Abstract
Accumulation of the DNA/RNA binding protein fused in sarcoma as cytoplasmic inclusions in neurons and glial cells is the pathological hallmark of all patients with amyotrophic lateral sclerosis with mutations in FUS as well as in several subtypes of frontotemporal lobar degeneration, which are not associated with FUS mutations. The mechanisms leading to inclusion formation and fused in sarcoma-associated neurodegeneration are only poorly understood. Because fused in sarcoma belongs to a family of proteins known as FET, which also includes Ewing’s sarcoma and TATA-binding protein-associated factor 15, we investigated the potential involvement of these other FET protein family members in the pathogenesis of fused in sarcoma proteinopathies. Immunohistochemical analysis of FET proteins revealed a striking difference among the various conditions, with pathology in amyotrophic lateral sclerosis with FUS mutations being labelled exclusively for fused in sarcoma, whereas fused in sarcoma-positive inclusions in subtypes of frontotemporal lobar degeneration also consistently immunostained for TATA-binding protein-associated factor 15 and variably for Ewing’s sarcoma. Immunoblot analysis of proteins extracted from post-mortem tissue of frontotemporal lobar degeneration with fused in sarcoma pathology demonstrated a relative shift of all FET proteins towards insoluble protein fractions, while genetic analysis of the TATA-binding protein-associated factor 15 and Ewing’s sarcoma gene did not identify any pathogenic variants. Cell culture experiments replicated the findings of amyotrophic lateral sclerosis with FUS mutations by confirming the absence of TATA-binding protein-associated factor 15 and Ewing’s sarcoma alterations upon expression of mutant fused in sarcoma. In contrast, all endogenous FET proteins were recruited into cytoplasmic stress granules upon general inhibition of Transportin-mediated nuclear import, mimicking the findings in frontotemporal lobar degeneration with fused in sarcoma pathology. These results allow a separation of fused in sarcoma proteinopathies caused by FUS mutations from those without a known genetic cause based on neuropathological features. More importantly, our data imply different pathological processes underlying inclusion formation and cell death between both conditions; the pathogenesis in amyotrophic lateral sclerosis with FUS mutations appears to be more restricted to dysfunction of fused in sarcoma, while a more global and complex dysregulation of all FET proteins is involved in the subtypes of frontotemporal lobar degeneration with fused in sarcoma pathology.
Keywords: FUS, TAF15, EWS, amyotrophic lateral sclerosis, frontotemporal dementia
Introduction
The identification of the DNA/RNA binding protein TAR-DNA binding protein 43 kDa (TDP-43) as the disease protein in most forms of amyotrophic lateral sclerosis (ALS) and in the most common form of frontotemporal lobar degeneration (FTLD), confirmed that these two neurodegenerative conditions belong to a clinicopathological spectrum of diseases and initiated the concept of RNA dysmetabolism as a crucial event in disease pathogenesis (Neumann et al., 2006; Mackenzie et al., 2010a). This idea was corroborated with the subsequent discovery of another DNA/RNA binding protein fused in sarcoma (FUS), as the pathological protein in many remaining TDP-43-negative cases with ALS and FTLD.
Briefly, the finding of mutations in the FUS gene as cause of familial ALS (Kwiatkowski et al., 2009; Vance et al., 2009) was rapidly confirmed in genetic screenings of large ALS cohorts throughout the world and were found to account for ∼3% of familial ALS and ∼1% of sporadic ALS (Mackenzie et al., 2010a). The majority of FUS mutations cluster in the C-terminus of the protein that encodes for a non-classical nuclear localization sequence (Lee et al., 2006; Dormann et al., 2010). FUS mutations have been shown to disrupt this motif, resulting in impaired Transportin-mediated nuclear import of FUS and increased concentrations of cytoplasmic FUS (Dormann et al., 2010; Ito et al., 2011; Kino et al., 2011). In line with the idea that altered nuclear import is a key event in disease pathogenesis, the neuropathology associated with ALS with FUS mutations (ALS-FUS) is characterized by abnormal cytoplasmic neuronal and glial inclusions that are immunoreactive for FUS (Kwiatkowski et al., 2009; Vance et al., 2009; Blair et al., 2010; Groen et al., 2010; Hewitt et al., 2010; Rademakers et al., 2010; Mackenzie et al., 2011b).
Subsequently, FUS was studied in other neurodegenerative diseases and identified as a component of the inclusions in several subtypes of FTLD, now subsumed as FTLD-FUS (Mackenzie et al., 2010b). This group includes cases initially designated as atypical FTLD with ubiquitin-positive inclusions (FTLD-U) (Neumann et al., 2009b), neuronal intermediate filament inclusion disease (NIFID) (Neumann et al., 2009a) and basophilic inclusion body disease (BIBD) (Munoz et al., 2009). In contrast to cases presenting with pure ALS, which are almost always associated with mutations in FUS, no genetic alterations of FUS have been reported to date for cases within the FTLD-FUS group (Neumann et al., 2009a, b; Rohrer et al., 2010; Urwin et al., 2010; Snowden et al., 2011). Thus, the mechanisms underlying FUS accumulation in FTLD-FUS as well as an explanation for the different patterns of FUS pathology in the distinct FTLD-FUS subtypes awaits further clarification (Mackenzie et al., 2011a).
FUS is a multifunctional DNA/RNA binding protein and belongs to the FET family of proteins that also includes Ewing’s sarcoma protein (EWS), TATA-binding protein-associated factor 15 (TAF15) and the Drosophila orthologue Cabeza (Law et al., 2006; Kovar 2011). The FET proteins were initially discovered as components of fusion oncogenes that cause human cancers. Their normal function is predicted to include roles in RNA transcription, processing, transport, microRNA processing and DNA repair (Law et al., 2006; Tan and Manley, 2009; Kovar, 2011). In most cell types, all of the FET proteins are predominantly localized to the nucleus, but they are able to continuously shuttle between the nucleus and cytoplasm (Zinszner et al., 1997; Zakaryan and Gehring 2006; Jobert et al., 2009). Protein-interaction studies have revealed that FET proteins are able to interact with each other, suggesting that they may form protein complexes (Pahlich et al., 2008; Kovar, 2011). This raises the possibility that alterations of TAF15 and EWS might also be involved in the pathogenesis of FUS-opathies.
In order to address this hypothesis we performed detailed immunohistochemical, biochemical and genetic analyses of TAF15 and EWS in a range of cases with FTLD-FUS and ALS-FUS that covers the complete spectrum of FUS-opathies. Our data revealed striking differences in FET protein alterations between ALS-FUS and FTLD-FUS, thereby strongly suggesting different disease mechanisms underlying these conditions.
Materials and methods
Case selection
Cases with FUS pathology, including atypical FTLD-U (n = 15), BIBD (n = 7), NIFID (n = 4) and ALS-FUS (n = 6), were selected from previous studies (Munoz et al., 2009; Neumann et al., 2009a, b; Mackenzie et al., 2011a, b). Detailed clinical and pathological description of each of the FTLD-FUS and ALS-FUS cases has been published previously and is summarized in Supplementary Table 1.
Neurological control cases for immunohistochemistry included FTLD with TDP-43 pathology [(n = 17); including sporadic subtype 1 (n = 3), subtype 2 (n = 2), subtype 3 (n = 6), according to Mackenzie et al. 2006, familial with GRN mutations (n = 2), familial with VCP mutations (n = 2) and familial linked to chromosome 9p (n = 2)], FTLD with tau pathology (n = 8; including two each of Pick’s disease, progressive supranuclear palsy, corticobasal degeneration and argyrophilic grain disease), FTLD with CHMP2B mutations (n = 2), sporadic ALS with TDP-43 pathology (n = 8), familial ALS with SOD1 mutations (n = 2), Alzheimer’s disease (n = 4), Lewy body disease (n = 4), multiple system atrophy (n = 2), Huntington’s disease (n = 2), spinocerebellar ataxia (n = 3) and neuronal intranuclear inclusion body disease (n = 1). Normal control tissue (n = 4) was from elderly patients with no history of neurological disease.
Antibodies
A number of commercially available anti-TAF15 and anti-EWS antibodies were tested by immunohistochemistry on formalin-fixed paraffin-embedded brain tissue and by immunoblot. Results are summarized in Supplementary Table 2. Three TAF15 antibodies revealed physiological staining in tissue sections. The polyclonal antibody TAF15-IHC-00094-1 (Bethyl) was used for staining of all cases and for immunofluorescence. TAF15-309A and 308A (Bethyl) were used for confirmation in selected sections and for immunoblots; TAF15-308A was used in cell culture experiments. For EWS, four antibodies revealed physiological staining in tissue sections. The monoclonal antibody EWS-G5 (Santa Cruz) was used for staining of all cases, immunofluorescence, immunoblotting and cell culture experiments. Selected sections were stained with EWS-IHC-00086 (Bethyl), EWS-3319-1 and EWS-3320-1 (Epitomics) for confirmation. Given the homology of FET proteins, possible cross-reactivity of the TAF15 and EWS antibodies with FUS was excluded by immunoblot analysis (Supplementary Fig. 1).
Other primary antibodies employed included polyclonal anti-FUS HPA008784 (Sigma-Aldrich, 1 : 2000), FUS-302A (Bethyl, 1 : 10 000), monoclonal anti-FUS (ProteintechGroup, 1 : 1000), monoclonal anti-α-internexin (Zymed, 1 : 500), monoclonal anti-haemagglutinin (Sigma, 1 : 500), and polyclonal anti-haemagglutinin (Sigma, 1 : 200).
Immunohistochemistry and immunofluorescence
Immunohistochemistry was performed on 5-µm thick paraffin sections using the Ventana BenchMark XT automated staining system (Ventana) and developed with aminoethylcarbizole or using the NovoLink™ Polymer Detection Kit and developed with 3,3′-diaminobenzidine. Microwave antigen retrieval was performed for all stainings.
FUS, TAF15 and EWS pathology was evaluated using a semi-quantitative grading system, similar to that used in previous studies in which the pathological lesions are scored as absent (−), rare (+), occasional (++), common (+++) or numerous (++++). A grading of ‘rare’ indicates that extensive survey of the tissue section is required for identification. ‘Occasional’ means that the lesions are easy to find but not present in every microscopic field. The pathology is considered ‘common’ when at least one example is present in most high-power fields. When many lesions are present in every high-power field, then the lesions are considered to be ‘numerous’.
Double-label immunofluorescence was performed on selected cases for FUS and TAF15 or EWS, and α-internexin and TAF15 or EWS. The secondary antibodies were Alexa Fluor 594 and Alexa Fluor 488 conjugated anti-mouse and anti-rabbit IgG (Invitrogen, 1 : 500). 4′-6-diamidino-2-phenylindol was used for nuclear counterstaining. Immunofluorescence images of brain sections were obtained by wide-field fluorescence microscopy (BX61 Olympus with digital camera F-view, Olympus).
Biochemical analysis
Fresh-frozen post-mortem frontal grey matter from atypical FTLD-U (n = 5), BIBD (n = 1), NIFID (n = 1), FTLD with TDP-43 pathology (n = 5), Alzheimer’s disease (n = 2) and normal controls (n = 4) was used for the sequential extraction of proteins with buffers of increasing stringency, using a protocol described previously (Neumann et al., 2009b). Briefly, grey matter was extracted at 2 ml/g (v/w) by repeated homogenization and centrifugation steps (120 000 g, 30 min, 4°C) with high-salt buffer (50 mM Tris–HCl, 750 mM NaCl, 10 mM NaF, 5 mM EDTA, pH 7.4), 1% Triton-X 100 in high-salt buffer, radioimmunoprecipitation assay buffer (50 mM Tris–HCl, 150 mM NaCl, 5 mM EDTA, 1% NP-40, 0.5 % sodium deoxycholate, 0.1% sodium dodecyl sulphate) and 2% sodium dodecyl sulphate buffer. To prevent carry over, each extraction step was performed twice. Supernatants from the first extraction steps were analysed while supernatants from the wash steps were discarded. The 2% sodium dodecyl sulphate insoluble pellet was extracted in 70% formic acid at 0.5 ml/g (v/w), evaporated in a SpeedVac system. The dried pellet was resuspended in sample buffer and the pH adjusted with NaOH. Protease inhibitors were added to all buffers prior to use. For immunoblot analysis, fractions were resolved by 7.5% sodium dodecyl sulphate–polyacrylamide gel electrophoresis and transferred to polyvinylidene difluoride membranes (Millipore). Membranes were blocked with Tris-buffered saline containing 3% powdered milk and probed with anti-FUS, anti-TAF-15 or anti-EWS antibodies. Primary antibodies were detected with horseradish peroxidase-conjugated anti-rabbit or anti-mouse IgG (Jackson ImmunoResearch), signals were visualized by a chemiluminescent reaction (Pierce) and the Chemiluminescence Imager Stella 3200 (Raytest). Quantification of band intensities was performed with AIDA software. The Mann–Whitney U-test was used for statistical analysis of insoluble/soluble ratios with significance level set as P < 0.05.
Cell culture experiments
HeLa cells were cultured in Dulbecco’s modified Eagle’s medium with Glutamax (Invitrogen) supplemented with 10% (v/v) foetal calf serum (Invitrogen) and penicillin/streptomycin. Transfection of HeLa cells was carried out with Fugene 6 (Roche) or Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. Expression vectors with haemagglutinin-tagged human FUS with the p.P525L mutation and with the Transportin-specific inhibitor peptide M9M fused to green fluorescent protein were generated as described previously (Dormann et al., 2010). In some experiments, cells were subjected to heat shock (1 h at 44°C) 24 h after transfection. For immunofluorescence, HeLa cells were fixed for 15 min in 4% paraformaldehyde in phosphate-buffered saline, permeabilized for 5 min in 0.2% Triton X-100 with 50 mM NH4Cl and subsequently blocked for 20–30 min in 5% goat serum. Cells were stained with the indicated primary and secondary antibodies, diluted in blocking buffer for 30 min. Alexa Fluor 488, 555 and 647 conjugated goat anti-mouse and goat anti-rabbit IgGs were used as secondary antibodies. To visualize nuclei, cells were stained with TO-PRO-3 iodide (Invitrogen) for 15 min. Confocal images were obtained with an inverted laser scanning confocal microscope (Zeiss Axiovert 200M).
Genetic analysis
DNA was available from six atypical FTLD-U, one NIFID and one BIBD case. EWS breakpoint region 1 (EWSR1) exons 1–18 and TAF15 exons 1–16 were polymerase chain reaction amplified using primers designed to flanking intronic sequences using Qiagen products (Qiagen). Polymerase chain reaction conditions and primer sequences available on request. Polymerase chain reaction products were purified using the Ampure system (Agencourt Bioscience Corporation) and sequenced using Big Dye terminator V.3.1 products (Applied Biosystems). Sequencing products were purified using the CleanSEQ method (Agencourt) and analysed on an ABI 3730 DNA analyser (Applied Biosystems). Sequence analysis was performed using Sequencher software (Gene Codes).
Results
Detailed clinical and pathological descriptions of each of the cases with FTLD-FUS and ALS-FUS have been published previously and are summarized in Supplementary Table 1. TAF15 and EWS pathology was evaluated in neuroanatomical regions previously shown to be most affected by FUS pathology in each condition and results are summarized in Table 1.
Table 1.
Summary of immunoreactivity for FET proteins in FTLD-FUS subtypes and ALS-FUS
| FUS |
TAF15 |
EWS |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| FC | HC | BG | BS | LMN | FC | HC | BG | BS | LMN | FC | HC | BG | BS | LMN | |
| Atypical FTLD-U | |||||||||||||||
| 1 | ++ | ++++ | +++ | +++ | ++ | +++ | ++++ | ND | ND | +++ | + | +++ | ND | ND | NA |
| 2 | +++ | NA | +++ | +++ | ++ | +++ | NA | ND | ND | ++ | +++ | NA | ND | ND | (+) |
| 3 | ++ | +++ | +++ | ++ | +++ | ++ | +++ | ND | ND | +++ | ++ | ++ | ND | ND | ++ |
| 4 | ++ | ++ | ++ | NA | NA | +++ | ++ | ND | NA | NA | ++ | (+) | ND | NA | NA |
| 5 | +++ | ++++ | +++ | +++ | +++ | +++ | ++++ | ND | ND | ++++ | ++ | +++ | ND | ND | (++) |
| 6 | +++ | ++++ | +++ | +++ | + | ++++ | ++++ | ND | ND | ++ | ++ | +++ | ND | ND | (+) |
| 7 | +++ | ++++ | NA | NA | + | +++ | ++++ | NA | NA | + | + | ++ | NA | NA | − |
| 8 | ++++ | ++++ | ++++ | ++ | ++ | ++++ | ++++ | ND | ND | ++ | + | +++ | ND | ND | + |
| 9 | ++ | +++ | NA | NA | ++ | ++ | NA | NA | NA | ++ | + | NA | NA | NA | + |
| 10 | ++ | +++ | ++ | ++ | ++ | ++ | +++ | ND | ND | ++ | + | + | ND | ND | + |
| 11 | ++ | ++ | ++ | ++ | + | ++ | NR | ND | ND | ++ | + | NR | ND | ND | + |
| 12 | ++ | ++ | ++ | + | ++ | +++ | ++ | ND | ND | ++ | + | (+) | ND | ND | + |
| 13 | +++ | ++ | ++ | ++ | ++ | +++ | ++ | ND | ND | ++ | + | + | ND | ND | + |
| 14 | +++ | ++++ | +++ | +++ | ++ | +++ | ++++ | ND | ND | +++ | + | + | ND | ND | + |
| 15 | + | ++ | +++ | + | + | + | ++ | ND | ND | + | (+) | ++ | ND | ND | + |
| Intensity NCI | Strong | Strong | Weak/Moderate | ||||||||||||
| NIFID | |||||||||||||||
| 1 | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ND | ++++ | ++++ | ++++ | ++++ | ND | +++ |
| 2 | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ND | ++++ | ++++ | ++++ | +++ | ND | +++ |
| 3 | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ND | ++++ | ++++ | ++++ | ++++ | ND | ++++ |
| 4 | ++++ | ++++ | ++++ | ++++ | + | ++++ | ++++ | ++++ | ND | + | ++++ | ++++ | ++++ | ND | NA |
| Intensity NCI | Strong | Strong | Strong | ||||||||||||
| BIBD | |||||||||||||||
| 1 | ++++ | ++++ | +++ | ++++ | +++ | ++++ | ++++ | +++ | ND | ++++ | (++++) | (+++) | (+++) | ND | (+) |
| 2 | ++++ | ++++ | ++++ | ++++ | ++ | ++++ | ++++ | ND | ++++ | +++ | (++) | (++) | ND | (+) | (+) |
| 3 | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | NR | ++++ | ND | NR | (++++) | NR | (+) | ND | (++) |
| 4 | ++++ | + | NA | ++++ | ++++ | ++++ | + | NA | ++++ | ++++ | ++++ | (+) | NA | (++++) | NR |
| 5 | ++++ | ++ | ++ | ++++ | ++++ | NR | + | +++ | +++ | ++++ | NR | NA | ND | (+++) | (+) |
| 6 | ++++ | +++ | +++ | ++++ | ++++ | +++ | +++ | +++ | ++++ | +++ | +++ | ++ | +++ | +++ | +++ |
| 7 | ++++ | +++ | ++++ | ++++ | ++ | ++++ | +++ | ++++ | ND | ++ | ++++ | +++ | ++++ | ND | + |
| Intensity NCI | Strong | Strong | Strong | ||||||||||||
| ALS-FUS | |||||||||||||||
| 1 p.R521C | + | − | ++ | + | +++ | − | ND | − | − | − | − | ND | − | − | − |
| 2 p.R521C | ++ | − | ++ | − | +++ | − | ND | − | NA | − | NA | ND | NA | NA | − |
| 3 p.R514S/E516V | +++ | − | +++ | ++ | +++ | − | ND | − | − | − | − | ND | − | − | − |
| 4 p.P525L | +++ | − | − | + | ++++ | NA | ND | − | − | − | NA | ND | − | − | − |
| 5 p.P525L | +++ | − | − | +++ | ++++ | − | ND | − | − | − | − | ND | − | − | − |
| 6 p.Q519lfsX9 | +++ | − | + | +++ | +++ | − | ND | − | − | − | − | ND | − | − | − |
| Intensity NCI | Strong | Absent | Absent | ||||||||||||
Semiquantitative grading: − = absent; + = rare; ++ = occasional; +++ = moderate; ++++ = numerous. Scores in parentheses indicate sections where the quality of the immunostaining makes the accuracy of the scoring uncertain. BG = basal ganglia; BS = upper brainstem; FC = frontal cortex (mid-frontal gyrus for atypical FTLD-U, NIFID and BIBD; precentral gyrus for ALS-FUS); HC = hippocampus; LMN = lower motor neurons of spinal cord or hypoglossal nucleus; NA = not available; NCI = neuronal cytoplasmic inclusion; ND = staining not done; NR = no immunoreactivity, neither physiological nor pathological in the entire section.
TAF15 and EWS pathology is present in all subtypes of frontotemporal lobar degeneration with FUS pathology
Immunohistochemistry for TAF15 revealed robust physiological staining of neuronal nuclei and weaker and more variable staining of glial nuclei in all cases and controls (Fig. 1A). All subtypes of FTLD-FUS showed strong TAF15 immunoreactivity in neuronal and glial inclusions that were of similar morphology, number and anatomical distribution as demonstrated with FUS antibodies (Fig. 1 and Table 1). Specifically, atypical FTLD-U cases showed TAF15-positive round neuronal cytoplasmic inclusions in hippocampus, neocortex and lower motor neurons, as well as vermiform or round neuronal intranuclear inclusions predominantly in the dentate gyrus (Fig. 1B–E). NIFID and BIBD cases were found to have numerous round or tangle-like inclusions throughout cortical, subcortical, brainstem and spinal cord regions (Fig. 1F–H). In addition to neuronal inclusions, all subtypes of FTLD-FUS revealed at least some TAF15-positive dystrophic neurites and glial cytoplasmic inclusions (Fig. 1I, J) that were more numerous in NIFID and BIBD than in atypical FTLD-U. Notably, most inclusion bearing cells in FTLD-FUS showed a striking reduction of the physiological nuclear staining for TAF15 (Fig. 1B and C). Similar results were observed using TAF15 antibodies recognizing different epitopes, including the mid-region and C-terminus.
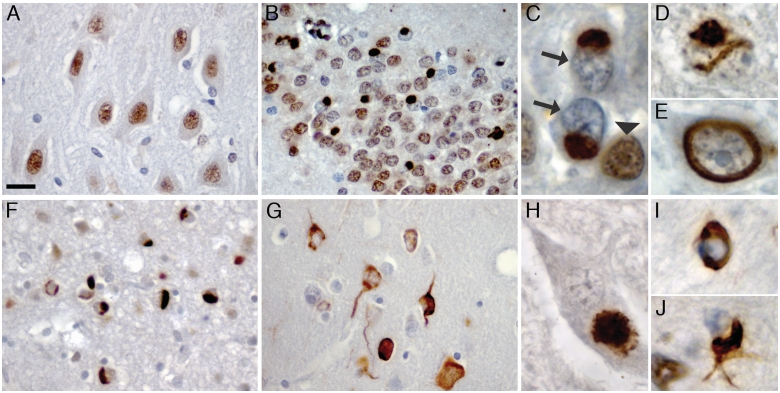
Figure 1.
TAF15 pathology in FTLD-FUS. TAF15 immunohistochemisty performed on sections of post-mortem brain tissue from normal control (A), atypical FTLD-U (B–E), NIFID (F) and BIBD (G–J). Normal physiological staining pattern, consisting of strong immunoreactivity of neuronal nuclei was seen in normal controls (A) and FTLD-FUS subjects (B). In atypical FTLD-U numerous round neuronal cytoplasmic inclusions were seen in the dentate granule cells (B and C). Note the dramatically reduced nuclear staining in inclusion bearing cells (arrows in C) compared with adjacent cells without inclusions (arrowhead in C). Neuronal intranuclear inclusions with vermiform (D) or ring-like morphology (E) were a consistent finding in the dentate granule and pyramidal cells of the hippocampus in all subjects with atypical FTLD-U. Numerous cytoplasmic inclusions with variable morphology ranging from round, crescentic, globular and tangle-like were present in neurons in NIFID (F) and BIBD (G) as shown here in frontal cortex. All FTLD-FUS cases revealed at least rare inclusions in lower motor neurons (H) as well as variable numbers of glial cytoplasmic inclusions in the white matter of affected brain regions (I, J). Scale bar: A, B, F and G = 25 µm; C–E, I and J = 5 µm; H = 10 µm.
Double-label immunofluorescence confirmed co-localization of FUS and TAF15 in almost all inclusions in FTLD-FUS cases (Fig. 2). There was a tendency for intranuclear inclusions in atypical FTLD-U and NIFID to be more strongly labelled for FUS compared with TAF15 and they were rarely found to be only FUS positive (Fig. 2B). In NIFID cases, TAF15 and α-internexin labelled discrete inclusions in the same neurons (Supplementary Fig. 2A), a finding similar to our previous results for FUS and α-internexin (Neumann et al., 2009a).
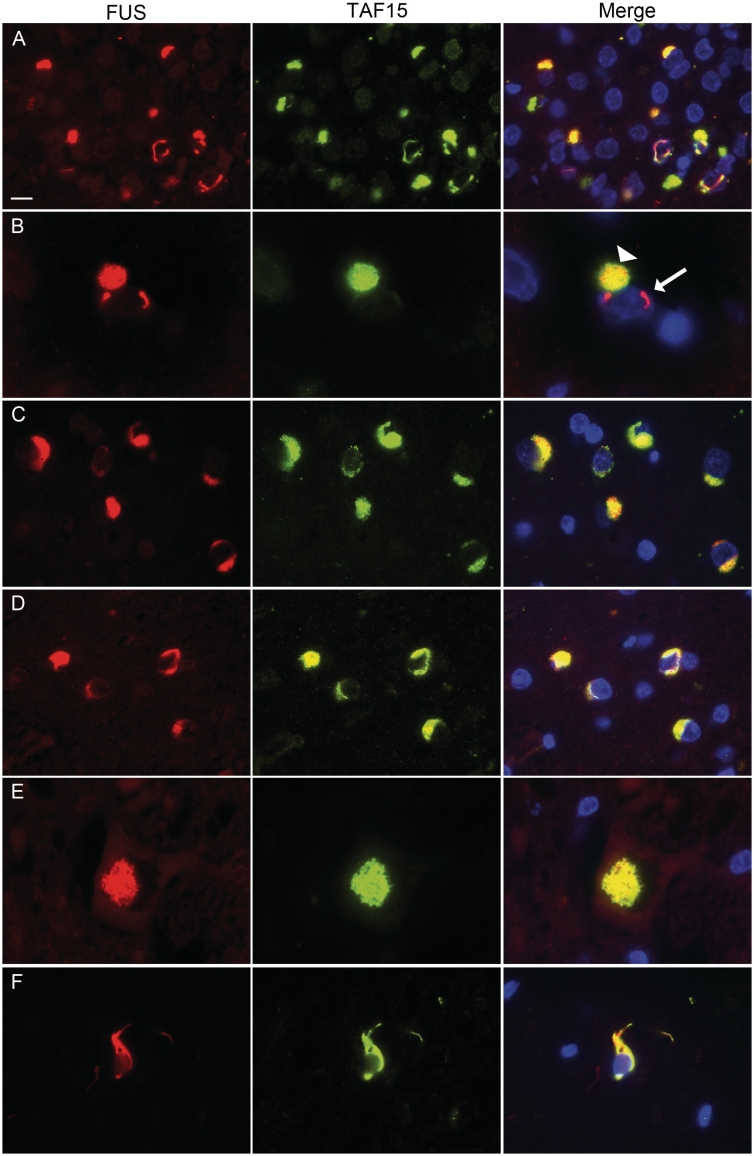
Figure 2.
Co-localization of TAF15 and FUS in FTLD-FUS inclusions. Double-label immunofluorescence for FUS (red) and TAF15 (green), with DAPI staining of nuclei in the merged images. (A) In atypical FTLD-U the vast majority of inclusions showed co-localization of FUS and TAF15. (B) However, note that single neuronal intranuclear inclusions in atypical FTLD-U were not labelled for TAF15 (arrow) while the cytoplasmic inclusion in the same cell shows co-localization (arrowhead). Consistent co-labelling for TAF15 was revealed for FUS pathology in NIFID (C) and BIBD (D). Inclusions in the lower motor neurons (E, atypical FTLD-U case) and glial cytoplasmic inclusions (F, BIBD case), also showed colocalization. Scale bar: A, C and D = 10 µm; B = 4 µm; E and F = 6.5 µm.
Antibodies against EWS revealed nuclear and diffuse cytoplasmic staining of neuronal and glial cells as the normal physiological staining pattern (Fig. 3A). However, EWS staining was more variable among cases compared with TAF15 with some sections completely lacking physiological staining while others revealed strong background staining making the scoring of EWS pathology in some cases more uncertain. Nevertheless, all subtypes of FTLD-FUS revealed at least some EWS-positive inclusions (Fig. 3 and Table 1) and similar results were obtained with four EWS antibodies recognizing different epitopes at the N-terminal and mid-region. Importantly, notable differences were observed between the distinct FTLD-FUS subtypes. In atypical FTLD-U, EWS-positive neuronal cytoplasmic and intranuclear inclusions were less numerous than those labelled with FUS, being rare to moderate in neocortical regions and lower motor neurons (Fig. 3B–D and G) and the staining intensity tended to be rather weak. In contrast, inclusions in NIFID and BIBD revealed a much more robust EWS staining intensity and the frequency of pathology in cortical, subcortical, brainstem and spinal cord regions was comparable with that seen with FUS (Fig. 3E, F and H). Due to the variability in staining intensity among cases, analysis of the normal physiological staining pattern of EWS was more difficult to assess; however, nuclear EWS staining was retained in at least some inclusion bearing cells.
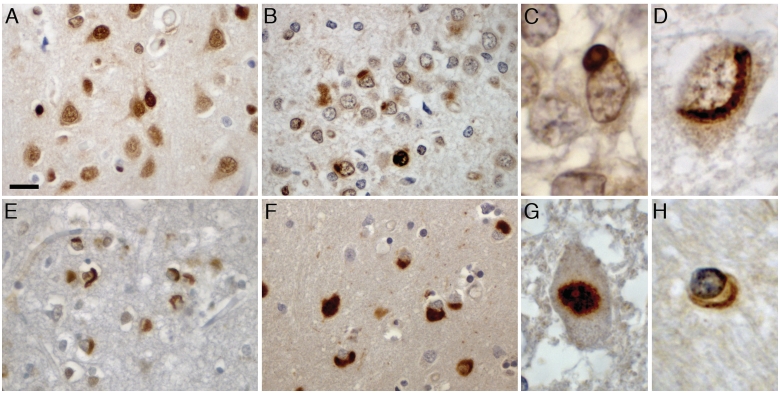
Figure 3.
EWS pathology in FTLD-FUS. EWS immunohistochemistry performed on sections of post-mortem brain tissue from normal control (A), atypical FTLD-U (B–D, G), NIFID (E) and BIBD (F and H). Normal physiological staining pattern of nuclei and diffuse cytoplasmic labelling (A). In atypical FTLD-U, round cytoplasmic and intranuclear inclusions were observed in the dentate granule cells with variable labelling intensity (B). Higher magnification of cytoplasmic (C) and vermiform intranuclear inclusion (D) in atypical FTLD-U. Numerous neuronal cytoplasmic inclusions with variable morphology including round, crescentic, globular and tangle-like showed strong immunoreactivity in NIFID (E) and BIBD (F) as shown here in frontal cortex. Most cases with FTLD-FUS revealed at least rare inclusions in lower motor neurons (G) as well as variable numbers of glial cytoplasmic inclusions in the white matter of affected brain regions (H). Scale bar: A, B, E and F = 25 µm; C, D and H = 5 µm; G = 10 µm.
Double-label immunofluorescence for EWS and FUS confirmed that in atypical FTLD-U only a subset of FUS-positive inclusions also labelled for EWS (Fig. 4A), while most FUS pathology in NIFID and BIBD revealed clear EWS co-localization (Fig. 4B–E). EWS and α-internexin labelled discrete inclusions in the same neurons in NIFID (Supplementary Fig. 2B) in accordance with the FUS and TAF15 results.
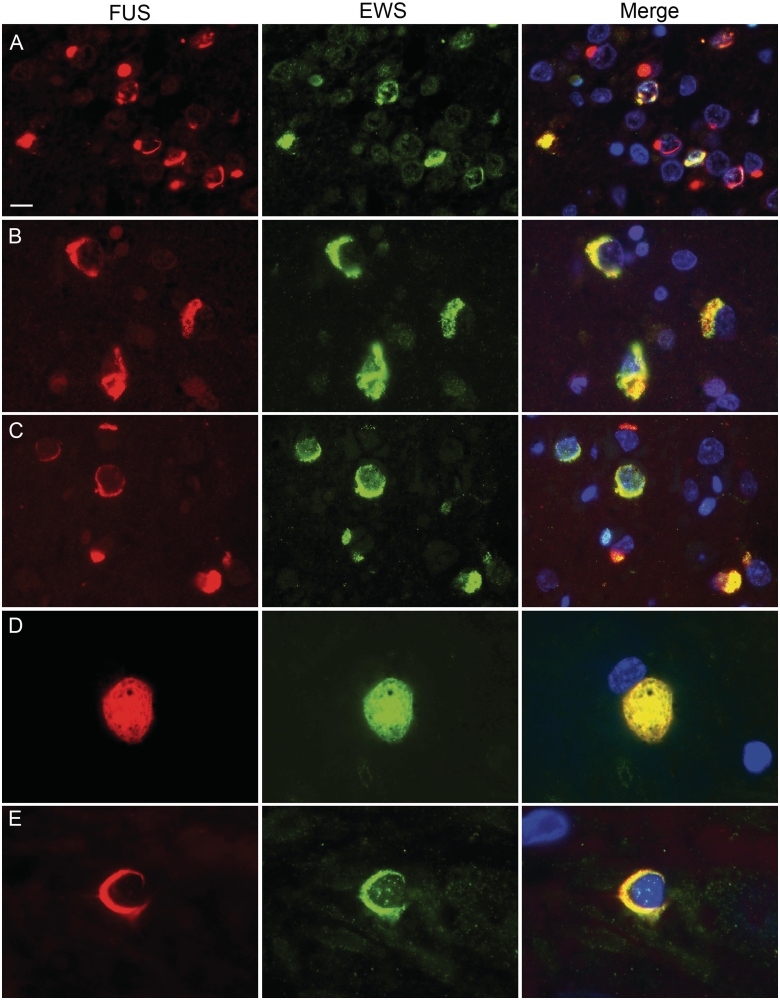
Figure 4.
Co-localization of EWS and FUS in FTLD-FUS inclusions. Double-label immunofluorescence for FUS (red) and EWS (green), with DAPI staining of nuclei in the merged images. In atypical FTLD-U, only a subset of FUS-positive neuronal cytoplasmic and intranuclear inclusions were stained for EWS (A). In contrast, robust co-labelling for EWS and FUS was observed in most inclusions in NIFID (B) and BIBD (C). Inclusions in the lower motor neurons (D, BIBD case) as well as glial cytoplasmic inclusions (E, BIBD case) also showed co-localization. Scale bar: A–C = 10 µm; D = 6.5 µm; E = 4 µm.
Absence of TAF15 and EWS pathology in amyotrophic lateral sclerosis with FUS mutations
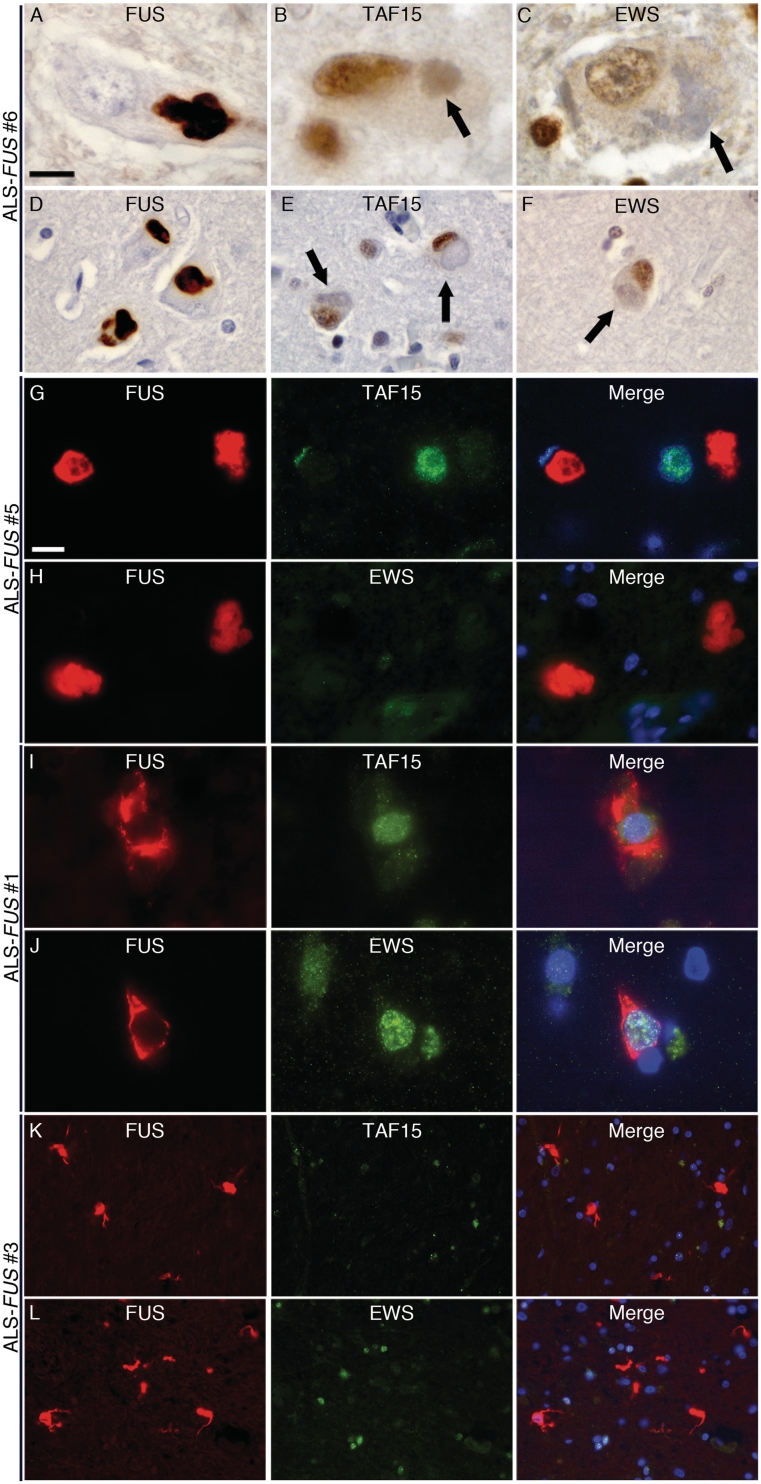
Next, we analysed the pattern of TAF15 and EWS staining in six ALS-FUS cases, which included four different FUS mutations. All cases showed robust FUS pathology, particularly in the spinal cord and motor cortex, with neuronal cytoplasmic inclusions (including basophilic inclusions) as well as variable presence of glial inclusions (Mackenzie et al., 2011b). Interestingly, and in striking contrast to FTLD-FUS, neither TAF15 nor EWS immunohistochemistry demonstrated any neuronal or glial inclusions in cortical, subcortical, brainstem or spinal cord regions in any of the ALS-FUS cases (Fig. 5). The absence of TAF15 and EWS immunoreactivity of FUS-positive inclusions in ALS-FUS was further confirmed by double-label immunofluorescence (Fig. 5G–L). Notably, cells with FUS-immunoreactive inclusions retained their physiological nuclear staining for TAF15 and EWS.
Figure 5.
Absence of TAF15 and EWS pathology in ALS-FUS. Lower (A) and upper (D) motor neurons in all ALS-FUS cases contained at least some cytoplasmic inclusions strongly labelled for FUS; however, no inclusions (including basophilic inclusions, arrows) were labelled for TAF15 (B, lower motor neuron; E, upper motor neuron) or EWS (C, lower motor neuron; F, upper motor neuron). Note the regular nuclear staining for both TAF15 (B and E) and EWS (C and F) in inclusion-bearing cells (arrows). The absence of TAF15 and EWS pathology in ALS-FUS was confirmed by double-label immunofluorescence that showed robust FUS-immunoreactivity of round and tangle-like neuronal inclusions in the spinal cord (red, G–J) that were not labelled for TAF15 (green in G and I) or EWS (green in H and J). In addition, FUS-positive glial cytoplasmic inclusions present in a subset of cases (red, K and L, basal ganglia) showed no co-localization for TAF15 (green, K) or EWS (green, L). Scale bar in A: A–C = 10 µm; D–F = 22 µm. Scale bar in G: G–J = 10 µm; K and L = 30 µm.
TAF15 and EWS immunoreactivity in neurological controls
The normal controls and the majority of neurological controls did not reveal any TAF or EWS pathology (Table 2). Specifically, there was no labelling of the characteristic inclusions in Alzheimer’s disease, Lewy body disease, FTLD with tau pathology, ALS with TDP-43 pathology or ALS due to SOD1 mutations. Inclusions in FTLD with TDP-43 pathology were negative, with the exception of one case that showed a small number of TAF15-positive cortical neurites and EWS staining of a minority of inclusions in the hippocampal dentate granule cells. Glial inclusions in multiple system atrophy were negative for FUS and TAF15; however, one case showed weak EWS labelling. Interestingly, intranuclear inclusions in spinocerebellar ataxia and Huntington’s disease, previously shown to be FUS positive (Doi et al., 2010; Woulfe et al., 2010), were consistently labelled for EWS but not TAF15, while the FUS-positive inclusions in neuronal intranuclear inclusion body disease were negative for both. These findings for intranuclear inclusions are noteworthy in suggesting that different combinations of FET proteins are involved in inclusion formation in a disease-specific fashion and that co-aggregation of all three FET proteins is a specific feature of FTLD-FUS.
Table 2.
Immunoreactivity for FET proteins in other neurodegenerative diseases
| Diagnosis | FUS | TAF15 | EWS |
|---|---|---|---|
| AD | 0/4 | 0/4 | 0/4 |
| FTLD-TDP | 0/17 | 1/17a | 1/17a |
| FTLD with CHMP2B | 0/2 | 0/2 | 0/2 |
| FTLD-tau | 0/8 | 0/8 | 0/8 |
| ALS-TDP | 0/8 | 0/8 | 0/8 |
| ALS with SOD1 | 0/2 | 0/2 | 0/2 |
| MSA | 0/2 | 0/2 | 1/2b |
| LBD | 0/2 | 0/2 | 0/2 |
| SCA | 3/3 | 0/3 | 3/3 |
| HD | 2/2 | 0/2 | 2/2 |
| NIIBD | 1/1 | 0/1 | 0/1 |
a One FTLD-TDP subtype 2 case [according to (Mackenzie et al., 2006)] with semantic dementia showed moderated EWS-immunoreactivity in a subset of neuronal cytoplasmic inclusions in the dentate gyrus and TAF15-immunoreactivity in a small proportion of long neurites.
b One case showed EWS-immunoreactivity in a small proportion of glial cytoplasmic inclusions.
AD = Alzheimer’s disease; ALS-TDP, amyotrophic lateral sclerosis with TDP-43 pathology; ALS with SOD1 = amyotrophic lateral sclerosis due to mutations in SOD1 gene; FTLD-TDP = frontotemporal lobar degeneration with TDP-43 pathology; FTLD-tau, frontotemporal lobar degeneration with tau pathology; FTLD with CHMP2B = frontotemporal lobar degeneration with mutations in CHMP2B gene; HD = Huntington’s disease; LBD = Lewy body disease; MSA = multiple system atrophy; NIIBD = neuronal intranuclear inclusion body disease; SCA = spinocerebellar ataxia.
Biochemical analysis of TAF15 and EWS in frontotemporal lobar degeneration with FUS pathology
A change in the solubility of FUS protein has previously been shown to be a consistent biochemical alteration in atypical FTLD-U (Neumann et al., 2009b) and NIFID (Page et al., 2011). To gain further insight into potential biochemical alterations of TAF15 and EWS, proteins were sequentially extracted from frozen brain tissue from FTLD-FUS, as well as normal and neurological controls, using a series of buffers containing detergents and acids with an increasing ability to solubilize proteins. Unfortunately, sufficient amounts of frozen tissue from ALS-FUS cases were not available for analysis.
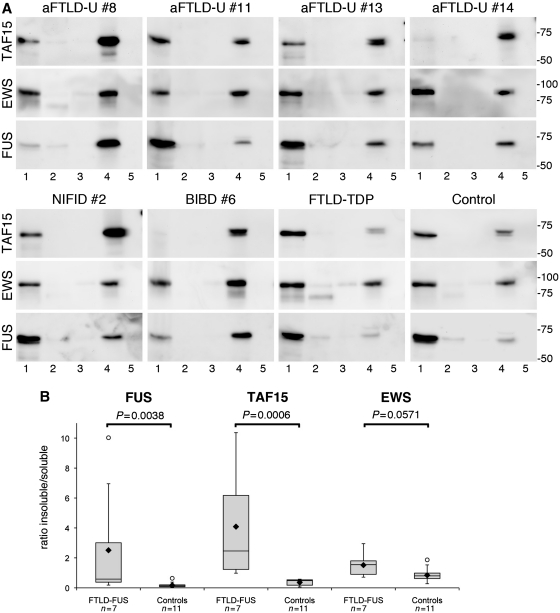
TAF15, EWS and FUS could be detected as major bands at the expected molecular mass of ∼75, ∼90 and ∼73 kDa, respectively, in the high salt (soluble proteins) and sodium dodecyl sulphate (enriched for insoluble proteins) fractions from FTLD-FUS, as well as controls (Fig. 6A). However, remarkable differences were observed in the amount of the proteins in the distinct fractions in FTLD-FUS compared with controls. In accordance with previous findings, a clear shift of FUS towards the insoluble fraction was seen in all FTLD-FUS cases resulting in a significantly higher insoluble : soluble ratio (median 0.58, mean 2.51 ± 3.54), compared with controls (median 0.13, mean 0.17 ± 0.17, P = 0.0038) (Fig. 6B). A similar change in solubility was observed for TAF15 with a significantly higher insoluble : soluble ratio for FTLD-FUS cases (median 2.47, mean 4.09 ± 3.70) compared with controls (median 0.50, mean 0.36 ± 0.21, P = 0.0006). Notably, in some FTLD-FUS cases the shift in solubility was even more pronounced for TAF15 than that observed for FUS (e.g. atypical FTLD-U Case 14 and NIFID Case 2). For EWS, there was a similar tendency for higher levels in the insoluble protein fraction in cases with FTLD-FUS (median 1.55, mean 1.5 ± 0.78) compared with controls (median 0.80, mean ratio = 0.8 ± 0.44); however, the difference did not reach significance.
Figure 6.
Biochemical analysis of FET proteins in FTLD-FUS. (A) Proteins were sequentially extracted from frontal cortex of atypical FTLD-U, NIFID, BIBD, normal as well as neurological controls. High salt (Lane 1), Triton-X-100 (Lane 2), radioimmunoprecipitation assay buffer (Lane 3), 2% sodium dodecyl sulphate (Lane 4) and formic acid (Lane 5) protein fractions were separated by sodium dodecyl sulphate–polyacrylamide gel electrophoresis and immunoblotted with anti-TAF15 (TAF15-309A), EWS (G5) and FUS (FUS-302A). All proteins were present in the soluble high salt fraction and sodium dodecyl sulphate fraction in each case as one major band at the expected molecular size for the full-length proteins. However, the amount of TAF15 and FUS in the sodium dodecyl sulphate fraction was much higher in FTLD-FUS compared with controls, while the shift towards the sodium dodecyl sulphate fraction was less obvious for EWS. (B) Densitometric quantification of band intensities of FUS, TAF15 and EWS in the soluble (high salt) and insoluble (sodium dodecyl sulphate) fraction was performed. Calculated insoluble/soluble ratios for each protein in the FTLD-FUS (n = 7) and control group (n = 11, including four normal controls, five FTLD with TDP-43 pathology and two cases with Alzheimer’s disease) are shown as box plot showing the range of values, with the box being subdivided by the median into the 25th and 75th percentiles. Filled rhombus represents the mean; circles represent outliers. aFTLD-U = atypical FTLD-U.
Despite the change in solubility, there was no evidence of other biochemical alterations of TAF15 and EWS, as indicated by abnormal molecular weight species, using antibodies specific for different TAF15 and EWS epitopes.
Genetic analysis of TAF15 and EWSR1 in frontotemporal lobar degeneration with FUS pathology
Sequence analyses of EWSR1 and TAF15 did not identify any novel coding variants in the eight FTLD-FUS cases with DNA available, with the exception of a 24 base pair deletion in TAF15 exon 15 in atypical FTLD-U Case 13 (c.1674_1697del), predicted to delete eight amino acids (p.G559_Y566del). This particular deletion has not been reported previously; however, similar deletions have been found in controls, suggesting it is likely a benign polymorphism (Ticozzi et al., 2011). Novel non-coding variants identified are summarized in Supplementary Table 3.
Characteristic features of human FUS-opathies are recapitulated in cultured cells
The strikingly different patterns of FET protein immunoreactivity in the pathology of FTLD-FUS versus ALS-FUS, suggest different mechanisms underlying inclusion body formation. To further address this issue we investigated whether the absence of TAF15 and EWS alterations seen in ALS-FUS would be recapitulated in cultured cells expressing mutant FUS.
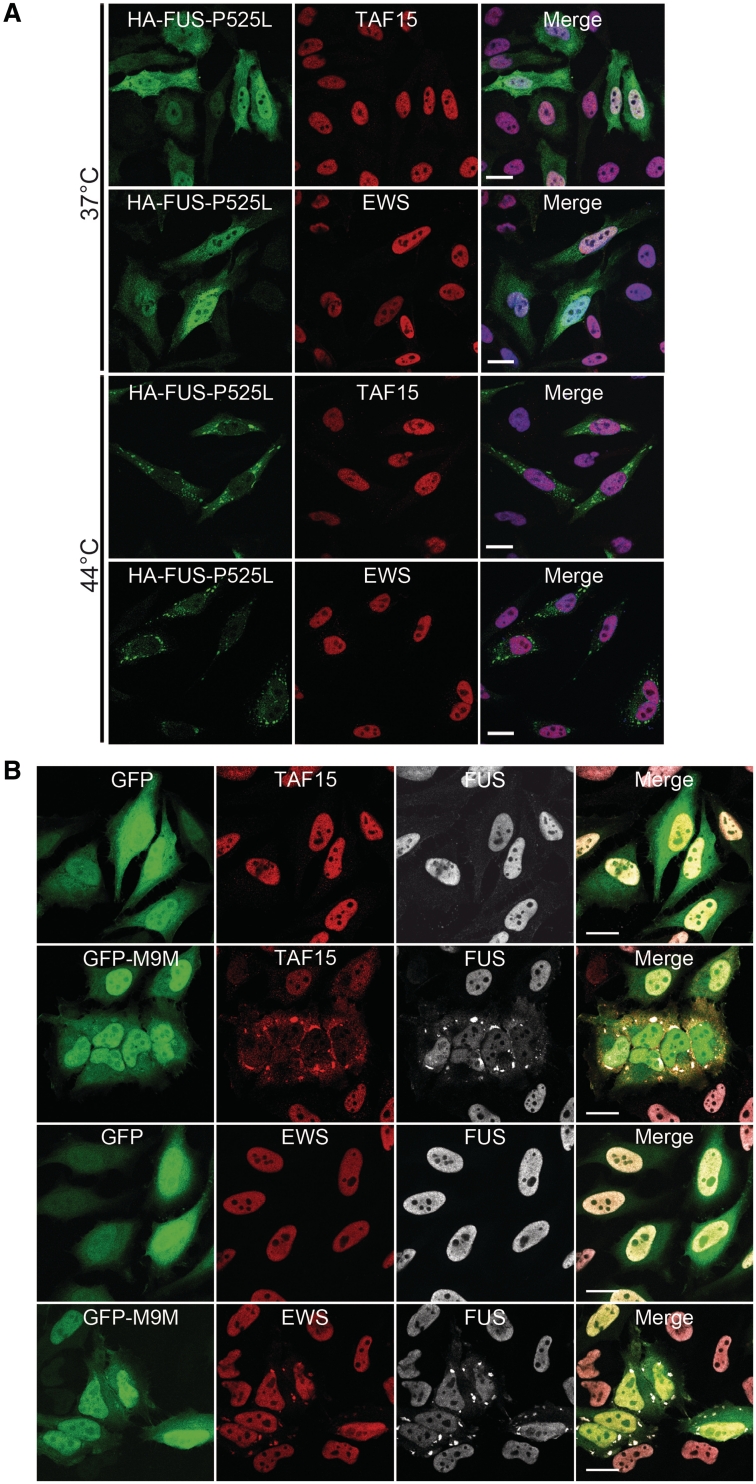
In accordance with previous results (Dormann et al., 2010), HeLa cells expressing FUS with the p.P525L mutation (a mutation present in two of our studied cases with ALS) showed a robust increase of cytoplasmic FUS compared with cells expressing wild-type FUS (Fig. 7A and Supplementary Fig. 3). Under stress conditions of heat shock, cells expressing mutant FUS showed recruitment of FUS into punctuate cytoplasmic structures, corresponding to stress granules. In contrast, these same conditions resulted in no changes in the subcellular distribution of endogenous TAF15 or EWS. Specifically, both proteins remained almost exclusively within the nucleus and there was no recruitment of TAF15 or EWS into FUS-positive stress granules. In this way, cells expressing an ALS-associated FUS mutation recapitulate our findings in human ALS-FUS, demonstrating the absence of other FET protein members in cytoplasmic FUS inclusions.
Figure 7.
Analysis of FET proteins in cell culture systems. (A) Cytoplasmically mislocalized mutant FUS does not sequester TAF15 or EWS into stress granules upon heat shock. HeLa cells transiently transfected with haemagglutinin-tagged human FUS with the P525L mutation (HA-FUS-P525L) were left untreated (37°C, top) or subjected to heat shock (1 h at 44°C, bottom) 24 h after transfection. Cells were stained with antibodies against haemagglutinin (green) and EWS (red) or TAF15 (red) and analysed by confocal microscopy. Under control conditions, HA-FUS-P525L is diffusely distributed in the cytoplasm, and endogenous TAF15 and EWS is localized in the nucleus. Upon heat shock, HA-FUS-P525L is recruited into cytoplasmic stress granules, while TAF15 and EWS remain predominantly nuclear and are not entrapped into FUS-positive stress granules. Scale bar = 20 µm. (B) Inhibition of the Transportin pathway leads to cytoplasmic mislocalization of TAF15, EWS and FUS into stress granules. The Transportin-specific peptide inhibitor M9M fused to green fluorescent protein (GFP-M9M, green) or GFP alone was expressed in HeLa cells for 24 h. Cells were stained with antibodies against TAF15, EWS (both shown in red) and FUS (white) and were analysed using confocal microscopy. Upon inhibition of Transportin-mediated nuclear import by the GFP-M9M peptide, TAF15 and EWS are recruited into cytoplasmic stress granules, where they co-localize with FUS. Note that EWS shows only mild cytoplasmic mislocalization, while FUS and especially TAF15 show a marked cytoplasmic redistribution with a nuclear depletion of these proteins. Scale bar = 20 µm.
To investigate whether the accumulation of all FET proteins in FTLD-FUS might reflect a more general problem of Transportin-mediated nuclear import, we studied the effect on TAF15 and EWS by transfecting HeLa cells with a Transportin-specific competitive inhibitor peptide (M9M) fused to green fluorescent protein (Fig. 7B). Similar to what has been shown previously for FUS (Dormann et al., 2010), a striking redistribution of endogenous TAF15 and EWS proteins to the cytoplasm was observed, that was associated with the formation of stress granules with co-localization of all FET proteins. Notably, recruitment of FUS and TAF15 into stress granules in this system seemed to be more efficient compared with EWS, based on staining intensities of stress granules, recapitulating the differences we observed in the staining intensities of inclusions for FET proteins in atypical FTLD-U. Furthermore, the most obvious reduction of normal nuclear protein levels was found for TAF15, similar to the dramatic decrease in nuclear staining intensity in inclusion bearing cells in FTLD-FUS. Thus, inhibition of Transportin-mediated nuclear import in cultured cells mimics characteristic alterations of FET proteins found in human FTLD-FUS.
Discussion
FUS accumulates in the pathological cellular inclusions that characterize all cases of ALS with FUS mutations and a variety of FTLD subtypes, collectively referred to as FTLD-FUS (Kwiatkowski et al., 2009; Munoz et al., 2009; Neumann et al., 2009a, b; Vance et al., 2009; Mackenzie et al., 2010b). Our knowledge of the underlying mechanisms leading to FUS accumulation and FUS-mediated cell death is still limited. So far, most insights come from studies analysing the functional consequences of FUS mutations. As demonstrated in cell culture experiments, pathogenic FUS mutations interfere with the Transportin-mediated nuclear import, leading to increased levels of cytoplasmic FUS where it is recruited into stress granules upon stress conditions (Dormann et al., 2010; Ito et al., 2011; Kino et al., 2011). Since stress granule markers have been found in FUS-positive inclusions in FTLD-FUS and ALS-FUS, it has been suggested that stress granules might be the precursors of pathological FUS-inclusions (Dormann et al., 2010; Dormann and Haass, 2011).
Although there is some clinical and pathological overlap between ALS-FUS and FTLD-FUS, the presence of significant differences in the phenotypes and the morphological patterns of FUS pathology (Mackenzie et al., 2011b) and the fact that no FTLD-FUS case has yet been associated with a FUS mutation (Neumann et al., 2009a, b; Rohrer et al., 2010; Urwin et al., 2010; Snowden et al., 2011), raise questions as to whether these conditions represent a clinicopathological spectrum of diseases with a shared pathomechanism or whether the pathogenic pathways triggered by FUS mutations may be different from those involved in FTLD-FUS.
In the present study, we performed a detailed analysis of the role of the FUS homologues TAF15 and EWS in the spectrum of FUS-opathies and identified remarkable differences in the protein composition of inclusions between FTLD-FUS and ALS-FUS. These findings strongly support the idea that the pathological processes underlying cell death in ALS-FUS might be different from those in FTLD-FUS.
None of the ALS-FUS cases investigated, including six cases with four different FUS mutations, showed any alteration in the subcellular distribution of TAF15 or EWS and no evidence of co-accumulation of these proteins in the FUS-positive pathological inclusions. Importantly, we confirmed retention of the normal physiological staining pattern and the absence of TAF15 and EWS co-localization in the cytoplasmic FUS pathology (i.e. stress granules) that develops in cultured cells expressing ALS-associated FUS mutations (Dormann et al., 2010). Thus, cytoplasmic accumulation of FUS per se does not trigger an alteration in the subcellular distribution of its homologues and does not lead to sequestration of TAF15 and EWS into FUS inclusions as a secondary phenomenon. This strongly implies that the pathological processes in ALS-FUS are restricted to dysfunctions of FUS. Since the ALS-FUS cases we studied do not cover the entire spectrum of reported FUS mutations, we cannot exclude the possibility that other FUS mutations, particularly those reported in exons 3, 5 or 6 (Mackenzie et al., 2010a) might be associated with TAF15 and/or EWS pathology. However, since our analysis did include two cases with the most common FUS mutation (p.R521C), this is unlikely to be a frequent finding.
In sharp contrast to ALS-FUS, abnormal co-accumulation of all three FET proteins into pathological inclusions was a consistent and specific feature of all subtypes of FTLD-FUS. This finding further extends the similarities between the various subtypes of FTLD-FUS, thereby strongly supporting the idea, that atypical FTLD-U, NIFID and BIBD are closely related disease entities (Mackenzie et al., 2011a). However, our results also suggest some important differences among distinct FET family members in the different FTLD-FUS subtypes. While antibodies against TAF15 robustly labelled virtually all FUS pathology in atypical FTLD-U, NIFID and BIBD, subtle disease-specific differences were observed for EWS. Only a proportion of inclusions in atypical FTLD-U cases labelled for EWS and the staining intensity was often weak. In contrast, inclusions in NIFID and BIBD were more consistently and robustly labelled for EWS. Because the quality of immunostaining obtained with the commercial EWS antibodies employed was not felt to be optimal in all sections, we are cautious in interpreting these results. However, they raise the possibility of subtle differences in the pathogenic pathways involved in the different FTLD-FUS subtypes, that may underlie the distinct clinico-pathological phenotypes previously described (Mackenzie et al., 2011a).
Another difference in the pattern of immunostaining among the FET proteins in FTLD-FUS is worth noting for its potential functional significance. Whereas inclusion bearing cells often demonstrated at least partial retention of nuclear FUS and EWS localization, a dramatic and consistent reduction of physiological nuclear staining was observed for TAF15, suggesting a possible loss-of-function mechanism.
The mechanisms leading to the accumulation of all FET proteins in FTLD-FUS remain unclear. The results in human ALS-FUS and in cultured cells expressing mutant FUS indicates that other FET proteins are not secondarily entrapped within FUS inclusions. An alternate mechanism is suggested by our cell culture data in which inhibition of Transportin-mediated nuclear import resulted in recruitment and co-localization of all FET proteins into stress granules. This favours a scenario in which a broader nuclear import defect in FTLD-FUS leads to increased cytoplasmic levels of all FET proteins (and possibly other proteins), which then predisposes to their abnormal accumulation. Although the underlying defect in nuclear import could reflect a direct dysfunction of the Transportin import machinery, preliminary studies in which we found no alterations in the subcellular distribution of other Transportin cargos, such as hnRNPA1, makes this mechanism more unlikely. Alternatively, altered post-translational modifications of FET proteins, such as phosphorylation or arginine methylation, might affect their subcellular localization and nuclear import in FTLD-FUS (Tan and Manley, 2009; Kovar, 2011). While biochemical analysis has so far revealed only a relative change in solubility for FET proteins (Neumann et al., 2009b and this study), the presence of potential disease-associated post-translational modifications as well as alterations of the transportin machinery requires further studies.
Our findings in FTLD-FUS add TAF15 and EWS to the growing list of DNA/RNA binding proteins involved in neurodegenerative diseases. Despite the fact that we have not detected any pathogenic mutations in TAF15 and EWSR1 in our FTLD-FUS cases, both genes are considered promising candidates for genetic screens in FTLD and ALS and a very recent report has described coding variants in TAF15 in ALS, although their pathogenicity remains to be confirmed (Ticozzi et al., 2011).
In summary, this study demonstrates the co-accumulation of all members of the FET protein family in the characteristic inclusions as specific feature of FTLD-FUS but not of ALS-FUS, thus allowing a clear separation between genetic and non-genetic forms of FUS-opathies by neuropathological features. More importantly, these findings imply that different pathomechanisms underlie inclusion body formation and cell death in ALS-FUS versus FTLD-FUS. Our data indicate that neurodegeneration associated with FUS mutations is probably the result of a restricted dysfunction of FUS, whereas a more complex dysregulation of all FET family members seems to be involved in FTLD-FUS pathogenesis. While the relative roles of the different FET proteins in the disease pathogenesis of FTLD-FUS remain to be determined in future studies, our data suggest that the conditions currently subsumed within the FTLD-FUS molecular subgroup might be more appropriately designated as FTLD-FET, in accordance with the recently proposed system of FTLD nomenclature (Mackenzie et al., 2009).
Supplementary material
Supplementary material is available at Brain online.
Funding
The Swiss National Science Foundation (31003A-132864, M.N.); the Synapsis Foundation (M.N.); the Canadian Institutes of Health Research (74580, I.R.A.M.); the Pacific Alzheimer Research Foundation (I.R.A.M.); the National Institutes of Health (R01 AG26251; R.R.); the ALS Association (R.R.); the Center for Integrated Protein Science Munich (CIPSM; C.H.), the German Federal Ministry of Education and Research (01GI1005B; C.H., M.N.), the Sonderforschungsbereich Molecular Mechanisms of Neurodegeneration (SFB 596; C.H.); and the National Institute for Health Research, Oxford Biomedical Research Centre (O.A.).
Supplementary Material
Acknowledgements
We thank Margaret Luk and Mareike Schroff for their excellent technical assistance.
Glossary
Abbreviations
- ALS
amyotrophic lateral sclerosis
- BIBD
basophilic inclusion body disease
- EWS
Ewing’s sarcoma protein
- FUS
fused in sarcoma
- FTLD
frontotemporal lobar degeneration
- FTLD-U
frontotemporal lobar degeneration with ubiquitin-positive inclusions
- NIFID
neuronal intermediate filament inclusion body disease
- TAF15
TATA-binding protein-associated factor 15
- TDP-43
TAR-DNA binding protein 43 kDa
References
- Blair IP, Williams KL, Warraich ST, Durnall JC, Thoeng AD, Manavis J, et al. FUS mutations in amyotrophic lateral sclerosis: clinical, pathological, neurophysiological and genetic analysis. J Neurol Neurosurg Psychiatry. 2010;81:639–45. doi: 10.1136/jnnp.2009.194399. [DOI] [PubMed] [Google Scholar]
- Doi H, Koyano S, Suzuki Y, Nukina N, Kuroiwa Y. The RNA-binding protein FUS/TLS is a common aggregate-interacting protein in polyglutamine diseases. Neurosci Res. 2010;66:131–3. doi: 10.1016/j.neures.2009.10.004. [DOI] [PubMed] [Google Scholar]
- Dormann D, Rodde R, Edbauer D, Bentmann E, Fischer I, Hruscha A, et al. ALS-associated fused in sarcoma (FUS) mutations disrupt Transportin-mediated nuclear import. EMBO J. 2010;29:2841–57. doi: 10.1038/emboj.2010.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dormann D, Haass C. TDP-43 and FUS – a nuclear affair. Trends Neuroscience. 2011;34:339–48. doi: 10.1016/j.tins.2011.05.002. [DOI] [PubMed] [Google Scholar]
- Groen EJ, van Es MA, van Vught PW, Spliet WG, van Engelen-Lee J, de Visser M, et al. FUS mutations in familial amyotrophic lateral sclerosis in the Netherlands. Arch Neurol. 2010;67:224–30. doi: 10.1001/archneurol.2009.329. [DOI] [PubMed] [Google Scholar]
- Hewitt C, Kirby J, Highley JR, Hartley JA, Hibberd R, Hollinger HC, et al. Novel FUS/TLS mutations and pathology in familial and sporadic amyotrophic lateral sclerosis. Arch Neurol. 2010;67:455–61. doi: 10.1001/archneurol.2010.52. [DOI] [PubMed] [Google Scholar]
- Ito D, Seki M, Tsunoda Y, Uchiyama H, Suzuki N. Nuclear transport impairment of amyotrophic lateral sclerosis-linked mutations in FUS/TLS. Ann Neurol. 2011;69:152–62. doi: 10.1002/ana.22246. [DOI] [PubMed] [Google Scholar]
- Jobert L, Argentini M, Tora L. PRMT1 mediated methylation of TAF15 is required for its positive gene regulatory function. Exp Cell Res. 2009;315:1273–86. doi: 10.1016/j.yexcr.2008.12.008. [DOI] [PubMed] [Google Scholar]
- Kino Y, Washizu C, Aquilanti E, Okuno M, Kurosawa M, Yamada M, et al. Intracellular localization and splicing regulation of FUS/TLS are variably affected by amyotrophic lateral sclerosis-linked mutations. Nucleic Acids Res. 2011;39:2781–98. doi: 10.1093/nar/gkq1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovar H. Dr. Jekyll and Mr. Hyde: the two faces of the FUS/EWS/TAF15 protein family. Sarcoma. 2011;2011:837474. doi: 10.1155/2011/837474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwiatkowski TJ, Jr, Bosco DA, Leclerc AL, Tamrazian E, Vanderburg CR, Russ C, et al. Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science. 2009;323:1205–8. doi: 10.1126/science.1166066. [DOI] [PubMed] [Google Scholar]
- Law WJ, Cann KL, Hicks GG. TLS, EWS and TAF15: a model for transcriptional integration of gene expression. Brief Funct Genomic Proteomic. 2006;5:8–14. doi: 10.1093/bfgp/ell015. [DOI] [PubMed] [Google Scholar]
- Lee BJ, Cansizoglu AE, Suel KE, Louis TH, Zhang Z, Chook YM. Rules for nuclear localization sequence recognition by karyopherin beta 2. Cell. 2006;126:543–58. doi: 10.1016/j.cell.2006.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie IR, Baborie A, Pickering-Brown S, Plessis DD, Jaros E, Perry RH, et al. Heterogeneity of ubiquitin pathology in frontotemporal lobar degeneration: classification and relation to clinical phenotype. Acta Neuropathol. 2006;112:539–49. doi: 10.1007/s00401-006-0138-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie IR, Neumann M, Bigio EH, Cairns NJ, Alafuzoff I, Kril J, et al. Nomenclature for neuropathologic subtypes of frontotemporal lobar degeneration: consensus recommendations. Acta Neuropathol. 2009;117:15–8. doi: 10.1007/s00401-008-0460-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie IR, Rademakers R, Neumann M. TDP-43 and FUS in amyotrophic lateral sclerosis and frontotemporal dementia. Lancet Neurol. 2010a;9:995–1007. doi: 10.1016/S1474-4422(10)70195-2. [DOI] [PubMed] [Google Scholar]
- Mackenzie IR, Neumann M, Bigio EH, Cairns NJ, Alafuzoff I, Kril J, et al. Nomenclature and nosology for neuropathologic subtypes of frontotemporal lobar degeneration: an update. Acta Neuropathol. 2010b;119:1–4. doi: 10.1007/s00401-009-0612-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie IR, Munoz DG, Kusaka H, Yokota O, Ishihara K, Roeber S, et al. Distinct pathological subtypes of FTLD-FUS. Acta Neuropathol. 2011a;121:207–18. doi: 10.1007/s00401-010-0764-0. [DOI] [PubMed] [Google Scholar]
- Mackenzie IRA, Ansorge O, Strong M, Bilbao J, Zinman L, Ang LC, et al. Pathological heterogeneity in amyotrophic lateral sclerosis with FUS mutations: two distinct patterns correlating with disease severity and mutation. Acta Neuropathol. 2011b;122:87–98. doi: 10.1007/s00401-011-0838-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz DG, Neumann M, Kusaka H, Yokota O, Ishihara K, Terada S, et al. FUS pathology in basophilic inclusion body disease. Acta Neuropathol. 2009;118:617–27. doi: 10.1007/s00401-009-0598-9. [DOI] [PubMed] [Google Scholar]
- Neumann M, Sampathu DM, Kwong LK, Truax AC, Micsenyi MC, Chou TT, et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314:130–3. doi: 10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- Neumann M, Roeber S, Kretzschmar HA, Rademakers R, Baker M, Mackenzie IR. Abundant FUS-immunoreactive pathology in neuronal intermediate filament inclusion disease. Acta Neuropathol. 2009a;118:605–16. doi: 10.1007/s00401-009-0581-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann M, Rademakers R, Roeber S, Baker M, Kretzschmar HA, Mackenzie IR. A new subtype of frontotemporal lobar degeneration with FUS pathology. Brain. 2009b;132:2922–31. doi: 10.1093/brain/awp214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page T, Gitcho MA, Mosaheb M, Carter D, Chakraverty S, Perry RH, et al. FUS immunogold labelling TEM analysis of the neuronal cytoplasmic inclusions of neuronal intermediate filament inclusion disease: a frontotemporal lobar degeneration with FUS proteinopathy. J Mol Neurosci. 2011 doi: 10.1007/s12031-011-9549-8. [Epub ahead of print] advance access date: May 21, doi:10.1007/s12031-011-9549-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pahlich S, Zakaryan RP, Gehring H. Identification of proteins interacting with protein arginine methyltransferase 8: the Ewing sarcoma (EWS) protein binds independent of its methylation state. Proteins. 2008;72:1125–37. doi: 10.1002/prot.22004. [DOI] [PubMed] [Google Scholar]
- Rademakers R, Stewart H, DeJesus-Hernandez M, Krieger C, Graff-Radford N, Fabros M, et al. FUS gene mutations in familial and sporadic amyotrophic lateral sclerosis. Muscle Nerve. 2010;42:170–6. doi: 10.1002/mus.21665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohrer JD, Lashley T, Holton J, Revesz T, Urwin H, Isaacs AM, et al. The clinical and neuroanatomical phenotype of FUS associated frontotemporal lobar degeneration. J Neurol Neurosurg Psychiatry. 2010 doi: 10.1136/jnnp.2010.214437. .[Epub ahead of print] advance access date: July 16, doi:10.1136/jnnp.2010.214437. [DOI] [PubMed] [Google Scholar]
- Snowden JS, Hu Q, Rollinson S, Halliwell N, Robinson A, Davidson YS, et al. The most common type of FTLD-FUS (aFTLD-U) is associated with a distinct clinical form of frontotemporal dementia but is not related to mutations in the FUS gene. Acta Neuropathol. 2011;122:99–110. doi: 10.1007/s00401-011-0816-0. [DOI] [PubMed] [Google Scholar]
- Tan AY, Manley JL. The TET family of proteins: functions and roles in disease. J Mol Cell Biol. 2009;1:82–92. doi: 10.1093/jmcb/mjp025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ticozzi N, Vance C, Leclerc AL, Keagle P, Glass JD, McKenna-Yasek D, et al. Mutational analysis reveals the FUS homolog TAF15 as a candidate gene for familial amyotrophic lateral sclerosis. Am J Med Genet B Neuropsychiatr Genet. 2011;156:285–90. doi: 10.1002/ajmg.b.31158. [DOI] [PubMed] [Google Scholar]
- Urwin H, Josephs KA, Rohrer JD, Mackenzie IR, Neumann M, Authier A, et al. FUS pathology defines the majority of tau- and TDP-43-negative frontotemporal lobar degeneration. Acta Neuropathol. 2010;120:33–41. doi: 10.1007/s00401-010-0698-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance C, Rogelj B, Hortobagyi T, De Vos KJ, Nishimura AL, Sreedharan J, et al. Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science. 2009;323:1208–11. doi: 10.1126/science.1165942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woulfe J, Gray DA, Mackenzie IR. FUS-immunoreactive intranuclear inclusions in neurodegenerative disease. Brain Pathol. 2010;20:589–97. doi: 10.1111/j.1750-3639.2009.00337.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakaryan RP, Gehring H. Identification and characterization of the nuclear localization/retention signal in the EWS proto-oncoprotein. J Mol Biol. 2006;363:27–38. doi: 10.1016/j.jmb.2006.08.018. [DOI] [PubMed] [Google Scholar]
- Zinszner H, Sok J, Immanuel D, Yin Y, Ron D. TLS (FUS) binds RNA in vivo and engages in nucleo-cytoplasmic shuttling. J Cell Sci. 1997;110:1741–50. doi: 10.1242/jcs.110.15.1741. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.