Abstract
Using methods that conserve nuclear architecture, we have reanalyzed the spatial organization of the initiation of mammalian DNA synthesis. Contrary to the commonly held view that replication begins at hundreds of dispersed nuclear sites, primary fibroblasts initiate synthesis in a limited number of foci that contain replication proteins, surround the nucleolus, and overlap with previously identified internal lamin A/C structures. These foci are established in early G1-phase and also contain members of the retinoblastoma protein family. Later, in S-phase, DNA replication sites distribute to regions located throughout the nucleus. As this progression occurs, association with the lamin structure and pRB family members is lost. A similar temporal progression is found in all the primary cells we have examined but not in most established cell lines, indicating that the immortalization process modifies spatial control of DNA replication. These findings indicate that in normal mammalian cells, the onset of DNA synthesis is coordinately regulated at a small number of previously unrecognized perinucleolar sites that are selected in early G1-phase.
Keywords: DNA replication, nuclear structure, lamin, perinucleolar foci, retinoblastoma protein
Several previous studies have examined the spatial and temporal patterns of DNA synthesis that occur during S-phase progression. A general consensus has emerged from this work suggesting that DNA synthesis initiates in numerous foci distributed throughout the nucleus (Nakamura et al. 1986; Nakayasu and Berezney 1989; Ma et al. 1998). Later in S-phase, a transition occurs and replication sites are found to cluster in regions of the nucleus that contain heterochromatin (Nakayasu and Berezney 1989). An important suggestion from these studies is that the nuclear organization of DNA replication is uniform in all mammalian cells (Nakamura et al. 1986; Nakayasu and Berezney 1989; van Dierendonck et al. 1989; O'Keefe et al. 1992; Rizzoli et al. 1992; Hassan and Cook 1993; Ma et al. 1998; Berezney et al. 2000). A further inference based on these findings is that the spatial organization of DNA replication is tightly controlled and thus unaffected by physiologic settings or by genetic changes seen in immortalization or transformation.
Very little is known about how the sites of DNA replication are established. Although many of the components of the DNA replication machinery have been identified, the proteins that mediate spatial control of DNA replication remain unknown. It also remains unclear how replication proteins are established in a particular pattern or how shifts from one pattern to another are regulated. Complicating this issue further, the sequence properties of a mammalian origin of replication have not been identified.
Numerous studies have suggested that there must be an organizing structure such as the nuclear matrix that helps coordinate spatial regulation of DNA synthesis (Dijkwel et al. 1979; Berezney and Buchholtz 1981; Hunt and Vogelstein 1981; Carri et al. 1986; Collins and Chu 1987; Vaughn et al. 1990; Neri et al. 1992; Hozak et al. 1993). Components of the nuclear substructure that have been implicated in establishing the pattern of DNA replication sites are the nuclear lamins. Nuclear lamins, which are intermediate filament proteins that form lattices around the nuclear envelope (Gerace et al. 1978) and also localize to regions in the nuclear interior (Bridger et al. 1993), have been implicated in the organization of a number of nuclear processes (Brylawski et al. 2000; Moir et al. 2000b; Wilson 2000). In Xenopus laevis egg extracts, disruption of lamin structure by introduction of dominant negative versions of the protein delays or impairs the initiation of DNA synthesis (Ellis et al. 1997; Spann et al. 1997; Moir et al. 2000a), indicating that these proteins may be important in organizing DNA replication. Moreover, lamin B has been found to localize to late S-phase sites of DNA synthesis in immortalized mouse cells (Moir et al. 1994). To date, lamins have not been found at early S-phase replication sites, however, and it remains unclear whether they play a direct role in the organization of replication sites.
DNA replication patterns have been observed generally using one of two methods. In the most common approach, cells are pulsed with the nucleotide analog bromodeoxyuridine (BrdU; Nakamura et al. 1986; Nakayasu and Berezney 1989). After fixation, sites of incorporation are determined by indirect immunofluorescence using antibodies specific for the analog. Importantly, these antibodies are unable to recognize BrdU incorporated into double-stranded DNA, and consequently, DNA must be denatured before immunofluorescence to expose single-stranded regions. Various denaturation methods have been used, but the consequences of these methods have not been carefully compared. In a second approach to study sites of DNA replication, cells are first permeabilized, then replication is allowed to proceed in the presence of biotin-labeled nucleotides (Nakayasu and Berezney 1989; Kill et al. 1991). Incorporation sites are later detected using avidin-bound fluorescent probes. Again, the changes in nuclear structure that might occur during permeabilization have not been clearly established.
Another common feature of the studies of DNA replication patterns has been the use of immortalized or transformed cell lines such as mouse 3T3 (Ma et al. 1998), hamster CHO (O'Keefe et al. 1992), or human HeLa cells (Hassan and Cook 1993; Jackson and Pombo 1998). We chose to reexamine replication patterns during S-phase progression in primary cells, as mutations involved in cell immortalization might alter patterns of DNA synthesis. Using BrdU-labeling and DNA denaturation methods that preserve nuclear structure, we find that primary cells initiate replication in a limited number of foci (five to 20) that surround the nucleolus. These sites contain proteins that are known to be involved in DNA replication, as well as members of the retinoblastoma (pRB) protein family, and are located within intranuclear lamin A/C foci. These replication structures are apparent in G1 and become dissociated as cells progress through mid-S-phase. In contrast, immortalized cells initiate replication in a more dispersed pattern with sites distributed throughout the nucleus. These findings challenge the notion that replication initiates in sites distributed throughout the nucleus and contradict the theory that replication patterns are invariant among different cells. Rather, we propose that mutations arising during the process of immortalization lead to changes in the nuclear organization of DNA replication.
Results
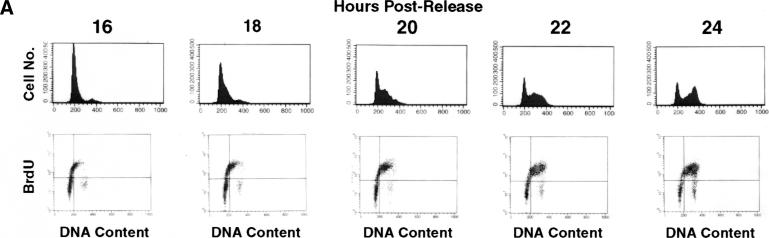
Human diploid fibroblasts (WI38) were arrested in G1-phase by contact inhibition and then replated at low density to achieve synchronous entry into S-phase (Figs. 1 and 2). Under these conditions, we find that up to 60% of cells move synchronously through S-phase (Fig. 2A). The remaining cells either stay in G0 or are significantly delayed in their entry into S-phase. At hourly intervals after release, cells were exposed to BrdU for 10-min pulses, and immunofluorescence was performed to determine sites of DNA synthesis using low concentrations of DNaseI to expose the BrdU epitopes (Krude 1995). To determine the relative frequency of each observed replication pattern, 100 cells from each time point were counted using standard immunofluorescence microscopy (Fig. 2B). Images throughout the report are generated using CCD and deconvolution microscopy (Fig. 1); however, unprocessed images generated using standard microscopy show identical patterns (Fig. 2C), and all quantitative data presented in the manuscript were generated by counting cells using standard microscopy.
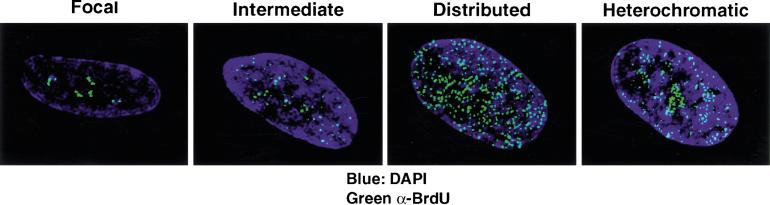
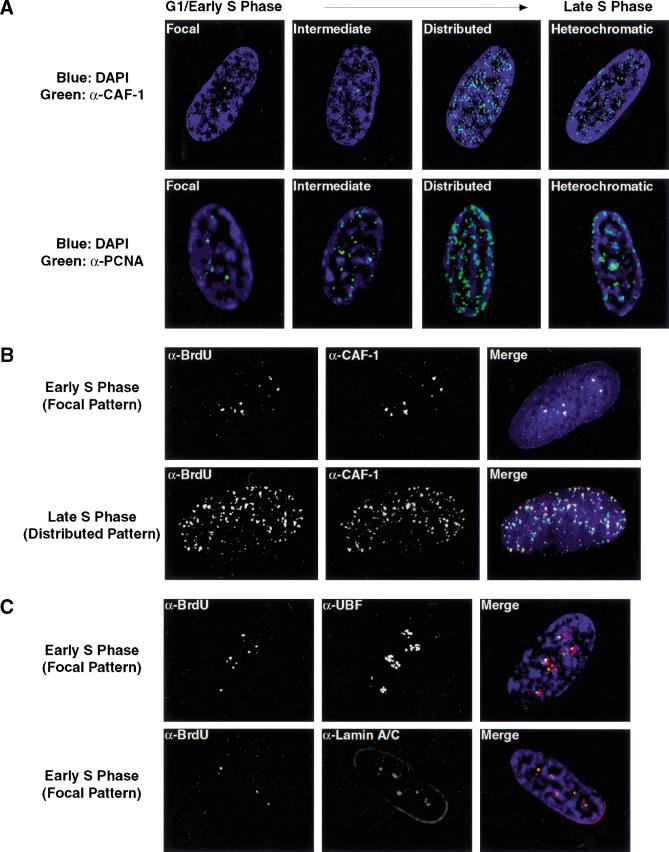
Figure 1.
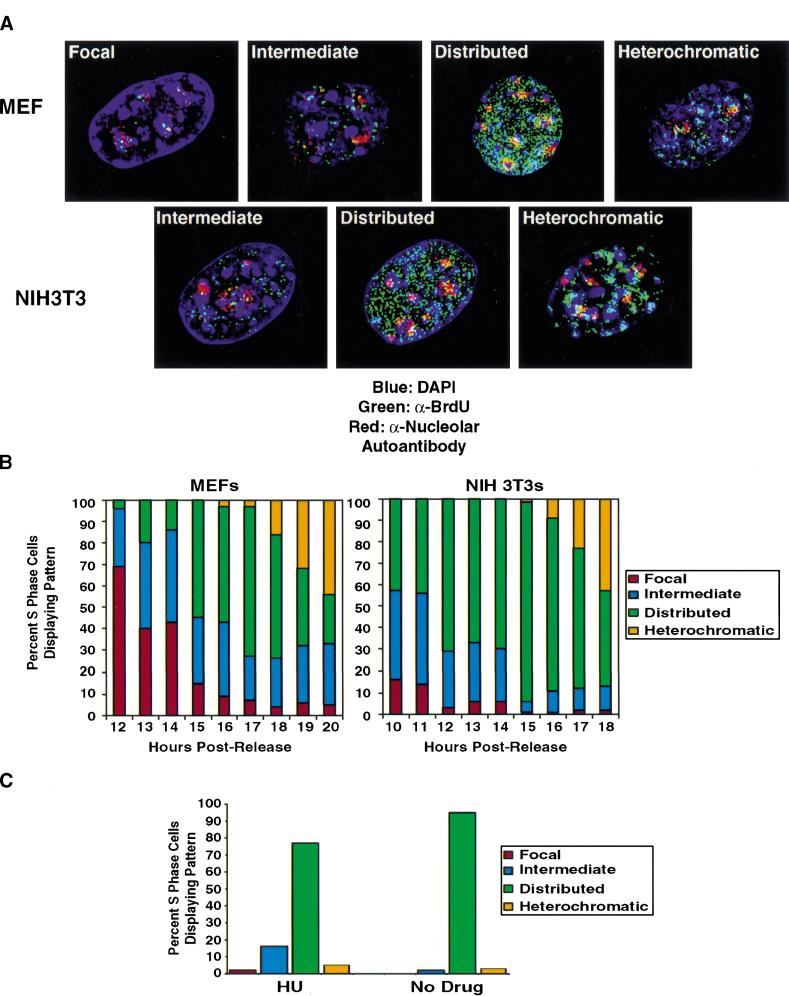
Images of different replication patterns during S-phase progression. Panels from left to right reflect the progression of replication patterns during S-phase progression. Sites of BrdU incorporation were detected by an anti-BrdU antibody containing DNaseI (green, Amersham) and overlaid on DAPI staining of the nucleus (blue). The pattern labeled intermediate likely reflects a transition phase between focal and distributed patterns. All processed images were recompiled from images on several sections throughout the nucleus and, thus, reflect the staining pattern of the entire nucleus compressed into a two-dimensional image. The intensity of individual foci in the focal pattern is stronger than in the distributed pattern, although this is not apparent as a result of the imaging process.
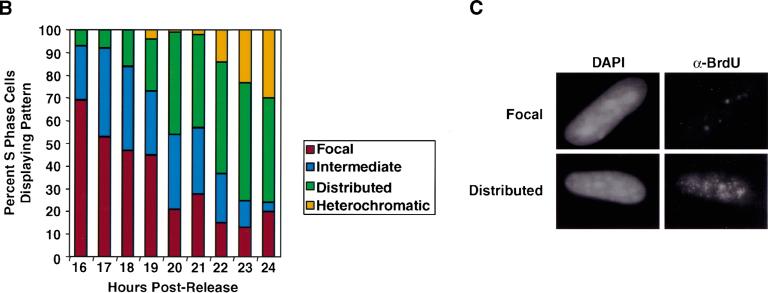
Figure 2.
DNA replication patterns in primary cells during S-phase progression. (A) FACS analysis was performed on WI38 cells released from contact inhibition. Upper panels plot cell number versus DNA content. Lower panels plot BrdU versus DNA content. Cells enter S-phase at ∼16 h postrelease and are in late S-phase by 24 h. (B) Quantitation of different replication patterns during S-phase progression. Only cells actively incorporating BrdU were included. (D) Images depict sites of BrdU incorporation either in the focal pattern (top) or the distributed pattern (bottom). These images were generated using standard immunofluorescence microscopy, which was used to generate all quantitative data.
At the onset of S-phase, the vast majority of cells exhibit DNA replication in only five to 20 foci (Figs. 1,2C). This focal replication pattern predominates for the first 2–3 h of S-phase, indicating that focal replication occurs for a significant fraction of the S-phase window in primary diploid fibroblasts. As S-phase progresses, replication sites distribute throughout the nucleus in several hundred foci. The relative intensity of one spot from a cell exhibiting the early S-phase focal pattern is higher than that of a spot from a cell exhibiting the distributed pattern. This observation is not reflected in processed images, where levels are not comparable from one image to another. By the end of S-phase, replication is observed in regions of the nucleus containing heterochromatin, consistent with previous reports (Nakayasu and Berezney 1989).
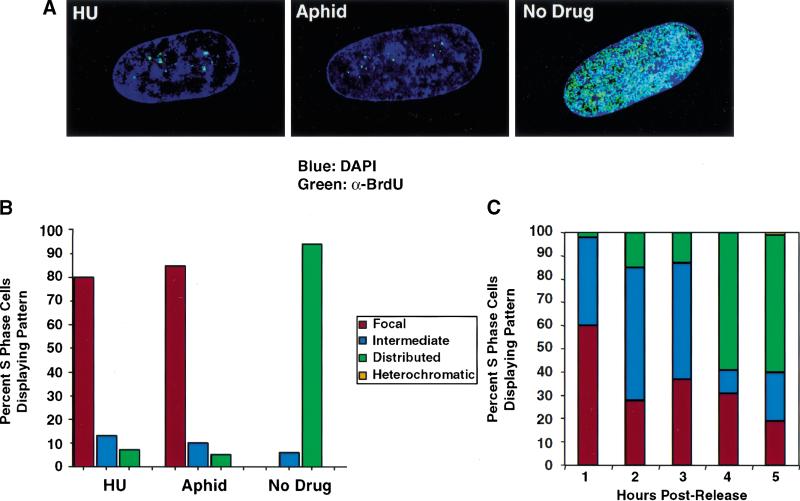
To confirm the observation that WI38 cells initiate DNA synthesis in a few foci, we treated G1 cells with either hydroxyurea or aphidicolin at concentrations known to generate arrest in early S-phase.Cells were released from arrest by contact inhibition into media in the presence of BrdU and containing hydroxyurea, aphidicolin, or no drug. After 24 h, cells were fixed for immunofluorescence. Cells that are not exposed to either drug do exhibit labeling throughout the nucleus. However, those arrested in early S-phase by either drug incorporate BrdU specifically in the focal pattern (Fig. 3A,B). In further experiments, cells were arrested with hydroxyurea and then allowed to progress synchronously through S-phase by washing out the drug. As they transgress S-phase, they exhibit a sequence of replication patterns identical to that observed with cells synchronized by contact inhibition (Fig. 3C).
Figure 3.
Synchronization in early S-phase by hydroxyurea or aphidicolin results in the focal replication pattern. (A) WI-38 cells were released from contact inhibition into media containing BrdU and either 1.5 mM hydroxyurea, 2 μg/mL aphidicolin, or no drug. Twenty-four hours after release, cells were fixed, and immunofluorescence was performed. BrdU was detected by an anti-BrdU antibody containing DNaseI (Amersham, green). When cells are arrested in early S-phase by either hydroxyurea or aphidicolin, incorporation of BrdU is found only in perinucleolar foci. In contrast, when cells are allowed to progress through S-phase in the absence of drug, BrdU is incorporated throughout the nucleus. (B) Quantitation of patterns, comparing cells treated either with hydroxyurea, aphidicolin, or no drug. (C) WI-38 cells were released from contact inhibition into media containing 1.5 mM hydroxyurea. Twenty-four hours after release, the cells were washed in PBS and were restimulated into S-phase by replacement with drug-free media. Cells were pulse-labeled with BrdU for 10 min at hourly intervals after release. At 1 h, a majority of cells continue to display focal replication patterns. By 3–4 h after release, significant distribution of replication sites has occurred.
In experiments using other methods of synchronization including serum starvation and nocodazole arrest, we find that cells exhibit the same sequence of replication patterns as they proceed through S-phase (data not shown). These results suggest that the appearance of perinucleolar foci is independent of the method of synchronization. Given that these findings do not agree with the generally held view of how DNA replication is organized in mammalian cells, we went on to characterize these structures and compare our results with those of previous studies.
Replication proteins and nuclear lamins also localize to the early S-phase sites of DNA synthesis
If sites of BrdU incorporation and detection accurately reflect DNA synthesis in primary cells, then replication proteins should exhibit similar patterns of localization during S-phase progression. Importantly, identification of these proteins by immunofluorescence does not require denaturation of DNA, so no artifacts could be introduced by the methods that are needed to expose the BrdU epitopes. We examined the localization of p150 (CAF-1; Smith and Stillman 1989; Kaufman et al. 1995) and PCNA (Bravo et al. 1987; Prelich et al. 1987) across the cell cycle in WI-38 cells, as both proteins have previously been shown to localize to sites of active DNA synthesis (Kill et al. 1991; Krude 1995). Both CAF-1 and PCNA are found in a limited number of foci in G1- and early S-phase of the cell cycle (Fig. 4A). As expected, we detect an increase in the intensity of PCNA staining concurrent with entry into S-phase (Bravo 1986). Later in S-phase, coincident with the distribution of BrdU incorporation sites, PCNA and CAF-1 relocalize to hundreds of foci throughout the nucleus.
Figure 4.
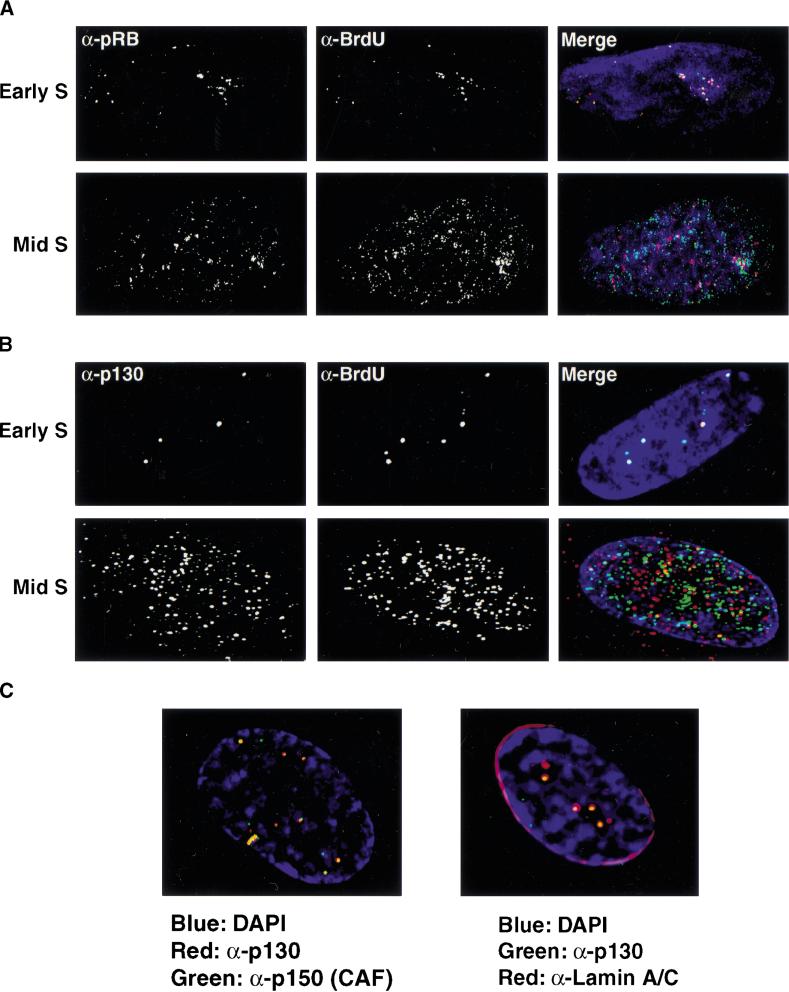
BrdU foci contain replication proteins and are associated with internal nuclear lamin A/C structures. (A) The localization of p150 (CAF-1; upper panels, green) and PCNA (lower panels, green) was determined during G1- and S-phase progression. At the G1-S boundary, both CAF-1 and PCNA foci increase in intensity. The mouse monoclonal antibody against p150 (MAB1) was kindly provided by B. Stillman. Similar results were seen with antibodies to the p60 subunit of CAF-1. PCNA was visualized with a rabbit polyclonal antibody (Santa Cruz). (B) Localization of BrdU was compared to that of p150 (red) in S-phase cells. DNaseI was used to expose BrdU. For this experiment, replication sites were detected using a rat anti-BrdU antibody (Harlan/Sera-Lab, green). (C) Early S-phase BrdU patterns were compared to that of UBF (upper panels, red), a nucleolar transcription factor, and nuclear lamins A/C (lower panels, red). The rabbit polyclonal anti-UBF antibody was kindly provided by L. Rothblum. Lamins A/C were visualized with a mouse monoclonal antibody (636, Santa Cruz). In all images, DNA is visualized by DAPI staining (blue).
In a separate experiment, we directly compared the localization of replication proteins with sites of BrdU incorporation. CAF-1 and PCNA colocalize with BrdU in both the focal early S-phase and distributed late S-phase replication patterns (Fig. 4B; data not shown). It is significant to note that the DNaseI conditions used to expose BrdU give identical patterns to those without nuclease and do not alter CAF-1 or PCNA localization (Fig. 4, cf. A and B).
As the early S-phase focal replication sites are found in distinct regions of the nucleus, we examined their localization relative to other defined nuclear structures. The foci do not align with coiled bodies, SC-35, PML, centromeres, telomeres, or α-satellite DNA (data not shown). They do, however, cluster in regions bordering the nucleolus (Fig. 4C). Strikingly, they also overlay internal nuclear lamin A/C structures, which have been previously shown to appear in G1 and early S-phase (Bridger et al. 1993). Later in S-phase, the distributed replication sites do not align with either the perinucleolar compartment or the internal lamins. These findings suggest that lamin A/C may be important for the establishment of early S-phase replication structures.
A comparison of methods used to detect sites of DNA synthesis
Our results indicate that replication initiates in a limited number of perinucleolar foci at the beginning of S-phase. This finding differs dramatically from those presented in previous reports, where replication is observed to begin in a dispersed pattern throughout the nucleus (Nakamura et al. 1986; Nakayasu and Berezney 1989; van Dierendonck et al. 1989; O'Keefe et al. 1992; Rizzoli et al. 1992; Hassan and Cook 1993; Ma et al. 1998; Berezney et al. 2000). By comparing our methods to those utilized in previous reports, we sought to identify the reasons for this discrepancy. We find that the identification of BrdU incorporation sites is not affected by the source of BrdU, the choice of anti-BrdU antibody, or the method of microscopy; standard immunofluorescence, confocal, and deconvolution microscopy all lead to identical patterns. However, there are two principal differences that account for the disparate results: the method of DNA denaturation and the cell type examined (discussed below).
For exposure of BrdU epitopes we have used DNaseI to generate regions of single-stranded DNA for detection with anti-BrdU antibodies (Krude 1995). In contrast, most previous studies denature DNA by treatment with 1–4N HCl generally for a period of 30 min (Nakamura et al. 1986; Nakayasu and Berezney 1989; van Dierendonck et al. 1989; O'Keefe et al. 1992; Rizzoli et al. 1992; Ma et al. 1998). We find that when HCl was used instead of DNaseI to expose the BrdU epitopes, sites of DNA synthesis appear distributed throughout the nucleus in early S-phase as previously reported (see Fig. 7A, below).
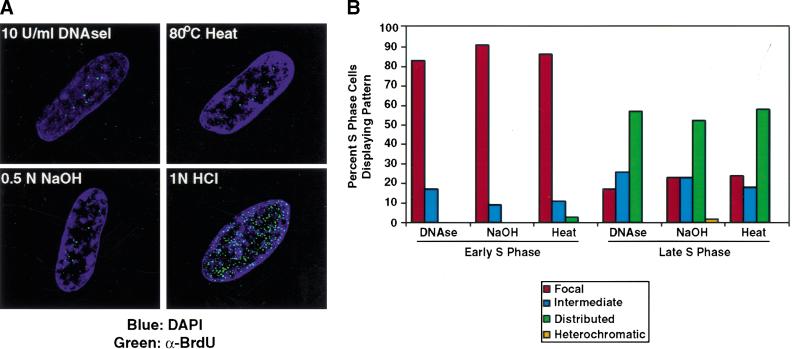
Figure 7.
Early S-phase replication appears focal using other methods to denature DNA. (A) Early S-phase BrdU incorporation sites (green) were compared using different methods to denature DNA. Using either heat exposure or base denaturation, early S-phase replication patterns appear focal. (B) Quantitation of replication patterns. Later in S-phase, denaturation by NaOH or heat yields a distributed pattern similar to that generated by DNaseI.
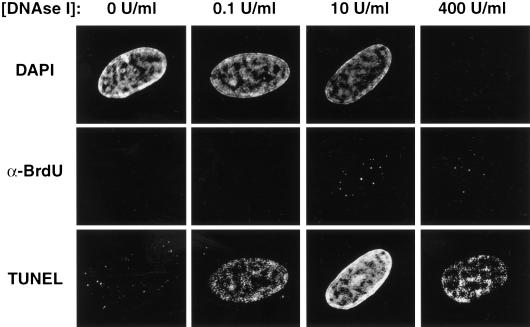
As denaturation of DNA by HCl yields different results than exposure to nuclease, we performed a series of experiments to determine which method permits the observation of authentic sites of DNA synthesis. One possibility was that the nuclear accessibility of DNaseI is nonuniform, such that perinucleolar BrdU epitopes are preferentially exposed early in S-phase. To test this possibility, we performed DAPI, BrdU, and TUNEL staining of early S-phase cells exposed to a variety of DNaseI concentrations. TUNEL staining, which recognizes free 3′ DNA ends, coincides with sites of nuclease activity in this assay. At the DNaseI concentration typically used (10 U/mL), BrdU incorporation patterns are focal, yet TUNEL staining occurs throughout the nucleus and is strongest in regions of highest DNA content (as judged by DAPI intensity; Fig. 5). This suggests that DNA is uniformly available for nuclease attack and indicates that many regions of the genome are not targeted by anti-BrdU antibodies even though they have single-stranded, accessible DNA. When DNaseI levels are increased 40-fold, perinucleolar replication patterns remain evident. Under these conditions, the DAPI signal is nearly absent, indicating a drastic reduction in double-stranded DNA needed for the intercalation of the dye and demonstrating that the nuclease has encountered nearly all of the nuclear DNA. DNA is still present at this concentration, as indicated by positive TUNEL staining. Finally, at the minimal DNaseI concentration required to generate a TUNEL signal (0.1 U/mL), the staining still does not favor perinucleolar regions. Therefore, we conclude that the DNaseI treatment used to expose BrdU does not specifically target perinucleolar regions.
Figure 5.
The early S-phase focal replication pattern does not result from differential access of DNaseI. Each column reflects a single cell stained for DAPI (upper panels), BrdU incorporation (middle panels), or TUNEL (lower panels, Intergen) at the indicated DNaseI concentration. Cells were treated with DNaseI (Sigma) and stained with an anti-BrdU antibody (Becton Dickinson). After secondary antibody application, cells were processed for TUNEL staining.
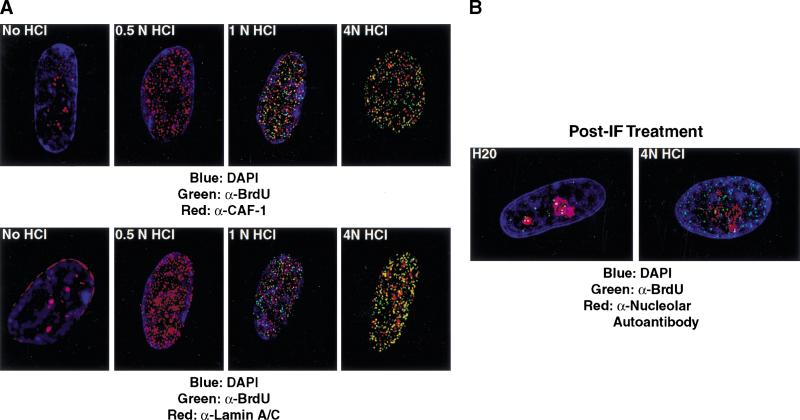
We next examined the effects of acid treatment on nuclear structure. It has been previously recognized that protein localization can be perturbed by HCl treatment (Kill et al. 1991). We tested this directly with antibodies to replication proteins and to markers of prominent nuclear structures. As shown above (Fig. 4A), the normal pattern of localization for CAF-1 and PCNA in early S-phase primary cells is perinucleolar foci, and these structures are unaffected by DNaseI treatment (Fig. 4B; data not shown). With increasing acid concentration, however, we observe a redistribution of epitopes for these antibodies such that the proteins appear dispersed throughout the nucleus in early S-phase cells (Fig. 6A, upper panels; data not shown). These results suggest that acid treatment either exposed new epitopes for these antibodies or that the CAF-1 and PCNA proteins relocalize following acid treatment. On the basis of the following experiments, we believe that the latter is occurring. First, staining for lamins A/C also becomes dispersed in response to acid treatment, suggesting that the nuclear framework may be disordered (Fig. 6A, lower panels). Second, dispersed CAF-1 and PCNA granules do not coincide well with the acid-generated BrdU pattern (Fig. 6A, data not shown). Thus untreated or DNaseI-treated nuclei show colocalization of between-replication proteins and sites of BrdU incorporation, whereas acid-treated nuclei show little or no overlap. Consistent with these findings, one previous report has suggested that nucleolar structures are highly sensitive to HCl treatment even after paraformaldehyde fixation (Smetana et al. 1998).
Figure 6.
HCl treatment is disruptive to nuclear structure. (A) Top panels compare sites of BrdU incorporation (Harlan/Sera-Lab, green) to localization p150 (CAF-1; red) with increasing acid concentration. No DNaseI is used in this experiment. At concentrations <1 N HCl, BrdU is not sufficiently exposed for detection. Lower panels depict nuclear lamins A/C localization (red) in increasing HCl concentration. (B) Cells were stained for BrdU with DNaseI and then exposed to HCl. BrdU incorporation sites (green, Becton Dickinson) are compared to the nucleolus as defined by staining with a human nucleolar autoantibody (red, Sigma; C).
Third, we tested directly whether chromosomal sites of recent replication can be relocalized by acid treatment. Cells were first treated with DNaseI and anti-BrdU antibodies and then exposed them to 4 N HCl for a period of 10 sec just before microscopic examination. When cells are treated with water as a control, the perinucleolar pattern remains evident (Fig. 6B). In HCl-exposed cells, however, BrdU staining is redistributed throughout the nucleus. From these experiments, we conclude that treatment of cells with HCl, even after fixation with paraformaldehyde, causes destruction of many nuclear structures and can lead to redistribution of recent sites of DNA synthesis. We believe that this technical difference explains the variation between our results and those arrived at in previous studies.
To confirm further the authenticity of our findings, we used either heat (80°C, 10 min in glycerol) or base (0.05 N NaOH) to denature DNA before treatment with primary antibody. Similar to treatment with DNaseI, denaturation of DNA by these methods in early S-phase cells results in perinucleolar replication foci and an intact nuclear structure (Fig. 7; data not shown). Taken together, these findings indicate that DNA replication is initiated in perinucleolar foci in primary cells.
Patterns of DNA synthesis are changed by immortalization of cells in culture
We next examined the conclusion, drawn largely from studies of acid-treated nuclei, that DNA replication patterns are invariant between different cells. Our attention centered on the analysis of immortalized cell lines in part because this is the experimental system most frequently used in previous studies to examine DNA patterns of replication. We compared sites of BrdU incorporation using DNaseI treatment in primary mouse embryo fibroblasts (MEFs) and immortalized mouse fibroblasts (NIH3T3). NIH3T3 cells are shown here, but we have also examined a number of other 3T3 lines with similar results. Again, cells were synchronized in G1 by contact inhibition and then released into the cell cycle by replating at low density. As expected, NIH3T3 cells entered S-phase ∼2 h earlier than primary MEFs as judged by FACS analysis (data not shown). When cells from the early stages of S-phase were compared, different patterns were seen. MEFs initiate DNA synthesis in perinucleolar foci, consistent with the results obtained from primary human cells (Fig. 8A). However, NIH3T3 cells exhibit a more distributed pattern of DNA synthesis on S-phase entry.
Figure 8.
Immortalization alters the nuclear organization of DNA replication in early S-phase. (A) DNA replication patterns in MEFs during S-phase progression. BrdU incorporation sites (green, Becton Dickinson) are compared to the nucleolus as defined by staining with a human nucleolar autoantibody (red, Sigma). Ten units per milliliter DNaseI was used to visualize BrdU. (B) Quantitation of replication patterns for MEFs and NIH3T3 cells. FACS analysis (data not shown) demonstrated that NIH3T3 cells entered S-phase at 10 h, whereas MEFs entered at 12 h postrelease from contact inhibition. (C) Replication patterns in NIH3T3 cells arrested in early S-phase by hydroxyurea.
To compare carefully the kinetics of pattern change throughout S-phase, 100 cells from each time point were counted to determine the most prominent replication pattern. The patterns of change in the MEFs were analogous to the changes we had seen earlier in human WI38 cells (cf. Fig. 2B with Fig. 8B). DNA synthesis begins in the perinucleolar pattern and by mid-S-phase the distributed patterns become more prominent. The 3T3 cells show more distributed patterns at the beginning of S-phase, and no focal stage is seen. As the later patterns of DNA synthesis are indistinguishable in both settings and resemble what is seen in human cells, we hypothesize that 3T3 cells have just skipped the early S-phase stage and begun DNA synthesis using more distributed patterns. To confirm this finding, we synchronized NIH3T3 cells in G1 by contact inhibition and then released them synchronously into the cycle by replating at low density with BrdU in the presence or absence of hydroxyurea (Fig. 8C). BrdU incorporation appears intermediate or distributed in these early S-phase-arrested cells, confirming that DNA synthesis initiates in a more distributed pattern in these cells.
When analysis of sites of DNA synthesis was extended to other primary fibroblasts and keratinocytes, as well as other immortalized cells, we found similar results; primary cells initiate DNA synthesis in perinucleolar foci, whereas, immortalized cells initiated in a more distributed nuclear pattern (data not shown). This observation is reflected by a comparative analysis of replication patterns in asynchronously cell culture (Table 1). A significant fraction of primary cells in culture (WI38, IMR90, MRC5, and MEFs) undergo replication in the focal pattern, whereas this pattern is not observed in cells from immortalized lines of various origin (U2OS, NIH3T3, and 10T1/2). We conclude that patterns of DNA synthesis are not invariant; rather, the immortalization process, presumably driven by mutations that promote continued proliferation in culture, leads to changes in the nuclear organization of DNA replication. These results indicate that initiation of DNA synthesis in the perinucleolar foci is a regulated event that depends on either the direct or the indirect action of proteins controlling mammalian proliferation.
Table 1.
Replication patterns in asynchronous primary cells and established cell lines
| Pattern
|
Focal
|
Intermediate
|
Distributed
|
Heterochromatic
|
|
|---|---|---|---|---|---|
| Primary cells | WI38 | 23 | 46 | 25 | 6 |
| IMR90 | 21 | 36 | 35 | 8 | |
| MRC5 | 20 | 49 | 24 | 7 | |
| MEFs | 15 | 55 | 23 | 7 | |
| Cell lines | NIH3T3 | 3 | 24 | 65 | 8 |
| 10T 1/2 | 2 | 18 | 70 | 10 | |
| U2OS | 1 | 18 | 75 | 6 | |
Asynchronous populations of cells at low density were pulsed with BrdU and analyzed for replication patterns. One hundred BrdU positive cells were scored in all cases.
Early S-phase perinucleolar replication foci contain retinoblastoma protein family members
During the course of these experiments, we examined the localization pattern of pRB, as well as the two other members of this family of proteins (p107 and p130). We find that pRB, p107, and p130 localize to perinucleolar foci in G1- and early S-phase and that they overlap with the intranuclear lamin structures (data not shown). These foci appeared remarkably similar to the perinucleolar replication foci described above. Therefore, we directly compared the localization of pRB family members to sites of DNA synthesis in both early and late S-phase. In early S-phase, we observe strong overlap between the two patterns indicating that pRB family members are associated with early S-phase replication sites (Fig. 9A,B). In contrast, the distributed replication pattern seen later in S-phase does not coincide well with the localization pattern of pRB, even though pRB has become more distributed in the nucleus at this point as well. p130 displays a nuclear and cytoplasmic distribution in late S-phase and also does not coincide with distributed sites of DNA synthesis. From these findings, we conclude that pRB family members are associated specifically with early S-phase DNA replication in primary cells.
Figure 9.
pRB family members localize to early S-phase sites of DNA synthesis. Synchronized cells were pulsed with BrdU in either early S-phase (upper panels) or mid-to-late S-phase (lower panels). Sites of DNA synthesis were determined in by indirect immunofluorescence (red) and compared to the localization pattern of pRB (green; A) or p130 (green; B). Overlapping signals appear yellow. (C) The localization patterns of replication proteins were compared to those of pRB family members in cells arrested in G1 by contact inhibition. Left panel: p130 (red), p150 (CAF-1; green). Right panel: p130 (green), Lamin A/C (red).
The localization of pRB family members to perinucleolar, lamin-associated foci is apparent in cells arrested by contact inhibition in G1 (Fig. 9C, right panel). We therefore tested whether replication proteins are targeted to these same sites in early G1. We examined p150 (CAF-1) localization in cells arrested by contact inhibition. p150 localizes to perinucleolar foci in contact-inhibited cells and throughout the G1-phase of the cell cycle and colocalizes with p130 during this window of the cell cycle (Fig. 9C, left panel). Similar results were observed with antibodies to PCNA (data not shown). This indicates that initial sites of replication are established in G1 and that pRB family members have access to these sites throughout G1 when they are thought to be active. These observations show that in primary human cells there is spatial coordination between the organization of the G1 nucleus and S-phase initiation. The results also raise the possibility that late G1 regulatory events may be physically linked to the initiation of DNA synthesis.
Discussion
Our results suggest several changes in the currently held view of spatial organization of replication in mammalian cells. First, we report that in normal primary fibroblasts grown in culture, DNA synthesis initiates in a relatively small number of distinct foci that surround the nucleolus. We do not yet understand how these foci are structured or how they are established. However, they do coincide with previously identified but still poorly characterized intranuclear lamin A/C structures (Bridger et al. 1993), and it is intriguing to speculate that the formation of these foci may be linked to these nuclear structural elements. Second, we find that the use of these structures for DNA synthesis is transient. By mid-S-phase, DNA replication moves to a large number of dispersed sites that exhibit less intense BrdU staining. Third, we report that the use of the perinucleolar sites is not mandatory but, instead, regulated. Replication patterns have been widely believed to be invariant in all cell lines and, thus, not changed by physiological events or mutations. Our findings invalidate this view. The immortalized cell lines that we have tested initiate replication in sites more distributed throughout the nucleus. Thus, the process of immortalization can induce changes in the spatial control of mammalian DNA replication.
Constitution of the perinucleolar replication foci
The intensity of the early S-phase replication foci is much higher than that of the later distributed sites, implying that the rate of DNA synthesis is higher in the perinucleolar foci. One possibility is that the bright, perinucleolar replication sites contain the same number of bidirectional forks as the comparatively dimmer distributed foci but that the rate of elongation is much higher in the focal sites. This could, in theory, be caused by a concentration of replication factors or an increased availability of free nucleotides. However, given that hundreds of independent foci become apparent after HCl treatment of early S-phase cells, we favor a model whereby each focus represents a large number of clustered origins. If so, these could be tandem origins on the same chromosome or origins from different regions throughout the genome. At present, this question cannot be addressed given that the regions of the genome that replicate first have not yet been identified.
In addition to the nuclear periphery, lamin A/C has been reported to concentrate in intranuclear foci (Bridger et al. 1993). Its function at these sites has remained mysterious. The observation that replication initiates at sites embedded within the lamin A/C structures suggests that the lamins may have an organizing role for early S-phase replication sites. Disruption of lamin structures has been shown to impair replication initiation in Xenopus egg extracts (Ellis et al. 1997; Spann et al. 1997; Moir et al. 2000a). Moreover, B-type lamins have been reported to localize to late S-phase heterochromatic replication sites in mouse cells (Moir et al. 1994). We have not compared lamin B localization to early S-phase replication foci in primary cells, although the reported localization pattern for this protein in other cell types does not coincide with the nucleolus. Finally, a number of reports have suggested that sites of DNA replication are associated with genomic matrix attachment regions (Dingman 1974; Pardoll et al. 1980; Amati and Gasser 1990; Jackson and Pombo 1998; Pemov et al. 1998).
Focal sites of DNA replication
The use of a limited number of sites within the nucleus to organize the initiation of DNA synthesis raises a number of interesting questions. For regulatory control, the focal architecture suggests a mechanism in which the decision to initiate DNA synthesis is not taken at a large number of sites simultaneously but, instead, is limited to a small number of sites that are organized around the nucleolus. Without knowing anything about the regulatory mechanisms, one can only suggest that this organization would lead to better control of initiation timing and to a more efficient method of responding to potential damage signals from the cell cycle checkpoints signaling system. The observation that pRB, a protein known to be required for DNA damage–induced G1 arrest (Harrington et al. 1998; Brugarolas et al. 1999), targets the perinucleolar foci prior to DNA synthesis indicates that it could be poised to directly inhibit the onset of DNA synthesis in response to arrest signals.
On a strictly structural basis, the use of a small number of replication foci to begin S-phase implies a physical hierarchy to the selection of origins. The distinction between early and late firing origins has been known for some time. However, it has not been clear how specific origins would be singled out for use from the entire list of potential origins. The discovery of a structure to organize these sequences may provide an explanation as to how they are chosen. We demonstrate that these foci are established early in G1 and that these structures also contain transcription regulatory proteins such as pRB that are needed for late G1 control. This observation, and the finding that E2F subunits also localize to these foci (B.K. Kennedy et al., unpubl.), suggests that the perinucleolar foci might contain chromosomal regions corresponding to E2F target genes. Physical association between late G1 transcriptional regulatory proteins and the initiation of DNA synthesis would be consistent with the view of research on a number of organisms that transcriptional activation is linked to the initiation of DNA replication (DeVilliers et al. 1984; Prives et al. 1987; Guo et al. 1989; Mul et al. 1990; Cheng et al. 1992; Ohba et al. 1996; Murakami and Ito 1999). It may also provide a rationale for why origin selection could be different in distinct cell types; that is, the transcriptional profile of a cell in G1 may influence which origins are ready for firing.
Our findings also add to the growing evidence for a sophisticated architecture that links nuclear substructure to specific nuclear functions. This type of organizational structure is well studied and deeply appreciated in work on the architecture of the eukaryotic cytoplasm. Nuclear substructure has been studied in detail by many groups (Strouboulis and Wolffe 1996; Misteli and Spector 1998; Brown 1999; Leonhardt et al. 1999), but in general, this work has been underappreciated. The demonstration of a structure that initiates DNA synthesis could provide a link between structure and function that will be assailable by modern molecular biology.
The observation that the use of early S-phase replication foci can be changed presumably by mutations associated with immortalization raises the question of whether the replication timing of individual genes is altered by the immortalization process. This issue has not been addressed directly. However, it was reported recently that the replication timing of homologous loci is more asynchronous in samples derived from malignant cells relative to their normal counterparts (Amiel et al. 1998). Finally, our preliminary results indicate that replication patterns can be altered changes in physiological conditions (D.A. Barbie et al., unpubl.), and that the organization of DNA replication may be regulated in other unanticipated ways. Clearly, much more work will be needed to understand how the nucleus is organized to coordinate higher-order regulation of DNA synthesis.
Materials and methods
Cell synchronization and immunofluorescence
Early passage cells were arrested in G1 by contact inhibition. After 2 d cells were replated at low density on coverslips and allowed to progress through the cell cycle. Cells were pulsed with BrdU labeling reagent (Amersham), and either harvested for FACS analysis or fixed for 10 min in 1× PBS, 4% paraformaldehyde, and then permeabilized in 1× PBS, 0.5% Triton X-100. Indirect immunofluorescence was performed using primary antibodies against BrdU supplemented with 10 U/mL DNaseI (Amersham), followed by exposure to secondary antibodies (Vector).
For cell synchronization in early S-phase, cells were released from contact inhibition into media containing BrdU and either 1.5 mM hydroxyurea, 2 μg/mL aphidicolin, or no drug. Twenty-four hours after release, cells were fixed and immunofluorescence was performed. For release for hydroxyurea arrest, cells were arrested as above except with no BrdU present, then washed in 1× PBS several times and allowed to proceed through S-phase by addition of drug-free media. At hourly intervals, cells were pulsed with BrdU for 10 min and immunofluorescence was performed using antibodies to BrdU (Amersham).
All quantitative data were collected using standard fluorescence microscopy. Images were generated using CCD and deconvolution microscopy (Scanalytics). The images reflect the localization of the proteins but do not portray relative intensities between individual foci. All processed images were recompiled from images on several sections throughout the nucleus and, thus, reflect the staining pattern of the entire nucleus compressed into a two-dimensional image. Antibodies to PCNA (Santa Cruz, Zymed), ANA-N (Sigma), pRB (Pharmingen), and p130 (Santa Cruz) were used to detect the locations of these antigens. Antibodies to CAF-1 were kindly supplied by B. Stillman (Cold Spring Harbor Laboratory, NY). UBF antibodies were kindly supplied by L. Rothblum (Weis Center for Research, PA).
BrdU labeling reagents and antibodies
Two BrdU labeling reagents (Amersham, Boehringer Mannheim) were tested, as well as CldU and IdU (Sigma). Three mouse anti-BrdU antibodies (Amersham, Becton Dickinson, Boehringer Mannheim) and one rat anti-Brdu antibody (Harlan/Sera) were tested. Standard fluorescence microscopy, confocal microscopy, and CCD/deconvolution microscopy-generated images were compared. Similar results were obtained with any combination of these reagents.
Methods to denature DNA for identification of BrdU incorporation sites
To generate denatured DNA, cells were exposed to DNaseI concomitant with treatment with the primary anti-BrdU antibody. The Amersham anti-BrdU antibody is supplied with DNaseI. In all other cases, we added DNaseI (Sigma) exogenously. The DNaseI concentration used was 10 U/mL unless otherwise listed. For acid denaturation, cells were exposed to 1–4 N HCl for 10–30 min before addition of antibodies.
For sequential denaturation experiments, the following procedure was used. Indirect immunofluorescence was performed on early S-phase cells as described previously using 10U/mL DNaseI to denature DNA. Then cells were treated in water (control) or 4 N HCl for a period of 10 sec and imaging was performed. Short-term exposure to HCl would seem to preclude the possibility that antibodies detach from their epitope and associate with new ones in other regions of the nucleus. To further rule out this possibility, we performed mock experiments in which no primary anti-BrdU antibodies were used after DNaseI treatment. Tenfold excess primary antibody was added with 4 N HCl for 20 sec, followed by secondary antibodies. Under these conditions, we detect no fluorescent staining (data not shown), indicating that new antibody epitope interactions cannot occur during short-term exposure to HCl.
In other experiments, DNA denaturation was achieved by either 10-min incubation at 80°C in 80% glycerol or 30-min exposure to 0.05 N NaOH. It should be noted that incubation of cells at higher temperatures or for prolonged periods often caused destruction of replication foci as visualized with antibodies to BrdU, replication factors, or lamin A/C. Higher base concentrations caused cells to become detached from coverslips.
Acknowledgments
We thank S. Bell, L. Guarente, T. Ha, A. Lai, J. Settleman, and J. Zhao for scientific advice and editorial comments on the manuscript and C. Srokowski for technical assistance. We thank L. Rothblum, J. Sylvester, B. Stillman for supplying reagents. Finally, we thank F. Dick and E. Harrington for generation of MEFs. Deconvolution microscopy was performed at the Whitehead Institute Microscopy Facility. B.K.K. is supported by a Leukemia Society of America Fellowship and D.A.B. by a Karen Grunebaum Cancer Research Fellowship and a Howard Hughes Medical Research Training Fellowship. This work was funded by an NIH grant to E.H. E.H. is an American Cancer Society Research Professor.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL kennedy@helix.mgh.harvard.edu; FAX (617) 726-7808.
Article and publication are at www.genesdev.org/cgi/doi/10.1101/gad.842600.
References
- Amati B, Gasser SM. Drosophila scaffold attached regions bind nuclear scaffolds and can function as ARS elements in both budding and fission yeasts. Mol Cell Biol. 1990;10:5442–5454. doi: 10.1128/mcb.10.10.5442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amiel A, Litmanovitch T, Lishner M, Mor A, Gaber E, Tangi I, Fejgin M, Avivi L. Temporal differences in replication timing of homologous loci in malignant cells derived from CML and lymphoma patients. Genes Chromosomes Cancer. 1998;22:225–231. doi: 10.1002/(sici)1098-2264(199807)22:3<225::aid-gcc8>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- Berezney R, Buchholtz LA. Dynamic association of replicating DNA fragments with the nuclear matrix of regenerating liver. Exp Cell Res. 1981;132:1–13. doi: 10.1016/0014-4827(81)90076-8. [DOI] [PubMed] [Google Scholar]
- Berezney R, Dubey DD, Huberman JA. Heterogeneity of eukaryotic replicons, replicon clusters, and replication foci. Chromosoma. 2000;108:471–484. doi: 10.1007/s004120050399. [DOI] [PubMed] [Google Scholar]
- Bravo R. Synthesis of the nuclear protein cyclin (PCNA) and its relationship with DNA replication. Exp Cell Res. 1986;163:287–293. doi: 10.1016/0014-4827(86)90059-5. [DOI] [PubMed] [Google Scholar]
- Bravo R, Frank R, Blundell PA, Macdonald-Bravo H. Cyclin/PCNA is the auxiliary protein of DNA polymerase-delta. Nature. 1987;326:515–517. doi: 10.1038/326515a0. [DOI] [PubMed] [Google Scholar]
- Bridger JM, Kill IR, O'Farrell M, Hutchison CJ. Internal lamin structures within G1 nuclei of human dermal fibroblasts. J Cell Sci. 1993;104:297–306. doi: 10.1242/jcs.104.2.297. [DOI] [PubMed] [Google Scholar]
- Brown K. Nuclear structure, gene expression and development. Crit Rev Eukaryot Gene Expr. 1999;9:203–212. doi: 10.1615/critreveukargeneexpr.v9.i3-4.50. [DOI] [PubMed] [Google Scholar]
- Brugarolas J, Moberg K, Boyd SD, Taya Y, Jacks T, Lees JA. Inhibition of cyclin-dependent kinase 2 by p21 is necessary for retinoblastoma protein-mediated G1 arrest after γ-irradiation. Proc Natl Acad Sci. 1999;96:1002–1007. doi: 10.1073/pnas.96.3.1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brylawski BP, Cohen SM, Cordeiro-Stone M, Schell MJ, Kaufman DG. On the relationship of matrix association and DNA replication. Crit Rev Eukaryot Gene Expr. 2000;10:91–99. [PubMed] [Google Scholar]
- Carri MT, Micheli G, Graziano E, Pace T, Buongiorno-Nardelli M. The relationship between chromosomal origins of replication and the nucelar matrix during the cell cycle. Exp Cell Res. 1986;164:426–436. doi: 10.1016/0014-4827(86)90041-8. [DOI] [PubMed] [Google Scholar]
- Cheng LZ, Workman JL, Kingston RE, Kelly TJ. Regulation of DNA replication in vitro by the transcriptional activation domain of GAL4–VP16. Proc Natl Acad Sci. 1992;89:589–593. doi: 10.1073/pnas.89.2.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins JM, Chu AK. Binding of the DNA polymerase alpha-DNA primase complex to the nuclear matrix in HeLA cells. Biochemistry. 1987;26:5600–5607. doi: 10.1021/bi00392a004. [DOI] [PubMed] [Google Scholar]
- DeVilliers J, Schaffner W, Tyndall C, Lupton S, Kamen R. Polyoma virus DNA replication requires an enhancer. Nature. 1984;312:242–246. doi: 10.1038/312242a0. [DOI] [PubMed] [Google Scholar]
- Dijkwel PA, Mullenders LH, Wanka F. Analysis of the attachment of replicating DNA to a nuclear matrix in mammalian interphase nuclei. Nucleic Acids Res. 1979;6:219–230. doi: 10.1093/nar/6.1.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dingman CW. Bidirectional chromosome replication: Some topological considerations. J Theor Biol. 1974;43:187–195. doi: 10.1016/s0022-5193(74)80052-4. [DOI] [PubMed] [Google Scholar]
- Ellis DJ, Jenkins H, Whitfield WG, Hutchison CJ. GST-lamin fusion proteins act as dominant negative mutants in Xenopus egg extract and reveal the function of the lamina in DNA replication. J Cell Sci. 1997;110:2507–2518. doi: 10.1242/jcs.110.20.2507. [DOI] [PubMed] [Google Scholar]
- Gerace L, Blum A, Blobel G. Immunocytochemical localization of the major polypeptides of the nuclear pore complex lamina fraction: Interphase and mitotic distribution. J Cell Biol. 1978;79:546–566. doi: 10.1083/jcb.79.2.546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z-S, Gutierrez C, Heine V, Sogo JM, DePamphilis ML. Origin auxiliary sequences can facilitate initiation of simian virus DNA replication in vitro as they do in vivo. Mol Cell Biol. 1989;9:3593–3602. doi: 10.1128/mcb.9.9.3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington EA, Bruce JL, Harlow E, Dyson N. pRB plays an essential role in cell cycle arrest induced by DNA damage. Proc Natl Acad Sci. 1998;95:11945–11950. doi: 10.1073/pnas.95.20.11945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan AB, Cook PR. Visualization of replication sites in unfixed human cells. J Cell Sci. 1993;105:541–550. doi: 10.1242/jcs.105.2.541. [DOI] [PubMed] [Google Scholar]
- Hozak P, Hassan AB, Jackson DA, Cook PR. Visualization of replication factories attached to the nucleoskeleton. Cell. 1993;73:361–373. doi: 10.1016/0092-8674(93)90235-i. [DOI] [PubMed] [Google Scholar]
- Hunt BF, Vogelstein B. Association of newly replicated DNA with the nuclear matrix of Physarum polycephalum. Nucleic Acids Res. 1981;9:349–363. doi: 10.1093/nar/9.2.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson DA, Pombo A. Replicon clusters are stable units of chromosome structure: Evidence that nuclear organization contributes to the efficient activationand propagation of S-phase in human cells. J Cell Biol. 1998;140:1285–1295. doi: 10.1083/jcb.140.6.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman PD, Kobayashi R, Kessler N, Stillman B. The p150 and p60 subunits of chromatin assembly factor I: A molecular link between newly synthesized histones and DNA replication. Cell. 1995;81:1105–1114. doi: 10.1016/s0092-8674(05)80015-7. [DOI] [PubMed] [Google Scholar]
- Kill JR, Bridger JM, Campbell KH, Maldonado-Codina G, Hutchison CJ. The timing of the formation and usage of replicase clusters in S-phase nuclei of human diploid fibroblasts. J Cell Sci. 1991;100:869–876. doi: 10.1242/jcs.100.4.869. [DOI] [PubMed] [Google Scholar]
- Krude T. Chromatin assembly factor I (CAF-1) colocalizes with replication foci in HeLa cell nuclei. Exp Cell Res. 1995;220:304–311. doi: 10.1006/excr.1995.1320. [DOI] [PubMed] [Google Scholar]
- Leonhardt H, Rahn HP, Cardoso MC. Functional links between nuclear structure, gene expression, DNA replication, and methylation. Crit Rev Eukaryot Gene Expr. 1999;9:345–351. doi: 10.1615/critreveukargeneexpr.v9.i3-4.190. [DOI] [PubMed] [Google Scholar]
- Ma H, Samarabandu J, Devdhar RS, Acharya R, Cheng PC, Meng C, Berezney R. Spatial and temporal dynamics of DNA replication sites in mammalian cells. J Cell Biol. 1998;143:1415–1425. doi: 10.1083/jcb.143.6.1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli T, Spector DL. The cellular organization of gene expression. Curr Opin Cell Biol. 1998;10:323–331. doi: 10.1016/s0955-0674(98)80007-0. [DOI] [PubMed] [Google Scholar]
- Moir RD, Spann TP, Goldman RD. Dynamic properties of nuclear lamins: Lamin B is associated with sites of DNA replication. J Cell Biol. 1994;125:1201–1212. doi: 10.1083/jcb.125.6.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moir RD, Spann TP, Herrmann H, Goldman RD. Disruption of nuclear lamin organization blocks the elongation phase of DNA replication. J Cell Biol. 2000a;149:1179–1192. doi: 10.1083/jcb.149.6.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moir RD, Spann TP, Lopez-Soler RI, Yoon M, Goldman AE, Khoun S, Goldman RD. The dynamics of the nuclear lamins during the cell cycle–relationship between structure and function. J Struct Biol. 2000b;129:324–334. doi: 10.1006/jsbi.2000.4251. [DOI] [PubMed] [Google Scholar]
- Mul YM, Cerrijzer CP, van der Vliet PC. Transcription factors NFI and NFIII/Oct-1 function independently, employing different mechanisms to enhance adenovirus DNA replication. J Virol. 1990;64:5510–5518. doi: 10.1128/jvi.64.11.5510-5518.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami Y, Ito Y. Transcription factors in DNA replication. Front Biosci. 1999;4:D824–D833. doi: 10.2741/murakami. [DOI] [PubMed] [Google Scholar]
- Nakamura H, Morita T, Sato C. Structural organizations of replicon domains during DNA synthetic phase in the mammalian nucleus. Exp Cell Res. 1986;165:291–297. doi: 10.1016/0014-4827(86)90583-5. [DOI] [PubMed] [Google Scholar]
- Nakayasu H, Berezney R. Mapping replicational sites in the eucaryotic cell nucleus. J Cell Biol. 1989;108:1–11. doi: 10.1083/jcb.108.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neri LM, Mazzotti G, Capitani S, Maraldi NM, Cinti C, Baldini N, Rana R, Martelli AM. Nuclear matrix-bound replicational sites detected in situ by 5-bromodeoxyuridine. Histochemistry. 1992;98:19–32. doi: 10.1007/BF00716934. [DOI] [PubMed] [Google Scholar]
- Ohba R, Matsumoto K, Ishimi Y. Induction of DNA replication by transcription in the region upstream of the human c-myc gene in a model replication system. Mol Cell Biol. 1996;16:5754–5763. doi: 10.1128/mcb.16.10.5754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Keefe RT, Henderson SC, Spector DL. Dynamic organization of DNA replication in mammalian cell nuclei: Spatially and temporally defined replication of chromosome-specific α-satellite DNA sequences. J Cell Biol. 1992;116:1095–1110. doi: 10.1083/jcb.116.5.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardoll DM, Vogelstein B, Coffey DS. A fixed site of DNA replication in eucaryotic cells. Cell. 1980;19:527–536. doi: 10.1016/0092-8674(80)90527-9. [DOI] [PubMed] [Google Scholar]
- Pemov A, Bavykin S, Hamlin JL. Attachment to the nuclear matrix mediates specific alterations in chromatin structure. Proc Natl Acad Sci. 1998;95:14757–14762. doi: 10.1073/pnas.95.25.14757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prelich G, Tan C-K, Kostura M, Mathews MB, So AG, Downey KM, Stillman B. Functional identity of proliferating cell nuclear antigen and a DNA polymerase-δ auxiliary protein. Nature. 1987;326:517–520. doi: 10.1038/326517a0. [DOI] [PubMed] [Google Scholar]
- Prives C, Murakami Y, Kem FG, Folc W, Basilico C, Hurwitz J. DNA sequence requirements for replication of polyomavirus DNA in vivo and in vitro. Mol Cell Biol. 1987;7:3694–3704. doi: 10.1128/mcb.7.10.3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzoli R, Baratta B, Maraldi NM, Falconi M, Galanzi A, Papa S, Vitale M, Rizzi F, Manzoli L, Mazzotti G. DNA synthesis progression in 3T3 synchronized fibroblasts: A high resolution approach. Histochemistry. 1992;97:181–187. doi: 10.1007/BF00267309. [DOI] [PubMed] [Google Scholar]
- Smetana K, Jiraskova I, Sedlackova M, Dvorak R, Spatova M, Hovak P. Preferential silver reaction of nucleolar regions adjacent to fibrillar centers in ring shaped nucleoli of leukemic lymphocytes. Acta Histochem. 1998;100:257–270. doi: 10.1016/s0065-1281(98)80012-3. [DOI] [PubMed] [Google Scholar]
- Smith S, Stillman B. Purification and characterization of CAF-I, a human cell factor required for chromatin assembly during DNA replication in vitro. Cell. 1989;58:15–25. doi: 10.1016/0092-8674(89)90398-x. [DOI] [PubMed] [Google Scholar]
- Spann TP, Moir RD, Goldman AE, Stick R, Goldman RD. Disruption of nuclear lamin organization alters the distribution of replication factors and inhibits DNA synthesis. J Cell Biol. 1997;136:1201–1212. doi: 10.1083/jcb.136.6.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strouboulis J, Wolffe A. Functional compartmentalization of the nucleus. J Cell Sci. 1996;109:1991–2000. doi: 10.1242/jcs.109.8.1991. [DOI] [PubMed] [Google Scholar]
- van Dierendonck JH, Keyzer R, van de Velde CJ, Cornelisse CJ. Subdivision of S-phase by analysis of nuclear 5-bromodeoxyuridine staining patterns. Cytometry. 1989;10:143–150. doi: 10.1002/cyto.990100205. [DOI] [PubMed] [Google Scholar]
- Vaughn JP, Dijkwel PA, Mullenders LH, Hamlin JL. Replication forks are associated with the nuclear matrix. Nucleic Acids Res. 1990;18:1965–1969. doi: 10.1093/nar/18.8.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson KL. The nuclear envelope, muscular dystrophy and gene expression. Trends Cell Biol. 2000;10:125–129. doi: 10.1016/s0962-8924(99)01708-0. [DOI] [PubMed] [Google Scholar]