Abstract
In an effort to control the variability of transgene expression in plants, we used Cre-lox mediated recombination to insert a gus reporter gene precisely and reproducibly into different target loci. Each integrant line chosen for analysis harbors a single copy of the transgene at the designated target site. At any given target site, nearly half of the insertions give a full spatial pattern of transgene expression. The absolute level of expression, however, showed target site dependency that varied up to 10-fold. This substantiates the view that the chromosome position can affect the level of gene expression. An unexpected finding was that nearly half of the insertions at any given target site failed to give a full spatial pattern of transgene expression. These partial patterns of expression appear to be attributable to gene silencing, as low gus expression correlates with DNA methylation and low transcription. The methylation is specific for the newly integrated DNA. Methylation changes are not found outside of the newly inserted DNA. Both the full and the partial expression states are meiotically heritable. The silencing of the introduced transgenes may be a stochastic event that occurs during transformation.
Keywords: Epigenetics, gene silencing, methylation, Cre-lox, site-specific recombination
Different levels of gene expression from independent transformants are common with plant or animal transformation systems. The variation in transgene expression has been attributed to several factors, including differences with respect to chromosome location, copy number, and transgene construct fidelity (Matzke and Matzke 1998; Kooter et al. 1999). Similar epigenetic phenomena are also associated with nontransgenic loci, such as position-effect variegation (PEV) and paramutation (Hollick et al. 1997; Wakimoto 1998).
Chromosome locations that promote transgene expression are not well characterized but are thought to be transcriptionally active regions of euchromatin. The heterochromatin state is thought to be inaccessible to transcription factors and often correlates with cytosine hypermethylation and histone hypoacetylation (Ng and Bird 1999). A consequence of this chromosome architecture is that transgenes that randomly integrate in the proximity of heterochromatin can show variable levels of expression (Dobie et al. 1996; Iglesias et al. 1997). For both transgenic and nontransgenic loci, repeat sequences have been implicated in gene silencing (Selker 1999). In Drosophila, plants, and mice, experiments with variable transgene repeats at the same locus support this hypothesis (Assaad et al. 1993; Sabl and Henikoff 1996; Garrick et al. 1998). A proposed role for the repeat sequences is to induce DNA–DNA interactions that lead to changes in chromatin conformation and epigenetic silencing (Matzke and Matzke 1998).
The effect of repeat sequences on transgene expression in plants has led to the term homology-dependent gene silencing (HDGS) (Kooter et al. 1999). Similar phenomena have also been reported in Drosophila (Pal-Bhadra et al. 1997), mice (Garrick et al. 1998), and fungi (Cogoni and Macino 1999). HDGS can occur at the level of transcription (Meyer et al. 1993; Park et al. 1996) or post-transcription (English et al. 1997). Transcriptional HDGS (TGS) is associated with homology in promoter regions (Park et al. 1996). In post-transcriptional HDGS (PTGS), mRNA transcripts are destabilized by dsRNA formed from complimentary RNA (cRNA) of the silenced gene (Waterhouse et al. 1998; Grant 1999). The cRNA might be produced by endogenous promoters adjacent to the transgene or read-through transcription of inverted repeats in a complex transgene (Kooter et al. 1999). Alternatively, aberrant transcripts may be produced that stimulate the production of cRNA by RNA-dependent RNA polymerase (Jorgensen et al. 1999). In Caenorhabditis elegans and Drosophila, the descriptions of RNA interference (RNAi) have substantiated the view that dsRNA can act as a potent and specific inhibitor of gene expression (Sharp 1999).
In recent years, the integration of DNA through use of recombinase-mediated site-specific integration systems has been described (Fukushige and Sauer 1992; Albert et al. 1995; Golic et al. 1997). Common features of these systems are that (1) the DNA inserts into a designated site in the genome; (2) the DNA inserts often as a single copy; and (3) the integrated molecule is usually free of rearrangements often seen with random DNA insertions. Hence, a commonly held belief is that recombinase-mediated DNA integration would produce consistent expression of the introduced DNA.
In this study, we find that a full spatial pattern of transgene expression can be obtained reproducibly through specific integration into a designated genomic site. Moreover, the absolute level of full expression is dependent on the site of integration. Contrary to expectation, however, we also recover a large percentage of integration events that give a partial spatial pattern of transgene expression. These events are not site dependent and appear to be due to a heritable form of gene silencing.
Results
Site-specific placement of a reporter gene
The experimental design was to insert pEL1 into 35S-lox-cre plant lines (Fig. 1A), in which 35S is the 35S RNA promoter from cauliflower mosaic virus, cre is the coding region for Cre recombinase, and lox is the 34-bp recombination site incorporated into the leader sequence of the transcript. This was achieved by transforming leaf mesophyll protoplasts, derived from 35S-lox-cre plants, with the pEL1 plasmid that contains lox-hpt and Cp-gus, in which hpt is the hygromycin phosphotransferase coding region, gus is the β-glucuronidase coding region, and Cp is the commelina yellow mottle virus promoter (Medberry et al. 1992). Cre-mediated integration of pEL1 into the genomic 35S-lox-cre target confers a hygromycin resistant (HygR) phenotype through a 35S-lox-hpt linkage, and abolishes cre transcription through displacement of the 35S promoter. Selection is not imposed on the linked Cp-gus reporter gene. Table 1 lists the five hemizygous 35S-lox-cre plant lines used as target lines.
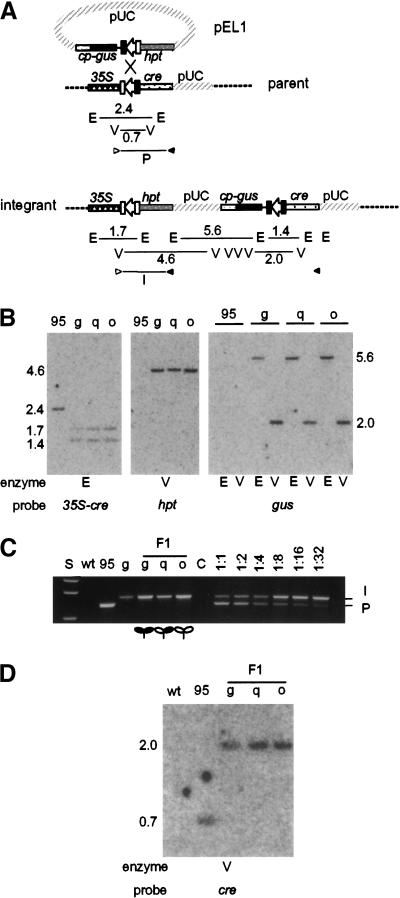
Figure 1.
Precise and stable integration of transgene. (A) Schematic map of the genomic target and the structure produced through integration of pEL1. The lox site is depicted by an open arrow with flanking rectangles that represent the 8-bp core and the flanking 13-bp Cre-binding sites, respectively. (Solid rectangle) Mutant sequence; when present on both sides of the core sequence, the site recombines less efficiently (Albert et al. 1995). Not shown are nos3′ terminator sequences of cre, hpt, and gus. Endonuclease sites shown: E, EcoRI; V, EcoRV. Fragment sizes, in kilobases, correspond to those next to the gels in (B,D). (B) Representative Southern blots of parent 95 and integrant T0 plants, 95g, 95q, and 95o, that show single precise copy pEL1 integration. (C) PCR products specific to parental (P) or F1 integrant (I) DNA detected from individual seedlings. Primers corresponding to 35S (open arrowhead) and nos terminator (solid arrowhead) are shown in A. Primer-only control lane is C. Lanes after C are reconstruction experiments of 95 and 95g DNA mixed, respectively, at ratios from 1:1 to 1:32. Expression of gus is depicted pictorially; solid and open leaves represent active and inactive gus expression, respectively. (D) Southern blot showing stable integration of pEL1 in integrant lines.
Table 1.
Expression of site-specific integrants
| Parent line
|
Transformation date
|
Integrant line
|
GUS pattern in leaves (0%–100%, n = 10)
|
GUS activity in leaves (pmole/min/mg, n = 3)
|
|---|---|---|---|---|
| nt35S-loxP-cre.5 | 4/26/95 | 95g | 100 | 81 |
| 95ea | 100 | 98 | ||
| 95fa | 100 | 67 | ||
| 5/5/95 | 95h | 100 | 98 | |
| 95sa | 100 | 27 | ||
| 5/23/95 | 95o | 0 | N.D. | |
| 95q | 10 | N.D. | ||
| 95pa | 5 | N.D. | ||
| nt35S-loxP-cre.11 | 5/5/95 | 911e | 100 | 533 |
| 5/23/95 | 911c | 30 | N.D. | |
| 911h | 0 | N.D. | ||
| 911Ia | 30 | N.D. | ||
| 911ba | 0 | N.D. | ||
| 6/2/95 | 911d | 5 | N.D. | |
| nt35-loxP-cre3 | 6/8/95 | 93b | 10 | N.D. |
| 93a | 0 | N.D. | ||
| 9/7/95 | 93g | 100 | 111 | |
| 93fa | 100 | 49 | ||
| nt35S-lox66-cre.5 | 8/31/95 | 75c | 5 | N.D. |
| 75d | 45 | N.D. | ||
| nt35S-lox76.cre.A | 8/31/95 | 85b | 100 | 845 |
| 85da | 100 | 743 |
N.D., not determined.
Potential sibling of another line from the same transformation.
Molecular analysis of integrant plants
A number of putative integrants from each experiment were analyzed by PCR for the presence of the expected 35S-lox-hpt junction. This junction was found in 77 of 81 candidates. The four plants that failed to yield a PCR product may be due to hpt fusion to an endogenous promoter in the genome. The 77 plants were analyzed by Southern hybridization. DNA treated with EcoRI was probed with a 35S-cre fragment. An intact left and right junction was confirmed in 73 of the 77 plants through detection of the 1.7-kb 35S-lox-hpt and the 1.4-kb gus-lox-cre band, and the loss of the 2.4-kb parental 35-lox-cre fragment (Fig. 1B). With EcoRV-treated DNA, a gus-specific probe hybridized to only one 2.0-kb gus-lox-cre fragment in 35 of the 73 plants (Fig. 1B). The presence of one or more additional bands in the other plants indicated more than a single copy of Cp-gus DNA, whether situated in tandem at the same site, or elsewhere in the genome.
These 35 plants were examined by hpt-specific hybridization to EcoRV-treated DNA that should detect a 4.6-kb 35S-lox-hpt-pUC backbone fragment (Fig. 1B). They were also examined by gus-specific hybridization to EcoRI-cleaved DNA that should reveal a 5.6-kb pUC-Cp-gus fragment (Fig. 1B). Fragments of the predicted sizes were found in 22 of the 35 single-copy integrant plants. Together, the detection of all five (three EcoRI and two EcoRV) overlapping fragments shows a contiguous arrangement of the pEL1 insertion as illustrated in Figure 1A. The failure to detect fragments of the expected sizes could be attributable to either of two events: (1) random integration of pEL1 followed by recombination between the two lox sites; in these occurrences, the 35S-lox-hpt and gus-lox-cre junctions are found, but they are not contiguous; and (2) site-specific integration of pEL1, followed by DNA rearrangement, such as insertion or deletion between the two lox junctions, such that the expected-size bands are not found.
Reporter gene expression pattern
Table 1 lists the 22 plants found with a precise single-copy integration of pEL1. For the plants from the same transformation experiment, each was regenerated from a different cluster of immobilized cells. The chances are remote for any two plants to be derived from the same protoplast during the brief incubation period before imbedding the protoplasts in agarose. Nonetheless, this possibility could not be entirely ruled out. Therefore, although we do not believe this could be the case, we yield to the possibility that eight lines might be sibling clones of the other 14 representative lines (Table 1, as indicated).
The primary transformants (T0) of the lines listed in Table 1 were crossed to wild-type tobacco to generate F1 progeny hemizygous for Cp-gus. Seedlings were germinated in the presence of hygromycin, and GUS enzyme activity was examined at the two-leaf stage by histochemical staining. Among the progeny of any given plant line, the staining pattern is highly consistent; hence, sibling seedlings are alike. The seedlings of different integrant lines, however, differ considerably. In four of the target sites, one to two integrant lines show the expression pattern previously reported for the Cp promoter, namely, expression throughout the plant in vascular tissue (Medberry et al. 1992). These lines have been designated as having a 100% value with regard to the pattern of Cp-gus expression. When quantified by enzyme activity, however, each locus has a distinct GUS activity level associated with the 100% gus pattern (Table 1). In other integrant lines, seedlings showed staining in only some tissue, as though the reporter gene has been silenced. These lines have been assigned a numeric value to indicate the approximate percentage of blue staining observed (Table 1). Figure 2 shows the typical staining patterns of three independent integrants at the same locus. The three lines 95g, 95q, and 95o have served as representative lines for detailed analysis.
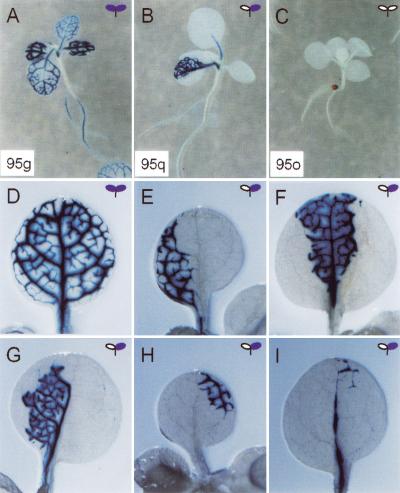
Figure 2.
Independent integrants at the same locus show different spatial patterns of gus expression. All seedlings are stained for GUS activity at the two-leaf stage. At the 95 locus, the 95g line has a vascular specific expression characteristic of the Cp promoter (A,D). The 95q line shows sectoring of gus expression (B,E–I), and the 95o line rarely shows gus expression (C).
In the silenced lines 95q and 95o, the blue patches of staining have sharp boundaries that define blue sectors. Typical sectors have boundaries that either divide a leaf in half (Fig. 2B) or follow a boundary between the midvein and the margin (Fig. 2E,F). The smaller sectors are still quite large (Fig. 2H,I), suggesting that the reversion of Cp-gus silencing occurs early during leaf development. These sectors are similar to clonal L2 sectors because they extend to the leaf margin. Less common L3-like sectors are also seen (Fig. 2G). These patterns suggest a clonal origin as they are similar to clonal sectors in tobacco (Poethig 1987).
When F1 seedlings are grown with or without selection for hpt or nptII, the degree of GUS expression does not change (data not shown). This indicates that the silencing phenomenon is not related to expression of linked marker genes. In F2 hemizygous seedlings, obtained from outcross to wild type as either male or female donor, the sectoring patterns are very similar to the F1 seedlings. The only minor difference was that in 95o, small blue sectors can be seen in ∼25% of the F2 cotyledons, whereas this was not seen with the F1 cotyledons unless the F1 seedlings were treated with 5-azacytidine (see below). This is an indication that there is some, but not significant, reactivation of Cp-gus in the F2 generation. Siblings that show an all-blue pattern (as in 95g) were not observed among more than 100 F2 seedlings of 95q or 95o. This can be interpreted to indicate that either cells representing the blue sectors in 95q and 95o are not progenitors of the germ line, or that the transgene is reset to the “off” state in the gamete. Because a limited number of flowers were outcrossed to produce seed, we cannot rule out the possibility that flowers with “blue” germ line sectors existed but were not selected for outcrossing. The observations made with F2 seedlings were also found in the F3 and F4 generation. Therefore, the silencing phenomenon is meiotically transmissible at high fidelity, through either male or female gametes.
The integrated DNA is stable
With 95q and 95o, the mosaic patterns of expression suggest that the Cp-gus transgene is active or inactive, respectively, in the blue or white sectors. The possibility that this phenotype could be due to instability of the integrated DNA was a consideration. Excision may be caused, for example, by residual Cre activity. To determine if this could be the cause, we used PCR to assay for the presence of the integration (35S-lox-hpt) or excision (35S-lox-cre) junctions. Both hpt and cre transcripts use the terminator sequence of the nopaline synthase gene (nos3′). Primers corresponding to 35S and nos3′ can amplify specific 35S-lox-hpt and 35S-lox-cre junction products. The sensitivity of the PCR assay is shown in Figure 1C. When genomic DNA from the parent and the T0 integrant lines were mixed in known ratios, the parental junction was visible in as low as 1 parent to 32 integrant DNA. Should the colorless phenotype observed in 95o be caused by excision of pEL1 from the genome, the assay would detect the excision-specific parental junction. Of the five representative F1 plants of 95g, 95q, or 95o analyzed by PCR, none showed the band representing the excision junction. In contrast, all showed the band representing the integration junction.
Genomic DNA prepared from pools of 10 F1 seedlings was also subjected to Southern analysis with a cre-specific probe (Fig. 1D). Although the gus-lox-cre integration junction was detected in all three lines, a band corresponding to the 35S-lox-cre excision junction was not observed. Both the PCR and the Southern data support the conclusion that the integrated DNA is stable. Therefore, the inactivity of the Cp-gus transgene is not attributable to excision of pEL1 from the target site.
Activation and developmental maintenance of silenced transgene
The pattern of small blue sectors seen in large colorless areas but not vice versa suggested that the blue sectors represent an activation of gene expression from an initial inactive state. To examine if the blue sectors were caused by the switching on of the transgene, as opposed to switching off the transgene in the colorless sectors, we stained the F1 seedlings at different stages after germination. The 95g line showed staining in the embryo a day after germination and the blue staining was restricted to the vascular tissue by day 11 (Fig. 3A–C3). In contrast, lines 95q and 95o did not show Cp-gus expression in the seedlings until days 9 to 11. This is when early leaf sectors in 95q and the first sign of blue staining near the shoot apical meristem in 95o could be seen (Fig. 3D,E). This is consistent with the interpretation that the Cp-gus transgene in the silenced lines was not initially expressed but was switched on later in development.
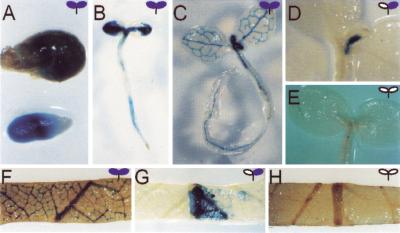
Figure 3.
Expression pattern during early stages of seedling development (A–E) and in mature leaves (F–H). Expression in 95g is observed at 3 (A), 7 (B), and 11 (C) days after germination. By day 11, expression is detected in the leaves of 95q (D), but not 95o (E). Expression patterns in mature leaves (adjacent to the inflorescence) of 95g (F), 95q (G), and 95o (H) are consistent with those observed in seedlings.
To examine if the expression pattern is maintained through development, we stained the second to last leaves before the inflorescence of fully grown plants for GUS activity. The expression pattern was similar to those seen at the two-leaf seedling stage (Fig. 3F–H). It appears that each line not only has a particular frequency at which Cp-gus expression switches from an off to an on state, but that the frequency is maintained through organismal development.
Steady state mRNA and reporter gene activity
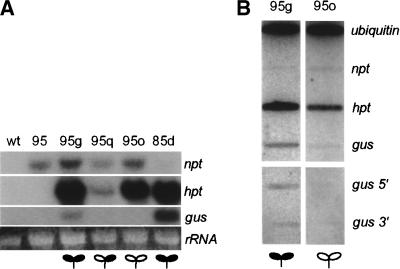
We examined if the silencing of Cp-gus in 95o and 95q is related to the steady state level of Cp-gus transcripts. RNA was prepared from a pool of 10 F1 seedlings of 95g, 95q, and 95o grown on hygromycin selection, and from the 95 parent line grown on kanamycin selection. Indeed, the level of Cp-gus mRNA correlated to the amount of GUS staining. 95g stained blue and Cp-gus transcript was detected by Northern analysis, whereas, 95q and 95o showed little GUS staining and Cp-gus transcripts were not detected (Fig. 4A). This correlation can be extended between loci because the level of gus transcript for 85d is significantly higher than 95g, similar to the GUS enzyme activity measured for the two lines (Table 1).
Figure 4.
Gene silencing acts at the level of transcription. (A) Steady state mRNA of parental and integrant plants. Probes for nptII, hpt, and gus mRNA were used in Northern blots. Loading control is large ribosomal RNA stained with ethidium bromide. (B) Run-on transcription from leaf nuclei of active and inactive lines. Radioactive transcripts were used to probe target DNA specific for the full-length ubiquitin, nptII, hpt, and gus genes. In an independent experiment, 5′- and 3′-specific regions of the gus gene were used as target DNA (bottom). The run-on transcripts from the 95g and 95o nuclei were hybridized to the target DNA and the signals from each filter were normalized relative to the ubiquitin signal.
To determine whether this observation is Cp-gus specific, or is characteristic of the locus, we measured the transcript levels of other transgenes of the integration locus. The nptII gene was used as the selectable marker for the first parental (35S-lox-cre) transgene. It therefore is present in both the parent and integrant lines. The hpt and gus genes are present only in the integrant lines as they were introduced through pEL1. The transcript levels of nptII are rather similar among the integrant and parent lines, whereas large differences are observed for hpt (Fig. 4A). The steady state level of hpt mRNA in 95q is lower than in 95g, consistent with that observed for Cp-gus. However, in the 95o line, the hpt transcript level is similar to that found in 95g, whereas the Cp-gus transcript is not detectable. Therefore, the data for nptII and hpt expression levels are inconsistent with a locus-wide silencing effect.
Nuclear run-on experiments were conducted to examine the rate of transcription (Fig. 4B). Compared with 95g, the transcription initiation rates of the gus and hpt genes in 95o are reduced by 10- and 3-fold, respectively, when compared with the ubiquitin gene standard. There is relatively low transcription from nptII, but the two lines appear to show similar rates. This supports an interpretation that the silencing phenomenon observed is attributable to an inhibition of transcription.
Gene dosage and allelic interactions
Genetic interactions between alleles of a locus that show HDGS have been reported (English et al. 1997; Vaucheret et al. 1997). In those instances, a positive gene dosage from an allelic or a nonallelic locus often correlates inversely with gene activity. In addition, it is common in HDGS for a silencing locus to act in trans to inactivate an otherwise actively expressing homologous gene (English et al. 1997).
When gus staining patterns of the self-fertilized T1 seedlings were examined, a third of the HygR 95g seedlings showed a more rapid rate of staining. Because this is the proportion of HygR seedlings expected to be homozygous for Cp-gus, this suggests that a higher level of expression may be attributable to the higher transgene dosage. When hemizygous 95g was crossed to hemizygous 95o, the HygR progeny pool should be comprised of the genotypes 95g/+, 95o/+, and 95g/95o in a ratio of 1 : 1 : 1. When the HygR progeny were analyzed, 85% (n = 15) had the all-blue phenotype characteristic of 95g. Similarly, in the cross between hemizygous 95g and hemizygous 95q, 76% (n = 49) had the 95g phenotype. A few of the blue plants do have paler leaves, so we cannot entirely rule out that a limited interaction can occur between the alleles. To examine for possible nonallelic interactions, we also crossed hemizygous 95g to hemizygous 911d. Of the HygR progeny, 60% (n = 20) showed the all-blue phenotype. In sum, the data do not suggest that the silenced alleles can exert a dominant negative effect on the nonsilenced alleles.
Cp-gus methylation correlates with silencing
Methylation sensitive and insensitive enzymes were used to probe the extent of DNA methylation in active and silenced Cp-gus transgenes at the different loci. The genomic DNA was first cleaved with methylation insensitive enzymes EcoRV and EcoR1 into defined 5′ (880-bp) and 3′ (1080-bp) fragments, and then further cleaved with methylation-sensitive enzymes HpaII, Sau3A, or BglII (Fig. 5). A methylation signature of small bands is detected by Southern blotting with either the Cp-gus 5′ fragment or a gus 3′ fragment. Analysis of the different size bands infers the methylation state of specific cytosines in the target sequence.
Figure 5.
Methylation analysis of active and inactive integrant lines at the 95, 93, and 911 target sites. (A) Predicted DNA fragments, in basepairs, of Cp-gus from cleavage with EcoRV and EcoRI and either BglII, Sau3A, or HpaII. Southern blots reveal the fragments hybridizing to a Cp-gus 5′ fragment probe (for BglII and Sau3A panels) or a gus 3′ fragment probe (for HpaII panel). Leftmost control lanes were not cleaved with BglII, Sau3A, or HpaII. Numbers next to autoradiograms correspond to sizes of predicted fragments (bp). Endonuclease sites shown are as follows: B, BglII; E, EcoRI; H, HpaII; S, Sau3A; V, EcoRV. (B) Predicted DNA fragments, in basepairs, of lox-cre after cleavage with EcoRV, EcoRI, and HpaII and detected by hybridization to a cre probe. Outermost control lanes were not cleaved with HpaII.
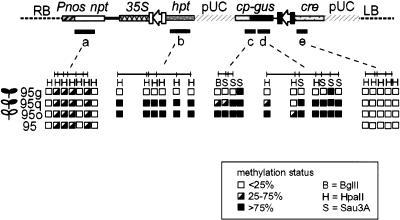
The 95o line is almost fully methylated at the restriction sites we assayed including sites in the promoter, coding region, and the 3′ untranslated region (UTR) (Figs. 5,6). Very few of the sites are methylated in the 95g line. The 95q line, with sectors of GUS expression, was partially methylated in the promoter and the coding region but was fully methylated in the 3′ UTR. Analysis of the promoter and coding region of Cp-gus was extended to the 93 and 911 loci. Lines that show active (93g and 911e) or relatively inactive (93b and 911b) Cp-gus expression also have a correlation between cytosine methylation in Cp-gus and low GUS activity (Fig. 5). For the 911 locus, hypermethylation in the Cp promoter and the gus coding region was detected in the 911b silenced allele. At the 93 locus, the Cp promoter and 5′ region of the gus gene are partially methylated in both 93 alleles, but differences were detected in the middle of the gus gene. In this region, the silenced allele 93b showed complete methylation whereas the expressed allele 93g is unmethylated. The analysis of three different loci all correlate hypermethylation to a silenced state, especially at the middle of the gus coding region.
Figure 6.
Methylation patterns in parental and integrant lines. Summary of methylation analysis at specific restriction sites in 95 and integrant lines 95g, 95q, and 95o. Specific probes (a, b, c, d, e) represent nptII, hpt, Cp-gus 5′ fragment, gus 3′ fragment and cre, respectively. (RB and LB) the positions of the T-DNA right border and left border, respectively. Endonuclease sites are as follows: S, Sau3A; B, BglII; H, HpaII.
Methylation is restricted to integrant DNA
To determine the extent of methylation beyond Cp-gus, we used probes for nptII, hpt, and cre to compare the methylation state throughout the 95 locus. Despite a positive correlation between hypermethylation and Cp-gus silencing, the hpt steady state mRNA data are inconsistent. When hpt was used to probe the methylation of HpaII restriction sites in 95q and 95o, those sites were fully methylated (Fig. 6). As the plants are HygR, both lines express hpt, but the 95o line accumulates a much higher level of hpt steady state mRNA (Fig. 4A). A lower level of methylation might have been expected for the 95o hpt gene, but there are examples of genes that express normally despite high levels of methylation (Luff et al. 1999).
For the nptII and cre probes, we also analyzed the 95 parental line to determine the level of methylation at the locus before transgene integration. Few of the HpaII restriction sites were methylated in the cre (Figs. 5,6) or nptII (Fig. 6) gene in either the parental or the integrant lines. This suggests that the low level of methylation in the 95 parent is maintained in the new transgenic lines, despite the variability of methylation seen among the newly introduced DNA.
Chromatin and methylation inhibitors partially activate expression
To test whether silenced states can reverse by manipulating the extent of DNA methylation, we treated the silenced 95o line with 5-azacytidine. This drug causes the genome to become hypomethylated. Plants were grown on a nontoxic concentration of 5-azacytidine and then stained for GUS activity. Approximately 18% of the total leaf area stained for GUS activity in sectoring patterns similar to 95q (Fig. 7B–E). This suggests that by hypomethylating the genome it is possible to reactivate Cp-gus from its silenced state. We also tested sodium butyrate, a deacetylase inhibitor, which perturbs the formation of chromatin. This inhibitor was also found to reactivate the Cp-gus, although not as well as 5-azacytidine. Again, sectoring was seen to increase and the area of leaves that stained was approximately 4% (Fig. 7F–H). It is possible that the reactivation of Cp-gus is not efficient due to insufficient application of the drugs. Higher concentrations, however, could not be used due to their toxicity to the tobacco seedlings. Although we did not see complete reactivation of Cp-gus, these experiments suggest that cytosine methylation and histone acetylation have a role in the silencing phenomenon. This substantiates the view that the correlation of methylation with gene silencing is not coincidental.
Figure 7.
Reactivation of Cp-gus expression in 95o by DNA methylation and acetylation inhibitors. (A) GUS activity from untreated cotyledons from the active line 95g. (B) Na butyrate–treated cotyledons from the inactive line 95o shows slight reactivation of expression. (C–H) Leaves of 95o treated with 5-azacytidine (C–E) or Na butyrate (F–H). Seedlings were grown on agar with 0.6 mM Na butyrate or 19 mg/L 5-azacytidine.
Discussion
This study was designed to test two hypotheses. First, we sought to determine whether placing a transgene into the same genomic target site would yield a reproducible level of expression. Second, we wanted to know if each target site might confer a distinct level of expression for independent integrant lines. The experimental strategy was designed to remove many of the variables that have been implicated to cause gene silencing in other systems, namely, the generation of inverted repeats, multiple transgene copies, and variable chromosome locations.
The data are consistent with both hypotheses. In three target loci in which two or more full spatial pattern lines were found (loci 95, 93, and 85), their levels of Cp-gus expression, as measured by enzyme activity, are quite similar to each other. In four target loci in which at least one full spatial pattern line was found, the Cp-gus expression is distinct for each target loci. Full expression at the 85 locus is close to 10-fold higher than that of the 95 locus. Therefore, the chromosome position is important for the level of transgene expression, and that reproducible insertion into the same chromosome position is capable of generating the same expression level.
What is most surprising is that gene silencing can also be obtained at a high frequency, resulting in a mosaic pattern of gene expression. This was not reported in other Cre-lox site-specific targeting experiments using mammalian cell lines (Fukushige and Sauer 1992; Feng et al. 1999). These groups assayed a marker gene at two and six genomic targets, respectively, and reported that transgene expression is reproducible at each site. Although the silencing effect we observed could reflect differences between plants and animals, it is also quite possible that silencing mechanisms in cultured cell lines behave quite differently from those in a differentiated whole organism. Several studies in mice have targeted transgenes to either the hprt or pim-1 loci by homologous recombination (Jasin et al. 1996). In some cases, variable expression of the transgene was observed, and this was postulated to be a position effect attributable to promoter occlusion. Further studies in transgenic mice have led to targeted integration with more stable expression, although few integration events have been studied in detail and variable expression from independent insertions remains a possibility (Jasin et al. 1996; Wallace et al 2000).
Genetic changes unlikely
The behavior of the silenced loci is inconsistent with a simple mutation theory. If a mutation had occurred to render the gene nonfunctional, then one would have to postulate that each silenced line has a distinct reversion rate. For example, the 95q and 95o lines would each have a unique and heritable reversion rate that occurs at a particular time during leaf development. A temporally nonrandom reversion rate that is also heritable places a severe limitation on a genetic change theory.
Cp-gus silencing compared with other examples of HDGS
The silencing of the Cp-gus reporter gene shares some of the characteristics of HDGS (Kooter et al. 1999). It correlates with hypermethylation of the transgene, there is meiotic transmission of the phenotype, and clonal sectors appear stochastically reminiscent of metastable alleles. Both PTGS and TGS can produce metastable alleles (Meyer et al. 1993; Davies et al. 1997), but in the case of PTGS, the sectors are nonclonal (Jorgensen 1995). In some cases, a silenced nonclonal sector can be transmitted throughout the plant by a systemic mechanism (Palauqui et al. 1997; Voinnet et al. 1998). Because the phenomenon we describe has clonal sectors, and that the nuclear run-on experiments showed reduced Cp-gus transcription, it is most likely TGS.
Nearly all TGS phenomena are attributed to an interaction between gene copies. Nonetheless, there are a few examples of a TGS-like phenomenon with single-copy transgenes. In one instance, the silencing of a single-copy transgene was attributed to its recognition as a nonplant sequence (Meyer and Heidmann 1994). In a second example, the transgene silencing was attributed to a position effect that influences chromatin structure (Iglesias et al. 1997).
The transgene in this study is a single hemizygous copy maintained by outcrossing to wild-type plants and thus would fall into this rare class of single-copy TGS.
Methylation confined to integrant DNA
The nonplant sequence model proposes a cellular scanning mechanism that recognizes foreign motifs in a transgene such as GC-rich, bacterial-like or unusual transgene/host boundary sequences (Matzke and Matzke 1998). This model is difficult to reconcile with our data. The Cp-gus is inserted precisely into a target transgene; therefore, no new plant/plasmid junctions are created from the integration. Moreover, only the new transgene DNA, and not the old transgene DNA, is subjected to hypermethylation. This is particularly striking when the silenced alleles of Cp-gus are highly methylated, and yet the adjacent cre gene does not change in methylation state. If a coordinated scanning mechanism can identify foreign DNA, either it does not achieve this by recognizing nonplant sequences or the nonplant sequences in our lox target lines are masked, perhaps by chromatin.
The position-effect model is similar to the stochastic silencing of PEV that is attributable either to the spread of heterochromatin into the gene or to the locus being looped into a heterochromatic complex (Wakimoto 1998). Either effect can silence other genes in the region as they are also incorporated into the heterochromatin (Dorer and Henikoff 1997). This model does not adequately explain the variegation we observe in our tobacco loci for two reasons. First, a spread of heterochromatin into the transgene should correlate with methylation across the 95 locus. Contrary to expectation, we observe an island of methylation that is specific to the integrant DNA. The possibility that integrant DNA has looped into a heterochromatic complex would also be unlikely because hpt and nptII are expressed. Note that nptII had not been used for selection after pEL1 integration. Second, because we observed gene silencing in most loci, the chromosome position-induced effect would have to be nearly ubiquitous throughout the genome.
Silenced alleles do not trans-inactivate
Epigenetic trans-inactivation and paramutation have been described in Drosophila and in maize (Dorer and Henikoff 1997; Hollick et al. 1997). In these instances, a silencer allele converts an active homolog to an inactive state, and the modification is meiotically stable after segregation of the trans-acting allele. Transgene analysis in both Drosophila and plants reveals that some silenced transgenes also behave in this manner. The transmission of the silenced state from one allele to another is believed to be facilitated by DNA pairing or chromatin interaction and are promoted by repetitive DNA (Hollick et al. 1997; Kooter et al. 1999). Most of the silencer alleles have gene duplications or complex structures (Jakowitsch et al. 1999; Luff et al. 1999). For the rare single-copy silencers, nearby repetitive sequences have been suggested to be the cause. In the case of the single-copy Pl′-mah gene in maize, its paramutagenic effect may be attributable to adjacent sequences of a transposable element (Hollick et al. 1997). Similarly, for the single-copy maize A1 transgene in petunia (Meyer et al. 1993), its ability to trans-inactivate may be explained by adjacent genomic elements. In this study, the silenced Cp-gus alleles 95o, 95q, or 911d did not affect the expression of the active 95g allele. This may suggest insufficient repetitive DNA elements that can cause silencing either adjacent to, or within, the transgenic loci.
The nos-nptII, 35S-hpt, Cp-gus, and cre genes are unique in the integrant loci and the tobacco genome; however, there are some direct repeat sequences in the locus. These include four nos terminator sequences 3′ of the nptII, hpt, gus, and cre genes and two pUC sequences, one in the integrant pEL1 plasmid and one 3′ of the cre gene in the target site. Potentially, these repetitive DNA might account for the silencing phenomenon we observe; however, there are several reasons why we do not consider this a likely interpretation. First, we do not see evidence for trans-inactivation that is commonly attributed to DNA–DNA interactions. Second, methylation and silencing induced by somatic pairing is usually restricted to sequences with homology (Luff et al. 1999; Melquist et al. 1999). In this study, the gus and hpt genes can be highly methylated despite being a single-copy sequence. If the nos or pUC repeats can induce methylation in hpt and gus, then cre should also be susceptible. However, this is not the case. Third, if the nos and pUC repeats can induce silencing of Cp-gus, we would expect the Cp-gus lines that show full expression to be susceptible to self-induced silencing. However, somatic or germinal silencing of the alleles that show full spatial patterns of expression was not observed. Therefore, all of the above suggest that the direct repeats do not play a key role in gene silencing at these loci.
The cre DNA is not associated with gene silencing
The strategy used in this study relies on integrating pEL1 between 35S and cre. We found no evidence of excision of the integrated molecule, which is consistent with the intended inactivation of cre transcription. Nonetheless, the presence of the cre coding region at the target construct is a common denominator in the integrant lines. The possibility of a cre effect on gene silencing, however, has been ruled out in a different set of integration experiments. One aspect of these experiments results in pEL1 integration into a number or 35S-lox-luc target lines, in which luc is the firefly luciferase gene (H. Albert et al., in prep.). The Cre recombinase was provided by transient expression, and cre DNA was not incorporated into the genome. F1 hemizygous seedlings with a single, precise integration of pEL1 at the target site likewise showed patterns of gene silencing as those described in Table 1. Therefore, the presence of cre DNA is not associated with the Cp-gus silencing phenomenon.
Silenced alleles meiotically heritable
Cubas et al. (1999) described a natural epiallele affecting the flower morphology of toadflax that is stable over many generations. The methylation pattern is not fixed because some revertant flowers are detected and these are shown to have lost their methylation. The role of methylation during development is still poorly understood, but it is clear that methylation patterns can become stable. The silenced Cp-gus alleles are relatively stable and can be maintained at least to the F4 generations. Occasionally, we see reversions at the somatic level for each of our silenced alleles. The frequency of reversion is comparable to epialleles of the SUPERMAN and PAI genes in Arabidopsis (Jacobsen 1999).
Possible mechanisms for specific methylation of new DNA
To account for the silencing of Cp-gus, a mechanism must be compatible with three observations. First, the silencing is independent of the integration locus. Second, the degree of methylation differs among independent transformation events. Third, the new methylation states are confined to the newly introduced DNA.
We propose that there is an initial imprinting step for newly introduced DNA. This imprinting is transient and inefficient, leading to a variety of methylation patterns on the new DNA. Because active alleles are not susceptible to silencing, we believe the methyltransferase activity that maintains a methylation state during development and meiosis is distinct from this initial imprinting process. We present several models that fit the criteria, although our data do not distinguish among them.
Model 1
The imprinting activity may be induced by environmental stress associated with tissue culture growth. Some somaclonal variants derived from tissue culture are associated with changes in DNA methylation (Phillips et al. 1994). The integrant DNA may be more susceptible to this methyltransferase activity before it incorporates into chromatin.
Model 2
Extrachromosomal DNA may mimic viral DNA and induce a defense response during transformation. The DNA virus CaMV can be recognized and methylated in tobacco cells as an extrachromosomal entity (Tang and Leisner 1998). Similarly, some plasmid molecules may be methylated before integration, resulting in a silenced allele.
Model 3
DNA cruciform junctions, similar to those formed during transposon and site-specific recombination, enhance animal DNA methytransferase activity. During DNA recombination into the genome, the integration intermediate may be a more susceptible target for de novo methylation (Bestor and Tycko 1996).
Model 4
A fourth scenario is an RNA–DNA interaction instigated by transient expression from the episomal plasmid. Transiently produced RNA may induce de novo methylation of plasmid DNA that has integrated into the genome. This mechanism is similar to a model of RNA-induced TGS proposed by Wassenegger and Pelissier (1998) and can account for both the specificity and transient period of the imprinting activity. Further evidence for such a mechanism is that a 35S-Pnos transgene producing RNA homologous to the nos promoter can specifically silence another gene driven by the nos promoter (Mette et al. 1999).
To conclude, our results show that a transgene can be delivered into a specific chromosome position and be expressed at a predicted level. It seems likely that target sites can be found such that the chromosomal context will yield a higher and more consistent level of transgene expression. However, regardless of the chromosome position, there is the probability of methylation-associated silencing. A future challenge is to design a transformation strategy that is not susceptible to transgene-specific imprinting during the transformation process.
Materials and methods
Plant transformation
Plasmid pEL1 was made by ligating a SalI to BamHI Cp-gus fragment from pCoYMV-GUS (Medberry et al. 1992) to EcoRI- and BamHI-cleaved pED112 (Albert et al. 1995), with SalI and EcoRI ends filled by DNA polymerase I (Klenow fragment). Polyethylene glycol–mediated tranformation of tobacco protoplasts and the four target lines nt35S-loxP-cre.5, nt35S-loxP-cre.3, nt35S-lox66-cre.5, and nt35S-lox76-cre.A have been described previously (Albert et al. 1995). A new line used in this study, nt35S-loxP-cre.11, likewise contains a single copy of the target site (data not shown).
Plant analyses
Tobacco seedlings were grown on 1/2 Murashige-Skoog medium with 3% sucrose, and when necessary with either 20 μg/mL hygromycin or 50 μg/mL kanamycin. The seedlings were selected at the two-leaf stage and stained for GUS enzyme activity in 2.5 mg/mL X-glucuronidase, 100 mM phosphate buffer (pH 7.4), 1.0 mM EDTA, 10% Triton X-100, and 5 mM ferri/ferrocyanide for 24 h. Tissues were then cleared in 70% ethanol and photographed using a Nikon SMZ-2T microscope. Gus activity was assayed using standard procedures.
Primers for PCR were as follows: 35S, 5′-GTTCATTTCA TTTGGAGAGG-3′ and nos 3′, 5′-CAAATGTTTGAACGATC TGC-3′. Probes for hybridization were amplified by PCR and purified from agarose with the Geneclean II kit (BIO101). Sources for the probes were nptII from pBIN19; hpt, Cp-gus 5′ fragment, and gus 3′ fragment from pEL1; and 35S-cre and cre 5′ fragment from pMM23. Primers used were as follows: nptII, 5′-ATGATTGAACAAGATGG-3′ and 5′-CTCGATGCGATG TTTCGCTT-3′ or 5′-CGCTCAGAAGAACTCGA-3′; hpt, 5′-ACCGCGACGTCTGTCGAGAA-3′ and 5′-CAAATGTTTGA ACGATCTGC-3′; Cp-gus 5′ fragment, 5′-GTCATCAATGA CATCATCAC-3′ and 5′-TGATATCGTCCACCCAG-3′, gus 3′ fragment, 5′-GGTCAGTGGCAGTGAAG-3′ and 5′-TGTAGC CGACGATGGTG-3′; 35S-cre, 5′-AGATCACCATGGACGAC 3′ and 5′-CTAATCGCCATCTTCCAGCA-3′; cre 5′ fragment, 5′-ATGTCCAATTTACTGACCGT-3′ and 5′-CTTTAACCCT GATCCTG-3′.
Standard conditions were used for PCR and hybridization analyses. Tobacco genomic DNA was isolated from two leaf seedlings or from the top leaves of a mature tobacco plant. Ten micrograms of DNA was cleaved with endonucleases, separated by agarose electrophoresis, and transferred to a positively charged nylon membrane. Membranes were hybridized to probes made radioactive by the random primed DNA labeling kit (Boehringer Mannheim) at 65°C in a rotating hybridization oven as recommended by the manufacturer (250 mM NaPO4 at pH 7.4, 1 mM EDTA, 7% SDS). For methylation analysis, the DNA fragments were separated by a 1.5% agarose gel. RNA was isolated by grinding plant tissue with liquid nitrogen. Extraction buffer (100 mM Tris-HCl at pH 8.5, 100 mM NaCl, 20 mM EDTA, 1% w/v sarkosyl) was added, and the solution was cleaned with phenol/chloroform extraction, ethanol precipitation, and resuspended in water. An equal volume of 4 M LiCl was added followed by an incubation on ice for 3 h and centrifuged. The pellet containing the RNA was washed with 70% ethanol and resuspended in water. Five micrograms of RNA was separated by a 1.3% agarose gel with 0.25 M formaldehyde and 1× MOPS buffer (pH 7.0). The RNA was blotted to nylon membrane and hybridized using the same conditions as for Southern analysis.
Run-on transcription
Nuclei were isolated from tobacco leaves (Vorst et al. 1993) except for the following modifications: (1) Dark-grown plants were used to reduce starch granules; (2) grinding of the leaf tissue in liquid nitrogen was omitted; (3) nuclei were passed through a discontinuous Percoll gradient after the first wash at 3000g for 20 min. The nuclei that partitioned in the 80% Percoll and sucrose cushion were used. Run-on experiments were conducted with 5 × 106 nuclei in 100 μL of buffer containing 25 mM Tris-HCl (pH 7.5), 75 mM NH4Cl2, 7.5 mM MgCl2, 0.5 mM each of ATP, CTP, and GTP, 1 mM DTT, 40 U RNA guard, and 150 μCi UTP (3000 Ci/mmole). The reaction was mixed every 5 min for a total incubation of 30 min at 30°C. DNA was then degraded for 10 min at 37°C with 10 U RNase free DNase, followed by 15 min at 37°C with 10 μg of proteinase K, 1% SDS, 10 mM EDTA (pH 8.0). Radioactive transcripts were purified twice by phenol:chloroform (1 : 1) extraction, precipitated with ethanol, and resuspended in hybridization buffer.
Hybridization of radioactive nuclear run-on transcripts to test DNA on slot blot filters was performed as described (Vorst et al. 1993), and the signals were quantified using a Molecular Dynamics STORM 860 detection system. The test DNA was produced by PCR using the same primers described above, and the ubiquitin gene DNA was made using PCR primers (Universal -20), (Reverse Primer), from the plasmid pSKUB, a gift from P. Quail. The test DNA was quantified by UV spectrophotometry, denatured in 500 mM NaOH, 1.5 M NaCl for 10 min at 37°C and 1 min at 100°C, and applied to nylon membrane as 2 μg per slot.
Acknowledgments
This work was supported by NRICGP grant 98-01816 and ARS CRIS Project 5335-21000-014-00D.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL ow@pgec.ars.usda.gov; FAX (510) 559-5678.
Article and publication are at www.genesdev.org/cgi/doi/10.1101/gad.849600.
References
- Albert H, Dale EC, Lee E, Ow DW. Site-specific integration of DNA into wild-type and mutant lox sites placed in the plant genome. Plant J. 1995;7:649–659. doi: 10.1046/j.1365-313x.1995.7040649.x. [DOI] [PubMed] [Google Scholar]
- Assaad FF, Tucker KL, Signer ER. Epigenetic repeat-induced gene silencing RIGS in Arabidopsis. Plant Mol Biol. 1993;22:1067–1085. doi: 10.1007/BF00028978. [DOI] [PubMed] [Google Scholar]
- Bestor TH, Tycko B. Creation of genomic methylation patterns. Nat Genet. 1996;12:363–367. doi: 10.1038/ng0496-363. [DOI] [PubMed] [Google Scholar]
- Cogoni C, Macino G. Homology-dependent gene silencing in plants and fungi: A number of variations on the same theme. Curr Opin Microbiol. 1999;2:657–662. doi: 10.1016/s1369-5274(99)00041-7. [DOI] [PubMed] [Google Scholar]
- Cubas P, Vincent C, Coen E. An epigenetic mutation responsible for natural variation in floral symmetry. Nature. 1999;401:157–161. doi: 10.1038/43657. [DOI] [PubMed] [Google Scholar]
- Davies GJ, Sheikh MA, Ratcliffe OJ, Coupland G, Furner IJ. Genetics of homology-dependent gene silencing in Arabidopsis: A role for methylation. Plant J. 1997;12:791–804. doi: 10.1046/j.1365-313x.1997.12040791.x. [DOI] [PubMed] [Google Scholar]
- Dobie KW, Lee M, Fantes JA, Graham E, Clark AJ, Springbett A, Lathe R, McClenaghan M. Variegated transgene expression in mouse mammary gland is determined by the transgene integration locus. Proc Natl Acad Sci. 1996;93:6659–6664. doi: 10.1073/pnas.93.13.6659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorer DR, Henikoff S. Transgene repeat arrays interact with distant heterochromatin and cause silencing in cis and trans. Genetics. 1997;147:1181–1190. doi: 10.1093/genetics/147.3.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- English J, Davenport G, Elmayan T, Vaucheret H, Baulcombe D. Requirement of sense transcription for homology dependent virus resistance and trans-inactivation. Plant J. 1997;12:597–603. [Google Scholar]
- Feng YQ, Seibler J, Alami R, Eisen A, Westerman KA, Leboulch P, Fiering S, Bouhassira EE. Site-specific chromosomal integration in mammalian cells: Highly efficient CRE recombinase-mediated cassette exchange. J Mol Biol. 1999;292:779–785. doi: 10.1006/jmbi.1999.3113. [DOI] [PubMed] [Google Scholar]
- Fukushige S, Sauer B. Genomic targeting with a positive-selection lox integration vector allows highly reproducible gene expression in mammalian cells. Proc Natl Acad Sci. 1992;89:7905–7909. doi: 10.1073/pnas.89.17.7905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrick D, Fiering S, Martin DI, Whitelaw E. Repeat-induced gene silencing in mammals. Nat Genet. 1998;18:56–59. doi: 10.1038/ng0198-56. [DOI] [PubMed] [Google Scholar]
- Golic MM, Rong YS, Petersen RB, Lindquist SL, Golic KG. FLP-mediated DNA mobilization to specific target sites in Drosophila chromosomes. Nucleic Acids Res. 1997;25:3665–3671. doi: 10.1093/nar/25.18.3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant SR. Dissecting the mechanisms of posttranscriptional gene silencing: Divide and conquer. Cell. 1999;96:303–306. doi: 10.1016/s0092-8674(00)80541-3. [DOI] [PubMed] [Google Scholar]
- Hollick JB, Dorweiler JE, Chandler VL. Paramutation and related allelic interactions. Trends Genet. 1997;13:302–308. doi: 10.1016/s0168-9525(97)01184-0. [DOI] [PubMed] [Google Scholar]
- Iglesias VA, Moscone EA, Papp I, Neuhuber F, Michalowski S, Phelan T, Spiker S, Matzke M, Matzke AJ. Molecular and cytogenetic analyses of stably and unstably expressed transgene loci in tobacco. Plant Cell. 1997;9:1251–1264. doi: 10.1105/tpc.9.8.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen SE. Gene silencing: Maintaining methylation patterns. Curr Biol. 1999;9:R617–619. doi: 10.1016/s0960-9822(99)80388-1. [DOI] [PubMed] [Google Scholar]
- Jakowitsch J, Papp I, Moscone EA, van der Winden J, Matzke M, Matzke AJ. Molecular and cytogenetic characterization of a transgene locus that induces silencing and methylation of homologous promoters in trans. Plant J. 1999;17:131–140. doi: 10.1046/j.1365-313x.1999.00357.x. [DOI] [PubMed] [Google Scholar]
- Jasin M, Moynahan ME, Richardson C. Targeted transgenesis. Proc Natl Acad Sci. 1996;93:8804–8808. doi: 10.1073/pnas.93.17.8804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen R. Cosuppression, flower color patterns, and metastable gene expression states. Science. 1995;268:686–691. doi: 10.1126/science.268.5211.686. [DOI] [PubMed] [Google Scholar]
- Jorgensen RA, Que Q, Stam M. Do unintended antisense transcripts contribute to sense cosuppression in plants? Trends Genet. 1999;15:11–12. doi: 10.1016/s0168-9525(98)01651-5. [DOI] [PubMed] [Google Scholar]
- Kooter JM, Matzke MA, Meyer P. Listening to the silent genes: Transgene silencing, gene regulation and pathogen control. Trends Plant Sci. 1999;4:340–347. doi: 10.1016/s1360-1385(99)01467-3. [DOI] [PubMed] [Google Scholar]
- Luff B, Pawlowski L, Bender J. An inverted repeat triggers cytosine methylation of identical sequences in Arabidopsis. Mol Cell. 1999;3:505–511. doi: 10.1016/s1097-2765(00)80478-5. [DOI] [PubMed] [Google Scholar]
- Matzke AJ, Matzke MA. Position effects and epigenetic silencing of plant transgenes. Curr Opin Plant Biol. 1998;1:142–148. doi: 10.1016/s1369-5266(98)80016-2. [DOI] [PubMed] [Google Scholar]
- Medberry SL, Lockhart BE, Olszewski NE. The Commelina yellow mottle virus promoter is a strong promoter in vascular and reproductive tissues. Plant Cell. 1992;4:185–192. doi: 10.1105/tpc.4.2.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melquist S, Luff B, Bender J. Arabidopsis PAI gene arrangements, cytosine methylation and expression. Genetics. 1999;153:401–413. doi: 10.1093/genetics/153.1.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mette MF, van der Winden J, Matzke MA, Matzke AJ. Production of aberrant promoter transcripts contributes to methylation and silencing of unlinked homologous promoters in trans. EMBO J. 1999;18:241–248. doi: 10.1093/emboj/18.1.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer P, Heidmann I. Epigenetic variants of a transgenic petunia line show hypermethylation in transgene DNA: An indication for specific recognition of foreign DNA in transgenic plants. Mol Gen Genet. 1994;243:390–399. doi: 10.1007/BF00280469. [DOI] [PubMed] [Google Scholar]
- Meyer P, Heidmann I, Niedenhof I. Differences in DNA-methylation are associated with a paramutation phenomenon in transgenic petunia. Plant J. 1993;4:89–100. doi: 10.1046/j.1365-313x.1993.04010089.x. [DOI] [PubMed] [Google Scholar]
- Ng H-H, Bird AP. DNA Methylation and chromatin modification. Curr Opin Genet Dev. 1999;9:158–163. doi: 10.1016/s0959-437x(99)80024-0. [DOI] [PubMed] [Google Scholar]
- Palauqui JC, Elmayan T, Pollien JM, Vaucheret H. Systemic acquired silencing: Transgene-specific post-transcriptional silencing is transmitted by grafting from silenced stocks to non-silenced scions. EMBO J. 1997;16:4738–4745. doi: 10.1093/emboj/16.15.4738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal-Bhadra M, Bhadra U, Birchler JA. Cosuppression in Drosophila: Gene silencing of alcohol dehydrogenase by white-Adh transgenes is polycomb dependent. Cell. 1997;90:479–490. doi: 10.1016/s0092-8674(00)80508-5. [DOI] [PubMed] [Google Scholar]
- Park YD, Papp I, Moscone EA, Iglesias VA, Vaucheret H, Matzke AJ, Matzke M A. Gene silencing mediated by promoter homology occurs at the level of transcription and results in meiotically heritable alterations in methylation and gene activity. Plant J. 1996;9:183–194. doi: 10.1046/j.1365-313x.1996.09020183.x. [DOI] [PubMed] [Google Scholar]
- Poethig RS. Clonal analysis of cell lineage patterns in plant development. Am J Bot. 1987;74:581–594. [Google Scholar]
- Phillips RL, Kaeppler SM, Olhoft P. Genetic instability of plant tissue cultures: Breakdown of normal controls. Proc Natl Acad Sci. 1994;91:5222–5226. doi: 10.1073/pnas.91.12.5222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabl JF, Henikoff S. Copy number and orientation determine the susceptibility of a gene to silencing by nearby heterochromatin in Drosophila. Genetics. 1996;142:447–458. doi: 10.1093/genetics/142.2.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selker EU. Gene silencing: Repeats that count. Cell. 1999;16:157–160. doi: 10.1016/s0092-8674(00)80725-4. [DOI] [PubMed] [Google Scholar]
- Sharp PA. RNAi and double-strand RNA. Genes & Dev. 1999;13:139–141. [PubMed] [Google Scholar]
- Tang W, Leisner S. Methylation of nonintegrated multiple copy DNA in plants. Biochem Biophys Res Commun. 1998;245:403–406. doi: 10.1006/bbrc.1998.8435. [DOI] [PubMed] [Google Scholar]
- Vaucheret H, Nussaume L, Palauqui J-C, Quilléré I, Elmayan T. A transcriptionally active state is required for post-transcriptional silencing (cosuppression) of nitrate reductase host genes and transgenes. Plant Cell. 1997;9:1495–1504. doi: 10.1105/tpc.9.8.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voinnet O, Vain P, Angell S, Baulcombe DC. Systemic spread of sequence-specific transgene RNA degradation in plants is initiated by localized introduction of ectopic promoterless DNA. Cell. 1998;95:177–187. doi: 10.1016/s0092-8674(00)81749-3. [DOI] [PubMed] [Google Scholar]
- Vorst O, van Dam F, Weisbeek P, Smeekens S. Light-regulated expression of the Arabidopsis thaliana ferredoxin A gene involves both transcriptional and post-transcriptional processes. Plant J. 1993;3:793–803. doi: 10.1111/j.1365-313x.1993.00793.x. [DOI] [PubMed] [Google Scholar]
- Wakimoto BT. Beyond the nucleosome: Epigenetic aspects of position-effect variegation in Drosophila. Cell. 1998;93:321–324. doi: 10.1016/s0092-8674(00)81159-9. [DOI] [PubMed] [Google Scholar]
- Wallace H, Ansell R, Clark J, McWhir J. Pre-selection of integration sites imparts repeatable transgene expression. Nucleic Acids Res. 2000;28:1455–1464. doi: 10.1093/nar/28.6.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassenegger M, Pelissier T. A model for RNA-mediated gene silencing in higher plants. Plant Mol Biol. 1998;37:349–362. doi: 10.1023/a:1005946720438. [DOI] [PubMed] [Google Scholar]
- Waterhouse PM, Graham MW, Wang M-B. Virus resistance and gene silencing in plants can be induced by simultaneous expression of sense and antisense RNA. Proc Natl Acad Sci. 1998;95:13959–13964. doi: 10.1073/pnas.95.23.13959. [DOI] [PMC free article] [PubMed] [Google Scholar]