Abstract
Many questions remain concerning the role of TFIID TBP-associated factors (TAFs) in transcription, including whether TAFs are required at most or only a small subset of promoters. It was shown previously that three histone-like TAFs are broadly required for transcription, but interpretation of this observation is complicated because these proteins are components of both TFIID and the SAGA histone acetyltransferase complex. Here we show that mutations in the yeast TFIID-specific protein Taf40 lead to a general cessation of transcription, even in the presence of excess TBP, suggesting that the TFIID complex is required at most promoters in vivo.
Keywords: TAFS, promoters, TFIID, TAF40, transcription
RNA polymerase II (Pol II) is positioned on a promoter by a set of accessory basal transcription factors. One of these factors, TFIID, binds the consensus promoter sequence TATAA and nucleates the assembly of other transcription factors. One subunit of TFIID, the TATA-binding protein (TBP), is necessary and sufficient for this activity in vitro. However, TBP in vivo is associated with several multisubunit complexes that mediate transcription by RNA polymerase I, II, or III (Hernandez 1993). The TBP-containing complexes implicated in Pol II transcription are TFIID, the TBP–MOT1 complex (Auble and Hahn 1993), and the SNAPc complex in higher eukaryotes (Hernandez 1993). In addition, the SAGA histone acetyltransferase complex can associate with TBP transiently (Eisenmann et al. 1992; Sterner et al. 1999).
The TFIID complex consists of TBP and 10–12 TBP-associated factors (TAFs). The essential role of TBP in Pol II-mediated transcription has been demonstrated both in vivo and in vitro (Hampsey 1998). The functions of TAFs in transcription are less clear (for review, see Verrijzer and Tjian 1996; Hoffmann et al. 1997; Grant and Workman 1998; Hahn 1998). In vitro experiments in mammalian, Drosophila, or yeast systems have indicated a requirement for TAFs in responding to transcriptional activators but not for basal transcription (Kokubo et al. 1993; Chen et al. 1994; Reese et al. 1994). However, activated transcription in vitro has also been reported in a yeast system apparently lacking TAFs, with activation being mediated by the Pol II-associated mediator/SRB proteins (Kim et al. 1994; Koleske and Young 1994). TAF-independent activation has also been reported in mammalian in vitro transcription systems (Oelgeschlager et al. 1998; Wu et al. 1998). Therefore, there may be both TAF-dependent and -independent activation mechanisms that can be emphasized by the particular choice of in vitro transcription system. In vivo experiments are therefore essential for testing the physiological relevance of in vitro results.
Several other roles for TAFs have been documented in addition to their proposed function as transcriptional coactivators. TFIID makes extensive contacts with core promoter elements in addition to the TATA element, and these contacts are made by the TAFs (for review, see Hoffmann et al. 1997). Furthermore, the largest TAF can inhibit TBP binding to the TATA element in vitro (Kokubo et al. 1994; Bai et al. 1997). Therefore, TAFs contribute to recognition of and affinity for basal promoters. The TFIID large subunit exhibits both protein kinase and histone acetyltransferase activities (Dikstein et al. 1996; Mizzen et al. 1996), suggesting that some TAFs have enzymatic functions relevant to transcription and/or chromatin structure.
Because TAFs are essential for viability and are usually required for activated transcription in vitro, it was predicted that they would be generally required for transcription in vivo. This view was challenged by observations that depletion or temperature inactivation of many TAFs did not cause an overall defect in Pol II-mediated transcription in vivo (Moqtaderi et al. 1996a, 1998; Walker et al. 1996). This observation suggested that TFIID is not required at most promoters for either activated or basal transcription in vivo and that gene expression is supported by TBP alone or other TBP-containing complexes. The fact that most TAFs are required for viability was explained by their need at promoters of selected genes, whose products are, in turn, essential. Individual promoters could be identified that had more stringent requirements for certain TAFs (Holstege et al. 1998).
Interpretation of TAF depletion experiments has been complicated by several recent findings. The histone H3-like Taf17 (Apone et al. 1998; Michel et al. 1998; Moqtaderi et al. 1998), as well as the histone H4-like Taf60 and H2B-like TAF61/68 (Michel et al. 1998; Natarajan et al. 1998) are required for the majority of transcriptions. Similar findings have been reported for Taf23/25 (Sanders et al. 1999). Therefore, some TAFs may be generally required while others function at only a few promoters.
Another complication is that some components of TFIID (Taf90, Taf23/25, and the histone-like TAFs Taf17, Taf60, and Taf61/68) are also subunits of the SAGA histone acetyltransferase complex (Grant et al. 1998). A similar sharing of identical or homologous subunits has been reported for mammalian TFIID and the P/CAF histone acetyltransferase complex (Ogryzko et al. 1998). Because the histone-like TAFs and Taf23/25 are components of at least two protein complexes involved in RNA polymerase II transcription, it was unclear if the observed general cessation of transcription was due to loss of TFIID, SAGA, or a redundant function carried out by both complexes (Grant and Workman 1998; Hahn 1998). It seems unlikely that the transcriptional deficiency in histone-like TAF mutants could arise solely from SAGA complex disruption because SAGA-specific proteins are not essential for viability and yeast strains lacking functional SAGA complex do not show a similar effect on Pol II-mediated transcription (Michel et al. 1998).
To determine the contribution of a TFIID subunit to transcription independent of the SAGA complex, we analyzed conditional alleles of TAF40, a TAF present in TFIID but not SAGA (Moqtaderi et al. 1996b; Klebanow et al. 1997; Grant et al. 1998). The mammalian homolog of Taf40 (TAFII28) interacts in vitro with the homolog of Taf19 (TAFII18) via a histone-fold motif, and the dimer may be structurally and functionally similar to the Spt3 protein within the SAGA complex (Birck et al. 1998). Consistent with this, we present genetic evidence for the interaction between TAF40 and TAF19 in vivo. Furthermore, we find that Taf40 is generally required for transcription by Pol II, even when TBP is overexpressed. These findings suggest that TFIID is the complex responsible for delivering TBP to most promoters.
Results
Isolation and characterization of TAF40 temperature-sensitive alleles
Temperature-sensitive alleles of Taf40 were generated by error-prone PCR and selected using plasmid shuffling as described (Michel et al. 1998). All taf40 alleles isolated were recessive (data not shown). Despite similar phenotypes displayed by the taf40 mutants when incubated on plates at 37°C, differences between various alleles were noticeable. Some strains resume growth when shifted back to permissive temperature (30°C) after two days of incubation at 37°C. Generally, these strains do not completely stop dividing when shifted to 37°C in liquid media, but instead show very long doubling times. Based on our previous experience with Taf60, 61/68, and 17, we selected strains that did not resume growth when returned from restrictive to permissive conditions and that exhibited a complete growth arrest at 37°C in liquid media (Michel et al. 1998).
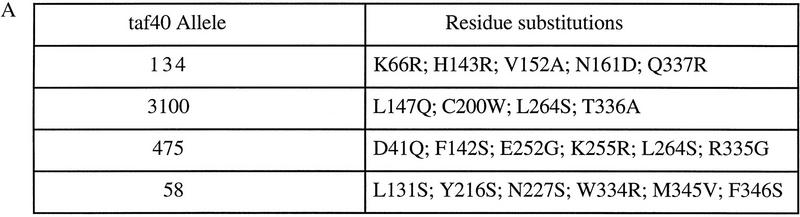
Four independently generated temperature-sensitive taf40 alleles were sequenced and the mutations are summarized in Figure 1. All alleles contain multiple missense mutations. One region mutated in all four alleles is the very carboxyl terminus of the protein (Fig. 1). A second region around amino acids 142–148 is mutated in three out of four taf40 alleles. In the homologous mammalian TAFIII28/TAFII18 crystal structure (Birck et al. 1998), this region is located amino-terminal to the histone fold and participates in the interaction with TAFII18. The residue in human TAFII28 corresponding to amino acid 147 in yeast Taf40 contacts the TAFII18 amino-terminal amino group. Human TAFII28 residues corresponding to yeast Taf40 positions 142 and 143 participate in the stabilization of the αN helix, which makes several important contacts with TAFII18 (Birck et al. 1998).
Figure 1.
Amino acid substitutions in temperature-sensitive taf40 alleles. (A) Substitutions are listed in the single-letter amino acid code. Wild-type amino acid and residue number are followed by the mutant amino acid. (B) Schematic diagram of Taf40 structure is shown, based on the alignment in Birck et al. (1998). Predicted α-helices are shown as shaded boxes; numbers are the Taf40 residues. Helices α1, α2, and α3 comprise the histone-fold for interaction with Taf19. LN, L1, and L2 are linker regions between helices.
Taf19 overexpression suppresses temperature sensitivity of Taf40 mutants
We tested for genetic interactions between TFIID subunits by seeing whether the Taf40 mutant phenotypes could be suppressed by overexpressing other TAFs. Of eight different TAFs tested, only Taf19 could suppress the temperature sensitivity of strains carrying mutant temperature-sensitive alleles of Taf40 (Fig. 2). Furthermore, overexpression of Spt3 does not suppress (data not shown). These results provide in vivo support for the finding that human homologs of Taf40 and Taf19 (TAFII28 and TAFII18, respectively) interact in vitro via their histone fold domains (Birck et al. 1998) and suggest that temperature-sensitive alleles of Taf40 are partially defective in interaction with Taf19 (Fig. 1).
Figure 2.
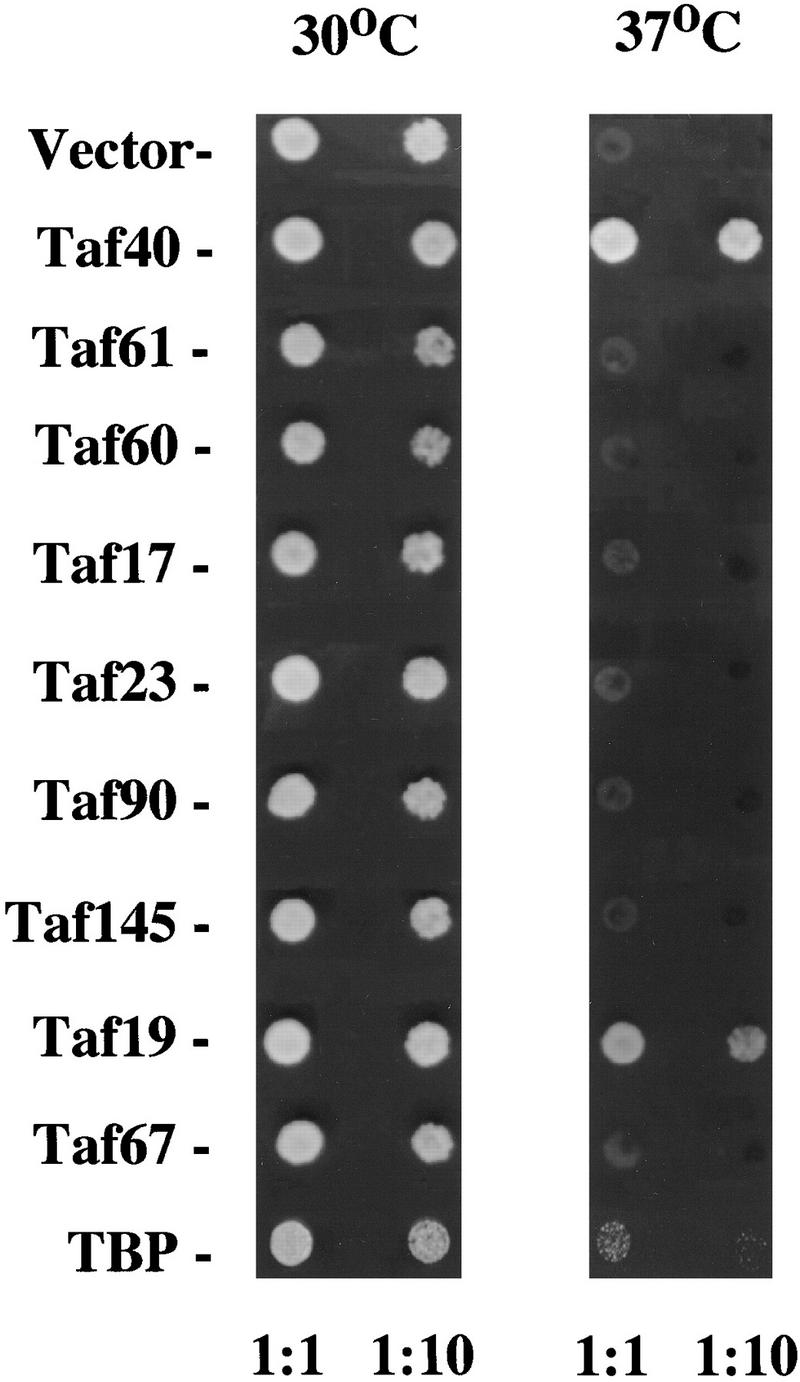
High copy suppression of a taf40 mutant phenotype. A strain carrying the temperature-sensitive taf40-3100 allele was transformed with high copy plasmids carrying the indicated TAF genes and dilutions were spotted at 30°C and 37°C. TBP overexpression was from a PGK1 promoter driving TBP expression (Jackson-Fisher et al. 1999). Photographs were taken after 60 hr of growth. The photograph is a composite of several plates (due to different markers used in some of the overexpressing plasmids).
Overexpression of TBP from a PGK1 promoter (see below) was also tested for suppression and did allow some growth of the taf40 mutant on plates but not in liquid culture (Fig. 2; data not shown). Suppression was much weaker than that seen for Taf19 and appeared to occur in only a small percentage of cells carrying the plasmid. Furthermore, the TAF40 gene was still essential in the presence of overexpressed TBP (data not shown). Therefore, TBP overexpression suppresses the mutant Taf40 phenotype weakly at best, probably by allowing a few extra divisions before cell death.
Taf40 is generally required for RNA polymerase II-mediated transcription
To determine the effect of Taf40 depletion/inactivation on Pol II-mediated transcription, strains carrying the wild-type or conditional alleles of Taf40 were shifted to nonpermissive temperature. Total RNA from these strains was isolated and analyzed. Specific mRNAs were quantified using an S1 nuclease protection assay, and total mRNA levels were determined using a poly(dT) probe as described in Materials and Methods.
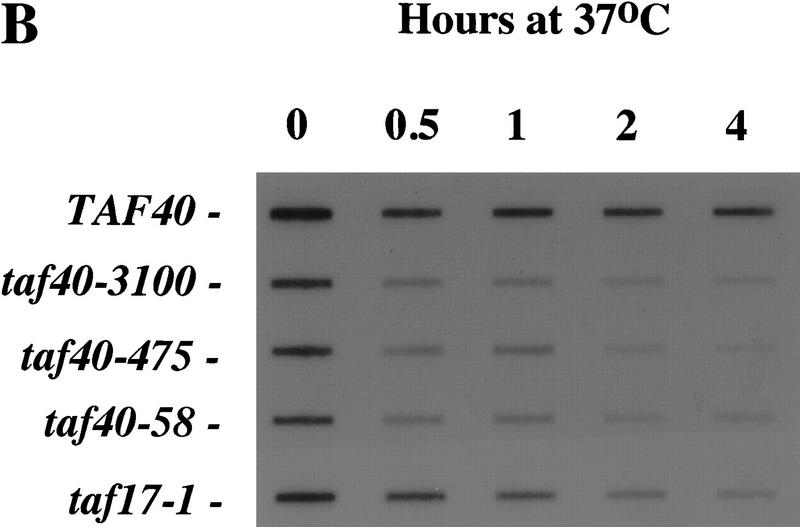
As shown in Figure 3A, mRNA levels of RPS4, DED1, HTA2, and ACT1 genes rapidly drop upon temperature shift in a strain carrying the taf40-3100 allele. The effect of Taf40 inactivation on the mRNA levels of those genes is readily detectable by 1 hr postshift. The kinetics of mRNA decrease differs among genes tested but parallels those observed with mutants of Taf60, Taf61/68, and Taf17 (Michel et al. 1998). Thus, we consider it unlikely that this kinetic difference reflects dependence of a specific gene on an individual TAF, but rather, may be a reflection of mRNA half-lives.
Figure 3.
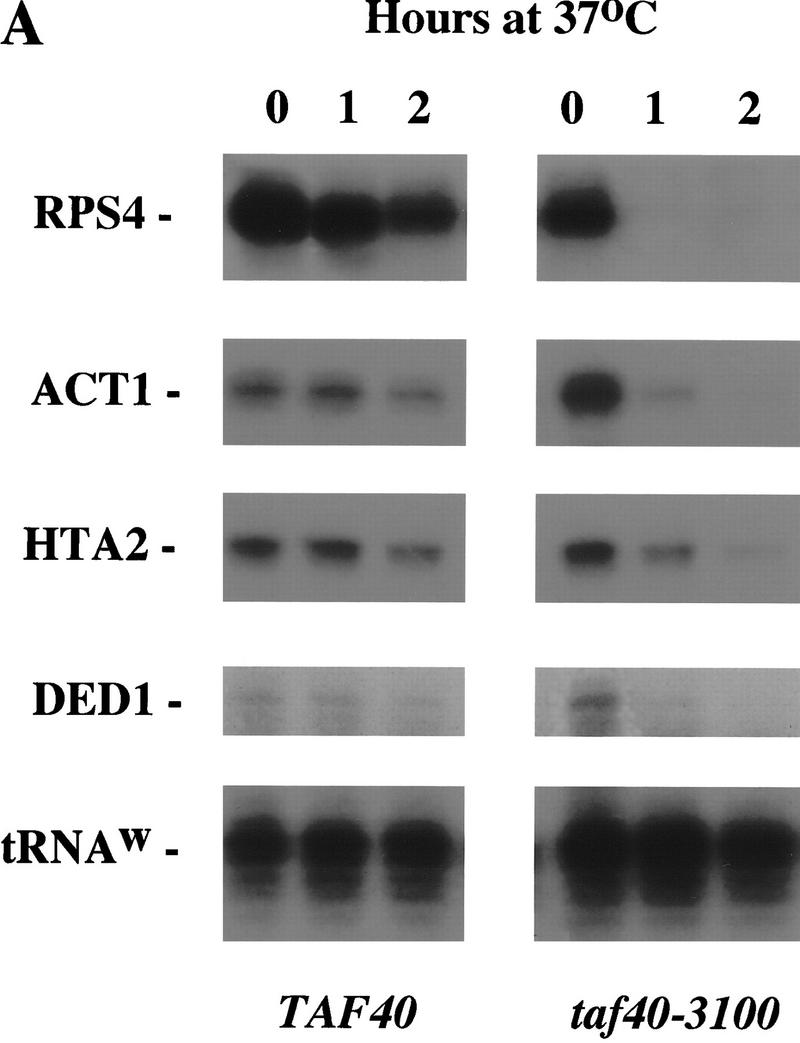
Taf40 is generally required for in vivo transcription by Pol II. (A) S1 nuclease protection assay of mRNAs in the taf40-3100 mutant strain. RNA was isolated from wild-type or mutant strains incubated at 37°C for the indicated amount of time. Probes for the RPS4, DED1, HTA2, and ACT1 genes were used to monitor levels of those transcripts. A probe for Pol III-transcribed tRNAW was used as an internal control for RNA normalization. (B) Total poly(A)+ levels in strains carrying either the indicated wild-type or the mutant TAF40 alleles. Total RNAs from wild-type or the indicated mutant TAF strains were isolated as described in A and slot-blotted to a membrane. Poly(A)+ levels were assayed by hybridization with a radioactive oligo(dT) probe.
Total poly(A)+ mRNA levels decrease detectably by 30 min of incubation at 37°C and after 4 hr are reduced to the background level (Fig. 3B). The total mRNA level decrease was observed in four independently isolated tight alleles. The extent and kinetics of total mRNA decrease upon Taf40 inactivation are similar to that seen upon Taf17 inactivation. Taf17 has been shown previously by us and others to be broadly required for Pol II-mediated transcription (Apone et al. 1998; Michel et al. 1998; Moqtaderi et al. 1998). The strong effect of Taf40 inactivation indicates that this TFIID-specific TAF is also generally required for Pol II-mediated transcription.
Taf40 temperature inactivation specifically affects TFIID complex components
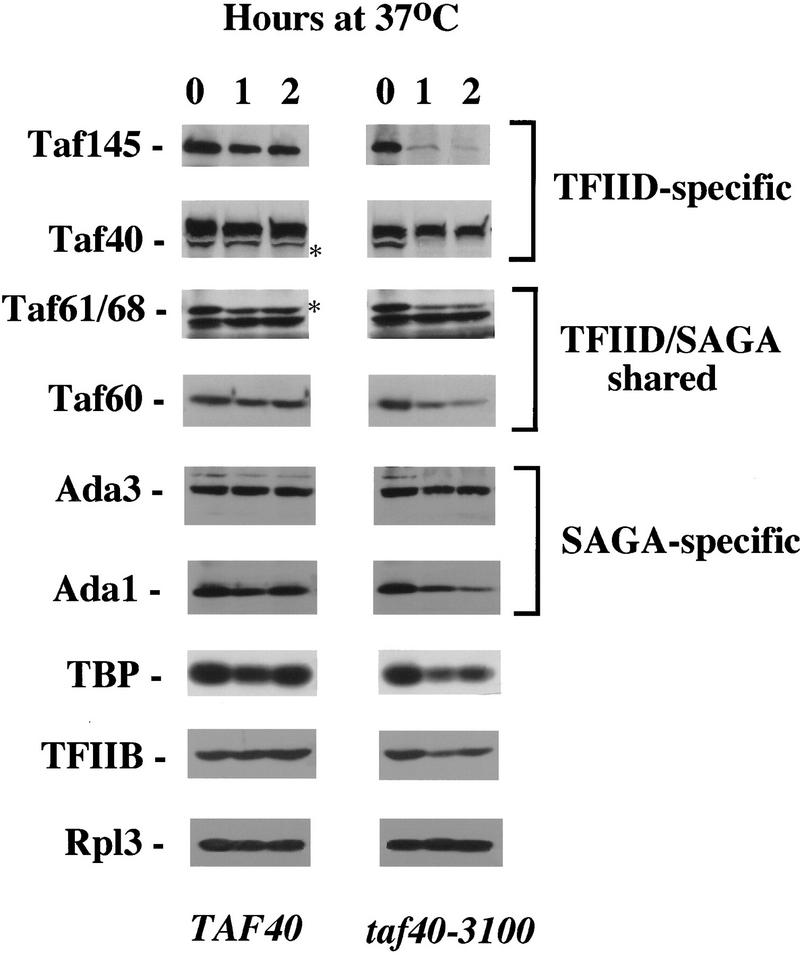
Because Taf40 is a component of TFIID but not the SAGA complex, it was of interest to study the effects of Taf40 temperature-sensitive alleles on the level of proteins constituting either complex. As reported previously, inactivation of the shared Tafs 60, 61/68, and 17 led to a dramatic decrease in the levels of proteins comprising both TFIID and SAGA (Michel et al. 1998). As shown in Figure 4, mutant Taf40 protein is rapidly degraded after 1 hr at 37°C. Similarly, levels of the TFIID-specific Taf145 protein are rapidly reduced after temperature shift. Taf60 and Taf61/68, present in both TFIID and SAGA complexes, are reduced but to a lesser extent than Taf145. The level of TBP diminished by approximately half. The SAGA-specific Ada3 protein level does not change for at least 2 hr, by which time transcriptional effects are readily detectable, and Ada1 protein levels show a very slight decrease by 2 hr. The levels of transcription factor TFIIB and ribosomal protein Rpl3 were monitored as a control for indirect effects on protein level. The extent of TFIIB decrease in the Taf40 temperature-sensitive strain is similar to that of TBP and SAGA complex components and significantly less dramatic than that of Taf145. These results indicate that upon Taf40 temperature inactivation the TFIID complex is rapidly depleted, but SAGA complex remains relatively unaffected.
Figure 4.
Effect of Taf40 temperature inactivation on the levels of TFIID and SAGA components. Aliquots of wild-type or taf40-3100 mutant strains were taken after the indicated time at 30°C or 37°C. Total protein was blotted and probed with antibodies to the indicated proteins. In the case of Taf40 and Taf61/68 a background band was seen nearby; the actual TAF band is indicated by an asterisk. Other taf40 temperature-sensitive alleles had similar effects on protein levels at the nonpermissive temperature (data not shown).
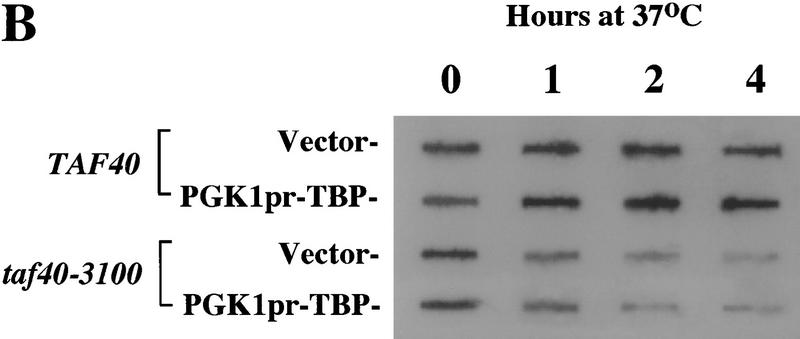
Loss of transcription in Taf40 mutants is not caused by limiting TBP
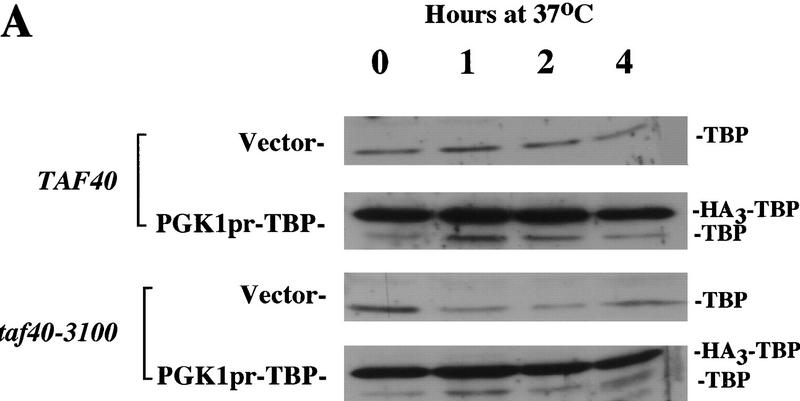
Because TBP levels are reduced upon shift to nonpermissive temperature in taf40 and histone-like taf mutant strains (Fig. 4; Michel et al. 1998), it was important to test whether the effect of TAF depletion on transcription was merely because the TAFs were required to stabilize TBP. If so, overexpression of TBP in the mutant strain should restore normal transcription levels at 37°C. Appoximately 15-fold overexpression of TBP was obtained using a construct in which an epitope-tagged TBP is driven by the PGK1 promoter (Fig. 5A; Jackson-Fisher et al. 1999). Even with high levels of TBP, poly(A)+ RNA was rapidly lost in the taf40 mutant strain at the restrictive temperature (Fig. 5B). Several individual genes were also monitored by RNase protection analysis and found to be rapidly lost even in the presence of overexpressed TBP (data not shown). These results indicate that the defect in transcription is not simply due to a decrease in TBP levels and that TBP alone cannot functionally substitute for TFIID in vivo.
Figure 5.
TBP overexpression does not restore transcription to a taf40-3100 mutant strain. (A) Samples of a wild-type (TAF40) and mutant strain (taf40-3100) carrying either vector or an epitope-tagged TBP overexpression construct (PGK1pr–TBP) were taken after the indicated time at 37°C. Total protein was blotted and probed with antibodies to TBP. The positions of the endogenous (TBP) and overexpressed, tagged TBP (HA3–TBP) are indicated. (B) Total RNA was isolated from the cultures used in A and analyzed for total poly(A)+ RNA as described in Fig. 3.
Discussion
In vitro studies have suggested that TFIID is the complex that delivers the TBP a typical Pol II-transcribed promoter. In many in vitro systems, the presence of TAFs is required to observe response to upstream activators. Activators can quantitatively and qualitatively change the DNase I footprint of TFIID on some promoters, and correlations between activator-TAF interactions and stimulation of transcription have been made (for reviews, see Verrijzer and Tjian 1996; Hoffmann et al. 1997). Furthermore, specific contacts between certain TAFs and basal promoter elements other than the TATA element have been documented. However, only a small fraction of Pol II promoters have actually been analyzed in vitro. A key question needs to be addressed in vivo: Is the TFIID complex utilized at all promoters, or do individual promoters require, at most, a small subset of TAFs?
It has now been shown that five TFIID TAF proteins are generally required for transcription in vivo. Conditional mutants in any of the three histone-like TAFs or Taf23/25 result in a rapid loss of poly(A)+ mRNA (Michel et al. 1998; Sanders et al. 1999). However, these four proteins are components of both TFIID and the SAGA complex. Deletions of SAGA-specific components such as Spt20 are not lethal (for review, see Hampsey 1998) and do not cause a similar loss of transcription. Thus, TAF dependence of transcription is not uniquely due to SAGA (Michel et al. 1998). However, shared TAFs raise the possibility that TFIID and SAGA have a redundant function that is required for transcription (Grant and Workman 1998; Hahn 1998). To test this hypothesis, we analyzed conditional alleles of the TFIID-specific Taf40. Taf40 inactivation results in TFIID depletion and a rapid loss of Pol II-mediated transcription. Transcription cannot be rescued by overexpression of TBP alone. Therefore, the TFIID complex appears to be required for the majority of transcription.
Several in vivo studies in yeast have observed ongoing transcription and response to activators in the apparent absence of TAFs. In some of these cases, TAFs other than those analyzed here were inactivated, raising the possibility that Taf40 and the histone-like TAFs could be components of an essential transcription complex other than TFIID, or that some TAFs can be removed from TFIID without disrupting the integrity or function of the entire complex. The second model seems unlikely, as it is generally observed that degradation of any of the Taf proteins leads to degradation of the other TFIID subunits (Walker et al. 1996; Michel et al. 1998; Moqtaderi et al. 1998). There is currently no evidence for additional TAF complexes other than TFIID and SAGA in yeast, but heterogeneity of TAF complexes has been observed in mammalian extracts (for review, see Bell and Tora 1999). However, the mammalian homolog of Taf40 has not been found in any complexes other than TFIID (Bell and Tora 1999), and we have thus far failed to observe any evidence for multiple complexes containing Taf40 using gel filtration chromatography (data not shown).
Earlier studies failed to observe a general requirement for Taf60, Taf61/68, or Taf40, all of which we find to be essential for transcription in vivo. We have observed that many temperature-sensitive TAF alleles have residual activity at the nonpermissive temperature; only the tightest alleles exhibit a general effect on transcription (Michel et al. 1998; data not shown). Therefore, many promoters in the cell may have sufficient in vivo affinities for TFIID such that they can function even at significantly reduced TAF levels.
Interestingly, earlier TAF depletion studies used the CUP1 and SSA4 heat shock-inducible promoters to monitor response to activators (Moqtaderi et al. 1996a, 1998; Walker et al. 1996). These promoters are highly inducible in the apparent absence of TAFs, but also in the apparent absence of the Pol II carboxy-terminal domain, the TFIIH kinase subunit Kin28, or the essential mediator component Srb4 (Lee and Lis 1998; McNeil et al. 1998). Like the histone-like TAFs and Taf40, Kin28 and Srb4 are generally required for transcription at the vast majority of promoters (Thompson and Young 1995; Holstege et al. 1998). Because CUP1 and SSA4 have evolved to be strongly transcribed under stress conditions that inhibit transcription of most genes, they may use an unusual transcription mechanism that is not representative of most promoters.
Our results are explained most simply by the hypothesis that TFIID, rather than free TBP or SAGA, is required at most Pol II-transcribed promoters in vivo. A general role for TFIID is supported by immunolocalization studies of Drosophila polytene chromosomes that find TFIID subunits at nearly every locus (Rabenstein et al. 1999; Soldatov et al. 1999). It remains to be determined what the functions of individual TAFs are and whether each function is required at every promoter. This will be difficult, as individual subunits are required for complex integrity in vivo. However, it may be possible to identify mutants that disrupt function without causing protein degradation and to reconstitute partial TFIID complexes for in vitro studies.
Materials and methods
Isolation and suppression of conditional alleles in Taf40
The TAF40 gene was randomly mutagenized by PCR-based misincorporation as described (Michel et al. 1998). Primers Taf40-A (5′-GGGAATTCCCTGTTCTAGAGCCCAACAA-3′) and pBS/RS–PvuI (5′-GGGAAGGGCGATCGGTGCGGGCCTCTTCGC-3′) were used for PCR amplification of 2307-bp DNA from the pRS313–Taf40 template. The amplified fragment contained the Taf40 ORF as well as 650 bp of upstream and 618 bp of the downstream sequence. The mutagenized PCR product was cotransformed into yeast strain YSB373 [MATα, ura3-52, leu2, his3Δ200, taf40Δ::LEU2 (pJA73-TAF40a)] with a 5290-bp NcoI–BamHI- gapped vector fragment from pRS313–Taf40. In vivo recombination was mediated by 354-bp overlap with the upstream end and 186-bp overlap with the downstream end of the PCR product. Transformants were selected on synthetic complete medium lacking histidine and then replica plated onto medium containing 5-fluoro-orotic acid (5-FOA) to shuffle out the wild-type TAF40 plasmid marked with URA3. Temperature-sensitive alleles in TAF40 were identified by replica plating 5-FOA-resistant cells at 30°C and 37°C. Plasmids were recovered and retransformed into YSB373 to confirm plasmid linkage of temperature-sensitive mutants. To test for high-copy suppression, 2μ plasmids carrying the indicated genes were transformed into YSB681 [MATa, ura3-52, leu2, trp1Δ1, his3Δ200, taf40Δ::LEU2 (pRS313-taf40-3100)] and the resulting strains were plated at 30°C and 37°C.
Protein analysis
Whole cell extracts were prepared by glass bead disruption in lysis buffer (10 mm Tris-HCl at pH 7.4, 1 mm EDTA, 0.5% SDS) supplemented with 1 mm PMSF. Immunoblotting was done using standard methods. Fifty micrograms of protein from each sample was subjected to SDS-PAGE and blotted to Immobilon-P PVDF membrane (Millipore). Blots were incubated with rabbit polyclonal serum directed against the indicated proteins, followed by detection with goat anti-rabbit IgG conjugated with horseradish peroxidase and chemiluminescent detection.
RNA analysis
Yeast was grown to early log phase at 30°C, briefly heated to prevent heat shock artifacts, and after an additional 30 min at 30°C shifted to 37°C. Equal numbers of cells were harvested at the indicated times and washed once with cold water. RNA was isolated using hot acid phenol extraction (Ausubel et al. 1991) and quantitated by measuring A260. The integrity and normalization of the RNA was confirmed by ethidium bromide staining of RNA in agarose gels.
To monitor specific genes, S1 nuclease protection assays were carried out with 35μg of RNA and 0.1 pmole oligonucleotide probes as described (Cormack and Struhl 1992). Oligonucleotides DED1, RPS4, and tRNAW (Cormack and Struhl 1992) and ACT1 (Thompson and Young 1995) were as described. HTA2 oligonucleotide sequence is 5′-GCAGCTGAACCAGCTTTACCACCTTTACCACCGGACATTATATATTAAATTTGCTCTTGTTCTGTACTT-3′.
To monitor overall Pol II transcription, slot blot analysis of total poly(A)+ RNA was performed (Thompson and Young 1995) using 2μg of RNA for each time point. Poly (dT) labeling and hybridization was performed as described (Michel et al. 1998).
Acknowledgments
We are extremely grateful to J. Reese, M. Green, and P.A. Weil for providing TAF antibodies; to L. Guarente for ADA antibodies; E. Elion for Rpl1 antibodies; and F. Pugh for the PGK1-TBP construct. We thank V. Podolny for technical assistance and M. Keogh for critical reading of the manuscript. This work was supported by grant GM46498 from the National Institutes of Health. S.B. is a Pew Scholar in the Biomedical Sciences and a Leukemia Society of America Scholar.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked ‘advertisement’ in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL steveb@hms.harvard.edu; FAX (617) 738-0516.
References
- Apone LM, Virbasius CA, Holstege FC, Wang J, Young RA, Green MR. Broad, but not universal, transcriptional requirement for yTAFII17, a histone H3-like TAFII present in TFIID and SAGA. Mol Cell. 1998;2:653–661. doi: 10.1016/s1097-2765(00)80163-x. [DOI] [PubMed] [Google Scholar]
- Auble DT, Hahn S. An ATP-dependent inhibitor of TBP binding to DNA. Genes & Dev. 1993;7:844–856. doi: 10.1101/gad.7.5.844. [DOI] [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current protocols in molecular biology. New York, NY: John Wiley and Sons; 1991. [Google Scholar]
- Bai Y, Perez GM, Beechem JM, Weil PA. Structure-function analysis of TAF130: Identification and characterization of a high-affinity TATA-binding protein interaction domain in the N terminus of yeast TAFII130. Mol Cell Biol. 1997;17:3081–3093. doi: 10.1128/mcb.17.6.3081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell B, Tora L. Regulation of gene expression by multiple forms of TFIID and other novel TAFII-containing complexes. Exp Cell Res. 1999;246:11–19. doi: 10.1006/excr.1998.4294. [DOI] [PubMed] [Google Scholar]
- Birck C, Poch O, Romier C, Ruff M, Mengus G, Lavigne AC, Davidson I, Moras D. Human TAFII28 and TAFII18 interact through a histone fold encoded by atypical evolutionary conserved motifs also found in the SPT3 family. Cell. 1998;94:239–249. doi: 10.1016/s0092-8674(00)81423-3. [DOI] [PubMed] [Google Scholar]
- Chen JL, Attardi LD, Verrijzer CP, Yokomori K, Tjian R. Assembly of recombinant TFIID reveals differential coactivator requirements for distinct transcriptional activators. Cell. 1994;79:93–105. doi: 10.1016/0092-8674(94)90403-0. [DOI] [PubMed] [Google Scholar]
- Cormack BP, Struhl K. The TATA-binding protein is required for transcription by all three nuclear RNA polymerases in yeast cells. Cell. 1992;69:685–696. doi: 10.1016/0092-8674(92)90232-2. [DOI] [PubMed] [Google Scholar]
- Dikstein R, Ruppert S, Tjian R. TAFII250 is a bipartite protein kinase that phosphorylates the basal transcription factor RAP74. Cell. 1996;84:781–790. doi: 10.1016/s0092-8674(00)81055-7. [DOI] [PubMed] [Google Scholar]
- Eisenmann DM, Arndt KM, Ricupero SL, Rooney JW, Winston F. SPT3 interacts with TFIID to allow normal transcription in Saccharomyces cerevisiae. Genes & Dev. 1992;6:1319–1331. doi: 10.1101/gad.6.7.1319. [DOI] [PubMed] [Google Scholar]
- Grant PA, Workman JL. Transcription. A lesson in sharing? Nature. 1998;396:410–411. doi: 10.1038/24723. [DOI] [PubMed] [Google Scholar]
- Grant PA, Schieltz D, Pray-Grant MG, Steger DJ, Reese JC, Yates JRI, Workman JL. A subset of TAFs are integral components of the SAGA complex required for nucleosome acetylation and transcriptional stimulation. Cell. 1998;94:45–53. doi: 10.1016/s0092-8674(00)81220-9. [DOI] [PubMed] [Google Scholar]
- Hahn S. The role of TAFs in RNA polymerase II transcription. Cell. 1998;95:579–582. doi: 10.1016/s0092-8674(00)81625-6. [DOI] [PubMed] [Google Scholar]
- Hampsey M. Molecular genetics of the RNA polymerase II general transcriptional machinery. Microbiol Mol Biol Rev. 1998;62:465–503. doi: 10.1128/mmbr.62.2.465-503.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez N. TBP, a universal eukaryotic transcription factor? Genes & Dev. 1993;7:1291–1308. doi: 10.1101/gad.7.7b.1291. [DOI] [PubMed] [Google Scholar]
- Hoffmann A, Oelgeschlager T, Roeder R G. Considerations of transcriptional control mechanisms: do TFIID-core promoter complexes recapitulate nucleosome-like functions? Proc Natl Acad Sci. 1997;94:8928–8935. doi: 10.1073/pnas.94.17.8928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holstege FC, Jennings EG, Wyrick JJ, Lee TI, Hengartner CJ, Green M R, Golub TR, Lander ES, Young RA. Dissecting the regulatory circuitry of a eukaryotic genome. Cell. 1998;95:717–728. doi: 10.1016/s0092-8674(00)81641-4. [DOI] [PubMed] [Google Scholar]
- Jackson-Fisher AJ, Chitikila C, Mitra M, Pugh BF. A role for TBP dimerization in preventing unregulated gene expression. Mol Cell. 1999;3:717–727. doi: 10.1016/s1097-2765(01)80004-6. [DOI] [PubMed] [Google Scholar]
- Kim YJ, Bjorklund S, Li Y, Sayre MH, Kornberg RD. A multiprotein mediator of transcriptional activation and its interaction with the C-terminal repeat domain of RNA polymerase II. Cell. 1994;77:599–608. doi: 10.1016/0092-8674(94)90221-6. [DOI] [PubMed] [Google Scholar]
- Klebanow ER, Poon D, Zhou S, Weil PA. Cloning and characterization of an essential Saccharomyces cerevisiae gene, TAF40, which encodes yTAFII40, an RNA polymerase II-specific TATA-binding protein-associated factor. J Biol Chem. 1997;272:9436–9442. doi: 10.1074/jbc.272.14.9436. [DOI] [PubMed] [Google Scholar]
- Kokubo T, Takada R, Yamashita S, Gong DW, Roeder RG, Horikoshi M, Nakatani Y. Identification of TFIID components required for transcriptional activation by upstream stimulatory factor. J Biol Chem. 1993;268:17554–17558. [PubMed] [Google Scholar]
- Kokubo T, Yamashita S, Horikoshi M, Roeder RG, Nakatani Y. Interaction between the N-terminal domain of the 230-kDa subunit and the TATA box-binding subunit of TFIID negatively regulates TATA-box binding. Proc Natl Acad Sci. 1994;91:3520–3524. doi: 10.1073/pnas.91.9.3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koleske AJ, Young RA. An RNA polymerase II holoenzyme responsive to activators. Nature. 1994;368:466–469. doi: 10.1038/368466a0. [DOI] [PubMed] [Google Scholar]
- Lee D, Lis JT. Transcriptional activation independent of TFIIH kinase and the RNA polymerase II mediator in vivo. Nature. 1998;393:389–392. doi: 10.1038/30770. [DOI] [PubMed] [Google Scholar]
- McNeil JB, Agah H, Bentley D. Activated transcription independent of the RNA polymerase II holoenzyme. Mol Cell Biol. 1998;19:2510–2521. doi: 10.1101/gad.12.16.2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel B, Komarnitsky P, Buratowski S. Histone-like TAFs are essential for transcription in vivo. Mol Cell. 1998;2:663–673. doi: 10.1016/s1097-2765(00)80164-1. [DOI] [PubMed] [Google Scholar]
- Mizzen CA, Yang XJ, Kokubo T, Brownell JE, Bannister AJ, Owen-Hughes T, Workman J, Wang L, Berger SL, Kouzarides T, Nakatani Y, Allis CD. The TAFII250 subunit of TFIID has histone acetyltransferase activity. Cell. 1996;87:1261–1270. doi: 10.1016/s0092-8674(00)81821-8. [DOI] [PubMed] [Google Scholar]
- Moqtaderi Z, Bai Y, Poon D, Weil PA, Struhl K. TBP-associated factors are not generally required for transcriptional activation in yeast. Nature. 1996a;383:188–191. doi: 10.1038/383188a0. [DOI] [PubMed] [Google Scholar]
- Moqtaderi Z, Yale JD, Struhl K, Buratowski S. Yeast homologues of higher eukaryotic TFIID subunits. Proc Natl Acad Sci. 1996b;93:14654–14658. doi: 10.1073/pnas.93.25.14654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moqtaderi Z, Keaveney M, Struhl K. The histone H3-like TAF is broadly required for transcription in yeast. Mol Cell. 1998;2:675–682. doi: 10.1016/s1097-2765(00)80165-3. [DOI] [PubMed] [Google Scholar]
- Natarajan K, Jackson BM, Rhee E, Hinnebusch AG. yTAFII61 has a general role in RNA polymerase II transcription and is required by Gcn4p to recruit the SAGA coactivator complex. Mol Cell. 1998;2:683–692. doi: 10.1016/s1097-2765(00)80166-5. [DOI] [PubMed] [Google Scholar]
- Oelgeschlager T, Tao Y, Kang Y K, Roeder R G. Transcription activation via enhanced preinitiation complex assembly in a human cell-free system lacking TAFIIs. Mol Cell. 1998;1:925–931. doi: 10.1016/s1097-2765(00)80092-1. [DOI] [PubMed] [Google Scholar]
- Ogryzko VV, Kotani T, Zhang X, Schlitz RL, Howard T, Yang XJ, Howard BH, Qin J, Nakatani Y. Histone-like TAFs within the PCAF histone acetylase complex. Cell. 1998;94:35–44. doi: 10.1016/s0092-8674(00)81219-2. [DOI] [PubMed] [Google Scholar]
- Rabenstein MD, Zhou S, Lis JT, Tjian RT. TATA box-binding protein (TBP)-related factor 2 (TRF2), a third member of the TBP family. Proc Natl Acad Sci. 1999;96:4791–4796. doi: 10.1073/pnas.96.9.4791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reese JC, Apone L, Walker SS, Griffin LA, Green MR. Yeast TAFIIs in a multisubunit complex required for activated transcription. Nature. 1994;371:523–527. doi: 10.1038/371523a0. [DOI] [PubMed] [Google Scholar]
- Sanders SL, Klebanow ER, Weil PA. TAF25p, a non-histone-like subunit of TFIID and SAGA complexes, is essential for total mRNA gene transcription in vivo. J Biol Chem. 1999;274:18847–18850. doi: 10.1074/jbc.274.27.18847. [DOI] [PubMed] [Google Scholar]
- Soldatov A, Nabirochkina E, Georgieva S, Belenkaja T, Georgiev P. TAFII40 protein is encoded by the e(y)1 gene: Biological consequences of mutations. Mol Cell Biol. 1999;19:3769–3778. doi: 10.1128/mcb.19.5.3769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterner DE, Grant PA, Roberts SM, Duggan LJ, Belotserkovskaya R, Pacella LA, Winston F, Workman J L, Berger SL. Functional organization of the yeast SAGA complex: Distinct components involved in structural integrity, nucleosome acetylation, and TATA-binding protein interaction. Mol Cell Biol. 1999;19:86–98. doi: 10.1128/mcb.19.1.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson CM, Young RA. General requirement for RNA polymerase II holoenzymes in vivo. Proc Natl Acad Sci. 1995;92:4587–4590. doi: 10.1073/pnas.92.10.4587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verrijzer CP, Tjian R. TAFs mediate transcriptional activation and promoter selectivity. Trends Biochem Sci. 1996;21:338–342. [PubMed] [Google Scholar]
- Walker SS, Reese JC, Apone LM, Green MR. Transcription activation in cells lacking TAFIIs. Nature. 1996;383:185–188. doi: 10.1038/383185a0. [DOI] [PubMed] [Google Scholar]
- Wu SY, Kershnar E, Chiang CM. TAFII-independent activation mediated by human TBP in the presence of the positive cofactor PC4. EMBO J. 1998;17:4478–4490. doi: 10.1093/emboj/17.15.4478. [DOI] [PMC free article] [PubMed] [Google Scholar]