Abstract
Sugar transport is critical for normal plant development and stress responses. However, functional evidence for the roles of monosaccharide transporters in rice (Oryza sativa) has not previously been presented. In this study, reversed genetics was used to identify OsGMST1 as a member of the monosaccharide transporter family in rice. The predicted 481 amino acid protein has the typical features of a sugar transporter in the plastid glucose transporter subfamily consistent with reduced monosaccharide accumulation in plants with reduced OsGMST1 expression. OsGMST1-green fluorescent protein is localized to the Golgi apparatus. OsGMST1 expression is induced by salt treatment and reduced expression confers hypersensitivity to salt stress in rice. OsGMST1 may play a direct or an indirect role in tolerance to salt stress in rice.
Keywords: Golgi, monosaccharide transporter, NaCl stress, Oryza sativa L., rice, SGB1
Introduction
In higher plants, sugars function as an energy source, signal molecules, building blocks of polysaccharide structures, and osmotic regulators. Sugars are synthesized in the phototrophic tissues (source) of the plant and then exported to heterotrophic tissues (sink), and this process requires several transport steps across membranes (Lalonde et al., 1999; Williams et al., 2000). Sugar transporters play key roles in sugar allocation at both the subcellular level and in long-distance transport via the phloem (Buttner, 2007). Long-distance transport is required for sinks to import photosynthetic assimilates from sources in the form of sucrose (Sauer, 2007). In sink tissues, released sucrose travels to sink cells via plasmodesmata and/or sucrose transporters (SUCs) or via monosaccharide transporters (MSTs) after hydrolysis to glucose and fructose by cell wall-bound invertases (Williams et al., 2000; Weschke et al., 2003). As such, the process of long-distance transport involves both cell-to-cell transport as well as to different organelles in a single cell. The sugar transport mode is dependent on the plant developmental stage and the sugar location (Williams et al., 2000). SUCs and MSTs are the main sugar transporters in plants.
MSTs mediate the transport of a wide range of monosaccharides including glucose, fructose, maltose, raffinose, and sugar alcohols (Turgeon and Medville, 2004; Klepek et al., 2005). In Arabidopsis, 53 monosaccharide transporter (-like) genes group into seven subfamilies: (i) STP (plasma membrane monosaccharide transporter), (ii) ERD-like, (iii) VGT (vacuolar glucose transporter), (iv) PLT (polyol transporter), (v) INT (inositol transporter), (vi) TMT (tonoplast monosaccharide transporter), and (vii) pGlcT (plastid glucose transporter) (Johnson et al., 2006; Buttner, 2007). Most MSTs have 12 membrane-spanning domains separated by a cytoplasmic loop between transmembrane helices 6 and 7. In addition to transport across the plasma membrane, monosaccharides are also transported across organelle membranes, such as the inner plastid envelope (Weber et al., 2000; Niittyla et al., 2004) and tonoplast (Wormit et al., 2006). MSTs on the Golgi apparatus (Wang et al., 2006) and mitochondria (Szarka et al., 2004) have also been reported. The rice genome encodes a total of 65 MST genes that group into the seven known subfamilies as defined by the Arabidopsis clades (Johnson and Thomas, 2007). The localization of a few of these transporters are known but, to date, nothing is known about their physiological functions (Toyofuku et al., 2000; Ngampanya et al., 2003; Mamun et al., 2006; Oliver et al., 2007; Wang et al., 2007, 2008; Cho et al., 2010).
In plants, sugars are signalling molecules that regulate plant growth and development and responses to biotic and abiotic stresses (Moore and Sheen, 1999; Rolland et al., 2002, 2006; Gibson, 2005). There is evidence that abiotic stresses affect the expression of plant monosaccharide transporters. The transcript level of TMT1 increased in response to salt, drought, and cold treatment in Arabidopsis, and TMT1 is involved in vacuolar monosaccharide accumulation during cold stress (Wormit et al., 2006). The expression of ERD6, which encoded a putative sugar transporter of Arabidopsis, was up-regulated by dehydration and cold treatment (Kiyosue et al., 1998). AtESL1 was also an abiotic stress-inducible monosaccharide transporter gene in Arabidopsis (Yamada et al., 2010). In rice, expression of OsMST7 and OsMST8 were affected by cold treatment during male gametophyte development (Mamun et al., 2006; Oliver et al., 2007). Therefore, some MSTs may play a pivotal role during stress responses.
The Golgi-localized hexose transporter SGB1 (Wang et al., 2006) belongs to the pGlcT subfamily in Arabidopsis. The gain-of-function mutant sgb1-1D is a suppressor of agb1, a null mutant of the β subunit of G protein. sgb1 and agb1 exhibited similar phenotypes with shorter hypocotyls and open hooks. SGB1 may transport glucose for cell wall biosynthesis in Arabidopsis during cell division (Wang et al., 2006). To date, SGB1 is the only known monosaccharide transporter localized in the Golgi apparatus in plants.
OsGMST1, encoded by locus Os02g17500 with unknown function, showed the highest sequence identity (61%) with SGB1 by a BLASTP search. The function of OsGMST1, a Golgi-localized monosaccharide transporter in rice, is described here.
Materials and methods
Plant materials
Rice plants (Oryza sativa L. ssp. Japonica cv. Zhonghua 10) were grown under field conditions or in a greenhouse with a 16/8 h, 30/28 °C, light/dark photoperiod.
Molecular cloning, vector construction, and the generation of transgenic rice
The cDNA fragment of OsGMST1 was cloned by reverse transcription PCR (RT-PCR) using specific primers (forward primer 5′-ATGCGGTGGAAGCTTAAGTC-3′ and reverse primer 5′-TCGCCCTTACTGGTCACA-3′), and then it was ligated into the binary vector pUN1301 digested with BamHI and KpnI in an antisense orientation to enable expression to be driven by the maize (Zea mays) ubiquitin promoter. This constructed vector (Ubi::antiOsGMST1) was genetically transformed into rice mediated by Agrobacterium tumefaciens EHA105 as previously described by Ge et al. (2004). The T2 generations of transgenic plants were used for further functional characterization.
Heterologous expression of OsGMST1 in yeast
The complete open reading frame of OsGMST1 was ligated with vector pEX-Tag (Meyer et al., 2000) digested by EcoRI in both the sense and antisense orientations, and then the recombinant vectors or empty vector were transformed into the yeast hexose transporter-deficient mutant EBY. VW4000 (Wieczorke et al., 1999), to create strains S1, S2, AS1, and Vector. Transformed cells were pregrown in YEPM medium (1% yeast extract, 2% peptone, 2% maltose) to OD600 of 1.0. Serial dilutions of cell suspensions were streaked on uracil-deficient solid medium containing 0.67% yeast nitrogen base with ammonium sulphate, supplemented with leucine, tryptophan, histidine, and various sugars, such as maltose, glucose, fructose, mannose, galactose, xylose, and ribose, and were incubated at 30 °C for 2 or 3 d.
Subcellular localization of the OsGMST1-GFP fusion protein
A transient expression vector 35S::OsGMST1-GFP was constructed, in which OsGMST1 was fused with the 5′-end of GFP driven by the 35S promoter. Protoplast transformation was as previously described (Sheen, 2001; Bart et al., 2006). Briefly, leaf sheaths from 10-d-old etiolated rice seedlings were cut and digested by cellulase RS and macerozyme R-10 (Yakult, Japan), and the released protoplast cells were transformed by polyethylene glycol (PEG) mediation and then were incubated in darkness overnight. To observe the auto fluorescence of chloroplasts, transformed protoplast cells were incubated under dim light. Mitochondria were stained with 20 nM fluorescent dye MitoTracker Red CMXRos (Molecular Probes) for 2 min, and then visualized at an excitation wavelength of 543 nm and an emission wavelength of 599 nm. Chlorophyll autofluorescence was used as the chloroplast marker. Then, cells were observed under confocal microscope (LSM 510 Meta, Carl Zeiss).
Quantitation of mRNA steady-state levels
Total RNA was prepared from seedlings using Trizol reagent (Invitrogen, USA) and first strand cDNA was synthesized with M-MLV reverse transcriptase (Promega, USA) according to the manufacturer's instructions, and PCR was used to clone genes by specific primers. Real-time PCR was performed by using SYBR Green Master Mix PCR reagent (Toyobo, Japan) and the MX3000P detector (Stratagene, USA). In the analysis of relative transcript level, gene-specific primers of OsGMST1 were used with the rice α-TUBULIN gene (Accession EF575922) as the internal control.
Tissue expression pattern analysis
Total RNA was extracted from different tissues, and the OsGMST1 transcript level was analysed by real-time PCR. OsGMST1::GUS transgenic plants were generated, and GUS staining of various tissues was performed by the histochemical method as previously described by Jefferson et al. (1987). Different organs of OsGMST1::GUS transgenic seedlings were incubated in 5-bromo-4-chloro-3-indolyl-β-glucuronic acid buffer at 37 °C for 12 h. After staining, the tissues were destained in 70% ethanol several times until chlorophyll was removed and images were visualized on an Olympus SZX9 microscope.
Southern blot
Genomic DNA was extracted from 14-d-old seedlings of wild-type and transgenic rice. 20 μg of DNA was digested with EcoRI or HindIII, electrophoresed on a 0.7% agarose gel, and transferred to a nylon membrane (Hybond N+; Amersham Pharmacia Biotech, Buckinghamshire, UK) under alkaline conditions. [α-32P]dCTP-labelled GUS amplified from pUN1301 was used as a probe for hybridization. Membrane was stored in –70 °C for 2–4 d, and then exposed to X-ray film (Eastern Kodak, Rochester NY).
Expression pattern analysis with stress treatment
To detect the transcript level of OsGMST1, seedlings of wild-type rice plants were used. Two-week-old seedlings grown in Kimura B nutrient solution (Kato-Noguchi and Ino, 2005; Chen et al., 2006) were treated with 200 mM NaCl or 18% PEG4000, followed by sampling at 0, 3, 5, 7, 12, 24, and 36 h. For the cold stress treatment, soil-grown seedlings were transferred to a chamber at 4 °C with a 12/12 h light/dark photoperiod, and sampled at the same time as the two stresses above.
Stress tolerance assays
Seeds of wild-type or transgenic rice plants were soak in water at 28 °C for 2 d, and germinated seeds were sown in the 96-well plates from which the bottoms were removed. The plate was floated in Kimura B nutrient solution, and seedlings were incubated in a growth chamber with a 30/28 °C, 16/8 h light/dark photoperiod. For the NaCl treatment, 14-d-old or 21-d-old seedlings were treated in culture solution containing 150 mM or 200 mM NaCl for 9–11 d, and the treated seedlings were recovered for 7 or 8 d followed by survival rate analysis. For PEG treatment, 3-week-old seedlings were transferred to culture solutions containing 20% (w/v) PEG4000. For cold treatment, 3-week-old seedlings were incubated at 4 °C in a growth chamber for 4 d, and then removed to normal growth conditions for 7 d. For the seed germination assay, T2 homozygous transgenic seeds were germinated on 1/2 MS medium supplemented with 0, 150, and 200 mM NaCl for 4–6 d, and were scored for germination based on whether the shoot length exceeded half that of the seed length.
Measurement of glucose/fructose/sucrose levels in leaves
Two-week-old seedlings of wild-type and transgenic rice were treated with 0 mM and 150 mM NaCl for 24 h, and then leaves were cut and weighed for fresh weight determination. Sugar extraction from the leaves was performed as described with modifications (Strand et al., 1999). Briefly, leaves were ground to powder in liquid nitrogen, and then extracted with 80% ethanol at 80 °C for 40 min, and this extraction was repeated once. Extracts were evaporated to dryness then dissolved in sterile water. Spectroscopic quantification and calculation of the sugar contents were performed according to the protocol of D-glucose/D-fructose/sucrose kits (Biosentec, France).
Results
OsGMST1 encodes a putative monosaccharide transporter
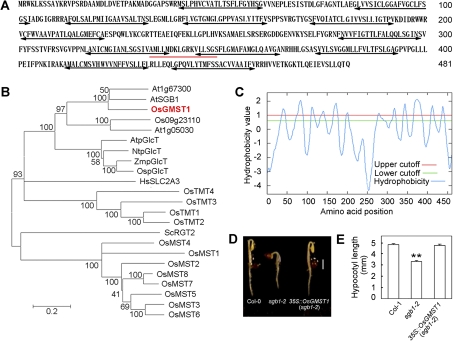
The OsGMST1 full-length cDNA is 2196 bp, with an opening reading frame of 1446 bp, encoding a protein of 481 amino acids (Fig. 1A) with a predicted molecular mass of 52 kDa and an isoelectric point of 8.45. It is a putative monosaccharide transporter, and has the sugar transport proteins signature 1 (Fig. 1A, underlined sequence) by the ExPASy (Expert Protein Analysis System) proteomics server (http://ca.expasy.org/prosite/). The plant membrane protein database Aramemnon (http://aramemnon.botanik.uni-koeln.de/) predicted 10–12 transmembrane domains, and TopPred (http://mobyle.pasteur.fr/cgi-bin/portal.py?form=toppred) predicted 12 hydrophobic transmembrane domains with a hydrophobicity value above 1.0 (Fig. 1A, double-arrow lines) arranged in two sets separated by a central hydrophilic loop (Fig. 1C). The transmembrane topology structure is consistent with the monosaccharide transporters in plants (Bush, 1999), yeast (Bisson et al., 1993), and mammals (Bell et al., 1993).
Fig. 1.
Sequence characterization of OsGMST1 and OsGMST1 complements the short hypocotyl phenotype of sgb1-2. (A) Deduced amino acid sequence of OsGMST1; numbers on the right refer to the positions of amino acid residues. Sugar transporter signature 1 predicted for OsGMST1 is indicated by the red underline. Double-headed arrows show the transmembrane regions with a high hydrophobicity value. (B) Phylogenic analysis of OsGMST1 homologues from rice and other organisms by MEGA 4.0. The predicted protein sequence is compared with OsMST1-8, Os09g23110, OspGlcT, and OsTMT1-4 from Oryza sativa, AtSGB1, At1g67300, At1g05030, and AtpGlcT from Arabidopsis thaliana, ScRGT2 from Saccharomyces cerevisiae, HsSLC2A3 from Homo sapiens, NtpGlcT from Nicotiana tabacum, and ZmpGlcT from Zea mays. (C) OsGMST1 is a membrane protein predicted by the TopPred program, showing 12 putative transmembrane helix domains. The red line and the green line indicate upper and lower cutoff of hydrophobicity, respectively. (D) Etiolated seedlings of Col-0 (WT), sgb1-2, and a transgenic line over-expressing OsGMST1 in the sgb1-2 mutant background. Bar=2 mm. (E) The hypocotyl length of etiolated seedlings. A total of 36 hypocotyls were measured for each line (wild-type, mutant, and transgenic). Error bars represent SE. ** indicates P <0.01 by Student's t test. (This figure is available in colour at JXB online.)
Phylogenetic analysis (Fig. 1B) shows that the putative Arabidopsis paralogues to OsGMST1 are encoded by At1g67300, SGB1, At1g05030, and AtpGlcT and the proteins have 67%, 65%, 44%, and 43% identity to OsGMST1, respectively. These four MST proteins are in the pGlcT subfamily (Buttner, 2007). SGB1 (suppressor of G protein beta1) was identified as a Golgi-localized hexose transporter in this subfamily (Wang et al., 2006). Wang and coworkers showed that SGB1 genetically complemented the phenotype of the agb1 mutant lacking the β subunit of G protein and that the transcript-null mutant, sgb1-2, mimicked the agb1 2 d etiolated phenotypes with shorter hypocotyls and open hooks (Wang et al., 2006). It has been proposed that, in rice (Japonica), OsGMST1, Os09g23110, and Os01g04190 (OspGlcT) belong to the pGlcT subfamily (Johnson and Thomas, 2007) with sequence identity approximately 36% (see Supplementary Fig. S1 at JXB online). Tobacco pGlcT and maize pGlcT belong to the pGlcT subfamily and are also closely related to OsGMST1 (36.33% and 36.16% identity, respectively). Yeast RGT2 and human SCL2A are similar to OsGMST1.
Expression of OsGMST1 in sgb1-2 mutants rescued the short hypocotyl phenotype to the wild type (P=0.634) (Fig. 1D, E), indicating that OsGMST1 is paralogous to SGB1. As was the case for SGB1 (Wang et al., 2006), OsGMST1 did not rescue the growth phenotype of a yeast strain EBY.VW4000 (Wieczorke et al., 1999) which lacked 22 monosaccharide transporter genes, HXT1-17, GAL2, STL1, AGT1, MPH2, and MPH3 (see Supplementary Fig. S2 at JXB online).
OsGMST1 is localized on the Golgi apparatus
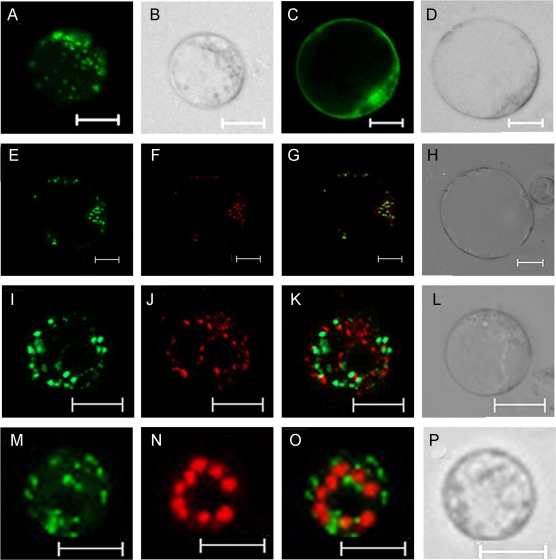
To identify the subcellular localization of OsGMST1, a transient expression vector harbouring OsGMST1-GFP fusion protein driven by the 35S promoter (35S::OsGMST1-GFP) was constructed. Protoplast cells were prepared from leaf sheaths of 7–10 d dark grown rice seedlings, and then transiently transformed with recombinant or empty vector as a control. Fluorescence images were observed under the confocal microscope. OsGMST1-GFP fluorescence was distributed in cells in a punctate pattern (Fig. 2A, B), but GFP alone was distributed throughout the cytoplasm (Fig. 2C, D). To test the hypothesis that OsGMST1 is localized to the Golgi as is SGB1 (Wang et al., 2006), OsGMST1-GFP was co-expressed with the Golgi marker sialyltransferase (ST) tagged red fluorescence protein (RFP). Most of the red and green fluorescence merged (Fig. 2E, F, G, H), indicating that OsGMST1 was in the Golgi body. By contrast, OsGMST1-GFP fluorescence did not co-localize with the mitochondrion fluorescence dye MitoTracker Red (Fig. 2I, J, K, L) or with chlorophyll autofluorescence (Fig. 2M, N, O, P).
Fig. 2.
Subcellular localization of OsGMST1-GFP transiently expressed in rice protoplasts. (A, B) A rice protoplast cell expressing OsGMST1-GFP (A) and its DIC image (B), showing a punctuate expression pattern. Bars=10 μm. (C, D) A rice protoplast cell expressing GFP (C) as control and its DIC image (D), showing its distribution in nucleus, membrane, and cytoplasm. Bars=10 μm. (E–H) A rice protoplast cell expressing OsGMST1-GFP (E), Golgi localized ST-RFP (F) ,a merged image (G), and its DIC image (H). Bars=10 μm. (I–L) A rice protoplast expressing OsGMST1-GFP (I), stained by mitochondria dye MitoTracker Red (J), a merged image (K), and its DIC image (L). Bars=10 μm. (M–P) A rice protoplast cell expressing OsGMST1-GFP (M), the chlorophyll autofluorescence (N), a merged image (O), and its DIC image (P). Bars=10 μm. (This figure is available in colour at JXB online.)
OsGMST1 is ubiquitiously expressed
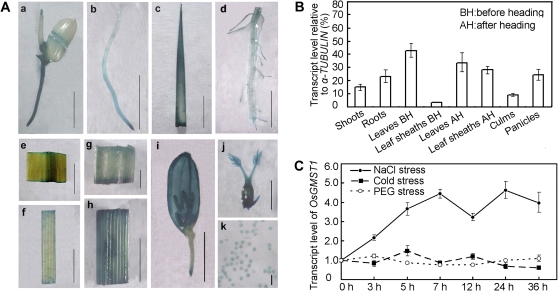
Transgenic rice plants expressing β-glucoronidase gene driven by the OsGMST1 promoter showed that OsGMST1 was expressed in all developmental stages from germination to flowering (Fig. 3A). Furthermore, quantitative PCR confirmed high levels of OsGMST1 mRNA in all the organs tested, such as the shoot, root, culm, panicle, leaf, and leaf sheath before and after heading (Fig. 3B), suggesting that OsGMST1 is ubiquitously expressed.
Fig. 3.
Expression pattern analysis of OsGMST1. (A) GUS histochemical staining in OsGMST1-promoter::GUS transgenic rice plants, showing a germinating seedling (a), young root (b), young leaf (c), mature root (d), mature leaf (e), leaf sheath (f), node (g), internode (h), panicle (i), pistil (j), and pollen gains (k). (a–f, h, i) Bars=5 mm; (g, j) 1 mm; (k) 0.1 mm. (B) Quantitative real-time PCR analysis of relative OsGMST1 transcript levels in shoots, roots, leaves, leaf sheaths, culms, and panicles, and data are the percentage of α-TUBULIN expression. The results are means ±SD of triplicate assays. (C) Real time PCR analysis of OsGMST1 transcript level under salt stress, PEG, and cold treatment with different time-courses. Error bars indicates SD of three replicates. (This figure is available in colour at JXB online.)
OsGMST1 expression is salt responsive
The OsGMST1 promoter sequence (1500 bp upstream of the start codon ATG) contains stress-responsive related cis-elements, such as ABRE, a MYB recognition site, a MYC recognition site, and a GCC box (see Supplementary Fig. S3 at JXB online) based on predictions using the PLACE database (http://www.dna.affrc.go.jp/PLACE). As shown in Fig. 3C, the transcript level of OsGMST1 in seedlings was induced by NaCl treatment but unaffected by low temperature (4 °C) and PEG4000. Salt treatment conferred a sustained increase in the steady-state level of the OsGMST1 mRNA (Fig. 3C).
Reduced expression of OsGMST1 confers hypersensitivity to NaCl
Reduced expression of OsGMST1 by antisense technology was performed (see Supplementary Fig. S4A at JXB online; see the Materials and methods). Three independent transgenic rice lines, AS-L4, L12, L18 (see Supplementary Fig. S4B at JXB online) were confirmed by quantitative PCR to have reduced expression (see Supplementary Fig. S4C at JXB online). In AS-L12 and AS-L18, the expression of OspGlcT (Os01g04190) and Os09g23110, which have the highest nucleic acid sequence identity (46.8% and 39.8%) with OsGMST1 were not affected (see Supplementary Fig. S4D at JXB online). In addition, the expression of other genes might not be affected in the antisense transgenic lines, as sequence similarity of OsGMST1 with other genes is lower than 30%, and there is no gene that has high similarity with OsGMST1 in a short sequence stretch by BLAST search in rice. The development and morphology of the T2 progeny was no different from the wild type (WT) under normal growing conditions (see Supplementary Fig. S4E at JXB online).
There were no differences in the seed germination rate between WT and the transgenic lines without NaCl stress (Fig. 4A). Treatment with 150 mM and 200 mM NaCl, however, reduced the germination rate of AS-L12 and AS-L18 (Fig. 4A, B). At 200 mM NaCl, the germination rate of the wild type was 90%, while the germination rates of the AS-L12 and AS-L18 lines were reduced to 40% and 50%, respectively (P <0.01) (Fig. 4B), and this difference was maintained over several days (P <0.01) (Fig. 4C).
Fig. 4.
Salt tolerance test of OsGMST1 antisense transgenic rice plants. (A) Seed germination of WT and OsGMST1 knockdown transgenic rice plants on MS medium containing 0 mM and 200 mM NaCl after 6 d. (B) Germination rate of WT and OsGMST1 knockdown transgenic rice seeds with 0, 150, and 200 mM NaCl treatment for 6 d. Data are means ±SD of three repeats, 20 seeds in each repeat. * and ** indicate P <0.05 and P <0.01, respectively by Student's t test. (C) Seed germination rate of WT and OsGMST1 knockdown transgenic rice seeds with 200 mM NaCl treatment in 0–6 d. Data are means ±SD of three repeats, 20 seeds in each repeat. ** indicate P <0.01 by Student's t test. (D) Growth of WT and transgenic seedlings before (left) and after (right) NaCl treatment. 2-week-old seedlings were treated with 200 mM NaCl for 9 d, and then recovered for 8 d. Bars=5 cm. (E) Survival rate of WT and transgenic seedlings after being treated with 150 mM and 200 mM NaCl. 15 seedlings at 2 weeks old were used in each repeat. Error bars are SE of three replicates. ** indicates P <0.01 by Student's t test. (F) PEG stress tolerance test of WT and OsGMST1 knockdown seedlings. Three-week-old seedlings were treated with 20% PEG for 15 d, and then recovered for 8 d. Bars=5 cm. (G) Cold stress tolerance test of WT and knockdown seedlings. 3-week-old seedlings were treated with 4 C for 4 d, and then recovered for 7 d. Bars=5 cm. (This figure is available in colour at JXB online.)
Two-week-old rice seedlings of WT, AS-L12, and AS-L18 were treated with 200 mM NaCl as a supplement in Kimura B nutrient solution (see the Materials and methods) for 9 d, and then recovered in Kimura B nutrient solution alone for 8 d. Reduced expression of OsGMST1 also conferred reduced tolerance to salt stress in young seedlings (Fig. 4D). The survival rate of AS-L12 (40% and 20% in 150 mM NaCl and 200 mM NaCl, respectively) and AS-L18 (50% and 30%) were lower than the WT (70% and 50%) with treatment of 150 mM and 200 mM NaCl (P <0.01) (Fig. 4E). By contrast, the tolerance of OsGMST1 antisense plants to PEG and cold stress was the same as that of WT plants (Fig. 4F, G). Just as for SGB1, increased expression of OsGMST1 had no effect (see Supplementary Fig. S5 at JXB online). The observation that OsGMST1 overexpression in rice seedlings did not confer increased salt tolerance suggests that OsGMST1 is not rate limiting.
Knockdown expression of OsGMST1 reduced glucose and fructose content in leaves
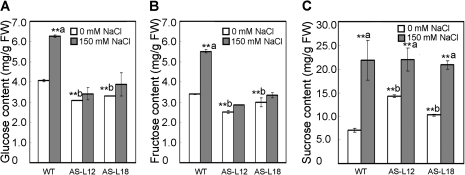
Because OsGMST1 is a putative monosaccharide transporter and its transcript is up-regulated by salt stress, the leaf sugar contents were determined in normal and salt stress conditions. As shown in Fig. 5A, under normal growth conditions, the glucose content was 4.0 mg g−1 fresh weight in the WT, and 3.0 and 3.3 mg g−1 fresh weight in the antisense transgenic plants, representing 25% and 20% reduced glucose content in knockdown plants (P <0.01). Fructose content was reduced by 25% and 12% of the WT (P <0.01) (Fig. 5B), whereas sucrose levels in transgenic rice plants were 1.5–2-fold of that in the WT, 10.0–14.0 versus 7.0 mg g−1 fresh weight, respectively (P <0.01) (Fig. 5C). The data indicated that sugar levels were interrupted in OsGMST1 knockdown rice, although they showed no visibly different phenotype compared with WT plants.
Fig. 5.
Effect of NaCl treatment on sugar content in rice leaves. Glucose (A), fructose (B), and sucrose level (C) in the leaves of WT and OsGMST1 knockdown transgenic rice plants under 0 mM and 150 mM NaCl treatment. Two-week-old seedlings were treated with 0 mM and 150 mM NaCl for 24 h, and then the sugar levels of the leaves (mg g−1 FW) were measured as described in the Materials and methods. Twelve plants were used in each experiment. Data are the means of three independent repeats ±SD. The confidence level for significant differences in sugar content between 0 mM and 150 mM NaCl is indicated by **a (P <0.01); and between WT and knockdown plants under non-salt stress treatment (0 mM NaCl) confidence is indicated by **b (P <0.01).
To analyse the differences in sugar accumulation in plants under NaCl stress further, the sugar levels were quantified after the wild-type and OsGMST1 knockdown rice were treated with 150 mM NaCl for 24 h. In leaves of WT plants, salt stress caused glucose and fructose contents to increase 50% and 60% compared with that of the untreated control leaves, respectively (P < 0.01) (Fig. 5A, B), and sucrose rose to 3-fold that of the untreated leaves (P <0.01) (Fig. 5C). In the antisense transgenic lines, glucose and fructose levels were not statistically increased by salt stress (Fig. 5A, B). In addition, sucrose contents in antisense plants accumulated only 0.5–1.0-fold higher than the untreated leaves in contrast to the 2-fold accumulation of sucrose in WT plants (P <0.01) (Fig. 5C). These results indicate that accumulation of sugars is, in part, by OsGMST1.
Discussion
OsGMST1 is a Golgi-localized monosaccharide transporter in rice
Monosaccharide transporters play a pivotal role in the translocation and distribution of monosaccharides throughout the plant. In rice, 12 MSTs, OsMST1–8 and OsTMT1–4 have been reported. Nevertheless, functional evidence for their roles in rice is not presented. In this study, we identified OsGMST1 as a Golgi-localized monosaccharide transporter in rice. Although the transport activity of OsGMST1 has not been characterized, we speculate that it is a sugar transporter based on its sequence characteristics (Fig. 1) and the effect on the sugar content when this gene expression is reduced in rice (Fig. 5). The orthologue of OsGMST1 in Arabidopsis is SGB1 (Wang et al., 2006), which is a Golgi-localized hexose transporter. Furthermore, expression of OsGMST1 in the sgb1-2 mutant can rescue the hypocotyl phenotype (Fig. 1). Therefore, we conclude that the biochemical function of OsGMST1 is the same or similar to SGB1 function. Some monosaccharide transporter genes are only expressed in source tissues (Buttner et al., 2000), and in sink tissues (Schneidereit et al., 2003, 2005; Scholz-Starke et al., 2003; Mamun et al., 2006; Oliver et al., 2007), or in both (Wormit et al., 2006; Wang et al., 2007). OsGMST1 is expressed ubiquitously (Fig. 3A, B), suggesting that it may be involved in sugar translocation in both source and sink tissues of rice.
OsGMST1 may play a role in salt-stress tolerance
Under abiotic stress conditions, soluble sugars derived from starch breakdown accumulate in plants to increase stress tolerance (Yano et al., 2005; Lee et al., 2009; Yamada et al., 2010). The Arabidopsis sex1 mutant, which was unable to accumulate sugars when exposed to freezing stress, displayed impaired freezing tolerance (Yano et al., 2005). OsGMST1 knockdown rice seedlings are hypersensitive to NaCl stress compared with the WT (Fig. 4), as the NaCl-induced accumulation of glucose and fructose is impaired in knockdown plants, and the accumulation amount of sucrose is also different from the WT (Fig. 5). Sugars generate adenosine 5'-triphosphate (ATP) and other important metabolites needed for biosynthesis and growth during O2 deficiency stress of rice (Lee et al., 2009). Sugars can also function as signals that affect gene expression under stress conditions (Hanson and Smeekens, 2009; Hey et al., 2010), so there is cross-talk between sugar signalling and stress. The interaction of stress and sugar signalling is essential for plants to tolerate stress. Sucrose non-fermenting-1-related protein kinases (SnRKs) play a central role in the interaction of the two signalling pathways, which serve as metabolic sensors to adjust energy homeostasis under stress (Halford et al., 2003; Halford and Hey, 2009). In rice, the SnRK1-dependent sugar-sensing cascade regulates sugar and energy production in order to enable rice growth under flooding stress (Lee et al., 2009). In OsGMST1 knockdown rice plants, the accumulation of sugars are affected under NaCl stress, suggesting that sugar signal transduction may be interrupted. Consequently, the expression of genes regulated by sugars may not be activated (or depressed) to provide energy for rice.
Expression of tonoplast localized TMT1 and TMT2 is induced by cold treatment, and glucose uptake into isolated leaf mesophyll vacuoles of WT plants is promoted by cold stress. In addition, tmt knockout lines accumulated less glucose and fructose compared with the WT plants (Wormit et al., 2006), analogous to results found with reduced expression of OsGMST1.
OsGMST1 resides in the Golgi apparatus (Fig. 2) and contributes to monosaccharide transport, and NaCl stress up-regulates the expression of OsGMST1 transcriptionally (Fig. 3C). One possible explanation is that NaCl stress stimulates the monosaccharide transport activity of OsGMST1 and, consequently, the activated OsGMST1 upon salt stress import sugars into the Golgi or export them out of the organelle to keep cytosolic sugar at homeostasis. In conclusion, it is hypothesized that sugar homeostasis is affected in OsGMST1 knockdown transgenic rice, which putatively results in altered sugar contents, so the seedlings show a NaCl hypersensitive phenotype.
In summary, OsGMST1 is a novel Golgi-localized monosaccharide transporter that may play a role in salt stress tolerance in rice.
Supplementary data
Supplementary data can be found at JXB online.
Supplementary Fig. S1. Alignment of OsGMST1 with other members of the rice pGlcT (plastid glucose transporter) subfamily.
Supplementary Fig. S2. Growth of hexose transporter-deficient yeast cells transformed with sense and antisense OsGMST1 on plates supplemented with sugars.
Supplementary Fig. S3. Distribution of stress related cis-elements in the OsGMST1 promoter region.
Supplementary Fig. S4. Molecular identification and phenotype observation of OsGMST1 knockdown transgenic rice plants.
Supplementary Fig. S5. Molecular identification and phenotype observation of OsGMST1 overexpressed transgenic rice plants.
Acknowledgments
The authors thank Professor Norbert Sauer of Germany for providing the vector pEX-Tag and Professor Eckhard Boles of Germany for providing the yeast strain EBY.VW4000. This work was supported by the Foundation for Innovative Research Groups of the National Natural Science Foundation of China (30821007). Work in the Jones laboratory was supported by grants to AMJ from the NIGMS (R01GM065989), DOE (DE-FG02-05er15671), and NSF (MCB-0723515 and MCB-0718202).
References
- Bart R, Chern M, Park CJ, Bartley L, Ronald PC. A novel system for gene silencing using siRNAs in rice leaf and stem-derived protoplasts. Plant Methods. 2006;2:13–21. doi: 10.1186/1746-4811-2-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell GI, Burant CF, Takeda J, Gould GW. Structure and function of mammalian facilitative sugar transporters. Journal of Biological Chemistry. 1993;268:19161–19164. [PubMed] [Google Scholar]
- Bisson LF, Coons DM, Kruckeberg AL, Lewis DA. Yeast sugar transporters. Critical Reviews in Biochemistry and Molecular Biology. 1993;28:259–308. doi: 10.3109/10409239309078437. [DOI] [PubMed] [Google Scholar]
- Bush DR. Sugar transporters in plant biology. Current Opinion in Plant Biology. 1999;2:187–191. doi: 10.1016/S1369-5266(99)80034-X. [DOI] [PubMed] [Google Scholar]
- Buttner M. The monosaccharide transporter(-like) gene family in Arabidopsis. FEBS Letters. 2007;581:2318–2324. doi: 10.1016/j.febslet.2007.03.016. [DOI] [PubMed] [Google Scholar]
- Buttner M, Truernit E, Baier K, Scholz-Starke J, Sontheim M, Lauterbach C, Huss VAR, Sauer N. AtSTP3, a green leaf-specific, low affinity monosaccharide-H+ symporter of Arabidopsis thaliana. Plant and Cell Physiology. 2000;23:175–184. [Google Scholar]
- Chen RF, Shen RF, Gu P, Dong XY, Du CW, Ma JF. Response of rice (Oryza sativa) with root surface iron plaque under aluminium stress. Annals of Botany. 2006;98:389–395. doi: 10.1093/aob/mcl110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho JI, Burla B, Lee DW, et al. Expression analysis and functional characterization of the monosaccharide transporters, OsTMTs, involving vacuolar sugar transport in rice (Oryza sativa. New Phytologist. 2010;186:657–668. doi: 10.1111/j.1469-8137.2010.03194.x. [DOI] [PubMed] [Google Scholar]
- Ge L, Chen H, Jiang JF, Zhao Y, Xu ML, Xu YY, Tan KH, Xu ZH, Chong K. Overexpression of OsRAA1 causes pleiotropic phenotypes in transgenic rice plants, including altered leaf, flower, and root development and root response to gravity. Plant Physiology. 2004;135:1502–1513. doi: 10.1104/pp.104.041996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson SI. Control of plant development and gene expression by sugar signaling. Current Opinion in Plant Biology. 2005;8:93–102. doi: 10.1016/j.pbi.2004.11.003. [DOI] [PubMed] [Google Scholar]
- Halford NG, Hey S, Jhurreea D, Laurie S, McKibbin RS, Paul M, Zhang Y. Metabolic signalling and carbon partitioning: role of Snf1-related (SnRK1) protein kinase. Journal of Experimental Botany. 2003;54:467–475. doi: 10.1093/jxb/erg038. [DOI] [PubMed] [Google Scholar]
- Halford NG, Hey SJ. Snf1-related protein kinases (SnRKs) act within an intricate network that links metabolic and stress signalling in plants. Biochemical Journal. 2009;419:247–259. doi: 10.1042/BJ20082408. [DOI] [PubMed] [Google Scholar]
- Hanson J, Smeekens S. Sugar perception and signaling: an update. Current Opinion in Plant Biology. 2009;12:562–567. doi: 10.1016/j.pbi.2009.07.014. [DOI] [PubMed] [Google Scholar]
- Hey SJ, Byrne E, Halford NG. The interface between metabolic and stress signalling. Annals of Botany. 2010;105:197–203. doi: 10.1093/aob/mcp285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. The EMBO Journal. 1987;6:3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DA, Hill JP, Thomas MA. The monosaccharide transporter gene family in land plants is ancient and shows differential subfamily expression and expansion across lineages. BMC Evolutionary Biology. 2006;6:64–83. doi: 10.1186/1471-2148-6-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DA, Thomas MA. The monosaccharide transporter gene family in Arabidopsis and rice: a history of duplications, adaptive evolution, and functional divergence. Molecular Biology and Evolution. 2007;24:2412–2423. doi: 10.1093/molbev/msm184. [DOI] [PubMed] [Google Scholar]
- Kato-Noguchi H, Ino T. Possible involvement of momilactone B in rice allelopathy. Journal of Plant Physiology. 2005;162:718–721. doi: 10.1016/j.jplph.2004.11.009. [DOI] [PubMed] [Google Scholar]
- Kiyosue T, Abe H, Yamaguchi-Shinozaki K, Shinozaki K. ERD6, a cDNA clone for an early dehydration-induced gene of Arabidopsis, encodes a putative sugar transporter. Biochimica et Biophysica Acta. 1998;1370:187–191. doi: 10.1016/s0005-2736(98)00007-8. [DOI] [PubMed] [Google Scholar]
- Klepek YS, Geiger D, Stadler R, Klebl F, Landouar-Arsivaud L, Lemoine R, Hedrich R, Sauer N. Arabidopsis POLYOL TRANSPORTER5, a new member of the monosaccharide transporter-like superfamily, mediates H+-symport of numerous substrates, including myo-inositol, glycerol, and ribose. The Plant Cell. 2005;17:204–218. doi: 10.1105/tpc.104.026641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalonde S, Boles E, Hellmann H, Barker L, Patrick JW, Frommer WB, Ward JM. The dual function of sugar carriers. Transport and sugar sensing. The Plant Cell. 1999;11:707–726. doi: 10.1105/tpc.11.4.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KW, Chen PW, Lu CA, Chen S, Ho TH, Yu SM. Coordinated responses to oxygen and sugar deficiency allow rice seedlings to tolerate flooding. Science Signaling. 2009;2:ra61. doi: 10.1126/scisignal.2000333. [DOI] [PubMed] [Google Scholar]
- Mamun EA, Alfred S, Cantrill LC, Overall RL, Sutton BG. Effects of chilling on male gametophyte development in rice. Cell Biology International. 2006;30:583–591. doi: 10.1016/j.cellbi.2006.03.004. [DOI] [PubMed] [Google Scholar]
- Meyer S, Melzer M, Truernit E, Hummer C, Besenbeck R, Stadler R, Sauer N. AtSUC3, a gene encoding a new Arabidopsis sucrose transporter, is expressed in cells adjacent to the vascular tissue and in a carpel cell layer. The Plant Journal. 2000;24:869–882. doi: 10.1046/j.1365-313x.2000.00934.x. [DOI] [PubMed] [Google Scholar]
- Moore B, Sheen J. Plant sugar sensing and signaling: a complex reality. Trends in Plant Science. 1999;4:250–250. doi: 10.1016/s1360-1385(99)01433-8. [DOI] [PubMed] [Google Scholar]
- Ngampanya B, Sobolewska A, Takeda T, Toyofuku K, Narangajavana J, Ikeda A, Yamaguchi J. Characterization of rice functional monosaccharide transporter, OsMST5. Bioscience, Biotechnology, and Biochemistry. 2003;67:556–562. doi: 10.1271/bbb.67.556. [DOI] [PubMed] [Google Scholar]
- Niittyla T, Messerli G, Trevisan M, Chen J, Smith AM, Zeeman SC. A previously unknown maltose transporter essential for starch degradation in leaves. Science. 2004;303:87–89. doi: 10.1126/science.1091811. [DOI] [PubMed] [Google Scholar]
- Oliver SN, Dennis ES, Dolferus R. ABA regulates apoplastic sugar transport and is a potential signal for cold-induced pollen sterility in rice. Plant and Cell Physiology. 2007;48:1319–1330. doi: 10.1093/pcp/pcm100. [DOI] [PubMed] [Google Scholar]
- Rolland F, Baena-Gonzalez E, Sheen J. Sugar sensing and signaling in plants: conserved and novel mechanisms. Annual Review of Plant Biology. 2006;57:675–709. doi: 10.1146/annurev.arplant.57.032905.105441. [DOI] [PubMed] [Google Scholar]
- Rolland F, Moore B, Sheen J. Sugar sensing and signaling in plants. The Plant Cell. 2002;14(Supplement):S185–S205. doi: 10.1105/tpc.010455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer N. Molecular physiology of higher plant sucrose transporters. FEBS Letters. 2007;581:2309–2317. doi: 10.1016/j.febslet.2007.03.048. [DOI] [PubMed] [Google Scholar]
- Schneidereit A, Scholz-Starke J, Buttner M. Functional characterization and expression analyses of the glucose-specific AtSTP9 monosaccharide transporter in pollen of Arabidopsis. Plant Physiology. 2003;133:182–190. doi: 10.1104/pp.103.026674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneidereit A, Scholz-Starke J, Sauer N, Buttner M. AtSTP11, a pollen tube-specific monosaccharide transporter in Arabidopsis. Planta. 2005;221:48–55. doi: 10.1007/s00425-004-1420-5. [DOI] [PubMed] [Google Scholar]
- Scholz-Starke J, Buttner M, Sauer N. AtSTP6, a new pollen-specific H+-monosaccharide symporter from Arabidopsis. Plant Physiology. 2003;131:70–77. doi: 10.1104/pp.012666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheen J. Signal transduction in maize and Arabidopsis mesophyll protoplasts. Plant Physiology. 2001;127:1466–1475. [PMC free article] [PubMed] [Google Scholar]
- Strand A, Hurry V, Henkes S, Huner N, Gustafsson P, Gardestrom P, Stitt M. Acclimation of Arabidopsis leaves developing at low temperatures. Increasing cytoplasmic volume accompanies increased activities of enzymes in the Calvin cycle and in the sucrose-biosynthesis pathway. Plant Physiology. 1999;119:1387–1398. doi: 10.1104/pp.119.4.1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szarka A, Horemans N, Banhegyi G, Asard H. Facilitated glucose and dehydroascorbate transport in plant mitochondria. Archives of Biochemistry and Biophysics. 2004;428:73–80. doi: 10.1016/j.abb.2004.05.011. [DOI] [PubMed] [Google Scholar]
- Toyofuku K, Kasahara M, Yamaguchi J. Characterization and expression of monosaccharide transporters (OsMSTs) in rice. Plant and Cell Physiology. 2000;41:940–947. doi: 10.1093/pcp/pcd016. [DOI] [PubMed] [Google Scholar]
- Turgeon R, Medville R. Phloem loading. A reevaluation of the relationship between plasmodesmatal frequencies and loading strategies. Plant Physiology. 2004;136:3795–3803. doi: 10.1104/pp.104.042036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HX, Weerasinghe RR, Perdue TD, Cakmakci NG, Taylor JP, Marzluff WF, Jones AM. A Golgi-localized hexose transporter is involved in heterotrimeric g protein-mediated early development in Arabidopsis. Molecular Biology of the Cell. 2006;17:4257–4269. doi: 10.1091/mbc.E06-01-0046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Xiao Y, Zhang Y, Chai C, Wei G, Wei X, Xu H, Wang M, Ouwerkerk PB, Zhu Z. Molecular cloning, functional characterization and expression analysis of a novel monosaccharide transporter gene OsMST6 from rice (Oryza sativa L.) Planta. 2008;228:525–535. doi: 10.1007/s00425-008-0755-8. [DOI] [PubMed] [Google Scholar]
- Wang Y, Xu H, Wei X, et al. Molecular cloning and expression analysis of a monosaccharide transporter gene OsMST4 from rice (Oryza sativa L.) Plant Molecular Biology. 2007;65:439–451. doi: 10.1007/s11103-007-9228-x. [DOI] [PubMed] [Google Scholar]
- Weber A, Servaites JC, Geiger DR, Kofler H, Hille D, Groner F, Hebbeker U, Flugge UI. Identification, purification, and molecular cloning of a putative plastidic glucose translocator. The Plant Cell. 2000;12:787–802. doi: 10.1105/tpc.12.5.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weschke W, Panitz R, Gubatz S, Wang Q, Radchuk R, Weber H, Wobus U. The role of invertases and hexose transporters in controlling sugar ratios in maternal and filial tissues of barley caryopses during early development. The Plant Journal. 2003;33:395–411. doi: 10.1046/j.1365-313x.2003.01633.x. [DOI] [PubMed] [Google Scholar]
- Wieczorke R, Krampe S, Weierstall T, Freidel K, Hollenberg CP, Boles E. Concurrent knock-out of at least 20 transporter genes is required to block uptake of hexoses in Saccharomyces cerevisiae. FEBS Letters. 1999;464:123–128. doi: 10.1016/s0014-5793(99)01698-1. [DOI] [PubMed] [Google Scholar]
- Williams LE, Lemoine R, Sauer N. Sugar transporters in higher plants: a diversity of roles and complex regulation. Trends in Plant Science. 2000;5:283–290. doi: 10.1016/s1360-1385(00)01681-2. [DOI] [PubMed] [Google Scholar]
- Wormit A, Trentmann O, Feifer I, Lohr C, Tjaden J, Meyer S, Schmidt U, Martinoia E, Neuhaus HE. Molecular identification and physiological characterization of a novel monosaccharide transporter from Arabidopsis involved in vacuolar sugar transport. The Plant Cell. 2006;18:3476–3490. doi: 10.1105/tpc.106.047290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K, Osakabe Y, Mizoi J, Nakashima K, Fujita Y, Shinozaki K, Yamaguchi-Shinozaki K. Functional analysis of an Arabidopsis thaliana abiotic stress-inducible facilitated diffusion transporter for monosaccharides. Journal of Biological Chemistry. 2010;285:1138–1146. doi: 10.1074/jbc.M109.054288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano R, Nakamura M, Yoneyama T, Nishida I. Starch-related alpha-glucan/water dikinase is involved in the cold-induced development of freezing tolerance in Arabidopsis. Plant Physiology. 2005;138:837–846. doi: 10.1104/pp.104.056374. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.