Abstract
In this report, a rapeseed (Brassica napus) haem oxygenase-1 gene BnHO1 was cloned and sequenced. It shared high homology with Arabidopsis HY1 proteins, and encodes a 32.6 kDa protein with a 54-amino-acid transit peptide, predicting the mature protein of 25.1 kDa. The mature BnHO1 expressed in Escherichia coli exhibits haem oxygenase (HO) activity. Furthermore, the application of lower doses of NaCl (10 mM) and polyethylene glycol (PEG) (2%) mimicked the inducible effects of naphthylacetic acid and the HO-1 inducer haemin on the up-regulation of BnHO1 and subsequent lateral root (LR) formation. Contrasting effects were observed when a higher dose of NaCl or PEG was applied. The above inducible and inhibitory responses were blocked significantly when the HO-1 inhibitor zinc protoporphyrin IX (ZnPPIX) or haemin was applied, both of which were reversed by the application of carbon monoxide or ZnPPIX, respectively. Moreover, the addition of ZnPPIX at different time points during LR formation indicated that BnHO1 might be involved in the early stages of LR formation. The auxin response factor transcripts and the auxin content in seedling roots were clearly induced by lower doses of salinity or osmotic stress. However, treatment with the inhibitor of polar auxin transport N-1-naphthylphthalamic acid prevented the above inducible responses conferred by lower doses of NaCl and PEG, which were further rescued when the treatments were combined with haemin. Taken together, these results suggested a novel role of the rapeseed HO-1 gene in salinity and osmotic stress-induced LR formation, with a possible interaction with auxin signalling.
Keywords: BnHO1, Brassica napus, lateral root formation, osmotic stress, salinity stress
Introduction
Abiotic stress is a worldwide problem limiting plant growth and yield. In response to abiotic stress, plants have gradually evolved a complete set of stress acclimation mechanisms after long-term natural selection. For example, plants are able to regulate ion homeostasis to adapt to salt stress (Zhu, 2001, 2003), whereas osmoprotectants secreted by plant cells protect them against osmotic stress (Zhu, 2002; Sairam and Tyagi, 2004). Besides the physiological and biochemical responses mentioned above, morphogenic responses, including increased root hair formation, enhanced formation of lateral or adventitious roots, and the inhibition of root elongation, also occur. The phenotype is referred to as a stress-induced morphogenic response (SIMR) (Pasternak et al., 2005; Potters et al., 2007).
Using morphogenic responses, plants can accommodate to and grow out of the environmental stresses. It is further postulated that the similarities in the SIMR phenotypes induced by distinct stresses reflect common molecular processes such as increased production of reactive oxygen species (ROS) and altered phytohormone transport and/or metabolism. For instance, there is a close link between altered auxin metabolism and stress-induced lateral root (LR) formation, a key component of SIMR. When Arabidopsis thaliana seedlings were exposed to salt stress (He et al., 2005), heavy metal (Cd) (Potters et al., 2007), low phosphorus (López-Bucio et al., 2005; Pérez-Torres et al., 2008), and sulphur starvation (Kutz et al., 2002), auxin signalling was involved. In fact, auxin has been established to play an important role in LR formation, because its polar transport inhibitor N-1-naphthylphthalamic acid (NPA) blocks LR initiation (Casimiro et al., 2001), and mutations that render the plants less sensitive to auxin exhibit a reduction in LR numbers. Thus, LR development induced by various stresses is not only of agronomic significance, but it also acts as a useful model for the investigation of hormonal signals and interaction(s) modulating root organogenesis.
In animals, carbon monoxide (CO) synthesized mainly by haem oxygenase (HO, EC 1.14.99.3) has been confirmed to function as a signal and cytotoxic agent in many physiological and immunological processes. There are three known isoforms of HO: HO-1, an inducible form, and HO-2/3, two constitutively expressed forms. Related HO genes have been described in many species including mammals, plants, red algae, cryptophytes, cyanobacteria, and pathogenic bacteria (Ryter et al., 2002; Dulak and Józkowicz, 2003). In A. thaliana, for example, a small family of four HO members clusters into two subfamilies: HY1 (HO1), HO3, and HO4 belong to the HO-1 subfamily (Emborg et al., 2006), whereas HO2 is the only member of the HO-2 subfamily; interestingly, HO2 was recently reported to be unable to bind or degrade haem and is therefore not a true HO (Gisk et al., 2010). Other publications illustrated that either HO-1 or its catalytic product CO was involved in plant responses to salinity stress (Xie et al., 2008, 2011), heavy metals (Noriega et al., 2004; Balestrasse et al., 2006; Han et al., 2008; Cui et al., 2011), UV radiation (Yannarelli et al., 2006), programmed cell death (Ling et al., 2009; Wu et al., 2011), stomatal movement (Cao et al., 2007a), LR formation (Cao et al., 2007b), and adventitious root development (Xuan et al., 2008b). However, previous research on HO genes in plants has focused primarily on a few plants such as Arabidopsis (Muramoto et al., 1999, 2002; Kohchi et al., 2001; Gisk et al., 2010), rice (Izawa et al., 2000), tomato (Terry and Kendrick, 1996), pine (Davis et al., 2001), pea (Linley et al., 2006), and alfalfa (Fu et al., 2011). Thus, further investigation of the functions of HO genes from other plant species is required to understand the default and unique physiological mechanism of plant development and responses to various stresses.
In this study, a cDNA encoding an HO-1 protein of Brassica napus (named BnHO1) was cloned and the enzymic properties of a mature recombinant protein derived from this clone were carefully characterized. The enzymic properties of this recombinant protein were compared with those of Arabidopsis HY1 (Muramoto et al., 2002). We subsequently discovered that lower doses of salinity and osmotic stress triggered LR development in B. napus seedlings. Since CO, the catalytic product of HO, is a critical molecule implicated in the regulation of plant development and responses to various abiotic stresses (Cao et al., 2007a, b; Xuan et al., 2008a), a working hypothesis was developed that BnHO1 may be required for salinity and osmotic stress-induced LR formation. Further pharmacological, physiological, and molecular evidence supported this view, suggesting that BnHO1 is a newly identified component of the signal transduction pathway in LR formation triggered by salinity and osmotic stress. Finally, its relationship with auxin signalling was also investigated.
Materials and methods
Chemicals
All chemicals were obtained from Sigma (St Louis, MO, USA) unless stated otherwise. Haemin (H), an HO-1 inducer applied in animal and plant research (Lamar et al., 1996; Xuan et al., 2008b), was used at 1.0 μM or indicated concentrations. Bilirubin (BR), FeSO4, and CO, were used at 1.0 μM, 1.0 μM, and 10% saturated aqueous solution, respectively. The compound zinc protoporphyrin IX (ZnPPIX), an inhibitor of HO-1, was used at 50 μM (Lang et al., 2005; Hirai et al., 2006; Xuan et al., 2008b; Wu et al., 2011). Naphthylacetic acid (NAA), the well-known LR inducer, was used at 100 nM (Correa-Aragunde et al., 2004; 2006). NPA, from Chem Service (West Chester, PA, USA), was used as the auxin transport inhibitor at 2 μM. Other reagents, including NaCl and polyethylene glycol (PEG), were domestic products.
Preparation of CO aqueous solution
CO gas was prepared by heating formic acid (HCOOH) with concentrated sulphuric acid (H2SO4) according to the reaction H2SO4 (l) + HCOOH (l) → CO (g) + H2SO4·H2O (l). The aqueous solution of CO was further obtained by bubbling the CO gas gently through a glass tube into 50 ml of distilled water in an open bottle for at least 20 min, a period of time long enough to saturate the solution with CO gas. Then, the saturated solution was diluted immediately with distilled water to the concentration required.
Plant material and growth conditions
Rapeseed (B. napus ‘Yangyou 6’) seeds were kindly supplied by State Key Laboratory of Crop Genetics and Germplasm Enhancement, Nanjing Agricultural University, China. Seeds were surface-sterilized with 2% NaClO for 10 min, rinsed extensively and germinated in distilled water at 25±1 °C in the dark for 3 d. Seedlings with 2- to 3-mm radicles were selected and then transferred to Petri dishes (90 mm diameter) containing filter paper soaked with 6 ml of treatment solution as indicated. During experiments, seedlings were grown in an illuminating incubator at 25±1 °C with a 14-h photoperiod at 200 μmol m−2 s−1 intensity. At least three replicates were performed for each treatment, and 30 samples were included in each replicate. All solutions were renewed every day to maintain identical concentrations. Meanwhile, Petri dishes were sealed to avoid interference between different treatments.
After various treatments, the total length (>1 mm) and number of LRs per seedling was calculated and photographs were then taken. Additionally, LR primordia (LRP) were observed after 1 d of treatment by root squash preparations and quantified by a light microscope (model Stemi 2000-C; Carl Zeiss, Germany). For tissue-specific expression analysis, roots (R), stems (S), and young leaves (L) were collected from 6-d-old seedlings under normal growth condition. Blooming flowers (F) and dry seeds (DS) were also sampled. All samples harvested at the indicated time were immediately used or frozen in liquid nitrogen, and stored at –80 °C until further analysis.
Cloning of BnHO1, sequence alignment, and phylogenetic analysis
The mRNA sequence of A. thaliana HY1 gene (AB021858) was used as a query probe to search B. napus expressed sequence tags (ESTs) in the dbEST database of GenBank. Sixteen ESTs (EV078426, EE466369, EE475109, FG564197, FG557573, EE449513, EV221523, CN736027, CD814686, EV160257, ES908030, EV133255, EV221293, CX190816, DY023946, and EV167101) having homology with the probe were used to assemble into one contig, and then B. napus HO-1 (BnHO1) gene-specific primers were designed based on this contig. Additionally, since there are HO2, HO3, and HO4 in Arabidopsis, corresponding genes were also used as query sequence to search B. napus dbEST division in GenBank. However, no sequence sharing homology with query probe HO2, HO3, or HO4 was identified.
Two primers (forward, 5′-ACTGGTACCATGGCTTACTCAGCTCCC-3′, and reverse, 5′-TTGAAGCTTTCAGGACAATATGAGACG-3′) were employed to clone BnHO1 by RT-PCR. Total RNA from rapeseed seedling roots was extracted using Trizol reagent (Invitrogen, Gaithersburg, MD, USA) according to the manufacturer's instructions. First-strand cDNAs were synthesized from 2 μg of total RNA (pre-treated with DNase I) with AMV reverse transcriptase (TaKaRa) according to the manufacturer's protocol. The PCR conditions for amplifying BnHO1 were as follows: a pre-denaturation of 5 min at 94 °C, 35 cycles of 1 min at 94 °C, 1 min at 47 °C, 1 min at 72 °C, and a final extension for 10 min at 72 °C. The PCR product was gel-purified with AxyPrep Gel DNA Extraction Kit (Axygen, Hangzhou, China) according to the manufacturer's protocol. The purified product was then cloned into the pMD-19T vector (TaKaRa) for sequencing (Invitrogen, Shanghai, China).
Alignment of deduced BnHO1 or other plant HO protein sequences was performed with DNAMAN 6.0.40 software. The phylogenetic tree of mature plant HO proteins was constructed with the MEGA program (ver 4.0) by the neighbor-joining (NJ) method. The parameters pairwise deletion and p-distance model were used. Bootstrap test of phylogeny was performed with 1000 replicates.
DNA extraction and BnHO1 genomic sequence amplification
A CTAB-based method for genomic DNA extraction from rapeseed seedlings was carried out according to the previous method (Doyle and Doyle, 1990). The PCR reaction, using the same two primers as in BnHO1 cDNA amplification, runs as follows: an initial denaturation at 94 °C for 10 min; 35 cycles at 94 °C for 1 min, 47 °C for 1 min, 72 °C for 2 min, and a final extension for 10 min at 72 °C. The PCR product was gel-purified, and cloned into pMD-19T vector (TaKaRa) for sequencing (Invitrogen, Shanghai, China).
Prokaryotic expression and purification of recombinant His-tagged BnHO1
mBnHO1, the mature BnHO1 gene without the predicted transit peptide, was subcloned into the Escherichia coli expression vector pET-30a (Novagen) to produce pET-30a-mBnHO1. mBnHO1 was amplified using the primers 5′-GTGGGTACCGCGACGGCGGCGGAGAAG-3′ (forward) and 5′-TTGAAGCTTTCAGGACAATATGAGACG-3′ (reverse), which contained KpnI and HindIII sites (underlined), respectively, and cloned into KpnI/HindIII-digested pET-30a vector to construct pET-30a-mBnHO1. The integrity of the construct was verified by restriction analysis and complete DNA sequencing of the insert (Invitrogen, Shanghai, China). The constructed vector was transferred into E. coli strain BL21. The mBnHO1 protein was induced by 1 mM IPTG at 28 °C for 12 h, based on the manufacturer's instructions (Novagen), and purified through Ni-NTA column and Sephadex G-25 chromatography. After centrifugation at 20,000×g in a rotor (model Avanti J-25; Beckman) for 30 min, the purified mBnHO1 protein was used for the following biochemical experiments.
HO activity determination
HO activity was analysed using the method described in previous reports (Han et al., 2008; Xuan et al., 2008b). In the HO activity test, the concentration of biliverdin IX (BV) was estimated using a molar absorption coefficient at 680 nm of 6.25 mM−1 cm−1 in 0.1 M HEPES–NaOH buffer (pH 7.2). One unit of activity (U) was calculated by taking the quantity of enzyme to produce 1 nmol of BV per 30 min. Protein concentration was determined by the method of Bradford (1976) using bovine serum albumin as the standard. Km and Vmax values were calculated for mBnHO1 using a Lineweaver–Burk plot. For haemin, the data were obtained using the standard assay, with the haemin concentrations varied between 1 μM and 20 μM. The effects of temperature and pH were determined using the standard assay conditions as described above.
Western blot analysis for BnHO1
Rabbit polyclonal antibody was made against the mature BnHO1 protein expressed in E. coli, with a molecular mass of 25.1 kDa. Homogenates obtained for the HO activity assays were also analysed by western blot. Fifty micrograms of protein from homogenates were subjected to SDS-PAGE using a 12.5% acrylamide resolving gel (Mini Protean II System; Bio-Rad). Separated proteins were then transferred to polyvinylidene difluoride membranes, and non-specific binding of antibodies was blocked with 5% non-fat dried milk in phosphate-buffered saline (pH 7.4) for 4 h at room temperature. Membranes were then incubated overnight at 4 °C, with primary antibodies diluted 1:3000 in the same buffer plus 1% non-fat dried milk. Immune complexes were detected using horseradish peroxidase-conjugated goat anti-rabbit immunoglobulin G. The colour was developed with a solution containing 3, 3'-diaminobenzidine tetrahydrochloride as horseradish peroxidase substrate.
RNA extraction and semi-quantitative RT-PCR analysis
Total RNA was isolated from 100 mg of fresh-weight samples by grinding with mortar and pestle in liquid nitrogen until a fine powder appeared and using Trizol reagent according to the manufacturer's instructions. DNA-free total RNA (5 μg) from different treatments was used for first-strand cDNA synthesis in a 20-μl reaction volume containing 2.5 U of AMV reverse transcriptase XL (TaKaRa) and 1 μM oligo(dT) primer. PCR reactions were performed using 2 μl of a 2-fold dilution of the cDNA, 10 pmol of each oligonucleotide primer, and 1 U of Taq polymerase (TaKaRa) in a 25-μl reaction volume.
Primers used were as follows: for BnHO1 (accession number GU390397), forward (5′-TCTCCATCCCTATCCTCC-3′) and reverse (5′-ACCAACTTGCTATCCACC-3′), amplifying a 353-bp fragment; for actin (accession number AF111812), forward (5′-CTGGAATGGTGAAGGCTGGGTT-3′) and reverse (5′-CGGAGGATAGCGTGAGGAAGAG-3′), amplifying a 490-bp fragment. To standardize the results, the relative abundance of actin was determined and used as the internal standard.
The number of cycles of PCR was adjusted for each gene to obtain visible bands on agarose gels. Aliquots from the PCR were loaded on 1.2% agarose gels with the use of ethidium bromide (EB). Specific amplification products of the expected size were observed, and their identities were confirmed by sequencing. EB-stained gels were scanned and analysed using Quantity One software.
Real-time quantitative RT-PCR analysis
Quantitative PCR was performed with a Mastercycler® ep realplex real-time PCR system (Eppendorf, Hamburg, Germany) with SYBR® Premix Ex Taq™ (TaKaRa Bio, Inc., Otsu, Shiga, Japan) according to the manufacturer's instructions. The cDNA was amplified by real time PCR using the following primers: for BnHO1, forward 5′-AAGGTGAGAAAGAGACAAAA-3′ and reverse 5′-GAAAGTGGACTGACCAATAA-3′; for auxin response factor (accession number AJ716227), forward 5′-GGAGAATTACGTGTGGGTGT-3′ and reverse 5′-TGGTTCCAGTTGAAATAGCG-3′; and for actin, forward 5′-CTAAGGCTAACAGGGAAAAA-3′ and reverse 5′-AGCGTAGAGAGAAAGAACAG-3′. Gene-specific primers were designed with the software tool Primer Express (Applied Biosystems, Foster city, CA, USA). The relative expression level was calculated from three replicates using the comparative Ct method after normalization to an actin control.
IAA determination
Roots of seedlings treated for 6 h were harvested and IAA was quantified using HPLC as described previously (Edlund et al., 1995).
Data analysis
Where indicated, results are expressed as the means ±SE of at least three independent experiments. Statistical analysis was performed using SPSS 10.0 software. For statistical analysis, t test (P<0.05 or P<0.01) or Duncan's multiple test (P<0.05) was chosen as appropriate.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers GU390397, AJ716227, and AF111812.
Results
Isolation of the haem oxygenase-1 gene from rapeseed
The Arabidopsis HY1 mRNA (GenBank accession number AB021858) was used as a query sequence to search the B. napus dbEST division in GenBank. Sixteen rapeseed ESTs with high homology to the query were obtained and assembled into one longer contig of 1643 bp. A full-length cDNA of 849 bp was amplified from the rapeseed seedling roots with two specific primers based on this contig. The results of nucleotide sequence alignment illustrated that this contig was identical to AtHO1 (HY1) by 83.6%. This sequence was designated BnHO1 and was deposited in GenBank with the accession number GU390397.
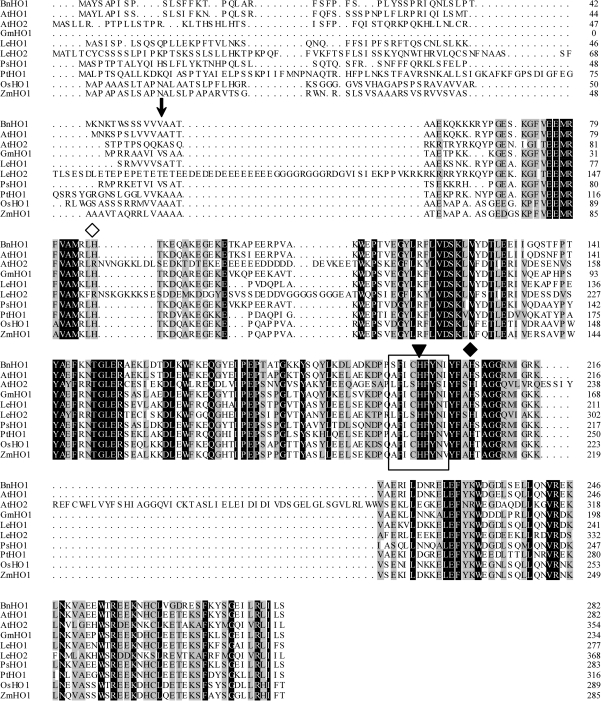
The sequence of complete deduced amino acids is shown in Fig. 1. Interestingly, the BnHO1 protein is similar to HO-1 in both structure and size. For example, the modelled structure using SWISS-MODLE analysis (see Supplementary Fig. S1 available at JXB online) is mostly α-helical and quite similar to PsHO1 (Linley et al., 2006). The BnHO1 gene encodes a single open reading frame of 282 amino acids with a predicted molecular mass of 32.6 kDa. This includes a 54-amino-acid transit peptide identified by the ChloroP algorithm (Emanuelsson et al., 1999), suggesting that the mature protein (mBnHO1) is 25.1 kDa after the cleavage of the transit peptide. The deduced amino acid sequence shows strong similarity to well-known plant HO-1 proteins (Fig. 1). Indeed, the HO signature sequence AFICHFYNI (amino acid residues 194–202 in Arabidopsis HY1), which is highly conserved among HOs of several plant species (Ito-Maki et al., 1995; Matera et al., 1997; Muramoto et al., 1999; Linley et al., 2006), is conserved in BnHO1 (Fig. 1, boxed). Interestingly, H-132 in the rat HO-1 signature sequence, which is thought to play a structural role in stabilizing the HO protein (Matera et al., 1997), is also conserved in BnHO1 (H-198, reverse triangle). In addition, H-86 (white diamond) in BnHO1 and HY1, which corresponds to H-25 of HO-1 in rat, is conserved (Muramoto et al., 1999). In fact, H-25 is the axial haem iron ligand and is therefore a vital residue for HO enzymic activity (Ito-Maki et al., 1995). The H-206 residue (Fig. 1, black diamond) of BnHO1 might be involved in ascorbic acid binding (Linley et al., 2006). Together, the conservation of these important residues implies that the sequence similarity of BnHO1 to plant and animal HO-1 protein is significant. Subsequently, a phylogenetic tree of plant mature HO was constructed (Fig. 2A). From the phylogenetic tree, the BnHO1 identified in this study clearly groups with the HO1 family, further supporting its designation as BnHO1.
Fig. 1.
Alignment of BnHO1 with other plant HO-1/2 proteins. Dark shading with white letters and gray shading with black letters reflect 100% and 75% sequence conservation, respectively. Database accession numbers are GU390397 for BnHO1 (B. napus), AB021858 for AtHO1 (A. thaliana), AF132477 for AtHO2 (A. thaliana), AF320024 for GmHO1 (Glycine max), AF320028 for LeHO1 (Lycopersicon esculentum), AF320029 for LeHO2 (L. esculentum), AF276228 for PsHO1 (Pisum sativum), AF320030 for PtHO1 (Pinus taeda), EU781632 for OsHO1 (Oryza sativa), and EU962994 for ZmHO1 (Zea mays), respectively. Solid frame means signature sequence. The arrow identifies the predicted cleavage site between the transit peptide and mature protein. The conserved histidine residue involved in haem–iron binding and catalysis is shown by a white diamond. The conserved histidine residue for protein stability is illustrated by a reverse triangle. The conserved histidine residue involved in ascorbic acid binding is also shown by a black diamond.
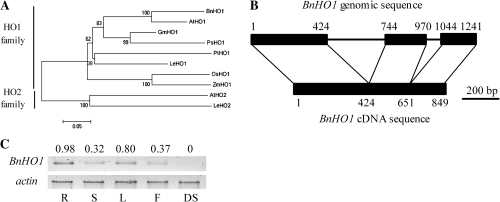
Fig. 2.
Phylogenetic properties, structure, and expression pattern of BnHO1. The phylogenetic tree of corresponding mature HO-1/2 proteins in plants (A). Database accession numbers are shown in Fig. 1. The NJ tree was constructed with MEGA 4.0. The scale bar corresponds to 0.05 estimated amino acid substitutions per site. Diagrammatic alignment of the genomic DNA and cDNA sequences of BnHO1 indicating the location of intron–exon boundaries (B). Black boxes and lines denote coding regions and introns, respectively. Numbers refer to the first and last nucleotides of the exons, respectively. This illustration is drawn to scale. mRNA expression of BnHO1 (C) in roots (R), stems (S), leaves (L), flowers (F), and dry seeds (DS). The actin amplification band is shown to confirm equal loading of RNA and RT efficiency. The PCR products were run on 1.2% agarose gel and visualized under UV light. The number above the band indicates relative abundance of the corresponding gene with respect to the loading control actin.
Using the same primers in BnHO1 cDNA amplication, a 1241-bp sequence was obtained from rapeseed genomic DNA. The corresponding BnHO1 genomic sequence contained three exons, and two introns of 319 bp and 73 bp (Fig. 2B), like HY1 (in other plants, HO1 contains three introns) (Davis et al., 2001). Together, these data clearly demonstrated that BnHO1 is a member of the HO1 family. Additionally, the expression pattern of BnHO1 mRNA in various tissues was determined by semi-quantitative RT-PCR (Fig. 2C). Under the experimental conditions used here, the BnHO1 transcripts were highly expressed in seedling roots and leaves, exhibited substantially lower levels in stems and flowers, and were almost negligible in dry seeds.
Expression, purification, and biochemical analysis of the recombinant protein
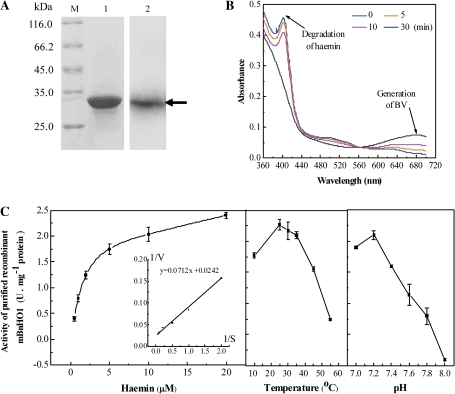
To confirm that BnHO1 encodes an HO protein and to further characterize its catalytic properties, mBnHO1 was expressed as a fusion protein in E. coli. The His-tagged fusion protein was obtained after IPTG induction. Then, the fusion protein was purified to homogeneity by Ni-affinity chromatography and used for further biochemical studies. In the subsequent western blot analysis, the His-mBnHO1 recombinant protein exhibited an apparent molecular mass of 32.7 kDa (Fig. 3A), which is slightly higher than the predicted weight (Niu and Guiltinan, 1994).
Fig. 3.
Expression and biochemical characterization of purified recombinant His-tagged mBnHO1 protein in E. coli. Expressed protein purified by Ni-affinity chromatography and its western blot analysis, 50 μg protein/well (A). Lane M, marker proteins; lane 1, purified protein; lane 2, western blot analysis of purified protein developed with the polyclonal antiserum against the mature rapeseed HO-1 (mBnHO1). Arrow indicates the position of the fusion protein of mBnHO1 with a molecular mass of 32.7 kDa. Time-dependent absorbance changes were determined during the mBnHO1 reaction with spectra taken at 0, 5, 10, and 30 min after the addition of NADPH (B). Arrows indicate the direction of absorbance changes during the incubation. Michaelis–Menten plot (left), temperature and pH dependence (right) of the mBnHO1 (C). Data shown are the means ±SE from three independent measurements.
We performed a biochemical experiment by measuring the HO activity of mBnHO1. The absorbance was monitored between 300 and 800 nm (only the range 360–700 nm is illustrated in Fig. 3B) by determining the conversion of haem to BV spectrophotometrically, because this showed the strong degradation of haemin at 405 nm and generation of BV at 680 nm. The bound haem peak decreases substantially over 30 min of incubation, and a concomitant rise in the BV absorbance maxima was observed.
Kinetic constants for the mBnHO1 reaction were also investigated (Fig. 3C). Under the experimental conditions reported here, the Vmax was 98.4 nmol BV h−1 nmol−1 protein with an apparent Km value for haemin of 2.52 μM. These values are comparable to those previously reported for HY1 in Arabidopsis (Muramoto et al., 2002). The corresponding Vmax and apparent Km values of HY1 were 156 nmol BV h−1 nmol−1 protein and 1.3 μM. The rate of the BnHO1 reaction increased to a temperature of 25 °C and declined thereafter. Similar to those of the Arabidopsis HY1 (Muramoto et al., 2002), alfalfa MsHO1 (Fu et al., 2011), and chick liver HO (Bonkovshy et al., 1990), the optimum pH of BnHO1 was 7.2, and dropped sharply either side of this value (Fig. 3C).
Lower doses of NaCl and PEG promote LR formation and induce BnHO1 expression
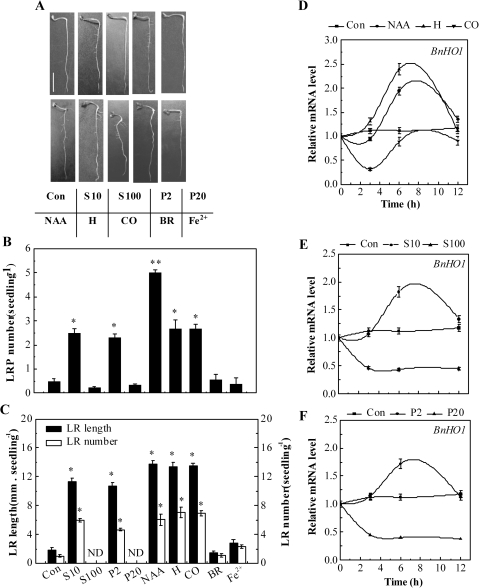
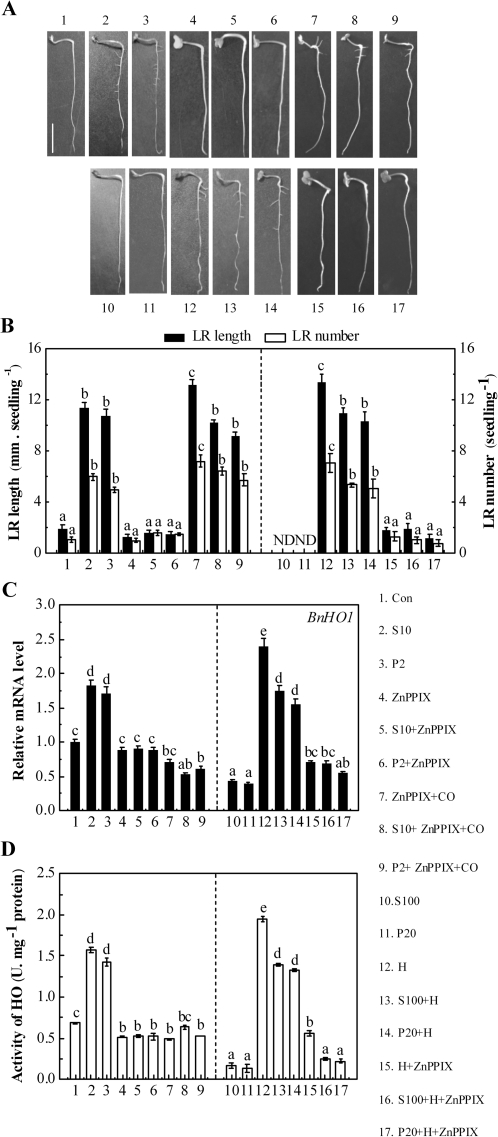
SIMR is typically induced by relatively low, chronic doses of stress, rather than by high phytotoxic doses (Potters et al., 2007; Zolla et al., 2010). Our further goal was to investigate whether the nature of salinity and osmotic stress-induced LR formation is related to doses of stress. Figure 4B, C shows the number of LRP as well as the length and number of emerged LRs after 1 d or 3 d of treatment, respectively. As expected, in comparison with the control sample (Con), lower doses of NaCl (10 mM, S10) and PEG (2%, P2) obviously increased LRP and LR formation (Fig. 4A–C), mimicking the inducing effects of 100 nM NAA, 1 μM haemin, and 10% saturated CO aqueous solution (P<0.05; Correa-Aragunde et al., 2004, 2006; Cao et al., 2007b). In contrast, higher doses of NaCl (100 mM, S100) and PEG (20%, P20) inhibited LR development. Additionally, no significant changes were observed when either bilirubin (BR) or Fe2+ was applied.
Fig. 4.
Effects of NaCl, PEG, NAA, haemin, CO, BR, and Fe2+ on LR formation and BnHO1 transcripts. Three-day-old rapeseed seedlings were treated with distilled water (Con), 10 mM or 100 mM NaCl (S10 or S100), 2% or 20% PEG (P2 or P20), 100 nM NAA, 1 μM haemin (H), 10% saturated CO aqueous solution (CO), 1 μM BR, or 1 μM FeSO4 (Fe2+) for another 3 d. Representative photos were then taken. Bar, 1 cm (A). The number of LRP per seedling was calculated after treatment for 1 d (n=30, B). The length (>1 mm) and number of LRs per seedling were determined after 3 d of treatment (n=90, C). Data are the means ±SE of at least three independent experiments. Bars with asterisks were significantly different with respect to Con at the P<0.05 and P<0.01 level according to t-test (B, C). ND, none detected. Time-course analysis of BnHO1 transcripts in seedling roots after various treatments (D–F). BnHO1 expression was analysed by real-time quantitative RT-PCR at the indicated time points, and presented relative to those of the corresponding control samples at 0 h.
In addition, BnHO1 transcripts were investigated by quantitative RT-PCR. Our time-course analysis of gene expression (Fig. 4D–F) showed that BnHO1 transcript levels were modulated differentially over 12 h of treatment. For example, BnHO1 transcripts were gradually up-regulated by NAA and haemin, reaching a peak after 6 h, and then declined. However, as expected (Han et al., 2008), the expression of BnHO1 was rapidly down-regulated by CO aqueous solution after 3 h but recovered later. Interestingly, changes in BnHO1 transcripts treated with different doses of NaCl and PEG were also detected (Fig. 4E, F). Among these, lower doses of NaCl (10 mM) and PEG (2%) increased BnHO1 transcripts to a peak after 6 h, but the transcripts then gradually decreased; both of these responses mimicked the pattern modulated by NAA and haemin. The trend of HO activity after 6 h of various treatments displayed similar tendencies (see Supplementary Fig. S2 at JXB online). We also noted that the inducible responses of BnHO1 triggered by lower doses of NaCl and PEG apparently preceded LR formation, suggesting a possible interrelationship between the up-regulation of BnHO1 and LR formation. However, high doses of NaCl and PEG decreased BnHO1 transcripts.
Up- or down-regulation of BnHO1 expression contributes to the induction or inhibition of LR formation
In order to assess the requirement of BnHO1 expression during NaCl- or PEG-induced LR formation, the potent HO-1 inhibitor ZnPPIX was added. Figure 5A, B showed that the 50 μM ZnPPIX plus 10 mM NaCl treatment produced a significant reduction in the length and number of LRs (P<0.05), in comparison with the NaCl stress alone sample. Similarly, ZnPPIX caused considerable inhibition in 2% PEG-stressed samples. Meanwhile, ZnPPIX-inhibited BnHO1 transcripts and HO activity in rapeseed seedling roots were significantly lower than those of samples when 10 mM NaCl or 2% PEG was applied alone (P<0.05; Fig. 5C, D). However, when a 10% saturated aqueous solution of CO was applied together with ZnPPIX in a 10 mM NaCl or 2% PEG-stressed sample, the inhibition of LR formation was relieved (Fig. 5A, B). We also observed that the application of CO alone (see Supplementary Fig. S2 at JXB online) or with other reagents remarkably decreased BnHO1 transcript levels or HO activity (Figs 4D, 5C, D). In addition, the application of ZnPPIX alone produced a slight reduction in BnHO1 transcripts and LR formation but significantly decreased HO activity.
Fig. 5.
Pharmacological and molecular evidence showing that BnHO1 is required for salinity and osmotic stress-induced LR formation. Three-day-old rapeseed seedlings were treated with distilled water (Con), 10 mM or 100 mM NaCl (S10 or S100), 2% or 20% PEG (P2 or P20), 50 μM ZnPPIX, 10% saturated CO aqueous solution (CO), 1 μM haemin alone, or the combination treatments for another 3 d. Representative photos were then taken. Bar, 1 cm (A). Meanwhile, the length (>1 mm) and number of LRs per seedling were determined after 3 d of treatment (n=90, B). BnHO1 transcripts (C) and HO activity (D) were analysed after 6 h of various treatments. Data are the means ±SE of at least three independent experiments. Within each set of experiments, bars denoted by the same letter did not differ significantly at the P<0.05 level according to Duncan's multiple test. ND, none detected.
Our previous report found that the exogenous HO-1 inducer haemin induced LR formation of rapeseed seedlings (Cao et al., 2007b). To further confirm the possible role of BnHO1 in NaCl- or PEG-induced LR formation, 1 μM haemin was applied. As expected, haemin was not only able to significantly block the inhibition of LR development induced by 100 mM NaCl or 20% PEG treatment (P<0.05, Fig. 5A, B), but it also induced the up-regulation of BnHO1 transcripts and HO activity (Fig. 5C, D), respectively. When haemin was used alone, BnHO1 was significantly up-regulated, and the HO activity and LR formation responses were induced (Fig. 5). However, the haemin-blocked inhibition of LR formation was reversed by exogenous 50 μM ZnPPIX and declined to a similar extent to that appearing in rapeseed seedlings incubated in 50 μM ZnPPIX alone (Fig. 5A, B). Furthermore, the recovery of the BnHO1 transcript and HO activity induced by haemin was clearly inhibited by ZnPPIX as well (Fig. 5C, D). Taken together, these results suggested that BnHO1 up-regulation may be required for LR formation triggered by lower doses of NaCl and PEG.
BnHO1 might be required in the early stages of LR formation
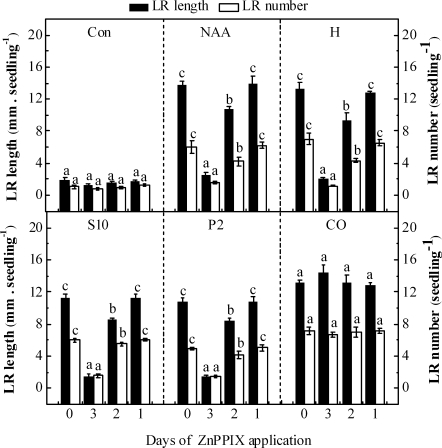
In order to further assess the BnHO1 requirement during the formation and/or elongation of the salinity and osmotic stress-induced LR formation, 3-d-old rapeseed seedlings were incubated in distilled water (Con), 100 nM NAA, 1 μM haemin, 10 mM NaCl, 2% PEG, and 10% saturated CO aqueous solution for 3 d. Meanwhile, the HO-1 inhibitor ZnPPIX was added after 0, 1, or 2 d of the various treatments. The results in Fig. 6 show that ZnPPIX, when applied with various treatments (except for CO treatment and control sample) for 3 d, led to a remarkable decrease (>3-fold) in LR formation. In contrast, only a slight effect could be observed in response to the application of ZnPPIX after 1 d or 2 d of various treatments. However, ZnPPIX had no apparent effects on CO-induced LR formation, which was reported previously (Cao et al., 2007b). Additionally, only a slight but not significant decrease in LR formation was observed in the ZnPPIX-treated control sample for 3 d. Thus, the events involving BnHO1 may occur within the first 1 d of 10 mM NaCl, 2% PEG treatment. These findings suggested that BnHO1 might be required in the early stages of LR formation.
Fig. 6.
BnHO1 is required during the early events that trigger LR formation. Three-day-old rapeseed seedlings were incubated in distilled water (Con), 100 nM NAA, 1 μM haemin (H), 10 mM NaCl (S10), 2% PEG (P2), and 10% saturated CO aqueous solution (CO) alone for 3 d, or in the presence of 50 μM ZnPPIX for 3, 2, or 1 d, respectively. The length (>1 mm) and number of LRs per seedling were determined after 3 d of treatments. Data are the means ±SE of at least three independent experiments (n=30). Within each set of experiments, bars denoted by the same letter did not differ significantly at the P<0.05 level according to Duncan's multiple test.
Possible auxin signalling
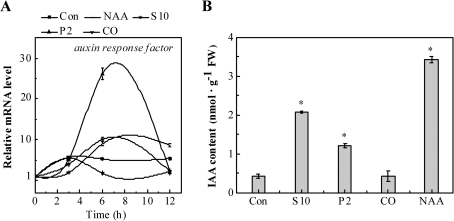
Given the key role of auxin in LR development (Casimiro et al., 2001), the changes in the auxin response gene and IAA content in rapeseed seedling roots upon different treatments was analysed. The time-dependent analysis indicated that the auxin response factor (Schruff et al., 2006) transcripts were up-regulated gradually by 100 nM NAA, 10 mM NaCl, and especially 2% PEG, reaching a peak after 6 h, and then declined (Fig. 7A). We observed that the CO-treated sample exhibited a slight increase in auxin response factor transcript after 3 h of treatment and later returned to its basal level. The transcripts increased gently when treated with distilled water over 12 h. Besides, the IAA content was increased in rapeseed seedling roots upon NaCl, PEG, and NAA treatment (especially, Fig. 7B), when compared with the control sample. No significant change in the IAA level was observed in the CO-treated sample. Combined with previous results (Xuan et al., 2008b), it was deduced that there was a possible link between auxin signalling and the up-regulation of BnHO1 during LR formation triggered by lower doses of NaCl and PEG.
Fig. 7.
Changes of auxin response factor transcripts (A) and IAA content (B) in rapeseed seedling roots. Three-day-old rapeseed seedlings were treated with distilled water (Con), 100 nM NAA, 10 mM NaCl (S10), 2% PEG (P2), or 10% saturated CO aqueous solution (CO), respectively. Time-course analysis of auxin response factor transcripts (A). IAA content was determined using HPLC after 6 h of various treatments (B). Data are the means ±SE of at least three independent experiments. Bars with asterisks were significantly different with respect to Con at P<0.05 level according to t-test.
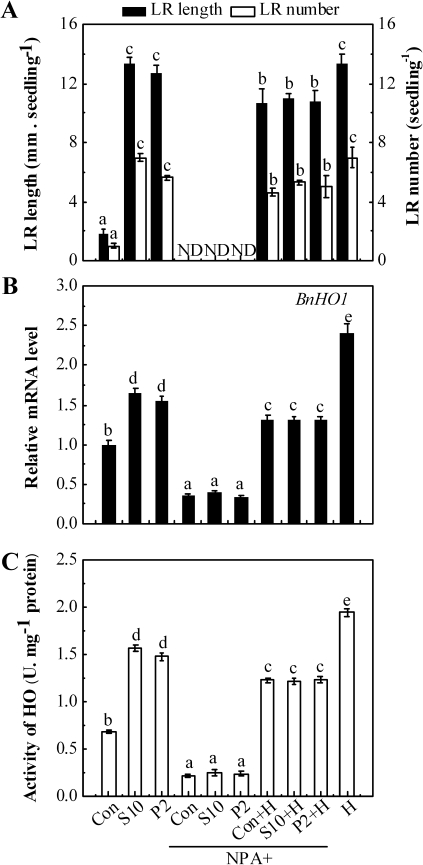
Previous results (Reed et al., 1998) showed that the inhibitor of polar auxin transport, NPA, generates reduced levels of auxin in the root. In this study, it was observed that the application of 2 μM NPA combined with distilled water (Con), 10 mM NaCl, or 2% PEG resulted in the almost complete abolition of LR emergence (P<0.05, Fig. 8A). A clearly decreased BnHO1 transcript level and HO activity (Fig. 8B, C) were also observed. In contrast, the addition of haemin not only resulted in the up-regulation of BnHO1 gene expression and enhanced HO activity, but it also significantly alleviated the inhibition of LR formation in rapeseed seedlings treated with NPA and 10 mM NaCl, or 2% PEG. These findings provide preliminary evidence for the involvement of endogenous auxin transport and/or metabolism in the salinity or osmotic stress-induced up-regulation of BnHO1 and subsequent LR formation.
Fig. 8.
Effects of NaCl, PEG, NPA, and haemin on LR formation, BnHO1 transcripts, and HO activity in rapeseed seedlings. Three-day-old rapeseed seedlings were treated with distilled water (Con), 10 mM NaCl (S10), or 2% PEG (P2) in the presence or absence of 2 μM NPA with or without 1 μM haemin (H) for another 3 d. The length (>1 mm) and number of LRs per seedling were determined (n=90, A). BnHO1 transcripts (B) and HO activity (C) were analysed after 6 h of various treatments. Data are the means ±SE of at least three independent experiments. Within each set of experiments, bars denoted by the same letter did not differ significantly at the P<0.05 level according to Duncan's multiple test. ND, none detected.
Discussion
To date, several HO-1 genes have been isolated or characterized from different plant species, and some of them were demonstrated to play important roles in various plant developmental and stress responses, especially in light signalling (Izawa et al., 2000; Muramoto et al., 2002; Linley et al., 2006). More recently, three rapeseed HO genes were isolated (nominated BnHO-1 to BnHO-3; Shen et al., 2011), and one of them, BnHO-1, confers plant tolerance to mercury toxicity. However, the biochemical and physiological characterization of these HO genes has not been fully investigated. This report describes the cloning and expression of BnHO1, an HO1-like gene in rapeseed similar to the BnHO-1 gene reported previously (Shen et al., 2011), and it is shown that BnHO1 was induced differentially by lower doses of salinity and osmotic stress, whereas the opposite response was caused by higher doses of stress. Both pharmacological and biochemical evidence supports the idea that lower levels of salinity and osmotic stress-induced BnHO1 expression and HO activity were required for stress-mediated LR formation (especially in the early stage), an important component of SIMR in plants, which might interact with auxin signalling.
BnHO1 in rapeseed is a counterpart of Arabidopsis HY1
Like the SE5 gene of rice (Izawa et al., 2000), several lines of evidence showed that BnHO1 of rapeseed is a counterpart of the Arabidopsis HY1 gene. First, the deduced amino acids of mBnHO1 share >88% identity with the HY1 product, in comparison with 48%, 84%, and 72% when compared with the products of HO2, HO3, and HO4 in Arabidopsis (data not shown). Secondly, the conserved HO signature sequence, histidine residue, intron numbers and positions in BnHO1 and HY1, and the existence of a putative transit peptide in both BnHO1 and HY1 (Muramoto et al., 1999) also support this idea (Figs 1, 2B). It was also noticed that intron positions of BnHO1 (Fig. 2B) are similar to those of BnHO-3 rather than BnHO-1 (Shen et al., 2011). Phylogenetic tree analysis (Fig. 2A) illustrated that BnHO1 belongs to the HO1 family. Furthermore, it is shown that the purified recombinant mBnHO1 exhibits HO activity. In fact, the degradation activity of the substrate–protein complex (absorbance peak at 405 nm; Yoshida and Kikuchi, 1978) concomitant with the generation of BV (absorbance peak at 680 nm) was confirmed in a time-course manner (Fig. 3B; Muramoto et al., 2002). The kinetic parameters for the BnHO1 reaction were also approximately similar to those previously reported for HY1 (Muramoto et al., 2002) and MsHO1 (Fu et al., 2011). For example, BnHO1 had a Km value for haemin of 2.52 μM (Fig. 3C) compared with 1.3 μM for recombinant HY1. However, semi-quantitative RT-PCR demonstrated that BnHO1 was very strongly expressed in root tissues as well as seedling leaves (Fig. 2C). Comparatively, promoter–GUS fusions showed that HY1 is actively expressed in the shoot apex, cotyledons, vascular tissue, and hypocotyl–root junction (Davis et al., 2001).
BnHO1: an endogenous modulator of salinity and osmotic stress-induced LR formation
In animals, HO-1 was able to modulate haemoprotein levels and maintain cellular haem homeostasis. HO-1 is also regarded as an important cellular defence enzyme, because its expression can alleviate oxidative stress injury and has been shown to have anti-inflammatory, anti-apoptotic, and anti-proliferative effects in cardiovascular, kidney, liver, and lung tissues (Ryter et al., 2002). Recently in plants, evidence has accumulated showing that the expression of HO-1 was triggered by diverse stress-inducing stimuli including salinity stress (Xie et al., 2008), osmotic stress (Liu et al., 2010), heavy metal exposure (Noriega et al., 2004; Han et al., 2008; Cui et al., 2011), haem compounds (Xuan et al., 2008b; Liu et al., 2010), UV radiation (Yannarelli et al., 2006), reactive oxygen species such as hydrogen peroxide (Yannarelli et al., 2006; Chen et al., 2009), and nitric oxide (Noriega et al., 2007). Meanwhile, inducible responses of HO expression were also observed when auxin (Xuan et al., 2008b) and abscisic acid (ABA) (Cao et al., 2007a) were applied. Between them, auxin is considered the prime candidate to control SIMR and therefore adventitious rooting and LR formation (Potters et al., 2007); ABA is an important signal against various stresses. Consequently, the biological functions of HO-1 in plants are probably associated with a fundamental adaptive and defensive response against oxidative stress and cellular stress as well as developmental processes (Shekhawat and Verma, 2010). These results were confirmed when it was further discovered that NAA and haemin not only stimulated LR and LRP formation, an important component of SIMR in plants, but also induced BnHO1 expression in a time-dependent fashion (Fig. 4). We also noticed that BnHO1 up-regulation preceded the LR formation triggered by NAA and haemin.
Previous results indicated that both salinity and osmotic stress cause increased LR formation in Arabidopsis and drought-sensitive rice (Yang et al., 2004; He et al., 2005; Kolbert et al., 2010). Furthermore, it was discovered that lower doses of NaCl and PEG mimicked the inducible effects of NAA and the HO-1 inducer haemin on the up-regulating BnHO1 transcripts and subsequently rapeseed LR formation (Fig. 4); the latter may benefit the salinity or osmotic acclimation response (He et al., 2005; Potters et al., 2007). Furthermore, it was discovered that only CO mimicked the above responses of lower doses of NaCl and PEG, but no significant responses of BR and Fe2+ were observed. Thus, it was speculated that BnHO1 induction triggered by NaCl and PEG would be beneficial not only by protecting auxins from oxidation, but also by enhancing the release of CO, a signalling molecule responsible for LR formation in rapeseed and tomato seedlings (Cao et al., 2007b; Xu et al., 2011). In contrast, the opposite effects occurred when higher doses of NaCl and PEG were applied (Fig. 5). Similarly, when searching results from the Genevestigator DNA microarray database (Zimmermann et al., 2004), HY1 mRNA in root tissues was induced significantly in Arabidopsis seedlings upon mild osmotic and salinity stress. Thus, it was further deduced that BnHO1 might be involved in plant tolerance to abiotic stresses, and that an interrelationship between BnHO1 and stress-modulated LR formation might exist.
We subsequently found strong correlations between the modulation of BnHO1 transcript, HO activity, and corresponding responses of LR formation modulated by NaCl and PEG treatments. For example, the specific inhibitor of HO-1 ZnPPIX caused a reduction in LR formation (P<0.05) as well as the down-regulation of the BnHO1 transcript and HO activity in plants stressed with a lower dose of NaCl (S10) and PEG (P2) as well as in control samples (Fig. 5). Meanwhile, CO, which promoted LR formation in rapeseed (Cao et al., 2007b), reversed the inhibition of LR formation caused by ZnPPIX, although it down-regulated BnHO1 transcripts and had no obvious effect on HO activity (Fig. 5). However, increased BnHO1 gene expression, HO activity, and induction of LR formation was demonstrated in response to haemin together with a higher dose of NaCl (S100) or PEG (P20) (Fig. 5). These findings were consistent with previous work, which reported that haemin could show a time- and dose-dependent increase in monocytic HO-1 mRNA and protein levels (Lang et al., 2005). Conversely, the up-regulation of BnHO1 transcripts, enhancement of HO activity, and increase in LR formation were restrained when ZnPPIX was added (Fig. 5). Previously, reductions or increases in HO-1 transcript and/or its protein level, and HO activity caused by ZnPPIX or haemin treatment were reported in both animals (Lang et al., 2005; Hirai et al., 2006; Tsoyi et al., 2009) and plants (Xuan et al., 2008b; Wu et al., 2011; Xie et al., 2011). Therefore, these results suggested that the up-regulation of BnHO1 expression is closely associated with the lower-dose NaCl or PEG stress-induced LR formation. Meanwhile, it has been shown that ZnPPIX is able to inhibit the heme carrier protein 1 (HCP1)-dependent uptake of haemin in cultured HeLa cells and astrocytes (Shayeghi et al., 2005; Dang et al., 2010). Thus, the possibility of antagonistic uptake of ZnPPIX and haemin in plant tissues should be investigated.
LR formation is known to be a very complex process of organogenesis, which is divided into several distinct stages and regulated by a series of signals (Malamy and Benfey, 1997; Casimiro et al., 2003; Péret et al., 2009). The results in this study suggest the involvement of BnHO1 in LR formation. However, it is not clear at exactly which stage BnHO1 is required. Himanen et al. (2002) developed a system using the depletion of endogenous auxin in Arabidopsis for studying the auxin signal transduction pathways modulating LR formation. As a result, a pharmacological and histological method analysed LR developmental events in detail. In this study, the use of another system, based on the inhibition of HO activity and/or its gene expression in rapeseed seedling roots by the application of HO-1 inhibitor ZnPPIX at different time points after various treatments is reported. The early stages of LR development in rapeseed have been shown to be similar to those described in Arabidopsis and tomato (Malamy and Benfey, 1997; Correa-Aragunde et al., 2006). Therefore, it is proposed that a ZnPPIX-dependent inducible system would be useful to study the early events leading to LR development. Based on this analogy, it is suggested that BnHO1 might be required in the early stage of LR formation (Fig. 6).
Relationship between auxin signalling and BnHO1-mediated LR formation
The alteration of auxin content, response, biosynthesis and distribution, or mutations in the corresponding signalling pathways is known to influence LR formation, especially in a variety of abiotic stresses (Zolla et al., 2010). For example, there is a good correlation between Cd-induced auxin redistribution and LR formation (Potters et al., 2007). When the root system of 6-d-old Arabidopsis seedlings was exposed to 50 μM Cd, the auxin levels increased in the root middle and upper zones, but the levels decreased near the root tip, and this coincides with the auxin-induced development of LPR. Additional data also suggest a possible link involving BnHO-mediated LR formation and stress-altered auxin homeostasis. This deduction is based on several pieces of evidence. (i) NAA not only induced LR formation but also resulted in the up-regulation of BnHO1 transcript in a time-dependent fashion (Fig. 4A–D). We also noted that the inducible responses of BnHO1 apparently preceded LR formation. (ii) The auxin response factor transcripts and auxin content in rapeseed seedling roots upon lower doses of salinity or osmotic stress were clearly induced (Fig. 7). (iii) The up-regulation of BnHO1 transcripts and enhanced HO activity are early responses to lower doses of salinity or osmotic stress, which are sensitive to the auxin polar transport inhibitor NPA (Fig. 8B, C). (iv) In IAA-depleted seedlings, lower doses of salinity or osmotic stress failed to induce subsequent LR development (Fig. 8A). (v) The abovementioned IAA depletion-induced responses could be alleviated by the addition of the HO-1 inducer haemin (Fig. 8). Therefore, these results were consistent with previously reported findings showing that auxin rapidly activates HO activity and that the product of the HO reaction, CO, then triggers the signal transduction events that lead to the auxin response of adventitious root formation in cucumber (Xuan et al., 2008b). In advanced stages of LR formation, the primordia have been suggested to develop a functional meristem from the pericycle of the primary root probably by synthesizing their own auxin to trigger the formation of LR, because young LRP cannot continue to divide and emerge when they are excised from the parent root (Laskowski et al., 1995). Thus, the possibility of changes in auxin sensitivity and content accounting for the increase in LR formation observed in salinity or osmotically stressed rapeseed seedlings cannot be ruled out. Additionally, it was previously reported that phytochrome could regulate the emergence of LRs, at least partly by manipuating auxin distribution within the Arabidopsis seedlings (Salisbury et al., 2007). Combined with the fact that HY1 in Arabidopsis is required for the synthesis of the phytochrome chromophore (Muramoto et al., 1999; Davis et al., 2001), the possible roles of BnHO1 in the formation of phytochrome chromophore and thereafter involvement in rapeseed LR development should be investigated.
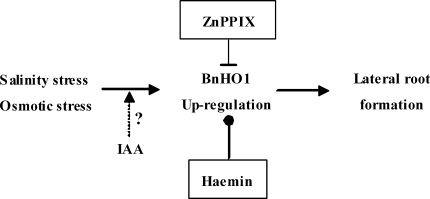
Overall, it has been demonstrated that lower doses of salinity and osmotic stress promoted LR formation, an important component of SIMR in plants (Fig. 9). We also suggested that stress-induced BnHO1 expression and HO activity, which are required in the LR establishment stage, were dependent on auxin signalling leading to LR formation (Fig. 9). Thus, BnHO1, as well as its catalytic product CO (Cao et al., 2007b; Xuan et al., 2008b), might represent a candidate that can incorporate environmental and/or endogenous hormone factors into plant developmental processes.
Fig. 9.
Schematic representation of the signalling pathway induced by salinity or osmotic stress during LR formation in rapeseed. The dashed line denotes possible involvement of IAA. The inhibitor or inducer of HO-1 applied in this study is boxed (top or bottom, respectively).
Supplementary Material
Acknowledgments
This work was supported by the National Basic Research Program of China (973 Programme, grant no. 2011CB109300), the Project for the Ministry of Agriculture Key Laboratory of Crop Nutrition and Fertilization (grant no. 10-08), Fundamental Research Funds for the Central Universities (grants no. KYJ200912 and KYZ200905), the Priority Academic Program Development of Jiangsu Higher Education Institutions, the Technology Support Program in Jiangsu Province (grant no. BE2010382), and the Program for New Century Excellent Talents in University (grant no. NCET-07-0441). We also thank Dr J. Liu from HeFei University of Technology, China, and Dr Evan Evans from the University of Tasmania, Australia, for their kind help in writing the manuscript.
References
- Balestrasse KB, Noriega GO, Batlle A, Tomaro ML. Heme oxygenase activity and oxidative stress signaling in soybean leaves. Plant Science. 2006;170:339–346. [Google Scholar]
- Bonkovsky HL, Healey JF, Pohl J. Purification and characterization of heme oxygenase from chick liver. European Journal of Biochemistry. 1990;189:155–166. doi: 10.1111/j.1432-1033.1990.tb15472.x. [DOI] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Analytical Biochemistry. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Cao ZY, Huang BK, Wang QY, Xuan W, Ling TF, Zhang B, Chen X, Nie L, Shen WB. Involvement of carbon monoxide produced by heme oxygenase in ABA-induced stomatal closure in Vicia faba and its proposed signal transduction pathway. Chinese Science Bulletin. 2007a;52:2365–2373. [Google Scholar]
- Cao ZY, Xuan W, Liu ZY, Li XN, Zhao N, Xu P, Wang Z, Guan RZ, Shen WB. Carbon monoxide promotes lateral root formation in rapeseed. Journal of Integrative Plant Biology. 2007b;49:1070–1079. [Google Scholar]
- Casimiro I, Beeckman T, Graham N, Bhalerao R, Zhang H, Casero P, Sandberg G, Bennett MJ. Dissecting Arabidopsis lateral root development. Trends in Plant Science. 2003;8:165–171. doi: 10.1016/S1360-1385(03)00051-7. [DOI] [PubMed] [Google Scholar]
- Casimiro I, Marchant A, Bhalerao RP, et al. Auxin transport promotes Arabidopsis lateral root initiation. The Plant Cell. 2001;13:843–852. doi: 10.1105/tpc.13.4.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen XY, Ding X, Xu S, Wang R, Xuan W, Cao ZY, Chen J, Wu HH, Ye MB, Shen WB. Endogenous hydrogen peroxide plays a positive role in the upregulation of heme oxygenase and acclimation to oxidative stress in wheat seedling leaves. Journal of Integrative Plant Biology. 2009;51:951–960. doi: 10.1111/j.1744-7909.2009.00869.x. [DOI] [PubMed] [Google Scholar]
- Correa-Aragunde N, Graziano M, Chevalier C, Lamattina L. Nitric oxide modulates the expression of cell cycle regulatory genes during lateral root formation in tomato. Journal of Experimental Botany. 2006;57:581–588. doi: 10.1093/jxb/erj045. [DOI] [PubMed] [Google Scholar]
- Correa-Aragunde N, Graziano M, Lamattina L. Nitric oxide plays a central role in determining lateral root development in tomato. Planta. 2004;218:900–905. doi: 10.1007/s00425-003-1172-7. [DOI] [PubMed] [Google Scholar]
- Cui W, Fu G, Wu H, Shen W. Cadmium-induced heme oxygenase-1 gene expression is associated with the depletion of glutathione in the roots of Medicago sativa. Biometals. 2011;24:93–103. doi: 10.1007/s10534-010-9377-2. [DOI] [PubMed] [Google Scholar]
- Dang T, Bishop GM, Dringen R, Robinson S. The putative heme transporter HCP1 is expressed in cultured astrocytes and contributes to the uptake of hemin. Glia. 2010;58:55–65. doi: 10.1002/glia.20901. [DOI] [PubMed] [Google Scholar]
- Davis SJ, Bhoo SH, Durski AM, Walker JM, Vierstra RD. The heme-oxygenase family required for phytochrome chromophore biosynthesis is necessary for proper photomorphogenesis in higher plants. Plant Physiology. 2001;126:656–669. doi: 10.1104/pp.126.2.656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle JJ, Doyle JL. Isolation of plant DNA from fresh tissue. Focus. 1990;12:13–15. [Google Scholar]
- Dulak J, Józkowicz A. Carbon monoxide – a ‘new’ gaseous modulator of gene expression. Acta Biochimica Polonica. 2003;50:31–47. [PubMed] [Google Scholar]
- Edlund A, Eklöf S, Sundberg B, Moritz T, Sanberg G. A microscale technique for gas chromatography-mass sepectrometry measurements of picogram amounts of indole-3-acetic acid in plant tissues. Plant Physiology. 1995;108:1043–1047. doi: 10.1104/pp.108.3.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, von Heijne G. ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Science. 1999;8:978–984. doi: 10.1110/ps.8.5.978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emborg TJ, Walker JM, Noh B, Vierstra RD. Multiple heme oxygenase family members contribute to the biosynthesis of the phytochrome chromophore in Arabidopsis. Plant Physiology. 2006;140:856–868. doi: 10.1104/pp.105.074211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu GQ, Xu S, Xie YJ, Han B, Nie L, Shen WB, Wang R. Molecular cloning, characterization, and expression of an alfalfa (Medicago sativa L.) heme oxygenase-1 gene, MsHO1, which is pro-oxidants-regulated. Plant Physiology and Biochemistry. 2011;49:792–799. doi: 10.1016/j.plaphy.2011.01.018. [DOI] [PubMed] [Google Scholar]
- Gisk B, Yasui Y, Kohchi T, Frankenberg-Dinkel N. Characterization of the haem oxygenase protein family in Arabidopsis thaliana reveals a diversity of functions. Biochemical Journal. 2010;425:425–434. doi: 10.1042/BJ20090775. [DOI] [PubMed] [Google Scholar]
- Han Y, Zhang J, Chen X, Gao Z, Xuan W, Xu S, Ding X, Shen WB. Carbon monoxide alleviates cadmium-induced oxidative damage by modulating glutathione metabolism in the roots of Medicago sativa. New Phytologist. 2008;177:155–166. doi: 10.1111/j.1469-8137.2007.02251.x. [DOI] [PubMed] [Google Scholar]
- He XJ, Mu RL, Cao WH, Zhang ZG, Zhang JS, Chen SY. AtNAC2, a transcription factor downstream of ethylene and auxin signaling pathways, is involved in salt stress response and lateral root development. The Plant Journal. 2005;44:903–916. doi: 10.1111/j.1365-313X.2005.02575.x. [DOI] [PubMed] [Google Scholar]
- Himanen K, Boucheron E, Vanneste S, de Almeida Engler J, Inzé D, Beeckman T. Auxin-mediated cell cycle activation during early lateral root initiation. The Plant Cell. 2002;14:2339–2351. doi: 10.1105/tpc.004960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirai K, Sasahira T, Ohmori H, Fujii K, Kuniyasu H. Inhibition of heme oxygenase-1 by zinc protoporphyrin IX reduces tumor growth of LL/2 lung cancer in C57BL mice. International Journal of Cancer. 2006;120:500–505. doi: 10.1002/ijc.22287. [DOI] [PubMed] [Google Scholar]
- Ito-Maki M, Ishikawa K, Matera KM, Sato M, Ikeda-Saito M, Yoshida T. Demonstration that histidine 25, but not 132, is the axial heme ligand in rat heme oxygenase-1. Archives of Biochemistry and Biophysics. 1995;317:253–258. doi: 10.1006/abbi.1995.1160. [DOI] [PubMed] [Google Scholar]
- Izawa T, Oikawa T, Tokutomi S, Okuno K, Shimamoto K. Phytochromes confer the photoperiodic control of flowering in rice (a short-day plant) The Plant Journal. 2000;22:391–399. doi: 10.1046/j.1365-313x.2000.00753.x. [DOI] [PubMed] [Google Scholar]
- Kohchi T, Mukougawa K, Frankenberg N, Masuda M, Yokota A, Lagarias JC. The Arabidopsis HY2 gene encodes phytochromobilin synthase, a ferredoxin-dependent biliverdin reductase. The Plant Cell. 2001;13:425–436. doi: 10.1105/tpc.13.2.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolbert Z, Ortega L, Erdei L. Involvement of nitrate reductase (NR) in osmotic stress-induced NO generation of Arabidopsis thaliana L. roots. Journal of Plant Physiology. 2010;167:77–80. doi: 10.1016/j.jplph.2009.08.013. [DOI] [PubMed] [Google Scholar]
- Kutz A, Müller A, Hennig P, Kaiser WM, Piotrowski M, Weiler EW. A role for nitrilase 3 in the regulation of root morphology in sulphur-starving Arabidopsis thaliana. The Plant Journal. 2002;30:95–106. doi: 10.1046/j.1365-313x.2002.01271.x. [DOI] [PubMed] [Google Scholar]
- Lamar CA, Mahesh VB, Brann DW. Regulation of gonadotrophin-releasing hormone (GnRH) secretion by heme molecules: a regulatory role for carbon monoxide? Endocrinology. 1996;137:790–793. doi: 10.1210/endo.137.2.8593832. [DOI] [PubMed] [Google Scholar]
- Lang D, Reuter S, Buzescu T, August C, Heidenreich S. Heme-induced heme oxygenase-1 (HO-1) in human monocytes inhibits apoptosis despite caspase-3 up-regulation. International Immunology. 2005;17:155–165. doi: 10.1093/intimm/dxh196. [DOI] [PubMed] [Google Scholar]
- Laskowski MJ, Williams ME, Nusbaum HC, Sussex IM. Formation of lateral root meristems is a two-stage process. Development. 1995;121:3303–3310. doi: 10.1242/dev.121.10.3303. [DOI] [PubMed] [Google Scholar]
- Ling T, Zhang B, Cui W, Wu M, Lin J, Zhou W, Huang J, Shen W. Carbon monoxide mitigates salt-induced inhibition of root growth and suppresses programmed cell death in wheat primary roots by inhibiting superoxide anion overproduction. Plant Science. 2009;177:331–340. [Google Scholar]
- Linley PJ, Landsberger M, Kohchi T, Cooper JB, Terry MJ. The molecular basis of heme oxygenase deficiency in the pcd1 mutant of pea. FEBS Journal. 2006;273:2594–2606. doi: 10.1111/j.1742-4658.2006.05264.x. [DOI] [PubMed] [Google Scholar]
- Liu YH, Xu S, Ling TF, Xu LL, Shen WB. Heme oxygenase/carbon monoxide system participates in regulating wheat seed germination under osmotic stress involving the nitric oxide pathway. Journal of Plant Physiology. 2010;167:1371–1379. doi: 10.1016/j.jplph.2010.05.021. [DOI] [PubMed] [Google Scholar]
- López-Bucio J, Hernández-Abreu E, Sánchez-Calderón L, Pérez-Torres A, Rampey RA, Bartel B, Herrera-Estrella L. An auxin transport independent pathway is involved in phosphate stress-induced root architectural alterations in Arabidopsis. Identification of BIG as a mediator of auxin in pericycle cell activation. Plant Physiology. 2005;137:681–691. doi: 10.1104/pp.104.049577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malamy JE, Benfey PN. Organization and cell differentiation in lateral roots of Arabidopsis thaliana. Development. 1997;124:33–44. doi: 10.1242/dev.124.1.33. [DOI] [PubMed] [Google Scholar]
- Matera KM, Zhou H, Migita CT, Hobert SE, Ishikawa K, Katakura K, Maeshima H, Yoshida T, Ikeda-Saito M. Histidine-132 does not stabilize a distal water ligand and is not an important residue for the enzyme activity in heme oxygenase-1. Biochemistry. 1997;36:4909–4915. doi: 10.1021/bi962321m. [DOI] [PubMed] [Google Scholar]
- Muramoto T, Kohchi T, Yokota A, Hwang I, Goodman HM. The Arabidopsis photomorphogenic mutant hy1 is deficient in phytochrome chromophore biosynthesis as a result of a mutation in a plastid heme oxygenase. The Plant Cell. 1999;11:335–348. doi: 10.1105/tpc.11.3.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muramoto T, Tsurui N, Terry MJ, Yokota A, Kohchi T. Expression and biochemical properties of a ferredoxin-dependent heme oxygenase required for phytochrome chromophore synthesis. Plant Physiology. 2002;130:1958–1966. doi: 10.1104/pp.008128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu X, Guiltinan MJ. DNA binding specificity of the wheat bZIP protein EmBP-1. Nucleic Acids Research. 1994;22:4969–4978. doi: 10.1093/nar/22.23.4969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noriega GO, Balestrasse KB, Batlle A, Tomaro ML. Heme oxygenase exerts a protective role against oxidative stress in soybean leaves. Biochemical and Biophysical Research Communications. 2004;323:1003–1008. doi: 10.1016/j.bbrc.2004.08.199. [DOI] [PubMed] [Google Scholar]
- Noriega GO, Yannarelli GG, Balestrasse KB, Batlle A, Tomaro ML. The effect of nitric oxide on heme oxygenase gene expression in soybean leaves. Planta. 2007;226:1155–1163. doi: 10.1007/s00425-007-0561-8. [DOI] [PubMed] [Google Scholar]
- Pasternak T, Rudas V, Potters G, Jansen MAK. Morphogenic effects of abiotic stress: reorientation of growth in Arabidopsis thaliana seedlings. Environmental and Experimental Botany. 2005;53:299–314. [Google Scholar]
- Péret B, De Rybel B, Casimiro I, Benková E, Swarup R, Laplaze L, Beeckman T, Bennett MJ. Arabidopsis lateral root development: an emerging story. Trends in Plant Science. 2009;14:399–408. doi: 10.1016/j.tplants.2009.05.002. [DOI] [PubMed] [Google Scholar]
- Pérez-Torres CA, López-Bucio J, Cruz-Ramírez A, Ibarra-Laclette E, Dharmasiri S, Estelle M, Herrera-Estrella L. Phosphate availability alters lateral root development in Arabidopsis by modulating auxin sensitivity via a mechanism involving the TIR1 auxin receptor. The Plant Cell. 2008;20:3258–3272. doi: 10.1105/tpc.108.058719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potters G, Pasternak TP, Guisez Y, Palme KJ, Jansen MAK. Stress-induced morphogenic responses: growing out of trouble? Trends in Plant Science. 2007;12:98–105. doi: 10.1016/j.tplants.2007.01.004. [DOI] [PubMed] [Google Scholar]
- Reed RC, Brady SR, Muday GK. Inhibition of auxin movement from the shoot into the root inhibits lateral root development in Arabidopsis. Plant Physiology. 1998;118:1369–1378. doi: 10.1104/pp.118.4.1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryter SW, Otterbein LE, Morse D, Choi AMK. Heme oxygenase/carbon monoxide signaling pathways: regulation and functional significance. Molecular and Cellular Biochemistry. 2002;234/235:249–263. doi: 10.1023/A:1015957026924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sairam RK, Tyagi A. Physiology and molecular biology of salinity stress tolerance in plants. Current Science. 2004;86:407–421. [Google Scholar]
- Salisbury FJ, Hall A, Grierson CS, Halliday KJ. Phytochrome coordinates Arabidopsis shoot and root development. The Plant Journal. 2007;50:429–438. doi: 10.1111/j.1365-313X.2007.03059.x. [DOI] [PubMed] [Google Scholar]
- Schruff MC, Spielman M, Tiwari S, Adams S, Fenby N, Scott RJ. The AUXIN RESPONSE FACTOR 2 gene of Arabidopsis links auxin signaling, cell division, and the size of seeds and other organs. Development. 2006;133:251–261. doi: 10.1242/dev.02194. [DOI] [PubMed] [Google Scholar]
- Shayeghi M, Latunde-Dada GO, Oakhill JS, et al. Identification of an intestinal heme transporter. Cell. 2005;122:789–801. doi: 10.1016/j.cell.2005.06.025. [DOI] [PubMed] [Google Scholar]
- Shekhawat GS, Verma K. Haem oxygenase (HO): an overlooked enzyme of plant metabolism and defence. Journal of Experimental Botany. 2010;61:2255–2270. doi: 10.1093/jxb/erq074. [DOI] [PubMed] [Google Scholar]
- Shen Q, Jiang M, Li H, Che LL, Yang ZM. Expression of a Brassica napus heme oxygenase confers plant tolerance to mercury toxicity. Plant, Cell and Environment. 2011;34:752–763. doi: 10.1111/j.1365-3040.2011.02279.x. [DOI] [PubMed] [Google Scholar]
- Terry MJ, Kendrick RE. The aurea and yellow-green-2 mutants of tomato are deficient in phytochrome chromophore synthesis. Journal of Biological Chemistry. 1996;271:21681–21686. doi: 10.1074/jbc.271.35.21681. [DOI] [PubMed] [Google Scholar]
- Tsoyi K, Lee TY, Lee YS, Kim HJ, Seo HG, Lee JH, Chang KC. Heme-oxygenase-1 induction and carbon monoxide-releasing molecule inhibit lipopolysaccharide (LPS)-induced high-mobility group box 1 release in vitro and improve survival of mice in LPS- and cecal ligation and puncture-induced sepsis model in vivo. Molecular Pharmacology. 2009;76:173–182. doi: 10.1124/mol.109.055137. [DOI] [PubMed] [Google Scholar]
- Wu MZ, Huang JJ, Xu S, Ling TF, Xie YJ, Shen WB. Haem oxygenase delays programmed cell death in wheat aleurone layers by modulation of hydrogen peroxide metabolism. Journal of Experimental Botany. 2011;62:235–248. doi: 10.1093/jxb/erq261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie YJ, Ling TF, Han Y, et al. Carbon monoxide enhances salt tolerance by nitric oxide-mediated maintenance of ion homeostasis and up-regulation of antioxidant defence in wheat seedling roots. Plant, Cell and Environment. 2008;31:1864–1881. doi: 10.1111/j.1365-3040.2008.01888.x. [DOI] [PubMed] [Google Scholar]
- Xie YJ, Xu S, Han B, Wu MZ, Yuan XX, Han Y, Gu Q, Xu DQ, Yang Q, Shen WB. Evidence of Arabidopsis salt acclimation induced by up-regulation of HY1 and the regulatory role of RbohD-derived reactive oxygen species synthesis. The Plant Journal. 2011;66:280–292. doi: 10.1111/j.1365-313X.2011.04488.x. [DOI] [PubMed] [Google Scholar]
- Xu S, Zhang B, Cao ZY, Ling TF, Shen WB. Heme oxygenase is involved in cobalt chloride-induced lateral root development in tomato. Biometals. 2011;24:181–191. doi: 10.1007/s10534-010-9386-1. [DOI] [PubMed] [Google Scholar]
- Xuan W, Xu S, Yuan XX, Shen WB. Carbon monoxide: a novel and pivotal signal molecule in plants? Plant Signaling and Behavior. 2008a;3:381–382. doi: 10.4161/psb.3.6.5374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xuan W, Zhu FY, Xu S, Huang BK, Ling TF, Qi JY, Ye MB, Shen WB. The heme oxygenase/carbon monoxide system is involved in the auxin-induced cucumber adventitious rooting process. Plant Physiology. 2008b;148:881–893. doi: 10.1104/pp.108.125567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Zheng B, Mao C, Qi X, Liu F, Wu P. Analysis of transcripts that are differentially expressed in three sectors of the rice root system under water deficit. Molecular Genetics and Genomics. 2004;272:433–442. doi: 10.1007/s00438-004-1066-9. [DOI] [PubMed] [Google Scholar]
- Yannarelli GG, Noriega GO, Batlle A, Tomaro ML. Heme oxygenase up-regulation in ultraviolet-B irradiated soybean plants involves reactive oxygen species. Planta. 2006;224:1154–1162. doi: 10.1007/s00425-006-0297-x. [DOI] [PubMed] [Google Scholar]
- Yoshida T, Kikuchi G. Purification and properties of heme oxygenase from pig spleen microsomes. Journal of Biological Chemistry. 1978;253:4224–4229. [PubMed] [Google Scholar]
- Zhu JK. Plant salt tolerance. Trends in Plant Science. 2001;6:66–71. doi: 10.1016/s1360-1385(00)01838-0. [DOI] [PubMed] [Google Scholar]
- Zhu JK. Salt and drought stress signal transduction in plants. Annual Review of Plant Biology. 2002;53:247–273. doi: 10.1146/annurev.arplant.53.091401.143329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu JK. Regulation of ion homeostasis under salt stress. Current Opinion in Plant Biology. 2003;6:441–445. doi: 10.1016/s1369-5266(03)00085-2. [DOI] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W. GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiology. 2004;136:2621–2632. doi: 10.1104/pp.104.046367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolla G, Heimer YM, Barak S. Mild salinity stimulates a stress-induced morphogenic response in Arabidopsis thaliana roots. Journal of Experimental Botany. 2010;61:211–224. doi: 10.1093/jxb/erp290. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.