Abstract
The liver X receptors (LXRs) are members of the nuclear hormone receptor superfamily that are bound and activated by oxysterols. These receptors serve as sterol sensors to regulate the transcription of gene products that control intracellular cholesterol homeostasis through catabolism and transport. In this report, we describe a novel LXR target, the sterol regulatory element-binding protein-1c gene (SREBP-1c), which encodes a membrane-bound transcription factor of the basic helix-loop-helix-leucine zipper family. SREBP-1c expression was markedly increased in mouse tissues in an LXR-dependent manner by dietary cholesterol and synthetic agonists for both LXR and its heterodimer partner, the retinoid X receptor (RXR). Expression of the related gene products, SREBP-1a and SREBP-2, were not increased. Analysis of the mouse SREBP-1c gene promoter revealed an RXR/LXR DNA-binding site that is essential for this regulation. The transcriptional increase in SREBP-1c mRNA by RXR/LXR was accompanied by a similar increase in the level of the nuclear, active form of the SREBP-1c protein and an increase in fatty acid synthesis. Because this active form of SREBP-1c controls the transcription of genes involved in fatty acid biosynthesis, our results reveal a unique regulatory interplay between cholesterol and fatty acid metabolism.
Keywords: LXR, RXR, cholesterol, fatty acids, SREBP-1, nuclear receptor
The liver X receptors (LXRs) were originally identified as orphan members of the nuclear hormone receptor superfamily of transcription factors, as their cognate ligands were unknown. LXRα (NR1H3, also known as RLD-1) is highly expressed in liver, intestine, kidney, and adipose (Willy et al. 1995; Repa and Mangelsdorf 2000). LXRβ (NR1H2, also known as UR, NER1, OR-1, and RIP15) is found in nearly all tissues examined (Teboul et al. 1995; Repa and Mangelsdorf 2000). Early studies revealed that LXRs bind DNA as obligate heterodimers with retinoid X receptors (RXRs), a family of nuclear receptors that are activated by their endogenous ligand 9-cis retinoic acid and highly specific synthetic agonists, called rexinoids (Boehm et al. 1995). The RXR/LXR heterodimer belongs to a subclass of nuclear receptor heterodimers that are permissive to ligand-induced transactivation of target genes by the RXR partner, which facilitated the early characterization of this orphan receptor heterodimer (Willy and Mangelsdorf 1997). RXR/LXR binds with high affinity to a DNA sequence comprised of two direct repeats of a hexanucleotide motif (AGGTCA) separated by four nucleotides, now commonly referred to as an LXRE of the DR4 type.
A major advance in the study of LXRs came with the identification of their ligands as a specific class of naturally occurring oxysterols, most arising as metabolic derivatives of cholesterol (Janowski et al. 1996, 1999; Lehmann et al. 1997). The most potent physiologic ligands include 24(S),25-epoxycholesterol, which is present in liver; 22(R)-hydroxycholesterol, which is found in abundance in adrenal gland; and 24(S)-hydroxycholesterol, also known as cerebrosterol because of its high levels in brain. In a variety of assays, these ligands were found to bind and activate LXRα and β in a specific, saturable manner at concentrations consistent with those found in tissues in which cholesterol metabolism and LXR expression are high (Janowski et al. 1999; Repa and Mangelsdorf 2000; Schroepfer 2000). Recently, a synthetic nonsteroidal LXR agonist has also been described (Repa et al. 2000; Schultz et al. 2000).
The pattern of expression of LXRs and their oxysterol ligands first suggested a role for LXRs in cholesterol metabolism. Indeed, the first target gene identified for LXRα was cholesterol 7α-hydroxylase (CYP7A1), which encodes the rate-limiting enzyme in the classic pathway of bile acid biosynthesis (Lehmann et al. 1997; Peet et al. 1998). Mice subjected to high cholesterol diets or synthetic agonists of LXR showed marked increases in CYP7A1 expression, resulting in increased production and fecal elimination of cholesterol-derived bile acids (Peet et al. 1998; Repa et al. 2000). Recently, the ATP-binding cassette transporters, ABC1 and ABC8, which are implicated in the flux of cellular free cholesterol, have been found to be under the transcriptional regulation of the RXR/LXR heterodimer (Costet et al. 2000; Repa et al. 2000; Venkateswaran et al. 2000). Cholesterol ester transfer protein (CETP), which has been implicated in reverse cholesterol transport has also been reported to be a direct LXR target gene (Luo and Tall 2000). These findings suggest that LXRs may serve as sterol sensors by responding to elevated oxysterol concentrations and up-regulating the expression of key genes responsible for enhancing cholesterol catabolism and transport.
The identification of oxysterol-regulated genes dependent on RXR/LXR has been greatly facilitated by studies in mouse strains devoid of LXRα and/or LXRβ (Peet et al. 1998; Repa et al. 2000; Venkateswaran et al. 2000), and the availability of synthetic RXR and LXR agonists (Boehm et al. 1995; Repa et al. 2000). The initial characterization of the LXRα-null mouse strain had previously shown that liver mRNA levels for several genes involved in fatty acid synthesis are reduced compared with levels in control mice (Peet et al. 1998). In addition, we and others have noted that one of the major pharmacologic responses of animals treated with rexinoids or LXR agonists is a significant increase in serum triglyceride levels (Miller et al. 1997; Schultz et al. 2000).
In this report, we reveal the mechanism responsible for the RXR/LXR-dependent regulation of lipogenesis in mice. We show that sterol regulatory element-binding protein-1c (SREBP-1c), a transcription factor known to regulate the expression of lipogenic enzymes (Kim and Spiegelman 1996; Shimano et al. 1997a; Horton and Shimomura 1999; Shimano et al. 1999), is a direct target gene of RXR/LXR. In mice lacking the LXR genes, the basal expression level of liver SREBP-1c mRNA is significantly reduced and, in parallel, so are the mRNA levels of lipogenic enzymes known to be affected by SREBP-1c. In addition, feeding diets containing high cholesterol, rexinoids or LXR agonists results in an increase in SREBP-1c mRNA and protein expression in wild-type mice, but not in Lxrα/β−/− mice. Finally examination of the 5′-flanking region of the mouse SREBP-1c gene revealed an LXRE motif necessary for the RXR/LXR-mediated up-regulation of expression. These studies establish mouse SREBP-1c as a target gene of the oxysterol receptor, LXR, and suggest a novel convergence of homeostatic mechanisms for cholesterol and fatty acid metabolism.
Results
SREBP-1 expression and fatty acid synthesis in LXR-deficient mice
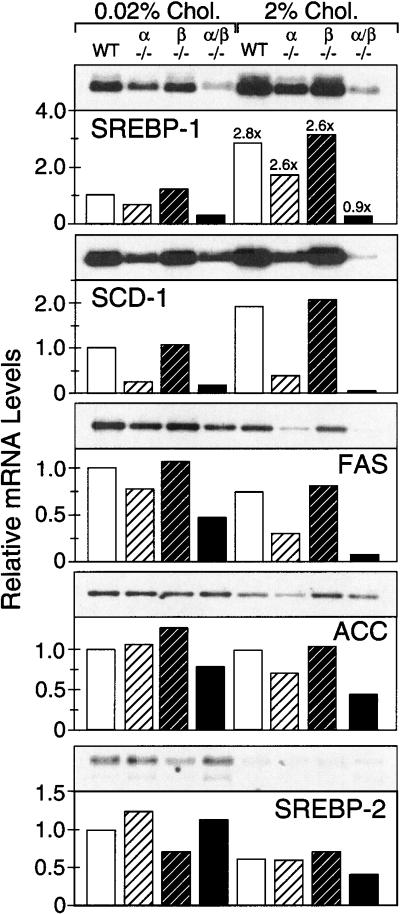
As observed previously, the expression of liver SREBP-1 mRNA was reduced in LXRα null mice fed a standard rodent diet (Fig.1; Peet et al. 1998). No such change was seen in mice lacking only the LXRβ gene, but a profound reduction in SREBP-1 mRNA was shown by Lxrα/β−/− mice (Fig. 1). When fed a high cholesterol diet for seven days, mice harboring at least one LXR gene showed a nearly threefold increase in SREBP-1 mRNA relative to chow-fed animals of a similar genotype. A modest increase in liver SREBP-1 mRNA levels was also observed within the first day of feeding wild-type mice a 2% cholesterol diet (Y. Zhang and D.J. Mangelsdorf, unpubl.). Mice lacking both LXRα and LXRβ, however, failed to show this dietary cholesterol-mediated induction in SREBP-1 expression.
Figure 1.
Effect of dietary cholesterol on the liver expression of SREBPs and lipogenic enzymes in wild-type and Lxr-knockout mice. Mixed strain (A129/C57Bl6) male mice of various Lxr genotypes were fed basal (0.02% cholesterol) or high cholesterol (2% cholesterol) diets for seven days. Liver mRNA was pooled from six animals per treatment group and Northern analysis performed using probes for SREBP-1 and -2, SCD-1, FAS, and ACC (Shimano et al. 1996). All RNA levels are expressed relative to wild-type mice on basal diet. Numbers above bars refer to relative fold changes in mRNA levels between animals of the same genotype after high cholesterol feeding. Genotypes denoted are: wild-type (WT); Lxrα−/− (α−/−); Lxrβ−/− (β−/−); and Lxrα/β−/− (α/β−/−).
Examination of several lipogenic target genes of SREBP-1 revealed similar changes in expression (Fig. 1). Stearoyl CoA desaturase (SCD) mRNA was coordinately reduced in Lxrα−/− and Lxrα/β−/− mice fed a basal diet and modestly induced by cholesterol feeding in all mice except the Lxr double-knockout strain. The SCD cDNA probe used in these northern analyses does not distinguish SCD-1 and SCD-2. However, liver from mice fed standard basal diets is reported to express SCD-1 exclusively (Kaestner et al. 1989), and only under conditions in which elevated levels of the nuclear, active SREBPs are present in mouse liver does SCD-2 mRNA become barely detectable (Korn et al. 1998). Therefore, the changes observed in SCD expression most likely reflect alterations in SCD-1 mRNA levels. The expression levels of fatty acid synthase (FAS) and acetyl CoA carboxylase (ACC) mRNA were not elevated by cholesterol feeding but continued to show a relative decrease in Lxrα−/− and Lxrα/β−/− mice fed either low- or high-cholesterol diets. In agreement with the observed changes in RNA levels of lipogenic enzymes, determination of fatty acid synthesis rates in organs of chow-fed Lxrα−/− mice and their wild-type littermates by a whole animal in vivo [3H]water incorporation technique (Shimano et al. 1996) revealed that fatty acid synthesis was reduced 75% in liver, 25% in kidney, and 35% in small intestine compared with wild-type mice. Furthermore, these studies showed that the incorporation of C18:1 and C16:1 fatty acids into phospholipids and triglycerides was significantly reduced (J.D. Horton, D.J. Peet, and D.J. Mangelsdorf, unpubl.). SREBP-2 mRNA levels were reduced by cholesterol feeding as previously reported (Shimomura et al. 1997a) and did not show any consistent alteration according to LXR genotype.
RXR and LXR agonists induce SREBP-1 expression in vivo
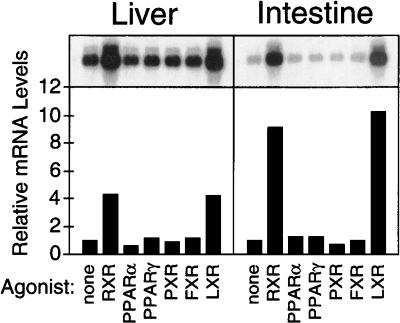
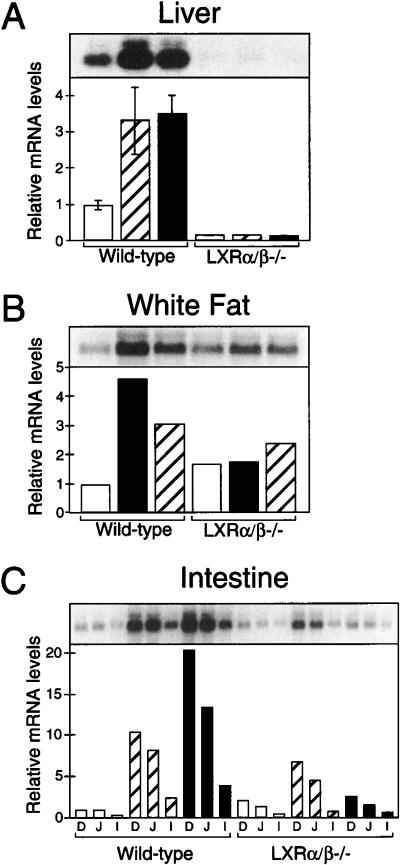
The dietary cholesterol-induced up-regulation of SREBP-1 expression that was observed only in mice harboring LXR strongly suggested that SREBP-1 is a primary target gene of the RXR/LXR heterodimer. To confirm this hypothesis, wild-type mice were treated with synthetic agonists of a variety of nuclear hormone receptors for 12 h (Fig. 2). This brief, oral administration of the RXR ligand LG268 (Boehm et al. 1995) or the LXR agonist T0901317 (Repa et al. 2000; Schultz et al. 2000) was sufficient to induce the expression of SREBP-1 mRNA in liver (greater than fourfold) and intestine (10-fold). Up-regulation of SREBP-1 expression was also observed in intestine within 6 h of administering LG268 (data not shown). No other nuclear receptor agonists elicited significant changes in SREBP-1 expression, even when these compounds were supplied at doses previously shown to alter expression of their respective nuclear receptor target genes (Repa et al. 2000). To confirm that LG268 and T0901317 were acting through an RXR/LXR-dependent pathway, similar experiments were performed in wild-type and Lxrα/β−/− mice (Fig. 3). Again, increased SREBP-1 mRNA levels were observed in the liver, white adipose, and intestine of wild-type mice receiving these agonists. However, LG268 and T0901317 were ineffective in inducing SREBP-1 in the Lxr double-knockout mice, except for a modest increase by LG268 in the small intestine. Interestingly, the basal level of SREBP-1 mRNA in Lxrα/β−/− mice receiving the vehicle control was reduced in liver but slightly elevated in adipose and intestine.
Figure 2.
Effect of nuclear receptor agonists on SREBP-1 mRNA expression. Male mice were fed diets containing 0.2% cholesterol plus vehicle or the following agonists for 12 h: 30 mpk LG268 (RXR), 1000 mpk fenofibrate (PPARα), 150 mpk troglitazone (PPARγ), 100 mpk pregnenolone α-carbonitrile (PXR), 1000 mpk chenodeoxycholic acid (FXR), or 50 mpk T0901317 (LXR). Northern analysis was performed on mRNA from liver or duodenal mucosa (n = 4 animals/group) using a SREBP-1 probe.
Figure 3.
Regulation of SREBP-1 mRNA by RXR and LXR agonists in liver (A), white adipose tissue (B), and small intestine (C). Male wild-type and Lxrα/β−/− mice were fed diets containing 0.2% cholesterol plus vehicle (open bars), 30 mpk LG268 (hatched bars), or 50 mpk T0901317 (black bars) for 12 h. Northern analysis was performed on mRNA individually (A) or as a pooled sample consisting of three to four animals per group (B,C). Relative mRNA levels in liver (A) are expressed as the mean ± SEM, and a representative Northern blot is shown above. Small intestine mRNA was isolated from the mucosa of the duodenum (D), jejunum (J), and ileum (I).
SREBP-1c mRNA and protein are regulated by RXR/LXR
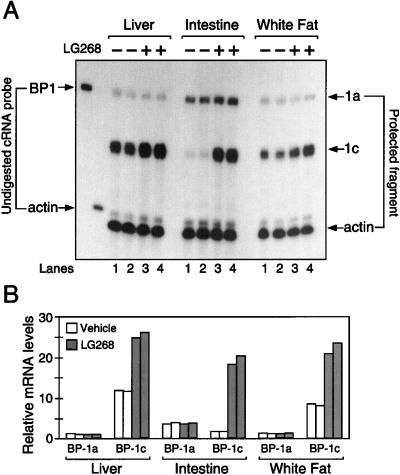
Two SREBP-1 isoforms are derived from a single gene by the use of two distinct transcriptional start sites under the control of separate promoters and are alternatively spliced to yield the SREBP-1a isoform (containing 29 amino acids derived from exon 1) or the SREBP-1c isoform (containing only five amino acids encoded by exon 1) (Yokoyama et al. 1993; Hua et al. 1995), which is also called ADD1 (Tontonoz et al. 1993). Northern analyses such as those depicted in Figures 1 through 3 do not distinguish SREBP-1a and SREBP-1c. Therefore, a previously described RNase protection assay (Shimomura et al. 1997b) was used to differentiate between the two mouse SREBP-1 mRNA isoforms (Fig. 4A; see Materials and Methods). Oral administration of the RXR agonist for 16 to 18 h resulted in the induction of only SREBP-1c expression in liver, intestine, and white fat of wild-type mice (Fig. 4A,B) to a degree similar to that observed previously by Northern analysis. No change in the mRNA levels of SREBP-1a were detected. Kidney SREBP-1c mRNA was also increased by LG268 or T0901317 by threefold in wild-type mice, but not in Lxrα/β−/− mice (data not shown).
Figure 4.
Expression of SREBP-1c mRNA is regulated by rexinoid. (A) Three mice per group were fed diets containing 0.2% cholesterol plus vehicle (−) or 30 mpk LG268 (+) for 16–18 h. Pooled RNA was isolated from liver, intestine, and epididymal white adipose depots from each group. Aliquots of the pooled RNA (20μg) were subjected to RNase protection assays using a 32P-labeled cRNA probe that can distinguish between 1a and 1c isoforms of SREBP-1 (BP1). RNA samples from two independent studies were examined in the same blot. Lanes 1 and 3 contain the samples from one experiment; lanes 2 and 4 are from an independent experiment. (B) Data in A were quantified as described in Materials and Methods and normalized relative to the β-actin signal. The data for SREBP-1a and SREBP-1c mRNA are plotted as the fold change relative to the SREBP1a mRNA level in the liver of the first vehicle group. Autoradiography was performed for 8 h at −80°C on Kodak X-Omat (XB-1) film with an intensifying screen.
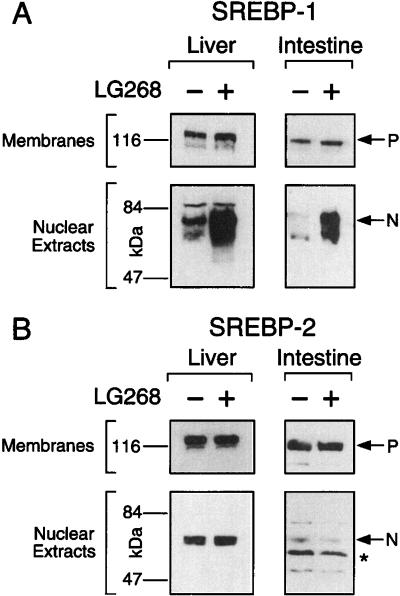
SREBPs are synthesized as ∼120 kD precursor proteins that are bound to the endoplasmic reticulum and nuclear envelope (Brown and Goldstein 1997). Under low cellular sterol conditions, two sequential proteolytic cleavage reactions occur to liberate an ∼70-kD amino-terminal fragment that translocates to the nucleus to activate gene transcription (Brown and Goldstein 1999). Immunoblot analyses of membrane and nuclear extracts of liver and intestine of LG268-treated mice revealed that the increase in SREBP-1c mRNA by the RXR/LXR heterodimer is reflected in the amount of the active nuclear form of SREBP-1c protein that serves as a transcription factor (Fig. 5A). This elevation in the nuclear active form of SREBP-1c would account for the up-regulation of expression for lipogenic enzymes regulated by SREBP-1 (Fig. 1). The relative increase in the nuclear form of SREBP-1 protein mirrored the change observed previously in the mRNA levels in these tissues from mice receiving rexinoids as measured by Northern analysis. The similar increase in the levels of SREBP-1 mRNA and the processed, nuclear form of SREBP-1 protein suggests that the enzymatic activities of the SREBP proteases are not rate-limiting under these experimental conditions, since the amount of the unprocessed, precursor form of SREBP-1 does not change. SREBP-2 protein levels were unaffected in these mice (Fig. 5B).
Figure 5.
Regulation of SREBP-1 protein expression by rexinoid. Immunoblot analysis of SREBP-1 and SREBP-2 was performed on fractionated cell extracts from liver and intestine of wild-type male mice fed diets containing 0.2% cholesterol plus vehicle (−) or 30 mpk LG268 (+) for 18 h. P and N denote the precursor and cleaved nuclear forms of SREBP, respectively, and the asterisk indicates a nonspecific band. The filters for liver and intestine were exposed to film at room temperature for 15 sec and 45 sec, respectively.
Identification of an LXRE in the mouse SREBP-1c promoter
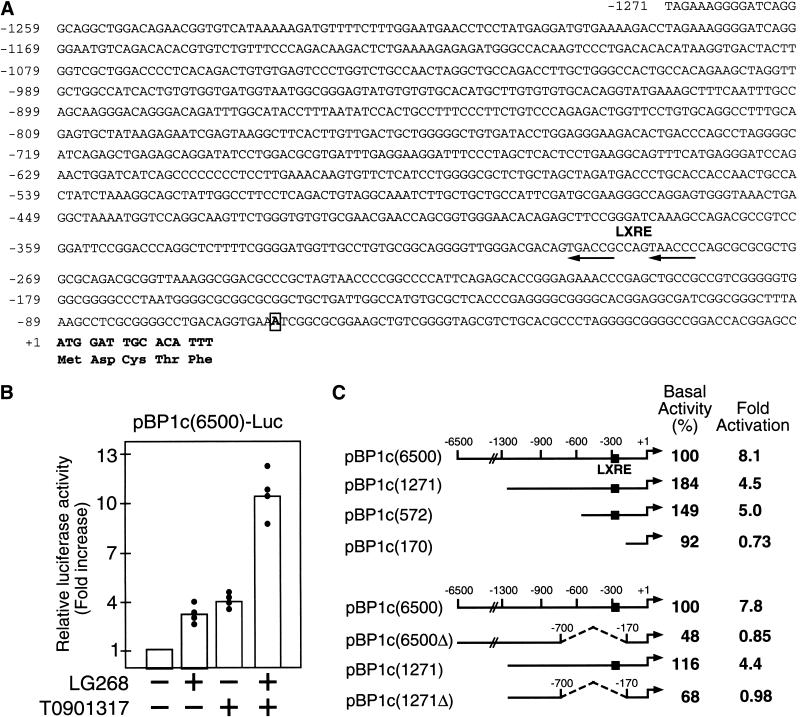
To investigate the molecular mechanism of RXR/LXR-mediated regulation of mouse SREBP-1c the 5′-flanking region of this gene was sequenced and analyzed (Fig. 6A). A 6.5-kb genomic fragment of the mouse SREBP-1c promoter, including the transcription start site, was placed before a luciferase reporter gene to test for RXR/LXR responsiveness in a cell-based reporter assay (Fig 6B). When this reporter plasmid was introduced into HEK293 cells, luciferase activities were increased three- to fourfold on incubating the cells with either LG268 or T0901317 and greater than 10-fold when both synthetic agonists were present. Similar results were observed in primary hepatocytes transfected with the pBP1c6500 luciferase reporter and treated with LG268 and T0901317 (data not shown). Serial 5′ truncations of the SREBP-1c promoter revealed that the majority of ligand-induced activation required sequences between nucleotides −572 and −170 (Fig. 6C). Deletion analyses confirmed the importance of this region for RXR/LXR regulation in the context of the full promoter (6.5 kb) and the shorter version depicted in Figure 6A (1.3 kb). Inspection of this region of the mouse SREBP-1c promoter revealed a putative LXRE of the DR4 type (arrows in Fig. 6A).
Figure 6.
RXR and LXR agonists activate mouse SREBP-1c promoter. (A) The sequence of the mouse SREBP-1c 5′-flanking region (∼1.3 kb) is shown with the DR4 LXRE motif denoted by arrows. The putative transcription start site at −63 is boxed. (B) RXR and LXR agonists (LG268 and T0901317) synergistically activate a luciferase reporter driven by the SREBP-1c promoter. A 6.5-kb fragment of the SREBP-1c promoter was linked to pGL3 basic luciferase reporter. The resulting plasmid pBP1c(6500)-Luc was cotransfected into HEK-293 cells with a control reporter plasmid pCMV-βGal as described in Materials and Methods. Twenty hours after transfection, cells were treated with vehicle (DMSO and/or ethanol), LG268 (1 μM), T0901317 (10 μM), or LG268 (1 μM) plus T0901317 (10 μM). Normalized luciferase activity of cells treated with vehicle is arbitrarily defined as 1, and relative luciferase activities from different treatments are shown as “fold increase”. Each value represents data from four independent transfection experiments (each in duplicate). (C) Deletions of the ∼6500 kb mouse SREBP-1c promoter were assayed as described in B. Data shown are the average of three independent transfection experiments (each in duplicate) in the upper panel and two independent experiments in the lower panel. The 100% value for basal activity corresponds to the normalized luciferase activity obtained with the pBP1c(6500)-Luc promoter construct in the absence of LG268 and T0901317. Individual values for fold-activations were: upper panel 8.1 (4.9, 7.4, 12), 4.5 (3.8, 4.8, 5.0), 5.0 (2.4, 5.5, 7.0), 0.73 (0.4, 0.8, 1.0); lower panel 7.8 (5.1, 10.5), 0.85 (0.6, 1.1), 4.4 (4, 4.8), 0.98 (0.95, 1.0).
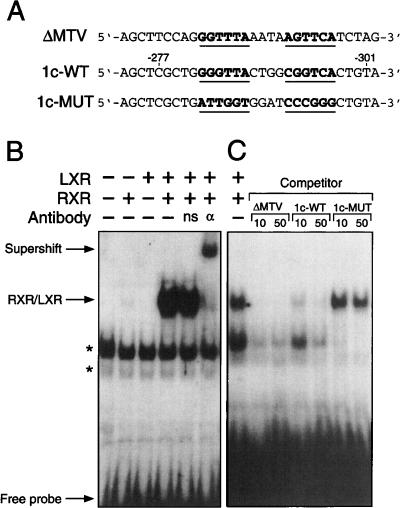
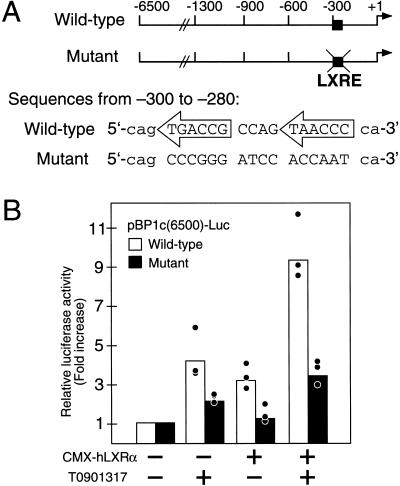
The ability of RXR/LXR to specifically bind to the SREBP-1c LXRE was tested by electrophoretic mobility shift assay (Fig. 7). The wild-type LXRE (1c-WT; Fig. 7A) was radiolabeled with [32P]dCTP and incubated with RXRα and/or LXRα, then separated from free 1c-WT probe by gel electrophoresis. Only in the presence of both receptors was a specific complex of retarded mobility observed (Fig. 7B). The presence of RXRα was confirmed by the supershift of this complex following the addition of an antibody against RXRα to the incubation mixture (Fig. 7B). The specificity of binding was shown by the ability of a well-characterized nonradiolabeled LXRE (ΔMTV; Fig. 7A; Willy et al. 1995) or the SREBP-1c LXRE (1c-WT) added in excess to compete for binding with the radiolabeled probe, thereby eliminating the detectable shifted complex (Fig. 7C). A mutated form of the SREBP-1c LXRE (1c-MUT; Fig. 7A), in which the hexanucleotides recognized by RXR and LXR were randomly scrambled, was ineffective in competing for binding with the radiolabeled 1c-WT probe (Fig. 7C). This same mutation of the SREBP-1c LXRE in the context of the full 6.5-kb promoter (Fig. 8A) eliminated nearly all RXR/LXR-mediated up-regulation of luciferase expression in the cell reporter assay (Fig. 8B). This effect was more pronounced when additional LXRα was provided to the HEK293 cells (Fig. 8B). These in vitro assays (Figs. 6–8) in conjunction with the whole animal analyses (Figs. 1–5) show that mouse SREBP-1c is a direct target gene of the RXR/LXR heterodimer.
Figure 7.
The LXRE in mouse SREBP-1c promoter is a high affinity binding site for RXR/LXR heterodimer. (A) Sequences of the LXRE derived from the mouse mammary tumor virus LTR (ΔMTV-LXRE), wild-type mouse SREBP-1c promoter region (1c-WT), and mutant mouse SREBP-1c promoter region (1c-MUT). For comparison, the LXRE sequences of the bottom strands of 1c-WT and 1c-MUT are aligned with the top strand sequence of ΔMTV. (B) Electrophoretic-mobility shift assays were performed as described in Materials and Methods. The 32P-labeled 1c-WT LXRE probe was incubated with in vitro synthesized human LXRα and RXRα proteins as indicated. For antibody supershift experiments, anti-RXRα antibody (αRXR) or a non-specific antibody (ns) were used. Free probe, RXR/LXR complex, and antibody-supershifted RXR/LXR complex are denoted by arrows. Nonspecific bands are indicated by asterisks. (C) Competition gel mobility-shift assays were performed as described in Materials and Methods using 32P-labeled 1c-WT LXRE as input probe and unlabeled oligonucleotides as competitors at 10- and 50-fold molar excesses. Autoradiography was performed for 16 h at −80°C with an intensifying screen.
Figure 8.
Mutation of the LXRE sequence abolishes activation of mouse SREBP-1c promoter by RXR/LXR. (A) Schematic diagrams of pBP1c(6500)-Luc wild-type and mutant reporter plasmids. Differences between wild-type and mutant plasmids (nucleotides −300 to −280) are shown for comparison. (B) Activation of wild-type, but not mutant, LXRE reporter plasmids. Wild-type and mutant pBP1c(6500)-Luc were cotransfected into HEK-293 cells. For each 60 mm dish 0.5 μg pBP1c-Luc, 25 ng, CMV-βgal and CMX-LXRα or pCDNA3 were used. Three hours after transfection, cells were switched to medium A containing vehicle (DMSO) or 10 μM T0901317, and assayed as described in Figure 6. Data represent three independent transfection experiments (each in duplicate).
Discussion
The work presented here describes the first report of nutrient-mediated regulation of SREBP-1c transcription defined at a molecular level: Cholesterol-derived oxysterols activate the RXR/LXR heterodimer to induce expression of the mouse SREBP-1c gene through an LXRE located in its proximal promoter. This finding was confirmed by experiments using RXR- and LXR-specific synthetic agonists and mouse strains of defined Lxr genotype, and by the identification and characterization of the LXRE in the 5′-flanking region of the mouse SREBP-1c gene responsible for this transcriptional regulation.
SREBP-1c regulation of fatty acids
Numerous reports of nutrient regulation of SREBP-1c have appeared in the recent literature (for review, see Osborne 2000). Investigators have identified a reduction in SREBP-1c mRNA in mouse liver and adipose with fasting and a restoration or overshoot of SREBP-1c mRNA levels on refeeding mice a high-carbohydrate diet (Horton et al. 1998; Kim et al. 1998). This overshoot in SREBP-1c expression may be caused by a recently identified SRE in the promoter of mouse SREBP-1c that would allow for a feedforward autoregulation of this gene on SREBP-1c induction (Amemiya-Kudo et al. 2000). Interestingly, when Lxrα/β−/− and wild-type mice are subjected to this fasting/refeeding regimen, SREBP-1c mRNA levels are similarly affected in both mouse strains, indicating that LXRs are not involved in this regulatory pathway (data not shown).
Insulin/glucose have been strongly implicated as the mediators of the refeeding-induced up-regulation of SREBP-1c transcription, although their precise molecular mechanism of action has not yet been defined. When isolated rat adipocytes or hepatocytes are treated with insulin, a transcriptional increase in SREBP-1c/ADD1 is observed (Kim et al. 1998; Foretz et al. 1999). Likewise, when rats are made diabetic by the administration of streptozotocin, liver SREBP-1c mRNA levels are reduced and can be restored with insulin administration (Shimomura et al. 1999b).
Specific polyunsaturated fatty acids or fish oil have also been used in animal feeding studies, with variable results, suggesting that these nutrients may reduce SREBP-1 mRNA levels or affect SREBP-1 protein processing (Kim et al. 1999; Yahagi et al. 1999). These latter data have recently been confirmed by demonstrations that unsaturated fatty acids repress both SREBP-1 transcription and proteolytic processing in HEK293 cells (V.C. Hannah, J. Ou, A. Luong, J. Goldstein, and M.S. Brown, in prep.).
Hypertriglyceridemia and RXR/LXR agonists: the role of SREBP-1c
The consequence of SREBP-1c up-regulation has been extensively investigated in cells and mice that overexpress the active, nuclear form of this transcription factor (Kim and Spiegelman 1996; Shimano et al. 1997a; Horton et al. 1998b; Pai et al. 1998). Increased production of either the SREBP-1a or SREBP-1c transcription factor results in the enhanced expression of genes encoding enzymes involved in lipogenesis (for review, see Horton and Shimomura 1999; Osborne 2000). SREBP-1c, which contains a shorter, less negatively charged activation domain, is not as active as SREBP-1a (Shimano et al. 1997a); however, SREBP-1c is the predominant isoform found in most tissues (Shimomura et al. 1997b). Ultimately, the result of SREBP-1c production is an increase in fatty acid synthesis and development of a fatty liver (Shimano et al. 1997a; Shimomura et al. 1999a).
Further evidence that SREBP-1c is integral to increased liver fatty acid synthesis was shown by the failure of SREBP1−/− mice to up-regulate the expression of lipogenic enzymes when subjected to a fasting/refeeding treatment (Shimano et al. 1999). Consistent with an LXR-mediated up-regulation of SREBP-1c, liver triglyceride concentrations are elevated in normal mice following long-term cholesterol feedings or when treated with rexinoids and LXR agonists (Schultz et al. 2000). This accumulation of triglyceride by the feeding of cholesterol or RXR/LXR acting drugs is absent in mice lacking the LXRs (Peet et al. 1998; Schultz et al. 2000) or mice that harbor a hepatocyte-specific deletion of RXRα (Wan et al. 2000). Thus, the RXR/LXR-mediated up-regulation of fatty acid synthesis through SREBP-1c provides an explanation for the hypertriglyceridemia seen in animals and patients receiving rexinoid therapy (Miller et al. 1997). Interestingly, in one study in hamsters (Shimomura et al. 1997a) dietary cholesterol did not lead to an increase in SREBP-1, suggesting that the response to endogenous LXR ligands is weak or absent in this species. Consistent with this hypothesis we note that the hamster also fails to up-regulate CYP7A1, another known LXR target gene, in response to dietary cholesterol (Horton et al. 1995). Nevertheless, administration of synthetic RXR or LXR agonists to hamsters results in potent hypertriglyceridemia, suggesting that the LXR-mediated SREBP-1 induction of lipogenesis is conserved in this species (Schultz et al. 2000). The basis for this species variation is currently under investigation.
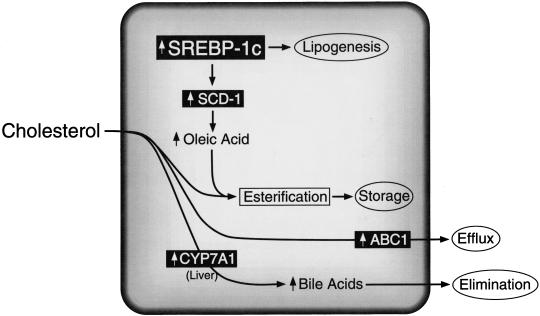
Model of LXR action as a cholesterol sensor
Stearoyl CoA desaturase-1 is a target gene of SREBP-1c (Shimano et al. 1997a; Tabor et al. 1999). This enzyme is responsible for the Δ9-cis desaturation of stearoyl-CoA and palmitoyl-CoA, converting them to oleoyl-CoA and palmitoleoyl-CoA, respectively. Early studies of cholesterol-fed rats suggested that SCD expression was enhanced by an indirect means (Garg et al. 1986). The current studies would suggest that cholesterol feeding results in LXR-mediated up-regulation of SREBP-1c and subsequent SREBP-1c-directed induction of SCD-1. The role of SCD-1 in cholesterol homeostasis was recently shown in mice harboring a null mutation in this gene. SCD1−/− mice showed impaired production of oleic acid and a diminished capacity to esterify cholesterol for hepatic storage and packaging into VLDL for export to other tissues (Miyazaki et al. 2000). This notion is consistent with unpublished data demonstrating C18:1 fatty acid synthesis is significantly reduced in LXR knockout mice (J.D. Horton, D.J Peet, and D.J. Mangelsdorf, unpubl.). The benefit of up-regulating SCD-1 would be to increase oleoyl-CoA, the preferred substrate for ACAT-mediated cholesterol esterification (Landau et al. 1997). Thus, under conditions of high cholesterol, LXR would indirectly promote the esterification of free cholesterol to protect the cell from its harmful effects (Fig. 9). In addition, the increase in fatty acid synthesis under these conditions would promote a more appropriate ratio of cholesterol to other lipids to maintain cell membrane integrity (Fig. 9).
Figure 9.
Model depicting LXR-activated target genes that regulate cellular cholesterol metabolism. Cholesterol-derived oxysterols function as LXR ligands to regulate the expression of RXR/LXR target genes (shown in black boxes), which, in turn, modulate intracellular free cholesterol levels through increased elimination, efflux, and storage. The identification of SREBP-1c as an RXR/LXR-regulated gene links cholesterol and fatty acid metabolism, perhaps as a means for the cell to achieve the appropriate ratio of cholesterol to other lipids and thereby maintain cellular membrane integrity. LXR-regulated genes include cholesterol 7α-hydroxylase (CYP7A1; Peet et al. 1998), the ATP-binding cassette transporter-1 (ABC1; Repa et al. 2000), and sterol regulatory element binding protein 1c (SREBP-1c), which in turn governs the expression of stearoyl CoA desaturase (SCD-1).
In summary, the identification of SREBP-1c as a direct target of LXR action further solidifies the role of LXRs as sterol sensors that function to ameliorate the effects of high free cholesterol levels (Fig. 9). In the liver, LXR up-regulates the cholesterol 7α-hydroxylase (CYP7A1) gene to promote production and elimination of bile acids (Peet et al. 1998). In macrophages and enterocytes, LXR-mediated expression of ABC1 facilitates the efflux of free cholesterol from the cell (Repa et al. 2000). Finally, in this report it is shown that oxysterol/LXR-mediated up-regulation of SREBP-1c occurs in numerous cell types to promote lipid synthesis to coordinate a homeostatic balance between fatty acids and sterols.
Materials and methods
Animals and diets
All experiments were performed on age-matched, male mice of 2.5 to 4 months of age. Lxr knockout mice were generated as described (Peet et al.1998; Lobaccaro and Mangelsdorf, unpubl.), and genotypes were confirmed by Southern and Northern analyses. Animals were housed in temperature-controlled rooms (22°C) with 12-hr light/dark cycles. Mice were fed ad libitum a cereal-based powdered diet (Teklad 7001, Harlan Teklad) supplemented with any of the following: cholesterol (ICN Biomedicals); chenodeoxycholic acid, fenofibrate, or pregnenolone α-carbonitrile (Sigma), LG268 (Ligand Pharmaceuticals) provided in a vehicle containing 0.9% carboxy methyl cellulose, 9% polyethylene glycol 400, and 0.05% Tween 80; T0901317 (Repa et al. 2000); or troglitazone (kindly provided by Dr. Roger Unger, University of Texas Southwestern Medical Center, Dallas). Diets were supplemented with these agents at a level sufficient to provide the appropriate miligrams per kiligram body weight (mpk) dose on consumption of 5 g diet by a 25 g mouse per day. Experiments were performed with the approval of the Institutional Animal Care and Research Advisory Committee at the University of Texas Southwestern Medical Center.
Northern blotting
At the completion of feeding studies, mice were anesthetized and exsanguinated. Liver and epididymal adipose samples were flash-frozen in liquid nitrogen and stored at −80°C until RNA was prepared. Small intestine was removed and flushed with cold phosphate-buffered saline; divided into three equal lengths designated duodenum (proximal), jejunum (medial), and ileum (distal); and the mucosa was carefully removed and placed in a tube for freezing and storage prior to RNA preparation. Total RNA was extracted using RNA STAT-60 (Tel-Test), and mRNA was obtained using oligo(dT)-cellulose columns (Pharmacia Biotech). mRNA was prepared by pooling equal amounts of total RNA from three to six mice per treatment group or from individual mouse liver total RNA samples. mRNA (5 μg/lane) was size fractionated on 1% formaldehyde agarose gels and transferred to nylon membrane (Zetaprobe, BioRad) for probing with 32P-labeled cDNAs (Shimano et al. 1996) as previously described (Peet et al. 1998). Signals were quantitated by PhosphorImager (Molecular Dynamics) and normalized against actin.
RNase protection assay
The cRNA probe to compare the amounts of mRNA for mouse SREBP-1a and SREBP-1c was generated as described (Shimomura et al. 1999b). The cDNA fragment for mouse SREBP-1a was amplified by RT–PCR from first-strand cDNA using mouse liver poly(A)+ RNA as a template and primers derived from the mouse SREBP-1a sequence: 5′ primer, 5′-GCCGGCGCCATGGACGAGCTGGCC-3′ and 3′ primer, 5′-CAGGAAGGCTTCCAGAGAGGAGGC-3′ (Shimomura et al. 1997b). The ends of the 5′- and 3′-primers contain HindIII and EcoRI sites, respectively. The amplified fragment contains exon 1a (specific for SREBP-1a) and part of exon 2 (common to SREBP-1a and SREBP-1c). The cDNA fragment was subcloned into the pGEM-3Zf(+) vector (Promega). After linearization of plasmid DNA with HindIII, antisense cRNA probe was generated using bacteriophage T7 RNA polymerase (Ambion) in the presence of [α-32P]CTP (20 mCi/mL).
Aliquots of total RNA (20 μg) from each sample were subjected to the RNase protection assay with a HybSpeed RPA kit (Ambion). Each assay tube contained a cRNA probe for the mRNA of SREBP-1a plus cRNA probe for β-actin. In preparing the probes, the specific activity of the [α-32P]CTP was adjusted to give an actin signal comparable to the SREBP-1 mRNAs. After digestion by RNase A/T1, protected fragments were separated on 8 M urea/4.8 % polyacrylamide gels and subjected to autoradiography using reflection film and intensifying screens (Dupont). Protected fragments corresponding to mouse SREBP-1a (262 bp) and SREBP-1c (168 bp) were analyzed quantitatively with a Fuji PhosphorImager. Levels of β-actin in each RNA sample were used to normalize signals obtained for the SREBP-1 mRNAs. For comparison of mouse SREBP-1a and SREBP-1c mRNA levels, the results were corrected for the difference in number of 32P-labeled CTP molecules in the protected fragments of mouse SREBP-1a and SREBP-1c (72 and 39 cytidines, respectively).
Immunoblotting
Rabbit polyclonal antibodies against mouse SREBP-1 and –2 were prepared as described (Shimano et al. 1996). Mouse liver or intestinal mucosa was immediately Dounce homogenized; nuclear extracts and membrane fractions (105g pellet) were prepared; and 30μg aliquots of protein were size fractionated by 8% SDS-PAGE and transferred to Hybond C extra membrane (Amersham) as described (Shimano et al. 1996). The membrane was incubated with 5 μg/mL of rabbit antimouse SREBP-1 IgG or 5 μg/mL rabbit antimouse SREBP-2 IgG. Immunoblot analysis was performed with the Enhanced Chemiluminescence (ECL) western blotting kit (Amersham), using a horseradish peroxidase-conjugated donkey anti-rabbit secondary IgG (Amersham).
Plasmids
Human LXRα and RXRα expression vectors pCMX-hLXRα and pCMX-hRXRα have been described (Willy et al. 1995). Isolation of genomic DNA containing mouse SREBP-1 gene has been described (Shimano et al. 1997b). An XhoI/NcoI fragment (1.3 kb) containing the mSREBP-1c promoter (from −1271 to ATG of mSREBP-1c) was cloned into pGL3 basic vector (Promega) to generate pBP1c(1271)-Luc. The adjacent upstream 5.2-kb SacI/XhoI fragment containing SREBP-1c promoter from −6500 to −1271 was then cloned into pBP1c(1271)-Luc to generate pBP1c(6500)-Luc. To generate pBP1c(572)-Luc, pBP1c(1271)Luc was digested with NheI to release a 700-bp fragment, and the remaining 5.4-kb fragment was relegated to create pBP1c(572)-Luc. To generate pBP1c(170)-Luc, pBP1c(572)-Luc was digested with NheI/ApaI to release a 400-bp fragment, the remaining 5-kb fragment was then treated with Mung Bean nuclease and relegated to create pBP1c(170)-Luc. To generate pBP1c(1271Δ)-Luc, pBP1c(1271)-Luc was first digested with EcoRV/ApaI to release the 530-bp fragment containing SREBP-1c promoter from −700 to −170. The remaining 5.5-kb fragment was then treated with Mung Bean nuclease and relegated to create pBP1c(1271Δ)-Luc. The 5.2-kb SacI/XhoI fragment containing the SREBP-1c promoter region from −6500 to −1271 was cloned into pBP1c(1271Δ)-Luc to generate pBP1c(6500Δ)-Luc. pBP1c (6500)-Luc(mut) is the same as pBP1c(6500)-Luc(wt) except that the BP1c-LXRE was mutated from 5′-TGACCGCCAGTAACCC-3′ to 5′-CCCGGGATCCACCAAT-3′ using QuickChange Site-Directed Mutagenesis Kit (Stratagene). Complementary primers used in the mutagenesis reaction were 5′GGGTTGGGACGACAGCCCGGGA TCCACCAATCAGCGCGCGCTGGC-3′ and 5′-GCCAGCGC GCGCTGATTGGTGGATCCCGGGCTGTCGTCCCAAC CC-3′.Mutagenesis was performed using pBP1c(1271)-Luc as a template to create pBP1c(1271)-Luc (mut) first. The 5.2-kb SacI/XhoI fragment containing the SREBP-1c promoter from −6500 to −1271 was then cloned into pBP1c(1271)-Luc(mut) to generate pBP1c(6500)-Luc(mut). All constructs were confirmed by sequencing.
Cell culture and transfection assays
Human embryonic kidney-293 (HEK-293) cells were maintained at 37°C in an atmosphere of 8.8% CO2 in medium A (DMEM containing 100 U/mL penicillin and 100 μg/mL streptomycin sulfate) supplemented with 10% fetal calf serum (FCS). On day 0, HEK-293 cells were plated at 4 × 105 cells per 60-mm dish in medium A. On day 2, the medium was switched to medium A supplemented with 6% MBS (Stratagene) and cotransfected with 0.5 μg of the indicated pBP1c-Luc reporter plasmid and 25 ng CMV-βGal using the MBS kit (Stratagene) according to the manufacturer's protocol. Three hours after transfection, the medium was switched to medium A supplemented with 10% FCS except for the experiment depicted in Figure 8 (see legend). After incubation for 20 h, the medium was changed to medium A containing LG268 and/or T0901317 as indicated. T0901317 and LG268 were added to cells at 103-fold dilution in DMSO and ethanol, respectively. Cells were harvested 24 h after addition of ligand and analyzed for luciferase and β-galactosidase activity using commercial kits (Promega and Clontech, respectively). β-galactosidase activity was used to normalize the luciferase activity.
Electrophoretic mobility-shift assays
hLXRα and hRXRα proteins were generated from expression vectors (Willy et al. 1995), using a coupled in vitro transcription/translation system (Promega). Sequences of double-stranded oligonucleotides shown in Figure 7 were synthesized with HindIII overhangs. Oligonucleotide probes were labeled by end-filling, using AMV reverse transcriptase (Boehringer) and Redivue [α-32P]dCTP (6000 Ci/mmol, Amersham Pharmacia). Binding reactions were performed in a total volume of 20 μL consisting of 75 mM of KCl, 20 of mM Hepes (pH 7.4), 2 mM of dithiothreitol, 7.5% (v/v) glycerol, 0.1% (v/v) NP-40, 2 μg of poly[d(I-C)] (Amersham Pharmacia), and 2 μL of nuclear receptor lysates or unprogrammed lysates. Reactions containing lysates were incubated for 10 min at room temperature, followed by the addition of 40 fmol of 32P-labeled probe, and further incubated for 20 min. Samples were then loaded onto 5% polyacryamide gels (1:40 crosslink) and run in 0.5× TBE buffer (200 V; 3 h for 15 cm × 20 cm gel at 4°C). For antibody supershift experiments, 1 μL of anti-hRXRα antibody (Santa Cruz) or nonspecific antibody (anti-Myc from Invitrogen) was added to the reaction after the addition of probe and incubated at room temperature for another 10 min before electrophoresis. For oligonucleotide competition experiments, competing cold oligonucleotides were added to the reaction with the radiolabeled probe at 10- or 50-fold molar excesses. After electrophoresis, the gels were dried and autoradiographed with intensifying screens at −80°C for 16 h.
Acknowledgments
We acknowledge the excellent technical assistance provided by Heather Lawrence, Chuanzhen Wu, Lisa Beatty, and Jill Abadia and the invaluable help of Jeff Cormier and Lorena Avila with DNA sequencing. We thank Dr. Rich Heyman for LG268. We thank Drs. D.J. Peet and J.D. Horton for sharing unpublished data on liver fatty acid composition in mice. D.J.M. is an Associate Investigator, and J.J.R. and J.M.L. are Research Associates of the Howard Hughes Medical Institute (HHMI). This work was funded by HHMI and grants from the Human Frontier Science Program and the Robert A. Welch Foundation (D.J.M.) and the National Institutes of Health (GM20948; M.S.B. and J.L.G.).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL davo.mango@utsouthwestern.edu; FAX (214) 648-5419.
Article and publication are at www.genesdev.org/cgi/doi/10.1101/gad.844900.
References
- Amemiya-Kudo M, Shimano H, Yoshikawa T, Yahagi N, Hasty AH, Okazaki H, Tamura Y, Shionoiri F, Iizuka Y, Ohashi K, et al. Promoter analysis of the mouse sterol regulatory element-binding protein-1c gene. J Biol Chem. 2000;275:31078–31085. doi: 10.1074/jbc.M005353200. [DOI] [PubMed] [Google Scholar]
- Boehm MF, Zhang L, McClurg MR, Berger E, Wagoner M, Mais DE, Suto CM, Davies PJA, Heyman RA, Nadzan AM. Design and synthesis of potent retinoid X receptor selective ligands that induce apoptosis in leukemia cells. J Med Chem. 1995;38:3146–3155. doi: 10.1021/jm00016a018. [DOI] [PubMed] [Google Scholar]
- Brown MS, Goldstein JL. The SREBP pathway: Regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell. 1997;89:331–340. doi: 10.1016/s0092-8674(00)80213-5. [DOI] [PubMed] [Google Scholar]
- ————— A proteolytic pathway that controls the cholesterol content of membranes, cells, and blood. Proc Natl Acad Sci. 1999;96:11041–11048. doi: 10.1073/pnas.96.20.11041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costet P, Luo Y, Wang N, Tall AR. Sterol-dependent transactivation of the ABC1 promoter by LXR/RXR. J Biol Chem. 2000;275:28240–28245. doi: 10.1074/jbc.M003337200. [DOI] [PubMed] [Google Scholar]
- Foretz M, Guichard C, Ferré P, Foufelle F. Sterol regulatory element binding protein-1c is a major mediator of insulin action on the hepatic expression of glucokinase and lipogenesis-related genes. Proc Natl Acad Sci. 1999;96:12737–12742. doi: 10.1073/pnas.96.22.12737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg ML, Snoswell AM, Sabine JR. Influence of dietary cholesterol on desaturase enzymes of rat liver microsomes. Prog Lipid Res. 1986;25:639–644. doi: 10.1016/0163-7827(86)90131-1. [DOI] [PubMed] [Google Scholar]
- Horton JD, Cuthbert JA, Spady DK. Regulation of hepatic 7α-hydroxylase expression and response to dietary cholesterol in the rat and hamster. J Biol Chem. 1995;270:5381–5387. doi: 10.1074/jbc.270.10.5381. [DOI] [PubMed] [Google Scholar]
- Horton JD, Bashmakov Y, Shimomura I, Shimano H. Regulation of sterol regulatory element binding proteins in livers of fasted and refed mice. Proc Natl Acad Sci. 1998a;95:5987–5992. doi: 10.1073/pnas.95.11.5987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton JD, Shimomura I, Brown MS, Hammer RE, Goldstein JL, Shimano H. Activation of cholesterol synthesis in preference to fatty acid synthesis in liver and adipose tissue of transgenic mice overproducing sterol regulatory element-binding protein-2. J Clin Invest. 1998b;101:2331–2339. doi: 10.1172/JCI2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton JD, Shimomura I. Sterol regulatory element-binding proteins: Activators of cholesterol and fatty acid biosynthesis. Curr Opin Lipidol. 1999;10:143–150. doi: 10.1097/00041433-199904000-00008. [DOI] [PubMed] [Google Scholar]
- Hua X, Wu J, Goldstein JL, Brown MS, Hobbs HH. Structure of the human gene encoding sterol regulatory element binding protein-1 (SREBF1) and localization of SREBF1 and SREBF2 to chromosomes 17p11.2 and 22q13. Genomics. 1995;25:667–673. doi: 10.1016/0888-7543(95)80009-b. [DOI] [PubMed] [Google Scholar]
- Janowski BA, Willy PJ, Devi TR, Falck JR, Mangelsdorf DJ. An oxysterol signalling pathway mediated by the nuclear receptor LXRα. Nature. 1996;383:728–731. doi: 10.1038/383728a0. [DOI] [PubMed] [Google Scholar]
- Janowski BA, Grogan MJ, Jones SA, Wisely GB, Kliewer SA, Corey EJ, Mangelsdorf DJ. Structural requirements of ligands for the oxysterol liver X receptors LXRα and LXRβ. Proc Natl Acad Sci. 1999;96:266–271. doi: 10.1073/pnas.96.1.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaestner KH, Ntambi JM, Kelly TJ, Lane MD. Differentiation-induced gene expression in 3T3-L1 preadipocytes. A second differentially expressed gene encoding stearoyl-CoA desaturase. J Biol Chem. 1989;264:14755–14761. [PubMed] [Google Scholar]
- Kim H-J, Takahashi M, Ezaki O. Fish oil feeding decreases mature sterol regulatory element-binding protein 1 (SREBP-1) by down-regulation of SREBP-1c mRNA in mouse liver. J Biol Chem. 1999;274:25892–25898. doi: 10.1074/jbc.274.36.25892. [DOI] [PubMed] [Google Scholar]
- Kim JB, Spiegelman BM. ADD1/SREBP1 promotes adipocyte differentiation and gene expression linked to fatty acid metabolism. Genes & Dev. 1996;10:1096–1107. doi: 10.1101/gad.10.9.1096. [DOI] [PubMed] [Google Scholar]
- Kim JB, Sarraf P, Wright M, Yao KM, Mueller E, Solanes G, Lowell BB, Spiegelman BM. Nutritional and insulin regulation of fatty acid synthetase and leptin gene expression through ADD1/SREBP1. J Clin Invest. 1998;101:1–9. doi: 10.1172/JCI1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korn BS, Shimomura I, Bashmakov Y, Hammer RE, Horton JD, Goldstein JL, Brown MS. Blunted feedback suppression of SREBP processing by dietary cholesterol in transgenic mice expressing sterol-resistant SCAP(D443N) J Clin Invest. 1998;102:2050–2060. doi: 10.1172/JCI5341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landau JM, Sekowski A, Hamm MW. Dietary cholesterol and the activity of stearoyl CoA desaturase in rats: Evidence for an indirect regulatory effect. Biochim Biophys Acta. 1997;1345:349–357. doi: 10.1016/s0005-2760(97)00010-6. [DOI] [PubMed] [Google Scholar]
- Lehmann JM, Kliewer SA, Moore LB, Smith-Oliver TA, Oliver BB, Su J-L, Sundseth SS, Winegar DA, Blanchard DE, Spencer TA, et al. Activation of the nuclear receptor LXR by oxysterols defines a new hormone response pathway. J Biol Chem. 1997;272:3137–3140. doi: 10.1074/jbc.272.6.3137. [DOI] [PubMed] [Google Scholar]
- Luo Y, Tall AR. Sterol upregulation of human CETP expression in vitro and in transgenic mice by an LXR element. J Clin Invest. 2000;105:513–520. doi: 10.1172/JCI8573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller VA, Benedetti FM, Rigas JR, Verret AL, Pfister DG, Straus D, Kris MG, Crisp M, Heyman R, Loewen GR, et al. Initial clinical trial of a selective retinoid X receptor ligand, LGD1069. J Clin Oncol. 1997;15:790–795. doi: 10.1200/JCO.1997.15.2.790. [DOI] [PubMed] [Google Scholar]
- Miyazaki M, Kim Y-C, Keller MP, Attie AD, Ntambi JM. The biosynthesis of hepatic cholesterol esters and triglycerides is impaired in mice with a disruption of the gene for stearoyl-CoA desaturase I. J Biol Chem. 2000;275:30132–30138. doi: 10.1074/jbc.M005488200. [DOI] [PubMed] [Google Scholar]
- Osborne TF. Sterol regulatory element binding proteins (SREBPs): Key regulators of nutritional homeostasis and insulin action. J Biol Chem. 2000;276:32379–32382. doi: 10.1074/jbc.R000017200. [DOI] [PubMed] [Google Scholar]
- Pai J-T, Guryev O, Brown MS, Goldstein JL. Differential stimulation of cholesterol and unsaturated fatty acid biosynthesis in cells expressing individual nuclear sterol regulatory element-binding proteins. J Biol Chem. 1998;273:26138–26148. doi: 10.1074/jbc.273.40.26138. [DOI] [PubMed] [Google Scholar]
- Peet DJ, Turley SD, Ma W, Janowski BA, Lobaccaro J-MA, Hammer RE, Mangelsdorf DJ. Cholesterol and bile acid metabolism are impaired in mice lacking the nuclear oxysterol receptor LXRα. Cell. 1998;93:693–704. doi: 10.1016/s0092-8674(00)81432-4. [DOI] [PubMed] [Google Scholar]
- Repa JJ, Mangelsdorf DJ. The role of orphan nuclear receptors in the regulation of cholesterol homeostasis. Ann Rev Cell Dev Biol. 2000;16:459–481. doi: 10.1146/annurev.cellbio.16.1.459. [DOI] [PubMed] [Google Scholar]
- Repa JJ, Turley SD, Lobaccaro J-MA, Medina J, Li L, Lustig K, Shan B, Heyman RA, Dietschy JM, Mangelsdorf DJ. Regulation of absorption and ABC1-mediated efflux of cholesterol by RXR heterodimers. Science. 2000;289:1524–1529. doi: 10.1126/science.289.5484.1524. [DOI] [PubMed] [Google Scholar]
- Schroepfer GJ. Oxysterols: Modulators of cholesterol metabolism and other processes. Physiol Rev. 2000;80:361–554. doi: 10.1152/physrev.2000.80.1.361. [DOI] [PubMed] [Google Scholar]
- Schultz JR, Tu H, Luk A, Repa JJ, Medina JC, Li L, Schwender S, Wang S, Thoolen M, Mangelsdorf DJ, Lustig KD, Shan B. Role of LXRs in control of lipogenesis. Genes & Dev. 2000;14:2831–2838. doi: 10.1101/gad.850400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimano H, Horton JD, Hammer RE, Shimomura I, Brown MS, Goldstein JL. Overproduction of cholesterol and fatty acids causes massive liver enlargement in transgenic mice expressing truncated SREBP-1a. J Clin Invest. 1996;98:1575–1584. doi: 10.1172/JCI118951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimano H, Horton JD, Shimomura I, Hammer RE, Brown MS, Goldstein JL. Isoform 1c of sterol regulatory element binding protein is less active than isoform 1a in livers of transgenic mice and in cultured cells. J Clin Invest. 1997a;99:846–854. doi: 10.1172/JCI119248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimano H, Shimomura I, Hammer RE, Herz J, Goldstein JL, Brown MS, Horton JD. Elevated levels of SREBP-2 and cholesterol synthesis in livers of mice homozygous for a targeted disruption of the SREBP-1 gene. J Clin Invest. 1997b;100:2115–2124. doi: 10.1172/JCI119746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimano H, Yahagi N, Amemiya-Kudo M, Hasty AH, Osuga J-I, Tamura Y, Shionoiri F, Iizuka Y, Ohashi K, Harada K, et al. Sterol regulatory element-binding protein-1 as a key transcription factor for nutritional induction of lipogenic enzyme genes. J Biol Chem. 1999;274:35832–35839. doi: 10.1074/jbc.274.50.35832. [DOI] [PubMed] [Google Scholar]
- Shimomura I, Bashmakov Y, Horton JD. Increased levels of nuclear SREBP-1c associated with fatty livers in two mouse models of diabetes mellitus. J Biol Chem. 1999a;274:30028–30032. doi: 10.1074/jbc.274.42.30028. [DOI] [PubMed] [Google Scholar]
- Shimomura I, Bashmakov Y, Ikemoto S, Horton JD, Brown MS, Goldstein JL. Insulin selectively increases SREBP-1c mRNA in the livers of rats with streptozotocin-induced diabetes. Proc Natl Acad Sci. 1999b;96:13656–13661. doi: 10.1073/pnas.96.24.13656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimomura I, Bashmakov Y, Shimano H, Horton JD, Goldstein JL, Brown MS. Cholesterol feeding reduces nuclear forms of sterol regulatory element binding proteins in hamster liver. Proc Natl Acad Sci. 1997a;94:12354–12359. doi: 10.1073/pnas.94.23.12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimomura I, Shimano H, Horton JD, Goldstein JL, Brown MS. Differential expression of exons 1a and 1c in mRNAs for sterol regulatory element binding protein-1 in human and mouse organs and cultured cells. J Clin Invest. 1997b;99:838–845. doi: 10.1172/JCI119247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabor DE, Kim JB, Spiegelman BM, Edwards PA. Identification of conserved cis-elements and transcription factors required for sterol-regulated transcription of stearoyl-CoA desaturase 1 and 2. J Biol Chem. 1999;274:20603–20610. doi: 10.1074/jbc.274.29.20603. [DOI] [PubMed] [Google Scholar]
- Teboul, M., Enmark, E., Li, Q., Wikström, A.C., Pelto-Huikko, M., and Gustafsson, J.-Å. OR-1, a member of the nuclear receptor superfamily that interacts with the 9-cis-retinoc acid receptor. Proc. Natl. Acad. Sci. 92: 2096–2100. [DOI] [PMC free article] [PubMed]
- Tontonoz P, Kim JB, Graves RA, Spiegelman BM. ADD1: A novel helix–loop–helix transcription factor associated with adipocyte determination and differentiation. Mol Cell Biol. 1993;13:4753–4759. doi: 10.1128/mcb.13.8.4753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkateswaran A, Repa JJ, Lobaccaro J-MA, Bronson A, Mangelsdorf DJ, Edwards PA. Human White/murine ABC8 mRNA levels are highly induced in lipid-loaded macrophages: A transcriptional role for specific oxysterols. J Biol Chem. 2000;275:14700–14707. doi: 10.1074/jbc.275.19.14700. [DOI] [PubMed] [Google Scholar]
- Wan Y-JY, An D, Cai Y, Repa JJ, Chen TH-P, Flores M, Postic C, Magnuson MA, Chen J, Chien KR, et al. Hepatocyte-specific mutation establishes retinoid X receptor α as a heterodimeric integrator of multiple physiological processes in the liver. Mol Cell Biol. 2000;20:4436–4444. doi: 10.1128/mcb.20.12.4436-4444.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willy PJ, Mangelsdorf DJ. Unique requirements for retinoid-dependent transcriptional activation by the orphan receptor LXR. Genes & Dev. 1997;11:289–298. doi: 10.1101/gad.11.3.289. [DOI] [PubMed] [Google Scholar]
- Willy PJ, Umesono K, Ong ES, Evans RM, Heyman RA, Mangelsdorf DJ. LXR, a nuclear receptor that defines a distinct retinoid response pathway. Genes & Dev. 1995;9:1033–1045. doi: 10.1101/gad.9.9.1033. [DOI] [PubMed] [Google Scholar]
- Yahagi N, Shimano H, Hasty AH, Amemiya-Kudo M, Okazaki H, Tamura Y, Iizuka Y, Shionoiri F, Ohashi K, Osuga J-I, et al. A crucial role of sterol regulatory element-binding protein-1 in the regulation of lipogenic gene expression by polyunsaturated fatty acids. J Biol Chem. 1999;274:35840–35844. doi: 10.1074/jbc.274.50.35840. [DOI] [PubMed] [Google Scholar]
- Yokoyama C, Wang X, Briggs MR, Admon A, Wu J, Hua X, Goldstein J L, Brown MS. SREBP-1, a basic-helix–loop–helix-leucine zipper protein that controls transcription of the low density lipoprotein receptor gene. Cell. 1993;75:187–197. [PubMed] [Google Scholar]