Abstract
Telomeres are specialized DNA/protein structures that act as protective caps to prevent end fusion events and to distinguish the chromosome ends from double-strand breaks. We report that TRF1 and Ku form a complex at the telomere. The Ku and TRF1 complex is a specific high-affinity interaction, as demonstrated by several in vitro methods, and exists in human cells as determined by coimmunoprecipitation experiments. Ku does not bind telomeric DNA directly but localizes to telomeric repeats via its interaction with TRF1. Primary mouse embryonic fibroblasts that are deficient for Ku80 accumulated a large percentage of telomere fusions, establishing that Ku plays a critical role in telomere capping in mammalian cells. We propose that Ku localizes to internal regions of the telomere via a high-affinity interaction with TRF1. Therefore, Ku acts in a unique way at the telomere to prevent end joining.
Keywords: Telomere, TRF1, Ku
Telomeres, composed of repetitive DNA sequences bound by telomere protein complexes, function to protect the chromosome termini from fusion events and promote chromosomal end replication (Blackburn 1999). Telomere maintenance is a critical determinant of cellular senescence (Bodnar et al. 1998), and telomerase activity is altered in most cancers (Artandi and DePinho 2000). Telomere maintenance requires a homeostatic balance between addition of telomeric sequences by telomerase, repression of telomerase, and persistent capping activity by telomeric proteins and structures (Blackburn 1997; Shore 1998; Griffith et al. 1999). To understand telomere maintenance, the components of the telomere and how they interact need to be defined.
Several proteins that were originally identified by their important roles in DNA repair have recently been shown to play additional roles in telomere maintenance (Boulton and Jackson 1998; d'Adda di Fagagna et al. 1999; Hande et al. 1999). One example is the Ku heterodimer (composed of ∼70- and ∼80-kD subunits; denoted here as Ku70 and Ku80), shown to be crucial for nonhomologous DNA double-strand break repair (Taccioli et al. 1994; Critchlow and Jackson 1998; Kanaar et al. 1998), and that, in addition, binds site specifically to particular DNA sequences (Giffin et al. 1996; Ludwig et al. 1997; Galande and Kohwi-Shigematsu 1999), functions in site-specific recombination of V(D)J gene segments (Nussenzweig et al. 1996), and plays an important role at the telomere (Boulton and Jackson 1996; Porter et al. 1996; Bailey et al. 1999; Hsu et al. 1999; Gasser 2000). During the repair of double-strand breaks, Ku binds nonspecifically to DNA ends with high affinity (Mimori et al. 1986; Paillard and Strauss 1991; Cary et al. 1997; Dynan and Yoo 1998). However, telomeric ends are capped or bound by specific telomere proteins that serve to conceal and disguise the telomeric DNA end, thereby preventing end fusion events and preventing cellular DNA damage signaling. We, along with our collaborators, recently found that Ku is in close proximity to the mammalian telomere and that loss of Ku, in virally transformed Ku-deficient mouse cell lines, resulted in telomere fusion events (Bailey et al. 1999; Hsu et al. 1999). However, the exact role that Ku plays at the telomere and how it localizes to the telomere is not yet known.
The mammalian telomere binding protein TRF1 localizes at telomeres by binding specifically to telomeric DNA and plays a role in telomere length regulation (Zhong et al. 1992; van Steensel and de Lange 1997). Here we report that Ku forms a high-affinity protein/protein interaction with TRF1 to localize to internal regions of telomeric DNA. In addition, we present evidence that Ku provides an essential telomere capping function in primary mammalian cells to prevent telomere fusions. Therefore, Ku functions in a different way at the telomere than during joining nonhomologous DNA double-stranded breaks.
Results and Discussion
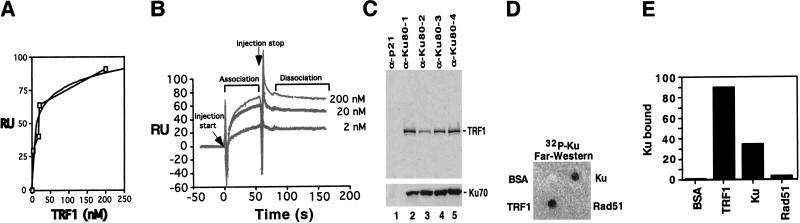
We quantitated the affinity of the Ku and TRF1 interaction using surface plasmon resonance on a Biacore 2000 (Malmqvist and Karlsson 1997). Recombinant Ku or a control protein, α-Ku80 Fab fragment, were covalently attached to the surface of a CM5 chip, and binding was monitored after injection of TRF1 or BSA. TRF1 did not bind significantly (<5 resonance units) to the control, and no binding was observed for any concentration of BSA to either the Ku or the control protein surface. As the concentration of TRF1 was increased from 2 nM to 200 nM, increasing amounts of TRF1 bound to the Ku-coupled surface, generating a typical hyperbolic saturation curve (Fig. 1A). The fit produced an apparent ka of 2.1 × 106 (1/Ms) and kd of 1.01 × 10−3, giving an equilibrium dissociation constant, KD, of 0.4 nM. Since the extremely slow dissociation rate of the TRF1/Ku complex prohibited confirmation of the apparent KD by Biacore steady state analysis, we can only state that the KD of this interaction appears to be within the low nanomolar range (Fig. 1B). The specificity and calculated rate constants for the interaction were similar when TRF1 was immobilized on the chip and various concentrations of Ku were passed over the surface (data not shown). Therefore, TRF1 binds with high affinity to Ku and the Ku/TRF1 complex is very stable, as evidenced by the extremely slow dissociation rate.
Figure 1.
Ku binds TRF1 with high affinity in vitro. (A) Graph of relative response levels obtained for specific TRF1 concentrations after injection was stopped for 10 sec (during dissociation phase) with a best fit curve (Cricket-Graph). RU, Resonance units. (B) Three representative sensorgrams obtained by flowing the indicated concentrations of TRF1 over Ku70/Ku80 covalently attached to the surface of a Biacore CM-5 chip. Curves were aligned on the X- and Y-axes and subtracted against a negative control using BiaEvaluation software. Injection start indicates the point at which TRF1 begins to flow over the Ku-coupled surface, while injection stop indicates the point at which buffer alone begins to flow over the surface. The TRF1 that remained associated with Ku after a 2–3-min dissociation period was removed in subsequent regeneration steps. Injections of each TRF1 concentration were repeated a minimum of three times using two different Ku-coupled CM-5 chips. (C) In vitro translated [35S]methionine labeled His-tagged TRF1 was preincubated with baculovirally expressed purified Ku70/Ku80 heterodimer. The mixtures were immunoprecipitated by p21 antibody (α-p21, lane 1) and by various Ku80 monoclonal antibodies (α-Ku80, lanes 2–5). Immunoprecipitates were resolved on SDS-PAGE followed by autoradiography. The translated His-TRF1 migrated as a 64-kD protein. (C, lower panel) Western analysis of coimmunoprecipitated Ku70 protein by the Ku80 antibodies. (D) Far-Western analysis TRF1 interaction with Ku. BSA (1 μg), TRF1 (200 ng), Ku70/Ku80 (200 ng), and Rad51 (200 ng) proteins were placed on a nitrocellulose membrane, and this membrane was incubated with a 32P-labeled Ku70/Ku80 probe. (E) PhosphorImager quantitation of 32P-Ku bound on the membrane. The relative level of Ku bound to these proteins was normalized to the amount of proteins used with BSA set at 1 U.
We further characterized the Ku/TRF1 complex using an in vitro immunoprecitation-binding assay. Radiolabeled TRF1 produced by in vitro translation was incubated with purified recombinant Ku, followed by immunoprecipitation using several Ku80 monoclonal antibodies (mAbs). All Ku80 mAbs efficiently coimmunoprecipitated a Ku/TRF1 complex (Fig. 1C, lanes 2–5), with up to 19% of the input TRF1 present in the immune complex (Fig. 1C, lanes 2,5). The p21 mAb control did not immunoprecipitate detectable levels of TRF1 (Fig. 1C, lane 1). Therefore, under these in vitro conditions with Ku and TRF1 free in solution, we observe a stable Ku/TRF1 complex.
In a far-Western assay, TRF1, Ku, Rad51, and BSA were immobilized on a nitrocellulose membrane and incubated with radiolabeled Ku. Immobilized TRF1 protein interacted strongly with the labeled Ku probe (Fig. 1D). In addition, labeled Ku hybridized with the immobilized Ku, presumably because of dissociation and reassociation of the Ku heterodimer components. Control proteins BSA and Rad51 displayed no significant interaction with Ku. Therefore, TRF1 bound the greatest amount of radiolabeled Ku; Ku bound 30%–40% as much radiolabeled Ku, whereas BSA and Rad51 bound barely detectable levels (Fig. 1E).
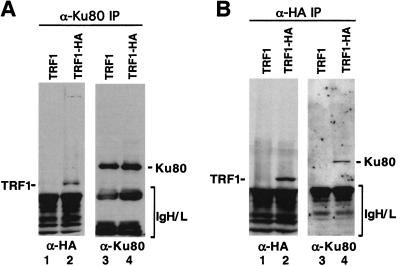
We tested for the presence of a Ku/TRF1 complex in human HT1080 fibrosarcoma cells expressing a haemagglutinin-tagged TRF1 (HA-TRF1; Kim et al. 1999). Protein extracts were analyzed for a Ku/TRF1 complex by coimmunoprecipitation and Western analysis using α-Ku80 or α-HA mAbs (Fig. 2). When extracts were immunoprecipitated with Ku80 mAb, HA-TRF1 coprecipitated (Fig. 2A, lane 2). The Ku80 antibody precipitated equal amounts of Ku protein from lysates of HA-TRF1-expressing HT1080 cells or control HT1080 cells (Fig. 2A, lanes 3,4). In reciprocal experiments, we observed that Ku80 was present in α-HA immunoprecipitates from cells expressing HA-TRF1 (Fig. 2B, lane 4), whereas no Ku80 was immunoprecipitated from the control cell line (Fig. 2B, lane 3). Taken together, these results indicate that Ku interacts with TRF1 with high affinity and specificity under several in vitro conditions and in human cells.
Figure 2.
Ku and TRF1 form a complex in vivo. Cell lysates from HT1080 infected with HA-tagged TRF1 (HA-TRF1) retrovirus or control retrovirus alone (TRF1) were immunoprecipitated with α-Ku80 or α-HA monoclonal antibodies. The immunoprecipitates were resolved by SDS-PAGE and probed with α-Ku80 (A) or α-HA (B).
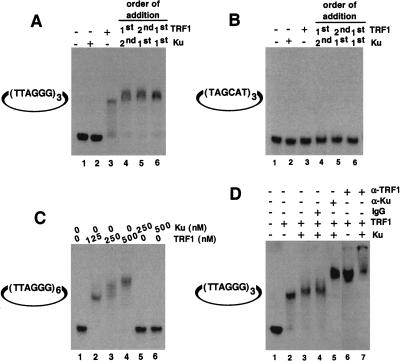
Ku displays two DNA binding activities: nonspecific end binding (Mimori et al. 1986; Dynan and Yoo 1998) and site-specific internal binding (Giffin et al. 1996; Ludwig et al. 1997; Galande and Kohwi-Shigematsu 1999). We tested whether Ku binds site specifically to telomeric DNA sequences. Using a gel shift assay, we tested whether microcircular DNAs (∼200 bps) with internal telomeric repeats showed site-specific Ku binding. Microcircular DNA substrates were used to eliminate background from nonspecific Ku end-binding activity. We found that Ku alone did not bind microcircles containing either three (Fig. 3A, lane 2) or six internal telomeric repeats (Fig. 3C, lanes 5,6). Addition of TRF1 to microcircles with three internal telomeric repeats (a single TRF1-binding site; Zhong et al. 1992) produced a single mobility-shift band (Fig. 3A, lane 3; Fig. 3D, lane 2). Microcircles containing six internal telomeric repeats also showed a band at lower concentrations of TRF1 consistent with occupation of one TRF1 binding site (Fig. 3C, lane 2). In addition, with increasing concentrations of TRF1, a higher shifted band appeared consistent with occupation of two TRF1 binding sites (Fig. 3C, lanes 3,4). When both Ku and TRF1 were added to the telomeric microcircular DNA, we observed a slight band shift compared with TRF1 alone (Fig. 3A, lane 3, cf. lanes 4–6; Fig. 3D, cf. lane 2 with lanes 3,4). This increase in mobility shift was seen regardless of the order of addition of the individual components (Fig. 3A, lanes 4,5) or the addition of a preformed Ku/TRF1 complex (Fig. 3A, lane 6). We observed no band shift of microcircles containing three nontelomeric repeats, (TAGCAT)3, under these same conditions (Fig. 3B). Confirming that both Ku and TRF1 were bound simultaneously to the telomeric microcircles, we observed a super-shift on addition of TRF1 antibody (Fig. 3D, lane 7) and an additional super-shift on addition of Ku antibody (Fig. 3D, lane 5). No super-shifted bands appeared on addition of normal mouse IgG (Fig. 3D, lane 4). Our results indicate that Ku does not bind specifically to telomeric DNA but requires and employs TRF1 to localize to telomeric DNA.
Figure 3.
Telomeric DNA binding analysis by EMSA. (A) Ku does not bind directly to internal telomeric repeats. Binding reactions were performed as described above using a mirocircle containing (TTAGGG)3 repeats. Lane 1, no protein added; lane 2, 250 nM Ku70/Ku80 only; lane 3, 125 nM TRF1 alone; lane 4, 250 nM Ku was incubated with substrate DNA first, followed by 125 nM TRF1; lane 5, 125 nM TRF1 was incubated with substrate DNA first, followed by 250 nM Ku; lane 6, a preformed complex containing 250 nM Ku and 125 nM TRF1 was incubated with substrate DNA. (B) Microcircles containing random sequence repeats were not bound by both TRF1 and Ku. Reaction mixtures are same as A except that the substrate used was a microcircle containing a random sequence (TAGCAT)3 repeat. (C) Titration of binding sites. Binding studies were performed using a microcircle containing (TTAGGG)6 repeats. Lane 1, no protein; lane 2, 125 nM TRF1; lane 3, 250 nM TRF1; lane 4, 500 nM TRF1; lane 5, 250 nM Ku; lane 6, 500 nM Ku. (D) Antibody mediated super-shift. The telomeric microcircle DNA containing (TTAGGG)3 repeats was incubated with either TRF1 alone (lane 2) or as a preformed complex of TRF1 with Ku70/Ku80 (lanes 3–5,7) as described above, except that the concentration of the poly (dI/dC) was reduced to 0.1 mg/mL. Appropriate antibody was then added to individual reaction mixtures, and the resulting DNA–protein complexes were resolved as described in methods. As a control, mouse IgG2a was added to one of the reactions (lane 4). Rabbit α-TRF1 antibody (lanes 6,7) or mouse α-Ku70/Ku80 (lane 5) antibodies were used.
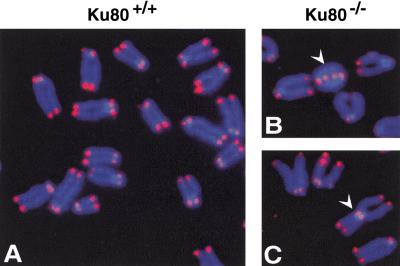
To determine whether Ku functions to cap telomeres in mammals, we analyzed primary mouse embryonic fibroblasts (MEFs) deficient for Ku80 (Fig. 4; Nussenzweig et al. 1996). Ku80-deficient MEF metaphase chromosome spreads contained high levels of telomere fusions (31 telomere fusion in 50 cells) compared with wild-type MEFs (one telomere fusion in 50 cells; Fig. 4D). The large accumulation of telomere fusions in Ku80-deficient MEFs indicates that Ku plays an important telomere capping function to prevent end fusions. Previously, it was shown that both Ku and TRF1 localize at telomeres (van Steensel and de Lange 1997; Hsu et al. 1999), with essentially all the cellular TRF1 localized to telomeres (van Steensel and de Lange 1997), and that a Ku/TRF1 complex exists in human cells (Fig. 2). Taken together, these results strongly suggest that the Ku/TRF1 complex must be localized to telomeres, which we find critical for telomere maintenance.
Figure 4.
FISH analysis of metaphase chromosomal spreads from wild-type and Ku80−/− mouse embryonic fibroblasts. (A) A metaphase chromosomes from Ku80+/+ mouse embryonic fibroblast (MEF). Chromosomal DNA was stained with DAPI (blue), and telomeres were hybridized with Cy3-labeled telomere probe (red). (B–C) Chromosome aberrations detected in metaphase spreads from Ku80−/− MEF. (B) White arrow points to a dicentric chromosome ring; (C) White arrow points to a Robertsonian fusion-like configuration with telomeres at the fusion point. (D) Compilation of the chromosomal analysis of wild-type and Ku80−/− MEF. Percentages indicate the number of specific chromosomal aberrations per metaphase chromosome set.
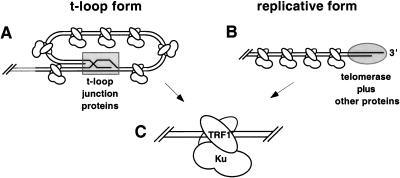
It is likely that the structure of the telomere changes conformations depending on the developmental stage or phase of the cell cycle. The 3′ telomeric DNA single-stranded overhang is apparently buried in the t-loop form with telomeric proteins bound to stabilize and conceal the t-loop junction (Fig. 5A). However, during telomeric DNA replication, the 3′ end must be exposed to allow telomerase access to anneal for telomeric DNA replication (Fig. 5B). It is possible that Ku could load onto telomeric DNA, using its nonspecific end-binding activity at this stage; however, even as the 3′ telomeric DNA overhang is unwound, it is likely to be associated with specific proteins and eventually bound by telomerase (Fig. 5B). Given that the telomeric DNA end is capped or blocked in either case, Ku would not be physically capable of loading onto the telomeric termini. As Ku and TRF1 form a high-affinity complex in human cell and essentially all TRF1 localizes to the telomere, our results give a possible mechanism for Ku localization to the capped telomere via complex formation with TRF1 (Fig. 5C).
Figure 5.
Model for Ku localization to the telomere via TRF1. (A) Schematic representation of the 3′ telomeric overhang duplexed within the t-loop (Griffith et al. 1999). Shaded rectangle represents hindrance by specialized t-loop junction proteins such as TRF2. (B) Unwound 3′ telomeric tail during telomeric DNA replication. Shaded oval represents hindrance by telomerase and specialized 3′ overhang telomeric proteins (LaBranche et al. 1998). (C) The TRF1 homodimer bound to internal telomeric tracts with Ku bound to TRF1. Note that Ku is not directly bound to telomeric DNA.
Recently, several DNA repair proteins, which function to join DNA double-stranded breaks, have been shown to play important roles at the telomere (Gasser 2000). One perplexing question that has arisen from this finding is: Why are proteins that function to join broken DNA ends found at the telomere? We present evidence that Ku, originally identified as being critical for joining nonhomologous DNA double-stranded breaks, provides essential telomere capping function in mammals to prevent telomere fusions. Importantly, we find that Ku acts uniquely at the telomere by forming a high-affinity protein/protein interaction with TRF1 to localize to internal regions of the telomere. Therefore, Ku functions at the telomere differently than during joining nonhomologous DNA double-stranded breaks. This suggests that once Ku forms a complex with TRF1, Ku-specific DNA repair domains required to tether broken DNA ends and recruit other DNA repair proteins may be obscured to prevent DNA end-joining activity at the telomere (Cary et al. 1997; Nick McElhinny et al. 2000). The exact role that Ku and other DNA repair proteins are performing at the telomere will be important to decipher for our continued understanding of telomere function and maintenance.
Materials and methods
Cell culture
HeLa cells were cultured in RPMI 1640 medium (GIBCO BRL) supplemented with 10% fetal bovine serum (HyClone). Human HT1080 fibrosarcoma cells with and without HA-TRF1 expressed from a pLXSN retroviral vector (Kim et al. 1999) were cultured in RPMI 1640 medium containing 400 μg/mL Geneticin (GIBCO BRL).
Vector construction and recombinant protein purification
His-tagged recombinant TRF1 was expressed in Escherichia coli. Culture were cooled to 18°C and induced with 0.1 mM IPTG (Sigma) for 16 h. Cells were lysed by sonication and the lysates cleared by centrifugation. The recombinant His-tagged TRF1 was purified from the soluble fraction using 1 mL Ni-NTA agarose (QIAGEN). The Ni-NTA beads were washed (3×, 10 mL each time) with PBS plus 25 mM imidazole. The His-TRF1 protein was eluted with 250 mM imidazole. The His-TRF1 protein was purified to near homogeneity as evidenced by a single coomassie-stained band on SDS-PAGE.
Purification of recombinant Ku70/Ku80 heterodimer from insect cells
Human genes encoding full-length Ku70 and Ku80 were cloned into the baculoviral shuttle vector pBluebacII (Invitrogen). Recombinant Ku70/Ku80 was purified as described previously (Cary et al. 1997). The Ku70 and Ku80 proteins were purified to near homogeneity as evidence by two coomassie stain bands on SDS-PAGE (Cary et al. 1997).
Surface plasmon resonance
Surface plasmon resonance (SPR) experiments were performed on a Biacore 2000 (Biacore) using a flow rate of 10–20 μL/min. Ligands (1800 and 1600 resonance units of Ku70/Ku80 or α-Ku80 Fab fragment) were coupled to individual flow cells of a CM5 chip using amine coupling chemistry (Malmqvist and Karlsson 1997). Two flow cells on the same chip were activated and deactivated with the coupling reagents to provide background values for subtraction throughout the binding experiments. The running HBS buffer consisted of 10 mM HEPES (pH 7.4), 150 mM NaCl, 3.4 mM EDTA, and 0.005% surfactant P20, and regeneration was carried out using one 30-sec pulse of 10 mM glycine followed by a 30-sec pulse of 100 mM HCl and 20 mM EDTA. TRF1 and BSA were diluted in HBS before injection. Injections were performed three times for each protein concentration, varying the association times from 30 sec to 1 min and the dissociation times from 1 to 5 min. We obtained an acceptable fit for one TRF1 concentration series to a 1 : 1 binding (Langmurir) model after increasing the injection times to 1 min. Sensorgrams were evaluated kinetically using Biacore Evaluation Software (Biacore).
In vitro transcription/translation and binding assay
To prepare 35S-methionine labeled TRF1 protein, a rabbit reticulocyte lysate TNT system was used (Promega). TNT reaction mixtures were labeled with [35S]methionine (Amersham) at 30°C for 2 h according to the manufacturer's instructions. In vitro translated TRF1 protein was incubated with recombinant Ku70/Ku80, in a 50-μL total volume of buffer containing 0.5% NP-40, 1 mM EDTA, 1 mM EGTA, 150 mM NaCl, 5 mM MgCl2, and 50 mM Tris-HCl (pH 8.0), for 4 h at 4°C. Monoclonal antibodies against Ku80 or p21 (Calbiochem) were added to reactions for 4 h at 4°C with occasional rotation. After incubation, mixture of protein A and G beads (Pharmacia Biotech, Boehringer Mannheim) was added for 1 h and then washed six times in 1 mL buffer A (150 mM NaCl, 0.05% Tween 20, 50 mM Tris-HCl at pH 7.4) at 4°C for 10 min each. Bound proteins were eluted by SDS sample buffer and analyzed by 4%–12% SDS-PAGE (Novex).
Far-Western analysis
Purified His-TRF1, His-Rad51, recombinant Ku70/Ku80, and BSA proteins were spotted onto a nitrocellulose membrane for far-Western analysis (Ishiguro et al. 1998). Filters were blocked in buffer A (150 mM NaCl, 0.05% Tween 20, 50 mM Tris-HCl at pH 7.4) containing 5% nonfat dry milk for 1 h, and were incubated with 32P-labeled Ku in buffer A containing 1% milk for 2 h at room temperature. Blots were washed four times for 10 min in buffer A without milk, dried, and analyzed by PhosphorImager (Molecular Dynamics) and ImageQuant software.
Immunoprecipitation
Mixture of protein A/G beads suspended in NET buffer (0.5% NP-40, 1 mM EDTA, 1 mM EGTA, 150 mM NaCl, 5 mM MgCl2, 50 mM Tris-HCl at pH 8.0) were incubated overnight with mAbs at 4°C. HeLa lysates were immunoprecipitated with α-Ku80 mAb or α-HA mAb (Sigma) coupled protein A/G sepharose beads. After incubation for 4 h at 4°C, the beads were collected by centrifugation and washed six times with NET buffer. The immune complexes were released by 40 μL of SDS sample buffer and subjected to SDS-PAGE analysis.
Western blot analysis
Triton X-100 treated cell extracts and immunoprecipitated proteins were separated on 4%–12% SDS-PAGE and transferred to nitrocellulose membranes. The membranes were probed with goat α-Ku70 Ab (Santa Cruz Biotechnology), mouse α-Ku80 Ab, or rabbit α-HA Ab (Santa Cruz Biotechnology), followed by incubation with horseradish peroxidase-conjugated, α-goat, α-mouse, or α-rabbit IgG secondary antibodies. The blots were developed using enhanced chemiluminescence (Amersham Pharmacia).
Preparation of microcircles
Telomeric [GAAGATCT(TTAGGG)nAAGATCTTC] or nontelomeric [GAAGATCT(TAGCAT)nAAGATCTTC] sequence-containing duplex oligonucleotides (n = 3 or n = 6) were digested with BglII and cloned into the BamHI site of pSP73 (Promega). The cloned sequences were excised by NdeI and XhoI digestion and were 32P-end labeled using Klenow polymerase (New England BioLabs). The labeled DNAs were circularized as described previously (Galande and Kohwi-Shigematsu 1999).
DNA binding analysis by electrophoretic mobility shift assay (EMSA)
Binding reactions contained 10 mM HEPES (pH 7.9), 50 mM KCl, 2.5 mM MgCl2, 1 mM DTT, 10% glycerol, 0.2 mg/mL BSA, and 0.25 mg/mL poly (dI/dC) (Pharmacia). Proteins were first incubated in the above reaction mixture and then microcircle DNA was added, followed by addition of a second protein and/or antibody. After a 15-min incubation at 25°C, the reaction mixtures were loaded directly onto 6% native polyacrylamide gels and electrophoresed for 4 h at 150 V, 4°C, in 0.57× Tris/borate/EDTA buffer. DNA was visualized by autoradiography of the dried gels.
Fluorescence in situ hybridization
Early passage Ku80+/+ and Ku80−/− mouse embryonic fibroblasts were treated with colcemid (0.1 μg/mL) for 4–5 h and were subsequently trypsinized and spun for 8 min at 120g. After hypotonic swelling in 30 mM sodium citrate for 25 min at 37°C, cells were fixed in methanol:acetic acid (3 : 1). After two to three additional changes in fixative, cell suspensions were dropped on wet, clean slides and dried overnight. FISH with Cy3-labeled (CCCTAA)3 peptide nucleic acid was performed as described (Hande et al. 1999). Cells were viewed with an Olympus BH2 microscope equipped with a CCD camera, and the images were acquired using a Cytovision software (Applied Imaging). The scoring criteria were as described elsewhere (Bailey et al. 1999; Hande et al. 1999). Fifty metaphases per sample were analyzed.
Acknowledgments
We thank Janice Pluth and Steven Yannone for helpful comments and Michael Murphy for valuable and timely help in the purification of the TRF1 protein. We thank Titia de Lange for kindly providing the TRF1 gene. This work was supported by the U.S. Department of Energy under contract DE-AC03–76SF00098 (D.J.C.) and NIH grants AG17709 (D.J.C.), CA50519 (D.J.C.), CA39681 (T.K.-S.), and AG11658 (J.C.).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL DJChen@LBL.gov; FAX (510) 486-6816.
Article and publication are at www.genesdev.org/cgi/doi/10.1101/gad.844000.
References
- Artandi SE, DePinho RA. A critical role for telomeres in suppressing and facilitating carcinogenesis. Curr Opin Genet Dev. 2000;10:39–46. doi: 10.1016/s0959-437x(99)00047-7. [DOI] [PubMed] [Google Scholar]
- Bailey SM, Meyne J, Chen DJ, Kurimasa A, Li GC, Lehnert BE, Goodwin EH. DNA double-strand break repair proteins are required to cap the ends of mammalian chromosomes. Proc Natl Acad Sci. 1999;96:14899–14904. doi: 10.1073/pnas.96.26.14899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackburn E. The telomere and telomerase: How do they interact? Mt Sinai J Med. 1999;66:292–300. [PubMed] [Google Scholar]
- Blackburn EH. The telomere and telomerase: Nucleic acid-protein complexes acting in a telomere homeostasis system. A review. Biochemistry. 1997;62:1196–1201. [PubMed] [Google Scholar]
- Bodnar AG, Ouellete M, Frolkis M, Holt SE, Chiu CP, Morin GB, Harley CB, Shay JW, Lichtsteiner S, Wright WE. Extension of life-span by introduction of telomerase into normal human cells. Science. 1998;279:349–352. doi: 10.1126/science.279.5349.349. [DOI] [PubMed] [Google Scholar]
- Boulton SJ, Jackson SP. Identification of a Saccharomyces cerevisiae Ku80 homologue: Roles in DNA double strand break rejoining and in telomeric maintenance. Nucleic Acids Res. 1996;24:4639–4648. doi: 10.1093/nar/24.23.4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Components of the Ku-dependent non-homologous end-joining pathway are involved in telomeric length maintenance and telomeric silencing. EMBO J. 1998;17:1819–1828. doi: 10.1093/emboj/17.6.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cary RB, Peterson SR, Wang J, Bear DG, Bradbury EM, Chen DJ. DNA looping by Ku and the DNA-dependent protein kinase. Proc Natl Acad Sci. 1997;94:4267–4272. doi: 10.1073/pnas.94.9.4267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Critchlow SE, Jackson SP. DNA end-joining: From yeast to man. Trends Biochem Sci. 1998;23:394–398. doi: 10.1016/s0968-0004(98)01284-5. [DOI] [PubMed] [Google Scholar]
- d'Adda di Fagagna F, Hande MP, Tong WM, Lansdorp PM, Wang ZQ, Jackson SP. Functions of poly(ADP-ribose) polymerase in controlling telomere length and chromosomal stability. Nat Genet. 1999;23:76–80. doi: 10.1038/12680. [DOI] [PubMed] [Google Scholar]
- Dynan WS, Yoo S. Interaction of Ku protein and DNA-dependent protein kinase catalytic subunit with nucleic acids. Nucleic Acids Res. 1998;26:1551–1559. doi: 10.1093/nar/26.7.1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galande S, Kohwi-Shigematsu T. Poly(ADP-ribose) polymerase and Ku autoantigen form a complex and synergistically bind to matrix attachment sequences. J Biol Chem. 1999;274:20521–20528. doi: 10.1074/jbc.274.29.20521. [DOI] [PubMed] [Google Scholar]
- Gasser SM. A sense of the end. Science. 2000;288:1377–1379. doi: 10.1126/science.288.5470.1377. [DOI] [PubMed] [Google Scholar]
- Giffin W, Torrance H, Rodda DJ, Prefontaine GG, Pope L, Hache RJ. Sequence-specific DNA binding by Ku autoantigen and its effects on transcription. Nature. 1996;380:265–268. doi: 10.1038/380265a0. [DOI] [PubMed] [Google Scholar]
- Griffith JD, Comeau L, Rosenfield S, Stansel RM, Bianchi A, Moss H, de Lange T. Mammalian telomeres end in a large duplex loop. Cell. 1999;97:503–514. doi: 10.1016/s0092-8674(00)80760-6. [DOI] [PubMed] [Google Scholar]
- Hande P, Slijepcevic P, Silver A, Bouffler S, van Buul P, Bryant P, Lansdorp P. Elongated telomeres in scid mice. Genomics. 1999;56:221–223. doi: 10.1006/geno.1998.5668. [DOI] [PubMed] [Google Scholar]
- Hsu HL, Gilley D, Blackburn EH, Chen DJ. Ku is associated with the telomere in mammals. Proc Natl Acad Sci. 1999;96:12454–12458. doi: 10.1073/pnas.96.22.12454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiguro A, Kimura M, Yasui K, Iwata A, Ueda S, Ishihama A. Two large subunits of the fission yeast RNA polymerase II provide platforms for the assembly of small subunits. J Mol Biol. 1998;279:703–712. doi: 10.1006/jmbi.1998.1823. [DOI] [PubMed] [Google Scholar]
- Kanaar R, Hoeijmakers JH, van Gent DC. Molecular mechanisms of DNA double strand break repair. Trends Cell Biol. 1998;8:483–489. doi: 10.1016/s0962-8924(98)01383-x. [DOI] [PubMed] [Google Scholar]
- Kim SH, Kaminker P, Campisi J. TIN2, a new regulator of telomere length in human cells. Nat Genet. 1999;23:405–412. doi: 10.1038/70508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaBranche H, Dupuis S, Ben-David Y, Bani MR, Wellinger RJ, Chabot B. Telomere elongation by hnRNP A1 and a derivative that interacts with telomeric repeats and telomerase. Nat Genet. 1998;19:199–202. doi: 10.1038/575. [DOI] [PubMed] [Google Scholar]
- Ludwig DL, Chen F, Peterson SR, Nussenzweig A, Li GC, Chen DJ. Ku80 gene expression is Sp1-dependent and sensitive to CpG methylation within a novel cis element. Gene. 1997;199:181–194. doi: 10.1016/s0378-1119(97)00366-1. [DOI] [PubMed] [Google Scholar]
- Malmqvist M, Karlsson R. Biomolecular interaction analysis: Affinity biosensor technologies for functional analysis of proteins. Curr Opin Chem Biol. 1997;1:378–383. doi: 10.1016/s1367-5931(97)80077-4. [DOI] [PubMed] [Google Scholar]
- Mimori T, Hardin JA, Steitz JA. Characterization of the DNA-binding protein antigen Ku recognized by autoantibodies from patients with rheumatic disorders. J Biol Chem. 1986;261:2274–2278. [PubMed] [Google Scholar]
- Nick McElhinny SA, Snowden CM, McCarville J, Ramsden DA. Ku recruits the XRCC4-ligase IV complex to DNA ends. Mol Cell Biol. 2000;20:2996–3003. doi: 10.1128/mcb.20.9.2996-3003.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nussenzweig A, Chen C, da Costa Soares V, Sanchez M, Sokol K, Nussenzweig MC, Li GC. Requirement for Ku80 in growth and immunoglobulin V(D)J recombination. Nature. 1996;382:551–555. doi: 10.1038/382551a0. [DOI] [PubMed] [Google Scholar]
- Paillard S, Strauss F. Analysis of the mechanism of interaction of simian Ku protein with DNA. Nucleic Acids Res. 1991;19:5619–5624. doi: 10.1093/nar/19.20.5619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter SE, Greenwell PW, Ritchie KB, Petes TD. The DNA-binding protein Hdf1p (a putative Ku homologue) is required for maintaining normal telomere length in Saccharomyces cerevisiae. Nucleic Acids Res. 1996;24:582–585. doi: 10.1093/nar/24.4.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shore D. Telomeres—Unsticky ends. Science. 1998;281:1818–1819. doi: 10.1126/science.281.5384.1818. [DOI] [PubMed] [Google Scholar]
- Taccioli GE, Gottlieb TM, Blunt T, Priestley A, Demengeot J, Mizuta R, Lehmann AR, Alt FW, Jackson SP, Jeggo PA. Ku80: Product of the XRCC5 gene and its role in DNA repair and V(D)J recombination. Science. 1994;265:1442–1445. doi: 10.1126/science.8073286. [DOI] [PubMed] [Google Scholar]
- van Steensel B, de Lange T. Control of telomere length by the human telomeric protein TRF1. Nature. 1997;385:740–743. doi: 10.1038/385740a0. [DOI] [PubMed] [Google Scholar]
- Zhong Z, Shiue L, Kaplan S, de Lange T. A mammalian factor that binds telomeric TTAGGG repeats in vitro. Mol Cell Biol. 1992;12:4834–4843. doi: 10.1128/mcb.12.11.4834. [DOI] [PMC free article] [PubMed] [Google Scholar]