Abstract
Carnivory in plants is an adaptation strategy to nutrient-poor environments and soils. Carnivorous plants obtain some additional mineral nutrients by trapping and digesting prey; the genus Nepenthes is helped by its specialized pitcher traps. To make the nutrients available, the caught prey needs to be digested, a process that requires the concerted activity of several hydrolytic enzymes. To identify and investigate the various enzymes involved in this process, fluid from Nepenthes traps has been analysed in detail. In this study, a novel type of Nepenthes endochitinase was identified in the digestion fluid of closed pitchers. The encoding endochitinase genes have been cloned from eight different Nepenthes species. Among these, the deduced amino acid sequence similarity was at least 94.9%. The corresponding cDNA from N. rafflesiana was heterologously expressed, and the purified protein, NrChit1, was biochemically characterized. The enzyme, classified as a class III acid endochitinase belonging to family 18 of the glycoside hydrolases, is secreted into the pitcher fluid very probably due to the presence of an N-terminal signal peptide. Transcriptome analyses using real-time PCR indicated that the presence of prey in the pitcher up-regulates the endochitinase gene not only in the glands, which are responsible for enzyme secretion, but at an even higher level, in the glands’ surrounding tissue. These results suggest that in the pitchers’ tissues, the endochitinase as well as other proteins from the pitcher fluid might fulfil a different, primary function as pathogenesis-related proteins.
Keywords: Carnivorous plants, digestive fluid, endochitinase, Nepenthes, PR proteins, real-time PCR
Introduction
Nepenthaceae are a monotypic family of carnivorous plants scattered throughout the Old World tropics. Around 130 species of Nepenthes have been described. This number is rapidly increasing, with several new species each year (McPherson, 2010). Their leaf morphology is very similar and consists of a photosynthetic part of the leaf (enlarged leaf base) and a tendril that carries a pitfall trap. These so-called pitchers are divided into zones which include a lid and a peristome involved in attracting and trapping prey; a waxy zone for trapping and preventing prey from escaping (Gaume et al., 2002; Riedel et al., 2003; Scholz et al., 2010); and, at the bottom, a digestive zone. The inside of this part is covered with multicellular glands and often filled with a viscoelastic fluid to retain and digest caught prey, mainly insects (Gorb et al., 2004; Gaume and Forterre, 2007). The glands of the digestive zone fulfil various functions: the perception of chemical stimuli; the secretion of digestive enzymes; and nutrient absorption (Owen et al., 1999; Schulze et al., 1999). When visiting the pitchers, insects fall into the traps (Gaume et al., 2002; Bohn and Federle, 2004), drown, and can be digested by the enzyme cocktail of the pitcher fluid (Mithöfer, 2011). As Nepenthes plants grow in nutrient-poor environments, this is an effective way to obtain additional nutrients, mainly nitrogen, that are otherwise difficult to come by.
The presence of proteolytic activities in carnivorous plants has been known since the time of Charles Darwin (Darwin, 1875). In addition to proteases, other hydrolytic activities have been described in Nepenthes as well, such as RNase, esterase, phosphatase, and chitinase (Heslop-Harrison, 1975; Juniper et al., 1989; Eilenberg et al., 2006). The presence of additional enzymes has been suggested, such as a β-1,3-glucanase and a β-D-xylosidase (for an overview, see Hatano and Hamada, 2008; Mithöfer, 2011). All these hydrolytic enzymes are very probably employed in the digestion of the prey and help make nutrients available to the plant. Additionally, the acidification of the pitcher fluid by plasma membrane H+-ATPase is important for the digestion of prey and the absorption of nutrients (An et al., 2001; Moran et al., 2010).
Chitinases (EC 3.2.1.14) are particularly interesting plant enzymes because their substrate is not present in plant tissues per se. The induction of chitinases in plants upon fungal infection as well as the inhibition of fungal growth has been shown for several plant–pathogen interactions (Schlumbaum et al., 1986; Theis and Stahl, 2004; Van Loon et al., 2006). Thus, chitinases are thought to be necessary and involved as typical pathogenesis-related (PR) proteins in plant defence against pathogenic fungi, due to their ability to hydrolyse and degrade chitin (Theis and Stahl, 2004; Van Loon et al., 2006). Most often, chitinases act as endochitinases, producing chito-oligosaccharides of 2–6 N-acetylglucosamine units, but they also may act as exochitinases. This large group of antifungal proteins belongs to the group of O-glycoside hydrolases, which hydrolyse the glycosidic bond between two or more carbohydrates or between a carbohydrate and a non-carbohydrate moiety. Based on amino acid sequence similarities, such enzymes are classified in 85 families; chitinases belong to families 18 and 19 (Henrissat, 1991).
Since insects represent the major fraction of animals trapped by carnivorous plants, there may be a special role for chitinases in the process of prey digestion. However, only two reports identify chitinases from Nepenthes species. Four genes representing two subgroups of basic chitinases from class I, Nkchit1b and Nkchit2b, have been isolated from N. khasiana; these were further characterized in a heterologous system (Eilenberg et al., 2006). Recently, Hatano and Hamada (2008) were able to clone another chitinase from class IV from N. alata but did not show any enzymatic activities. Thus, the aim of this study was to investigate the digestive fluid of Nepenthes plants for the presence of additional hydrolytic enzymes; the study focused on chitinases. Using a proteomic approach in combination with molecular techniques, a novel chitinase was identified and heterologously expressed in Escherichia coli to enable its biochemical characterization. In order to analyse whether or not this enzyme is widespread in the genus Nepenthes, chitinase activity in the pitchers of various species was determined and a DNA-based phylogeny was calculated. Furthermore, the regulation of the enzyme was addressed using a real-time PCR approach. The effect of the presence of the chitinase on the transcript level in the tissues of different pitchers was studied and an attempt was made to unravel possible prey-mediated regulations.
Materials and methods
Chemicals and organisms
Nepenthes species (N. singalana, N. ventricosa, N. gracilis, N. thorelii, N. mirabilis, N. ampullaria, N. alata, N. rafflesiana, and the hybrid ‘Mizuho’) were grown in the greenhouses of the Botanical Gardens in Jena and Munich. All chemicals used, unless specified, were of analytical grade and purchased from Roth (Karlsruhe, Germany).
Protein analysis
A 15 ml aliquot of fluid was collected from 18 closed pitchers of various Nepenthes species (N. singalana, N. gracilis, N. mirabilis, N. alata, and N. rafflesiana) using a sterile syringe. A 50 μg aliquot of protein was precipitated using trichloroacetic acid (TCA) from the homogenized fluid and subjected to SDS–PAGE (10% separation gel). The limited amount of pitcher fluid proteins meant that one-dimensional SDS–PAGE with a higher protein loading capacity had to be used instead of the more frequently used two-dimensional isoelectric focusing/SDS–PAGE system, which has a higher resolution. For microsequencing, peptides obtained after in-gel digestion with trypsin (Shevchenko et al., 1996) were desalted and concentrated by ZipTip columns (C18-RP, Millipore, Schwalbach, Germany). Subsequently, electrospray ionization–tandem mass spectrometry (ESI-MS/MS) was performed on a Q-TOF 1.5 hybrid mass spectrometer (Micromass, Bremen, Germany) using ‘medium’ nano ESI capillaries according to Mithöfer et al. (2002). Data obtained were processed using MassLynx 3.5 (Micromass) and peptide sequences were calculated manually. For the identification of the proteins, databases were used by similarity or blast searches such as Swiss-Prot (http://www.expasy.ch/sprot/) and EMBL (http://www.ebi.ac.uk/Tools/fasta33/index.html). Search parameters were set as recommended by the database programs. Alignments and homology searches were carried out with Clustal X.
The putative signal peptide was predicted using the SignalP 3.0 server (http://www.cbs.dtu.dk/services/SignalP; Bendtsen et al., 2004). For homology modelling, a three-dimensional model of NrChit1 was built with the automated comparative modelling program SWISS-MODEL (http://swissmodel.expasy.org/workspace; Guex and Peitsch, 1997; Schwede et al., 2003; Arnold et al., 2006). As homologous protein template, the crystal structure of hevamine, an endochitinase from family 18 class III (Protein Data Bank entry, 1hvqA; Swiss Prot, P23472; sequence similarity with NrChit1, 65.4%, X-ray resolution, 2.2 Å; Terwisscha van Scheltinga et al., 1994) from Hevea brasiliensis, was used. Because of the high level of sequence similarities between the templates and NrChit1, the model was evaluated to be of a high quality.
Cloning, expression, and purification of NrChit1 endochitinase cDNA
Total RNA from N. rafflesiana pitchers was isolated using the Concert™ Plant RNA Reagent (Invitrogen, Carlsbad, CA, USA) following the manufacturer's protocol. RNA was purified to eliminate genomic DNA using the Qiagen RNeasy Plant RNA kit (Qiagen, Hilden, Germany) and DNA was digested by TURBO™ DNase (Applied Biosystems/Ambion, Darmstadt, Germany). First-strand cDNA was synthesized using SuperScript III Reverse Transcriptase (Invitrogen, Darmstadt, Germany), oligo(dT)20 primer, and 1 μg of total RNA at 50 °C for 55 min.
Degenerate primers, designed according to conserved protein sequences of known plant endochitinases (NCBI GenBank), were used to amplify a fragmental cDNA sequence. Cloning the 5′ and 3′ end of NrChit1 cDNA was accomplished by rapid amplification of cDNA ends (RACE) PCR using total RNA and the FirstChoice® RLM-RACE Kit (Applied Biosystems/Ambion) following the manufacturer's protocol. Primers were designed by using DNASTAR Lasergene® Software (GATC BIOTECH, Konstanz, Germany). The resulting amplified products were cloned into a pCR®-TOPO®-vector, and the resulting plasmid was subjected to nucleotide sequencing (Eurofins MWG Operon, Ebersberg, Germany). The complete NrChit1 cDNA sequence was amplified by PCR using Pfu DNA polymerase (Fermentas, St. Leon-Rot, Germany), and the primers: forward 5′-ATG AAG ACC CAT TAT TCA TCA GCA ATT C-3′ and reverse 5′-TTA AAC ACT ATC CTT GAT AGC TGA G-3′ (PCR: 3 min at 94 °C; 35 cycles of 30 s at 94 °C, 30 s at 60 °C, 60 s at 72 °C; and 10 min at 72 °C).
For functional identification, cDNA was amplified with primers for an open reading frame (ORF) lacking the signal peptide. The cDNA was subcloned into the pHIS8-3 expression vector (Jez et al., 2000). The recombinant vector was transformed into E. coli BL21(DE3) already transformed with the chaperone-coding plasmid pG-Tf2 (Takara Bio Europe S.A.S., Saint-Germain-en-Laye, France). The bacterial strain was grown to A600=0.6 at 37 °C in LB medium with kanamycin at 50 μg ml−1 and chloramphenicol at 20 μg ml−1. Cultures were induced with 1 mM isopropyl 1-thio-β-D-galactopyranoside (IPTG) for NrChit1 and with 10 ng ml−1 tetracycline for chaperone co-expression. Cultures were kept overnight at 16 °C while being shaken at 200 rpm. After expression, the protein was purified following the instructions of QIAexpressionist™ with a modification of the elution buffer (10 mM NaH2PO4, 300 mM NaCl, 20 mM imidazole, pH 8).
The respective endochitinase sequences from genomic DNA of seven additional Nepenthes species were also cloned. Therefore, genomic DNA was isolated from pitchers using the CTAB (cetyltrimethylammonium bromide) method (Doyle and Doyle, 1990). Amplification and cloning were done as described above.
Chitinase activity assays
The chinolytic activity was determined by using the Chitinase Assay Kit, Fluorometric assay from Sigma-Aldrich (Taufkirchen, Germany). Substrates tested were the dimer 4-methylumbelliferyl (4MU) β-D-N-acetylglucosaminide [(GlcNAc)1] for exochitinase, the trimer 4-methylumbelliferyl β-D-N,N′-diacetylchitobioside hydrate [4MU-(GlcNAc)2] for chitobiosidase, or the tetramer 4-methylumbelliferyl β-D-N,N′,N′′-triacetylchitotriose [4MU-(GlcNAc)3)] for endochitinase activity. Chitinase specificity was estimated by the cleavage of the β-1,4 bond that releases 4MU from the different oligomers. Chitinase reactions were done following the manufacturer's protocol. Fluorescence was measured by a fluorescence spectrophotometer (excitation at 360 nm, emission at 450 nm).
As a second substrate for endochitinase activity, the soluble polymeric substrate carboxymethyl-chitin-Remazol Brilliant Violet (CM-Chitin-RBV; LOEWE Biochemica, Sauerlach, Germany) was used. The reaction contained 50 μl of 50 mM sodium acetate buffer, pH 5, standard (chitinase from Streptomyces griseus, 10 μg ml−1, Sigma-Aldrich) or sample (expressed endochitinase or pitcher fluid), and 150 μl of CM-Chitin-RBV (2 mg ml−1). The reaction was stopped by adding 20 μl of 0.25 M HCl, and non-digested substrate was precipitated by incubation at –20 °C for 5 min and centrifugation at 4 °C for 5 min. Absorbance at 560 nm was measured by a Spectramax 250 microplate reader.
Real-time PCR
For real-time PCR analyses, total RNA was isolated from either whole pitchers, glands, or the epidermal tissue of the digestive zone of closed and open pitchers from N. mirabilis or N. alata. When pitchers were fed with Drosophila melanogaster (30 individuals each; three replicates), they were packed in a full-fashioned stocking to avoid contamination and harvested after 7 d. Fifty glands or surrounding epidermal tissue were isolated using the aureka platform (aura optik, Jena, Germany; Rottloff et al., 2009). NrChit1 expression levels were analysed and compared as follows: for total RNA isolation, the RNAqueous®-Micro kit (Applied Biosystems/Ambion) was used following the manufacturer's protocol. DNA digestion and the reverse transcription reaction were done as described above. Actin was chosen as the internal control gene; the sequence was isolated using the primers published by Van den Berg et al. (2004), actinF 5′-ACC GAA GCC CCT CTT AAC CC-3′ and actinR 5′-GTA TGG CTG ACA CCA TCA CC-3′, coding for a 180 bp fragment with 93% identity to an actin gene from Quercus robur (GQ339769) (PCR: 3 min at 94 °C; 35 cycles of 30 s at 94 °C, 30 s at 57 °C, 60 s at 72 °C; and 10 min at 72 °C).
Primers for real-time PCR were designed in order to obtain resulting PCR products of ∼100 bp using the Primer3Plus software (http://www.bioinformatics.nl/cgi-bin/primer3plus/primer3plus.cgi): for actin, 5′-CTC TTA ACC CCA AAG CAA ACA GG-3′ and 5′-GTG AGA GAA CAG CCT GGA TG-3′; and for endochitinase, 5′-AAG GGA TCA AGG TCC TCC TAT C-3′ and 5′- GAG GTA GTT ATT CCA AAG GTA AGC-3′.
Real-time PCR was done on a Mx3000P Real-Time PCR System (Stratagene, La Jolla, CA, USA). The process was performed with 25 μl of reaction mixture containing 12.5 μl of 2× Brilliant II SYBR® Green QPCR Master Mix (Stratagene), cDNA (20–75 ng), 400 nM of each primer, and 30 nM ROX as a passive reference dye. The following protocol was used: initial polymerase activation for 10 min at 95 °C, 40 cycles of 30 s at 95 °C, 60 s at 61 °C, and 60 s at 72 °C. Actin levels were equal in every reaction independently of tissue type. PCR conditions were determined by a non-reverse transcriptase template control and a non-template control for each primer pair. Relative RNA levels were calibrated and normalized with the level of actin mRNA by determining the efficiency of every single reaction using the method of Liu and Saint (2002). Calculation of expression ratios and statistical analyses were performed with the Relative Expression Software Tool (REST© 2009, http://www.gene-quantification.de/rest.html; Pfaffl et al., 2002). Data are from triplicates.
Phylogenetic analyses
To infer phylogenetic relationships, a maximum parsimony analysis of the endochitinase sequence data set that had been obtained from genomic DNA as described above was carried out in PAUP* 4.0b10 (Swofford, 2003) using the default exhaustive search settings. The 50% bootstrap majority rule consensus trees (Felsenstein, 1981) were calculated from 10 000 replicates under the default heuristic search settings with no maxtrees limit, and character states specified as unordered and equally weighted.
Additionally, a Bayesian analysis was conducted using the Markov-Chain-Monte-Carlo algorithm of MrBayes 3.1.2 (Huelsenbeck and Ronquist, 2001) under the assumption of the HKY model (Hasegawa et al., 1985) and invariable sites (HKY+I), which was determined to be the best-fit model of sequence evolution by the likelihood ratio tests implemented in MrModeltest 2.2 (Nylander, 2004). Four Markov chains were calculated simultaneously, according to MrBayes’ default setting. Analysis was terminated after 1 000 000 generations; trees were summarized in a 50% majority rule consensus tree after discarding burn-in trees yielded before reaching likelihood stationarity.
Results and Discussion
Protein identification
Based on the peptide sequence YYDNGYSSA(I/L)K that was determined from a 30 kDa protein band, high similarities of this protein with acid endochitinases from other plant species, Nicotiana tabacum (NCBI accession no. CAA77656), Vitis vinifera (BAC65326), Hevea brasiliensis (CAA09110), Malus domestica (AAG25709), and Beta vulgaris (AAB28479), were detected by blast searches. This was possible because this N-terminal-localized 12 amino acid long peptide was specific enough to suggest that the protein was very likely to be an acid endochitinase. In particular, the GYS tripeptide is a highly conserved motif which can be found all over these proteins (Supplementary Fig. S1 available at JXB online).
Cloning of NrChit1 from Nepenthes rafflesiana and its heterologous expression
In order to isolate the corresponding cDNA, degenerated primers based on the peptide sequence and on a conserved peptide stretch close to the N-terminus of the consensus sequence of the various endochitinases were synthesized and applied in a reverse transcription–PCR (RT-PCR) using RNA isolated from N. rafflesiana pitchers. However, no PCR fragment was amplified. Only the usage of oligonucleotides, which have been deduced from the consensus sequence of the cDNAs of the endochitinases mentioned above, resulted in the amplification of a 474 bp fragment. RACE PCR (Frohman et al., 1988) was further used to isolate the missing 5' and 3' regions of the cDNA, again by using RNA isolated from N. rafflesiana. The isolated full-length cDNA (GenBank accession no. GQ338257) encoding the putative endochitinase (NrChit1) showed an ORF of 879 bp corresponding to 292 amino acid residues (Fig. 1A). The deduced amino acid sequence comprised the sequenced peptide from position 279Y to 289K, indicating an isoleucine at position 288. Moreover, NrChit1 also contained the conserved sequence of the catalytic domain of family 18 glycoside hydrolases (Karlsson and Stenlid, 2009), LDGIDFDIE, from position 145 to 154 (Fig. 1A). Its primary structure suggested a class III chitinase (Collinge et al., 1993), and it shares significant homology with plant class III chitinases (Supplementary Fig. S1 at JXB online), including the presence of six cysteine residues at conserved positions in all class III chitinases (Kim et al., 1999) (Fig. 1A). This classification is strongly supported by further sequence analyses that predicted the presence of an N-terminal region representing a signal peptide; this peptide in turn directs the protein to the apoplasm (Bendtsen et al., 2004). These signal peptides are cleaved off the mature protein. Most probably, the cleavage site is between amino acid positions 26 and 27 (Fig. 1A) (Bendtsen et al., 2004).
Fig. 1.
(A) Deduced amino acid sequence of the endochitinase gene from Nepenthes rafflesiana. The predicted signal peptide (amino acids 1–26) is indicated in italics and underlined, the conserved sequence of the catalytic domain of family 18 glycoside hydrolases is underlined (amino acids 145–153), the sequenced peptide (amino acids 279–189) is indicated in bold and underlined, and conserved cysteines are in bold. (B) Coomassie-stained SDS–PAGE of recombinant endochitinase NrChit1 heterologously expressed in E. coli and isolated using His-Tag-based purification. M, broad range protein marker (Biorad); E1–6, different fractions eluted with imidazole.
In addition, the endochitinase sequences from genomic DNA of N. ventricosa (GenBank accession no. GQ338254), N. mirabilis (GQ338258), N. thorelii (GQ338255), N. ampullaria (GQ338261), Nepenthes hybrid ‘Mizuho’ (GQ338259), N. gracilis (GQ338260), and N. singalana (GQ338256) were also cloned. Interestingly, no introns could be found for any of them. The significance of single exon genes in plants is still unknown. Genes without introns are often retrogenes and formed by retroposition, a cellular process in which spliced mRNAs are reverse transcribed and inserted into new genomic positions. Typically, they become non-expressed pseudogenes, because of the lack of regulatory elements (Wang et al., 2006). However, many retrogenes were identified in Populus species and Arabidopsis thaliana, and most were functional (Wang et al., 2006; Zuh et al., 2009). Although some genes encoding PR proteins do not have introns, such as the PR-1 gene from Oryza sativa (Liu and Xue, 2006) and TLP from N. singalana (Rottloff et al., 2009), this is not a general phenomenon; such a general phenomenon would include, for example, four genes encoding basic chitinases class I (Nkchit1b-1, -2 and Nkchit2b-1, -2) of pitchers from N. khasiana which possess two introns (Eilenberg et al., 2006). However, it is conceivable that the single exon structure is typical for class III acidic chitinases, as genomic sequence analysis with other plant species coding for this enzyme [M. domestica (AF309514), Medicago truncatula (AY238969), and V. vinifera (AB105374)] also showed no introns in the coding region.
Biochemical characterization
The recombinant NrChit1 protein, which was heterologously expressed in E. coli under chaperone co-expression and purified using a His-tag of eight histidines (Fig. 1B), was further used to characterize catalytic activities (Table 1). Among the various compounds that have been tested to elucidate the substrate specificity—4MU-(GlcNAc)1 for exochitinase activity, 4MU-(GlcNAc)2 for chitobiosidase activity, and 4MU-(GlcNAc)3 and CM-chitin-RBV for endochitinase activity—only CM-chitin-RBV could be hydrolysed. Using this substrate, basic enzymatic properties have been determined (Table 1). However, the specific activity of NrChit1 was quite low, ΔE550: 0.08 AU (h μg protein)−1. Thus, a detailed kinetic analysis of the enzymatic reaction was not feasible because CM-chitin-RBV concentrations >1.5 μg ml−1 were not applicable and, as a consequence, substrate saturation was impossible to reach. In spite of this handicap, the endochitinase activity of NrChit1 could be demonstrated with the recombinant enzyme. Thus, clearly NrChit1 is able to hydrolyse longer chitin polymers occurring in the exoskeleton of arthropods or fungal cell walls. This might happen together with other endochitinases in the digestion fluid, described by Eilenberg et al. (2006). However, the question remains: why have no chitobiosidases or exochitinases been identified up to now? Such enzymes release and provide dimeric or even monomeric products which easily can be further metabolized by the plant.
Table 1.
Properties of the heterologously expressed endochitinase from Nepenthes rafflesiana
| Protein characteristics | Specificities |
| Enzyme activity | Endochitinase |
| Glycosyl hydrolase family | 18 |
| Classification | Class III |
| Substrate specificity | CM-chitin-RBV |
| Temperature optimum | 41 °C |
| pH optimum | 3–4 |
| Molecular mass | 31.1 kDa |
| Isoelectric point | 3.86 |
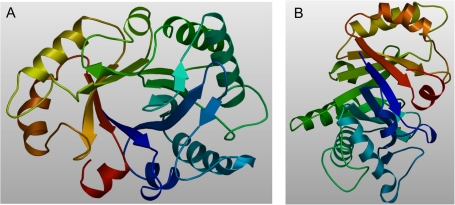
As several crystal structures were determined for chitinases from family 18 class III, the spatial structure of the NrChit1 was modelled by comparative modelling. The modelled residue range was from amino acids 1 to 266 (i.e. without a signal peptide). The sequence of NrChit1 was then structurally compatible with the fold of chitinases from class III. Similar to other chitinases from this class, the predicted NrChit1 structure was composed of a (βα)8-barrel folding motif, which is the typical architecture of chitinases from family 18 (Terwisscha van Scheltinga et al., 1994) and consists of an eight-stranded parallel β-barrel surrounded by eight α-helices. Two additional β-strands can be found outside the (βα)8-barrel (Fig. 2).
Fig. 2.
Calculated 3D structure of the endochitinase NrChit1 from Nepenthes rafflesiana. The structure was suggested by the SWISS-MODEL web server upon comparative modelling using an endochitinase sequence from Hevea brasiliensis (NCPI, CAA09110; Swiss Prot, P23472) as the homologous template structure. The modelled residue range is from amino acids 1 to 266. A and B represent different views of the modelled structure.
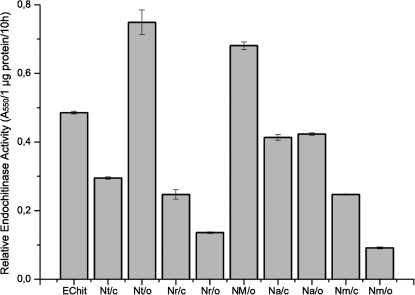
The widespread occurrence of this particular type of chitinase in the genus Nepenthes was demonstrated by the identification of its enzymatic activities in several species (Fig. 3). This demonstrates that in all tested Nepenthes species the encoding gene is indeed not a pseudogene but expresses an active enzyme. The endochitinase is detectable in closed pitchers, suggesting it is constitutively expressed. In N. rafflesiana and N. mirabilis, the activities in closed pitchers are even higher compared with the open ones. On the other hand, at least for N. thorelii, it may be that some proteins were induced. In any case, the enzymatic activities were calculated on the basis of protein concentration; thus dilution effects due to different volumes of fluid in the pitchers can be ruled out. The differences in the activities can be explained by a lack of information about the relative participation of the endochitinase with respect to the total amount of protein in the fluid of the particular species. Thus, the ratio of endochitinase to total protein might vary in different species.
Fig. 3.
Endochitinase activities in the pitcher fluid of various Nepenthes species and the heterologously expressed NrChit1 using CM-chitin-RBV as substrate. Echit, E. coli-expressed NrChit1 from N. rafflesiana; Nt, N. thorelli; Nr, N. rafflesiana; NM, N. hybrid ‘Mizuho’; Na: N. ampullaria; Nm, N. mirabilis; o, open pitchers; c, closed pitchers. Bars represent means calculated from n=3 pitchers ±SD.
Phylogenetic analyses
To investigate whether the encoding genes and proteins of the endochitinases from various Nepenthes species are homologues within this genus as well as compared with other species, a phylogenetic analysis was performed. A comparison of the whole amino acid sequence of NrChit1 with protein sequences of endochitinases from other plants (Supplementary Fig. S1 at JXB online) revealed a similarity, always in the range from ∼61.6% with signal peptide and 65.2% without signal peptide (V. vinifera) to 66.0% and 69.5% (B. vulgaris), respectively. Within the additional endochitinases of seven other Nepenthes species that have been cloned as well, the similarity on the amino acid level was never lower than 94.9%.
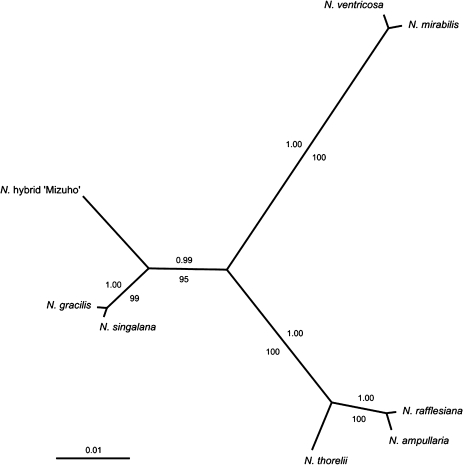
A maximum parsimony analysis of the corresponding DNA sequence data set resulted in one tree with a length of 78 steps (consistency index=0.95, retention index=0.95). The topology of the bootstrap 50% majority rule consensus tree (identical values) corresponded to the topology of the Bayesian consensus tree which was computed from 1 000 000 generations (burn-in: 10 000). This is shown in Fig. 4, including Bayesian posterior probabilities (PPs) and bootstrap support (BS) values from the parsimony analysis. The topology indicates three major lineages, each with high statistical support. One lineage consists of N. ventricosa and N. mirabilis (PP=1.00, BS=100) and the second (PP=1.00, BS=100) consists of N. thorelii and—as a sister group—a monophyly comprising N. rafflesiana and N. ampullaria (PP=1.00, BS=100). The third lineage (PP=0.99, BS=95) contains the hybrid ‘Mizuho’, which is a sister taxon of a monophyly that comprises N. gracilis and N. singalana (PP=1.00, BS=99).
Fig. 4.
The 50% majority rule consensus tree resulting from the Bayesian analysis of the DNA sequence data set, shown as a phylogram. For each node, posterior probabilities are given above the corresponding branch. Bootstrap values from 10 000 MP replicates are given below the branches.
A previous molecular study divided the genus Nepenthes into three major evolutionary lineages based on a phylogenetic reconstruction from chloroplast trnK intron sequence data (Meimberg et al., 2001). Six of the eight taxa investigated in the present study were present in the sampling of this chloroplast DNA (cpDNA) analysis; five were found to be members of the same lineage (N. ampullaria, N. gracilis, N. mirabilis, N. rafflesiana, and N. thorelii). Considering the moderate statistical support within the clades inferred by Meimberg et al. (2001), it cannot be said that there is a major conflict with the present phylogenetic reconstruction, but it is evident that the present data provide a significantly higher resolution among these five taxa. The sixth species, N. ventricosa, is nested within a different lineage in the cpDNA phylogeny; the results, however, indicate a close relationship to N. mirabilis with high statistical support (Fig. 4). This grouping receives support from phylogeographic considerations (McPherson, 2010), as both species occur in the Philippines. Biogeographic congruence is also true for two other indicated subgroups, N. gracilis and N. singalana (both are found in Sumatra), as well as N. amplullaria and N. rafflesiana (found in Borneo, Peninsular Malaysia, Singapore, Sulawesi, Sumatra, and Thailand).
The difference in resolution in comparison with cpDNA data and the inconsistency described above strongly indicate the crucial effect of the marker employed for phylogenetic analyses on the results. This suggests that a thorough re-examination of the phylogenetic relationships within Nepenthes, based on a multiple marker analysis or on fingerprinting methodology, might be rewarding.
Endochitinase transcript abundance
To gain insight into its molecular regulation, endochitinase gene expression was measured in pitchers of N. mirabilis. In order to investigate whether or not the presence of prey affects the transcript level, pitchers fed with fruit flies were analysed. As shown in Table 2, a slight but significant up-regulation was found (1.40-fold) in open pitchers due to the presence of the flies, suggesting that the endochitinase gene is regulated on the mRNA level. In addition, in the glands’ surrounding tissue, a significantly higher level of endochitinase transcripts was detected in open compared with closed pitchers (7.52-fold). This was not the case in the glands themselves, although a trend (not significant) towards this condition was observed [1.76-fold; P(H1): 0.16]. These results can be interpreted as an induction of the respective gene in the presence of prey. Recently, the injection of colloidal chitin into the trap of N. khasiana was shown to induce a type I basic chitinase, NkChit1b, which is very probably secreted into the pitcher fluid (Eilenberg et al., 2006). Analysing glands and their surrounding tissue in open pitchers fed with flies demonstrated that in the tissue, a striking higher level of endochitinase transcripts (10.04-fold) occurred compared with in the glands (Table 2). This result was somewhat surprising if it is assumed that the endochitinase functions in the digestion of prey and is therefore synthesized and secreted mainly by the glands. However, the results of Owen et al. (1999) showed that at least in N. alata, the glands of mature, open pitchers are less able to secrete a fluorescent dye, 6(5)-carboxyfluorescein, than glands of younger pitchers. Consequently, it might be that the reduced secretion ability diminishes transcript accumulation in glands but not in the surrounding tissues.
Table 2.
Real-time PCR analyses for the NrChit1 level in Nepenthes pitchers under various conditions
| Plant | Comparison | Expression ratio | P(H1)a |
| N. mirabilis | Open + Dm versus open | 1.40 | <0.001 |
| N. mirabilis, gl | Open + Dm versus closed | 1.76 | 0.160 |
| N. mirabilis, ts | Open + Dm versus closed | 7.52 | <0.001 |
| N. mirabilis | Open + Dm, tissue versus glands | 10.04 | <0.001 |
| N. mirabilis | Closed, tissue versus glands | 2.97 | <0.001 |
| N. alata | Closed, tissue versus glands | 2.98 | <0.001 |
Whole tissue of open or closed pitchers was investigated or the indicated parts: glands only (gl) and tissues surrounding the glands only (ts), respectively. Some pitchers were treated with prey (Drosophila melanogaster, Dm) for 7 d before harvesting.
The hypothesis test P(H1) represents the probability of the alternative hypothesis that the difference between the sample and control group is due only to chance. P(H1) <0.001 indicates significant differences between the groups compared.
Thus, to investigate further whether this distribution of endochitinase transcripts in glands and surrounding tissue is typical for Nepenthes and present even in developing traps, the abundance of endochitinase transcript in glands and in the tissue of closed pitchers of two species, N. mirabilis and N. alata, was analysed. In principle, higher levels of endochitinase transcripts have usually been found in N. alata compared with the respective part of N. mirabilis (whole pitcher, glands, and surrounding tissue). However, this difference (5.89-fold) was significant only for the tissue. However, even more interesting, the results obtained revealed significantly higher expression levels (∼3-fold) in the tissue than in glands for both plants (Table 2). Thus, given that reduced secretion ability is typical for mature but not developing pitchers, it is unlikely that the main site for transcript accumulation shifts from glands to their surrounding tissue. Although a fast transcript turnover in the glands could explain the findings, it is also conceivable that the glands are indeed responsible for protein secretion but not necessarily for their biosynthesis. All pitcher tissues might be employed in the generation of enzymes for the digestion fluid, but only the glands can secrete them. In any case, more experiments addressing these questions should be performed.
Conclusions
Several lines of evidence are consistent with the hypothesis that chitinases as PR proteins are an important component of plant defence systems against pathogenic fungi. However, the presence of such a hydrolytic activity in the digestion fluid of carnivorous plants also makes sense as it is employed to break down captured insect prey. That means the plant is taking a given enzyme and using it in a new, different context. This represents a sophisticated and highly efficient way of overcoming an environmental handicap. The fact that the endochitinase is inducible supports the hypothesis that this enzyme actually is a PR protein because inducibility is a typical property of PR proteins (Van Loon et al., 2006). Moreover, the occurrence of this protein not only in the pitcher fluid or the secreting glands but also in the tissue of the pitchers, as indicated by the detection of transcripts, suggests that the endochitinase might be employed in defence. It is hoped to prove this hypothesis by further experiments with non-pitcher tissues of Nepenthes. If a pathogenic infection causes the induction of the endochitinase in these parts of the plant, it is very likely to be a real PR protein. The signal peptide suggests that the endochitinase is located in the apoplasm. Similar findings have been reported for a thaumatin-like protein from Nepenthes pitchers (Rottloff et al., 2009). Further experiments will study the exact localization of such enzymes in the pitchers’ tissue as well as how their genes are regulated on the molecular level. Thus, although the activity of the enzyme remains the same, its function has shifted from defence to digestion as postulated recently (Mithöfer, 2011).
Supplementary data
Supplementary data are available at JXB online.
Figure S1. Amino acid sequence alignment of endochitinases from various species. The amino acid sequence alignment and the deduced consensus sequence were generated using Clustal X involving the plant species Pyrus pyrifolia, Lupinus albus, Cucumis sativus, Nepenthes rafflesiana, Beta vulgaris, Vitis vinifera, and Oryza sativa.
Acknowledgments
We thank the Botanical Gardens of the Friedrich Schiller University Jena and the Ludwig-Maximilians-Universität München, Germany, as well as Andreas Fleischmann for continuous support with Nepenthes plants, and the Max Planck Society for financial support. We also thank Emily Wheeler for improving the English.
References
- An CI, Fukosaki EI, Kobayashi A. Plasma-membrane H+-ATPases are expressed in pitchers of the carnivorous plant Nepenthes alata Blanco. Planta. 2001;212:547–555. doi: 10.1007/s004250000455. [DOI] [PubMed] [Google Scholar]
- Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL Workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: SignalP 3.0. Journal of Molecular Biology. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- Bohn HF, Federle W. Insect aquaplaning: Nepenthes pitcher plants capture prey with the peristome, a fully wettable water-lubricated anisotropic surface. Proceedings of the National Academy of Sciences, USA. 2004;101:14138–14143. doi: 10.1073/pnas.0405885101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collinge DB, Kragh KM, Mikkelsen JD, Nielsen KK, Rasmussen U, Vad K. Plant chitinases. The Plant Journal. 1993;3:31–40. doi: 10.1046/j.1365-313x.1993.t01-1-00999.x. [DOI] [PubMed] [Google Scholar]
- Darwin C. Insectivorous plants. London: John Murray; 1875. [Google Scholar]
- Doyle JJ, Doyle JL. A rapid total DNA preparation procedure for fresh plant tissue. Focus. 1990;12:13–15. [Google Scholar]
- Eilenberg H, Pnini-Cohen S, Schuster S, Movtchan A, Zilberstein A. Isolation and characterization of chitinase genes from pitchers of the carnivorous plant Nepenthes khasiana. Journal of Experimental Botany. 2006;57:2775–2784. doi: 10.1093/jxb/erl048. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Confidence limits on phylogenesis: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- Frohman MA, Dush MK, Martin GR. Rapid production of full-length cDNAs from rare transcripts: amplification using a single gene-specific oligonucleotide primer. Proceedings of the National Academy of Sciences, USA. 1988;85:8998–9002. doi: 10.1073/pnas.85.23.8998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaume L, Forterre Y. A viscoelastic deadly fluid in carnivorous pitcher plants. PLoS ONE. 2007;2:e1185. doi: 10.1371/journal.pone.0001185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaume L, Gorb S, Rowe N. Function of epidermal surfaces in the trapping efficiency of Nepenthes alata pitchers. New Phytologist. 2002;156:479–489. doi: 10.1046/j.1469-8137.2002.00530.x. [DOI] [PubMed] [Google Scholar]
- Gorb E, Kastner V, Peressadko A, Arzt E, Gaume L, Rowe N, Gorb S. Structure and properties of the glandular surfaces in the digestive zone of the pitcher in the carnivorous plant Nepenthes ventrata and its role in insect trapping and retention. Journal of Experimental Biology. 2004;207:2947–2963. doi: 10.1242/jeb.01128. [DOI] [PubMed] [Google Scholar]
- Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modelling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- Hasegawa M, Kishino H, Yano T. Dating the human–ape split by a molecular clock of mitochondrial DNA. Journal of Molecular Evolution. 1985;22:160–174. doi: 10.1007/BF02101694. [DOI] [PubMed] [Google Scholar]
- Hatano N, Hamada T. Proteome analysis of pitcher fluid of the carnivorous plant Nepenthes alata. Journal of Proteome Research. 2008;7:809–816. doi: 10.1021/pr700566d. [DOI] [PubMed] [Google Scholar]
- Henrissat B. A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochemical Journal. 1991;280:309–316. doi: 10.1042/bj2800309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heslop-Harrison Y. Enzyme release in carnivorous plants. Frontiers in Biology. 1975;43:525–578. [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogeny. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- Jez JM, Ferrer J, Bowman ME, Dixon RA, Noel JP. Dissection of malonyl-coenzyme A decarboxylation from polyketide formation in the reaction mechanism of a plant polyketide synthase. Biochemistry. 2000;39:890–902. doi: 10.1021/bi991489f. [DOI] [PubMed] [Google Scholar]
- Juniper BE, Robins RJ, Joel DH. The carnivorous plants. London: Academic Press; 1989. [Google Scholar]
- Karlsson M, Stenlid J. Evolution of family 18 glycoside hydrolases: diversity, domain structures and phylogenetic relationships. Journal of Molecular Microbiology and Biotechnology. 2009;16:208–223. doi: 10.1159/000151220. [DOI] [PubMed] [Google Scholar]
- Kim MG, Lee KO, Cheong NE, Choi YO, Jeong JH, Cho MJ, Kim SC, Lee SY. Molecular cloning and characterization of a class III chitinase in pumpkin leaves, which strongly binds to regenerated chitin affinity gel. Plant Science. 1999;147:157–163. [Google Scholar]
- Liu WH, Saint DH. A new quantitative method of real time reverse transcription polymerase chain reaction assay based on simulation of polymerase chain reaction kinetics. Analytical Biochemistry. 2002;302:52–59. doi: 10.1006/abio.2001.5530. [DOI] [PubMed] [Google Scholar]
- Liu Q, Xue Q. Computational identification of novel PR-1-type genes in Oryza sativa. Journal of Genetics. 2006;85:193–198. doi: 10.1007/BF02935330. [DOI] [PubMed] [Google Scholar]
- McPherson S. Carnivorous plants in their habitats. Vol. 1. Poole, UK: Redfern Natural History Productions; 2010. [Google Scholar]
- Meimberg H, Wistuba A, Dittrich P, Heubl G. Molecular phylogeny of Nepenthaceae based on cladistic analysis of plastid trnK intron sequence data. Plant Biology. 2001;3:164–175. [Google Scholar]
- Mithöfer A. Carnivorous pitcher plants: insights in an old topic. Phytochemistry. 2011 doi: 10.1016/j.phytochem.2010.11.024. in press. [DOI] [PubMed] [Google Scholar]
- Mithöfer A, Müller B, Wanner G, Eichacker LA. Identification of defence-related cell wall proteins in Phytophthora sojae-infected soybean roots by ESI-MS/MS. Molecular Plant Pathology. 2002;3:163–166. doi: 10.1046/j.1364-3703.2002.00109.x. [DOI] [PubMed] [Google Scholar]
- Moran JA, Hawkins BJ, Gowen BE, Robbins SL. Ion fluxes across the pitcher walls of three Bornean Nepenthes pitcher plant species: flux rates and gland distribution patterns reflect nitrogen sequestration strategies. Journal of Experimental Botany. 2010;61:1365–1374. doi: 10.1093/jxb/erq004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nylander JAA. MrModeltest v2. 2004. Program distributed by the author. Evolutionary Biology Centre, Uppsala University. [Google Scholar]
- Owen TP, Jr., Lennon KA, Santo MJ, Anderson AN. Pathways for nutrient transport in the pitchers of the carnivorous plant. Nepenthes alata. Annals of Botany. 1999;84:459–466. [Google Scholar]
- Pfaffl MW, Horgan GW, Dempfle L. Relative expression software tool (REST©) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Research. 2002;30:E36. doi: 10.1093/nar/30.9.e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedel M, Eichener A, Jetter R. Slippery surfaces of carnivorous plants: composition of epicuticular wax crystals in Nepenthes alata Blanco pitchers. Planta. 2003;218:87–97. doi: 10.1007/s00425-003-1075-7. [DOI] [PubMed] [Google Scholar]
- Rottloff S, Müller U, Kilper R, Mithöfer A. Micropreparation of single secretory glands from the carnivorous plant Nepenthes. Analytical Biochemistry. 2009;394:135–137. doi: 10.1016/j.ab.2009.07.013. [DOI] [PubMed] [Google Scholar]
- Schlumbaum A, Mauch F, Vogeli U, Boller T. Plant chitinases are potent inhibitors of fungal growth. Nature. 1986;324:365–367. [Google Scholar]
- Scholz I, Bückins M, Dolge L, et al. Slippery surfaces of pitcher plants: Nepenthes wax crystals minimize insect attachment via microscopic surface roughness. Journal of Experimental Biology. 2010;213:1115–1125. doi: 10.1242/jeb.035618. [DOI] [PubMed] [Google Scholar]
- Schulze W, Frommer WB, Ward JM. Transporters for ammonium, amino acids and peptides are expressed in pitchers of the carnivorous plant Nepenthes. The Plant Journal. 1999;17:637–646. doi: 10.1046/j.1365-313x.1999.00414.x. [DOI] [PubMed] [Google Scholar]
- Schwede T, Kopp J, Guex N, Peitsch MC. SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Research. 2003;31:3381–3385. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Analytical Chemistry. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- Swofford DL. 2003. PAUP: Phylogenetic Analysis Using Parsimony. Version 4.0b10 for 32-bit Microsoft Windows. Sunderland, MA Sinauer Associates. [Google Scholar]
- Theis T, Stahl U. Antifungal proteins: targets, mechanisms and prospective applications. Cellular and Molecular Life Sciences. 2004;61:437–455. doi: 10.1007/s00018-003-3231-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwisscha van Scheltinga AC, Kalk KH, Beintema JJ, Dijkstra BW. Crystal structures of hevamine, a plant defence protein with chitinase and lysozyme activity, and its complex with an inhibitor. Structure. 1994;2:1181–1189. doi: 10.1016/s0969-2126(94)00120-0. [DOI] [PubMed] [Google Scholar]
- Van den Berg N, Crampton BG, Hein I, Birch PRJ, Berger DK. High-throughput screening of suppression subtractive hybridization cDNA libraries using DNA microarray analysis. BioTechniques. 2004;37:818–824. doi: 10.2144/04375RR02. [DOI] [PubMed] [Google Scholar]
- Van Loon LC, Rep M, Pieterse CMJ. Significance of inducible defense-related proteins in infected plants. Annual Review of Phytopathology. 2006;44:135–162. doi: 10.1146/annurev.phyto.44.070505.143425. [DOI] [PubMed] [Google Scholar]
- Wang W, Zheng H, Fan C, et al. High rate of chimeric gene origination by retroposition in plant genoms. The Plant Cell. 2006;18:1791–1802. doi: 10.1105/tpc.106.041905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z, Zhang Y, Long M. Extensive structural renovation of retrogenes in the evolution of the Populus genome. Plant Physiology. 2009;151:1943–1951. doi: 10.1104/pp.109.142984. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.