Abstract
Background
Mycoplasma mycoides subsp. mycoides SC is the pathogenic agent of contagious bovine pleuropneumonia (CBPP), the most important disease of cattle in Africa causing significant economic losses. The re-emergence of CBPP in Europe in the 1980s and 1990s illustrates that it is still a threat also to countries that have successfully eradicated the disease in the past. Nowadays, probe-based real-time PCR techniques are among the most advanced tools for a reliable identification and a sensitive detection of many pathogens, but only few protocols have been published so far for CBPP diagnosis. Therefore we developed a novel TaqMan®-based real-time PCR assay comprising the amplification of two independent targets (MSC_0136 and MSC_1046) and an internal exogenous amplification control in a multiplex reaction and evaluated its diagnostic performance with clinical samples.
Results
The assays detected 49 MmmSC strains from diverse temporal and geographical origin, but did not amplify DNA from 82 isolates of 20 non-target species confirming a specificity of 100%. The detection limit was determined to be 10 fg DNA per reaction for the MSC_0136 assay and 100 fg per reaction for the MSC_1046 assay corresponding to 8 and 80 genome equivalents, respectively. The diagnostic performance of the assay was evaluated with clinical samples from 19 experimentally infected cattle and from 20 cattle without CBPP and compared to those of cultivation and a conventional PCR protocol. The two rt-PCR tests proved to be the most sensitive methods and identified all 19 infected animals. The different sample types used were not equally suitable for MmmSC detection. While 94.7% of lung samples from the infected cohort were positively tested in the MSC_0136 assay, only 81% of pulmonal lymph nodes, 31% of mediastinal lymph nodes and 25% of pleural fluid samples gave a positive result.
Conclusions
The developed multiplex rt-PCR assay is recommended as an efficient tool for rapid confirmation of a presumptive CBPP diagnosis in a well-equipped laboratory environment.
Background
Contagious bovine pleuropneumonia (CBPP) caused by Mycoplasma mycoides subsp. mycoides SC (MmmSC) is a disease of cattle affecting the respiratory tract of animals. It is manifested by fever, anorexia, cough, and dyspnoea. Macropathological examinations show gross lesions in the lung including marbling, thickened interlobular septa and sequestra. However, clinical and pathological signs are not always evident, and chronically infected animals might act as carriers and source of infections. Today the disease is mainly confined to sub-Saharan Africa where it causes severe economic losses. As a result of rigid control measures and a consequent stamping out policy, Europe has been CBPP free since the late 1990s. Even so, the risk of re-introducing CBPP remains high. In case of a CBPP outbreak in non-endemic regions caused by animals with inapparent infection or by re- introduction, rapid action such as setting up exclusion zones and culling of affected live stock is essential to efficiently prevent transmission and spread of disease. All these measures depend on an early identification of the pathogen. Two serological tests, the complement fixation test (CFT) and a competitive enzyme-linked immunosorbent assay (cELISA), are approved by the World Organization for Animal Health (OIE) [1], but they are recommended at herd level only [2-5]. Traditionally, mycoplasmas including MmmSC are isolated by culture and identified by biochemical and antigenic techniques [6-8]. Drawbacks of these methods include low sensitivity caused by bacterial contamination, low specificity due to cross-reactivity of antigenic determinants of closely related species and time- and labour-intensive laboratory procedures. Considerable improvements have been achieved by the introduction of PCR since 1994 [2,9,10] which provides a much quicker and more sensitive diagnosis of CBPP. However, due to the close phylogenetic relatedness among MmmSC and other members of the M. mycoides cluster including M. mycoides subsp. capri, M. capricolum subsp. capricolum, M. capricolum subsp. capripneumoniae and M. leachii, identification by conventional PCR protocols remains problematic [7]. Real time PCR (rt-PCR) formats using SYBR green detection of PCR products compensated some of the disadvantages connected to conventional PCR [11,12], but are less specific than real-time PCR assays that include specific probes [13,14].
In this study, we developed a new multiplex rt-PCR assay using TaqMan®-labelled locked nucleic acid (LNA) probes for the detection of MmmSC and validated it using samples from experimentally infected Boran cattle taken at necropsy. The assay specifically targets the gene lppQ (MSC_1046) and a gene encoding an uncharacterized lipoprotein (MSC_0136) close to the fba locus, and it includes an internal amplification control. The new protocol is intended for confirmatory diagnosis of clinically or pathologically suspected cases and is considered to contribute to further improvement of CBPP control in routine laboratory diagnosis.
Methods
Bacterial strains
The specificity of the assay was verified by testing DNA from 48 MmmSC field strains, from 46 other type and field strains of the M. mycoides cluster to exclude potential cross-reactivity due to close phylogenetic relatedness and from 36 isolates of 14 other relevant mollicute species occurring in cattle (Table 1). All mycoplasma strains were recovered from lyophilized stocks and cultivated in modified Hayflick medium [15] containing 20% horse serum at 37°C for 2-6 days. DNA extracts from the non-related bacterial pathogens Pasteurella multocida and Mannheimia haemolytica, that cause respiratory diseases in cattle, were also included in the specificity testing (Table 1). 10 ng of genomic DNA extracted from bacterial cultures were used in the specificity tests.
Table 1.
Specificity testing of rt-PCR assays with isolates of the M.mycoides cluster (A), mollicute strains frequently isolated from bovine lungs (B) and bacterial strains causing CBPP-like symptoms (C)
| Species | Strain | n | Host | Origin | Year | MSC_0136 rt-PCR | MSC_1046 rt-PCR | |
|---|---|---|---|---|---|---|---|---|
| A | M. mycoides | PG1T | cattle | ? | ? | + | + | |
| subsp. Mycoides SC | diverse | 19 | cattle | Italy | 1990-1992 | + | + | |
| diverse | 3 | cattle | Spain | unknown | + | + | ||
| P0 | cattle | France | 1967 | + | + | |||
| N6 | cattle | Botswana | 1996 | + | + | |||
| M375 | cattle | Botswana | 1995 | + | + | |||
| Afade | cattle | Cameroon | 1968 | + | + | |||
| PO2 | cattle | France | 1980 | + | + | |||
| L2 | cattle | Italy | 1993 | + | + | |||
| Fatick | cattle | Senegal | 1968 | + | + | |||
| C11 | cattle | Chad | 1962 | + | + | |||
| 95014 | cattle | Tanzania | 1995 | + | + | |||
| T1/44 | cattle | Tanzania | 1952 | + | + | |||
| Gladysdale | cattle | Australia | 1953 | + | + | |||
| Shawawa | cattle | Botswana | 1995 | + | + | |||
| 2091 | cattle | France | 1984 | + | + | |||
| 197 | cattle | Italy | 1992 | + | + | |||
| B103 | cattle | Portugal | 1986 | + | + | |||
| 6305 | cattle | Portugal | 1993 | + | + | |||
| 6526 | cattle | Portugal | 1993 | + | + | |||
| Astercous | cattle | Spain | 1987 | + | + | |||
| Madrid | cattle | Spain | 1993 | + | + | |||
| Tan8 | cattle | Tanzania | 1998 | + | + | |||
| Matapi | cattle | Namibia | 2004 | + | + | |||
| Mandigwan | cattle | Namibia | 2001 | + | + | |||
| V5 | cattle | Australia | 1936 | + | + | |||
| IS31 | cattle | Tanzania | 1998 | + | + | |||
| 2022 | cattle | France | 1984 | + | + | |||
| B237 | cattle | Kenya | 1987 | + | + | |||
| M. mycoides | PG3, NCTC 10137T | goat | Turkey | 1950 | - | - | ||
| subsp. capri | Y-Goat | goat | Australia | 1956 | - | - | ||
| 7302 | goat | Portugal | < 1994 | - | - | |||
| Kombolcha | goat | Ethiopia | 1975 | - | - | |||
| 7730 | goat | France | < 1997 | - | - | |||
| 9096-C9415 | goat | Nigeria | unknown | - | - | |||
| diverse | 6 | goat | Gran Canaria | 1993 | - | - | ||
| My-I | goat | Croatia | 1986 | - | - | |||
| D2482 | goat | Switzerland | 1991 | - | - | |||
| 95010 | goat | France | 1995 | - | - | |||
| 6443-90 | goat | France | 1990 | - | - | |||
| My-325 | goat | Croatia | 1986 | - | - | |||
| C260/4 | goat | Gran Canaria | 1993 | - | - | |||
| Wk354 | goat | Switzerland | 1980 | - | - | |||
| L | goat | France | 1975 | - | - | |||
| Wi18079 | goat | Germany | 2009 | - | - | |||
| diverse | 3 | Barbary sheep | Germany | 1994 | - | - | ||
| M-5 | goat | Croatia | 1988 | - | - | |||
| M-18 | goat | Croatia | 1988 | - | - | |||
| M-29 | goat | Croatia | 1988 | - | - | |||
| M. capricolum | Cal. kid, NCTC 10154T | goat | USA | 1954 | - | - | ||
| subsp. capricolum | C47 | sheep | Germany | < 1992 | - | - | ||
| 4146 | goat | France | 1980 | - | - | |||
| 7714 | goat | France | 1967 | - | - | |||
| 609-79 | goat | France | 1979 | - | - | |||
| 8086-1 | goat | France | 1980 | - | - | |||
| M. capricolum | F38, NCTC 10192T | goat | Kenya | 1976 | - | - | ||
| subsp. capripneumoniae | 87F05 | goat | Turkey | 2005 | - | - | ||
| diverse | 2 | gazelle | UAE | 2004 | - | - | ||
| 3535 | mouflon | Qatar | 2005 | - | - | |||
| GL97P | goat | Tunisia | 1980 | - | - | |||
| M. leachii | PG50, NCTC 10133T | cattle | Australia | 1963 | - | - | ||
| D318b | cattle | Germany | unknown | - | - | |||
| D424 | cattle | Germany | unknown | - | - | |||
| Calf1 | cattle | Nigeria | unknown | - | - | |||
| M.nov.spec. | 8756-13 | mountain goat | USA | 1987 | - | - | ||
| G1650 | capricorn | Germany | 1994 | - | - | |||
| G5847 | capricorn | Germany | 1993 | - | - | |||
| B | M. putrefaciens | KS1, NCTC 10155T | goat | USA | 1955 | - | - | |
| 7578.94 | goat | France | 1994 | - | - | |||
| Tours2 | goat | France | 1972 | - | - | |||
| M. bovis | PG45, NCTC 10131T | cattle | USA | 1962 | - | - | ||
| 363/88 | cattle | Germany | 1988 | - | - | |||
| 334/96 | cattle | Germany | 1996 | - | - | |||
| 54/10 | cattle | Germany | 2010 | - | - | |||
| M. bovirhinis | PG43, NCTC 10118T | cattle | UK | 1967 | - | - | ||
| 320/93 | cattle | Germany | 1993 | - | - | |||
| 19/94 | cattle | Germany | 1994 | - | - | |||
| 365/09 | cattle | Germany | 2009 | - | - | |||
| M. bovigenitalium | PG11, NCTC 10122T | cattle | UK | 1947 | - | - | ||
| 338/87 | cattle | Germany | 1987 | - | - | |||
| 18/92 | cattle | Germany | 1992 | - | - | |||
| 466/94 | cattle | Germany | 1994 | - | - | |||
| M. arginini | G230, NCTC 10129T | mouse | USA | 1968 | - | - | ||
| 162/81 | cattle | Germany | 1981 | - | - | |||
| 208/94 | Barbary sheep | Germany | 1994 | - | - | |||
| 23/05 | sheep | Germany | 2005 | - | - | |||
| M. californicum | St-6, NCTC 10189T | cattle | USA | 1981 | - | - | ||
| 456/83 | cattle | Germany | 1983 | - | - | |||
| 111/84 | cattle | Germany | 1984 | - | - | |||
| M. canadense | 275C, NCTC 10152T | cattle | Canada | 1974 | - | - | ||
| 380/94 | cattle | Germany | 1994 | - | - | |||
| M. verecundum | 107, NCTC 10145T | cattle | UK | 1974 | - | - | ||
| 49/94 | cattle | Germany | 1994 | - | - | |||
| M. alkalescense | PG51, NCTC 10135T | cattle | Australia | 1961 | - | - | ||
| 26/93 | cattle | Germany | 1993 | - | - | |||
| M. canis | PG14, NCTC 10146T | dog | UK | 1951 | - | - | ||
| 306/93 | dog | Germany | 1993 | - | - | |||
| A. laidlawii | PG8, NCTC 10116T | unknown | UK | 1967 | - | - | ||
| 54/99 | cattle | UK | 1999 | - | - | |||
| A. axanthum | S743, NCTC 10138T | tissue culture | UK | 1970 | - | - | ||
| 824/80 | cattle | Germany | 1980 | - | - | |||
| Ureaplasma diversum | A417, NCTC 10182T | cattle | UK | 1972 | - | - | ||
| C | Pasteurella multocida | ATCC 43137T | pig | - | - | |||
| Mannheimia haemolytica | ATCC 33396T | unknown | - | - | ||||
Clinical samples
Tissue samples from an animal infection trial performed according to the Kenyan national legislation for animal experimentation and approved by the ILRI Institutional Animal Care and Use Committee (IACUC reference number 2008.08) were obtained from the International Livestock Research Institute Nairobi, Kenya [16]. Twenty 14-16 months old castrated outbred Boran cattle (Bos indicus) were infected intratracheally with 5 × 1010 cfu of the strain MmmSC Afade in 50 ml culture broth. Six days post infection nine animals were depleted for CD4+ T lymphocytes using repeated injections of CD4-specific mouse monoclonal antibody. The CD4+ T cell depletion had no discernable effect on the infection status, clinical presentation, pathology or humoral response of the animals [16]. Out of the 20 infected cattle, 19 were available for sampling. Three of them (animals no. BD097, BD098, BD118) showed acute CBPP-typical clinical signs such as coughing, dyspnea and fever (> 39.4°C) for at least eight consecutive days. Because of the severity of the disease, these animals had to be euthanized 16 to 21 days post infection. Out of the 16 surviving animals, 13 showed clinical signs, and three animals (no. BD096, BD100, BD111) were asymptomatic. All were euthanized 28-30 days post infection.
Post mortem examination of lung tissue revealed typical pathological signs of pneumonia and respiratory disorder in all but one (no. BD102) individuals. Gross lesions of red hepatisation and marmorisation, necrosis and sequestra were seen in animals no. BD097, BD098 and BD118, whereas fibrous adhesions between pleura and chest wall and the production of massive amounts of pleural fluid (a typical symptom of acute CBPP) were only observed in animals no. BD097 and BD118. All other animals, with the exception of no. BD102, developed necrotic lesions, sequestra and areas of resolution of differing sizes. Their low morbidity together with the appearance of lung lesions indicated that these animals were in the chronic stage of infection. Despite the artificial infection and the constant contact with its infected companions, animal no. BD102 did not show any pathological changes in the lung, and also clinical signs were mild. All animals showed a specific antibody response detected by the complement fixation test (CFT) (data not shown). According to the case definition for CBPP given by the OIE [17], 18 animals are defined as MmmSC-positive and animal no. BD102 as MmmSC-negative. But since animal no. BD102 had MmmSC specific antibodies and mild clinical symptoms (fever) it was also included in the positive group.
Specimens of lungs and lymph nodes as well as pleural fluid were collected, immediately frozen at -80°C and used for DNA extraction and subsequent PCR.
Re-isolation of MmmSC was done by inoculation of 50-100 mg smashed tissue or 300 μl pleural fluid into 3 ml modified Hayflick broth containing 20% horse serum and 1000 IU/ml penicillin for 3-5 days. Subsequently, an aliquot was plated onto a modified Hayflick agar plate without penicillin to control for growth of mycoplasma colonies. Sub-cultures were grown in 3 ml of modified Hayflick broth without penicillin to identify the re-isolated species. DNA was extracted and conventional MmmSC-specific PCR was performed as described below.
20 negative control samples from cattle flocks in Germany (which is CBPP-free) were included in the analysis. Lothar Hoffmann (TLLV Bad Langensalza, Germany) kindly provided lung samples of cattle diagnosed with pneumonia (n = 3). Bovine lung samples from an experimental chlamydia infection (approved by the Thuringian State Office for Food Safety and Consumer Protection under registration no. 04-002/07) causing pneumonia in cattle (n = 8) and lung samples from uninfected animals (n = 9) were obtained from the animal facilities of the Friedrich Loeffler Institute.
DNA extraction
Cells from 3 ml broth cultures were harvested by centrifugation, washed twice in PBS, and DNA was extracted using the High Pure PCR Template Preparation Kit™ (Roche Diagnostics, Mannheim, Germany) according to the manufacturer's instructions. DNA extraction from animal tissue was performed following the protocol of the innuSPEED Tissue DNA Kit™ (Analytik Jena, Jena, Germany) and DNA was eluted with 100 μl of buffer.
Confirmation of strain identity
All strains presumably belonging to the M. mycoides cluster were characterized by amplification and sequencing of a 992 bp fragment of uvrC.
M. mycoides cluster strains other than MmmSC were additionally analyzed by amplification and sequencing of fragments of rpoB. Universal primers uvrC-F1 5'-GATCTATTTTATTAACACTACAACG-3' and uvrC-R2 5'-TCTTTAGCTGCATGAATTTG and universal rpoB-specific primers as described by Manso-Silvan et al. and Vilei et al. [18,19] were used at a concentration of 400 nM in a 25 ul PCR reaction volume with 1.5 mM MgCl2, 250 μM each dNTP, reaction buffer and 1 U InnuTaq Hot A DNA Polymerase (Analytik Jena, Jena, Germany). The temperature-time profile on a UNO II thermocycler (Biometra, Göttingen, Germany) was 95°C for 5 min and 35 times 95°C for 30 s, 50°C for 30s and 72°C for 60 s. Amplicons were purified by the DNA Clean and Concentrator Kit (Zymo Research, Irvine, USA) and subsequently sequenced by Eurofins MWG Operon (Ebersberg, Germany) using the respective forward primers. Sequences were edited by the software BioEdit™ version 7.0.9 and aligned and compared to reference sequences by the software tool Multalin™ [20].
The field strains of other mycoplasma species of bovine origin investigated in this study were isolated in our lab over the last decades and identified by an in-house indirect immunofluorescence test according to Rosendal and Black, [8].
Conventional MmmSC-specific PCR
Conventional PCR [10] was done on dilution series of quantified DNA from cultures and DNA extracted from clinical specimens using the primer pair SC3NEST1-L/SC3NEST1-R and Taq DNA Polymerase 5 Prime (VWR International, Darmstadt, Germany) on a UNO II thermocycler. 5 μl of each PCR product were analyzed by gelelectrophoresis and visualised by ethidiumbromide staining.
Oligonucleotide Primers and TaqMan® probes
MmmSC-specific primers and probes were designed using the software tool IDT OligoAnalyzer3.1 based on sequence comparisons between the respective genes of MmmSC field strains and type and field strains of the M. mycoides cluster performed by the software tool Multalin™ [20].
Locked nucleic acid (LNA)-containing dual-labelled probes, designed in cooperation with and produced by TIB Molbiol (Berlin, Germany), were applied to compensate the low GC-content of mycoplasma DNA and to ensure specific and sensitive probe hybridization despite the low Tm values of probe sequences.
The primers MSC_1046-S and MSC_1046-R2 obtained from MWG Operon (Ebersbach, Germany) (Table 2) target the gene of a well-characterized antigenic and specific MmmSC lipoprotein [21]. This gene occurs in two copies in type strain PG1, but only in one copy in many other MmmSC strains [22]. The respective probe MSC_1046-TM1 was designed to anneal to a highly MmmSC-specific segment exhibiting ten nucleotide mismatches to the homologous sequences from M. mycoides subsp. capri (GenBank accession number FQ377874) and M. leachii (CP002108).
Table 2.
Primers and TaqMan® probes
| Target | Primers/Probes | Sequence (5'-3') | Position2 |
|---|---|---|---|
| IppQ MSC_0136 IAC |
MSC_1046-S MSC_1046-R2 MSC_1046-TM1 MSC_0136-F MSC_0136-R MSC_0136-TM EGFP-1F EGFP-10R EGFP-Hex |
ATCAAGATATTTCGAGTTGAAATGTAAG TGTATATTTTTTAGATTTCAATCTGAAAGTG FAM-TTTCAGCTCGATAAAACATATTT-BBQ1 AACACAAAAAACCGAACAGCCT AGCTTATCAGGAACTTTTTTAACGTA Cy5-TGAGATAGTTCAAATTGGCTTT-BBQ1 GACCACTACCAGCAGAACAC CTTGTACAGCTCGTCCATGC HEX-AGCACCCAGTCCGCCCTGAGCA-BHQ1 |
1166686-1166713 1190443-1190470 1166797-1166767 1190554-1190524 1166754-1166732 1190511-1190489 159994-160015 1023331-1023310 160131-160106 1023194-1023219 160042-160063 1023283-1023262 |
1Locked nucleic acid (LNA) containing nucleotides are underlined,2Based on GenBank sequence BX293980
Primers MSC_0136-F and MSC_0136-R mediate the amplification of the target MSC_0136, which codes for a hypothetical transmembrane protein and is located directly adjacent upstream of the fba locus [23]. The PG1 genome also harbours a second copy of the gene in reverse orientation [24]. Amplification products were detected by hybridization and hydrolysis of probe MSC_0136-TM which was designed with at least 3 mismatches to sequences from other members of the M. mycoides cluster.
Internal amplification control
A 712 bp fragment of the enhanced green fluorescent protein (EGFP) gene was used as internal amplification control (IAC) template as described previously [25] which is also commercially available as Intype IC-DNA (Labor Diagnostik, Leipzig, Germany). A 177 bp amplicon was generated by amplification with primers EGFP-1F and EGFP-10R and detected by probe EGFP-HEX to demonstrate the absence of PCR inhibitory substances.
Multiplex real-time PCR
Amplifications were performed as duplex assays (MSC_0136 and MSC_1046 assay) with culture material and as triplex assays including also the IAC with tissue samples. The assays were run on a Mx3000P® thermocycler (Stratagene, Heidelberg, Germany) using the following cycling parameters: initial denaturation at 95°C for 5 min, 45 cycles of denaturation at 95°C for 20 s, primer annealing at 57°C for 45 s and extension at 68°C for 45 S. Optimal primer and probe concentrations were determined by empirical testing of different concentrations ranging from 200 nM to 800 nM final concentration. Finally, each 25 μl duplex reaction contained 12.5 μl 2 × QuantiTect® Multiplex PCR Master Mix (Qiagen, Hilden, Germany), 400 nM each of primers MSC_1046-S, MSC_1046-R2, MSC_0136-F and MSC_0136-R, 300 nM each of probes MSC_1046-TM1 and MSC_0136-TM and 1 μl of DNA template. Additionally, 800 nM of the EGFP-specific primers and 400 nM of probe EGFP-HEX as well as 0.25 μl IAC template DNA corresponding to 12,500 copies were added to reactions in the triplex assay.
Amplified products were analyzed using the Stratagene software MxPro™, and amplification plots were generated with an adaptive baseline. The threshold, i.e. the level of fluorescence signal that reflects a significant increase over the baseline signal, was manually adjusted at 0.2 for the MSC_0136 assay and at 0.15 for the MSC_1046 assay. The threshold cycles were calculated accordingly by the instrument.
Each sample was analysed by rt-PCR in duplicate. Each microtitre plate included a serial dilution of quantified standard DNA containing 10 fg to 10 ng.
Standard curve calculation
Standard curves were generated from duplicates of five tenfold dilution series of MmmSC PG1 genomic DNA quantified by UV-spectrophotometry (Nanodrop®, Thermo Scientific, Wilmington, USA) containing 1 fg - 100 ng. Linear regressions, efficiencies and correlation coefficients were calculated from the standard graphs of the Ct values plotted against the logarithm of the DNA amount, and intra-assay variability was determined. Additionally, five independent runs on different days were performed in duplicates to assess inter-assay variance and coefficients of variance were calculated.
Correlation of cfu and DNA content
To investigate the correlation between counts of colony forming units (cfu) and extracted DNA and to define an adequate reference value for pathogen quantification, a growth curve of MmmSC PG1 was recorded. Cfu counts and the respective quantities of total DNA determined using the newly developed rt-PCR assay were compared at different time points. Two-day cultures of MmmSC were diluted 1:100 into fresh liquid medium and incubated at 37°C. Every 6 to 12 h up to 48 h, 100 μl aliquots were removed, tenfold serial dilutions were prepared and 20 μl of each dilution were seeded on agar plates for counting cfus after incubation of the plates for 3 days. The doubling time of the strain was calculated during the logarithmic growth phase with Doubling Time Software v1.0.10 (Roth, 2006, http://www.doubling-time.com/compute.php).
Furthermore, aliquots were taken after 12, 18, 24 and 48 h of growth from which tenfold dilution series were prepared. DNA was extracted from 200 μl of each dilution sample as described above and quantified in duplex rt-PCR assays. Results were compared with standard curves obtained from tenfold serial dilutions of quantified MmmSC DNA. The calculated genome equivalents of the 10-1 dilution samples at the time points 12, 18, 24 and 48 h were related to the respective cfu counts.
Results
Specificity testing
In order to determine the ability of the MSC_1046 and MSC_0136 assays to detect MmmSC DNA, genomic DNA extracts of the type strain PG1 and of 48 MmmSC field strains were subjected to the duplex rt-PCR. All isolates were readily detected by both assays.
All non-MmmSC strains belonging to the M. mycoides cluster (46 strains) and 36 isolates of 16 other mollicute and bacterial species yielded a signal below a Ct value of 39 in any of the two amplification reactions. The results are summarized in Table 1 and confirm an analytical specificity of the developed duplex rt-PCR of 100%.
Detection limit and standard curves of MSC_1046 and MSC_0136 assay
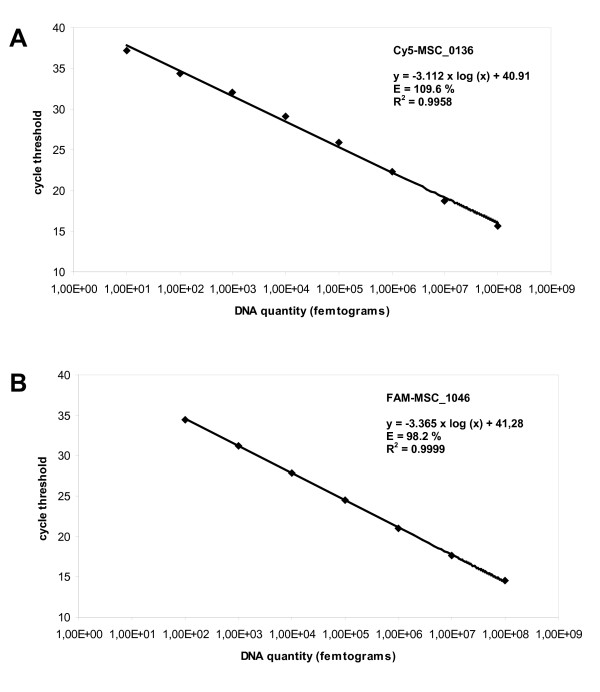
Five tenfold serial dilutions of genomic DNA covering a range from 1 fg to 100 ng were analysed in duplex assay to calculate detection limits (LOD) and standard curves for MSC_1046 and MSC_0136 amplifications (Figure 1). The LOD giving a reproducible signal (10 out of 10) was determined to be 10 fg DNA per reaction for the MSC_0136 assay and 100 fg per reaction for the MSC_1046 assay corresponding to 8 and 80 genome equivalents, respectively, based on the size of the published MmmSC genome sequence [24]. Conventional PCR performed with the same dilution series exhibited a detection limit of 1 pg, corresponding to 800 genome equivalents and thus, proved to be less sensitive by an order of 1-2 log units.
Figure 1.
Real-time quantification of DNA extracted from MmmSC cultures. DNA concentration was measured spectrophotometrically and adjusted to 100 ng μl-1. Five tenfold dilution series were prepared and applied in duplex rt-PCR assays comprising the amplification and detection of the targets MSC_0136 (A) and MSC_1046 (B) with the TaqMan® LNA probes FAM-MSC_1046 and Cy5-MSC_0136, respectively. The detection limits are 10 fg DNA and 100 fg DNA corresponding to approximately 8 genome equivalents and 80 genome equivalents for the MSC_0136 and MSC_1046 assay, respectively.
While the standard curve of the MSC_1046 reaction is linear over a range of 7 log units (from 100 fg to 100 ng), as illustrated by a correlation coefficient (R2) of 0.9999, the standard curve of the MSC_0136 reaction shows linearity over a range of 6 log units (from 1 pg to 100 ng) only and has a lower R2 value of 0.9958 for the entire detectable dilution range of 8 log units. The efficiency of amplification calculated from standard curve slopes was determined by the instruments software to be 109.6% for MSC_0136 and 98.2% for MSC_1046. Intra-assay variation coefficients varied between 1.1 and 2.6% for MSC_0136 and between 0.7 and 2.3% for MSC_1046. Inter-assay variation coefficients ranged from 1.1 to 2.4% for MSC_0136 and from 0.8 to 2.9% for MSC_1046 indicating a good reproducibility also between different runs.
When compared to the two simplex reactions, the duplex format did not shift Ct values and did in particular not compromise the sensitivity of the two assays. Also the presence of the IAC did not negatively influence the amplification of MmmSC DNA in triplex rt-PCR (data not shown).
Growth curve and MmmSC quantification
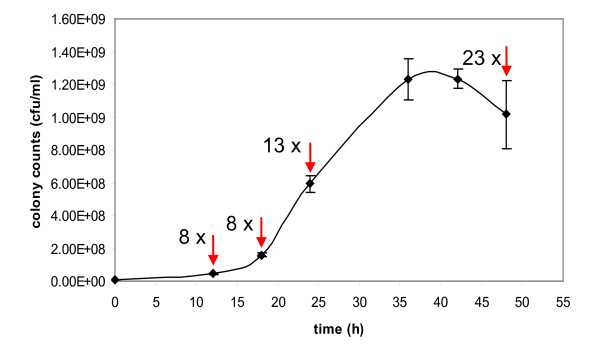
The growth curve in Figure 2 was obtained for MmmSC type strain PG1 after colony counting under cultivation conditions described above. The culture reached the stationary phase after 24 to 36 hours of cultivation. The doubling time during the logarithmic phase after 18 to 24 hours of cultivation was 188 min which is in the range reported for MmmSC strains [26]. In parallel, cultures at time points 12, 18, 24 and 48 h were examined through the quantification of extracted DNA by MSC_1046 rt-PCR assay. As expected, the total amount of deduced genome equivalents exceeded the number of viable mycoplasmas determined by colony counting. While in the lag phase (12 h) and early logarithmic phase (18 h), the total amount of DNA expressed in genome equivalents exceeded the cfu values by a factor of 8, this factor rose to 13 in the culture after 24 h of growth and to 23 after 48 h.
Figure 2.
Growth curve of Mycoplasma mycoides subsp. mycoides SC PG1 expressed in colony forming units (cfu/ml). Numbers next to arrows indicate factors by which colony counts differ from genome equivalents calculated after analysis of extracted DNA in the presented MSC_1046 rt-PCR assay.
Detection of MmmSC in clinical samples
Clinical samples were subjected to the triplex rt-PCR assay comprising the amplification of the two MmmSC-specific targets MSC_0136 and MSC_1046 and the IAC template. The IAC template was consistently amplified in all samples as illustrated by a Ct value of approximately 27 ruling out a PCR inhibition in the sample preparations.
Lung samples from cattle flocks in Germany with and without diagnosed pneumonia were used as negative control samples and examined in the multiplex rt-PCR and conventional PCR. All negative control samples yielded negative results as expected.
66 clinical specimens obtained from 19 animals of the CBPP infection trial were investigated by the multiplex rt-PCR, conventional PCR and cultivation (Table 3). 19 samples were positive in all tests, whereas 23 samples yielded negative results in all tests. 30 specimens (45.5%) were culture positive, but 41 samples (62.1%) were tested positive with the MSC_0136 assay and 37 samples (56.1%) with the MSC_1046 assay. 19 samples (28.8%) gave a positive result with the conventional SC3 PCR.
Table 3.
Detection of Mycoplasma mycoides subsp. mycoides SC in lung, lymph nodes and pleural fluid from experimentally infected Boran cattle by cultivation, MSC_0136 and MSC_1046 rt-PCR and conventional SC3-PCR assay
| Animal | Lung | Lymph nodes, pulmonal | Lymph nodes, mediastinal | Pleural fluid | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Culture | rt-PCR | PCR | Culture | rt-PCR | PCR | Culture | rt-PCR | PCR | Culture | rt-PCR | PCR | |||||
| MSC 0136 |
MSC 1046 |
SC3 | MSC 0136 |
MSC 1046 |
SC3 | MSC 0136 |
MSC 1046 |
SC3 | MSC 0136 |
MSC 1046 |
SC3 | |||||
| BD091 | + | + | + | + | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. |
| BD092 | + | + | + | - | - | + | + | - | + | - | - | - | - | - | - | - |
| BD093 | + | + | + | + | - | + | - | - | + | + | - | - | - | - | - | - |
| BD094 | + | + | + | + | - | + | + | - | - | - | - | - | - | - | - | - |
| BD095 | + | + | + | + | + | - | - | - | - | - | - | - | - | - | - | - |
| BD096 | - | + | + | + | - | - | - | - | - | - | - | - | - | - | - | - |
| BD097 | + | + | + | + | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | + | + | + | + |
| BD099 | + | + | + | - | - | - | - | - | - | - | - | - | - | - | - | - |
| BD100 | + | + | + | + | - | + | - | - | - | - | - | - | - | - | - | - |
| BD101 | + | + | + | + | + | + | + | - | + | + | - | - | - | + | + | - |
| BD102 | - | - | - | - | - | + | + | - | - | - | - | - | - | + | + | - |
| BD105 | + | + | + | + | - | + | + | - | n.d. | n.d. | n.d. | n.d. | - | - | - | - |
| BD106 | - | + | + | - | + | + | + | - | - | - | - | - | - | - | - | - |
| BD107 | + | + | + | + | + | + | + | - | - | + | + | - | - | - | - | - |
| BD111 | + | + | + | + | + | + | + | - | - | - | - | - | n.d. | n.d. | n.d. | n.d. |
| BD115 | + | + | + | + | - | + | + | - | - | + | + | - | n.d. | n.d. | n.d. | n.d. |
| BD116 | + | + | + | + | + | + | + | + | + | + | + | - | - | - | - | - |
| BD118 | + | + | + | + | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | + | + | + | + |
| BD119 | + | + | + | + | + | + | + | + | + | + | + | - | - | - | - | - |
| positive/total | 16/19 | 18/19 | 18/19 | 15/19 | 7/16 | 13/16 | 11/16 | 2/16 | 5/15 | 6/15 | 4/15 | 0/15 | 2/16 | 4/16 | 4/16 | 2/16 |
n.d.: not done, +: positive test, -: negative test, rt: real time.
All infected animals were correctly identified by the rt-PCR assays if results from different sample types were considered. Thus, calculation of test sensitivities based on diagnosis for individual animals resulted in sensitivities of 100% for the MSC_0136 and MSC_1046 rt-PCR, 78.9% for the SC3 PCR and 89.5% for culture (Table 4).
Table 4.
Detection rates of Mycoplasma mycoides subsp. mycoides SC in CBPP-infected animals after parallel investigation of lung, lymph nodes and pleural fluid by cultivation, MSC_0136 and MSC_1046 rt-PCR and conventional SC3-PCR assay
| Animal: n = 19, Condition: positive = 19 | ||||
|---|---|---|---|---|
| rt-PCR | PCR | |||
| Culture | MSC_0136 | MSC_1046 | SC3 | |
| Positives | 17 | 19 | 19 | 15 |
| Detection rate | 89.5% | 100% | 100% | 78.9% |
n = total number, rt: real time.
Table 5 presents test specificities and sensitivities based on data derived from the examination of lung specimens. No false positive results were obtained in any test resulting in specificities of 100% for the PCR assays. The two rt-PCR assays failed to identify one out of 19 lung samples as MmmSC-positive (sensitivity of 94,7%), but are more sensitive than detection by culture (84. 2%) and conventional SC3 PCR (78.9%).
Table 5.
Comparison of specificity and sensitivity of different tests for the detection of Mycoplasma mycoides subsp. mycoides SC in lung tissue
| Lung: n = 39, Condition: positive = 19, negative = 20 | ||||
|---|---|---|---|---|
| rt-PCR | PCR | |||
| Culture | MSC_0136 | MSC_1046 | SC3 | |
| True positives | 16 | 18 | 18 | 15 |
| False positives | 0 | 0 | 0 | 0 |
| False negatives | 3 | 1 | 1 | 4 |
| True negatives | - | 20 | 20 | 20 |
| Sensitivity | 84.2% | 94.7% | 94.7% | 78.9% |
| Specificity | - | 100% | 100% | 100% |
n = total number, rt: real time.
A strong difference was observed in rt-PCR test results obtained from lung samples, lymph nodes and pleural fluid samples (Table 3). While 94.7% of lung samples were positively tested in the MSC_0136 assay, 81% of pulmonal lymph nodes, 31% of mediastinal lymph nodes and 25% of pleural fluid samples gave a positive result.
Discussion
Sensitive, reliable and time-saving molecular diagnostic methods are indispensible tools for a successful control of contagious bovine pleuropneumonia. Here, we present the development and preliminary evaluation of a new TaqMan®-based real-time PCR assay for the detection of the CBPP agent Mycoplasma mycoides subsp. mycoides SC in clinical samples. The assay comprises the amplification of two independent targets, MSC_1046 and MSC_0136, as well as an internal exogenous amplification control in a multiplex reaction. Nowadays, PCR is an accepted alternative to time- and labour-intensive and often insensitive culture-based methods for the direct detection of MmmSC in infected animals to confirm a presumptive clinical or pathomorphological diagnosis [17]. In general, PCR is known as a sensitive, specific, straightforward and fast tool for pathogen detection and several conventional PCR tests have been proposed to detect MmmSC [2,9,10,27-29]. Despite the superiority of real time-PCR assays over conventional PCR and the importance and impact of the disease, only few rt-PCR protocols have been introduced so far and none of them was validated on more than five clinical samples. Gorton et al. [13] published three independent assays with MGB-TaqMan® probes with rather low amplification efficiencies and detection limits of 100 fg to 1 pg. Higher sensitivities were reached with a SYBR Green® rt-PCR assay [12], but the lack of specific amplicon detection by a probe might negatively influence the specificity of the assay. Recently, Vilei and Frey [14] reported on the development of two assays with TaqMan® probes intended for the detection of MmmSC in bronchoalveolar lavage fluid of cattle and for the discrimination of wild type strains from a mutant vaccine strain. The assays are based on the gene of the well-described antigenic surface lipoprotein LppQ [21,30] that is also used in the study presented here. However, in our study, we preferred LNA probes to Minor Grove Binding (MGB) probes as they were shown to be significantly more sensitive, have a low optimum concentration and low background fluorescence [31] and are also cheaper. Furthermore, we used a probe binding site downstream of the sites proposed by Vilei and Frey [14] which guaranteed a more specific detection due to a higher number of nucleotide mismatches to the homologous sequences from M. mycoides subsp. capri and M. leachii. The differentiation between MmmSC and its close relatives of the M. mycoides cluster is important and best performed by the use of variable genetic targets with high discriminatory potential such as MSC_1046 and MSC_0136. In fact, our duplex assay was able to detect DNA of all of the 48 tested MmmSC strains and did not cross-react with any non-target strain indicating excellent specificity. The analytical sensitivity and detection limits of the two single assays were in the range reported for rt-PCR tests. The lower LOD of the MSC_0136 assay compared to the MSC_1046 assay was also reflected in its higher diagnostic sensitivity. Although both assays are predominantly designed for qualitative pathogen detection, especially the MSC_1046 assay with a dynamic range of 7 log units may also be a useful tool for quantitative evaluation of pathogen load. Standard curve preparation and pathogen quantification by rt-PCR are often related to cfu counts assuming a strict correlation between numbers of viable cfu and extracted DNA [12,14]. We have tested this assumption and found that the total amount of detected DNA exceeded the counted cfu already in the lag and early exponential phase by eight times and that the ratio varies depending on the age of the culture (Figure 2). Thus, especially for slow growing fastidious organisms such as mycoplasmas, the use of cfu instead of DNA amount may lead to an overestimation of sensitivity when assessing analytical test performance since the contribution of non-viable genome equivalents to total DNA amount is not considered.
In accordance with recommendations after which the detection of MmmSC by PCR should be performed by the amplification of two independent targets [1,7], our new rt-PCR test was conceived as a multiplex assay including two targets and an internal control without loss of sensitivity. The multiplex format is advantageous over simplex assays as it is particularly economical in terms of cost of material (mastermix, consumables) and turnaround time and suitable for small volume samples and high-throughput.
The clinical usability of a PCR test is not solely dependent on its inherent specificity and sensitivity, but also influenced by the course and stage of infection, sampling strategy and sample preparation. To evaluate the ability of the new test to diagnose MmmSC infection in cattle, different specimens from CBPP infected animals and from cattle with non-MmmSC related pneumonia as well as from healthy animals were examined. It was evident that the extraction procedure efficiently removed DNA polymerase inhibitors from the tissue matrices since amplification of the IAC template was not impaired. However, DNA degradation was observed in some samples after long-time storage at -20°C possibly due to nucleases remaining in the samples after DNA preparation. Therefore, an immediate processing of samples after DNA extraction is advisable.
The rt-PCR assays' diagnostic specificity proved to be 100%. Diagnostic sensitivities were compared to those of MmmSC detection by culture and conventional PCR. The MSC_0136 and MSC_1046 assays were found to be most sensitive. Concerning the diagnosis for the individual animal, the sensitivity was 100% for both rt-PCR assays, compared to 89.5% for culture and 78.9% for SC3 PCR (Table 4). Thus, both rt-PCR assays reliably identified infected animals provided that different clinical specimens were investigated. If lung samples were exclusively considered, diagnostic sensitivity was 94.7% for the two rt-PCR assays, 84.2% for culture and 78.9% for the conventional PCR test (Table 5). In contrast, lymph nodes were obviously less appropriate to detect MmmSC in infected animals. This observation is congruent to findings of a study using tissue samples from a natural CBPP outbreak in Portugal [32]. As expected, pathogen detection was more often successful in pulmonal lymph nodes that directly drain the lungs than in the more distant mediastinal lumph nodes. Pleural fluid was previously recommended as the sample of choice for the diagnosis of CBPP [33]. However, it is not always present in clinical cases of CBPP. We could detect high titres of MmmSC (Ct values for MSC_0136 below 23 correlating to > 2.3E+08 GE/ml) in the voluminous pleural fluid samples from the two acutely infected animals no. BD097 and BD118, whereas the other animals accumulated only tiny amounts of pleural fluid of which only two tested positive in the rt-PCR assay with Ct values close to the detection limit (> 36.0). In conclusion, the apparent weak diagnostic performance in the investigation of single specimens has to be attributed to the used sample types being not all equally suitable.
Animal no. BD102 did not show any pathological modifications in the lung, a fact that did not allow targeted sampling. This may explain the negative rt-PCR result obtained for the animal's lung sample. However, MmmSC was detected in lymph node tissue and pleural fluid of the animal, which underlines the importance of a diversified sampling procedure, especially in animals with negative pathology, in order to prevent false negative results [34,35]. The frequently reported cases of detection of MmmSC by PCR in samples of asymptomatic animals without organic lesions and without successful cultivation ([36,34,32], present study) are not reflected by the OIE case definition of CBPP. However, those animals are nevertheless important as carriers and shedders.
Although the number of samples was limited and far from satisfying the Principles of Validation of the OIE Terrestrial Manual [1], the pre-validation of the new rt-PCR assay presented here is a first important step in assessing its diagnostic capabilities. In the future, well-characterized field samples from endemic areas are needed for further validation of the assay according to OIE guidelines.
Conclusions
1) The developed multiplex rt-PCR is more sensitive than cultivation and conventional PCR. All infected animals could be identified, but only a combined analysis of different sample types ensured 100% sensitivity of the test.
2) Lung samples collected from lesions at the interface between diseased and healthy tissue are most appropriate to detect MmmSC infections and should predominantly be considered in the analysis by rt-PCR. However, if resources allow, we propose a more comprehensive sampling strategy including also lymph node tissue and pleural fluid to clarify the infection status of the animal.
3) In conclusion, we recommend the duplex rt-PCR assay as an efficient tool for rapid confirmation of a presumptive CBPP diagnosis based on clinical suspicion or pathological observations especially in a laboratory setting of developed countries and for epidemic surveillance in CBPP-free countries. In the future, the test will be used in routine diagnosis of CBPP in the German Reference Laboratory.
Authors' contributions
CS planned and supervised microbiological and preparative laboratory work, designed and conducted the real-time PCR experiments and wrote the manuscript. MH coordinated the project, organized funding and helped in sample acquisition at the animal trial. JJ designed and carried out the animal infection experiment. HT assisted in data analysis and reviewed the manuscript. HN contributed to acquisition of funding, organisation of the project and reviewing the manuscript. All authors have read and approved the final manuscript.
Contributor Information
Christiane Schnee, Email: christiane.schnee@fli.bund.de.
Martin Heller, Email: martin.heller@fli.bund.de.
Jörg Jores, Email: j.jores@cgiar.org.
Herbert Tomaso, Email: herbert.tomaso@fli.bund.de.
Heinrich Neubauer, Email: heinrich.neubauer@fli.bund.de.
Acknowledgements
We wish to thank Francois Thiaucourt (CIRAD, Montpellier, France), Colin Churchward (VLA, Weybridge, UK) and Lothar Hoffmann (TLLV Bad Langensalza, Germany) for providing mycoplasma strains and clinical samples, as well as Susann Bahrmann for technical support. The German Federal Ministry for Education and Research funded this project in the framework of the program "Research for Civil Security", part of the High-Tech Strategy of the Federal Government of Germany. JJ was supported by the German Federal Ministry of Cooperation and Development.
References
- OIE. Manual of Diagnostic Tests and Vaccines for Terrestrial Animals. 2010. Chapters 1.1.4/5 and 2.4.9, Paris.
- Bashiruddin JB, Taylor TK, Gould AR. A PCR-based test for the specific identification of Mycoplasma mycoides subspecies mycoides SC. J Vet Diagn Invest. 1994;6:428–34. doi: 10.1177/104063879400600405. [DOI] [PubMed] [Google Scholar]
- Nicholas RA, Santini FG, Clark KM, Palmer NM, De Santis P, Bashiruddin JB. A comparison of serological tests and gross lung pathology for detecting contagious bovine pleuropneumonia in two groups of Italian cattle. Vet Rec. 1996;139:89–93. doi: 10.1136/vr.139.4.89. [DOI] [PubMed] [Google Scholar]
- Amanfu W, Sediadie S, Masupu KV, Raborokgwe MV, Benkirane A, Geiger R, Thiaucourt F. Comparison between c-ELISA and CFT in detecting antibodies to Mycoplasma mycoides mycoides biotype SC in cattle affected by CBPP in Botswana. Ann N Y Acad Sci. 2000;916:364–369. doi: 10.1111/j.1749-6632.2000.tb05314.x. [DOI] [PubMed] [Google Scholar]
- Marobela-Raborokgwe C, Nicholas RA, Ayling RD, Bashiruddin JB. Comparison of complement fixation test, immunoblotting, indirect ELISA, and competitive ELISA for detecting antibodies to Mycoplasma mycoides subspecies mycoides small colony (SC) in naturally infected cattle from the 1995 outbreak in Botswana. Onderstepoort J Vet Res. 2003;70:21–7. [PubMed] [Google Scholar]
- Rice P, Houshaymi BM, Nicholas RA, Miles RJ. A rapid biochemical test to aid identification of Mycoplasma mycoides subsp. mycoides small colony (SC) strains. Lett Appl Microbiol. 2000;30:70–4. doi: 10.1046/j.1472-765x.2000.00674.x. [DOI] [PubMed] [Google Scholar]
- Le Grand D, Saras E, Blond D, Solsona M, Poumarat F. Assessment of PCR for routine identification of species of the Mycoplasma mycoides cluster in ruminants. Vet Res. 2004;35:635–49. doi: 10.1051/vetres:2004037. [DOI] [PubMed] [Google Scholar]
- Rosendal S, Black FT. Direct and indirect immunofluorescence of unfixed and fixed mycoplasma colonies. Acta Path Microbiol Scand. 1972;80:615–22. doi: 10.1111/j.1699-0463.1972.tb00186.x. [DOI] [PubMed] [Google Scholar]
- Dedieu L, Mady V, Lefevre PC. Development of a selective polymerase chain-reaction Assay for the detection of Mycoplasma mycoides subsp mycoides SC (contagious bovine pleuropneumonia agent) Vet Microbiol. 1994;42:327–339. doi: 10.1016/0378-1135(94)90064-7. [DOI] [PubMed] [Google Scholar]
- Miserez R, Pilloud T, Cheng X, Nicolet J, Griot C, Frey J. Development of a sensitive nested PCR method for the specific detection of Mycoplasma mycoides subsp. mycoides SC. Mol Cell Probes. 1997;11:103–11. doi: 10.1006/mcpr.1996.0088. [DOI] [PubMed] [Google Scholar]
- Fitzmaurice J, Sewell M, Manso-Silván L, Thiaucourt F, McDonald WL, O'Keefe JS. Real-time polymerase chain reaction assays for the detection of members of the Mycoplasma mycoides cluster. N Z Vet J. 2008;56:40–7. doi: 10.1080/00480169.2008.36803. [DOI] [PubMed] [Google Scholar]
- Lorenzon S, Manso-Silvan L, Thiaucourt F. Specific real-time PCR assays for the detection and quantification of Mycoplasma mycoides subsp. mycoides SC and Mycoplasma capricolum subsp. capripneumoniae. Mol Cell Probes. 2008;22:324–8. doi: 10.1016/j.mcp.2008.07.003. [DOI] [PubMed] [Google Scholar]
- Gorton TS, Barnett MM, Gull T, French RA, Lu Z, Kutish GF, Adams LG, Geary SJ. Development of real-time diagnostic assays specific for Mycoplasma mycoides subspecies mycoides Small Colony. Vet Microbiol. 2005;111:51–8. doi: 10.1016/j.vetmic.2005.09.013. [DOI] [PubMed] [Google Scholar]
- Vilei EM, Frey J. Detection of Mycoplasma mycoides subsp. mycoides SC in bronchoalveolar lavage fluids of cows based on a TaqMan real-time PCR discriminating wild type strains from an lppQ-mutant vaccine strain used for DIVA-strategies. J Microbiol Methods. 2010;81:211–8. doi: 10.1016/j.mimet.2010.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freundt EA. In: Methods in mycoplasmology. Razin S, Tully JG, editor. I. New York Academic Press; 1983. Culture media for classic mycoplasmas; pp. 128–35. Mycoplasma characterization. [Google Scholar]
- Sacchini F, Naessens J, Awino E, Heller M, Hlinak A, Haider W, Sterner-Kock A, Jores J. A minor role of CD4+ T lymphocytes in the control of a primary infection of cattle with Mycoplasma mycoides subsp. mycoides. Vet Res. 2011;42:77. doi: 10.1186/1297-9716-42-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OIE. Terrestrial Animal Health Code. 2010. Chapter 11.8.1, Paris.
- Manso-Silvan L, Perrier X, Thiaucourt F. Phylogeny of the Mycoplasma mycoides cluster based on analysis of five conserved protein-coding sequences and possible implications for the taxonomy of the group. Int J Syst Evol Microbiol. 2007;57:2247–58. doi: 10.1099/ijs.0.64918-0. [DOI] [PubMed] [Google Scholar]
- Vilei EM, Korczak BM, Frey J. Mycoplasma mycoides subsp capri and Mycoplasma mycoides subsp mycoides LC can be grouped into a single subspecies. Vet Res. 2006;37:779–90. doi: 10.1051/vetres:2006037. [DOI] [PubMed] [Google Scholar]
- Corpet F. Multiple sequence alignment with hierarchical clustering. Nucl Acids Res. 1988;16:10881–90. doi: 10.1093/nar/16.22.10881. http://multalin.toulouse.inra.fr/multalin/multalin.html [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdo EM, Nicolet J, Frey J. Antigenic and genetic characterization of lipoprotein LppQ from Mycoplasma mycoides subsp mycoides SC. Clin Diagn Lab Immunol. 2000. pp. 588–595. [DOI] [PMC free article] [PubMed]
- Bischof DF, Vilei EM, Frey J. Genomic differences between type strain PG1 and field strains of Mycoplasma mycoides subsp. mycoides small-colony type. Genomics. 2006. pp. 633–41. [DOI] [PMC free article] [PubMed]
- Thiaucourt F, Lorenzon S, David A, Breard A. Phylogeny of the Mycoplasma mycoides cluster as shown by sequencing of a putative membrane protein gene. Veterinary Microbiology. 2000;72:251–68. doi: 10.1016/S0378-1135(99)00204-7. [DOI] [PubMed] [Google Scholar]
- Westberg J, Persson A, Holmberg A, Goesmann A, Lundeberg J, Johansson KE, Pettersson B, Uhlen M. The genome sequence of Mycoplasma mycoides subsp. mycoides SC type strain PG1T, the causative agent of contagious bovine pleuropneumonia (CBPP) Genome Res. 2004;14:221–7. doi: 10.1101/gr.1673304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann B, Depner K, Schirrmeier H, Beer M. A universal heterologous internal control system for duplex real-time RT-PCR assays used in a detection system for pestiviruses. J Virol Methods. 2006;136:200–9. doi: 10.1016/j.jviromet.2006.05.020. [DOI] [PubMed] [Google Scholar]
- March JB, Clark J, Brodlie M. Characterization of strains of Mycoplasma mycoides subsp. mycoides small colony type isolated from recent outbreaks of contagious bovine pleuropneumonia in Botswana and Tanzania: evidence for a new biotype. J Clin Microbiol. 2000. pp. 1419–25. [DOI] [PMC free article] [PubMed]
- Persson A, Pettersson B, Bolske G, Johansson KE. Diagnosis of contagious bovine pleuropneumonia by PCR-laser- induced fluorescence and PCR-restriction endonuclease analysis based on the 16S rRNA genes of Mycoplasma mycoides subsp. mycoides SC. J Clin Microbiol. 1999;37:3815–21. doi: 10.1128/jcm.37.12.3815-3821.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilei EM, Abdo EM, Nicolet J, Botelho A, Goncalves R, Frey J. Genomic and antigenic differences between the European and African/Australian clusters of Mycoplasma mycoides subsp. mycoides SC. Microbiology. 2000;146:477–86. doi: 10.1099/00221287-146-2-477. [DOI] [PubMed] [Google Scholar]
- Miles K, Churchward CP, McAuliffe L, Ayling RD, Nicholas RA. Identification and differentiation of European and African/Australian strains of Mycoplasma mycoides subspecies mycoides small-colony type using polymerase chain reaction analysis. J Vet Diagn Invest. 2006;18:168–71. doi: 10.1177/104063870601800205. [DOI] [PubMed] [Google Scholar]
- Bruderer U, Regalla J, Abdo el-M, Huebschle OJ, Frey J. Serodiagnosis and monitoring of contagious bovine pleuropneumonia (CBPP) with an indirect ELISA based on the specific lipoprotein LppQ of Mycoplasma mycoides subsp. mycoides SC. Vet Microbiol. 2002;84:195–205. doi: 10.1016/S0378-1135(01)00466-7. [DOI] [PubMed] [Google Scholar]
- Josefsen MH, Löfström C, Sommer MH, Hoorfar J. Diagnostic PCR: Comparative sensitivity of four probe chemistries. Mol Cell Probes. 2009;23:201–3. doi: 10.1016/j.mcp.2009.02.003. [DOI] [PubMed] [Google Scholar]
- de Santis P, Persson A, Ball H, Regalla J. Detection of Mycoplasma mycoides subspecies mycoides SC in bovine lung and lymph node tissues by culture, sandwich ELISA and polymerase chain reaction systems. Res Vet Sci. 2005;78:199–205. doi: 10.1016/j.rvsc.2004.09.004. [DOI] [PubMed] [Google Scholar]
- Nicholas RA, Ayling RD, McAuliffe L. Mycoplasma diseases of ruminants. Wallingford, CAB International; 2008. Contagious Bovine Pleuropneumonia; pp. 69–97. [Google Scholar]
- Bashiruddin JB, De Santis P, Vacciana A, Santini FG. Detection of Mycoplasma mycoides subspecies mycoides SC in clinical material by a rapid colorimetric PCR. Mol Cell Probes. 1999;13:23–8. doi: 10.1006/mcpr.1998.0206. [DOI] [PubMed] [Google Scholar]
- Perreau P. In: The Diagnosis of Contagious Bovine Pleuropneumonia and Other Infections with Mycoplasma mycoides subsp. mycoides. Hall SA, editor. Luxembourg, C.E.C. Publications EUR 8654; 1983. Isolation procedures for the diagnosis of contagious bovine pleuropneumonia; pp. 18–26. [Google Scholar]
- Nicholas RA, Palmer N. Contagious bovine pleuropneumonia in Europe. State Vet J. 1994;4:14–6. [Google Scholar]