Abstract
We describe evidence that a regulatory B subunit of protein phosphatase 2A (PP2A) positively regulates an RTK–Ras–MAP kinase signaling cascade during Caenorhabditis elegans vulval induction. Although reduction of sur-6 PP2A-B function causes few vulval induction defects in an otherwise wild-type background, sur-6 PP2A-B mutations suppress the Multivulva phenotype of an activated ras mutation and enhance the Vulvaless phenotype of mutations in lin-45 raf, sur-8, or mpk-1. Double mutant analysis suggests that sur-6 PP2A-B acts downstream or in parallel to ras, but likely upstream of raf, and functions with ksr-1 in a common pathway to positively regulate Ras signaling.
Keywords: Suppressor of ras, phosphatase 2A, vulval induction, KSR, Raf, SUR-8
The protein phosphatase 2A (PP2A) enzymes, one of four families of serine/threonine protein phosphatases, are key regulators of many cellular events controlled by protein phosphorylation (Shenolikar 1994; Millward et al. 1999). PP2A is a heterotrimer, composed of a catalytic (C) subunit, an associated (A) subunit, and a regulatory (B) subunit. The C and A subunits form a core complex to which one of several classes of B subunits can bind. This differential binding of regulatory B subunits can influence the catalytic activity and substrate specificity of the catalytic core in vitro (Mayer-Jaekel and Hemmings 1994). However, although in vitro studies have identified numerous potential PP2A substrates, the broad substrate specificity has limited the ability to identify physiological targets or to understand normal PP2A regulation (Shenolikar 1994).
Recent studies have shown that PP2A can regulate signal transduction pathways. For example, PP2A stably interacts with and inactivates Ca2+-calmodulin-dependent protein kinase IV (CaMKIV) (Westphal et al. 1998). PP2A has also been proposed to inactivate MEK and MAP kinase, because PP2A can dephosphorylate MEK and MAP kinase in vitro (for review, see Millward et al. 1999), and interference of PP2A activity by SV40 small t antigen results in activation of MAP kinase in vivo (Sontag et al. 1993). Genetic studies in Drosophila suggested that PP2A both positively and negatively affects Ras pathway signaling during R7 photoreceptor cell fate specification (Wassarman et al. 1996).
Here we demonstrate that a B regulatory subunit of PP2A promotes Ras signaling during Caenorhabditis elegans vulval development. In C. elegans, a ras-mediated signal transduction pathway in part controls the fates of six cells, the vulval precursor cells (VPCs). An inductive signal from the anchor cell activates an RTK–Ras–MAP kinase signal-transduction pathway to induce the three neighboring VPCs (P5.p, P6.p, and P7.p) to adopt vulval cell fates. These cells undergo three rounds of division followed by morphogenesis to form the vulval structure. The remaining three VPCs (P3.p, P4.p, and P8.p) adopt nonvulval cell fates and, instead, divide only once before fusing with the surrounding hypodermis (Horvitz and Sternberg 1991). Mutations that result in the mis-specification of vulval cell fates have defined many of the genes necessary for normal vulval differentiation (Kornfeld 1997; Sternberg and Han 1998). Loss-of-function mutations in positively acting components of this pathway can cause fewer than three VPCs to adopt vulval cell fates, leading to a Vulvaless (Vul) phenotype. A gain-of-function mutation in let-60 ras (n1046gf or G13E), causes more than three VPCs to adopt vulval cell fates, leading to a Multivulva (Muv) phenotype.
To identify genes that act downstream of let-60 ras, screens have been conducted for mutations that suppress the Muv phenotype of let-60 ras(n1046gf) mutants. These screens have identified mutations in components acting downstream of Ras (such as lin-45 raf, mek-2 MEK, and mpk-1 MAP kinase), as well as in some new components such as ksr-1 and sur-8 (Sternberg and Han 1998). KSR-1 is a conserved putative kinase (Kornfeld et al. 1995; Sundaram and Han 1995; Therrien et al. 1995), and SUR-8 is a conserved Ras-binding protein with leucine-rich repeats (Selfors et al. 1998; Sieburth et al. 1998). Both KSR-1 and SUR-8 appear to stimulate signaling at the level of Ras or Raf (Therrien et al. 1995; Sieburth et al. 1998; this work). Here we describe the identification and characterization of another positive regulator of Ras-mediated signaling, sur-6, which encodes a regulatory PR55/B subunit of PP2A (PP2A-B).
Results
We screened for mutations that suppress the Muv phenotype caused by an activated let-60 ras mutation, n1046gf (Sundaram and Han 1995) or that enhance the Vul phenotype caused by a hypomorphic lin-45 raf mutation, ku112 (M. Sundaram, unpubl.). These screens identified two mutations, ku123 and cs24, that define the gene sur-6 (suppressor of ras). These sur-6 mutations cause few or no vulval defects in an otherwise wild-type background; ku123 and cs24 mutants display an average vulval induction of 100% or 99%, respectively (Table 1). However, both sur-6 mutations suppress the Muv phenotype of let-60(n1046gf) animals (Table 2), and enhance the vulval induction defects and larval lethality caused by weak alleles of lin-45 raf (Table 1). These strong genetic interactions suggest that the sur-6 mutations reduce signaling by the Ras pathway at a point downstream or in parallel to let-60 ras.
Table 1.
sur-6 mutations enhance the vulval defects of lin-45 Raf, mpk-1 MAP kinase, or sur-8 but not ksr-1 mutants
| Genotypea
|
Percent
|
Average percent
|
Percent larval lethald (n)
|
|
|---|---|---|---|---|
| Vulb
|
inductionc
|
(No.)
|
||
| sur-6(ku123) | 0 | 100 | (26) | 0 (474) |
| sur-6(ku123)/qDf8 | 0 | 100 | (28) | 0 (531)e |
| sur-6(cs24) | 2 | 99 | (48) | 0 (195) |
| sur-6(cs24)/qDf8 | 8 | 97 | (26) | 0 (386)e |
| +/qDf8 | 0 | 100 | (16) | N.D. |
| sur-6(cs24)/+ | 0 | 100 | (35) | N.D. |
| lin-45(sy96) | 58 | 53 | (38) | 86 (290) |
| sur-6(ku123); lin-45(sy96) | 100 | 5 | (17) | 94 (228) |
| lin-45(ku112) | 0 | 100 | (26) | <1 (66) |
| sur-6(cs24); lin-45(ku112) | 87 | 50 | (24) | 80 (360) |
| mpk-1(ku1) | 17 | 97 | (29) | 7 (229) |
| sur-6(ku123); mpk-1(ku1) | 82 | 76 | (28) | 77 (263) |
| sur-8(ku167) | 0 | 100 | (18) | <1 (271) |
| sur-6(ku123); sur-8(ku167)f | 71 | 56 | (24) | <1 (263) |
| sur-6(cs24); sur-8(ku167) | 65 | 67 | (20) | 3 (198) |
| ksr-1(ku68) | 0 | 100 | (23) | 24 (257) |
| sur-6(ku123); ksr-1(ku68) | 3 | 99 | (40) | 17 (282) |
| sur-8(ku167); ksr-1(ku68)g | 100 | 4 | (19) | 85 (164) |
| ksr-1(n2526) | 1 | 99 | (68) | 2 (607) |
| sur-6(cs24); ksr-1(n2526) | 3 | 99 | (62) | 1 (88) |
| sur-8(ku167); ksr-1(n2526) | 67 | 64 | (24) | 54 (132) |
ksr-1(ku68) (R531H) is a strong loss-of-function, possibly dominant-negative allele (Sundaram and Han 1995). ksr-1(n2526) (W255stop) is a putative null allele (Kornfeld et al. 1995). lin-45(sy96) and lin-45(ku112) are hypomorphic (partial loss-of-function) alleles (Han et al. 1993; Sundaram and Han 1995); lin-45(sy96) contains a splice site mutation, and the lin-45(ku112) lesion is unknown. sur-8(ku167) (E430K) is a strong hypomorphic allele (Sieburth et al. 1998). mpk-1(ku1) (A38V) is a hypomorphic allele (Wu and Han 1994). qDf8 removes the flanking genes mec-8 and fog-3 and thus should remove sur-6 (R. Ellis, pers. comm.). For deficiency strains, sur-6 was marked with unc-13, and qDf8 was marked with ces-1. No other defects were observed in sur-6/qDf8 strains. lin-45(sy96) was linked to dpy-20, ksr-1(ku68) was linked to lon-2. lin-45(ku112) was linked to dpy-20 for double mutants with ksr-1. sur-6(ku123) was linked to unc-29 for double mutants with lin-45, ksr-1; and sur-8. All other strains were unmarked. The dpy-20, unc-29, and lon-2 markers were also tested in the control strains to show that they have no effects on vulval induction.
Percent of animals in which <3 VPCs adopted vulval fates, as scored under Nomarski optics (Sieburth et al. 1998). In some sur-6 strains, VPCs were occasionally undivided or absent and may have adopted 4° fates (Clark et al. 1993). Nondivision of VPC P5.p, P6.p, or P7.p occurs infrequently (4/24 ku123; ku167 animals, 3/23 cs24;ku167 animals, 3/65 cs24;n2526 animals). %Vul is calculated only for those animals in which P5.p, P6.p, and P7.p were present and divided at least once.
Average percent of VPCs adopting a vulval cell fate, as scored under Nomarski optics (Sieburth et al. 1998; 100% for wild type). Induction is calculated only for those animals in which P5.p, P6.p, and P7.p were present and divided at least once.
Percent of animals arresting in early larval stages with a clear, rod-like phenotype (Sieburth et al. 1998). (N.D.) Not determined.
For qDf8 experiments, n corresponds to the entire brood of sur-6/qDf8 mothers, half of which should also have been sur-6/qDf8 hemizygotes. No more than 25% of such broods arrested as embryos, suggesting that sur-6/qDf8 (like sur-6/sur-6) has no significant embryonic lethal phenotype.
sur-6(ku123); sur-8 double mutants were 37% embryonic lethal with an average brood size (live worms plus dead embryos) of 26 (n = 263).
Data from Sieburth et al. (1998).
Table 2.
Epistasis analysis of sur-6 and ksr-1 with Muv mutations
|
sur-6 or ksr-1 mutationa
|
Muv mutationb
|
Percent Muvc (n)
|
Percent average inductiond (n)
|
|---|---|---|---|
| +/+ | +/+ | 0 (many) | 100 (many) |
| +/+ | let-60(n1046gf) | 87 (276) | 154 (27) |
| sur-6(ku123) | let-60(n1046gf) | 6 (240) | 103 (26) |
| sur-6(cs24) | let-60(n1046gf) | 19 (107) | 104 (33) |
| sur-6(ku123)/sur-6(cs24) | let-60(n1046gf) | 3 (31) | N.D. |
| sur-6(ku123)/+ | let-60(n1046gf) | 22 (148) | 115 (31) |
| sur-6(cs24)/+ | let-60(n1046gf) | 40 (53) | N.D. |
| sur-6(ku123)/qDf8 | let-60(n1046gf) | 1 (209) | N.D. |
| +/qDf8 | let-60(n1046gf) | 5 (281) | N.D. |
| +/+ | HSP–raf(gf) | 47 (30) | 119 (30) |
| sur-6(ku123) | HSP–raf(gf) | 47 (17) | 125 (17) |
| +/+ | HSP–raf(gf) | 47 (17) | 118 (17) |
| ksr-1(ku68) | HSP–raf(gf) | 46 (28) | 117 (28) |
| +/+ | lin-1(e1275) | 99 (68) | N.D. |
| sur-6(ku123) | lin-1(e1275) | 100 (102) | N.D. |
| +/+ | lin-15(n765) | 100 (59) | 157 (28) |
| sur-6(ku123) | lin-15(n765) | 6 (351) | 102 (20) |
ku123 was marked with unc-29 in all strains except those with n1046. The full genotype of ku123/cs24 was n1046/sy130 dpy-20; ku123/cs24. The full genotype of ku123/+ was ku123/unc-29; n1046; him-5/+. The full genotype for cs24/+ was n1046/sy130 dpy-20; cs24/+. sy130 encodes the same let-60 ras(G13E) substitution as n1046. Full genotypes for deficiency analysis: ces-1 qDf8/unc-13 ku123; n1046 and ces1 qDf8/ unc-13 dyp-24; n1046. HSP–raf(gf); ksr-1(ku68) animals (and their paired controls) were marked with unc-24.
For HSP–raf(gf) description, see text and Sieburth et al. (1998). Also see Sieburth et al. (1998) for positive control results (suppression of HSP–raf(gf) by mek-2 and mpk-1 alleles). HSP–raf(gf) strains were heat shocked during the early-mid L3 stage for 80 min at 36°C. lin-15 strains were grown at 18°C.
Percent Muv was determined by examining adult hermaphrodites with a dissecting microscope for the presence of ectopic ventral protrusions [for let-60(gf), lin-1 and lin-15 experiments], or by examining L4 larvae under Nomarski optics [for HSP–raf(gf) experiments].
See Table 1 footnote. In 1/34 cs24; n1046gf animals, P4.p and P8.p were undivided and may have adopted 4° fates. Induction is calculated here only for animals in which P(4-8).p divided at least once.
sur-6(ku123) and sur-6(cs24) appear to strongly reduce (but not eliminate) sur-6 gene function. These sur-6 alleles and a deficiency of the sur-6 locus each semidominantly suppress the Muv phenotype of let-60(n1046gf) mutants (Table 2), suggesting the sur-6 locus is haplo insufficient. RNA-mediated inhibition of sur-6 also suppresses the let-60(n1046gf) Muv phenotype and causes a partial Vul phenotype in a wild-type background (Table 3), arguing that these are loss-of-function phenotypes. qDf8 fails to complement the weak Vul phenotype of sur-6(cs24) mutants, and sur-6/sur-6 homozygotes and sur-6/qDf8 hemizygotes display similar phenotypes (Tables 1 and 2), consistent with the sur-6 mutations strongly reducing sur-6 function. This notion is further supported by the fact that the suppressor phenotype of sur-6(ku123) can be rescued by injecting wild-type DNA containing the sur-6 gene (see below and Materials and Methods). Nevertheless, the sur-6 mutations are likely non-null, because RNA inhibition suggests that the sur-6 null phenotype is embryonic lethal (Table 3; see below). Because reducing sur-6 function reduces vulval induction in sensitized genetic backgrounds, we conclude that sur-6 normally plays a positive role in regulating Ras pathway signaling during vulval development.
Table 3.
RNA interference of sur-6 PP2A-B, PP2A-C, or PP2A-A
| Genotype
|
dsRNAa or transgenesb
|
Percent Vul or Muv (n)
|
Percent avg. induction (n)
|
|---|---|---|---|
| + | no dsRNA | 0 (many) | 100 (many) |
| + | sur-6 PP2A-B | 19 Vul (21) | 94 (21) |
| + | PP2A-C | 5 Vul (22) | 99 (22) |
| let-60(n1046gf) | no dsRNA | 95 Muv (19) | 173 (19) |
| let-60(n1046gf) | sur-6 PP2A-B | 3 Muv (29) | 101 (29) |
| let-60(n1046gf) | PP2A-C | 70 Muv (23) | 132 (23) |
| let-60(n1046gf) | sur-8 | 15 Muv (33) | 106 (33) |
| let-60(n1046gf) | no dsRNA | 93 Muv (59) | 143 (59) |
| let-60(n1046gf) | sur-6 PP2A-B | 7 Muv (15) | 103 (15) |
| let-60(n1046gf) | PP2A-A | 57 Muv (14) | 125 (14) |
| let-60(n1046gf) | no transgenes | 82 Muv (242) | 175 (23) |
| let-60(n1046gf) | sur-6 PP2A-B | 20 Muv (172) | 110 (48) |
| let-60(n1046gf) | PP2A-C | 65 Muv (82) | N.D. |
In the case of sur-6 PP2A-B, PP2A-C or PP2A-A (but not sur-8), nearly complete embryonic lethality was observed in progeny laid 8–24 hr after double-stranded (ds) RNA injection. Most surviving sur-6(RNAi) animals displayed a weakly Uncoordinated phenotype. Vulval induction was scored in surviving progeny laid at least 6 hr postinjection (for the wild-type background), or in progeny laid 6–8 hr postinjection, before the lethal period began [for the let-60(gf) background]. In the wild-type background, sur-6 (RNAi) caused variable vulval defects, including defects in vulval induction (2° to 3° or hybrid fate transformation; 5/31 animals), VPC generation (absence of P5.p, P6.p, or P7.p, or failure of those cells to divide; 10/31 animals), and vulval lineage execution (failure of P5.pxx to complete last round of vulval division; 1/31 animals). The VPC generation defects were almost never seen in the let-60(n1046gf) background. In the wild-type background, PP2A-C RNAi also caused occasional defects in vulval induction (P5.p executed a “hybrid” lineage, generating only 3 vulval descendants and 1 nonvulval descendant; 1/23 animals), VPC generation (absence of P5.p; 1/23 animals), and vulval lineage execution (failure of P5.pxx to complete last round of vulval division; 1/23 animals). %Vul (and induction) are calculated only for animals in which P5.p, P6.p, and P7.p were present and divided at least once. %Muv (and induction) are calculated only for animals in which P(4-8).p were present and divided at least once.
We used the col-10 promoter to coexpress both sense and antisense RNA fragments in the hypodermis (see Material and Methods).
sur-8 and ksr-1 are two other genes that display genetic interactions similar to those of sur-6. Strong loss-of-function mutations in sur-8 or ksr-1 cause few defects on their own, but strongly modify the phenotypes of other Ras pathway mutants (Kornfeld et al. 1995; Sundaram and Han 1995; Sieburth et al. 1998). We found that sur-6;sur-8 double mutants display a synthetic Vul phenotype (Table 1), consistent with these mutations potentially affecting different aspects of Ras pathway regulation. Interestingly, however, sur-6;ksr-1 double mutants resemble the sur-6 or ksr-1 single mutants (Table 1). Because reducing the function of either ksr-1 or sur-6 has the same effect as reducing the function of both, it is likely that sur-6 acts together with ksr-1 in a common signaling pathway to regulate Ras signaling.
raf genetically acts downstream of ras in C. elegans and in Drosophila (Dickson et al. 1992; Han et al. 1993), and mammalian Raf is a direct Ras effector (for review, see Katz and McCormick 1997). Drosophila ksr has been shown previously to function genetically downstream or in parallel to ras and upstream of raf (Therrien et al. 1995). We tested whether this was also true for C. elegans ksr-1 and sur-6. An activated raf transgene under the control of a heat shock promoter [HSP–raf(gf)] causes a Muv phenotype that can be suppressed by mutations in the downstream Ras pathway components mek-2 MEK or mpk-1 MAP kinase (Sieburth et al. 1998). In contrast, this HSP–raf(gf) Muv phenotype is not suppressed by ksr-1 or sur-6 mutations (Table 2). Therefore, ksr-1 and sur-6 likely act upstream of lin-45 raf. Consistent with this, sur-6(ku123) also fails to suppress the Muv phenotype caused by a relatively weak mutation of lin-1, which encodes an ETS domain transcription factor acting downstream of mpk-1 MAP kinase (Jacobs et al. 1998; Tan et al. 1998). The genetic placement of ksr-1 and sur-6 suggests that ksr-1 and sur-6 may act in a regulatory branch that modifies or cooperates with Ras/Raf.
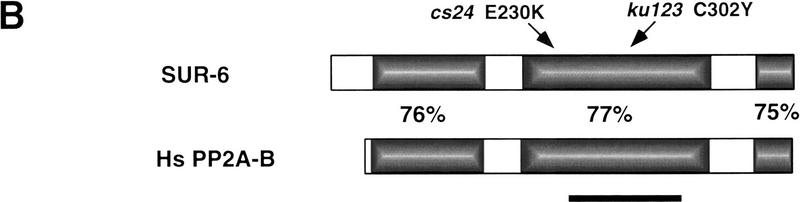
We cloned sur-6 by genetic mapping followed by transformation rescue (see Materials and Methods). An 11-kb fragment, pDS89, which contains sur-6(+) rescuing activity was predicted by the C. elegans genome sequencing consortium to contain a single gene (F26E4.1) that encodes a PR55 family regulatory B subunit of PP2A (PP2A-B, Fig. 1). The sur-6 alleles ku123 and cs24 each contain single G → A substitutions that introduce amino acid substitutions at highly conserved positions within PP2A-B (Fig. 1). The PP2A-B coding region, when expressed under the control of a heat shock promoter in transgenic animals, can rescue the sur-6(ku123) mutant (see Materials and Methods), and RNA interference of PP2A-B phenocopies the sur-6 suppression and partial Vul phenotypes (Table 3). Thus, we conclude that sur-6 encodes PP2A-B. SUR-6 shares >59% overall amino acid identity with PP2A-B from human or Drosophila, with three large stretches of at least 75% identity (Fig. 1B). In mammals there are three PR55/B isoforms that differ in spatial and temporal expression (Mayer-Jaekel and Hemmings 1994). SUR-6 is most similar to the mammalian Bα subtype.
Figure 1.

Sequence comparison between SUR-6 PP2AB and human PP2A-Bα. (A) Amino acid alignment of C. elegans SUR-6 PP2A-B (as predicted by cDNA analyses; see Materials and Methods, GenBank accession no. AF174643) with a human PP2A-Bα isoform (GenBank accession no. 231445; Mayer et al. 1991) and Drosophila Twins PP2A-B (GenBank accession no. 543716; Uemura et al. 1993). Identical amino acids are shaded. The position of the C302Y and E230K missense mutations identified in
sur-6(ku123) and sur-6(cs24) mutants, respectively, are denoted. C302 and E230 are conserved in PP2A-B from all species in which it has been identified, from human to yeast. (B) Structural comparison of sur-6 PP2A-B and human PP2A-Bα non-neuronal isoform. Percent amino acid identity is shown for shaded regions. Nonshaded regions share 20% or less amino acid identity. The overall amino acid identity is 59%. The bar denotes the region with similarity to the c-Abl proposed substrate-binding domain (amino acids 257–378).
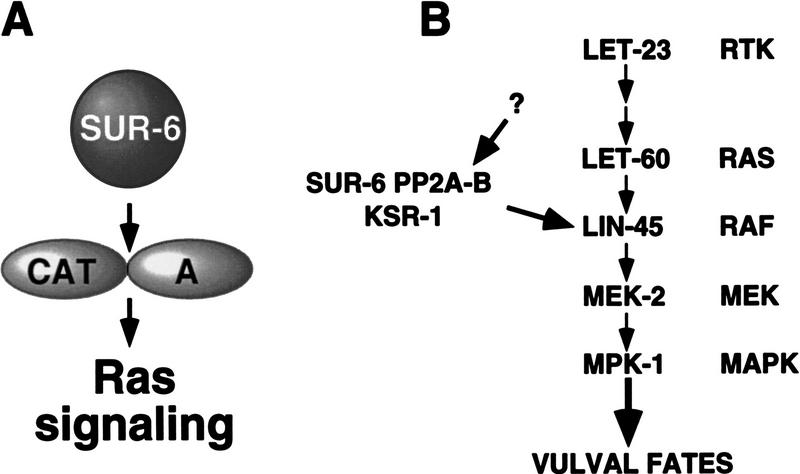
PP2A-B subunits modulate the activity and/or substrate specificity of the PP2A-A/C catalytic core (Mayer-Jaekel and Hemmings 1994). SUR-6 is the only predicted PR55/B family member encoded by the C. elegans genome, although other types of regulatory B subunits (such as PR56/B′) are also present. The C. elegans genome is predicted to encode a single PP2A-A subunit (F48E8.5) and a single PP2A-C subunit (F38H4.9), which share >90% amino acid identity with their mammalian counterparts. As expected, the PP2A-A subunit can bind to both SUR-6 PP2A-B and the PP2A-C subunit, as assayed by the yeast two-hybrid system (data not shown). Given the positive role of sur-6 PP2A-B defined by genetic analysis, sur-6 PP2A-B could either function to activate the catalytic core, which in turn would activate Ras pathway signaling, or it could function to relieve inhibition of Ras signaling by the core complex (Fig. 2A).
Figure 2.
Models for SUR-6 PP2A-B function in the vulval induction pathway. (A) SUR-6 PP2A-B is likely to regulate the activity of a PP2A core complex composed of a C and A subunit to influence Ras signaling. SUR-6 PP2A-B is a positive regulator of Ras signaling, so SUR-6 could function either to activate the PP2A core, which in turn activates Ras signaling, or to inhibit the PP2A core that normally inhibits Ras signaling. Potential targets for SUR-6 PP2A-B-regulated PP2A activity include LIN-45 Raf and KSR-1. (B) Model for the signal transduction pathway regulated by SUR-6. The receptor tyrosine kinase LET-23 is activated by an inductive signal that leads to the activation of LET-60 Ras. Ras–GTP recruits Raf kinase to the membrane and Raf phosphorylates and activates the MEK-2 MEK/MPK-1 MAPK kinase cascade. We showed that SUR-6 PP2A-B positively regulates signaling at a branch point at the level of Ras or Raf. KSR-1 and SUR-6 may work together to strengthen Ras pathway output during vulval cell fate specification.
To determine the requirements for PP2A during C. elegans development, we used RNA interference (RNAi) (Fire et al. 1998) to block sur-6 PP2A-B, PP2A-A, or PP2A-C expression. For each PP2A gene (but not for sur-8 or ksr-1), RNAi caused highly penetrant embryonic lethality in both wild-type and let-60(n1046gf) backgrounds. Embryos arrested at ∼100 cell stage, with widely variable cell sizes (data not shown). Thus, unlike ksr-1 and sur-8, PP2A appears to be absolutely required during embryonic development in addition to functioning later during vulval induction. Because the sur-6(ku123) and sur-6(cs24) mutations caused little or no embryonic lethality, even when hemizygous (Table 2), the two functions of sur-6 PP2A-B appear separable, with these sur-6 point mutations primarily affecting sur-6 PP2A-B function in vulval development but not in embryogenesis.
To avoid the PP2A(RNAi) lethality and test its effects on vulval development, we examined the last surviving progeny of RNA-injected mothers (Table 3). In this assay, sur-6(RNAi) caused a partial Vul phenotype in a wild-type background and efficiently suppressed the let-60(n1046gf) Muv phenotype. However, PP2A-C(RNAi) caused few vulval defects and only weakly suppressed the let-60(n1046gf) Muv phenotype. Similar results were obtained by use of a hypodermal-specific promoter to drive expression of sur-6 PP2A-B or PP2A-C RNAs (Table 3). Thus, it is still unclear whether PP2A catalytic activity promotes and/or inhibits Ras-mediated vulval induction.
Discussion
We have shown for the first time a positive and specific regulatory role for a PP2A regulatory PR55/B subunit in Ras-mediated signal transduction. We propose that sur-6 PP2A-B influences the catalytic activity of PP2A toward specific Ras pathway substrate(s), such as Raf or KSR, leading to the enhancement of Ras pathway signaling.
Genetic analysis shows that sur-6 PP2A-B positively regulates Ras pathway signaling during vulval induction. First, reducing sur-6 PP2A-B function by mutation or RNA inhibition suppresses the excess vulval cell fate specification caused by an activated let-60 ras allele (but not that caused by an activated raf transgene). Second, sur-6 PP2A-B mutations enhance defects in vulval cell-fate specification caused by weak mutations in the Ras pathway components lin-45 raf or mpk-1 MAP kinase. However, reducing sur-6 PP2A-B function by mutation or RNA inhibition only mildly reduces vulval induction in an otherwise wild-type background, suggesting that sur-6 PP2A-B activity might not be essential for vulval induction when other components are functioning normally.
sur-6 PP2A-B appears to regulate Ras pathway signaling in multiple tissues besides vulval precursor cells. For example, a sur-6 PP2A-B mutation suppresses the male mating defect caused by an activated ras mutation (data not shown) and enhances the rod-like larval lethality of lin-45 raf or mpk-1 MAP kinase mutants. However, sur-6 PP2A-B also appears to have functions that are currently not known to involve the Ras pathway. RNA inhibition of sur-6 PP2A-B function reveals an absolute requirement for sur-6 during embryogenesis. Additional roles of sur-6 PP2A-B in the generation or survival of VPCs, and in the development or function of muscles and/or motor neurons, are suggested by the fact that sur-6(cs24) animals and surviving sur-6(RNAi) animals sometimes lack one or more VPCs and are weakly uncoordinated. It would be interesting to know whether or not these defects also involve the Ras pathway.
SUR-6 PP2A-B likely functions by regulating the activity of the PP2A-A/C core complex. Mammalian PP2A-B regulatory subunits can either activate or inhibit core activity in vitro, depending on the substrate used (Mayer-Jaekel and Hemmings 1994). Genetic data in yeast support specific activating roles for PP2A regulatory subunits, because mutations in cdc55 (a PR55/B subunit) and rts1 (a PR56/B′ subunit) each cause a distinct subset of phenotypes associated with mutations in the PP2A catalytic core (Shu et al. 1997). If SUR-6 PP2A-B also functions to activate the PP2A catalytic core, this implies that PP2A positively regulates Ras signaling at the level of Ras or Raf (Fig. 2). Such a model would differ from previously proposed models, on the basis of experiments overexpressing SV40 small t antigen in cultured mammalian cells, that suggest PP2A inhibits MEK and MAP kinase activities (Sontag et al. 1993). However, it is likely that regulation of the Ras pathway by PP2A-C is complex, involving multiple positive and negative influences on different substrates (e.g., Wassarman et al. 1996). sur-6 PP2A-B may stimulate PP2A core activity toward a specific subset of these substrates, such as KSR and/or Raf.
Consistent with the idea that sur-6 PP2A-B acts by activating a PP2A core is the observation that the sur-6(ku123) allele affects an absolutely conserved cysteine residue within a region of PP2A-B that shares similarity to c-Abl kinase domains VI–X. This region of similarity is shared by non-kinase residues of c-Abl that may be involved in target specificity, and may thus define a region of PP2A-B involved in localization or target specificity (Mayer et al. 1991). We hypothesize that in sur-6(ku123) PP2A-B mutants, the PP2A core may be mislocalized or may fail to be targeted to the proper substrates. This model would be similar to the suggestion from a recent work in mammalian cells and Xenopus embryo explants that a PR56/B′ subunit of PP2A may interact with APC and direct PP2A to dephosphorylate specific components of the Wnt/β-catenin signaling pathway (Seeling et al. 1999).
Interestingly, our data suggest that sur-6 PP2A-B may function together with ksr-1. sur-6 PP2A-B and ksr-1 have the same epistatic relationship with respect to ras and raf, and sur-6; ksr-1 double mutants resemble sur-6 or ksr-1 single mutants. Taken together, these data are consistent with the idea that sur-6 PP2A-B and ksr-1 act in a common pathway to stimulate ras-mediated signaling at a branch point that feeds out of the pathway at the level of ras or into the pathway at the level of raf (Fig. 2B).
KSR proteins positively regulate Ras signaling in C. elegans and Drosophila (Kornfeld et al. 1995; Sundaram and Han 1995; Therrien et al. 1995), as well as in Xenopus oocytes and certain mammalian cells (Therrien et al. 1996). Murine KSR associates with several proteins in vivo, including Raf, MEK, and MAP kinase, and has been proposed to function as a scaffold protein involved in signal propagation through the Raf/MEK/MAP kinase cascade (Therrien et al. 1996; Stewart et al. 1999). Murine KSR is a phosphoprotein (Cacace et al. 1999); although the role of phosphorylation in KSR regulation is unclear. Thus, KSR-1 is a potential target for regulation by PP2A during vulval induction. Alternatively, KSR-1 may act to regulate PP2A-B function.
Another potential sur-6 PP2A-B-dependent PP2A target is LIN-45 Raf. The mechanism of Raf activation is still poorly understood, but there is evidence for both inhibitory and activating phosphates on Raf (Morrison and Cutler 1997). Whereas in vitro studies suggest that PP2A can dephosphorylate Raf, it is probably not the major phosphatase to remove activating phosphates (Dent et al. 1995). However, a role for PP2A in removing inhibitory phosphates has not been ruled out. The placement of a B regulatory subunit of PP2A as a positive regulator of the Ras pathway, and the unexpected finding that it acts together with KSR-1, should lead to a better understanding of PP2A regulation and its physiological substrates.
Materials and methods
Mutants were derived from the wild-type Bristol strain, N2, and grown under standard conditions (Brenner 1974) at 20°C unless otherwise indicated. Some strains were obtained from the Caenorhabditis Genetics Center. The alleles and deficiencies used are described in Riddle et al. (1997) unless otherwise indicated: LGI, unc-29(e1072, h1), ces-1(n703), unc-13(e1091), dpy-24(s71), qDf8, qDf5; LGIII, mpk-1(ku1), unc-119(ed3); LGIV, lin-1(e1275), sur-8(ku167) (Sieburth et al. 1998), unc-24(e138), lin-45(sy96), lin-45(ku112) (Sundaram and Han 1995), let-60(n1046gf), let-60(sy130gf), dpy-20(e1282); LGV, him-5(e1490); LGX, ksr-1(ku68), ksr-1(n2526), lin-15(n765).
sur-6 isolation and cloning
sur-6(ku123) was isolated as a dominant suppressor in screens for suppressors of the Muv phenotype of let-60 ras(n1046gf) homozygotes described previously (Wu and Han 1994; Sundaram and Han 1995). sur-6(cs24) was isolated in screens for enhancers of the lin-45 raf(ku112) Vul and lethal phenotypes (M. Sundaram, unpubl.). Both alleles were obtained after ethylmethanesulfonate mutagenesis.
sur-6 was first mapped between unc-29 and dpy-24 of linkage group I by standard three-point mapping. sur-6 was further mapped with unc-29(h1) hP6 dpy-24; let-60(n1046), derived from SP1726 (gift from E. Lundquist, University of Minnesota, St. Paul, MN). Unc non-Dpy recombinants were tested for ku123 by scoring the suppression phenotype and for the polymorphism hP6 by PCR with primers Tc1-1 and hP6-B (gift from D. Fitch, New York University, NY, NY). A total of 15 of 35 Unc-29 recombinants contained ku123 and of these, one was positive for hP6, placing sur-6 to the right of cosmid C03D6. sur-6 mapped to the left of cosmid F14G10, which is the left endpoint of the complementing deficiency qDf5 (R. Ellis, pers. comm.). Deficiency qDf8 uncovers sur-6 (Tables 1 and 2). sur-6(cs24) was mapped to an interval between dpy-5 and unc-101, and within two map units of unc-13 by standard mapping crosses.
Cosmids in the region were tested for sur-6(+) activity by assaying their ability to rescue the sur-6(ku123) suppressor phenotype. Cosmids were injected at 5–10 μg/ml together with 40 μg/ml unc-119(+) transformation marker pDP#MM016 (Maduro and Pilgrim 1995) and 10 μg/ml pBluescript into sur-6(ku123); unc-119; let-60 mutants. A single cosmid, K02A11 contained sur-6(+) rescuing activity, but an overlapping cosmid, F26E4, failed to rescue.
Northern analysis of mixed-stage RNA with a genomic fragment that spanned all predicted exons of F26E4.1 as a probe revealed the presence of an abundant 2.5-kb transcript and two minor transcripts of 2.3 and 1.8 kb. Approximately 1 million plaques were screened from a λgt11-mixed stage cDNA library (gift from P. Okkema, University of Illinois, Chicago, IL) with a 1.5-kb genomic probe, and 22 positive clones were isolated. One positive clone contained a partial SL1-spiced leader corresponding to base pair 19414 of cosmid K02A11. Positive clones analyzed differed in the length of the 3′ UTR because of differential use of transcription termination sites. The sequence of these clones confirmed the exon/intron structure predicted by the C. elegans genome sequencing consortium. A transgene (pDS54) that expresses the sur-6 PP2A-B cDNA under the control of the Hsp16-41 promoter could rescue the suppressor phenotype of sur-6(ku123). The sur-6(ku123); let-60(n1046gf) double mutant was used to host the transgene. When heat-shocked, 75% of the transgenic animals were Muv (average vulval induction is 138%, n = 12), compared with 0% Muv (100% vulval induction, n = 20) for non-heat-shocked controls.
The molecular lesion associated with sur-6 mutations was identified by PCR amplifying genomic DNA from lysates of sur-6 mutants and sequencing PCR products directly.
RNAi
RNAi with double-stranded RNA was performed essentially as described (Fire et al. 1998). PCR fragments containing >1 kb of the coding regions were used as templates for in vitro transcription reactions. RNA was injected in parallel into either let-60(n1046gf) or N2 hermaphrodites at a concentration of 0.5–1 mg/ml.
The col-10 promoter was used to coexpress both sense and antisense RNA fragments in hypodermis. The sur-6 PP2A-B transgenes are pDS94 and pDS95, which contain nucleotides 90-1488 of sur-6 cloned in opposite orientations into a col-10 promoter-containing vector. The PP2A-C transgenes are pDS96 and pDS97, which contain nucleotides 241-957 of PP2A-C cloned in opposite orientations into the col-10 promoter vector. Transgenes were coinjected with the marker pTG96 (Yochem et al. 1998).
Acknowledgments
We thank Y. Han for isolating the ku123 allele, A. Goldman for isolating the cs24 allele, R. Ellis for information on deficiencies, D. Fitch for hP6 mapping primers, V. Ambros for the col-10 promoter, P. Okkema for cDNA libraries, J. Brenner and C. Duffey for assistance, and K.L. Guan and G. Kao for comments on the manuscript. We thank Alan Coulson at the Sanger Centre for cosmids, and the Caenorhabditis Genetic Center, which is funded by the National Institutes for Health (NIH) National Center for Research Resources, for some strains used in this study. D.S.S. is funded with a predoctoral Breast Cancer Research Program Fellowship from the U.S. Army Medical Research and Material Command under DAMD17-96-6117. M.H. is a Howard Hughes Medical Institute Investigator. Supported by grants from the American Cancer Society (ACS) and NIH (GM47869) to M.H., and by grants from ACS, the McCabe Foundation, and the Penn-Hughes Developmental Biology Program to M.S.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked ‘advertisement’ in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL mhan@colorado.edu; FAX (303) 492-7744.
References
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cacace AM, Michaud NR, Therrien M, Mathes K, Rubin C T, GM, Morrison DK. Identification of constitutive and ras-inducible phosphorylation sites of KSR: Implications for 14-3-3 binding, mitogen-activated protein kinase binding, and KSR overexpression. Mol Cell Biol. 1999;19:229–240. doi: 10.1128/mcb.19.1.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark SG, Chisholm AD, Horvitz RH. Control of cell fates in the central body region of C. elegans by the homeobox gene lin-39. Cell. 1993;74:43–55. doi: 10.1016/0092-8674(93)90293-y. [DOI] [PubMed] [Google Scholar]
- Dent P, Jelinek T, Morrison DK, Weber MJ, Sturgill TW. Reversal of Raf-1 activation by purified and membrane-associated protein phosphatases. Science. 1995;268:1902–1905. doi: 10.1126/science.7604263. [DOI] [PubMed] [Google Scholar]
- Dickson B, Sprenger F, Morrison D, Hafen E. Raf functions downstream of Ras1 in the Sevenless signal transduction pathway. Nature. 1992;360:600–603. doi: 10.1038/360600a0. [DOI] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Han M, Golden A, Han Y, Sternberg PW. C. elegans lin-45 raf gene participates in let-60 ras-stimulated vulval differentiation. Nature. 1993;363:133–140. doi: 10.1038/363133a0. [DOI] [PubMed] [Google Scholar]
- Horvitz HR, Sternberg PW. Multiple intercellular signaling systems control the development of the C. elegans vulva. Nature. 1991;351:535–541. doi: 10.1038/351535a0. [DOI] [PubMed] [Google Scholar]
- Jacobs D, Beitel GJ, Clark SG, Horvitz HR, Kornfeld K. Gain-of-function mutations in the Caenorhabditis elegans lin-1 ETS gene identify a C-terminal regulatory domain phosphorylated by ERK MAP kinase. Genetics. 1998;149:1809–1822. doi: 10.1093/genetics/149.4.1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz ME, McCormick F. Signal transduction from multiple Ras effectors. Curr Opin Genet Dev. 1997;7:75–79. doi: 10.1016/s0959-437x(97)80112-8. [DOI] [PubMed] [Google Scholar]
- Kornfeld K. Vulval development in Caenorhabditis elegans. Trends Genet. 1997;13:55–61. doi: 10.1016/s0168-9525(97)01005-6. [DOI] [PubMed] [Google Scholar]
- Kornfeld K, Hom DB, Horvitz HR. The ksr-1 gene encodes a novel protein kinase involved in Ras-mediated signaling in Caenorhabditis elegans. Cell. 1995;83:902–913. doi: 10.1016/0092-8674(95)90206-6. [DOI] [PubMed] [Google Scholar]
- Maduro M, Pilgrim D. Identification and cloning of unc-119, a gene expressed in the Caenorhabditis elegans nervous system. Genetics. 1995;141:977–988. doi: 10.1093/genetics/141.3.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer RE, Hendrix P, Cron P, Matthies R, Stone SR, Goris J, Merlevede W, Hofsteenge J, Hemmings BA. Structure of the 55-kDa regulatory subunit of protein phosphatase 2A: Evidence for a neuronal-specific isoform. Biochemistry. 1991;30:3589–3597. doi: 10.1021/bi00229a001. [DOI] [PubMed] [Google Scholar]
- Mayer-Jaekel RE, Hemmings BA. Protein phosphatase 2A—a ‘menage a trois’. Trends Cell Biol. 1994;4:287–291. doi: 10.1016/0962-8924(94)90219-4. [DOI] [PubMed] [Google Scholar]
- Millward TA, Zolnierowicz S, Hemmings BA. Regulation of protein kinase cascades by protein phosphatase 2A. Trends Biochem Sci. 1999;24:186–191. doi: 10.1016/s0968-0004(99)01375-4. [DOI] [PubMed] [Google Scholar]
- Morrison DK, Cutler RE. The complexity of Raf-1 regulation. Curr Opin Cell Biol. 1997;9:174–179. doi: 10.1016/s0955-0674(97)80060-9. [DOI] [PubMed] [Google Scholar]
- Riddle DL, Blumenthal T, Meyer B, Preiss JR. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1997. C. elegans II. [PubMed] [Google Scholar]
- Seeling JM, Miller JR, Gil R, Moon RT, White R, Virshup DM. Regulation of beta-catenin signaling by the B56 subunit of protein phosphatase 2A. Science. 1999;283:2089–2091. doi: 10.1126/science.283.5410.2089. [DOI] [PubMed] [Google Scholar]
- Selfors LM, Schultzman JL, Borland CZ, Stern MJ. soc-2 encodes a leucine-rich repeat protein implicated in fibroblast growth factor receptor signaling. Proc Natl Acad Sci. 1998;95:6903–6908. doi: 10.1073/pnas.95.12.6903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shenolikar S. Protein serine/threonine phosphatases-new avenues for cell regulation. Annu Rev Cell Biol. 1994;10:55–86. doi: 10.1146/annurev.cb.10.110194.000415. [DOI] [PubMed] [Google Scholar]
- Shu Y, Yan H, Hallberg E, Hallberg R. Molecular genetic analysis of Rts1p, a B′ regulatory subunit of Saccharomyces cerevisiae protein phosphatase 2A. Mol Cell Biol. 1997;17:3242–3253. doi: 10.1128/mcb.17.6.3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieburth D, Sun Q, Han M. SUR-8, a conserved Ras-binding protein with leucine-rich repeats, positively regulates Ras-mediated signaling in C. elegans. Cell. 1998;94:119–130. doi: 10.1016/s0092-8674(00)81227-1. [DOI] [PubMed] [Google Scholar]
- Sontag E, Fedorov S, Kamibayashi C, Robins D, Cobb M, Mumby M. The interaction of SV40 small tumor anitgen with protein phosphatase 2A stimulates the Map kinase pathway and induces cell proliferation. Cell. 1993;75:887–897. doi: 10.1016/0092-8674(93)90533-v. [DOI] [PubMed] [Google Scholar]
- Sternberg PW, Han M. Genetics of Ras signaling in C. elegans. Trends Genet. 1998;14:466–472. doi: 10.1016/s0168-9525(98)01592-3. [DOI] [PubMed] [Google Scholar]
- Stewart S, Sundaram M, Zhang Y, Lee J, Han M, Guan K-L. Kinase Suppressor of Ras (KSR) forms a multi-protein signaling complex and modulates MEK localization. Mol Cell Biol. 1999;19:5523–5534. doi: 10.1128/mcb.19.8.5523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaram M, Han M. The C. elegans ksr-1 gene encodes a novel Raf-related kinase involved in Ras-mediated signal transduction. Cell. 1995;83:889–901. doi: 10.1016/0092-8674(95)90205-8. [DOI] [PubMed] [Google Scholar]
- Tan PB, Lackner MR, Kim SK. MAP kinase signaling specificity mediated by the LIN-1 Ets/LIN-31 WH transcription factor complex during C. elegans vulval induction. Cell. 1998;93:569–580. doi: 10.1016/s0092-8674(00)81186-1. [DOI] [PubMed] [Google Scholar]
- Therrien M, Chang HC, Solomon NM, Karim FD, Wassarman DA, Rubin GM. KSR, a novel protein kinase required for Ras signal transduction. Cell. 1995;83:879–887. doi: 10.1016/0092-8674(95)90204-x. [DOI] [PubMed] [Google Scholar]
- Therrien M, Michaud NR, Rubin GM, Morrison DK. KSR modulates signal propagation within the MAPK cascade. Genes & Dev. 1996;10:2684–2695. doi: 10.1101/gad.10.21.2684. [DOI] [PubMed] [Google Scholar]
- Uemura T, Shiomi K, Togashi S, Takeichi M. Mutation of twins encoding a regulator of protein phosphatase 2A leads to pattern duplication in Drosophila imaginal disks. Genes & Dev. 1993;7:429–440. doi: 10.1101/gad.7.3.429. [DOI] [PubMed] [Google Scholar]
- Wassarman DA, Solomon NM, Chang HC, Karim FD, Therrien M, Rubin GM. Protein phosphatase 2A positively and negatively regulates Ras-mediated photoreceptor development in Drosophila. Genes & Dev. 1996;10:272–278. doi: 10.1101/gad.10.3.272. [DOI] [PubMed] [Google Scholar]
- Westphal RS, Anderson KA, Means AR, Waszinski BE. A signaling complex of Ca2+-calmodulin-dependent protein kinase IV and protein phosphatase 2A. Science. 1998;280:1258–1261. doi: 10.1126/science.280.5367.1258. [DOI] [PubMed] [Google Scholar]
- Wu Y, Han M. Suppression of activated Let-60 Ras protein defines a role of Caenorhabditis elegans Sur-1 MAP kinase in vulval differentiation. Genes & Dev. 1994;8:147–159. doi: 10.1101/gad.8.2.147. [DOI] [PubMed] [Google Scholar]
- Yochem J, Gu T, Han M. A new marker for mosaic analysis of Caenorhabditis elegans indicates a fusion between hyp6 and hyp7, two major components of the hypodermis. Genetics. 1998;149:1323–1334. doi: 10.1093/genetics/149.3.1323. [DOI] [PMC free article] [PubMed] [Google Scholar]