particular, the cytoplasmic localization of EBER, which was not observed in any of the brain samples from cases with other inflammatory neurological diseases examined or in control lymphoid tissues (Serafini et al., 2007, 2010 and unpublished results), is not interpreted in such a dogmatic manner and as a necessarily non-specific finding. The coexistence of EBER+ cells with either a nuclear or cytoplasmic signal in the multiple sclerosis brain is rather seen as a feature that may be unique to multiple sclerosis and could be related to the survival and expansion of non-tumour EBV-infected cells in a chronically inflamed environment, with possible extracellular spread of EBER molecules, a condition that is fundamentally different from EBV-associated tumours or primary EBV infection. A careful re-analysis of EBV gene expression profiles in inflammatory conditions may thus be needed. Concerning the immunohistochemical findings, it may be necessary to more carefully consider the biology of EBV infection and its gene expression profiles in latency and lytic cycle as described above. The use of different protocols and of a non-overlapping panel of antibodies could account for the discrepancies observed and stresses the need for further standardization. It should also be considered that most protocols used to reveal EBV antigens in tumour biopsies need to be adapted for frozen or fixed frozen autopsy brain tissue. Finally, the distribution of EBV markers in the multiple sclerosis brain is compatible with a predominantly latent viral infection while reactivation events in plasma cells are restricted to active lesions and ectopic B cell follicles, namely to areas that can be found only after an extensive screening of multiple sclerosis brain samples (Aloisi et al., 2010). These areas may not have been included sufficiently in the brain samples analysed by the other groups.
Conclusion
Currently the evidence for the presence of EBV-infected cells in the brain of patients with multiple sclerosis remains controversial. Experience ranges widely between different groups, one seeing an expression in the vast majority of B cells in all inflamed brain samples (Rome) while the others finding no or only exceptional EBV-positive cells in very few patients (most groups present). After an extensive and detailed discussion of all the results, it was felt that independent confirmation of the presence of a multiple sclerosis-associated EBV infection in the CNS had so far not been achieved by any of the groups participating at the workshop. However, during the preparation of this manuscript Dr Luca Muzio at San Raffaele Scientific Institute in Milan analysed, in a blind manner, seven different fixed frozen brain samples of the UK multiple sclerosis cohort using a highly sensitive, radioactive in situ hybridization technique (Muzio et al., 2005; Centonze et al., 2009) with EBER1- and EBER2-specific probes (Niedobitek et al., 1991) that differ from those used by Serafini et al. (2007). The data obtained by Muzio were reported to confirm the presence of EBV-infected cells in multiple sclerosis lesions and to provide a picture that overlaps, in terms of localization and frequency of EBV-infected cells, to that obtained by Serafini et al. in adjacent brain sections using a non-radioactive EBER in situ hybridization protocol (L. Muzio and B. Serafini, unpublished results). However, the original slides have not been available for assessment. Therefore, while acknowledging that these results may be relevant, the working group is not able to provide a consensus opinion on this unpublished set of experiments.
There was agreement that an additional attempt should be made focusing on brain tissue samples that are regarded by the group in Rome as those with the highest level of EBV infection. This material should be sent to other groups for independent confirmation on all three levels (in situ hybridization, immunocytochemistry and polymerase chain reaction) using technical protocols on which agreement has been reached beforehand. These additional experiments may clarify whether or not there is a massive EBV infection in the CNS-infiltrating B cell population in patients with multiple sclerosis, as described by Serafini et al. (2007, 2010).
In case the presence of EBV-infected B cells in the majority of multiple sclerosis brains should be confirmed, investigations in other chronic inflammatory brain diseases will become of critical importance. Particularly interesting will be to study patients with chronic B cell activation and plasma cell infiltration in the CNS, associated with intrathecal immunoglobulin synthesis. This may help to exclude that EBV activation within the tissue is just a non-specific response to B cell activation in inflammatory brain lesions. Given the widely confirmed epidemiological and serological evidence linking EBV to multiple sclerosis and the growing literature on altered T cell responses to EBV in multiple sclerosis, it is mandatory that further work is done to better understand how altered virus–host interactions contribute to multiple sclerosis pathogenesis.
Funding
6th Framework Programme of the European Union, Project NeuroproMiSe (LSHM-CT-2005-01863) for the meeting held in Vienna on 14–15 July 2010.
Acknowledgements
We thank Wim Vos for expert EBV staining (Figs 3 and 4).
Glossary
Abbreviations
- EBV
Epstein–Barr virus
- EBNA1
Epstein–Barr nuclear antigen-1
Appendix I
The Neuropromise EBV working group
Sandra Amor: VU University Medical Centre, Amsterdam, The Netherlands; Jeffrey Bennett: University of Colorado Denver School of Medicine, Aurora, CO, USA; Jan Bauer: Medical University of Vienna, Vienna, Austria; Monika Bradl: Medical University of Vienna, Vienna, Austria; Wolfgang Brück: University Medical Centre, Georg August University, Göttingen, Germany; Eliana Coccia: Istituto Superiore di Sanità, Rome, Italy; Stefan Gattenlöhner: Medical University of Graz, Graz, Austria; Romana Höftberger: Medical University of Vienna, Vienna, Austria; Andreas Junker: University Medical Centre, Georg August University, Göttingen, Germany; Gulfaraz Khan: United Arab Emirates University, Abu Dhabi, UAE; Ute Meier: Queen Mary University London, Barts and the London School of Medicine and Dentistry, London, UK; Kevin O'Connor: Yale School of Medicine, New Haven, CT, USA; Greg Owens: University of Colorado Denver School of Medicine, Aurora, CO, USA; Richard Reynolds: Imperial College Faculty of Medicine, London, UK; Barbara Serafini: Istituto Superiore di Sanità, Rome, Italy; Christine Stadelmann: University Medical Centre, Georg August University, Göttingen, Germany; John Tzartos: Queen Mary University London, Barts and the London School of Medicine and Dentistry, London, UK; Paul Van der Valk: VU University Medical Centre, Amsterdam, The Netherlands.
References
- Aloisi F, Pujol-Borrell R. Lymphoid neogenesis in chronic inflammatory diseases. Nature Rev. 2006;6:205–17. doi: 10.1038/nri1786. [DOI] [PubMed] [Google Scholar]
- Aloisi F, Serafini B, Magliozzi R, Howell OW, Reynolds R. Detection of Epstein-Barr virus and B-cell follicles in the multiple sclerosis brain: what you find depends on how and where you look. Brain. 2010;133:e157. doi: 10.1093/brain/awq223. [DOI] [PubMed] [Google Scholar]
- Ascherio A, Munger KL. Epstein Barr virus infection and multiple sclerosis. J Neuroimmune Pharmacol. 2010;5:271–7. doi: 10.1007/s11481-010-9201-3. [DOI] [PubMed] [Google Scholar]
- Bonnet M, Guinebretiere JM, Kremmer E, Grunewald V, Benhamou E, Contesso G, et al. Detection of Epstein-Barr virus in invasive breast cancers. J Natl Cancer Inst. 1999;91:1376–81. doi: 10.1093/jnci/91.16.1376. [DOI] [PubMed] [Google Scholar]
- Brink AA, Dukers DF, van den Brule AJ, Oudejans JJ, Middeldorp JM, et al. Presence of Epstein-Barr virus latency type III at the single cell level in post-transplantation lymphoproliferative disorders and AIDS related lymphomas. J Clin Pathol. 1997;50:911–8. doi: 10.1136/jcp.50.11.911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldwell RG, Wilson JB, Anderson SJ, Longnecker R. Epstein-Barr virus LMP2A drives B cell development and survival in the absence of normal B cell receptor signals. Immunity. 1998;9:405–11. doi: 10.1016/s1074-7613(00)80623-8. [DOI] [PubMed] [Google Scholar]
- Casola S, Otipoby KL, Alimzhanov M, Humme S, Uytterspot N, Kutok JL, et al. B cell receptor signal strength determines B cell fate. Nat Immunol. 2004;5:317–27. doi: 10.1038/ni1036. [DOI] [PubMed] [Google Scholar]
- Centonze D, Muzio L, Rossi S, Cavasinni F, De Chiara V, Bergami A, et al. Inflammation triggers synaptic alteration and degeneration in experimental autoimmune encephalomyelitis. J Neurosci. 2009;29:3442–52. doi: 10.1523/JNEUROSCI.5804-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaganti S, Heath EM, Bergler W, Kuo M, Buettner M, Niedobitek G, et al. Epstein-Barr virus colonization of tonsillar and peripheral blood B-cell subsets in primary infection and persistence. Blood. 2009;113:6372–81. doi: 10.1182/blood-2008-08-175828. [DOI] [PubMed] [Google Scholar]
- Chen T, Hudnall SD. Anatomical mapping of human herpesvirus reservoirs of infection. Mod Pathol. 2006;19:726–37. doi: 10.1038/modpathol.3800584. [DOI] [PubMed] [Google Scholar]
- de Sanjosé S, Bosch R, Schouten T, Verkuijlen S, Nieters A, Foretova L, et al. Epstein-Barr virus infection and risk of lymphoma: immunoblot analysis of antibody responses against EBV-related proteins in a large series of lymphoma subjects and matched controls. Int J Cancer. 2007;121:1806–12. doi: 10.1002/ijc.22857. [DOI] [PubMed] [Google Scholar]
- Dukers DF, Meij P, Vervoort MB, Vos W, Scheper RJ, Meijer CJ, et al. Direct immunosuppressive effects of EBV-encoded latent membrane protein 1. J Immunol. 2000;165:663–70. doi: 10.4049/jimmunol.165.2.663. [DOI] [PubMed] [Google Scholar]
- Esiri MM. Immunoglobulin-containing cells in multiple-sclerosis plaques. Lancet. 1977;2:478. doi: 10.1016/s0140-6736(77)91603-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans AS, Niederman JC. Epstein-Barr virus. In: Evans AS, Kaslow RA, editors. Viral infections of humans: epidemiology and control. 3rd edn. Plenum Press; 1989. pp. 265–92. [Google Scholar]
- Flanagan J, Middeldorp J, Sculley T. Localization of the Epstein-Barr virus protein LMP 1 to exosomes. J Gen Virol. 2003;84:1871–9. doi: 10.1099/vir.0.18944-0. [DOI] [PubMed] [Google Scholar]
- Frangou P, Buettner M, Niedobitek G. Epstein-Barr virus (EBV) infection in epithelial cells in vivo: rare detection of EBV replication in tongue mucosa but not in salivary glands. J Infect Dis. 2005;191:238–42. doi: 10.1086/426823. [DOI] [PubMed] [Google Scholar]
- Frischer JM, Bramow S, Dal Bianco A, Lucchinetti C, Rauschka H, Schmidbauer M, et al. The relation between inflammation and neurodegeneration in multiple sclerosis. Brain. 2009;132:1175–89. doi: 10.1093/brain/awp070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilligan K, Rajadurai P, Resnick L, Raab-Traub N. Epstein-Barr virus small nuclear RNAs are not expressed in permissively infected cells in AIDS-associated leukoplakia. Proc Natl Acad Sci USA. 1990;87:8790–4. doi: 10.1073/pnas.87.22.8790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gires O, Zimber-Strobl U, Gonnella R, Ueffing M, Marschall G, Zeidler R, et al. Latent membrane protein 1 of Epstein-Barr virus mimics a constitutively active receptor molecule. EMBO J. 1997;16:6131–40. doi: 10.1093/emboj/16.20.6131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaser R, Padgett DA, Litsky ML, Baiocchi RA, Yang EV, Chen M, et al. Stress-associated changes in the steady-state expression of latent Epstein-Barr virus: implications for chronic fatigue syndrome and cancer. Brain Behav Immun. 2005;19:91–103. doi: 10.1016/j.bbi.2004.09.001. [DOI] [PubMed] [Google Scholar]
- Grässer FA, Murray PG, Kremmer E, Klein K, Remberger K, Feiden W, et al. Monoclonal antibodies directed against Epstein-Barr virus-encoded nuclear antigen 1 (EBNA1): immunohistologic detection of EBNA1 in the malignant cells of Hodgkin's disease. Blood. 1994;84:3792–8. [PubMed] [Google Scholar]
- Grimm t, Schneider S, Naschberger E, Huber J, Guenzi E, Kieser A, et al. EBV latent membrane protein-1 protects B-cells from apoptosis by inhibition of BAX. Blood. 2005;105:3263–9. doi: 10.1182/blood-2004-07-2752. [DOI] [PubMed] [Google Scholar]
- Grinstein S, Preciado MV, Gattuso P, Chabay PA, Warren PA, De Matteo E, et al. Demonstration of Epstein-Barr virus in carcinomas of various sites. Cancer Res. 2002;62:4876–8. [PubMed] [Google Scholar]
- Gruhne B, Kamranvar SA, Masucci MG, Sompallae R. EBV and genomic instability – a new look at the role of virus in the pathogenesis of Burkitt's lymphoma. Semin Cancer Biol. 2009;19:394–400. doi: 10.1016/j.semcancer.2009.07.005. [DOI] [PubMed] [Google Scholar]
- Gulley ML, Glaser SL, Craig FE, Borowitz M, Mann RB, Shema SJ, et al. Guidelines for interpreting EBER in situ hybridization and LMP1 immunohistochemical tests for detecting Epstein-Barr virus in Hodgkin lymphoma. Am J Clin Pathol. 2002;117:259–67. doi: 10.1309/MMAU-0QYH-7BHA-W8C2. [DOI] [PubMed] [Google Scholar]
- Gulley ML, Tang W. Laboratory assays for Epstein-Barr virus-related disease. J Mol Diagn. 2008;10:279–92. doi: 10.2353/jmoldx.2008.080023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadinoto V, Shapiro M, Sun CC, Thorley-Lawson DA. The dynamics of EBV shedding implicate a central role for epithelial cells in amplifying viral output. PLoS Pathog. 2009;5:e1000496. doi: 10.1371/journal.ppat.1000496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton-Dutoit SJ, Pallesen G. Detection of Epstein-Barr virus small RNAs in routine paraffin sections using non-isotype RNA/RNA in situ hybridization. Histopathology. 1994;25:101–11. doi: 10.1111/j.1365-2559.1994.tb01565.x. [DOI] [PubMed] [Google Scholar]
- Handel AE, Williamson AJ, Disanto G, Handunnetthi L, Giovannoni G, Ramagopalan SV. An up-dated metaanalysis of risk of multiple sclerosis following infectious mononucleosis. PLoS One. 2010;5:e12496. doi: 10.1371/journal.pone.0012496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He B, Raab-Traub N, Casali P, Cerutti A. EBV-encoded latent membrane protein 1 cooperates with BAFF/BLyS and APRIL to induce T cell-independent Ig heavy chain class switching. J Immunol. 2003;171:5215–24. doi: 10.4049/jimmunol.171.10.5215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennard C, Pfuhl T, Buettner M, Becker KF, Knöfel T, Middeldorp J, et al. The antibody 2B4 directed against the Epstein-Barr virus (EBV)-encoded nuclear antigen 1 (EBNA1) detects MAGE-4: implications for studies on the EBV association of human cancers. J Pathol. 2006;209:430–5. doi: 10.1002/path.1996. [DOI] [PubMed] [Google Scholar]
- Herbst H, Dallenbach F, Niedobitek G, Anagnostopoulos I, Hummel M, Finn T, et al. Expression of latent membrane proteins (LMP) od Epstein-Barr virus in malignant lymphomas. Verh Dtsch Ges Pathol. 1991;75:175–8. [PubMed] [Google Scholar]
- Herrmann K, Frangou P, Middeldorp J, Niedobitek G. Epstein-Barr virus replication in tongue epithelial cells. J Gen Virol. 2002;83:2995–8. doi: 10.1099/0022-1317-83-12-2995. [DOI] [PubMed] [Google Scholar]
- Heussinger N, Büttner M, Ott G, Brachtel E, Pilch BZ, Kremmer E, et al. Expression of the Epstein-Barr virus (EBV)-encoded latent membrane protein 2A (LMP2A) in EBV-associated nasopharyngeal carcinoma. J Pathol. 2004;203:696–9. doi: 10.1002/path.1569. [DOI] [PubMed] [Google Scholar]
- Hislop AD, Kuo M, Drake-Lee AB, Akbar AN, Bergler W, Hammerschmitt N, et al. Tonsillar homing of Epstein-Barr virus-specific CD8+ T cells and the virus-host balance. J Clin Invest. 2005;115:2546–55. doi: 10.1172/JCI24810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hislop AD, Palendira U, Leese AM, Arkwright PD, Rohrlich PS, Tangye SG, et al. Impaired Epstein-Barr virus-specific CD8+ T-cell function in X-linked lymphoproliferative disease is restricted to SLAM family-positive B-cell targets. Blood. 2010;116:3249–57. doi: 10.1182/blood-2009-09-238832. [DOI] [PubMed] [Google Scholar]
- Hislop AD, Taylor GS, Sauce D, Rickinson AB. Cellular responses to viral infection in humans: lessons from Epstein-Barr virus. Annu Rev Immunol. 2007;25:587–617. doi: 10.1146/annurev.immunol.25.022106.141553. [DOI] [PubMed] [Google Scholar]
- Hochberg D, Middeldorp JM, Catalina M, Sullivan JL, Luzuriaga K, Thorley-Lawson DA. Demonstration of the Burkitt's lymphoma Epstein-Barr virus phenotype in dividing latently infected memory cells in vivo. Proc Natl Acad Sci USA. 2004;101:239–44. doi: 10.1073/pnas.2237267100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohlfeld R, Wekerle H. Autoimmune concepts of multiple sclerosis as a basis for selective immunotherapy: from pipe dreams to (therapeutic) pipelines. Proc Natl Acad Sci USA. 2004;101(Suppl 2):14599–606. doi: 10.1073/pnas.0404874101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höllsberg P, Hansen HJ, Haahr S. Altered CD8+ T-cell responses to selected Epstein-Barr virus immunodominant epitopes in patients with multiple sclerosis. Clin Exp Immunol. 2003;132:137–43. doi: 10.1046/j.1365-2249.2003.02114.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmøy T, Kvale EØ, Vartdal F. Cerebrospinal fluid CD4+ T cells from a multiple sclerosis patient cross-recognize Epstein-Barr virus and myelin basic protein. J Neurovirol. 2004;10:278–83. doi: 10.1080/13550280490499524. [DOI] [PubMed] [Google Scholar]
- Houali K, Wang X, Shimizu Y, Djennaoui D, Nicholls J, Fiorini S, et al. A new diagnostic marker for secreted Epstein-Barr virus encoded LMP1 and BARF1 oncoproteins in the serum and saliva of patients with nasopharyngeal carcinoma. Clin Cancer Res. 2007;13:4993–5000. doi: 10.1158/1078-0432.CCR-06-2945. [DOI] [PubMed] [Google Scholar]
- Hudnall SD, Ge Y, Wei L, Yang NP, Wang HQ, Chen T. Distribution and phenotype of Epstein-Barr virus-infected cells in human pharyngeal tonsils. Mod Pathol. 2005;18:519–27. doi: 10.1038/modpathol.3800369. [DOI] [PubMed] [Google Scholar]
- Hutt-Fletcher LM. Epstein-Barr virus entry. J Virol. 2007;81:7825–32. doi: 10.1128/JVI.00445-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwakiri D, Zhou L, Samanta M, Matsumoto M, Ebihara T, Seya T, et al. Epstein-Barr virus (EBV)-encoded small RNA is released from EBV-infected cells and activates signaling from Toll-like receptor 3. J Exp Med. 2009;206:2091–9. doi: 10.1084/jem.20081761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jafari N, van Nierop GP, Verjans GM, Osterhaus AD, Middeldorp JM, Hintzen RQ. No evidence for intrathecal IgG synthesis to Epstein Barr virus nuclear antigen-1 in multiple sclerosis. J Clin Virol. 2010;49:26–31. doi: 10.1016/j.jcv.2010.06.007. [DOI] [PubMed] [Google Scholar]
- Jaquiery E, Jilek S, Schluep M, Meylan P, Lysandropoulos A, Pantaleo G, et al. Intrathecal immune responses to EBV in early MS. Eur J Immunol. 2010;40:878–87. doi: 10.1002/eji.200939761. [DOI] [PubMed] [Google Scholar]
- Jilek S, Schluep M, Meylan P, Vingerhoets F, Guignard L, Monney A, et al. Strong EBV-specific CD8+ T-cell response in patients with early multiple sclerosis. Brain. 2008;131:1712–21. doi: 10.1093/brain/awn108. [DOI] [PubMed] [Google Scholar]
- Kang MS, Hung SC, Kieff E. Epstein-Barr virus nuclear antigen 1 activates transcription from episomal but not integrated DNA and does not alter lymphocyte growth. Proc Natl Acad Sci USA. 2001;98:15223–8. doi: 10.1073/pnas.211556598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan G. Epstein-Barr virus and the germinal center B cells. Exp Hematol. 2006;34:695–6. doi: 10.1016/j.exphem.2006.02.021. [DOI] [PubMed] [Google Scholar]
- Khan G, Miyashita EM, Yang B, Babcock GJ, Thorley-Lawson DA. Is EBV persistence in vivo a model for B cell homeostasis? Immunity. 1996;5:173–9. doi: 10.1016/s1074-7613(00)80493-8. [DOI] [PubMed] [Google Scholar]
- Khan G, Coates PJ, Gupta RK, Kangro HO, Slavin G. Presence of Epstein-Barr virus in Hodgkin's disease is not exclusive to Reed-Sternberg cells. Am J Pathol. 1992;140:757–62. [PMC free article] [PubMed] [Google Scholar]
- Khan G, Coates PJ, Kangro HO, Slavin G. Epstein Barr virus (EBV) encoded small RNAs: targets for detection by in situ hybridisation with oligonucleotide probes. J Clin Pathol. 1992;45:616–20. doi: 10.1136/jcp.45.7.616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kis LL, Salamon D, Persson EK, Nagy N, Scheeren FA, Spits H, et al. IL-21 imposes a type II EBV gene expression on type III and type I B cells by the repression of C- and activation of LMP-1-promoter. Proc Natl Acad Sci USA. 2010;107:872–7. doi: 10.1073/pnas.0912920107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kis LL, Takahara M, Nagy N, Klein G, Klein E. Cytokine mediated induction of the major Epstein-Barr virus (EBV)-encoded transforming protein, LMP-1. Immunol Lett. 2006;104:83–8. doi: 10.1016/j.imlet.2005.11.003. [DOI] [PubMed] [Google Scholar]
- Kurth J, Spieker T, Wustrow J, Strickler GJ, Hansmann LM, Rajewsky K, et al. EBV-infected B cells in infectious mononucleosis: viral strategies for spreading in the B cell compartment and establishing latency. Immunity. 2000;13:485–95. doi: 10.1016/s1074-7613(00)00048-0. [DOI] [PubMed] [Google Scholar]
- Laichalk LL, Hochberg D, Babcock GJ, Freeman RB, Thorley-Lawson DA. The dispersal of mucosal memory B cells: evidence from persistent EBV infection. Immunity. 2002;16:745–54. doi: 10.1016/s1074-7613(02)00318-7. [DOI] [PubMed] [Google Scholar]
- Laichalk LL, Thorley-Lawson DA. Terminal differentiation into plasma cells initiates the replicative cycle of Epstein-Barr virus in vivo. J Virol. 2005;79:1296–307. doi: 10.1128/JVI.79.2.1296-1307.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang HL, Jacobsen H, Ikemizu S, Andersson C, Harlos K, Madsen L, et al. A functional and structural basis for TCR cross-reactivity in multiple sclerosis. Nat Immunol. 2002;3:940–3. doi: 10.1038/ni835. [DOI] [PubMed] [Google Scholar]
- Lassmann H, Brück W, Lucchinetti C. The immunopathology of multiple sclerosis: an overview. Brain Pathol. 2007;17:210–8. doi: 10.1111/j.1750-3639.2007.00064.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin LI, Munger KL, O'Reilly EJ, Falk KI, Ascherio A. Primary infection with Epstein-Barr virus and risk of multiple sclerosis. Ann Neurol. 2010;67:824–30. doi: 10.1002/ana.21978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovato L, Willis SN, Rodig SJ, Caron T, Almendinger SE, Howell OW, et al. Related B cell clones populate the meninges and parenchyma of patients with multiple sclerosis. Brain. 2011;134:534–41. doi: 10.1093/brain/awq350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Murakami M, Verma SC, Cai Q, Haldar S, Kaul R, et al. Epstein-Barr virus nuclear antigen 1 (EBNA1) confers resistance to apoptosis in EBV-positive B-lymphoma cells through upregulation of survivin. Virology. 2011;410:64–75. doi: 10.1016/j.virol.2010.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucchinetti C, Brück W, Parisi J, Scheithauer B, Rodriguez M, Lassmann H. Heterogeneity of multiple sclerosis lesions: Implications for the pathogenesis of demyelination. Ann Neurol. 2000;47:707–17. doi: 10.1002/1531-8249(200006)47:6<707::aid-ana3>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- Lünemann JD, Edwards N, Muraro PA, Hayashi S, Cohen JI, Münz C, et al. Increased frequency and broadened specificity of latent EBV nuclear antigen-1-specific T cells in multiple sclerosis. Brain. 2006;129:1493–506. doi: 10.1093/brain/awl067. [DOI] [PubMed] [Google Scholar]
- Lünemann JD, Jelcić I, Roberts S, Lutterotti A, Tackenberg B, Martin R, et al. EBNA1-specific T cells from patients with multiple sclerosis cross react with myelin antigens and co-produce IFN-gamma and IL-2. J Exp Med. 2008;205:1763–73. doi: 10.1084/jem.20072397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lünemann JD, Münz C. EBV in MS: guilty by association? Trends Immunol. 2009;30:243–8. doi: 10.1016/j.it.2009.03.007. [DOI] [PubMed] [Google Scholar]
- Maghzi AH, Marta M, Bosca I, Etemadifar M, Dobson R, Maggiore C, et al. Viral pathophysiology of multiple sclerosis: a role for Epstein-Barr virus infection? Pathophysiology. 2010;18:13–20. doi: 10.1016/j.pathophys.2010.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magliozzi R, Howell OW, Reeves C, Roncaroli F, Nicholas R, Serafini B, et al. A gradient of neuronal loss and meningeal inflammation in multiple sclerosis. Ann Neurol. 2010;68:477–93. doi: 10.1002/ana.22230. [DOI] [PubMed] [Google Scholar]
- Magliozzi R, Howell O, Vora A, Serafini B, Nicholas R, Puopolo M, et al. Meningeal B-cell follicles in secondary progressive multiple sclerosis associate with early onset of disease and severe cortical pathology. Brain. 2007;130:1089–104. doi: 10.1093/brain/awm038. [DOI] [PubMed] [Google Scholar]
- Middeldorp JM, Brink AA, van den Brule AJ, Meijer CJ. Pathogenic roles for Epstein-Barr virus (EBV) gene products in EBV-associated proliferative disorders. Crit Rev Oncol Hematol. 2003;45:1–36. doi: 10.1016/s1040-8428(02)00078-1. [DOI] [PubMed] [Google Scholar]
- Middeldorp JM, Pegtel DM. Multiple roles of LMP1 in Epstein-Barr virus induced immune escape. Semin Cancer Biol. 2008;18:388–96. doi: 10.1016/j.semcancer.2008.10.004. [DOI] [PubMed] [Google Scholar]
- Miller G, El-Guindy A, Countryman J, Ye J, Gradoville L. Lytic cycle switches of oncogenic human gammaherpesviruses. Adv Cancer Res. 2007;97:81–109. doi: 10.1016/S0065-230X(06)97004-3. [DOI] [PubMed] [Google Scholar]
- Morré SA, van Beek J, De Groot CJ, Killestein J, Meijer CJ, Polman CH, et al. Is Epstein-Barr virus present in the CNS of patients with MS? Neurology. 2001;56:692. doi: 10.1212/wnl.56.5.692. [DOI] [PubMed] [Google Scholar]
- Murray PG, Lissauer D, Junying J, Davies G, Moore S, Bell A, et al. Reactivity with a monoclonal antibody to Epstein-Barr virus (EBV) nuclear antigen 1 defines a subset of aggressive breast cancers in the absence of the EBV genome. Cancer Res. 2003;63:2338–43. [PubMed] [Google Scholar]
- Murray PG, Niedobitek G, Kremmer E, Grässer F, Reynolds GM, Cruchley A, et al. In situ detection of the Epstein-Barr virus-encoded nuclear antigen 1 in oral hairy leukoplakia and virus-associated carcinomas. J Pathol. 1996;178:44–7. doi: 10.1002/(SICI)1096-9896(199601)178:1<44::AID-PATH471>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- Muzio L, Soria JM, Pannese M, Piccolo S, Mallamaci A. A mutually stimulating loop involving emx2 and canonical wnt signalling specifically promotes expansion of occipital cortex and hippocampus. Cereb Cortex. 2005;15:2021–8. doi: 10.1093/cercor/bhi077. [DOI] [PubMed] [Google Scholar]
- Nicholls J, Kremmer E, Meseda CA, Mackett M, Hahn P, Gulley ML, et al. Comparative analysis of the expression of the Epstein-Barr virus (EBV) anti-apoptotic gene BHRF1 in nasopharyngeal carcinoma and EBV-related lymphoid diseases. J Med Virol. 2001;65:105–13. [PubMed] [Google Scholar]
- Niedobitek G, Agathanggelou A, Herbst H, Whitehead L, Wright DH, Young LS. Epstein-Barr virus (EBV) infection in infectious mononucleosis: virus latency, replication and phenotype of EBV-infected cells. J Pathol. 1997;182:151–9. doi: 10.1002/(SICI)1096-9896(199706)182:2<151::AID-PATH824>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Niedobitek G, Agathanggelou A, Rowe M, Jones EL, Jones DB, Turyaguma P, et al. Heterogeneous expression of Epstein-Barr virus latent protein in endemic Burkitt's lymphoma. Blood. 1995;86:659–65. [PubMed] [Google Scholar]
- Niedobitek G, Agathanggelou A, Steven N, Young LS. Epstein-Barr virus (EBV) in infectious mononucleosis: detection of the virus in tonsillar B lymphocytes but not in desquamated oropharyngeal epithelial cells. J Clin Pathol: Mol Pathol. 2000;53:37–42. doi: 10.1136/mp.53.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niedobitek G, Herbst H. In situ detection of Epstein-Barr virus and phenotype determination of EBV-infected cells. Methods Mol Biol. 2006;326:115–37. doi: 10.1385/1-59745-007-3:115. [DOI] [PubMed] [Google Scholar]
- Niedobitek G, Herbst H, Young LS, Brooks L, Masucci MG, Crocker J, et al. Patterns of Epstein-Barr virus infection in non-neoplastic lymphoid tissue. Blood. 1992;79:2520–6. [PubMed] [Google Scholar]
- Niedobitek G, Kremmer E, Herbst H, Whitehead L, Dawson CW, Niedobitek E, et al. Immunohistochemical detection of the Epstein-Barr virus-encoded latent membrane protein 2A in Hodgkin's disease and infectious mononucleosis. Blood. 1997;90:1664–72. [PubMed] [Google Scholar]
- Niedobitek G, Young LS, Lau R, Brooks L, Greenspan D, Greenspan JS, et al. Epstein-Barr virus infection in oral hairy leukoplakia: virus replication in the absence of a detectable latent phase. J Gen Virol. 1991;72:3035–46. doi: 10.1099/0022-1317-72-12-3035. [DOI] [PubMed] [Google Scholar]
- Niller HH, Wolf H, Minarovits J. Epigenetic dysregulation of the host cell genome in Epstein-Barr virus-associated neoplasia. Semin Cancer Biol. 2009;19:158–64. doi: 10.1016/j.semcancer.2009.02.012. [DOI] [PubMed] [Google Scholar]
- Obermeier B, Mentele R, Malotka J, Kellermann J, Kümpfel T, Wekerle H, et al. Matching of oligoclonal immunoglobulin transcriptomes and proteomes of cerebrospinal fluid in multiple sclerosis. Nat Med. 2008;14:686–93. doi: 10.1038/nm1714. [DOI] [PubMed] [Google Scholar]
- Owens GP, Bennett JL, Lassmann H, O'Connor KC, Ritchie AM, Shearer A, et al. Antibodies produced by clonally expanded plasma cells in multiple sclerosis cerebrospinal fluid. Ann Neurol. 2009;65:639–49. doi: 10.1002/ana.21641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panagopoulos D, Victoratos P, Alexiou M, Kollias G, Mosialos G. Comparative analysis of signal transduction by CD40 and the Epstein-Barr virus oncoprotein LMP1 in vivo. J Virol. 2004;78:13253–61. doi: 10.1128/JVI.78.23.13253-13261.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paschos K, Smith P, Anderton E, Middeldorp JM, White RE, Allday MJ. Epstein-Barr virus latency in B cells leads to epigenetic repression and CpG methylation of the tumour suppressor gene Bim. PLoS Pathog. 2009;5:e1000492. doi: 10.1371/journal.ppat.1000492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peferoen LA, Lamers F, Lodder LN, Gerritsen WH, Huitinga I, Melief J, et al. Epstein Barr virus is not a characteristic feature in the central nervous system in established multiple sclerosis. Brain. 2010;133:1–4. doi: 10.1093/brain/awp296. [DOI] [PubMed] [Google Scholar]
- Pegtel DM, Cosmopoulos K, Thorley-Lawson DA, van Eijndhoven MA, Hopmans ES, Lindenberg JL, et al. Functional delivery of viral miRNAs via exosomes. Proc Natl Acad Sci USA. 2010;107:6328–33. doi: 10.1073/pnas.0914843107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pegtel DM, Subramanian A, Sheen TS, Tsai CH, Golub TR, Thorley-Lawson DA. Epstein-Barr-virus-encoded LMP2A induces primary epithelial cell migration and invasion: possible role in nasopharyngeal carcinoma. J Virol. 2005;79:15430–42. doi: 10.1128/JVI.79.24.15430-15442.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pender MP. Infection of autoreactive B lymphocytes with EBV, causing chronic autoimmune disease. Trends Immunol. 2003;24:584–8. doi: 10.1016/j.it.2003.09.005. [DOI] [PubMed] [Google Scholar]
- Pender MP. The essential role of Epstein-Barr Virus in the pathogenesis of multiple sclerosis. Neuroscientist. 2011;17:351–367. doi: 10.1177/1073858410381531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portis T, Ikeda M, Longnecker R. Epstein-Barr virus LMP2A: regulatory cellular ubiquitination process for maintenance of viral latency? Trends Immunol. 2004;25:422–6. doi: 10.1016/j.it.2004.05.009. [DOI] [PubMed] [Google Scholar]
- Preciado MV, Chabay PA, De Matteo EN, Gonzales P, Grinstein S, Actis A, et al. Epstein-Barr virus in breast carcinoma in Argentinia. Arch Pathol Lab Med. 2005;129:377–81. doi: 10.5858/2005-129-377-EVIBCI. [DOI] [PubMed] [Google Scholar]
- Prineas JW, Wright RG. Macrophages, lymphocytes, and plasma cells in the perivascular compartment in chronic multiple sclerosis. Lab Invest. 1978;38:409–21. [PubMed] [Google Scholar]
- Ressing ME, Horst D, Griffin BD, Tellam J, Zuo J, Khanna R, et al. Epstein-Barr virus evasion of CD8(+) and CD4(+) T cell immunity via concerted actions of multiple gene products. Semin Cancer Biol. 2008;18:397–408. doi: 10.1016/j.semcancer.2008.10.008. [DOI] [PubMed] [Google Scholar]
- Roughan JE, Torgbor C, Thorley-Lawson DA. Germinal center B cells latently infected with Epstein-Barr virus proliferate extensively but do not increase in number. J Virol. 2010;84:1158–68. doi: 10.1128/JVI.01780-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvetti M, Giovannoni G, Aloisi F. Epstein-Barr virus and multiple sclerosis. Curr Opin Neurol. 2009;22:201–6. doi: 10.1097/WCO.0b013e32832b4c8d. [DOI] [PubMed] [Google Scholar]
- Sargsyan SA, Shearer AJ, Ritchie AM, Burgoon MP, Anderson S, Hemmer B, et al. Absence of Epstein-Barr virus in the brain and CSF of patients with multiple sclerosis. Neurology. 2010;74:1127–35. doi: 10.1212/WNL.0b013e3181d865a1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saridakis V, Sheng Y, Sarkari F, Holowaty MN, Shire K, Nguyen T, et al. Structure of the p53 binding domain of HAUSP/USP7 bound to Epstein-Barr nuclear antigen 1: implications for EBV-mediated immortalization. Mol Cell. 2005;18:25–36. doi: 10.1016/j.molcel.2005.02.029. [DOI] [PubMed] [Google Scholar]
- Sears J, Ujihara M, Wong S, Ott C, Middeldorp J, Aiyar A. The amino terminus of Epstein-Barr virus (EBV) nuclear antigen 1 contains AT hooks that facilitate the replication and partitioning of latent EBV genomes by tethering them to cellular chromosomes. J Virol. 2004;78:11487–505. doi: 10.1128/JVI.78.21.11487-11505.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serafini B, Severa M, Columba-Cabezas S, Rosicarelli B, Veroni C, Chiappetta G, et al. Epstein-Barr virus latent infection and BAFF expression in B-cells in the multiple sclerosis brain: implications for viral persistence and intrathecal B-cell activation. J Neuropath Exp Neurol. 2010;69:677–93. doi: 10.1097/NEN.0b013e3181e332ec. [DOI] [PubMed] [Google Scholar]
- Serafini B, Rosicarelli B, Franciotta D, Magliozzi R, Reynolds R, Cinque P, et al. Dysregulated Epstein Barr virus infection in the multiple sclerosis brain. J Exp Med. 2007;204:2899–912. doi: 10.1084/jem.20071030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serafini B, Rosicarelli B, Magliozzi R, Stigliano E, Aloisi F. Detection of ectopic B-cell follicles with germinal centers in the meninges of patients with secondary progressive multiple sclerosis. Brain Pathol. 2004;14:164–74. doi: 10.1111/j.1750-3639.2004.tb00049.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw KM, Stewart SE, Wei W, Woodman CB, O'Neil JD, Dawson CW, et al. The EBV-encoded latent membrane proteins, LMP2A and LMP2B, limit the actions of interferon by targeting interferon receptors for degradation. Oncogene. 2009;28:3903–14. doi: 10.1038/onc.2009.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon KC, van der Mei IA, Munger KL, Ponsonby A, Dickinson J, Dwyer T, et al. Combined effects of smoking, anti-EBNA antibodies, and HLA-DRB1*1501 on multiple sclerosis risk. Neurology. 2010;74:1365–71. doi: 10.1212/WNL.0b013e3181dad57e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivachandran N, Sarkari F, Frappier L. Epstein-Barr nuclear antigen 1 contributes to nasopharyngeal carcinoma through disruption of PML nuclear bodies. PLoS Pathog. 2008;4:e1000170. doi: 10.1371/journal.ppat.1000170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens SJ, Smits PH, Verkuijlen SA, Rockx DA, van Gorp EC, Mulder JW, et al. Aberrant Epstein-Barr virus persistence in HIV carriers is characterized by anti-Epstein-Barr virus IgA and high cellular viral loads with restricted transcription. AIDS. 2007;21:2141–9. doi: 10.1097/QAD.0b013e3282eeeba0. [DOI] [PubMed] [Google Scholar]
- Stevens SJ, Verschuuren EA, Verkuujlen SA, Van Den Brule AJ, Meijer CJ, Middeldorp JM. Role of Epstein-Barr virus DNA load monitoring in prevention and early detection of post-transplant lymphoproliferative disease. Leuk Lymphoma. 2002;43:831–40. doi: 10.1080/10428190290016971. [DOI] [PubMed] [Google Scholar]
- Sundström P, Nyström M, Ruuth K, Lundgren E. Antibodies to specific EBNA1 domains and HLA DRB1*1501 interact as risk factors for multiple sclerosis. J Neuroimmunol. 2009;215:102–7. doi: 10.1016/j.jneuroim.2009.08.004. [DOI] [PubMed] [Google Scholar]
- Swaminathan S. Noncoding RNAs produced by oncogenic human herpesviruses. J Cell Physiol. 2008;216:321–6. doi: 10.1002/jcp.21480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorley-Lawson DA. Epstein-Barr virus: exploiting the immune system. Nat Rev Immunol. 2001;1:75–82. doi: 10.1038/35095584. [DOI] [PubMed] [Google Scholar]
- Thorley-Lawson DA, Allday MJ. The curious case of the tumour virus: 50 years of Burkitt's lymphoma. Nat Rev Microbiol. 2008;6:913–24. doi: 10.1038/nrmicro2015. [DOI] [PubMed] [Google Scholar]
- Tobollik S, Meyer L, Buettner M, Klemmer S, Kempkes B, Kremmer E, et al. Epstein-Barr virus nuclear antigen 2 inhibits AID expression during EBV-driven B-cell growth. Blood. 2006;108:3859–64. doi: 10.1182/blood-2006-05-021303. [DOI] [PubMed] [Google Scholar]
- Torkildsen O, Stansberg C, Angelskar SM, Kooi EJ, Geurts JJ, van der Valk P, et al. Upregulation of immunoglobulin-related genes in cortical sections from multiple sclerosis patients. Brain Pathol. 2010;20:720–9. doi: 10.1111/j.1750-3639.2009.00343.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai CL, Li HP, Lu YJ, Hsueh C, Liang Y, Chen CL, et al. Activation of DNA methyltransferase 1 by EBV LMP1 involves c-jun NH(2)-terminal kinase signaling. Cancer Res. 2006;66:11668–76. doi: 10.1158/0008-5472.CAN-06-2194. [DOI] [PubMed] [Google Scholar]
- Ufret-Vincenty RL, Quigley L, Tresser N, Pak SH, Gado A, Hausmann S, et al. In vivo survival of viral antigen-specific T cells that induce experimental autoimmune encephalomyelitis. J Exp Med. 1998;188:1725–38. doi: 10.1084/jem.188.9.1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Nicholas MW, Conway KL, Sen P, Diz R, Tisch RM, et al. EBV latent membrane protein 2A induces autoreactive B cell activation and TLR hypersensitivity. J Immunol. 2006;177:2793–802. doi: 10.4049/jimmunol.177.5.2793. [DOI] [PubMed] [Google Scholar]
- Webster-Cyriaque J, Middeldorp J, Raab-Traub N. Hairy leukoplakia: an unusual combination of transforming and permissive Epstein-Barr virus infections. J Virol. 2000;74:7610–8. doi: 10.1128/jvi.74.16.7610-7618.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis SN, Stadelmann C, Rodig SJ, Caron T, Gattenlöhner S, Mallozzi SS, et al. Epstein-Barr virus infection is not a characteristic feature of multiple sclerosis brain. Brain. 2009;132:3318–28. doi: 10.1093/brain/awp200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates JL, Guan N. Epstein-Barr virus-derived plasmids replicate only once per cell cycle and are not amplified after entry into cells. J Virol. 1991;65:483–8. doi: 10.1128/jvi.65.1.483-488.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young LS, Rickinson AB. Epstein-Barr virus: 40 years on. Nat Rev Cancer. 2004;4:757–68. doi: 10.1038/nrc1452. [DOI] [PubMed] [Google Scholar]
- Zhang JX, Chen HL, Zong YS, Chan KH, Nicholls J, Middeldorp JM, et al. Epstein-Barr virus expression within keratinizing nasopharyngeal carcinoma. J Med Virol. 1998;55:227–33. doi: 10.1002/(sici)1096-9071(199807)55:3<227::aid-jmv8>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- Zeng Y, Middeldorp J, Madjar JJ, Ooka T. A major DNA binding protein encoded by BALF2 open reading frame of Epstein-Barr virus (EBV) forms a complex with other EBV DNA-binding proteins: DNAase, EA-D, and DNA polymerase. Virology. 1997;239:285–95. doi: 10.1006/viro.1997.8891. [DOI] [PubMed] [Google Scholar]