Abstract
CLE (CLAVATA3/ESR-related) peptides are developmental regulators that are secreted into the apoplast. Little is known about the role of the sequences that flank CLE peptides in terms of their biological activity or how they are targeted by proteases that are known to liberate the final active CLE peptides from their precursor sequences. The biological activity of Medicago truncatula CLE36, which possesses broadly conserved border sequences flanking the putative final active CLE36 peptide product, was assessed. Using in vitro root growth assays and an in vitro root and callus formation assay it is shown that CLE36 peptides of different lengths possess differential biological activities. Using mass spectrometry, Glycine max and Medicago extracellular fluids were each shown to possess an endoproteolytic activity that recognizes and cleaves at border sequences in a synthetic 31 amino acid CLE36 ‘propeptide bait’ to liberate biologically active peptide products. Inhibitor studies suggest that a subtilisin, in combination with a carboxypeptidase, liberated and trimmed CLE36, respectively, to form biologically relevant 11–15 amino acid cleavage products. The 15 amino acid cleavage product is more biologically potent on Arabidopsis than shorter or longer CLE peptides. In situ hybridization shows that the soybean orthologue of CLE36 (GmCLE34) is expressed in the provascular tissue. The results suggest that secreted subtilisins can specifically recognize the border sequences of CLE36 propeptides and liberate biologically active cleavage products. These secreted proteases may affect the stability and biological activity of CLE peptides in the apoplast or be involved in CLE36 processing.
Keywords: Arabidopsis, carboxypeptidase, CLE peptide, mass spectrometry, Medicago, proteomics, rice, secreted proteins, soybean, subtilisin
Introduction
Regulatory peptides are important developmental signal molecules in plants. They affect diverse plant processes including defence (e.g. systemins and Propep1; Huffaker et al., 2006), stem cell and vascular differentiation [CLAVATA3/ESR-related CLE peptides and tracheary element differentiation inhibitory factor (TDIF); Fiers et al., 2006; Kondo et al., 2006, 2011; Matsubayashi and Sakagami, 2006; Ni and Clark, 2006], and the local and systemic control of root nodulation (Mortier et al., 2010; Okamoto et al., 2009; Reid et al., 2011; Saur et al., 2011).
Most regulatory peptides are secreted into the apoplast where they act as short-range signals affecting nearby cell populations (DeYoung et al., 2006; Matsubayashi and Sakagami, 2006). The precursors of these regulatory peptides usually possess N-terminal secretion signals that facilitate their entry to the endoplasmic reticulum/Golgi secretion pathway before their release into the apoplast. The final active peptide is a smaller part of a predicted propeptide located at or near the propeptide's C-terminus (Matsubayashi and Sakagami, 2006) and there is a requirement for one or two endoproteolytic cleavages to liberate the final active product. Proteolytic processing of CLE propeptides is predicted, and recent evidence points to a role for subtilisins and carboxypeptidases in CLV3 processing (Ni et al., 2011).
The CLE peptide family has been extensively studied in Arabidopsis, rice, and Zinnia (Matsubayashi and Sakagami, 2006; Kinoshita et al., 2007), and the regulatory peptides encoded by this family play roles in many important developmental processes (Jun et al., 2010). CLV3 is an archetypal member of this large gene family that regulates shoot apical meristem (SAM) formation. The CLV3 ligand, thought to be a 12 or 13 amino acid peptide (Ohyama et al., 2009), interacts with membrane-bound receptors (Ogawa et al., 2008) to control the expression of the homeodomain transcription factor, WUSCHEL (WUS). Similarly, AtCLE40 regulates the expression of the homeodomain transcription factor WOX5 to control root stem cell differentiation (Stahl et al., 2009). In contrast, the AtCLE41/44 and AtCLE42 TDIF CLE peptides suppress Zinnia xylem cell development and promote cell division (Ito et al., 2006), but have no effect on the root apical meristem (RAM) or SAM. A subset of CLE peptides regulate protoxylem formation (Kondo et al., 2011). Commonly used approaches to assess biological activity include root growth inhibition bioassays (Ito et al., 2006; Kondo et al., 2006; Oelkers et al., 2008) and overexpression studies (Strabala et al., 2006; Mortier et al., 2010; Saur et al., 2011). For example, the addition of synthesized CLE peptides to Arabidopsis inhibits root growth at concentrations in the mid to high nanomolar range by reducing the proliferation of the stem cell daughters and altering the specification of certain cell types (Fiers et al., 2005).
Until recently, little was known about how CLE peptides are processed from larger precursors, which proteases are involved, what role bordering sequences play in processing or about the stability of CLE peptides in the apoplast. Sequences bordering the 12–13 amino acid final product of CLV3 may represent endoprotease cleavage sites (Ni et al., 2011). Regions of amino acid homology exist outside the conserved 12 amino acid consensus region (Oelkers et al., 2008; Okamoto et al., 2009) especially in orthologous sequences across divergent species, suggesting functionality for these border sequences. However, the significance of these broadly conserved CLE border sequences still requires investigation (Ni et al., 2011).
Hundreds of proteases exist in plants and they control a wide diversity of functions including development. A huge knowledge gap exists in identifying the in vivo substrates for these proteases (van der Hoorn, 2008). A distinct subset of plant proteases are secreted into the apoplast, but little information exists on their functional roles. Subtilisins are part of a large family (56 members in Arabidopsis) of proteases with functional redundancy (Rautengarten et al., 2005), and many subtilisins are secreted into the apoplast. Specific subtilisins, for example STOMATAL DENSITY AND DISTRIBUTION 1 (SDD1) and ABNORMAL LEAF SHAPE 1 (ALE1), have specific roles in plant development (Rautengarten et al., 2005; Srivastava et al., 2008; van der Hoorn, 2008).
In this study the function of the Medicago CLE36 peptide and its surrounding sequences was explored. MtCLE36 differs from other CLE types since it possesses extensive areas of protein sequence homology surrounding the putative 12 amino acid MtCLE36 peptide consensus sequence when compared with orthologues in other species and especially with CLE34 in soybean. Since the putative CLE36 peptide is near to but not at the predicted C-terminus, two endoproteolytic cleavages are required to liberate a functional CLE36 peptide from its predicted precursor. The function of the conserved border sequences was explored by synthesizing a series of CLE36 peptides of different lengths. The ability of these CLE36 peptides to inhibit root growth or to affect in vitro root and callus formation from leaf explants was assessed. In situ hybridization was used to determine the tissues where this gene is expressed. Using mass spectrometry, it was determined if the CLE36 border sequences were the targets for secreted proteases previously identified in Medicago and soybean such as subtilisins, aspartic proteases, and carboxypeptidases (Djordjevic et al., 2007; Kusumawati et al., 2008). To determine this, a ‘CLE36 31 amino acid propeptide bait’ sequence was utilized which incorporates the putative CLE36 12 amino acid consensus sequence as a target for the secreted proteases present in two extracellular fluids. It was determined whether the proteases specifically targeted the CLE36 border sequences in the bait sequence and if the breakdown products had biological relevance.
Materials and methods
Plant growth and root apical meristem inhibition assays
Seeds of Trifolium repens white clover (cv. Haifa), T. subterraneum subterranean clover (cv. Woogenellup (Morris and Djordjevic, 2006), or M. truncatula (A17) were surfaced sterilized with 50% hypochlorite and germinated on plates containing nitrogen-free Fåhraeus (F) medium (Morris and Djordjevic, 2001) solidified with 0.8% agar (Gelita, Beaudesert, Australia). After germination, seedlings with root radicals of ∼5–10 mm in length were transferred to fresh F plates containing CLE peptides at concentrations ranging from 100 pM to 10 μM or with no CLE peptide addition. Filter-sterilized CLE peptides were added to F medium after autoclaving. For Arabidopsis thaliana (ecotype Columbia), solidified MS medium (Sigma-Aldrich, St Louis, MO, USA) was used. Root lengths of the seedlings were marked at day 0 and subsequent growth measured each day over a period of 4–7 d. Plants were grown at 25 °C with a 16 h/8 h day/night cycle at 60–100 μE m−2 s−1.
The inhibition of RAM growth was assessed for all assays using a one-way Student t-test. Results were considered significant with a P-value of <0.05. In all root growth inhibition experiments, n was a minimum of eight and experiments were repeated independently. Two types of significant root growth inhibition responses were recorded: ‘arrested’ and ‘slowed’ (see Results). Arrested growth was defined as roots that failed to increase in length after the first effects of CLE-induced root growth inhibition were measured. Slowed growth was defined as roots that showed moderate root growth inhibition which failed to achieve the root length of the control. Both responses were significantly different from that of the controls.
Oryza sativa (rice) seeds (cv. Doongara) were germinated on F medium as above. Rice seedlings were transferred to platforms sitting in Magenta jars above 100 ml of F medium containing 10 μM CLE peptides. Root lengths were compared after 10 d exposure to peptides.
In vitro root and callus formation
Medicago truncatula cv Jemalong seed line 2HA was used for the leaf explant tissue culture (Nolan and Rose, 1998; Nolan et al., 2003). Plants were grown under controlled growth cabinet conditions with a 12 h photoperiod at 150 μmol m−2 s−1 and a 23 °C day temperature, a 19 °C night temperature, and a relative humidity of 80%. The basal medium used for the explant leaf culture was P4, which is based on Gamborg's B5 medium (Imin et al., 2007). Leaf explants were plated onto P4 medium containing 10 μM NAA (1-naphthaleneacetic acid; Sigma-Aldrich) with or without 4 μM BAP (6-benzylaminopurine; Sigma-Aldrich). CLE peptides were added at 10 μM. Cultures were incubated in the dark at 28 °C as described (Imin et al., 2007).
Peptide synthesis
Peptides were synthesized at the Biomolecular Resource Facility at the Australian National University according to methods in Saur et al. (2011). Peptides were synthesized with a C-terminal carboxamide, purified via reverse-phase HPLC, and the quality checked by matrix-assisted laser desorption ionization-time of flight/time of flight-mass spectrometry (MALDI-TOF/TOF-MS; see below). The peptides used are listed in Table 1.
Table 1.
Root growth inhibition activity of CLE36 peptides on three legumes, Arabidopsis, and rice
| Peptidea | Sequence | Mtb | Trb | Tsb | Atb | Osb |
| Control | N | N | N | N | N | |
| CLE34/36 (31aa)c | RAELDFNYMSKRRVPNGPDPIHNRRAGNSGR | S | A | A | A | A |
| CLE36 (15aa) | SKRRVPNGPDPIHNR | A | A | A | A | A |
| CLE36 (14aa) | SKRRVPNGPDPIHN | A | A | A | A | A |
| CLE36 (12aa) | RRVPNGPDPIHN | A | A | A | A | – |
| CLE36 (12aa) | RVPNGPDPIHNR | A | A | – | – | – |
| CLE36 (12aa) | KRRVPNGPDPIH | N | N | N | N | N |
| CLE36 (11aa) | RVPNGPDPIHN | A | A | S | A | S |
| CLE36 (10aa) | RVPNGPDPIH | N | N | N | N | N |
| CLV3 (14aa) | LRTVPSGPDPLHHH | S | A | N | A | A |
| CLV3 (12aa) | RTVPSGPDPLHH | S | A | A | A | A |
| CLV3 (12aa) | RTVPSGPDPLHH | – | – | A | – | – |
| CLV3 (10aa) | RTVPSGPDPL | N | N | N | – | – |
| CLE65/TDIF (14aa)d | AHEVPSGPNPISNR | N | N | N | N | N |
Nomenclature for CLE peptides as in Oelkers et al (2008). Peptides were added at 10μM. An underlined P indicates a hydroxyproline residue present in a CLV3 peptide derivative. ‘aa’, amino acid; the length the synthesized CLE peptide is indicated.
The plants used were M. truncatula (Mt), T. repens (Tr), T. subterraneum (Ts), A. thaliana (At), and O. sativa (Os). N is normal root growth, S is significantly reduced but slowed root growth, A is arrested root growth, and ‘–’ is not determined.
CLE34/36 refers to the conserved CLE36 in M. truncatula (TC131785) and CLE34 G. max (TC232036). The 31 amino acid peptide synthesized is CLE34.
CLE65 (TC 109337) is the putative MtTDIF (treachery element differentiation inhibitory factor); it is 100% homologous to the corresponding Arabidopsis and Zinnia TDIF peptide (Ito et al., 2006).
CLE peptide incubations in Medicago and soybean extracellular fluids
Soybean xylem sap was extracted from cultivar Bragg plants (Djordjevic et al., 2007). The CLE34/36 31 amino acid propeptide was added (10 mg ml−1) to the sap at a 1:1 (volume: volume) ratio and 1 μl aliquots removed at 0, 1, 3, 6, 24 and 48 h. Reactions were stopped by spotting a 1 μl aliquot onto a 374 spot MALDI target plate with 0.5 μl of α-cyano-4-hydroxy-cinnamic acid matrix and acidifying with 0.2 μl of 1% trifluoroacetic acid (TFA).
A cell suspension culture of the M. truncatula 2HA line was established (Kusumawati et al., 2008) and subcultured every 2 weeks by transferring 30 ml of culture into 50 ml of fresh Hildebrand and Schenk medium. The supernatants of suspension cultures were obtained (Kusumawati et al., 2008), freeze-dried, and resuspended in deionized water. Samples were incubated with the CLE34/36 31 amino acid propeptide for different times and aliquots assessed at 0 h and 3 h after addition.
Mass spectrometry analysis of CLE cleavage products
Peptide samples incubated in extracellular fluids were subjected to MS and MS/MS analysis in an ABI 4800 MALDI-TOF/TOF mass spectrometer (Applied Biosystems, Foster City, CA, USA) at the Biomolecular Resource Facility at the Australian National University. Spectra were obtained in positive ion reflectron mode (Zhang et al., 2006; Djordjevic et al., 2007; Miyahara et al., 2008). External calibration for MS was done using LaserBio Labs (Cedex, France) peptide calibration Mix 4; using angiotensin II [M+H]+ 1046.54, neurotensin [M+H] + 1672.92, adrenocorticotrophic hormone (ACTH; 18–39) [M+H] + 2465.20, and oxidized insulin B chain [M+H]+ 3494.65. External 10 point calibration for MS/MS was done using ACTH (18–39) [M+H] + 2465.20. Spectra were examined for the presence of the intact parent ions of the peptides and the corresponding cleavage products. Controls samples included the examination of peptide alone (without incubation with the extracellular fluids) or only extracellular fluids without peptide addition. The sequences of the cleavage products in peptide plus extracellular fluid samples were determined by comparisons with the theoretical m/z of the cleavage products calculated using the Peptide Mass algorithm at the ExPasy Server (http://au.expasy.org/tools/pi_tool.html). Most peaks were subjected to MS/MS analysis to confirm the sequence identify of the cleavage products. The cleavage products were found in independent experiments. For protease inhibition studies samples were treated with 1 mM phenylmethylsulphonyl fluoride (PMSF; Sigma) in isopropanol for 2 h at 4° C prior to incubation with the peptides or by adding pepstatin A to a final concentration of 150 nM. The reaction products were purified using a Zip Tip (Millipore) before MALDI-TOF/TOF analysis. Protease activity in the exudates was indirectly confirmed by boiling for 5 min to denature the proteases.
Partial purification of protease activities from soybean xylem sap
Proteins from 15 ml of soybean sap were concentrated by passage through a C18 column (500 mg/8 ml Alltech, NSW, Australia) and eluted with three 500 μl washes of 50% acetonitrile. The washes were combined and the volume reduced to 100 μl in a Speedivac. An aliquot was denatured and loaded onto an SDS–polyacrylamide gel (Invitrogen) to confirm that all the previously identified sap protein species (Djordjevic et al., 2007) eluted from the column. This was used for the western blotting. An aliquot of the remainder (5 μl) was loaded onto a TRIS-acetate 3–8% polyacrylamide gel using native gel running conditions and sample buffers according to the manufacturer's instructions (Invitrogen). A lane of the gel was stained with Coomassie blue and used as a guide to extract 10 segments of gel (5×1 mm) from an unstained lane. The gel segments were incubated separately with 10 μl of CLE34/36 31 amino acid (1 mg ml−1). Aliquots of 0.5 μl were removed at 2 h and 24 h, and acidified on a MALDI target plate as before for analysis by MALDI-TOF/TOF-MS.
Western blotting
Concentrated samples of Medicago and soybean extracellular fluids were run in duplicate on a one-dimensional poyacrylamide gel with protein size markers. One set of lanes was stained with Coomassie blue and the unstained gel portion was blotted to a polyvinylidene fluoride (PVDF) membrane before incubating with a subtilisin-specific antibody (Hamilton et al., 2003). The antibody was detected using donkey anti-rabbit IgG antibody (Amersham Biosciences, UK). The reaction was visualized with the Western Lightning Chemiluminescence Reagent ECL Plus (Amersham, UK) in the LAS 1000 Luminescence Image Analyser at the Biomolecular Resource Facility at the Australian National University.
RT-PCR analysis and in situ hybridization
The full-length cDNA sequence (Glyma01g04580.1) corresponding to TC267754, the Glycine max homologue of MtCLE36, was retrieved from Phytozome (http://www.phytozome.net). DNA primers were synthesized (Sigma, Castle Hill, NSW, Australia) to amplify a 143 bp fragment of the 3′-untranslated region (UTR) of the gene for quantitative RT-PCR analysis. The following forward and reverse primers were used: forward primer 5′-GAA AGT TAG ACA AGC TTC AGC AAC C-3′; reverse primer 5′-CAT GCA AGC ACT GAT CTC AAT TCC-3′. To determine the expression pattern of the gene encoding the G. max CLE34 peptide, an RNA template was prepared from leaf, stem, shoot apex, or root tissues of soybean using the RNAeasy Plant RNA kit (Qiagen, MD, USA). More than 30 samples were used to obtain tissues from the RAMs and SAMs, and the remaining samples were obtained from tissues pooled from at least five individual plants. A 1 μg aliquot of DNA-free RNA extract was converted into first-strand cDNA using the SuperScript III system for first-strand cDNA synthesis (Invitrogen) and oligo(dT)12–18. Subsequent quantitative RT-PCR was performed using 1 μl of cDNA template with SYBR® GreenER™ qPCR SuperMix Universal (Invitrogen) on a Mx3000P instrument (Stratagene) in a 20 μl reaction volume. ROX at a final concentration of 50 nM was used as a reference dye. A melting curve analysis was performed to check for specific product amplification. Gene expression was normalized against a housekeeping gene ELF1b (Jian et al., 2008) and relative expression was calculated using MxPro QPCR software v. 1.00 (Stratagene) according to the 2–ΔΔCT method (Livak and Schmittgen, 2001). The results were compared with those obtained from the soybean RNA-Seq Atlas (Severin et al., 2010) to ensure broad consistency before samples harvested in parallel were analysed for in situ hybridizations.
For in situ hybridization analysis of CLE34, probes were derived from a 400 bp 3′-UTR of CLE34 using the same primers that were used for the quantitative RT-PCR. Tissue fixation, probe labelling, and subsequent hybridization were carried out as described by the protocol at http://www.its.caltech.edu/∼plantlab/html/protocols.html.
Results
Putative CLE36 peptide sequences are conserved in diverse plant species
The M. truncatula gene product encoding the putative CLE36 peptide shows an extensive area of conservation across diverse plant species that extends outside the 12–14 amino acid ‘core region’ believed to represent the final active product for most CLE peptides (Fig. 1). For example, the M. truncatula CLE36 and soybean CLE34 C-terminal protein sequences are 100% homologous over 32 consecutive amino acids but diverge outside this region. In addition, a 15 amino acid region of CLE36 is 100% conserved in Medicago, rice, tomato, and poplar. The poplar CLE117 is 100% homologous to CLE36 over 21 consecutive amino acids (Fig. 1). The closest Arabidopsis homologue to CLE36 is AtCLE25 (Fig. 1), which is functionally distinct from CLV3 and one of the most effective CLE peptides regulating the size of the RAM (Kinoshita et al., 2007).
Fig. 1.
Sequence comparison of M. truncatula CLE36 orthologues in different species. The predicted CLE36 12 amino acid peptide, RRVPNGPDPIHN, is bracketed. The M. truncatula CLE36 and soybean CLE34 are 100% homologous over 32 consecutive amino acids at the C-terminal end but show sequence divergence outside this region. A 15 amino acid sequence (underlined) is 100% conserved in the species shown except the related Arabidopsis sequence (AtCLE25). Some CLE peptides, for example CLE34, CLE36, CLE163, and especially CLE117 (with 21 consecutive identical amino acids), show further amino acid conservation bordering the 15 amino acid sequence (underlined). MtCLE36 and GmCLE34 show more sequence homology in the border sequences than a comparison of the two M. truncatula sequences (CLE36 and CLE64). The left border residues of the three rice CLE36 homologues are 100% homologous to each other (DFKADDPFQD) but differ from the MtCLE36 and GmCLE34 left border sequences. Since all the CLE sequences related to CLE36 have C-terminal extensions, two endoproteolytic cleavages are required to liberate a bioactive CLE peptide centred on or near the predicted CLE36 12 amino acid consensus sequence. The CLE36 consensus is most similar to the Arabidopsis CLE25 sequence (15 of 17 residues are identical); CLE25 is incapable of complementing CLV3 in vivo. (This figure is available in colour at JXB online.)
CLE36 peptides of different lengths have differential biological activity
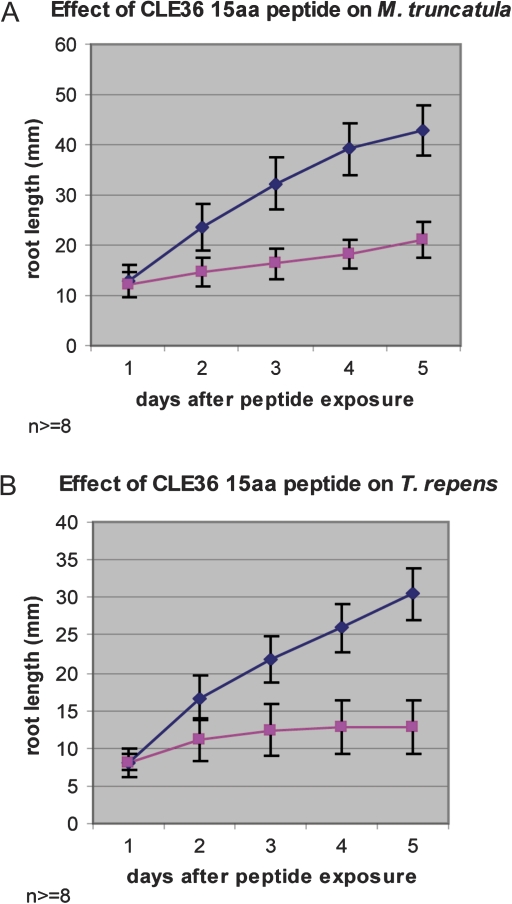
Given the high fidelity of C-terminal CLE36 sequence protein conservation across diverse species, the possibility that the homologous sequences bordering the putative CLE peptide were functionally relevant and that CLE36 had a conserved function in different plants was explored. The biological activity of synthetic CLE36 peptides of different sequence length and composition was assessed on several plants. The results showed that different length CLE36 peptides differentially affected primary root growth (Table 1). Several CLE36 derivatives of 31, 15, 14, 12, or 11 amino acids inhibited primary root growth within 2–3 d of exposure. Two primary root growth inhibition phenotypes were observed: arrested and slowed (Table 1, Fig. 2); some peptides gave no detectable response. Root growth was terminated rapidly and did not recover in plants showing the arrested phenotype, whereas root growth was significantly inhibited but continued at a reduced rate with the slowed phenotype (Fig. 2).
Fig. 2.
Examples of slowed and arrested root growth inhibition of a 15 amino acid CLE36 derivative on M. truncatula and T. repens. (A) The CLE36 15 amino acid peptide affects M. truncatula root growth from day 2 and growth is slowed thereafter. (B) The CLE36 15 amino acid peptide arrests white clover growth between day 2 and 3; no further growth occurs. Both peptides were added at 10 μM. Control plants with no peptide addition show linear growth. Both responses were significant (P <0.05; n >8). (This figure is available in colour at JXB online.)
Surprisingly, the 11 amino acid CLE36 derivative (RVPNGPDPIHN) inhibited root growth on all plants tested (Table 1), and this represents the shortest, broadly bioactive CLE peptide thus far discovered. The C-terminal asparagine of the putative CLE36 final product was critical for biological activity. Removal of this residue abolished the root growth inhibitory activity of 11 and 12 amino acid CLE36 derivatives (Table 1) and this asparagine residue most probably defines the right border of CLE36. TDIF and CLV3 peptides were used as controls (Table 1). MtCLE65 is identical to Arabidopsis and Zinnia TDIF and it shows no root growth inhibition activity, whereas CLV3 peptides with an intact C-terminus inhibited root growth as found previously (Fiers et al., 2005; Kondo et al., 2006). Similar to CLE36, deletion of the right boundary histidine residues of CLV3 peptides also generated inactive peptides (Table 1).
The biological activity of MtCLE36 peptides of different lengths (15, 12, and 11 amino acids) was also assessed on M. truncatula, T. repens, T. subterraneum, and A. thaliana using serial dilutions to determine the minimal inhibitory peptide concentration. The final size of the CLE36 peptide affected its biological activity considerably (Table 2; Supplementary Fig. S1.1–1.11 available at JXB online). For example, the CLE36 15 amino acid peptide (underlined in Fig. 1) inhibited A. thaliana primary root growth at 1 nM but the 12 and 11 amino acid CLE36 peptides required 100- and 10 000-fold higher concentrations, respectively (Table 2; Supplementary Fig. S1). In contrast, the roots of M. truncatula and T. subterraneum were most sensitive to the CLE36 12 amino acid peptide, whereas T. repens was equally responsive to each peptide (Table 2). Because of the diversity of responses to CLE36 peptides with different N-terminal amino acids, it was not possible to determine the amino acid that precisely defined the left border. However, it is likely to be the N-terminal arginine residue of the 11 amino acid CLE36 derivative (RVPNGPDPIHN) since its biological potency was greatly attenuated compared with the 15 amino derivative (SKRRVPNGPDPIHNR) on Arabidopsis.
Table 2.
Minimum inhibitory concentration (nM) of CLE36 peptide root growth inhibition on four plant species
| CLE36 peptide | A. thaliana | M. truncatula | T. subterraneum | T. repens |
| SKRRVPNGPDPIHNR | 1 | 1000 | 10 000 | 100 |
| RRVPNGPDPIHN | 100 | 100 | 10 | 100 |
| RVPNGPDPIHN | 10 000 | 10 000 | 10 000 | 100 |
Germinated seedlings with 5–10 mm root radicals were incubated with and without three CLE36 peptides (of 15, 12, and 11 amino acids in length, respectively) at concentrations ranging from 100 pM to 10 μM and grown over 4 d or 5 d (see Supplementary Fig. S1 at JXB online). Measurements were recorded daily. The minimum concentration that significantly inhibited root growth was recorded in nM for ease of comparison.
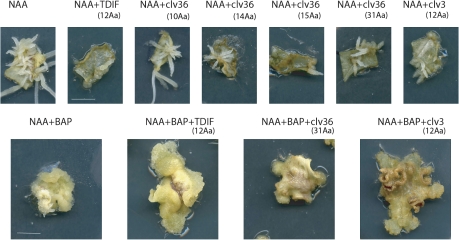
CLE36 derivatives inhibit in vitro callus and root formation
Since CLE peptides positively and negatively regulate cell proliferation and differentiation in meristems (Ito et al., 2006; Matsubayashi and Sakagami, 2006) and during vascular tissue formation (Hirakawa et al., 2008; Kondo et al., 2011), experiments were carried out to test whether the CLE36 peptides affected in vitro root and callus formation in M. truncatula. In M. truncatula, the exogenous application to leaf explants of auxin (as NAA) induces the formation of root stem cell niches that develop into root-like structures, whereas auxin and cytokinin (as BAP) addition inhibits root stem cell niche formation, and callus forms instead (Fig. 3) (Imin et al., 2007; Holmes et al., 2008). The exposure of the leaf explants to several CLE36 peptides (14, 15, or 31 amino acids) reduced the numbers of root-like structures formed on auxin-treated explants (Fig. 3). CLV3 peptide and, surprisingly, also the TDIF peptide inhibited in vitro root formation. CLE36 and CLV3 peptides, but not TDIF, inhibited in vitro callus formation in auxin- and cytokinin-treated explants. As expected, the CLE36 10 amino acid peptide (deleted for the important C-terminal asparagine residue) did not affect auxin-induced root formation (Fig. 3) and this was consistent with its lack of functionality using the root growth inhibition assay.
Fig. 3.
Effect of CLE peptides on in vitro callus and root formation in M. truncatula. Leaf explants from line 2HA were plated onto P4 medium containing 10 μM NAA with or without 4 μM BAP and with or without further addition of CLE peptides at 10 μM. All CLE36 derivatives inhibited root initiation and growth except the biologically inactive 10 amino acid derivative. In addition, TDIF and CLV3 also inhibited root initiation and growth from the explants. Callus formation was also inhibited by CLE36 and CLV3, but not TDIF. The identity of the peptides and length in parentheses are indicated. Scale bar=10 mm.
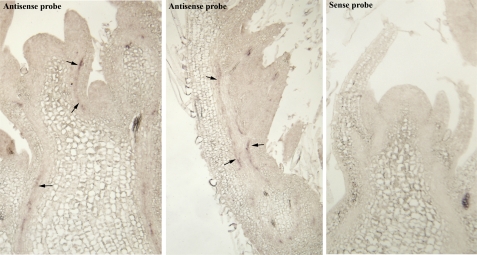
The in situ expression pattern of the CLE34 gene in soybean
The expression of the CLE36 homologue (CLE34) was examined in soybean using quantitative RT-PCR and in situ hybridization. CLE34 cDNA was amplified from the shoot apex, root, root tip, stem, and leaf tissues (Supplementary Fig 2A at JXB online). A single product was present at low levels (especially in the root tip). In situ hybridization showed that CLE34 was expressed in the provascular tissue and no expression was detected in the SAM or the meristems of lateral buds (Fig. 4). Two genes (Glyma01g04580 and Glyma02g02980) representing CLE34 were found in the RNA-Seq Atlas of G. max (Severin et al., 2010). Expression of these two genes is shown as the number of reads from Illumina deep sequencing data (Supplementary Fig. 2B). The quantitative RT-PCR results are in agreement with the RNA-Seq Atlas data. Similarly, examination of the Medicago expression atlas (http://mtgea.noble.org/v2/) showed that MtCLE36 (Affymetrix ID Mtr.42801.1.S1_at) is expressed at low levels in many tissue types (Supplementary Fig. 2C). Interestingly, MtCLE36 is repressed in root-forming culture but highly expressed in non-root-forming culture (Holmes et al., 2010; Supplementary Fig. 2C).
Fig. 4.
CLE34 expression in soybean. Signals associated with the expression of CLE34 (indicated by arrows) were observed in provascular tissues in young trifoliate leaves in the vicinity of the shoot apex. Sense probes showed no detectable expression.
Use of a CLE34/36 31 amino acid ‘propeptide bait’ to identify target sites for extracellular proteases using MALDI-TOF/TOF-MS
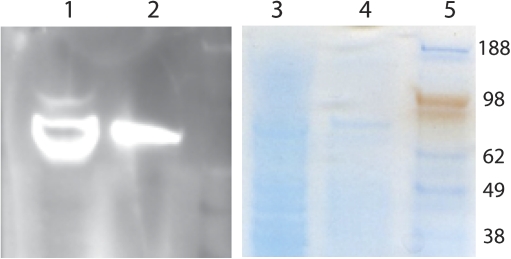
The regions of protein sequence homology surrounding the putative CLE36 peptide sequence could represent target sites for secreted proteases. To test the possible role of secreted proteases in this process, the 31 amino acid CLE34/36 sequence was used as a ‘propeptide bait’ target. This bait peptide was incubated with two extracellular fluids, soybean xylem sap (Djordjevic et al., 2007) and the culture medium of M. truncatula suspension cultures. Both fluids been shown to contain secreted proteases, (Kusumawati et al., 2008; Djordjevic et al. 2007). Previous proteomic analysis of soybean xylem fluid proteins had identified an aspartic protease amongst the identified protein species in the soybean xylem fluid (Djordjevic et al., 2007). However, previous data failed to identify the subtilisin at 80 kDa found by proteomic assessment of soybean xylem fluid by Subramanian et al. (2009). However, using western blotting (Fig. 5) the soybean xylem fluid used here as well as the M. truncatula suspension culture fluid were shown to contain a subtilisin (Fig. 5; consistent with the result of Subramanian et al., 2009).
Fig. 5.
Western blotting confirms the presence of subtilisin in Medicago and soybean extracellular fluids. Western blotting using a subtilisin-specific antibody (Hamilton et al., 2003) demonstrates the presence of a subtilisin in the Medicago (lanes 1 and 3) and soybean extracellular fluids (lanes 2 and 4) at ∼80 kDa, the expected size for many subtilisin species. Lanes 1 and 2 show specific binding of the antibody to protein species in the two extracellular fluids and lanes 3 and 4 the corresponding denaturing polyarylamde gel before transfer of the proteins to a membrane. The material run on the gel was first concentrated on a C18 reverse phase matrix before eluting with 50% acetonitrile. (This figure is available in colour at JXB online.)
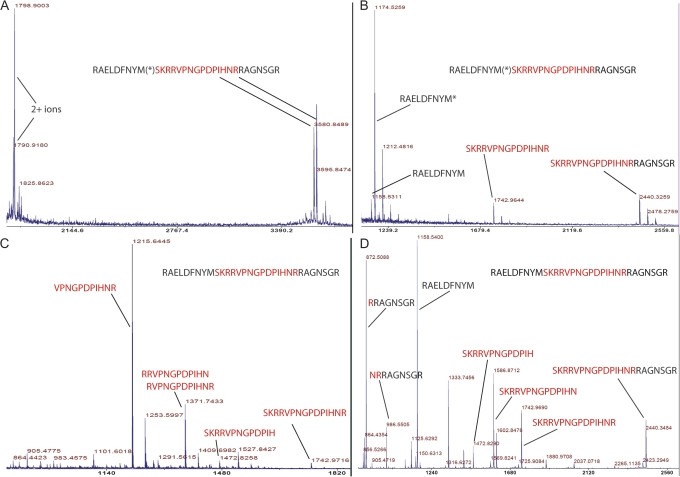
The cleavage products generated were determined by MS and their sequences confirmed by MS/MS (Fig. 6). When no extracellular fluid was added to the propeptide or if the extracellular fluid was boiled prior to addition, no cleavage of the CLE34/36 31 amino acid bait peptide occurred (Fig. 6A). However, incubation of the bait peptide in soybean xylem fluid resulted in two endoproteolytic cleavages liberating a 15 amino acid product with an m/z of 1742.96. This corresponded to the 15 amino acid conserved region of CLE36 (underlined in Fig. 1; Fig. 6B). The product at an m/z of 2440.32 corresponds to the centre and right border of the CLE34/36 propeptide (i.e. the CLE consensus and bordering C-terminal sequences to the end of the bait sequence); the products at 1158.53 and 1174.53 correspond to the left border of the CLE consensus (Fig. 6B).
Fig. 6.
MALDI-TOF/TOF spectra of the CLE34/36 31 amino acid ‘propeptide’ in the presence and absence of secreted proteases. (A) Spectrum of the CLE34/36 peptide in the absence of extracellular fluids. The expected molecular mass of the full-length product (m/z 3580.8) and its oxidized adduct (m/z 3596.8 Da) are present as well as the M/2+ ion and its oxidized adduct (1790.9 Da and 1798.9 Da). An identical spectrum resulted if the extracellular fluid was boiled prior to addition to the peptide or if PMSF was pre-incubated with the extracellular fluid prior to the addition of the peptide. (B) Digestion products after incubation of the 31 amino acid CLE34/36 propeptide in soybean xylem fluid. The parent ion is absent and instead ions at m/z 1158.6, 1174.5, and 1742.9 are present. The ions at m/z 1158.6 and 1174.5 correspond to the left border sequence (RAELDFNYM) without and with methionine oxidation (*). The ion at m/z 1742.9 corresponds to the 15 amino acid central region (underlined in Fig. 1). Potassium adducts of each ion are present (e.g. at m/z 2478.2). (C) Digestion products of CLE34/36 in M. truncatula suspension culture fluid. Ions at m/z 1215.6, 1371.7, 1472.8, and 1742.9 correspond to products ranging from 11, 12, 13, and 15 amino acids, respectively. Two peptide sequences assigned for the ion at 1371.7 are isomers and it was not possible to distinguish which was correct. Potassium adducts are apparent. (D) CLE34/36 after digestion for 2 h in non-denaturing gel band 5 (derived from soybean xylem sap). The ions at m/z 1158.5 and 2440.3 correspond to the left border (RAELDFNYM) and the middle 15 amino acids and the right border combined, respectively. The ions at m/z 1742.9, 1586.8, and 1472.8 represent 15, 14, and 13 amino acid digestion products. The peaks at 872.5 and 986.5 correspond to the sequences adjacent to the 14 amino acid and 13 amino acid peptides at 1586.8 and 1472.8, respectively, and therefore support endoproteolytic cleavages. Digestions for 24 h gave similar results except that the ion at 2440.3 was diminished or completely absent and the peaks corresponding to the 15, 14, and 13 amino acid products were proportionally elevated and a 12 amino acid product at m/z 1371.727 appeared (data not presented). The x-axis is m/z and the y-axis is relative abundance. (This figure is available in colour at JXB online.)
The addition of the CLE34/36 31 amino acid propeptide to the M. truncatula extracellular fluid resulted in a similar but more complex cleavage pattern. Endoproteolytic cleavages also liberated the same 15 amino acid product at an m/z of 1742.97 (Fig. 6C). However, additional cleavage products were also generated that were consistent with endoproteolytic cleavage at both ‘twin arginine sites’ or exoproteolytic trimming of larger products to yield smaller 13, 12, and 11 amino acid products (at m/z 1472.82, 1371.74, and 1215.64; Fig. 6C). Purification of the proteins present in the soybean extracellular fluid using non-denaturing gel electrophoresis enhanced the speed of cleavage (Fig. 6D) and supported endoproteolytic cleavage activity only.
Inhibitor studies suggested that the subtilisin species common to the Medicago and soybean extracellular fluids was responsible for the endoproteolytic cleavages. The serine protease inhibitor (PMSF) completely abolished the endoprotease cleavage of the bait peptide by both extracellular fluids, whereas the aspartic protease inhibitor (pepstatin A) did not affect the cleavage activity. Since carboxypeptidases are not capable of endoprotease cleavage, it is concluded that the subtilisin was most likely responsible for the cleavages observed.
Discussion
CLE peptide biological activity is determined by peptide size and sequence composition
Unlike most other CLE protein sequences, CLE36 shows areas of protein sequence homology surrounding the 12 amino acid CLE consensus sequence: RRVPNGPDPIHN. The present study was carried out to determine if these homologous border sequences were involved in the biological activity of CLE36 or represented target sites for proteases present in extracellular fluids. The results of assessing the biological activity of synthetic MtCLE36 peptides of different length and composition showed that these peptides differentially affect root growth on several plant species, with a 15 amino acid product being the most potent root growth inhibitor on Arabidopsis. This 15 amino acid product corresponds to the conserved core of most of the CLE36 homologues listed in Fig. 1, and it is possible that CLE36 acts as a 15 amino acid product. An 11 amino acid CLE36 product was broadly biologically active, but deletion of the C-terminal asparagine residue from CLE36 peptides rendered these peptides inactive.
The results of using other CLE peptide classes such as AtTDIF (CLE 65) and AtCLV3 also showed that peptide length and composition were important for root growth inhibition. MtTDIF was unable to inhibit root growth although it shares significant homology with CLV3 peptides that were active. The result for MtTDIF is consistent with its specific role in inhibiting tissue differentiation and promoting stem cell renewal in the vascular tissue in other species (Ito et al., 2006).
Several CLE peptides, including MtTDIF, were able to inhibit in vitro root formation from leaf explants. The repression of MtCLE36 in root-forming culture but not in non-root-forming culture (Supplementary Fig. 2C at JXB online) suggests a negative regulatory role for CLE36 peptide during in vitro root formation. There was also a very low level of CLE34 gene expression in the root tip. These results may suggest that CLE peptides of diverse function may interfere with in vitro root initiation or proliferation of root stem cells. It is possible that inappropriate levels of CLE peptides could negatively affect the formation of a root stem cell niche or its development into a functional root, and this requires further investigation. CLE65 did not affect in vitro callus formation, but the other CLE peptides tested did. Therefore, in vitro root and callus formation may provide alternative bioassays to screen for differential CLE peptide activities.
Two root growth inhibition phenotypes were recognized after daily measurement of root growth post-CLE peptide addition. A distinct delay in the apparent response of the root lasting 1–3 d was observed before root growth was either arrested (no further growth was recorded over the time period measured) or slowed. This apparent refractive period to CLE peptide addition is not understood and may vary between species. Nevertheless, the speed of growth inhibition on Medicago roots in this study was much faster than that recorded in other studies (Oelkers et al., 2008). The basis for slowed growth is also not understood as CLE addition would most probably lead to stem cell differentiation which should terminate root growth, leading to arrest. The arrest of root growth was most apparent in T. repens, whereas M. truncatula predominantly responded to the various added CLE peptides with slowed growth.
An extracellular protease likely to be a subtilisin targets CLE36 border sequences and may affect the stability and biological activity of CLE36 peptides
The results show that secreted proteases found in extracellular fluids of two legume species can target specific sequences bordering the putative CLE36 peptide, and it is possible that these proteases may affect the stability and biological activity of CLE peptides in vivo. There is evidence for specific endoproteolytic cleavages of the bait peptide when it was incubated with the extracellular fluids of Medicago or Glycine. A 15 amino acid CLE36 peptide cleavage product was generated after incubation of a propeptide bait sequence with the two extracellular fluids and this appeared to be a predominant product. It was shown that this 15 amino acid CLE36 peptide sequence is highly conserved across several plant species and as a peptide it appears to be more biologically potent on some plant species (e.g. Arabidopsis) than others. Smaller CLE cleavage products were also generated; from the results in Table 1 some would be predicted to be biologically active and others would be predicted to be inactive. The results of inhibitor studies suggest that a subtilisin species present in the soybean and Medicago extracellular fluids is most likely to be responsible for the endoproteolytic cleavages observed (Djordjevic et al., 2007; Kusumawati et al., 2008). The cleavage at the twin arginines on the right border of the 15 amino acid CLE34/36 consensus and the inhibition of activity by PMSF (but not by pepstatin) is consistent with subtilisin activity, which is a serine protease, but not consistent with aspartic protease activity (Ni et al., 2011). An aspartic protease is common to soybean and M. truncatula secreted fluids but it does not appear to recognize the bait peptide. However, it cannot be fully excluded that other PMSF-sensitive serine proteases at levels too low to be detected by MS may be responsible for the endoproteolytic cleavages seen and not the more abundant subtilisin found in the soybean and M. truncatula secreted fluids. The additional cleavages observed with the Medicago extracellular fluid could be due to the presence of the carboxypeptidase species present in the Medicago extracellular fluid (Kusumawati et al., 2008) but not the soybean xylem fluid (Djordjevic et al., 2007), and therefore the results of Ni et al. (2010) for CLV3 are consistent with the present results for CLE36. Carboxypeptidases are known to trim peptides from C-termini and are also involved in animal peptide hormone trimming (Seidah and Chrétien, 1997). Secreted carboxypeptidases have been implicated in developmental control in plants and in CLE-regulated responses (Casamitjana-Martınez et al., 2003). There was no evidence that the native protease inhibitors present in the two extracellular fluids examined affected proteolytic activity (Djordjevic et al., 2007; Kusumawati et al., 2008).
It is possible that the CLE36 border sequences might represent conserved proteolysis recognition sites. Endoproteases generally cut recognition sites of up to eight amino acids (e.g. P5-P4-P3-P2-P1—P'1-P'2-P'3), with cleavage occurring between P1 and P'1 (Turk et al., 2001). There is good sequence conservation surrounding a putative C-terminal cleavage site (PIHNR-RAX) in most of the putative CLE36 orthologues shown in Fig. 1. This putative C-terminal cleavage site shows 100% sequence homology (e.g. CLE36 and CLE34), similarity (CLE163, CLE117, AtCLE25, and CLE118), or divergence (CLE64 and the three rice CLEs; Fig. 1). In addition, CLE36 (M. truncatula) and CLE34 (soybean) show good N-terminal cleavage site conservation (DFNYM-SKR), but in CLE64 (M. truncatula) and other orthologous CLEs (e.g. those in rice) this potential cleavage site is different. This opens up the possibility that different protease activities could dictate orderly CLE processing (or destruction) both within and between species, and the activity of proteases may impose an additional layer of regulation to CLE peptide activity. This may explain why many proteases are specifically expressed in time and space and precisely directed to different subcellular compartments (van der Hoorn, 2008).
Glycine max CLE34 is expressed in a broad range of tissues including provascular tissue
In situ hybridization and quantitative RT-PCR studies in G. max combined with an examination of the M. truncatula expression atlas (http://mtgea.noble.org/v2/) suggest that CLE34 and CLE36 are expressed at low levels. Few expressed sequence tags exist for CLE34 and CLE36 (http://compbio.dfci.harvard.edu/tgi/plant.html). The G. max CLE34 gene shows expression in provascular tissue, but no expression was detected in the SAM or lateral bud meristems. Based on the precise location of CLE34 gene expression in provascular tissue, it may play a role in vascular differentiation. A subfamily of CLE peptides in Arabidopsis has recently been shown to affect protoxylem formation specifically, but this subfamily excluded AtCLE25 (Kondo et al., 2011), the most homologous Arabidopsis CLE peptide to CLE36 (Fig. 1). The putative CLE36 peptide region is homologous to AtCLE25 in 15 of 17 residues. Expression of CLE36 in the provascular tissue is consistent with the in situ expression pattern of AtCLE25 in Arabidopsis (Jun et al., 2010). These CLE peptides all share the -GPDPIHNR C-terminal residues, and this site may represent a recognition site for a specific subtilisin.
Evidence has been provided that the endoproteolytic cleavage of CLE36 peptides can be mediated by secreted subtilisins and the resulting product possibly trimmed by a carboxypeptidase. This is consistent with results obtained for CLV3 (Ni et al., 2011). The observations open up the possibility that secreted subtilisins may regulate CLE activity in the apoplast by processing a larger secreted peptide precursor or by inactivating or otherwise regulating the activity of extracellular CLE peptides. CLE bait peptides may serve as useful substrates to define the in vivo targets of secreted plant proteases.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. Inhibitory responses on plant root growth by external application of CLE36 peptides.
Figure S2. Indicative expression of the genes encoding CLE36 and CLE34 in various G. max and Medicago tissues.
Acknowledgments
We acknowledge our colleagues at the Australian Research Council Centre of Excellence for Integrative Legume Research (CILR), especially Karsten Oelkers, Jeremy Weinman, Ulrike Mathesius, and Peter Gresshoff for fruitful discussions. LK is grateful for financial support from the AUSAid Scheme. We acknowledge Isabel Saur for assistance with the tissue culture experiments. Angelia M. Hartono is acknowledged for assistance with root growth inhibition assays. Dr Peter Milburn of the Biomolecular Resource Facility at the ANU is thanked for operating the MALDI-TOF/TOF. ARA-12 antibody was supplied by Dr John Hamilton, School of Biological and Biomedical Sciences, University of Durham, Durham UK. This research was funded by the Australian Research Council's Centre of Excellence Scheme (CE0348212).
References
- Casamitjana-Martınez E, Hofhuis HF, Xu J, Liu C-M, Heidstra R, Scheres B. Root-specific CLE19 overexpression and the sol1/2 suppressors implicate a CLV-like pathway in the control of Arabidopsis root meristem maintenance. Current Biology. 2003;13:1435–1441. doi: 10.1016/s0960-9822(03)00533-5. [DOI] [PubMed] [Google Scholar]
- DeYoung BJ, Bickle KL, Schrage KJ, Muskett P, Patel K, Clark SE. The CLAVATA1-related BAM1, BAM2 and BAM3 receptor kinase-like proteins are required for meristem function in Arabidopsis. The Plant Journal. 2006;45:1–16. doi: 10.1111/j.1365-313X.2005.02592.x. [DOI] [PubMed] [Google Scholar]
- Djordjevic MA, Oakes M, Li DX, Hwang CH, Hocart CH, Gresshoff PM. The Glycine max xylem sap and apoplast proteome. Journal of Proteome Research. 2007;6:3771–3779. doi: 10.1021/pr0606833. [DOI] [PubMed] [Google Scholar]
- Fiers M, Golemiec E, van der Schors R, van der Geest L, Li KW, Stiekema WJ, Liu C- M. The CLAVATA3/ESR motif of CLAVATA3 is functionally independent from the nonconserved flanking sequences. Plant Physiology. 2006;141:1284–1292. doi: 10.1104/pp.106.080671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiers M, Golemiec E, Xu J, van der Geest L, Heidstra R, Stiekema W, Liu C- M. The 14-amino acid CLV3, CLE19, and CLE40 peptides trigger consumption of the root meristem in Arabidopsis through a CLAVATA2-dependent pathway. The Plant Cell. 2005;17:2542–2553. doi: 10.1105/tpc.105.034009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton JM, Simpson DJ, Hyman SC, Ndimba BK, Slabas AR. Ara12 subtilisin-like protease from Arabidopsis thaliana: purification, substrate specificity and tissue localization. Biochemical Journal. 2003;370:57–67. doi: 10.1042/BJ20021125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirakawa Y, Shinohara H, Kondo Y, Inoue A, Nakanomyo I, Ogawa M, Sawa S, Ohashi-Ito K, Matsubayashi Y, Fukuda H. Non-cell-autonomous control of vascular stem cell fate by a CLE peptide/receptor system. Proceedings of the National Academy of Sciences, USA. 2008;105:15208–15213. doi: 10.1073/pnas.0808444105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes P, Djordjevic MA, Imin N. Global gene expression analysis of in vitro root formation in Medicago truncatula. Functional Plant Biology. 2010;37:1117–1131. [Google Scholar]
- Holmes P, Goffard N, Weiller GF, Rolfe BG, Imin N. Transcriptional profiling of Medicago truncatula meristematic root cells. BMC Plant Biology. 2008;8:21. doi: 10.1186/1471-2229-8-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huffaker A, Pearce G, Ryan CA. An endogenous peptide signal in Arabidopsis activates components of the innate immune response. Proceedings of the National Academy of Sciences, USA. 2006;103:10098–10103. doi: 10.1073/pnas.0603727103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imin N, Nizamidin M, Wu T, Rolfe BG. Factors involved in root formation in Medicago truncatula. Journal of Experimental Botany. 2007;58:439–451. doi: 10.1093/jxb/erl224. [DOI] [PubMed] [Google Scholar]
- Ito Y, Nakanomyo I, Motose H, Iwamoto K, Sawa S, Dohmae N, Fukuda H. Dodeca-CLE peptides as suppressors of plant stem cell differentiation. Science. 2006;313:842–845. doi: 10.1126/science.1128436. [DOI] [PubMed] [Google Scholar]
- Jian B, Liu B, Bi Y, Hou W, Wu C, Han T. Validation of internal control for gene expression study in soybean by quantitative real-time PCR. BMC Molecular Biology. 2008;9:59. doi: 10.1186/1471-2199-9-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jun J, Fiume E, Roeder AH, et al. Comprehensive analysis of CLE polypeptide signaling gene expression and overexpression activity in Arabidopsis. Plant Physiology. 2010;154:1721–1736. doi: 10.1104/pp.110.163683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita A, Nakamura Y, Sasaki E, Kyozuka J, Fukuda H, Sawa S. Gain-of-function phenotypes of chemically synthetic CLAVATA3/ESR-related (CLE) peptides in Arabidopsis thaliana and Oryza sativa. Plant and Cell Physiology. 2007;48:1821–1825. doi: 10.1093/pcp/pcm154. [DOI] [PubMed] [Google Scholar]
- Kondo T, Sawa S, Kinoshita A, Mizuno S, Kakimoto T, Fukuda H, Sakagami Y. A plant peptide encoded by CLV3 identified by in situ MALDI-TOF MS analysis. Science. 2006;313:845–848. doi: 10.1126/science.1128439. [DOI] [PubMed] [Google Scholar]
- Kondo Y, Hirakawa Y, Kieber JJ, Fukuda H. CLE peptides can negatively regulate protoxylem vessel formation via cytokinin signaling. Plant and Cell Physiology. 2011;52:37–48. doi: 10.1093/pcp/pcq129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusumawati L, Imin N, Djordjevic MA. Characterization of the secretome of suspension cultures of Medicago species reveals proteins important for defense and development. Journal of Proteome Research. 2008;7:4508–4520. doi: 10.1021/pr800291z. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Matsubayashi Y, Sakagami Y. Peptide hormones in plants. Annual Review of Plant Biology. 2006;57:649–674. doi: 10.1146/annurev.arplant.56.032604.144204. [DOI] [PubMed] [Google Scholar]
- Miyahara A, Hirani TA, Oakes M, Kereszt A, Kobe B, Djordjevic MA, Gresshoff PM. Soybean nodule autoregulation receptor kinase phosphorylates two kinase-associated protein phosphatases in vitro. Journal of Biological Chemistry. 2008;283:25381–25391. doi: 10.1074/jbc.M800400200. [DOI] [PubMed] [Google Scholar]
- Morris AC, Djordjevic MA. Proteome analysis of cultivar-specific interactions between Rhizobium leguminosarum biovar trifolii and subterranean clover cultivar Woogenellup. Electrophoresis. 2001;22:586–598. doi: 10.1002/1522-2683(200102)22:3<586::AID-ELPS586>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Morris AC, Djordjevic MA. The Rhizobium leguminosarum biovar trifolii ANU794 induces novel developmental responses on the subterranean clover cultivar Woogenellup. Molecular Plant-Microbe Interactions. 2006;19:471–479. doi: 10.1094/MPMI-19-0471. [DOI] [PubMed] [Google Scholar]
- Mortier V, Den Herder G, Whitford R, Van de Velde W, Rombauts S, D'Haeseleer K, Holsters M, Goormachtig S. CLE peptides control Medicago truncatula nodulation locally and systemically. Plant Physiology. 2010;153:222–237. doi: 10.1104/pp.110.153718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni J, Clark SE. Evidence for functional conservation, sufficiency, and proteolytic processing of the CLAVATA3 CLE domain. Plant Physiology. 2006;140:726–733. doi: 10.1104/pp.105.072678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni J, Guo Y, Jin H, Hartsell J, Clark SE. Characterization of a CLE processing activity. Plant Molecular Biology. 2011;75:67–75. doi: 10.1007/s11103-010-9708-2. [DOI] [PubMed] [Google Scholar]
- Nolan KE, Irwanto RR, Rose RJ. Auxin up-regulates MtSERK1 expression in both Medicago truncatula root-forming and embryogenic cultures. Plant Physiology. 2003;133:218–230. doi: 10.1104/pp.103.020917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolan KE, Rose RJ. Plant regeneration from cultured Medicago truncatula with particular reference to abscisic acid and light treatments. Australian Journal of Botany. 1998;46:151–160. [Google Scholar]
- Oelkers K, Goffard N, Weiller GF, Gresshoff PM, Mathesius U, Frickey T. Bioinformatic analysis of the CLE signaling peptide family. BMC Plant Biology. 2008;8:1–15. doi: 10.1186/1471-2229-8-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa M, Shinohara H, Sakagami Y, Matsubayashi Y. Arabidopsis CLV3 peptide directly binds CLV1 ectodomain. Science. 2008;319:294. doi: 10.1126/science.1150083. [DOI] [PubMed] [Google Scholar]
- Ohyama K, Shinohara H, Ogawa-Ohnishi M, Matsubayashi Y. A glycopeptide regulating stem cell fate in Arabidopsis thaliana. Nature Chemical Biology. 2009;5:578–580. doi: 10.1038/nchembio.182. [DOI] [PubMed] [Google Scholar]
- Okamoto S, Ohnishi E, Sato S, Takahashi H, Nakazono M, Tabata S, Kawaguchi M. Nod factor/nitrate-induced CLE genes that drive HAR1-mediated systemic regulation of nodulation. Plant and Cell Physiology. 2009;50:67–77. doi: 10.1093/pcp/pcn194. [DOI] [PubMed] [Google Scholar]
- Rautengarten C, Steinhauser D, Bussis D, Stintzi A, Schaller A, Kopka J, Altmann T. Inferring hypotheses on functional relationships of genes: analysis of the Arabidopsis thaliana subtilase gene family. PLoS Computational Biology. 2005;1:e40. doi: 10.1371/journal.pcbi.0010040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid DE, Ferguson BJ, Gresshoff PM. Inoculation- and nitrate-induced CLE peptides of soybean control NARK-dependent nodule formation. Molecular Plant-Microbe Interactions. 2011;24:606–618. doi: 10.1094/MPMI-09-10-0207. [DOI] [PubMed] [Google Scholar]
- Saur I, Oakes M, Djordjevic MA, Imin N. Crosstalk between the nodulation signaling pathway and the autoregulation of nodulation in Medicago truncatula. New Phytologist. 2011;190:865–874. doi: 10.1111/j.1469-8137.2011.03738.x. [DOI] [PubMed] [Google Scholar]
- Seidah NG, Chrétien M. Eukaryotic protein processing: endoproteolysis of precursor proteins. Current Opinion in Biotechnology. 1997;8:602–607. doi: 10.1016/s0958-1669(97)80036-5. [DOI] [PubMed] [Google Scholar]
- Severin AJ, Woody JL, Bolon YT, et al. RNA-Seq atlas of Glycine max: a guide to the soybean transcriptome. BMC Plant Biology. 2010;10:160. doi: 10.1186/1471-2229-10-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava R, Liu JX, Howell SH. Proteolytic processing of a precursor protein for a growth-promoting peptide by a subtilisin serine protease in Arabidopsis. The Plant Journal. 2008;56:219–227. doi: 10.1111/j.1365-313X.2008.03598.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl Y, Wink RH, Ingram GC, Simon R. A signaling module controlling the stem cell niche in Arabidopsis root meristems. Current Biology. 2009;19:909–914. doi: 10.1016/j.cub.2009.03.060. [DOI] [PubMed] [Google Scholar]
- Strabala TJ, O'Donnell PJ, Smit AM, Ampomah-Dwamena C, Martin EJ, Netzler N, Nieuwenhuizen NJ, Quinn BD, Foote HC, Hudson KR. Gain-of-function phenotypes of many CLAVATA3/ESR genes, including four new family members, correlate with tandem variations in the conserved CLAVATA3/ESR domain. Plant Physiology. 2006;140:1331–1344. doi: 10.1104/pp.105.075515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian S, Cho UH, Keyes C, Yu O. Distinct changes in soybean xylem sap proteome in response to pathogenic and symbiotic microbe interactions. BMC Plant Biology. 2009;9:119. doi: 10.1186/1471-2229-9-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turk BE, Huang LL, Piro ET, Cantley LC. Determination of protease cleavage site motifs using mixture-based oriented peptide libraries. Nature Biotechnology. 2001;19:661–667. doi: 10.1038/90273. [DOI] [PubMed] [Google Scholar]
- van der Hoorn RA. Plant proteases: from phenotypes to molecular mechanisms. Annual Review of Plant Biology. 2008;59:191–223. doi: 10.1146/annurev.arplant.59.032607.092835. [DOI] [PubMed] [Google Scholar]
- Zhang KR, McKinlay C, Hocart CH, Djordjevic MA. The Medicago truncatula small protein proteome and peptidome. Journal of Proteome Research. 2006;5:3355–3367. doi: 10.1021/pr060336t. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.